Abstract
Background
In this study, we investigated the functional correlation between microRNA-210 (miR-210) and gene of F-box protein 31 (FBXO31) in regulating breast cancer.
Methods
Dual-luciferase assay and quantitative real-time polymerase chain reaction were used to investigate the binding of miR-210 with FBXO31 and their expression patterns in breast cancer. miR-210 was inhibited in breast cancer T47D and MCF-7 cells to assess its effect on cancer proliferation, cell cycle progression, and migration. FBXO31 was also downregulated in breast cancer cells to examine its effect on miR-210-mediated breast cancer regulation. The interaction between miR-210 and FBXO31 was further investigated by examining the effect of overexpressing miR-210 on FBXO31-induced suppression of breast cancer proliferation.
Results
FBXO31 was the downstream target gene of miR-210 in breast cancer. miR-210 and FBXO31 are inversely expressed in breast cancer cell lines. miR-210 downregulation reduced cancer progression, induced cell cycle arrest, and inhibited cancer migration in T47D and MCF-7 cells. Tumor suppression by miR-210 downregulation was reversed by downregulating FBXO31. In FBXO31-overexpressed breast cancer cells, upregulating miR-210 also reversed the tumor-suppressive effect of FBXO31 on breast cancer proliferation.
Conclusion
Our work demonstrated that the expression pattern and tumor regulatory functions of miR-210 and FBXO31 are inversely correlated in breast cancer.
Keywords: breast cancer, miR-210, FBXO31, cancer proliferation, cancer migration
Introduction
Breast carcinoma is one of the most common tumors among women worldwide.1,2 An estimated quarter million new cases of breast cancer and 41,000 breast cancer–related deaths are reported in the US annually.3 Thanks to the development of mammography and high awareness of early screening among female patients, the averaged 5-year survival for patients with breast cancer has gradually increased during the past decades.4,5 However, the exact molecular mechanisms contributing to carcinogenesis, development, and metastasis of breast cancer are largely unknown.
MicroRNAs (miRNAs) are families of short noncoding RNAs that bind to specific regions on the 3′-untranslated region (3′-UTR) of targeted messenger RNAs (mRNAs) to suppress gene transcription or protein translation in various cell types in both animals and humans.6–9 During the past decades, numerous studies have shown that miRNAs play an important role in breast cancer proliferation, maturation, apoptosis, and metastasis.10–12 For instance, miRNA-21 was overexpressed in breast carcinoma in human patients and closely correlated with the pathology, lymph node metastasis, and prognosis.13 Also, cluster of miR-211/222 was found to regulate the sensitivity of tamoxifen in breast cancer through the regulation of estrogen receptor and cell cycle inhibitor p27(Kip1).14,15 Furthermore, miRNA-146 was found to be overexpressed in highly metastatic human breast cancer, and inhibiting miRNA-146 resulted in reduced breast cancer metastasis by regulation of nuclear factor-κB.16 Specifically, miRNA-210, or miR-210, was found to be overexpressed in clinical samples of breast carcinoma and closely associated with the overall survival in patients with breast cancer.17,18 However, the exact downstream molecular complex associated with oncogenic miR-210 in breast cancer is yet to be elucidated.
Gene of F-box protein 31 (FBXO31), residing on human chromosome 16q24.3, has been identified as a tumor-suppressing gene in various human cancers, including breast cancer.19–23 In breast cancer cell lines, MCF-7, MCF-10A, and SKBR3 cells, FBXO31 was shown to be downregulated and the tumor-inhibitory effect of FBXO31 was suggested to be associated with a complex network of proteins such as Skp1, Roc-1, or Cullin-1.19 Interestingly, while much focus has been given on the downstream pathways of FBXO31, little is known about the upstream initiator of FBXO31 in breast cancer.
In the present study, we attempted to explore the functional correlation between oncogenic miR-210 and tumor suppressing FBXO31 in breast cancer. We firstly used dual-luciferase reporter assay to investigate whether miR-210 actually binds FBXO31 gene in breast cancer. We also used quantitative real-time polymerase chain reaction (qRT-PCR) to assess the reciprocal expression pattern of miR-210 and FBXO31 in breast cancer cell lines. We then inhibited endogenous miR-210 in breast cancer to investigate the mechanistic regulation of miR-210 on cancer proliferation, cell cycle progression, and migration. Subsequently, we inhibited the expression of FBXO31 in miR-210–downregulated breast cancer to examine the effect of FBXO31 on miR-210–mediated cancer regulation. Furthermore, in FBXO31-overexpressed breast cancer cells, we upregulated miR-210 to investigate whether reverse regulation of miR-210 on FBXO31 would also occur in breast cancer.
Materials and methods
Ethical statements
Approval for the study was granted by the Clinic Research and Ethics Committee at the First Affiliated Hospital, Sun Yat-Sen University in Guangzhou, People’s Republic of China. All experiments were performed in accordance with the Declaration of Helsinki.
Cell lines
In our study, six breast cancer cell lines, T47D, MCF-7, MDA-MB-231, BT549, SKBR3, and BT474, and two control normal breast epithelial cell lines, 184A1 and 184B5, were all obtained from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Wuhan, People’s Republic of China). They were maintained in Dulbecco’s Modified Eagle’s Medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific) and penicillin (100 U/mL) and streptomycin (100 μg/mL) (Sigma-Aldrich Co., St Louis, MO, USA) in a humidified tissue culture chamber at 37°C with 5% CO2.
miR-210 inhibition and upregulation in breast cancer cell lines
Human miRIDIAN miR-210 inhibitor and mimics (C-miR210-IN, miR210-mimics), and nonspecific miRIDIAN miRNA negative control (C-miRNA) were purchased from GE Health Dharmacon (Waltham, MA, USA). Breast cancer cell lines T47D and MCF-7 were transfected with C-miR210-IN, miR210-mimics, or C-miRNA using Lipofectamine 2000 reagent (Thermo Fisher Scientific) for 48 hours. Transfection efficiencies were then verified by qRT-PCR.
Dual-luciferase reporter assay
The 3′-UTR of the wild-type (wt) human FBXO31 gene, including the putative hsa-miR-210 binding site, was cloned from a human complementary DNA (cDNA) library and inserted into a dual-luciferase PmirGLO vector (Promega Corporation, Fitchburg, WI, USA). The hsa-miR-210 binding site was then mutated (mu) and the mutated FBXO31 3′-UTR was also inserted into the PmirGLO vector. T47D and MCF-7 cells were seeded in six-well plates overnight and cotransfected with luciferase vectors of FBXO31(wt), FBXO31(mu), or a negative control Renilla vector, and miR210-mimics. Forty-eight hours after cotransfection, relative luciferase activities were measured using a dual-luciferase reporter assay (Promega Corporation) according to the manufacturer’s instructions.
RNA extraction and qRT-PCR
Quantification of gene expressions of miR-210 and FBXO31 in breast cancer cell lines was performed according to the methods used in previous studies with slight modifications.17,19 Briefly, RNA extraction was carried out using an Ambion mirVana miRNA isolation kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The quality of RNA was verified using a NanoDrop ND-3000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Then, 5 ng of total RNA was used to synthesize cDNA using a QuantiTect Reverse Transcription Kit (Qiagen, Venlo, the Netherlands). qRT-PCR was performed using an iCycler IQ Detection System (Bio-Rad Laboratories Inc., Hercules, CA, USA). For detection of hsa-miR-210, a TaqMan miRNA assay (Thermo Fisher Scientific, Waltham, MA, USA) was used with the housekeeping gene of snRNA RNU43. For detection of FBXO31, a Brilliant SYBR Green II qRT-PCR kit (Stratagene, La Jolla, CA, USA) was used with the housekeeping gene of cyclophilin A to minimize gene expression variation.24
Proliferation assay
Breast cancer cell lines T47D and MCF-7 were seeded in 96-well plate (5,000 cells/well). A Vybrant® 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) Cell Proliferation Assay Kit (Molecular Probes, Eugene, OR, USA) was used for performing cell proliferation assay for 5 days according to the manufacturer’s instruction. Breast cancer cell proliferation was evaluated daily by reading MTT fluorescent absorbance, which was recorded at 490 nm by a V-5000 spectrophotometer reader (Yuanxi Instrument, Shanghai, People’s Republic of China).
Cell cycle assay
T47D and MCF-7 cells were quickly fixed with 70% methanol and incubated with a DNA-binding dye, propidium iodide (1 mM; Thermo Fisher Scientific), and 10 mg/mL RNAse (Sigma-Aldrich Co.) at 37°C for 30 minutes. DNA content was assessed with a two-laser BD Accuri C6 flow cytometer (Beckman Coulter, Miami, FL, USA). G0/G1, S, and M/G2 phases of the cell cycle distribution were then analyzed using a MultiCycle AV DNA Analysis software (Phoenix Flow Systems, San Diego, CA, USA).
Migration assay
In a transwell migration assay (Thermo Fisher Scientific), T47D and MCF-7 cells were seeded in the matrigel-coated inserts (5,000 cells/insert) in culture medium with 0.5% FBS. The lower chamber was filled with 10% FBS as the migration attractant. Cells were incubated at 37°C for 12 hours. After that, the inserts were discarded and the lower chambers were quickly fixed with 70% methanol and stained with 0.1% crystal violet. Images of the migrated cells in the lower chamber were visualized using an inverted microscope (XDS-1; Chongqing Optical & Electrical Instrument, Chongqing, People’s Republic of China).
FBXO31 inhibition and upregulation in breast cancer cell lines
For FBXO31 inhibition, small interfering RNA (siRNA) targeting human FBXO31 gene (Si-FBXO31) and a scrambled control siRNA (Si-C) were purchased from GE Health Dharmacon. T47D and MCF-7 cells were transfected with 100 nM Si-C or Si-FBXO31 using Lipofectamine 2000 reagent (Thermo Fisher Scientific) for 48 hours, following which qRT-PCR was performed to verify the downregulation efficiency. For BXO31 upregulation, the full length of human FBXO31 gene was cloned into a eukaryotic expression vector pcDNA3.1 (pcDNA-FBXO31; Promega Corporation). T47D and MCF-7 cells were transfected with pcDNA-FBXO31, or an empty pcDNA3.1 vector (pcDNA-C), also using Lipofectamine 2000 reagent for 48 hours. The efficiency of FBXO31 upregulation was also verified by qRT-PCR.
Statistical analysis
All assays were independently repeated for at least three times and data were expressed as mean ± standard error. Statistical analyses were carried out using Student’s t-test on Statistical Package for the Social Sciences software, version 13.0 (SPSS Inc., Chicago, IL, USA). Differences were considered significant if the P-value was <0.05.
Results
FBXO31 is the downstream target gene of miR-210 in breast cancer
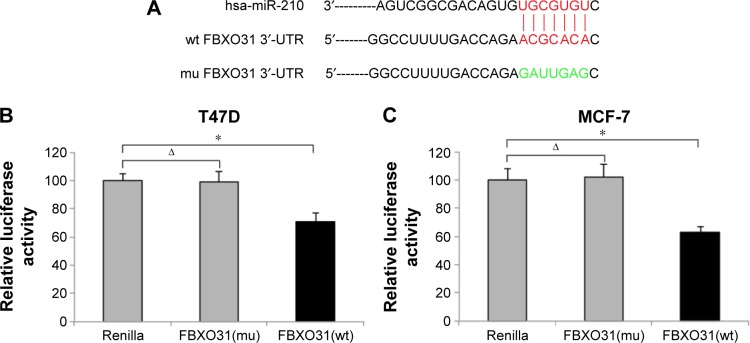
In a previous study, FBXO31 was shown to be a tumor suppressor in human breast cancer.19 However, the exact upstream regulator(s) of FBXO31 in breast cancer was never identified. In two recent reports, miR-210 was demonstrated to be highly expressed in breast cancer tumor samples and cancer cell lines with oncogenic effects.17,18 In this study, we investigated whether miR-210 was the upstream regulator of FBXO31, or vice versa, and FBXO31was the downstream target gene of miR-210 in human breast cancer. We used a dual-luciferase activity assay to test this hypothesis. Through several online services of miRNA-binding algorithms, we found that the 3′-UTR region of wt human FBXO31 gene included a DNA sequence complimentary to hsa-miR-210 (Figure 1A). We thus cloned this sequence into a PmirGLO vector to make luciferase FBXO31(wt) vector. In addition, we point-mutated the putative miR-210 binding sequence on 3′-UTR of FBXO31 (Figure 1A) and then cloned this mutant (mu) into PmirGLO to make FBXO31(mu) luciferase vector. Then, we selected two breast cancer cell lines, T47D and MCF-7, and cotransfected them with FBXO31(wt), FBXO31(mu), or control Renilla luciferase vector, and miR-210 mimics. Forty-eight hours after cotransfection, the breast cancer cells were examined by dual-luciferase activity assay. It showed that, in both T47D and MCF-7 cells, there were no significant differences in relative luciferase activities between the cells transfected with Renilla control and those transfected with FBXO31(mu) (Figure 1B, C) (ΔP>0.05). However, relative luciferase activities were much smaller in cells transfected with FBXO31 (mu) than the values in cells transfected with Renilla control (Figure 1B, C) (P<0.05). Thus, analysis of luciferase assay demonstrates that FBXO31 is the downstream target gene of miR-210 in breast cancer.
Figure 1.
miR-210 targets FBXO31 gene in breast cancer.
Notes: (A) In wt human FBXO31 gene, its 3′-UTR DNA sequence included a putative hsa-miR-210 binding site. In a mu FBXO31 3′-UTR, the hsa-miR-210 binding site was point-mutated to another amino acid to avoid binding. In T47D (B) and MCF-7 (C) cells, luciferase vectors of FBXO31(mu), FBXO31(wt), or Renilla control were cotransfected with hsa-miR-210 mimics for 48 hours. A dual-luciferase reporter assay was then carried out (ΔP>0.05, *P<0.05).
Abbreviations: FBXO31, F-box protein 31; miR-210, microRNA-210; mu, mutant; UTR, untranslated region; wt, wild type.
miR-210 and FBXO31 are reversely expressed in breast cancer
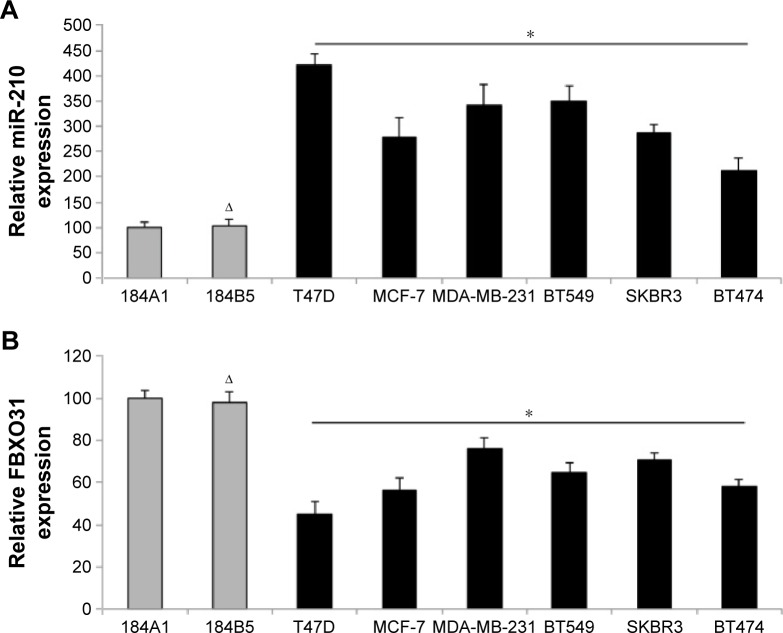
We then investigated the expression patterns of miR-210 and FBXO31 in breast cancer cell lines. We firstly used qRT-PCR to examine miR-210 expression in seven immortal breast cancer cell lines, T47D, MCF-7, MDA-MB-231, BT549, SKBR3, and BT474. The expression levels of miR-210 in those cancer cells were compared with the expression levels in two normal human breast epithelial cell lines, 184A1 and 184B5. The result showed that miR-210 was significantly upregulated in all breast cancer cell lines than in normal breast cells (Figure 2A) (*P<0.05). We also examined the expression pattern of FBXO31 gene. The qRT-PCR result showed that FBXO31 was significantly downregulated in all breast cancer cell lines than in normal breast cells (Figure 2B) (*P<0.05). In addition, we noticed that neither miR-210 nor FBXO31 differed much between the two normal breast cell lines, 184A1 and 184B5 (Figure 2A, B) (ΔP>0.05). Thus, our analysis of qRT-PCR demonstrates that the expression levels of miR-210 and FBXO31 are inversely related in breast cancer, supporting our previous findings that FBXO31 is the downstream target gene of miR-210.
Figure 2.
Expression levels of miR-210 and FBXO31 are inversely correlated in breast cancer.
Notes: (A) qRT-PCR was used to assess miR-210 expression in the breast cancer cell lines, T47D, MCF-7, MDA-MB-231, BT549, SKBR3, and BT474, and in normal human breast epithelial cell line, 184A1 (*P<0.05). miR-210 expression was also examined in another normal human breast epithelial cell line, 184B5 (ΔP<0.05). (B) qRT-PCR was also used to assess FBXO31 gene expression in breast cancer cell lines and normal human breast epithelial cells (ΔP<0.05, *P<0.05).
Abbreviations: FBXO31, F-box protein 31; miR-210, microRNA-210; qRT-PCR, quantitative real-time polymerase chain reaction.
Downregulation of miR-210 reduced cancer proliferation in breast cancer
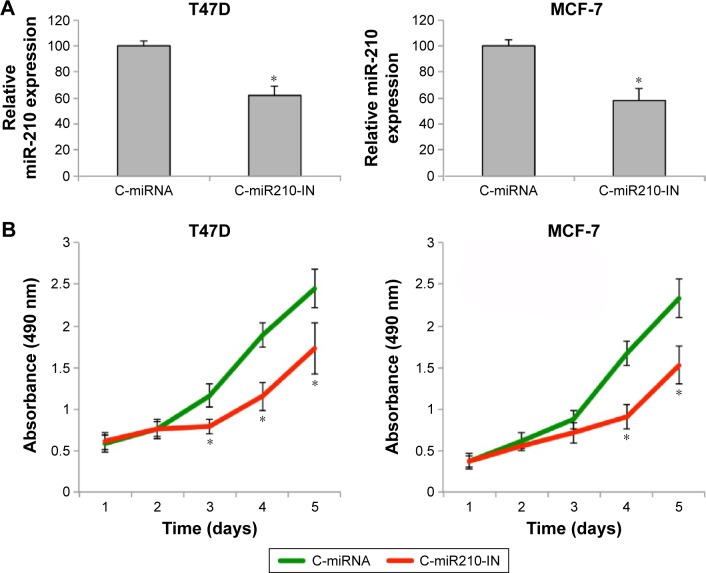
The breast cancer cell lines T47D and MCF-7 were transfected with miR-210 inhibitor (C-miR210-IN) or a control miRNA (C-miRNA) for 48 hours. qRT-PCR showed that C-miR210-IN was able to significantly downregulate miR-210 gene expression in breast cancer cells (Figure 3A) (*P<0.05). We then used those cells and performed a cancer proliferation assay. It showed that the cancer proliferating rates were significantly reduced in T47D and MCF-7 cells transfected with C-miR210-IN than in cells transfected with C-miRNA (Figure 3B) (*P<0.05).
Figure 3.
Inhibiting miR-210 suppressed cancer proliferation in breast cancer.
Notes: (A) Two breast cancer cell lines, T47D and MCF-7, were transfected with a nonspecific C-miRNA or a human miR-210 inhibitor (C-miR210-IN) for 48 hours. qRT-PCR was then performed to examine miR-210 expression between the cancer cells transfected with C-miRNA and the cells transfected with C-miR210-IN (*P<0.05). (B) After transfection, T47D and MCF-7 cells were seeded in 96-well plates and a proliferation assay was carried out for five consecutive days. MTT-induced luminescence was measured daily at an absorbance of 490 nm and compared between the cancer cells transfected with C-miRNA and the cells transfected with C-miR210-IN (*P<0.05).
Abbreviations: C-miRNA, control microRNA; miR-210, microRNA-210; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; qRT-PCR, quantitative real-time polymerase chain reaction.
Downregulation of miR-210 induced cell cycle arrest and reduced migration in breast cancer
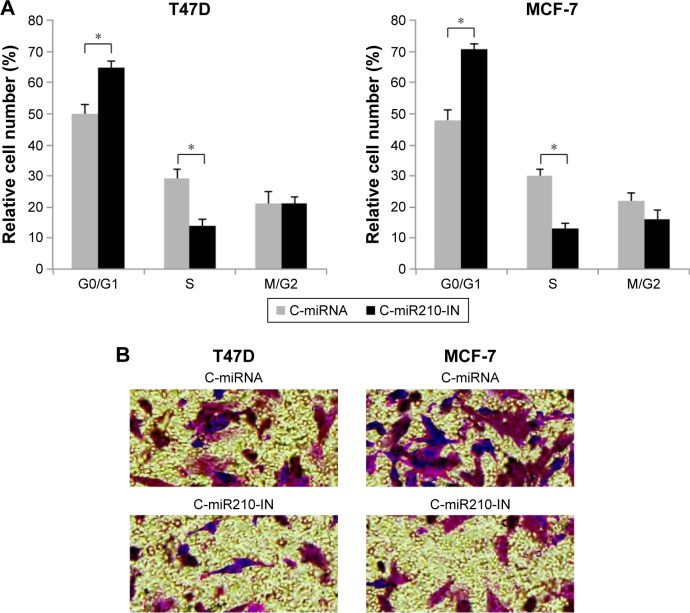
We then investigated other functional regulation by downregulating miR-210 in breast cancer. With cell cycle assay, we found that miR-210 downregulation induced G0/G1 phase arrest in both T47D and MCF-7 cells (Figure 4A) (*P<0.05). In addition, we examined the transfected breast cancer cells with a transwell migration assay. It showed that cancer migration was significantly reduced in T47D and MCF-7 cells transfected with C-miR210-IN than in cells transfected with C-miRNA (Figure 4B).
Figure 4.
Inhibiting miR-210 suppressed cancer migration and cell cycle progression in breast cancer.
Notes: (A) After transfection, T47D and MCF-7 cells were fixed and run through a cell cycle assay. It showed that the percentages of breast cancer cells at G0/G1 phase were significantly higher in cells transfected with C-miR210-IN (*P<0.05). (B) Transfected T47D and MCF-7 cells were also assessed through a transwell migration assay. Twelve hours after the cells were seeded, breast cancer cells migrated to lower chambers and were immunostained and examined under a microscope (40×). It revealed that significantly less T47D or MCF-7 cells invaded the lower chamber while they were transfected with C-miR210-IN.
Abbreviations: C-miRNA, control microRNA; miR-210, microRNA-210.
Inhibitory effect of miR-210 downregulation on breast cancer proliferation was reversed by downregulating FBXO31
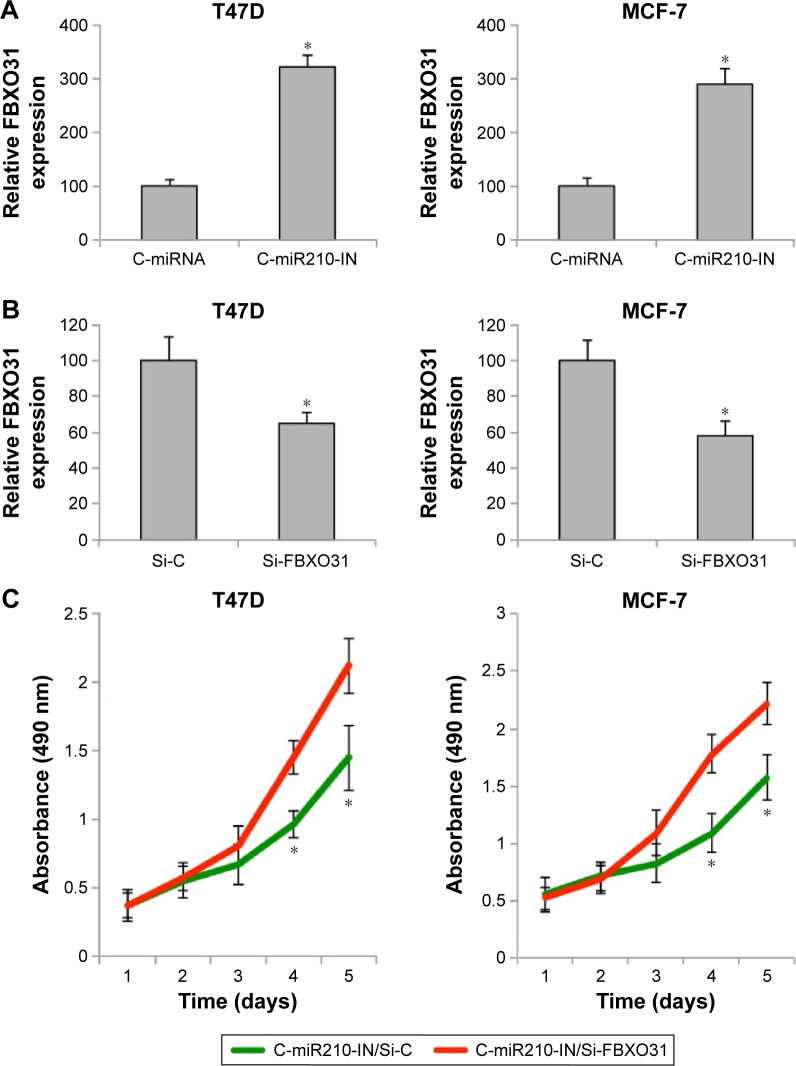
As we found that miR-210 downregulation had an inhibitory effect on breast cancer development, we wondered whether it also involved its downstream target gene of FBXO31. We firstly examined the dynamics of FBXO31 gene expression under the influence of miR-210 regulation. In transfected T47D and MCF-7 cells, we used qRT-PCR to examine FBXO31 expression. It showed that in both breast cancer cell lines, genes of FBXO31 were significantly upregulated by C-miR210-IN transfection, further confirming that FBXO31 was the downstream target of miR-210 (Figure 5A) (*P<0.05).
Figure 5.
FBXO31 reversely regulated miR-210–mediated cancer proliferation in breast cancer.
Notes: (A) After transfection, T47D and MCF-7 cells were examined by qRT-PCR to compare FBXO31 gene expression between the cells transfected with C-miRNA and the cells transfected with a human miR-210 inhibitor (C-miR210 IN) (*P<0.05). (B) In T47D and MCF-7 cells transfected with C-miR210-IN, second transfection with FBXO31-specific siRNA (Si-FBXO31) or nonspecific Si-C was performed for 48 hours. qRT-PCR was then performed to examine FBXO31 gene expression (*P<0.05). (C) In T47D and MCF-7 cells transfected with both C-miR210-IN and siRNAs (Si-C or Si-FBXO31), a proliferation assay was conducted for five consecutive days (*P<0.05).
Abbreviations: C-miRNA, control microRNA; FBXO31, F-box protein 31; miR-210, microRNA-210; qRT-PCR, quantitative real-time polymerase chain reaction; Si-C, control siRNA; SiRNA, small interfering RNA.
In order to manipulate FBXO31 gene expression in breast cancer cells, T47D and MCF-7 cells with downregulated miR-210 were further transfected with FBXO31-specific siRNA (Si-FBXO31) or Si-C for 48 hours. qRT-PCR confirmed that Si-FBXO31 transfection significantly downregulated FBXO31 gene expression, as compared to siRNA transfection (Si-C) (Figure 5B) (*P<0.05). These double-transfected breast cancer cells were then reseeded into 96-well plates and examined through a proliferation assay. The results showed that, in both T47D and MCF-7 cells, cancer proliferating rates were significantly increased by Si-FBXO31 transfection, rather than siRNA transfection (Figure 5C) (*P<0.05).
Inhibitory effect of miR-210 downregulation on breast cancer migration and cell cycle progression was reversed by downregulating FBXO31
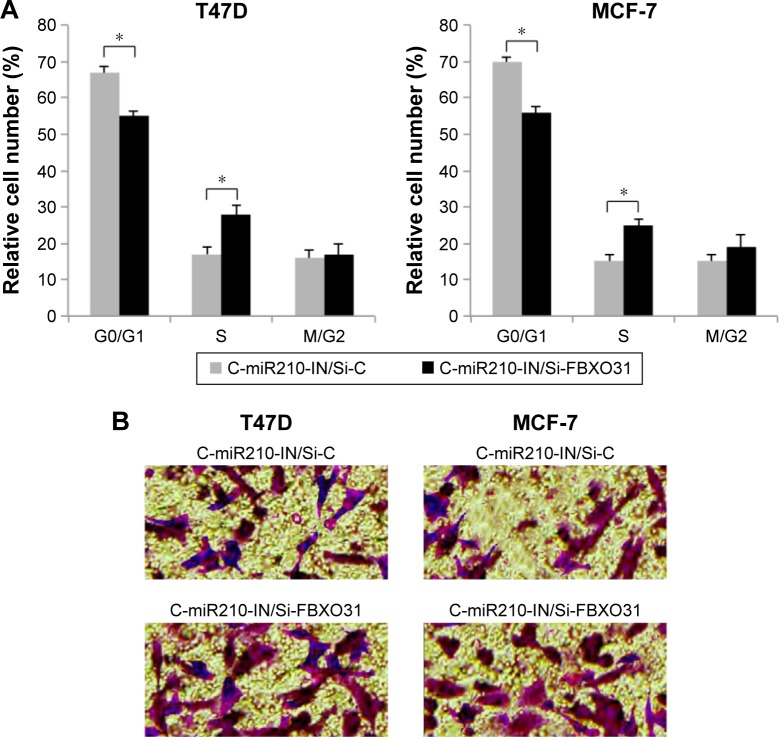
In double-transfected T47D and MCF-7 cells, we also performed a cell cycle assay. The results showed that in miR-210–downregulated breast cancer cells, Si-FBXO31 transfection significantly rescued G0/G1 phase arrest than Si-C transfection (Figure 6A) (*P<0.05). Furthermore, we examined double-transfected breast cancer cells with a transwell migration assay. The results showed that in miR-210–downregulated breast cancer cells, Si-FBXO31 transfection significantly increased cell migration, rather than Si-C transfection (Figure 6B).
Figure 6.
FBXO31 reversely regulated miR-210–mediated cancer cell cycle progression and migration in breast cancer.
Notes: (A) In T47D and MCF-7 cells transfected with both C-miR210-IN and siRNAs (Si-C or Si-FBXO31), a cell cycle assay was conducted. It showed that the percentages of breast cancer cells arrested at G0/G1 phase were significantly reduced in cells transfected with Si-FBXO31, rather than in cells transfected with Si-C (*P<0.05). (B) Double-transfected T47D and MCF-7 cells were also assessed by a transwell migration assay. It was found that significantly more T47D or MCF-7 cells invaded the lower chamber while they were transfected with Si-FBXO31 than with Si-C (40×).
Abbreviations: FBXO31, F-box protein 31; miR-210, microRNA-210; Si-C, control siRNA; SiRNA, small interfering RNA; C-miR210-IN, miR-210 inhibitor.
Suppression of breast cancer proliferation by FBXO31 was reversed by miR-210
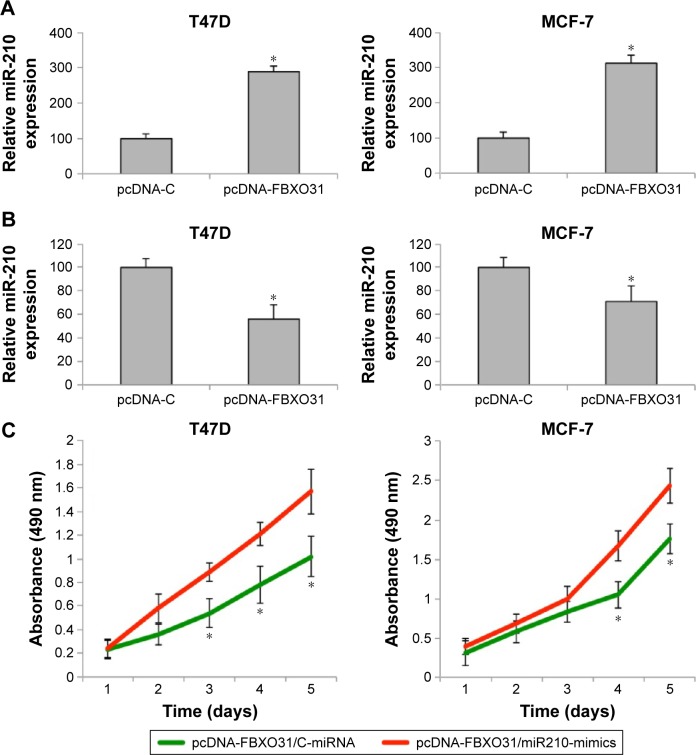
Finally, we investigated whether the tumor-suppressive effect by FBXO31, as shown in a previous study,19 would be affected by miR-210 in breast cancer. We firstly transfected T47D and MCF-7 cells with an FBXO31 expressing cassette (pcDNA-FBXO31) or an empty control expression cassette (pcDNA-C). qRT-PCR confirmed that endogenous FBXO31 was significantly upregulated by pcDBA-FBXO31 transfection (Figure 7A) (*P<0.05). Interestingly, we noticed that while FBXO31 was upregulated in breast cancer cells, endogenous miR-210 gene expression was subsequently downregulated (Figure 7B) (*P<0.05).
Figure 7.
Tumor-suppressive effect of FBXO31 was reversed by miR-210 in breast cancer.
Notes: (A) T47D and MCF-7 cells were transfected with an FBXO31 overexpression vector, pcDNA-FBXO31, or an empty control vector pcDNA-C for 48 hours. qRT-PCR was then performed to examine FBXO31 expression between the cancer cells transfected with pcDNA-C and the cells transfected with pcDNA-FBXO31 (*P<0.05). (B) qRT-PCR was also performed to examine miR-210 expression between the cancer cells transfected with pcDNA-C and the cells transfected with pcDNA-FBXO31 (*P<0.05). (C) FBXO31-overexpressed T47D and MCF-7 cells were transfected with either C-miRNA or miR210-mimics. A proliferation assay was then conducted for five consecutive days (*P<0.05).
Abbreviations: C-miRNA, control microRNA; FBXO31, F-box protein 31; miR-210, microRNA-210; qRT-PCR, quantitative real-time polymerase chain reaction; pcDNA-C, over extension vector DNA control.
Furthermore, we performed another double transfection in breast cancer cells by transfecting T47D and MCF-7 cells with pcDNA-FBXO31 first, followed by another transfection with either C-miRNA or miR210-mimics. After that, a proliferation assay was performed. It showed that in breast cancer cells with overexpressed FBXO31, miR-210 had significant oncogenic effect by promoting cancer cell proliferation (Figure 7C) (*P<0.05).
Discussion
Previous studies suggested that miR-210 is a tumor oncogene and FBXO31 a tumor suppressor in breast cancer.17–19 However, the possibly interactive relationship between miR-210 and FBXO31 had never been speculated. In this study, we firstly applied dual-luciferase activity assay to confirm that FBXO31 was indeed a downstream target gene of miR-210 in breast cancer. In addition, after examining the gene expression patterns of miR-210 and FBXO31 in several breast cancer cell lines, we discovered that miR-210 and FBXO31 were inversely expressed, as miR-210 was upregulated and FBXO31 downregulated in breast cancer. These results are in line with previous studies, supporting the notion that miR-210 is the oncogene and FBXO31 the tumor suppressor in breast cancer.
We then used a systemic method to investigate the functional role of miR-210 in breast cancer. As we found that miR-210 was predominantly overexpressed in breast cancer, we took the approach to downregulate miR-210 in breast cancer T47D and MCF-7 cells. After transfecting the breast cancer cells with miR-210 inhibitor, we applied several functional assays and found that miR-210 downregulation had significant tumor-suppressing effects by inhibiting cancer proliferation and migration and by inducing cell cycle arrest. In a previous study, similar manipulation of miR-210 resulted in an inhibitory effect on cancer proliferation in another breast cancer cell line, MDA-MB-231, which was characterized as a breast cancer cell line with high metastasis.18 Based on the data obtained in our study, it is very likely that downregulating miR-210 would have profound suppressing effect on various subtypes of breast carcinomas. It is worth noting that, the functional regulation of overexpressing miR-210 was also revealed in the breast cancer cell line MCF-7.18 However, as we focused on the interaction between miR-210 and FBXO31, we did not further pursue the functional assays with miR-210 overexpression.
Also, in our study, we performed double transfection in breast cancer cells to precisely measure the interactive effect of FBXO31 on miR-210–mediated breast cancer regulation. T47D and MCF-7 cells that were initially transfected with miR-210 inhibitor were further transfected with FBXO31-specific siRNA. After conducting functional assays, we found that the tumor-suppressive effects of miR-210 downregulation on breast cancer proliferation, cell cycle arrest, and migration were all reversed by the second downregulation of FBXO31, thus confirming our hypothesis that FBXO31 is the direct downstream target gene of miR-210, and miR-210–mediated functional regulation of breast cancer is predominantly through the reverse interaction with FBXO31.
Furthermore, we performed another double transfection in breast cancer cells by overexpressing FBXO31 first and then upregulating miR-210. In a previous study, ectopic expression of FBXO31 was shown to have significant tumor-inhibitory effect on breast cancer cells.19 This was confirmed by our qRT-PCR assay which showed that the oncogene miR-210 was downregulated by FBXO31 in breast cancer cells. Interestingly, in our study, while we subsequently upregulated miR-210 in those FBXO31-overexpressed breast cancer cells, we observed that, FBXO31 overexpression–induced cancer proliferation was greatly reversed by miR-210 upregulation. The most straightforward explanation would be that, miR-210 upregulation would have downregulated FBXO31, thus offsetting the gene overexpression of FBXO31. However, we cannot rule out the possibility that other interactively correlated signaling pathways of miR-210 or FBXO31 may contribute to the oncogenic effect of miR-210 in FBXO31-overexpressed breast cancer. Future experiments exploring the targeted genes or proteins of FBXO31-mediated cancer regulation in breast cancer may help to elucidate the full blueprint of miR-210/FBXO31 molecular complex in breast cancer.
Conclusion
In conclusion, our work demonstrates that the oncogene miR-210 and the tumor suppressor FBXO31 are inversely expressed in breast cancer, as well as have reciprocal functions in regulating breast cancer proliferation, cell cycle progression, and migration. The data obtained in our study may help to elucidate the molecular mechanisms contributing to breast cancer development and seek novel therapeutic targets for the diagnosis and treatment of breast cancer patients.
Acknowledgments
This work was supported by the Science and Technology Planning Project of Guangdong Province (2012B031800055) and the National Natural Science Foundation of China (81300421).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]; Adv Nutr. 2016;7(2):418–419. doi: 10.3945/an.116.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tao Z, Shi A, Lu C, Song T, Zhang Z, Zhao J. Breast cancer: epidemiology and etiology. Cell Biochem Biophys. 2015;72(2):333–338. doi: 10.1007/s12013-014-0459-6. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Nelson HD, Cantor A, Humphrey L, et al. Screening for Breast Cancer: A Systematic Review to Update the 2009 US Preventive Services Task Force Recommendation. Rockville, MD: Agency for Healthcare Research and Quality (US); 2016. edn. [PubMed] [Google Scholar]
- 5.Lo-Fo-Wong DN, Sitnikova K, Sprangers MA, de Haes HC. Predictors of health care use of women with breast cancer: a systematic review. Breast J. 2015;21(5):508–513. doi: 10.1111/tbj.12447. [DOI] [PubMed] [Google Scholar]
- 6.Niwa R, Slack FJ. The evolution of animal microRNA function. Curr Opin Genet Dev. 2007;17(2):145–150. doi: 10.1016/j.gde.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132(21):4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Khoshnaw SM, Green AR, Powe DG, Ellis IO. MicroRNA involvement in the pathogenesis and management of breast cancer. J Clin Pathol. 2009;62(5):422–428. doi: 10.1136/jcp.2008.060681. [DOI] [PubMed] [Google Scholar]
- 11.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 12.Sempere LF, Christensen M, Silahtaroglu A, et al. Altered microRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67(24):11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 13.Yan LX, Huang XF, Shao Q, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14(11):2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller TE, Ghoshal K, Ramaswamy B, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283(44):29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao JJ, Lin J, Yang H, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283(45):31079–31086. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;27(42):5643–5647. doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camps C, Buffa FM, Colella S, et al. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14(5):1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 18.Rothe F, Ignatiadis M, Chaboteaux C, et al. Global microRNA expression profiling identifies MiR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS One. 2011;6(6):e20980. doi: 10.1371/journal.pone.0020980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar R, Neilsen PM, Crawford J, et al. FBXO31 is the chromosome 16q24.3 senescence gene, a candidate breast tumor suppressor, and a component of an SCF complex. Cancer Res. 2005;65(24):11304–11313. doi: 10.1158/0008-5472.CAN-05-0936. [DOI] [PubMed] [Google Scholar]
- 20.Miller BJ, Wang D, Krahe R, Wright FA. Pooled analysis of loss of heterozygosity in breast cancer: a genome scan provides comparative evidence for multiple tumor suppressors and identifies novel candidate regions. Am J Hum Genet. 2003;73(4):748–767. doi: 10.1086/378522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin YW, Lee IN, Chen CH, et al. Deletion mapping of chromosome 16q24 in hepatocellular carcinoma in Taiwan and mutational analysis of the 17-beta-HSD gene localized to the region. Int J Cancer. 2001;93(1):74–79. doi: 10.1002/ijc.1287. [DOI] [PubMed] [Google Scholar]
- 22.Harkonen P, Kyllonen AP, Nordling S, Vihko P. Loss of heterozygosity in chromosomal region 16q24.3 associated with progression of prostate cancer. Prostate. 2005;62(3):267–274. doi: 10.1002/pros.20147. [DOI] [PubMed] [Google Scholar]
- 23.Huang HL, Zheng WL, Zhao R, Zhang B, Ma WL. FBXO31 is down-regulated and may function as a tumor suppressor in hepatocellular carcinoma. Oncol Rep. 2010;24(3):715–720. doi: 10.3892/or_00000912. [DOI] [PubMed] [Google Scholar]
- 24.Powell JA, Gardner AE, Bais AJ, et al. Sequencing, transcript identification, and quantitative gene expression profiling in the breast cancer loss of heterozygosity region 16q24.3 reveal three potential tumor-suppressor genes. Genomics. 2002;80(3):303–310. doi: 10.1006/geno.2002.6828. [DOI] [PubMed] [Google Scholar]