Abstract
RNA capping and decapping are thought to be distinctive features of eukaryotes. Recently, the redox cofactor NAD was discovered to be attached to small regulatory RNAs in bacteria in a cap-like manner, and Nudix hydrolase NudC was found to act as a NAD decapping enzyme in vitro and in vivo. Here, crystal structures of Escherichia coli NudC in complex with substrate NAD and with cleavage product NMN reveal the catalytic residues lining the binding pocket and principles underlying molecular recognition of substrate and product. Biochemical mutation analysis identifies the conserved Nudix motif as the catalytic center of the enzyme, which needs to be homodimeric as the catalytic pocket is composed of amino acids from both monomers. NudC is single-strand specific and has a purine preference for the 5’-terminal nucleotide. The enzyme strongly prefers NAD-RNA over NAD and binds to a diverse set of cellular RNAs in an unspecific manner.
RNA capping is considered a hallmark of eukaryotic gene expression, in which a 5′,5′-triphosphate-linked 7-methylguanosine protects mRNA from degradation and modulates maturation, localisation, and translation1. The m7G cap is removed by eukaryotic decapping enzymes, such as Dcp22, thereby initiating different RNA decay pathways. Recently, the ubiquitous redox cofactor nicotinamide adenine dinucleotide (NAD) has been found to be covalently linked to bacterial RNA3, and recent work from our laboratory discovered a subset of small regulatory RNAs (sRNAs) in the bacterium Escherichia coli to be specifically 5’-modified with NAD in a cap-like manner4. This finding provides a new and unexpected link between redox biology, metabolism, and RNA processing5, and represents the first description of a prokaryotic cap6,7. The NAD cap stabilizes the sRNAs in vitro against endonucleolytic processing by RNase E8, the major player of RNA decay in E. coli, and against 5’-end modification by RNA pyrophosphohydrolase RppH9, which converts 5’-triphosphate RNA into 5’-monophosphate RNA and thereby triggers endonucleolytic processing4. RppH is – like eukaryotic Dcp2 – a member of the class of Nudix phosphohydrolases, which are characterized by a signature Nudix motif10,11. Nudix hydrolases catalyze the hydrolysis of nucleoside diphosphates linked to different moieties and are known to hydrolyze nucleoside triphosphates, nucleotide sugars, dinucleoside polyphosphates, dinucleotide coenzymes and capped RNAs12. The Nudix motif in these enzymes functions as a versatile Mg2+-binding and catalytic site and forms a loop-helix-loop structure. The precise catalytic mechanisms of different Nudix hydrolases vary, typically requiring two or three conserved glutamate residues to coordinate a divalent metal ion crucial for catalysis13.
E. coli Nudix hydrolase NudC efficiently removes the NAD cap by hydrolyzing the pyrophosphate bond to produce nicotinamide mononucleotide (NMN) and 5’-monophosphate RNA4. NudC has been previously described as a NAD(H) pyrophosphohydrolase14, but any involvement in RNA processing was unknown. NAD capping may endow the bacterium with an additional RNA protection / degradation pathway that is orthogonal to the RppH / triphosphate RNA combination4.
Intrigued by this discovery of capping and decapping in prokaryotes and its far-reaching implications, we now set out to understand the molecular basis underlying NAD-RNA processing by NudC, as well as the properties of this bacterial decapping enzyme, by combining crystallographic and biochemical investigations. On the basis of two high-resolution crystal structures of a NudC/NAD substrate complex and a NudC/NMN product complex, which defined key residues lining the binding pocket in substrate and product states, we carried out extensive protein and RNA mutagenesis and analyzed the processing of NAD-capped E. coli RNAI and 5S rRNA. We found NudC to be a single-strand-specific RNA decapping enzyme with a strong preference for a purine as first nucleotide. In in vitro competition experiments, NudC preferred NAD-RNA over NAD(H) by several orders of magnitude, suggesting that NAD-RNA may be its primary biological substrate. NudC bound a diverse population of cellular RNAs in an unspecific, most likely electrostatic manner.
RESULTS
Structure of NudC in complex with NAD
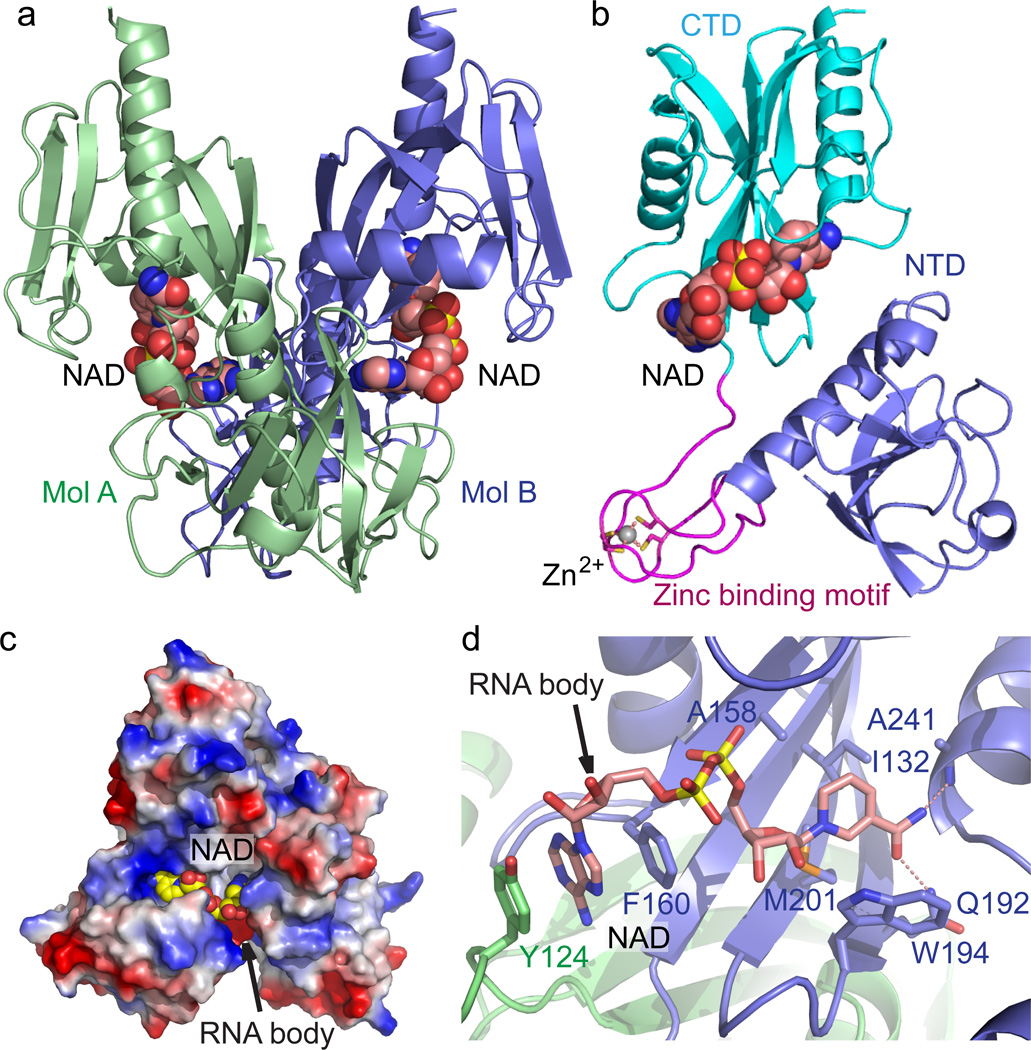
To investigate the molecular mechanism of the NudC dependent bacterial RNA decapping reaction, we carried out structural studies on E. coli NudC. Given the free form NudC structure is available in the PDB (PDB: 1VK6, 2GB5), we focused on the substrate- and product- bound form structures under Mg2+-containing conditions. Although we were unable to grow crystals of NudC in complex with NAD-capped RNA, we successfully obtained crystals of NudC in complex with NAD, which mimics the head group moiety of NudC substrate, NAD-RNA. The structure was determined with the molecular replacement method and refined to 2.6 Å resolution to represent its substrate bound form conformation (Fig. 1a and x-ray statistics in Supplementary Results, Supplementary Table 1). Two NudC-NAD complexes in the asymmetric unit form a dimer (Fig. 1a), which resembles the free-form NudC structure (PDB: 1VK6, 2GB5). The structure of each of the NudC monomers is composed of two tandem Nudix-fold domains, labeled N-terminal domain (NTD) and C-terminal domain (CTD) (Fig. 1b). Between these two domains, four Cys residues coordinate a Zn2+ ion to form an extended zinc-binding motif, which protrudes out from the NTD (Fig. 1b).
Figure 1. Crystal structure E. coli NudC in complex with bound NAD.
(a) Overall structure of the E. coli NudC dimer in complex with NAD. The two monomers of NudC (Mol A and Mol B) are colored in slate and light green, respectively. The bound NAD molecules are shown in space filling representation. (b) A detailed structural representation of a monomer of NudC-NAD complex. The NTD, zinc-binding motif, and CTD are colored in slate, magenta, and cyan, respectively. Zinc ion coordination is highlighted with dashed red lines. (c) An electrostatic surface view of the NAD binding pocket of NudC. The postulated NAD-RNA extension channel is highlighted with the arrow. In addition, a positively-charged surface patch locates at the entrance of the NAD binding pocket, suggesting a plausible role in RNA binding. (d) The specific recognition of NAD by NudC. The residues involved in NAD binding are shown in a stick representation. The hydrogen bonding interactions are shown as dashed red lines. The arrow indicates the postulated position of RNA body extension.
The structure of the overall complex is composed of a NudC dimer with each monomer containing a bound NAD ligand shown in a space-filling representation (Fig. 1a). The bound NAD molecules are positioned at the dimer interface with well-defined electron density (Supplementary Fig. 1a). The nicotinamide moiety of NAD is buried in a hydrophobic binding pocket of one monomer, with a directionality of the pyridine ring towards the bottom of this pocket and the phosphate group positioned near the entrance of the pocket (Fig. 1c,d). The AMP moiety of NAD extends into another hydrophobic pocket formed at the dimer interface, with a directionality of the adenine ring towards the bottom of this pocket and the ribose and phosphate group near the entrance (Fig. 1c,d). It is noteworthy that there are several positively charged residues clustered outside the entrance of the NAD-binding pocket (Fig. 1c), suggestive of a plausible pathway for specific recognition and anchoring of the sugar-phosphate backbone of the NAD-modified RNA. Several hydrophobic residues, including Ala158, Ile132, Met201, Trp194, Thr239 and Val240 contribute in forming a hydrophobic pocket to accommodate the hydrophobic pyridine ring (Fig. 1d). In addition to the hydrophobic interactions, Ala 241 and Gln192 form a hydrogen bond with the amide and carbonyl groups of the nicotinamide moiety, respectively (Fig. 1d). The adenine ring of NAD is stacked between the side chains of Phe160 from one NudC molecule and Tyr124 from the partner molecule in the dimer, revealing that dimerization of the NudC is essential for the substrate binding and recognition (Fig. 1d).
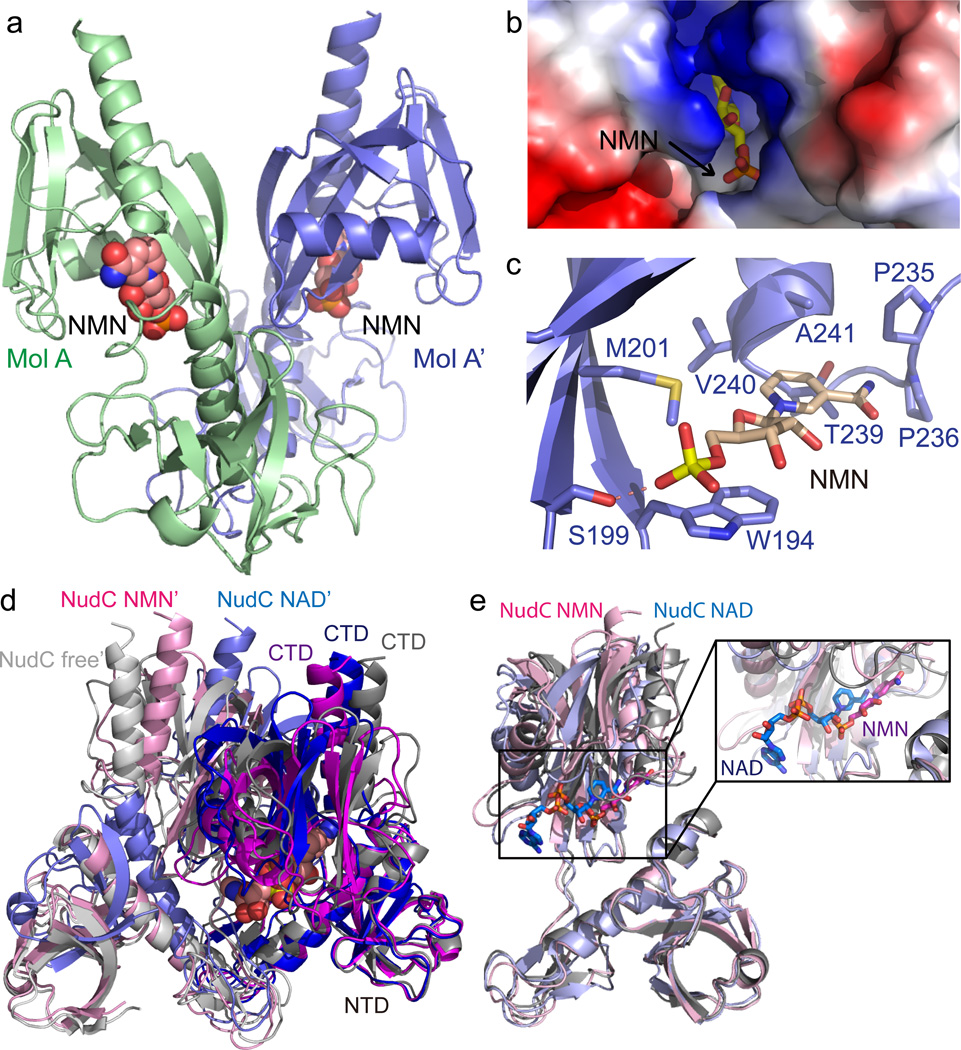
Structure of NudC in complex with NMN
In addition to the enzyme-substrate complex, we further determined the structure of NudC in complex with its cleavage product NMN at 2.7 Å resolution. Each NudC-NMN complex can form a biologically relevant symmetric dimer via the 2-fold crystallographic axis (Fig. 2a), resulting in a similar dimerization as in structures of the NudC-NAD complex (Fig. 1a) and free form NudC. The NMN molecule exhibits good electron density (Supplementary Fig. 1b), possesses an extended conformation and is deeply buried in a hydrophobic binding pocket between the NTD and CTD, with the pyridine ring directed towards the bottom of this pocket and the phosphate group positioned near the entrance of the pocket (Fig. 2b,c). Several hydrophobic residues, including Trp193, Met201, Pro235, Pro236, Thr239, Val240, and Ala241 of the CTD contribute to formation of the hydrophobic NMN-binding pocket (Fig. 2c) that accommodates both the hydrophobic pyridine ring and the sugar ring of bound NMN. Notably, the indole ring of Trp194 stacks with the sugar ring of NMN, thereby stabilizing the bound NMN (Fig. 2c). In addition to the hydrophobic interactions, Ser199 which is positioned near the entrance of the ligand-binding pocket, forms a hydrogen bond with the phosphate of NMN (Fig. 2c). Compared with the NAD position in the NudC-NAD complex, the pyridine ring of NMN makes a 90° turn in the NudC-NMN complex (Fig. 2d and zoomed in Fig. 2e). Notably, NMN is lifted away from the C-domain and the Nudix helix in the NudC-NMN complex compared to the position that NAD takes in the NudC-NAD complex, consistent with the product moving away from the catalytic center. At the same time, the N-terminal domain appears to close such that the bound NMN appears more buried than NAD, a feature that would prevent product release. These alignments suggest a plausible mechanism whereby the NMN binding pocket interacts with the NMN moiety of NAD and potentially facilitates positioning of the pyrophosphate in a conformation required for cleavage. Compared with the ligand-free structure of NudC (PDB code: 1VK6) and the NAD bound structure, the conformation of each monomer and the relative orientation between two monomers in the NMN bound structure undergo significant changes (Fig. 2e). When we superpose the NTDs of one monomer of NAD-bound, NMN-bound and ligand-free forms of NudC molecules, the CTDs exhibit significant orientation changes (Fig. 2e). Moreover, the relative orientation between the two monomers gradually moves a little closer upon binding NAD (Fig. 2d), indicating that the enzymatic activity process is not only influenced by the conformation of each monomer, but also involves dimeric arrangement movement.
Figure 2. Crystal structure of E. coli NudC in complex with bound NMN.
(a) Overall structure of the E. coli NudC-NMN complex with the NudC monomer in a light green ribbon (MolA) and its symmetry-related molecule in slate (MolA’). The bound NMN molecules are shown in space-filling representation. (b) The NMN binding pocket of NudC is shown in an electrostatistics surface representation and NMN in a stick representation. (c) Intermolecular interactions between NudC and NMN in the complex. Intermolecular hydrogen-bonding is shown by a dashed red line. (d) A superposition of the NTD of one monomer from the NudC-NAD dimer (blue and light blue), NudC-NMN dimer (magenta and pink), and free-form NudC dimer (gray and silver, PDB: 1VK6) shows a significant conformational change in the CTD on complex formation. In addition, the dimer interfaces exhibit significant orientation changes, indicating that the enzymatic activity not only involves the position of NTD relative to the CTD, but also the arrangement of the two monomers in the dimer. (e) Superposition of NudC-NAD (in light blue) and NudC-NMN (in pink) complexes reveals that the NMN binding site is much deeper in NudC-bound protein compared to the NAD-bound protein. The NAD/NMN binding pocket is highlighted in the expanded box.
The Nudix motif is the catalytic center of NudC
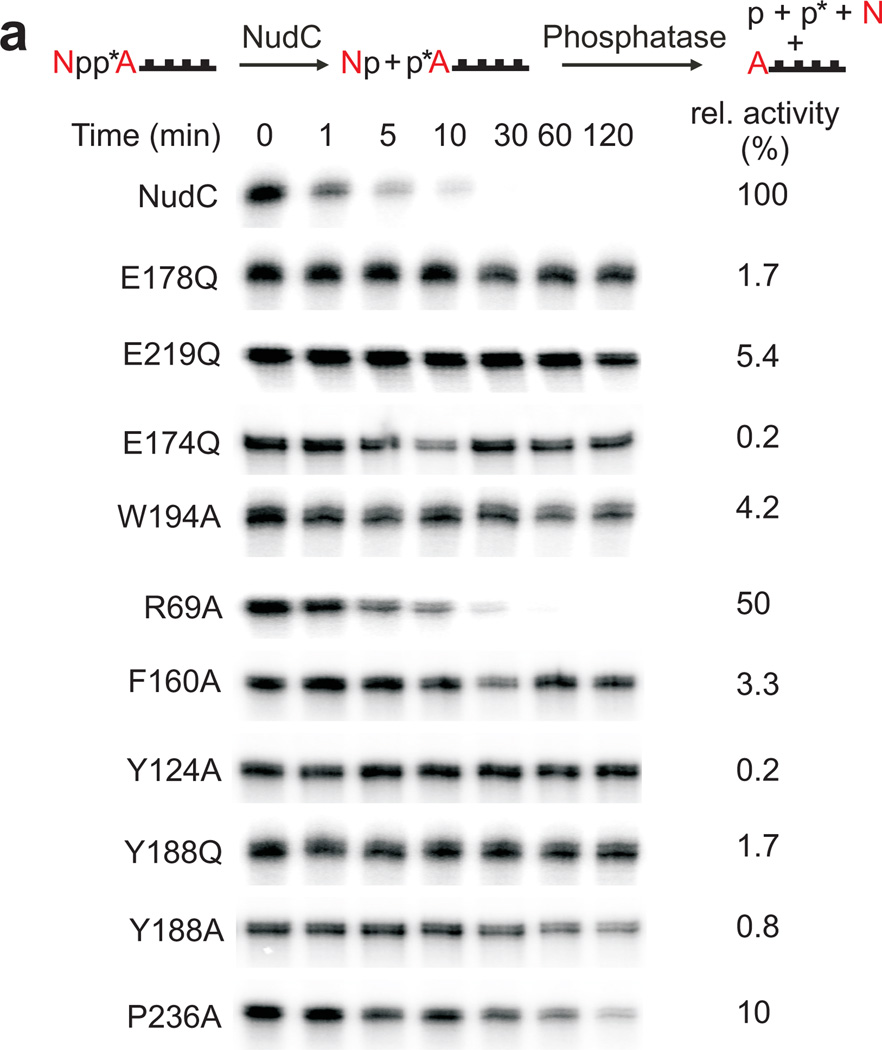
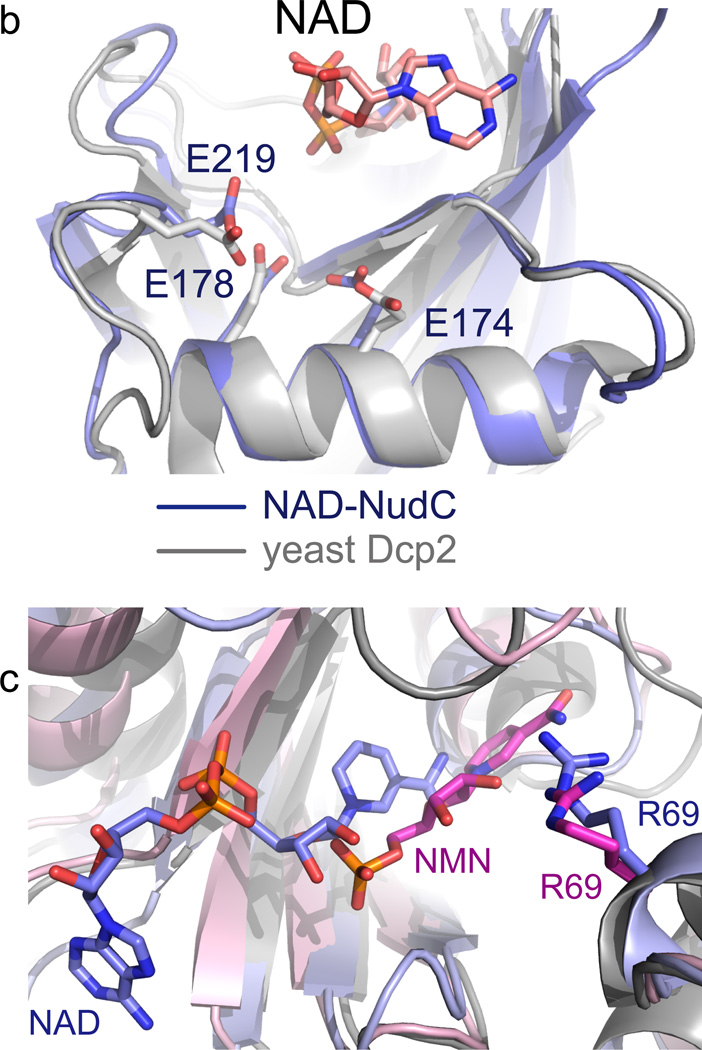
We probed the interactions observed in the crystal structures for functional relevance by activity determinations of NudC point mutants, analyzing the kinetics of hydrolysis of highly pure enzymatically prepared full-length NAD-RNAI (Fig. 3a), a sRNA known to be NAD-capped in E. coli (107 nucleotides, wild-type sequence)4. The canonical Nudix family share a conserved 23-amino acid Nudix motif (Nudix box), Gx5Ex5[UA]xREx2EExGU, where U is an aliphatic, hydrophobic residue. Our structural data showed that the negatively charged residues Glu174, Glu178 and Glu219 of NudC are located near the NAD (Fig. 3b) and are structurally conserved with their counterparts in the well-studied Nudix enzyme yeast Dcp215, which may reflect their involvement in catalytic activity. We found the conserved Nudix motif to be essential for function (Fig. 3a,b). Replacement of a single O atom by NH of central glutamates (E178Q, E219Q and E174Q) abolished activity (Fig. 3b). In addition to the Nudix box residues, some residues involved in NAD binding were also important for enzymatic activity. Trp194 interacted differently with the substrate and the product: With NAD, an imperfect stacking was observed with the nicotinamide moiety (Fig. 1d), while with NMN it interacted with the ribose ring (Fig. 2c). The inactivity of the W194A point mutant confirmed the importance of this residue (Fig. 3a). Arg69 appeared to be the main cause for the change in orientation of the bound NMN product compared to the NAD substrate (Fig. 3c). It was, however, not important for the catalytic mechanism, as indicated by the high activity of the R69A mutant (Fig. 3a).
Figure 3. Mutagenesis of NudC.
(a) Decapping of full-length E. coli NAD-RNAI (NppA = NAD; N: nicotinamide riboside, pp: pyrophosphate, A: adenosine) by NudC and single-mutants. NAD-RNA contains a site-specific 32P label at the pyrophosphate bridge (asterisk) which becomes accessible to excess phosphatase only upon hydrolysis of the pyrophosphate4, causing disappearance of the radioactive RNAI band. Aliquots taken at the indicated time points are separated on denaturing PAGE gels. Experiment was carried out three times. Full denaturing PAGE gels are shown in Supplementary Fig. 8a. (b) The superposition of the Nudix motif of NudC and yeast Dcp2 showing the conserved glutamates. (c) Superposition of NudC-NAD (in light blue) and NudC-NMN (in pink), relative to Arg69.
A most intriguing feature was the observed inter-subunit stacking of the NAD adenine ring between Phe160 from one NudC protomer and Tyr124 from the other (Fig. 1d). The functional role of this stacking interaction was supported by the catalytic inactivity of the F160A and Y124A mutants (Fig. 3a). An attempt to directly demonstrate the inactivity of monomeric NudC remained inconclusive (Fig. 3a), as mutations Y188A and Y188Q, designed to disrupt the aromatic Y188-Y188’ inter-subunit stacking interaction (Supplementary Fig. 2a,b), did not yield monomeric protein, similar to a report on homologous Arabidopsis Nudix hydrolase AtNudt716. Pro236 formed part of the binding pocket but does not interact with the bound NMN molecule in the crystal structure of the complex, consistent with this residue not being essential for catalytic activity (P236A mutant, 10% activity, Fig. 2c and 3a).
NudC is single-strand specific and prefers a 5’-purine
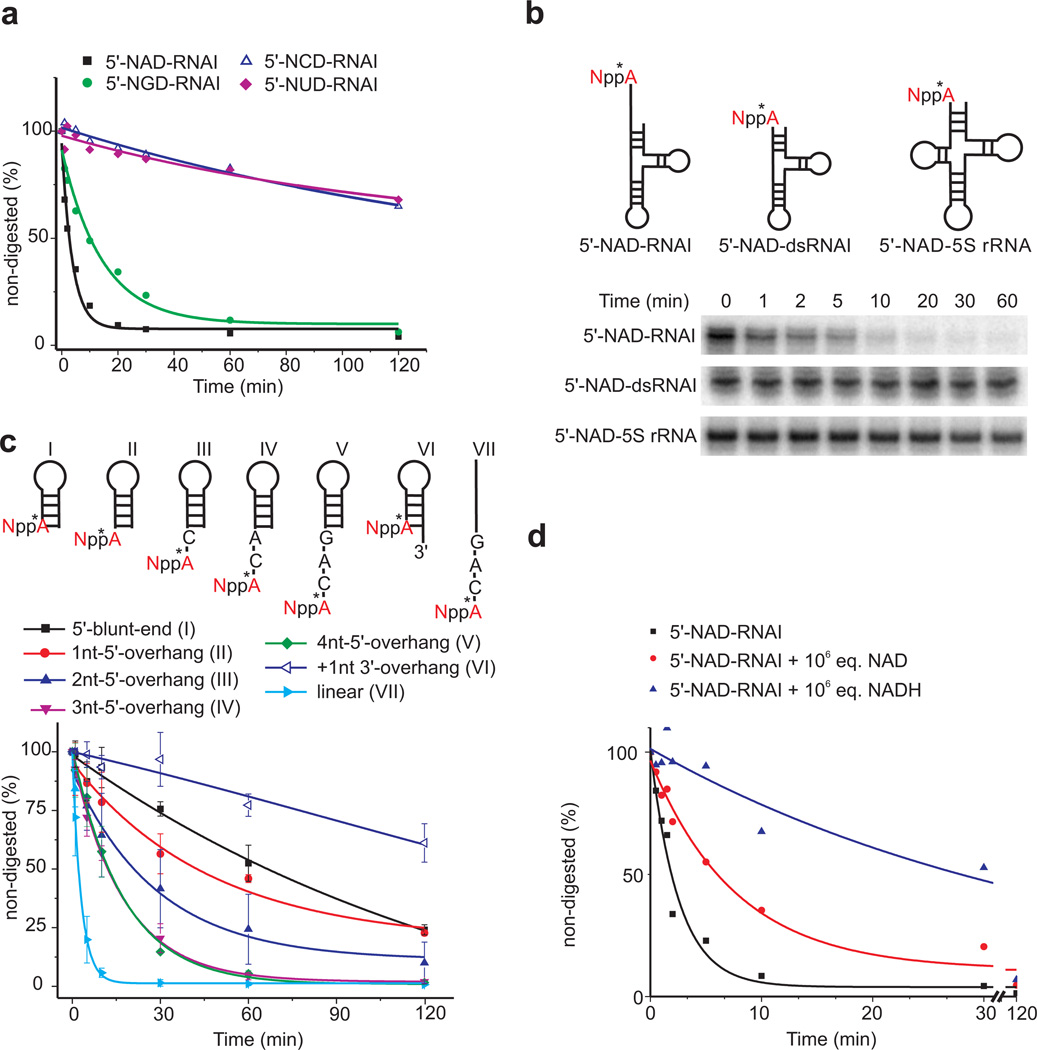
The F160-NAD adenine-Y124 intercalative interaction suggested a mechanism for the recognition of the first RNA base, which was adenosine in all RNAs enriched in NAD captureSeq4, namely by stacking with the aromatic side chains of Phe160 and Tyr124. To test this hypothesis, we prepared NAD-RNAI variants in which the NAD adenosine was replaced by guanosine (NGD), cytidine (NCD), and uridine (NUD). While the rate of decapping by NudC wild-type enzyme was only 3-fold reduced for NGD-RNAI, the pyrimidine substitutions exhibited 50-fold reduced enzymatic processing, revealing a strong purine preference of the enzyme for the nucleotide at the 5’-terminal position (Fig. 4a, Supplementary Fig. 3a).
Figure 4. RNA mutagenesis.
(a) Decapping kinetics of full-length E. coli NXD-RNAI depending on the nature of the 5’-terminal nucleotide. Data points represent mean ± standard deviation (s.d.), n=2. (b) Processing of native and a 5’-double-stranded mutant of E. coli RNAI, and of E. coli 5S rRNA. Full denaturing PAGE gels are shown in Supplementary Fig. 8b. (c) Analysis of the role of RNA secondary structure on NudC decapping. Assay as in Fig. 3a. For linear RNA (VII), NudC concentration was reduced 10-fold due to very fast decapping. Data points represent mean ± standard deviation (s.d.), n=3. (d) Decapping of 5 nM NAD-RNAI in the presence of 5 mM NAD+ or NADH. Assay as in Fig. 3a, except for the following differences: 0.5 µM total RNAI concentration, containing ~1% NAD-RNAI, was incubated with 5 mM NAD+ or NADH.
The narrow dimensions of the cleft above the adenosine ribose observed in the NudC-NAD co-crystal (7 Å width above the adenosine 3’-hydroxyl group, Supplementary Fig. 3b) led us to assume that it may only be accessible to NAD-RNAs with single-stranded ends, such as RNAI which has 8 unpaired 5’-nucleotides. In support of this assumption, an RNAI mutant with a paired 5’-end, as well as native E. coli 5S rRNA (also possessing a paired 5’-end) were resistant against NudC processing (Fig. 4b). To more thoroughly study the role of RNA secondary structure on NudC degradation, a series of short NAD-capped RNA hairpins with or without 3’- or 5’-overhangs as well as a linear 8-mer were analyzed (Fig. 4c, Supplementary Fig. 4a), confirming a strong preference for RNAs with at least 3 unpaired nt at their 5’-ends.
The pH-activity profile revealed – in agreement with data reported for other Nudix hydrolases15 – an optimum at 8 (Supplementary Fig. 4b), consistent with the use of a general base with near-neutral pKa to efficiently abstract a proton from water during catalysis. In Dcp2, E153 (equivalent to E178 in NudC) is known to serve this purpose15.
NAD-RNA is preferred as substrate over NAD in vitro
The published catalytic parameters14 indicate that NudC is not very efficient in hydrolyzing NAD. We therefore speculated that the primary biological substrate of NudC might be NAD-RNA, rather than NAD. To test this hypothesis in vitro, we analyzed the rate of NAD-RNA processing in the presence of an excess of NAD(H) (Fig. 4d, Supplementary Fig. 4c). Even a 1 million-fold excess (5 mM) slowed NAD-RNA processing only by a factor of 4 (NAD) or 20 (NADH), while under these conditions only a much smaller fraction of NAD (170-fold lower) or NADH (5-fold lower) was hydrolyzed per minute (Supplementary Fig. 4d), demonstrating the enzyme’s substrate preference in direct competition. The difference between the effects of NAD and NADH agreed with the reported affinities of the enzyme for these cofactors (KM, NAD+: 5.1 mM, KM, NADH: 0.11 mM)14. Structurally divergent E. coli Nudix hydrolase NudE17, by contrast, hydrolyzed NAD rapidly, but was inactive on NAD-RNAI (Supplementary Fig. 4e,f).
NudC is an unspecific RNA-binding protein
As already noted in 1995, NudC co-purifies with a population of tightly bound nucleic acids14. Isolation of this population and nuclease treatment revealed that these are various RNAs of different sizes (Supplementary Fig. 5a). NGS sequence analysis of the RNA bound to NudC identified a highly diverse population very similar in composition to a total RNA sample isolated from the same E. coli strain with very little enrichment or depletion of specific sequences (Supplementary Fig. 5b). RNAs bound to the catalytically inactive E178Q mutant were virtually indistinguishable from those bound to wild-type in amount, size, and sequence distribution (Supplementary Fig. 5a,b), indicating that RNA binding is not functionally connected to the decapping activity. However, the observation that a highly purified RNA-free NudC preparation prepared by chromatography in the presence of 1 M urea had a higher NAD-hydrolase activity than a crude RNA-containing NudC preparation (Supplementary Fig. 5c) suggests that RNA binding may interfere with NAD binding. We speculate that – although they do not constitute a known RNA binding motif – the extended positively charged surface around the entrance to the catalytic center (Fig. 1c) could potentially mediate RNA binding in a primarily electrostatic manner.
DISCUSSION
The crystallographic and biochemical data presented here support the role of NudC as a bacterial decapping enzyme. A wide variety of NAD-RNAs are processed by NudC, from highly structured long native RNAs with a single-stranded 5’-end down to short linear oligonucleotides. The nicotinamide riboside moiety and the 5’-terminal RNA nucleotide (typically adenosine) are specifically bound inside the catalytic pocket via stacking and hydrogen-bonding interactions. While most of the catalytic pocket is formed by one monomer, the other monomer contributes a tyrosine that is critical for binding the adenine of NAD. By contrast to the first nucleotide, it is likely that positioning further nucleotides takes place outside the catalytic pocket on the protein surface. The comparable rates by which RNAs of different sizes are processed suggest that only a small number of nucleotides potentially interact with NudC. Such an electrostatic mode has been proposed for the interaction of the eukaryotic decapping machinery with RNA substrates18,19 and would also partially explain our inability to date in obtaining diffracting co-crystals with NAD-RNAs of various sizes and sequences.
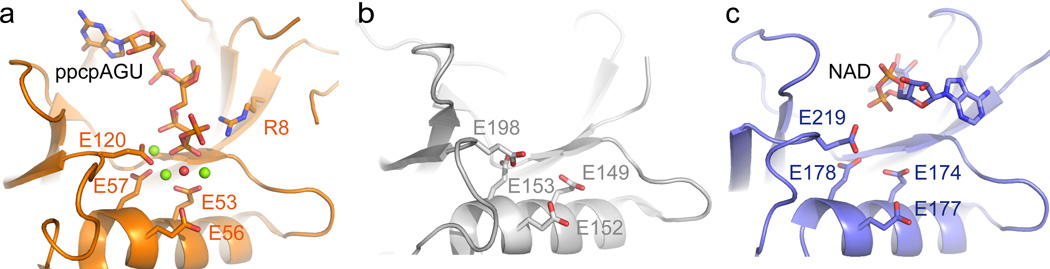
The catalytic activity undoubtedly resides in the Nudix domain and depends on conserved glutamate residues. In the bacterial Nudix family RNA pyrophosphohydrolase RppH, four glutamate residues can coordinate for interaction with three Mg2+ ions to drive a water molecule to attack a phosphorus atom which triggers hydrolysis of the pyrophosphate bond (Fig. 5a). An arginine residue interacts with the phosphate group and assists in stabilizing the leaving group20. In the eukaryotic decapping enzyme Dcp2, the four glutamate residues are present in a very similar arrangement, while the arginine residue is missing, indicating a different strategy for leaving group stabilization15 (Fig. 5b). Our NudC structure (Fig. 5c) is more similar to the latter case. In the structure of RppH in complex with Mg2+ ions and its substrate ppcpAGU RNA, the conserved glutamate residues locate near the Mg2+ ions, the important water molecule, and the substrate RNA. This conformation leads to in-line attack of phosphorus atom to trigger the reaction.20 In contrast, in our NudC-NAD complex structure, the NMN and AMP moieties of NAD are anchored in the protein (Fig. 1d) in an orientation that places the pyrophosphate bond to be cleaved quite distant from the conserved glutamates (Fig. 5c), suggesting that our NudC-NAD complex structure may represent a pre-reaction state. It appears that an additional conformational change will have to occur to get the pyrophosphate bond to approach the four glutamate region and further facilitate the enzymatic reaction.
Figure 5. Comparison of the active site conformation in decapping enzymes.
(a) The active site of RppH in complex with Mg2+ and ppcpAGU RNA (PDB: 4S2Y). RppH and substrate RNA are shown in golden ribbon and stick model. The Mg2+ ions and water molecule are shown as green and red balls. Conserved glutamate residues and the arginine residue are highlighted in stick model. The substrate RNA, the Mg2+ ions and the water molecule located close to catalytic glutamate residues (b) The active site of yeast decapping enzyme Dcp2 is shown in silver (PDB: 4K6E). The four conserved glutamate residues are highlighted in stick model. (c) The active site of NudC bound to substrate NAD is shown in slate ribbon and stick. The four conserved glutamate residues are highlighted in stick model. The pyrophosphate bond of NAD to be cleaved locates distant from the four glutamate region, indicating a pre-reaction state. We did not observe a stable bound Mg2+ ion in the NudC catalytic pocket despite the growth of the crystals in a Mg2+-containing buffer solution.
For many Nudix enzymes, the biological role is unclear. They were proposed to act as “housecleaning enzymes”, which can cleanse the cell of potentially deleterious endogenous metabolites and modulate the accumulation of biochemical intermediates21. The current study reveals that for NudC, the cleavage of NAD-capped RNA is carried out more efficiently than known small-molecule substrates NAD+ and NADH. Considering that E. coli has 13 Nudix enzymes, many of which with unclear function, it is conceivable that these might serve to process RNAs capped with other non-standard cap moieties, such as nucleotide sugars, dinucleoside polyphosphates, or other dinucleotide coenzymes5.
Capping and decapping have so far been considered exclusively eukaryotic phenomena. The discovery of NAD capping and the identification of NudC as the decapping enzyme suggest new pathways and strategies for RNA processing and decay in bacteria. NAD as a redox-active moiety may link the redox state to RNA processing. NudC may interact with other proteins (similar to the recently observed RppH-DapF interaction22) to trigger RNase E-dependent RNA decay in a NAD-dependent manner. The further exploration of these interactions in prokaryotes will reveal functional roles of NAD caps in bacterial physiology.
ONLINE METHODS
Cloning, expression and purification of NudC for crystallization
The E. coli NudC gene was inserted into the pET-Sumo expression vector (Invitrogen) which fuses a hexa-His tag plus a yeast sumo tag at the N-terminus. The plasmid was transformed into E. coli BL21 (DE3) strain (Stratagene). The cells were cultured at 37 °C until OD600 reached 0.8, and then the protein expression was induced with 0.2 mM IPTG at 18 °C overnight. The hexa-His-Sumo tagged protein was purified using a HisTrap FF column (GE Healthcare). The tag was cleaved by Ulp1 protease and further removed by a second step HisTrap FF column (GE Healthcare) purification. The target protein was further purified by a Hiload Superdex G200 16/60 column (GE Healthcare). The purified protein was of high purity (above 95%) as shown by SDS-PAGE.
Crystallization and diffraction data collection
The purified protein was concentrated to about 10 mg/ml in a storage buffer of 150 mM NaCl, 20 mM Tris, pH 7.5, 2 mM MgCl2, and 5 mM DTT. β–Nicotinamide mononucleotide (NMN) (Sigma) or nicotinamide adenine dinucleotide (NAD) was added with a fivefold excess molar ratio to NudC and incubated at 4 °C for 30 min. Crystallization was conducted at 20 °C using the hanging drop vapor diffusion method by mixing 1 µL protein solution and 1 µL reservoir solution. The NudC-NAD complex crystals were grown in a condition of 0.2 M sodium nitrate, 20% PEG3350 and the NudC-NMN complex crystals were grown in a condition of 1 M LiCl2, 0.1 M HEPES, pH 7.0, and 10% PEG6000. The crystals were soaked into the reservoir solution supplemented with 15% glycerol before flash-cooled into liquid nitrogen for diffraction data collection. The diffraction data were collected at 100 K with a wavelength of 1.075 Å at beamline X29A of Brookhaven National Laboratory, New York, and processed using the HKL200023. The statistics of the diffraction data are summarized in Supplementary Table 1.
Structure determination and refinement
The structure of NudC in complex with NAD was solved with the molecular replacement method as implemented in Phaser24 using the structure of apo NudC as the search model (PDB ID: 1VK6). In the initial difference Fourier maps, there was electron density for a bound NAD. The model building was carried out using Coot25 and the structure refinement was conducted using Phenix26. Throughout the refinement, a free R factor was calculated using 5% randomly chosen reflections. The stereochemistry of the structure model was analyzed using Procheck27 and MolProbity28. The NudC-NMN complex structure was determined using the same protocol. The final structural models are of good geometry with Ramanchandran analysis of 97% in favored and 3% in allowed regions for NudC-NAD complex, 94% in favored and 6% in allowed regions as calculated by MolProbity, respectively. The statistics of the refinement and the structure model are summarized in Supplementary Table 1. All the molecular graphics were generated using Pymol (DeLano Scientific LLC).
Plasmids and E. coli strains for functional analysis
The nudC and nudE gene were PCR-amplified from genomic DNA of E. coli K-12 (isolated via GenElute™ Bacterial Genomic DNA Kit, Sigma-Aldrich). XbaI/NotI (nudC) or XhoI/NcoI (nudE) restriction sites were introduced during amplification using the primers listed in Supplementary Table 2. The resulting PCR products were digested with XbaI/NotI or XhoI/NcoI and cloned into pET-28c vector (Merck Millipore). After Sanger sequencing, the resulting plasmids were transformed into E. coli One Shot BL21 Star (DE3) (Life Technologies). The NudC mutants were generated by site-directed mutagenesis as described4.
Purification and analytical characterization of NudC, NudC Mutants and NudE
E. coli One Shot BL21 Star (DE3) containing the respective plasmid were induced at OD600=0.8 with 1 mM IPTG for 3 h. Purification was performed as described4. All proteins yielded a single band on SDS page (Coomassie staining, Supplementary Fig. 6a). For preparation of RNA-free pure NudC, 1 M urea was added to His-Trap buffer A and His-Trap buffer B. To ensure dimer formation, all NudC variants were analyzed by size exclusion chromatography on a Superdex 200 10/300 column (50 mM Tris-HCl pH 7.5, 150 mM NaCl, Supplementary Fig. 6b–f). Folding of all mutants was confirmed by CD spectroscopy (J-810 Jasco spectropolarimeter, 0.2 mg/mL protein in 5 mM Tris-HCl pH 7.5, 5 mM MgSO4, 0.5 mM EDTA and 0.5 mM DTT, 1 nm bandwidth, 0.5 nm data pitch, 20 nm min−1 scanning speed, 1 s response time, Supplementary Fig. 7).
Isolation of total RNA and NudC co-purifying RNA
Total RNA from E. coli One Shot BL21 Star (DE3) pET-28c-NudC was isolated using RNAzol (Molecular Research Center). Bacteria (1 L) were grown at 37 °C in LB broth and protein expression induced at OD600=0.8 with 1 mM IPTG. After 3 h, 200 mL cells were lysed by sonication (30 s, 50% power, 5 times) in water. RNAzol (10 ml) was added to the lysate to isolate total RNA according to the supplier′s information. Finally, total RNA was phenol/ether extracted twice and ethanol precipitated. RNA co-purifying with NudC and NudC-E178Q was isolated from IMAC-purified NudC or NudC E178Q. Eluting fractions were pooled and phenol/ether extracted twice and ethanol precipitated. Isolated nucleic acids were digested with DNaseI (Roche) (10U/ 1 µg nucleic acids, 30 min 37°C) or RNase A/T1 mix (Thermo Scientific) (5 µL/1µg nucleic acids, 30 min 37°C). Digests were analysed by denaturing polyacrylamide gel electrophoresis and nucleic acids were visualized by Sybr gold staining (Life technologies) using a Typhoon 9400 imager (GE Healthcare).
cDNA library preparation
cDNA library preparation was performed by vertis Biotechnology AG, Germany. In brief, RNA samples were fragmented with ultrasound (four pulses of 30 s). A N6 randomized primer was used for cDNA synthesis. Subsequently, the Illumina TruSeq sequencing adapters were ligated to the 5' and 3' ends of the cDNA. The cDNA was purified using the Agencourt AMPure XP kit (Beckman Coulter Genomics) and analyzed by capillary electrophoresis. Finally, PCR, employing a high-fidelity DNA polymerase, was performed to amplify the cDNA pool (size range 300–500 bp) and to introduce barcodes for multiplexing of the samples. The primers used for PCR amplification were designed for TruSeq sequencing according to the instructions of Illumina.
The following adapter sequences flank the DNA insert:
TruSeq_Sense_primer
5´- AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3’
TruSeq_Antisense_ NNNNNN_primer (N6 = barcode)
5’-CAAGCAGAAGACGGCATACGAGAT-NNNNNN-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3’.
The samples were sequenced on an Illumina NextSeq500 instrument with a read-depth of 400 mio. reads in single-end read mode.
Analysis of the NGS data
Analysis of the NGS data was performed as previously described4. In brief, sequences were uploaded to the web-based sequence-analysis platform Geneprof29. At first, the reads were filtered according to quality using the Modify and Filter Sequence tool (Modify: cut after low quality 10; filter: cumulative quality score > 300; discard N-reads; sequence length > 12). Furthermore, adapter sequences were clipped (FASTX-Toolkit Clip Adapter Sequences). A custom reference set for Escherichia coli BL21 (DE3) was created using reference data from Ensembl Bacteria FTP30. The pET28-MHL plasmid sequence (GenBank accession EF456735.1) was manually added, whereby sacB was replaced by nudC. The clipped sequences were aligned to the custom reference set with Bowtie31 (v0.12.3) and further analysis conducted using the Quantitate Gene Expression tool. For each sample or control, read numbers as normalized read counts (in RPM) were calculated. Enrichment factors were determined as ratio of read counts of sample versus control. For each data set, relative abundance of different reads in the sample was plotted against relative abundance in the control. This analysis revealed no enriched sequences (≥ 4).
In vitro transcription and preparation of 5’-NAD capped RNA
The templates for in vitro transcriptions using T7 RNA polymerase (own lab stock 1 mg/mL) were amplified by PCR (sequences see Supplementary Table 1). 5’-NAD-RNA was prepared by in vitro transcription with NAD as initiator dinucleotide as described4.
5’-NUD, 5’-NGD or 5’-NCD-RNA were prepared by reaction of nicotinamide riboside phosphoimidazolide with 5’-32P-phophorylated RNA32. Remaining 5’-32P-RNA was removed by XRN-1 digest. To exclude effects of different synthesis procedures, 5’-NAD-RNA used in this experiment was prepared by the same method. RNA was purified by denaturing polyacrylamide gel electrophoresis and radiolabeled reaction products were visualized using storage phosphor screens (GE Healthcare) and a Typhoon 9400 imager (GE Healthcare).
NudC and NudE kinetic study
In vitro NudC cleavage assays were performed in 25 mM Tris-HCl pH 7.5, 50 mM NaCl, 50 mM KCl, 10 mM MgCl2, 1 mM DTT, using 0.5 µM site-specifically radiolabeled 5’-NAD-RNAI and 1 nM NudC / NudC E178Q, purified as described.4 Cleavage reactions were performed in the presence of 0.1 U/µl FastAP thermosensitive alkaline phosphatase (Thermo Scientific) in order to remove the 32P-phosphate that became accessible only after cleavage of the NAD pyrophosphate bond by NudC. pH dependency of NudC was tested in different buffers: buffer pH 6 (125 mM Bis-Tris-methane pH 6, 250 mM NaCl, 250 mM KCl, 50 mM MgCl2), buffer pH 6.5 (125 mM Bis-Tris-methane (6.5), 250 mM NaCl, 250 mM KCl, 50 mM MgCl2), buffer pH 7 (125 mM Tris-HCl pH 7, 250 mM NaCl, 250 mM KCl, 50 mM MgCl2), buffer pH 7.5 (125 mM Tris-HCl pH 7.5, 250 mM NaCl, 250 mM KCl, 50 mM MgCl2), buffer pH 8 (125 mM Tris-HCl pH 8, 250 mM NaCl, 250 mM KCl, 50 mM MgCl2), buffer pH 8.5 (125 mM Bis-Tris-propane pH 8.5, 250 mM NaCl, 250 mM KCl, 50 mM MgCl2), buffer pH 9 (125 mM Bis-Tris-propane pH 9, 250 mM NaCl, 250 mM KCl, 50 mM MgCl2).
Activity determination of NudE on NAD-RNA was performed exactly as for NudC, replacing NudC by the double amount of NudE.
Activity determination of NudC and NudE by thin layer chromatography
Hydrolysis of radioactively labelled NAD (32P, Perkin Elmer) by NudE or NudC was analyzed by TLC using 4:6 1 M ammonium acetate pH 5.0 : ethanol as mobile phase. Enzymatic reactions were stopped by freezing the samples at −20 °C. 1 µL samples were loaded onto TLC Silica Gel 60 F254 plates and run for approximately 6 h. Substrate hydrolysis was visualized using storage phosphor screens (GE Healthcare) and a Typhoon 9400 imager (GE Healthcare).
Supplementary Material
Acknowledgments
We are grateful to Laura Obenauer (Heidelberg University) for assistance with NGS data analysis, Leo Kiss for experimental assistance and to Dr. Wuxian Shi at beamline X29A at the Brookhaven National Laboratory for support in diffraction data collection. A.J. is supported by the German Research (DFG SPP 1784), grant # Ja 794/10-1 and by the Baden Württemberg Stiftung (BWST_NCRNA_045). The structural research was supported by funds from NIH grant GM104962 to D.J.P. and the Thousand Young Talent Program of China and the Chinese Academy of Sciences to J.D.
Footnotes
Accession codes
The structure of E. coli NudC in complex with NAD and NMN have been deposited in the Protein Data Bank under the accession codes 5IW4 and 5IW5, respectively.
Author Contributions
K.H., S.L., D.J.P. and A.J. designed research, K.H., S.L., F.A., J.F., J.S., J.G., J.D. performed research, whereby S.L. and J.D. performed the crystallographic investigation, and K.H., F.A., J.F., J.S., J.G., carried out the functional studies on NudC, all authors analyzed data, K.H., S.L., J.F., J.D., D.J.P. and A.J. wrote the paper.
Competing financial interests
The authors declare no competing financial interests.
References for main text
- 1.Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ. Cap and cap-binding proteins in the control of gene expression. Wiley interdisciplinary reviews. RNA. 2011;2:277–298. doi: 10.1002/wrna.52. [DOI] [PubMed] [Google Scholar]
- 2.Arribas-Layton M, Wu D, Lykke-Andersen J, Song H. Structural and functional control of the eukaryotic mRNA decapping machinery. Biochim. Biophys. Acta. 2013;1829:580–589. doi: 10.1016/j.bbagrm.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen YG, Kowtoniuk WE, Agarwal I, Shen Y, Liu DR. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat. Chem. Biol. 2009;5:879–881. doi: 10.1038/nchembio.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cahová H, Winz ML, Höfer K, Nübel G, Jäschke A. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature. 2015;519:374–377. doi: 10.1038/nature14020. [DOI] [PubMed] [Google Scholar]
- 5.Jäschke A, Höfer K, Nübel G, Frindert J. Cap-like structures in bacterial RNA and epitranscriptomic modification. Curr. Opin. Microbiol. 2016;30:44–49. doi: 10.1016/j.mib.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Luciano DJ, Belasco JG. NAD in RNA: unconventional headgear. Trends Biochem. Sci. 2015;40:245–247. doi: 10.1016/j.tibs.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marbaniang CN, Vogel J. Emerging roles of RNA modifications in bacteria. Curr. Opin. Microbiol. 2016;30:50–57. doi: 10.1016/j.mib.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Mackie GA. Ribonuclease E is a 5 '-end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- 9.Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5' pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 10.McLennan AG. The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 2006;63:123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLennan AG. Substrate ambiguity among the nudix hydrolases: biologically significant, evolutionary remnant, or both? Cell. Mol. Life Sci. 2013;70:373–385. doi: 10.1007/s00018-012-1210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song MG, Bail S, Kiledjian M. Multiple Nudix family proteins possess mRNA decapping activity. RNA. 2013;19:390–399. doi: 10.1261/rna.037309.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.She M, et al. Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe. Nat. Struct. Mol. Biol. 2006;13:63–70. doi: 10.1038/nsmb1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frick DN, Bessman MJ. Cloning, purification, and properties of a novel NADH pyrophosphatase. Evidence for a nucleotide pyrophosphatase catalytic domain in MutT-like enzymes. J. Biol. Chem. 1995;270:1529–1534. doi: 10.1074/jbc.270.4.1529. [DOI] [PubMed] [Google Scholar]
- 15.Aglietti RA, Floor SN, McClendon CL, Jacobson MP, Gross JD. Active site conformational dynamics are coupled to catalysis in the mRNA decapping enzyme Dcp2. Structure. 2013;21:1571–1580. doi: 10.1016/j.str.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olejnik K, et al. Mutational analysis of the AtNUDT7 Nudix hydrolase from Arabidopsis thaliana reveals residues required for protein quaternary structure formation and activity. Acta Biochim. Pol. 2009;56:291–300. [PubMed] [Google Scholar]
- 17.O'Handley SF, Frick DN, Dunn CA, Bessman MJ. Orf186 represents a new member of the Nudix hydrolases, active on adenosine(5')triphospho(5')adenosine, ADP-ribose, and NADH. J. Biol. Chem. 1998;273:3192–3197. doi: 10.1074/jbc.273.6.3192. [DOI] [PubMed] [Google Scholar]
- 18.Deshmukh MV, et al. mRNA decapping is promoted by an RNA-binding channel in Dcp2. Mol. Cell. 2008;29:324–336. doi: 10.1016/j.molcel.2007.11.027. [DOI] [PubMed] [Google Scholar]
- 19.She M, et al. Structural basis of dcp2 recognition and activation by dcp1. Mol. Cell. 2008;29:337–349. doi: 10.1016/j.molcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vasilyev N, Serganov A. Structures of RNA complexes with the Escherichia coli RNA pyrophosphohydrolase RppH unveil the basis for specific 5'-end-dependent mRNA decay. J. Biol. Chem. 2015;290:9487–9499. doi: 10.1074/jbc.M114.634824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bessman MJ, Frick DN, O'Handley SF. The MutT proteins or "Nudix" hydrolases, a family of versatile, widely distributed, "housecleaning" enzymes. J. Biol. Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 22.Lee CR, Kim M, Park YH, Kim YR, Seok YJ. RppH-dependent pyrophosphohydrolysis of mRNAs is regulated by direct interaction with DapF in Escherichia coli. Nucleic Acids Res. 2014;42:12746–12757. doi: 10.1093/nar/gku926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 23.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 28.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halbritter F, Vaidya HJ, Tomlinson SR. GeneProf: analysis of high-throughput sequencing experiments. Nat. Methods. 2011;9:7–8. doi: 10.1038/nmeth.1809. [DOI] [PubMed] [Google Scholar]
- 30.Kersey PJ, et al. Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res. 2016;44:D574–D580. doi: 10.1093/nar/gkv1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Höfer K, Abele F, Schlotthauer J, Jäschke A. Synthesis of 5'-NAD-Capped RNA. Bioconjug. Chem. 2016;27:874–877. doi: 10.1021/acs.bioconjchem.6b00072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.