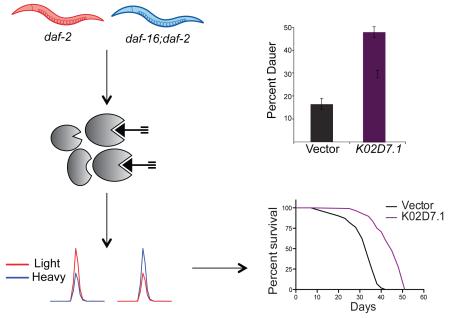
Summary
In the nematode, Caenorhabditis elegans, inactivating mutations in the insulin/IGF-1 receptor, DAF-2, result in a 2-fold increase in lifespan mediated by DAF-16, a FOXO-family transcription factor. Downstream protein activities that directly regulate longevity during impaired Insulin/IGF-1 signaling (IIS) are poorly characterized. Here, we use global cysteine-reactivity profiling to identify protein-activity changes during impaired IIS. Upon confirming that cysteine reactivity is a good predictor of functionality in C. elegans, we profiled cysteine-reactivity changes between daf-2 and daf-16;daf-2 mutants, and identified 40 proteins that display a >2-fold change. Subsequent RNAi-mediated knockdown studies revealed that lbp-3 and K02D7.1 knockdown caused significant increases in lifespan and dauer formation. The proteins encoded by these two genes, LBP-3 and K02D7.1, are implicated in intracellular fatty-acid transport and purine metabolism, respectively. These studies demonstrate that cysteine-reactivity profiling can be complementary to abundance-based transcriptomic and proteomic studies, serving to identify uncharacterized mediators of C. elegans longevity.
Graphical abstract
Introduction
The insulin/insulin-like growth factor (IGF) signaling (IIS) pathway has been shown to modulate lifespan across phylogeny. The life-extending properties of impaired IIS have been observed in organisms including Caenorhabditis elegans (C. elegans) (Kenyon et al., 1993), Drosophila melanogaster (Clancy et al., 2001; Tatar et al., 2001) and Saccharomyces cerevisiae (Kaeberlein et al., 2005). Furthermore, studies in humans revealed that genetic variations within components of the IIS pathway, in particular FOXO3A, were strongly associated with longevity (Anselmi et al., 2009; Flachsbart et al., 2009; Li et al., 2009; Willcox et al., 2008), implicating the IIS pathway as a common mediator of lifespan in both invertebrates and vertebrates.
C. elegans provides an ideal model organism for investigating the mechanistic basis of lifespan regulation due to their short and relatively invariant lifespan that enables rapid evaluation of genetic and environmental factors that affect longevity (Olsen et al., 2006). In C. elegans, the IIS pathway is initiated by activation of the single insulin/IGF-1-like tyrosine kinase receptor, DAF-2, which results in phosphorylation and activation of a series of downstream protein kinases including phosphoinositide 3-kinase (AGE-1), 3-phosphoinositide dependent protein kinase-1 (PDK-1), and AKT/protein kinase B (PKB). AKT/PKB phosphorylation ultimately results in inactivation of a forkhead box O (FOXO) transcription factor, DAF-16 (Mukhopadhyay et al., 2006). Inactivation of DAF-2, or other kinases in the IIS pathway, result in DAF-16 translocation to the nucleus where DAF-16 targets genes that mediate IIS-regulated processes including longevity, fat storage and dauer formation (Jensen et al., 2006; Oh et al., 2006; Ookuma et al., 2003). Dauer formation is a well-characterized alternative developmental pathway triggered by unfavorable growth conditions. Growth-arrested dauer larva exhibits increased stress resistance, altered metabolism, increased longevity, and has the ability to survive without food for several months (Riddle et al., 1997). Many of the components of the IIS pathway were originally identified in genetic screens for genes that modulate dauer formation (Kenyon et al., 1993). Therefore, reduction of function mutations in genes in the IIS pathway, such as daf-16 and daf-2, also modulate dauer diapause.
Inactivating mutations in the C. elegans daf-2 gene results in a 2.3-fold increase in lifespan compared to wild-type (WT) animals (Kenyon et al., 1993). All daf-2 phenotypes are suppressed when in combination with a reduction of function mutation in daf-16. Therefore, DAF-16 activity is critical to the observed longevity and daf-16;daf-2 double mutants are devoid of the long-lived phenotype (Kenyon et al., 1993). Downstream transcriptional targets of DAF-16 have been investigated using global transcriptomic and proteomic methods. Targeted transcriptomic studies have focused on genes involved in stress resistance due to the increased resistance to oxidative stress, heat, and heavy metals (Barsyte et al., 2001) that is characteristic of daf-2 mutants. These targeted studies identified the upregulation of transcripts for superoxide dismutase (sod-3), metallothionein (mtl-1), and heat-shock proteins (hsp-16) (Barsyte et al., 2001; Honda and Honda, 1999; Walker et al., 2001). Global transcriptomic analysis identified upregulated genes that encode proteins involved in metabolism, steroid and lipid synthesis, and dauer formation. Downregulated genes were involved in translation elongation, protein degradation, apolipoproteins, and peptide and lipid transport (Jensen et al., 2006; Lamitina and Strange, 2005; Murphy et al., 2003; Oh et al., 2006; Tullet, 2015; Yu et al., 2008). In addition to transcriptomic profiling, global proteomic analyses of daf-2 mutants using stable isotope labeling and quantitative mass spectrometry (MS) identified several protein-abundance changes that were not detected in the microarray analyses (Depuydt et al., 2013; Dong et al., 2007), underscoring the complementarity of different global profiling techniques such as transcriptomics and proteomics.
Due to a plethora of protein posttranslational modifications, protein-protein interactions and endogenous inhibitors, protein abundance is not a direct measure of activity state (Walsh et al., 2005). To complement abundance-based global profiling methods such as transcriptomics and proteomics, the field of activity-based protein profiling (ABPP) has evolved to directly measure protein activity in complex proteomes (Adam et al., 2002; Evans and Cravatt, 2006). Here, we apply the tools of ABPP, specifically reactivity-based proteomics, to profile changes associated with impaired IIS in C. elegans.
ABPP studies typically focus on a particular enzyme class of interest, and here, we focus on cysteine-mediated protein activities by applying cysteine-reactivity profiling, which monitors changes in cysteine reactivity across two or more proteomes (Deng et al., 2013; Pace and Weerapana, 2014; Wang et al., 2014; Weerapana et al., 2010). Cysteine residues on proteins serve critical functions in catalysis and regulation (Pace and Weerapana, 2013). Diverse protein families, including proteases, kinases and oxidoreductases, contain cysteine residues that are essential for protein function. These functional cysteines demonstrate elevated reactivity, allowing for enrichment of proteins with functional cysteines by using cysteine-reactive electrophilic probes (Weerapana et al., 2010). In C. elegans, proteins involved in stress resistance, such as heat-shock proteins (Hsu et al., 2003), oxidoreductases (e.g. peroxiredoxins) (Zarse et al., 2012), and detoxifying enzymes (e.g. glutathione S-transferases) (Ayyadevara et al., 2007), are regulated by the IIS pathway and rely on critical cysteine residues for function. Several of these and other cysteine-containing proteins are low abundant and intractable to standard abundance-based proteomic analyses. Cysteine-reactivity profiling can allow for monitoring abundance changes amongst these IIS-relevant proteins by enriching low-abundance proteins within this class for enhanced detection.
Dysregulated reactive oxygen species (ROS) is a characteristic feature of impaired IIS (Honda and Honda, 1999; Zarse et al., 2012). ROS can target cysteine residues within proteins {Couvertier, 2014 #125}, resulting in changes to the oxidation state and subsequent function of diverse cysteine-mediated proteins. The relationship between ROS and lifespan extension through IIS is complex and multifaceted. Acute inhibition of DAF-2 results in a transient increase in ROS levels due to an increase in metabolic rate to compensate for decreased glucose uptake; this spike in ROS then triggers the activation of a variety of antioxidant systems and the subsequent lowering of ROS levels (Zarse et al., 2012). Therefore, chronic inactivation of DAF-2 results in sustained increases in expression of antioxidant enzymes such as superoxide dismutase 3 (SOD3) and catalase that render lower ROS levels in daf-2 mutants (Honda and Honda, 1999). Due to the sensitivity of cysteine-mediated protein activities to changes in ROS, the abundance of these proteins is not a true representation of activity state. Cysteine-reactivity profiling can therefore serve to identify cysteine oxidation events that occur during IIS. Previous studies have applied redox-proteomic methods to identify C. elegans proteins that are oxidized upon exposure to peroxide (Kumsta et al., 2011), but similar studies have not been utilized to explore endogenous oxidative events associated with impaired IIS.
Importantly, comparing changes in cysteine reactivity across daf-2 and daf-16;daf-2 mutants allows identification of changes in protein abundance and/or oxidation driven by impaired IIS. We applied a promiscuous cysteine-reactive chemical probe, coupled with quantitative mass spectrometry (MS), (Qian et al., 2013; Weerapana et al., 2010) to globally quantify cysteine-reactivity changes between daf-2 and daf-16;daf-2 mutants. Our studies identified 40 cysteine-containing proteins that show a greater than two-fold change in cysteine reactivity upon impaired IIS. Subsequent RNAi-mediated knockdown of 17 genes identified lbp-3 and K02D7.1 as novel modulators of C. elegans lifespan and dauer formation. Importantly, our studies represent one of the first applications of the tools of ABPP in C. elegans and highlight the ability of chemical proteomics to complement traditional transcriptomic and proteomic methods used to study IIS.
Results
Reactive-cysteine profiling reveals functional cysteines in C. elegans
Cysteine is one of the most intrinsically nucleophilic amino acids, and this nucleophilicity can be modulated by the protein microenvironment to enable diverse biochemical functions (Giles et al., 2003; Pace and Weerapana, 2013). A global proteomic evaluation of cysteine reactivity demonstrated that functional cysteines involved in catalysis and regulation display elevated reactivity relative to non-functional cysteines in the proteome (Weerapana et al., 2010). In this previous study, the intrinsic reactivity of hundreds of cysteines in human proteomes was monitored using a promiscuous cysteine-reactive iodoacetamide-alkyne (IA) probe. Comparison of the extent of cysteine labeling as a function of time or IA concentration, revealed a subset of hyperreactive cysteines that saturated labeling at low time points, or low IA concentrations. This subset of hyperreactive cysteines was enriched in functional cysteines (Weerapana et al., 2010). To determine if a similar strategy would allow identification of functional cysteines in C. elegans lysates, a concentration-dependent analysis of cysteine labeling by the IA probe was performed.
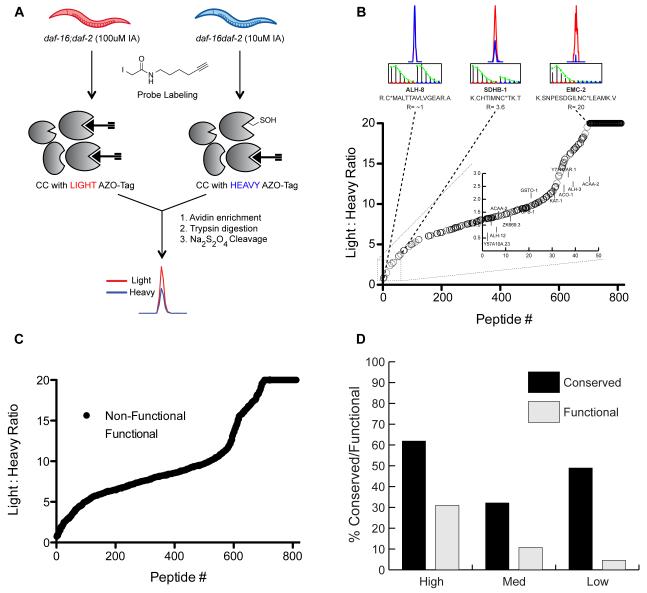
Lysates from daf-16;daf-2 mutants were used for these studies. These lysates were treated with either 10 μM or 100 μM IA probe prior to conjugation to isotopically labeled, chemically cleavable biotin tags (Azo-tags) (Qian et al., 2013) using copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC) (Rostovtsev et al., 2002). The Azo-tags are comprised of an azide for CuAAC-mediated conjugation to IA-labeled proteins, a biotin for enrichment of labeled proteins on streptavidin beads, a chemically cleavable azobenzene linker for selective release of probe-labeled peptides, and an isotopically light or heavy valine residue for quantitation of labeled peptides in two different proteomes. C. elegans lysates, treated with 10 μM and 100 μM IA, were conjugated to heavy and light Azo-tags, respectively. These samples were then combined and subjected to streptavidin enrichment, on-bead trypsin digestion and treatment with sodium dithionite to release the probe-labeled peptides for analysis by high-resolution LC/LC-MS/MS (Figure 1A). MS analysis identified 816 unique cysteine-containing peptides in the daf-16;daf-2 lysates with calculated light:heavy ratio (R) values in at least two out of four replicates (Table S1). The R values reflect the degree of cysteine labeling between the 10 μM and 100 μM IA-treated samples. A cysteine that is hyperreactive and saturates labeling at the low IA concentration will display R values of ~1, whereas less reactive cysteines will display R values >> 1. The 816 cysteine-containing peptides that were identified displayed a wide range of R values (Figure 1B and S1), with 46 cysteines demonstrating R values < 3 (Figure 1B; inset).
Figure 1. Identifying hyperreactive cysteines in C. elegans lysates.
(A) Workflow to identify hyperreactive cysteines in the daf-16;daf-2 mutant proteome. C. elegans lysates were treated with either 10 M or 100 M IA probe and conjugated to the heavy (10 M IA) or light (100 M IA) Azo-tags via click chemistry (CuAAC). The samples were combined and then enriched with streptavidin beads, subjected to on-bead trypsin digestion and Na2S2O4 treatment to release the probe-labeled peptides from the beads for quantitative LC/LC-MS/MS analysis. (B) 816 cysteine-containing peptides in order from high (light:heavy ratio < 3) to low (light:heavy ratio >>3) reactivity. Singleton peptides that are only detected in the Azo-L sample are designated an arbitrary R value of 20. Chromatograms and isotopic envelopes of representative peptides are shown above the plot with high (ALH-8, R=1.0), medium (SDHB-1, R=3.6), and low (EMC-2, R=20) cysteine reactivity. Chromatograms and isotopic envelopes for the light peptides are in red, the heavy peptides in blue. Inset displays 46 peptides with the lowest light:heavy ratio values representing the most reactive cysteines in the daf-2 mutant proteome. Cysteines with annotated biological function shown in Table 1 are labeled. (C) Cysteines with an annotated biological function in either C. elegans or the corresponding human homolog are highlighted in white along the ratio plot. (D) Cysteine-containing peptides were sorted into 3 groups of light:heavy ratios: R < 3 (hyperreactive), R = 3-6 (medium reactivity), R > 6 (low reactivity). The percentages of cysteines in each grouping that are conserved in humans (black) or have annotated biological function (gray) are shown.
To determine if those cysteines with low ratio values (R < 3) were enriched in known functional residues, the C. elegans UniProt database was mined for functional annotation of the identified cysteines as catalytic or regulatory residues. However, the C. elegans UniProt entries have poor annotation of residue and protein functions. Therefore, a BLAST search was performed for each identified C. elegans protein against the human UniProt database, identifying cysteine residues that were conserved in the corresponding human homolog and were functionally annotated to be involved in catalysis and regulation (Table S1). Comparing functional annotation with the observed R values for each cysteine demonstrated that cysteines with low R values (i.e. hyperreactive cysteines) were enriched in known functional residues (Figure 1C), similar to what was observed in human proteomes (Weerapana et al., 2010). Approximately 30% of all cysteines identified with ratios < 3 are known to be functional in either C. elegans or the corresponding human homolog (Figure 1D), in contrast to <5% of cysteines with R > 6 that were annotated to be functional. A similar trend in cysteine conservation was also observed (Figure 1D), albeit with lower enrichment amongst the hyperreactive subset of cysteines, suggesting that cysteine reactivity rather than conservation, is a more effective predictor of cysteine functionality. Functionally annotated hyperreactive cysteines in C. elegans include active-site cysteines in glutathione S-transferases, aldehyde dehydrogenase and acetyl-CoA acyltransferases (Table 1). These studies constitute the first evaluation of cysteine reactivity in C. elegans and provide a list of annotated and unannotated hyperreactive cysteines for future functional characterization.
Table 1. Functional cysteines identified in C. elegans.
Hyperreactive cysteines identified in the daf-2 mutant proteomes with an annotated biological function in either C. elegans or human Uniprot databases.
| Gene ID | Symbol | Description | Sequence | Average Ratio | Transcriptomic data |
Proteomic data |
|---|---|---|---|---|---|---|
| K07H8.6 | vit-6 | Vitellogenin-6 | VIC*PIAEVGTK | 0.185 ± 0.005 | decreased | decreased |
| TEEGLIC*R | 0.185 ± 0.025 | |||||
| EC*NEEQLEQIYR | 0.195 ± 0.005 | |||||
| SYANNESPC*EQTFSSR | 0.21 ± 0.01 | |||||
| NQFTPC*YSVLAK | 0.25 ± 0.02 | |||||
| C42D8.2 | vit-2 | Protein VIT-2, isoform b | VAIVC*SK | 0.215 ± 0.005 | decreased | not identified |
| C04F6.1 | vit-5 | Vitellogenin-5 | APLTTC*YSLVAK | 0.22 ± 0.01 | decreased | decreased |
| ZK228.3 | ZK228.3 | Protein ZK228.3 | DGVVYSVAC*STHQFV | 0.355 ± 0.015 | decreased | not identified |
| C17H12.13 | C17H12.13 | Protein C17H12.13, isoform b | DLVQDSLQC*SSTCVIR | 0.37 ± 0.01 | not identified |
|
| K02D7.1 | K02D7.1 | Protein K02D7.1 | ADLGIIC*GSGLGPIGDTVQDATILPYSK | 0.425 ± 0.025 | not identified |
|
| TVGADALGMSTC*HEVTVAR | 0.49 ± 0.05 | |||||
| R11H6.1 | pes-9 | Protein PES-9 | EGC*SIPITLTFQELTGK | 0.445 ± 0.035 | decreased | no change |
| F40F4.4 | lbp-3 | Fatty acid-binding protein homolog 3 | MVNNGITC*R | 0.465 ± 0.015 | decreased | not identified |
| F32D1.5 | F32D1.5 | Probable GMP reductase | SAC*TYTGAK | 0.49 ± 0.02 | decreased |
Human homologue of unannotated C. elegans proteins.
Chemical-proteomic analysis identifies changes in cysteine reactivity between daf-2 and daf-16;daf-2 mutants
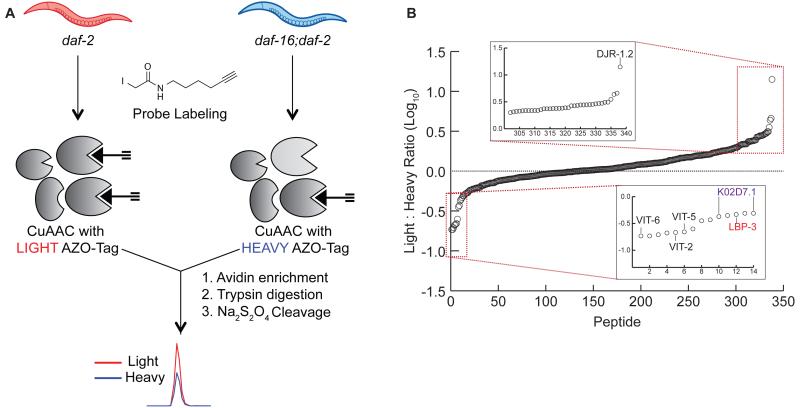
Given that the IA probe labels a large number of functionally relevant cysteine residues in C. elegans, cysteine reactivity was compared across daf-2 and daf-16;daf-2 mutants. These studies will reveal variations in protein abundance and cysteine posttranslational-modification state in the long-lived daf-2 mutants relative to the daf-16;daf-2 mutants. Initially, to confirm the longevity phenotype of the daf-2 mutants, a lifespan assay was performed on WT (N2), daf-2 and daf-16;daf-2 mutants cultured under identical conditions. As expected, daf-2 mutants demonstrated an almost 100% lifespan extension (Figure S2A) (Kenyon et al., 1993). To quantify cysteine-reactivity changes between daf-2 and daf-16;daf-2 mutants, lysates from daf-2 and daf-16;daf-2 animals were treated with 100 μM IA, appended to Azo-L and Azo-H, respectively, and subjected to the MS workflow utilized previously (Figure 2A). The higher concentration of IA (100 μM) was utilized for these comparative studies to maximize the number of cysteine residues identified, and is consistent with previous cysteine-reactivity profiling experiments in lysates treated with oxidants (Deng et al., 2013), Zn(II) ions (Pace and Weerapana, 2014), and lipid-derived electrophiles (Wang et al., 2014). MS analysis provided R-values for 338 cysteine-containing peptides on 254 proteins (Figure 2B and S2B; Table S2) from two biological replicates.
Figure 2. Identifying cysteine reactivity changes between daf-2 and daf-16;daf-2 mutants.
(A) Workflow to quantify cysteine reactivity changes between daf-2 and daf- 16;daf-2 mutants. Lysates from each mutant were treated with 100 M IA probe, conjugated to either the light (daf-2) or heavy (daf-16;daf-2) Azo-tags, and subjected to streptavidin enrichment, tryptic digest, and Na2S2O4 cleavage as before. (B) 338 cysteine-containing peptides were identified and the log10 value of each light;heavy ratio were plotted; log10 values less than 0 indicate cysteines that have decreased reactivity in daf-2 mutants, whereas log10 values greater than 0 indicate cysteines with increased reactivity in daf-2 mutants. Insets display peptides with cysteines that show at least a 2-fold increase (upper) or decrease (lower) in reactivity in daf-2 mutants. Proteins with a previously observed role in lifespan regulation (DJR-1.2, Vit-2,5,6) and those that we demonstrate to affect lifespan upon RNAi-mediated knockdown (LBP-3, K02D7.1) are specifically indicated.
High R-values are indicative of cysteines with increased reactivity in daf-2 mutants, whereas low R-values represent cysteines with decreased reactivity in daf-2 mutants. The majority of identified cysteines (84.9%) demonstrated R-values in the range 0.5 < R < 2, demonstrating less than 2-fold change across the daf-2 and daf-16;daf-2 proteomes (Table S2). In total, 50 cysteine residues on 40 proteins displayed a ≥ 2-fold change (Table 2 and Table 3), with 36 showing increases and 14 showing decreases in daf-2 mutants. These top 40 proteins were compared to transcriptomic data available for daf-2 and daf-16;daf-2 mutants (McElwee et al., 2003; McElwee et al., 2004), to determine if the changes we observed were also present at the transcript level. Transcriptomic data was available for 25 of the 40 proteins and in terms of the general trend (increase vs. decrease in daf-2 animals), all but one hit (protein Y39E4A.3) agreed with the previously reported transcriptomic data (Table 2 and Table 3). Further comparison of these 40 proteins to proteomic studies that measured protein abundance changes in daf-2 mutants {Depuydt, 2013 #119} demonstrated poor overlap between the proteins identified in unenriched proteomics compared to cysteine enrichment. Specifically, only 19 of the 40 (48%) were identified in these previous global proteomic studies {Depuydt, 2013 #119} (Table 2 and Table 3). This poor overlap underscores the ability of cysteine enrichment to promote the identification of low-abundant proteins that are typically suppressed in unenriched proteomic analyses. Of the 19 proteins identified in both cysteine enrichment and unenriched proteomic analyses, 13 showed the same changes (increase vs. decrease in daf-2 animals), whilst the remaining 6 (PES-9, RPS-12, RPS-17, DHS-25, HEL1, and INF-1) showed changes that were not statistically significant in the unenriched proteomic studies {Depuydt, 2013 #119}. Lastly, the cysteines identified in our study were also compared to a redox-proteomic study in wild-type C. elegans that utilized the OxiCAT method to identify sites of cysteine oxidation upon hydrogen peroxide exposure (Kumsta et al., 2011). Only 13 of the 338 cysteine residues from our study were identified in the OxiCAT data (Table S3), and only one of these cysteine residues (on RPS-17) were within the 40 proteins that demonstrated a ≥ 2-fold change between daf-2 and daf-16;daf-2 mutants.
Table 2. Cysteine residues identified with decreased reactivity (≥ 2-fold) in daf-2 relative to daf-16;daf-2 mutants.
The 14 cysteines on 9 proteins that demonstrated decreased reactivity in daf-2 (R ≤ 0.5) are shown. Transcriptomic changes (decreased or increased in daf-2) of the corresponding genes previously identified in microarray analysis of daf-2 and daf-16;daf-2 are shown {McElwee, 2003 #131;McElwee, 2004 #132}. Proteomic changes for these proteins in unenriched proteomic analyses (decreased, increased or no change in daf-2) are also indicated {Depuydt, 2013 #119}. “Not identified” indicates a protein that was not present in the unenriched proteomic datasets.
| Gene ID | Symbol | Transcriptomic data |
Proteomic data |
|||
|---|---|---|---|---|---|---|
| F54E7.2 | rps-12 | 40S ribosomal protein S12 | GLHETC*K | 2 ± 0.12 | no change | |
| Y39E4A.3 | Y39E4A.3 | Protein Y39E4A.3, isoform a | GYTMENFMNQC*YGNADDLGK | 2.06 ± 0.2 | decreased | not identified |
| C08B11.7 | ubh-4 | Probable ubiquitin carboxyl-terminal hydrolase ubh |
GHC*LSNSEEIR | 2.105 ± 0.135 | not identified | |
| R03D7.6 | gst-5 | Probable glutathione S-transferase 5 | ETC*AAPFGQLPFLEVDGK | 2.125 ± 0.025 | not identified | |
| F36H1.6 | alh-3 | Protein ALH-3 | GENC*IAAGR | 2.155 ± 0.115 | not identified | |
| Y54E10BR.6 | rpb-7 | Protein RPB-7 | LFNEVEGTC*TGK | 2.175 ± 0.335 | not identified | |
| T20G5.1 | chc-1 | Probable clathrin heavy chain 1 | AAIGQLC*EK | 2.175 ± 0.825 | increased | |
| K08E3.5 | K08E3.5 | Protein K08E3.5, isoform f | LNGGLGTSMGC*K | 2.18 ± 0.04 | not identified | |
| T08B2.10 | rps-17 | 40S ribosomal protein S17 | VC*DEVAIIGSK | 2.185 ± 0.105 | no change | |
| F09E10.3 | dhs-25 | Protein DHS-25 | TPMTEAMPPTVLAEIC*K | 2.19 ± 0.02 | no change | |
| K11H3.1 | gpdh-2 | Protein GPDH-2, isoform c | NVVAC*AAGFTDGLGYGDNTK | 2.26 ± 0.06 | not identified | |
| K12G11.3 | sodh-1 | Alcohol dehydrogenase 1 | LMNFNC*LNCEFCK | 2.35 ± 0.21 | increased | increased |
| LMNFNCLNC*EFCK | 2.35 ± 0.21 | |||||
| LMNFNCLNCEFC*K | 2.35 ± 0.21 | |||||
| DTNLAAAAPILC*AGVTVYK | 4.355 ± 0.135 | |||||
| C26D10.2 | hel-1 | Spliceosome RNA helicase DDX39B homolog |
YFVLDEC*DK | 2.38 ± 0.05 | no change | |
| W05G11.6 | pck-2 | Protein W05G11.6, isoform a | AELMNPAGIYIC*DGSQK | 2.385 ± 0.065 | increased | increased |
| TNAMAMESC*R | 2.405 ± 0.155 | |||||
| FIAAAFPSAC*GK | 3.53 ± 0.13 | |||||
| C36A4.9 | acs-19 | Protein ACS-19, isoform a | TNISYNC*LER | 2.41 ± 0.14 | increased | not identified |
| F55H12.4 | F55H12.4 | Protein F55H12.4 | GSTGHC*YK | 2.45 ± 0.18 | increased | increased |
| Y113G7B.23 | swsn-1 | Protein SWSN-1 | GVQAAAASC*LAAAAVK | 2.455 ± 0.245 | not identified | |
| ZK829.4 | gdh-1 | Glutamate dehydrogenase | CDIFVPAAC*EK | 2.67 ± 0.37 | not identified | |
| C05C10.6 | ufd-3 | Protein UFD-3, isoform b | ALAVTQGGC*LISGGR | 2.71 ± 0.04 | not identified | |
| K08F4.9 | dhs-12 | Protein DHS-12 | AAIVNIGSDC*ASQALNLR | 2.77 ± 0.17 | increased | increased |
| F01G10.1 | tkt-1 | Protein TKT-1 | ISSIEMTC*ASK | 2.775 ± 0.225 | increased | increased |
| B0286.3 | B0286.3 | Probable multifunctional protein ADE2 | MPNGIGC*TTVLDPSEAALAAAK | 2.785 ± 0.095 | increased | increased |
| F32A7.5 | F32A7.5 | Protein F32A7.5, isoform d | DISGEQLQAILC*GK | 2.805 ± 0.015 | not identified | |
| F20D6.11 | F20D6.11 | Protein F20D6.11 | FSGC*NQGSTK | 2.84 ± 0.48 | increased | not identified |
| F57B9.6 | inf-1 | Eukaryotic initiation factor 4A | AIVPC*TTGK | 2.905 ± 0.155 | no change | |
| B0035.5 | gspd-1 | Glucose-6-phosphate 1-dehydrogenase | SSC*ELSTHLAK | 2.95 ± 0.36 | increased | |
| ZK829.7 | ZK829.7 | Protein ZK829.7 | AILEVC*DPSSALDADQSGGVPIPAATSE | 2.98 ± 0.31 | increased | increased |
| K09A9.2 | rab-14 | Protein RAB-14 | AFAEENGLTFLEC*SAK | 3.11 ± 1.7 | not identified | |
| W01A11.6 | moc-2 | Protein MOC-2 | VCVITVSDTC*SAGTR | 3.125 ± 0.125 | not identified | |
| F20G2.2 | F20G2.2 | Protein F20G2.2 | SC*SIDLAK | 4.59 ± 0.55 | increased | |
| C49G7.11 | djr-1.2 | Protein DJR-1.2 | LAEC*PVIGELLK | 14.115 ± 1.505 | increased | not identified |
Table 3. Cysteine residues identified with increased reactivity (≥ 2-fold) in daf-2 relative to daf-16;daf-2 mutants.
The 36 cysteines on 31 proteins that demonstrated increased reactivity in daf-2 (R ≥ 2) are shown. Transcriptomic changes (decreased or increased in daf-2) of the corresponding genes previously identified in microarray analysis of daf-2 and daf-16;daf-2 are shown {McElwee, 2003 #131;McElwee, 2004 #132}. Proteomic changes for these proteins in unenriched proteomic analyses (decreased, increased or no change in daf-2) are also indicated {Depuydt, 2013 #119}. “Not identified” indicates a protein that was not present in the unenriched proteomic datasets.
| Gene Symbol |
Protein Name | Sequence | Ratio | Function |
|---|---|---|---|---|
| Y57A10A.23 | Thioredoxin domain- containing protein 12# |
K.SWC*HACK.A | 0.805 | Redox-active disulfide |
| alh-8 | Methylmalonate- semialdehyde dehydrogenase |
R.C*MALTTAVLVGEAR.A | 0.898 | Active site nucleophile |
| alh-12 | Aldehyde dehydrogenase# |
-AMLANFLNQGQVC*TNATR.V | 1.03 | Active Site Nucleophile |
| acaa-2 | Acetyl-CoA acyltransferase 2 |
R.LC*GSGFQAVVNAAQAIK.L | 1.15 | Acyl-thioester intermediate |
| ZK669.3 | Gamma-interferon- inducible lysosomal thiol reductase# |
R.C*SDTSYWMK.W | 1.44 | Redox-active disulfide |
| upb-1 | Ureidopropionase beta# | R.IGINIC*YGR.H | 1.92 | Active site nucleophile |
| gsto-1 | Glutathione transferase omega-1 |
R.FC*PWAER.A | 1.96 | Active Site Nucleophile |
| kat-1 | Acetyl-CoA acetyltransferase |
K.VC*SSGLK.A | 2.55 | Acyl-thioester intermediate |
| aco-1 | Aconitate hydratase | K.IGFNIAGYGC*MTCIGNSGP- | 2.66 | Iron-binding |
| Y71H2AR.1 | Thioredoxin domain- containing protein 17# |
K.ILTTGESWC*PDCVVAEPVV- | 2.76 | Active site nucleophile/ Redox-active disulfide |
| alh-3 | Formyltetrahydrofolate dehydrogenase |
K.GENC*IAAGR.V | 2.79 | Active site |
| acaa-2 | Acetyl-CoA acyltransferase 2 |
K.YGIGSAC*IGGGQGIAILFEK- | 2.98 | Active site |
Several of the protein targets we identified have been previously characterized using RNAi knockdown and phenotypic analysis, including the vitellogenins (VIT-2/5/6) and the DJR-1.2 glyoxalase (Fischer et al., 2013; Lee et al., 2013; Murphy et al., 2003; Yuan et al., 2012). The vitellogenins showed the largest decrease in daf-2 mutants, and DJR-1.2 showed the largest increase in daf-2 mutants according to our cysteine-profiling data (Table 2 and Table 3). Vitellogenesis is the process of egg yolk, or vitellogenin, production that provides the major nutrient source for developing embryos. Vitellogenesis is suppressed in the daf-2 mutant, possibly extending lifespan by using those resources to maintain somatic cells (DePina et al., 2011). Vitellogenins are among the most downregulated proteins in daf-2 mutants according to previous transcriptomic and proteomic analyses {Depuydt, 2013 #119;Dong, 2007 #120;McElwee, 2003 #131;McElwee, 2004 #132}, and RNAi-mediated knockdown lengthened the lifespan of daf-2 (+) animals (Murphy et al., 2003). DJR-1.2 is homologous to the human DJ-1 protein and defects in the dj-1 gene are a cause of autosomal recessive early-onset Parkinson’s disease (Bonifati et al., 2003). DJ-1 is a multifunctional redox-sensitive protein that plays roles in dampening mitochondrial oxidative stress and regulation of anti-apoptotic and anti-oxidant gene expression. DJ-1 also regulates toll-like receptor signaling, suggesting a role in innate immunity (Cornejo Castro et al., 2010). DJR-1.2 has been found to be upregulated in both daf-2 and dauer larva, and shows a DAF-16-dependent decrease in stress resistance and viability upon knockdown (Lee et al., 2013). These well-characterized lifespan-modulating effects for vitellogenins and DJR-1.2, the two most significantly changed proteins from our data, serves as validation of our platform to identify potential mediators of IIS and lifespan.
RNAi-mediated knockdown of lbp-3 and K02D7.1 results in modulation of lifespan and dauer formation
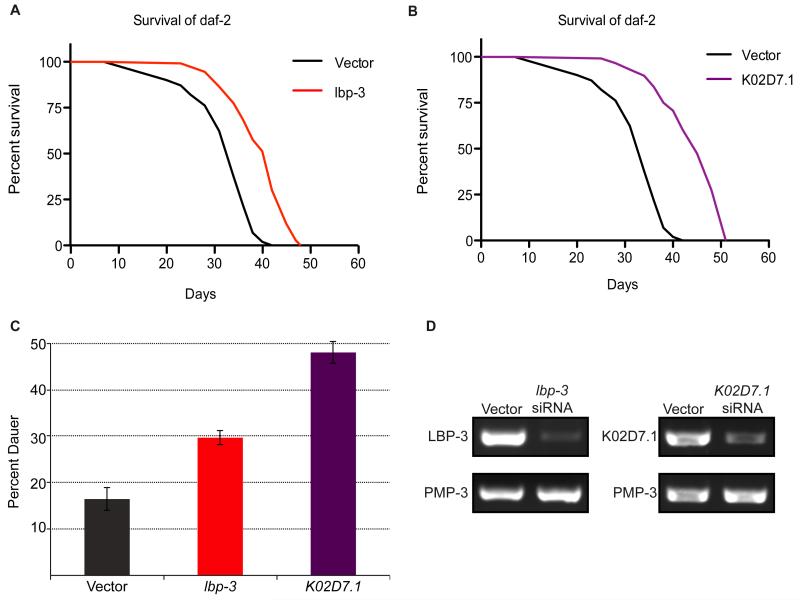
To determine if other proteins identified in our cysteine-reactivity profiling studies were implicated in IIS-mediated lifespan regulation, RNAi-mediated knockdown studies were performed followed by subsequent lifespan analyses. For these RNAi studies, we focused on a set of 20 genes, corresponding to the 10 proteins that showed the largest decrease (vit-6, vit-2, vit-5, ZK228.3, C17H12.13, K02D7.1, pes-9, lbp-3, F32D1.5, eef-2) and the 10 proteins with the largest increase (djr-1.2, F20G2.2, sodh-1, pck-1, moc-2, rab-14, ZK829.7, gspd-1, inf-1, F20D6.11) in daf-2 mutants, consistent across the two biological replicates from our cysteine-profiling data (Table S2). Of these 20 genes, in addition to the vitellogenin (vit-6/2/5) and djr-1.2 genes discussed previously, four other genes had already been subjected to RNAi and phenotypic analysis (sodh-1 (Murphy et al., 2003); eef-2 (Li et al., 2011); inf-1 (Curran and Ruvkun, 2007); and pck-1 (Yuan et al., 2012). The remaining 12 genes with no previous RNAi and phenotypic data related to lifespan were targeted for RNAi-mediated knockdown using bacterial strains from the Ahringer RNAi library (Kamath et al., 2003), and lifespan assays were performed with respect to vector-treated controls (Table S4, Figure S3). We observed a greater than 15% increase in lifespan for four of the genes tested (K02D7.1, pes-9, lbp-3 and gspd-1).
To determine if inactivation of these four genes would further augment dauer formation in daf-2 mutants, dauer-arrest assays were performed upon RNAi-mediated knockdown. Reduction of levels of both LBP-3 and K02D7.1 increased dauer formation by 23% and 37%, respectively (Figure 3C and Table S4). In contrast, knockdown of pes-9 and gspd-1 showed a decrease in dauer formation (Table S4). Therefore, these data implicate C. elegans LBP-3 and K02D7.1 proteins in both lifespan regulation (Figure 3A and B, Table S5) and entry into the dauer state (Figure 3C, Table S4). RT-PCR was used to confirm knockdown of lbp-3 and K02D7.1, and compared against the control gene pmp-3, which demonstrates unusually stable expression levels with little variation between adults, dauers, and L3 larvae, or between wild-type and daf-2 or daf-16 mutant adults (Figure 3D) (Hoogewijs et al., 2008). To determine if the observed effects on lifespan were dependent on the presence of functional DAF-2 and DAF-16, lifespan assays were repeated in the background of daf-16 mutants, daf-16;daf-2 double mutants, and WT (N2) animals (Figure S4). Knockdown of lbp-3 only affected the lifespan of daf-2 mutants, whereas K02D7.1 knockdown extended lifespan in all mutant backgrounds.
Figure 3. RNAi-mediated knockdown and phenotypic analysis.
Survival plots of daf-2 mutants treated with (A) lbp-3 siRNA or (B) K02D7.1 siRNA and compared to a vector-treated control. (C) Dauer-arrest assay comparing the percent dauer formation of daf-2 mutants with RNAi-mediated knockdown of lbp-3 and K02D7.1 compared to a vector-treated control. (D) RT-PCR of daf-2 mutants treated with lbp-3 or K02D7.1 siRNA using primers for lbp-3, K02D7.1, or pmp-3 as a control.
Discussion
Global profiling techniques, such as transcriptomics and proteomics, can provide valuable insight into changes in mRNA and protein across multiple biological samples. These studies are complemented by chemical-proteomic methods that apply chemical probes to enrich specific subsets of the proteome. Chemical-proteomic platforms, such as ABPP, can enrich low-abundant proteins that are intractable to whole-proteome analyses, and can additionally provide insight into posttranslational modifications that can activate or inactive proteins. Despite the application of chemical-proteomic methods such as ABPP to identify dysregulated protein activities in a variety of human, mouse and bacterial proteomes, these tools are yet to be applied in C. elegans. Here, we apply cysteine-reactivity profiling to uncover hyperreactive and functional cysteines in C. elegans and identify proteins dysregulated during impaired IIS. We identified a subset of hyperreactive cysteines in C. elegans and demonstrate that ~30% of these cysteines are known to perform essential roles in catalysis and regulation in the corresponding human homologs. Further studies into the unannotated hyperreactive cysteines that we identified are likely to reveal novel functional cysteine residues and serve to further annotate the C. elegans proteome.
Comparing cysteine-reactivity in daf-2 and daf-16;daf-2 proteomes revealed 40 proteins that displayed a >2-fold change. The majority of these changes have been previously reported in transcriptomic studies, serving to verify the accuracy of our platform to identify changes across daf-2 and daf-16:daf-2 proteomes. Importantly, several of these proteins, such as the vitellogenins and DJR-1.2, have also been functionally characterized to be important mediators of IIS and lifespan, further alluding to the promise of global profiling studies to reveal functional components of the IIS pathway. Importantly, many of the proteins identified were not detected in previous unenriched proteomic studies, alluding to the promise of ABPP and cysteine reactivity-profiling studies to enrich for low abundant proteins inaccessible to unenriched proteomics. To investigate previously uncharacterized proteins that were identified in our studies, we performed RNAi-mediated knockdown and subsequent phenotypic assays to monitor lifespan and dauer formation. These studies revealed that RNAi-mediated knockdown of lbp-3 and K02D7.1 results in significant increases in both lifespan and dauer formation, implicating these proteins as important components of the IIS pathway.
LBP-3 is an intracellular lipid chaperone in the fatty-acid-binding protein (FABP) family. FABPs are conserved from C. elegans to humans and are involved in fatty-acid uptake, transport and oxidation (Storch and Corsico, 2008). The C. elegans genome contains nine LBP genes and the exact functions of these individual LBPs remains unclear (Plenefisch et al., 2000). Of the C. elegans LBPs, LBP-5 appears to be the most extensively studied, whereby RNAi-mediated knockdown of lbp-5 results in fat accumulation in the intestine (Xu et al., 2011). The putative role of the C. elegans LBPs in fat accumulation is notable because a characteristic feature of the daf-2 mutants and long-lived dauers is increased fat content (Ogg et al., 1997). Previous studies have also shown that mutational inactivation of other lipid transport proteins, such as the intramembrane transporters, NDG-4 and NRF-5/6, increase stress resistance and lifespan through the IIS pathway (Brejning et al., 2014). These previous studies into the LBP family and related proteins supports a potential role for LBP-3 in C. elegans lipid metabolism and IIS.
Given the lack of functional characterization of the C. elegans LBPs, we probed the mammalian FABPs for insight into the function and regulation of this class of proteins. There are at least nine human FABPs and these have distinct tissue localization patterns. All FABPs are small intracellular proteins that can localize to the nucleus, (Furuhashi and Hotamisligil, 2008; Smathers and Petersen, 2011) and bind long-chain fatty acids, albeit with different binding affinities. In our cysteine-profiling studies, we identified Cys154 as labeled by the IA probe; this cysteine is not annotated as functional in the C. elegans UniProt database, but is conserved in several of the human FABPs (Figure S5A). C. elegans LBP-3 shares the most homology with human FABP-5 (27% sequence identity), which contains a disulfide bond between Cys120 and Cys127 that regulates protein structure and function under reducing/oxidizing conditions (Hohoff et al., 1999). In fact, several FABPs have been shown to be oxidized, glutathionylated and/or modified by oxidized lipid species such as 4-hydroxynonenal (HNE) at cysteine with effects on the lipid-binding ability and proteolytic stability (Bennaars-Eiden et al., 2002; Smathers et al., 2012; Smathers et al., 2013; Yang et al., 2014). It is therefore likely that Cys154 in C. elegans LBP-3 is similarly regulated through redox modifications. Interestingly, the 2.2-fold decrease in IA-labeled LBP-3 that we observed in the daf-2 animals is significantly less than the change reported for lbp-3 using transcriptomic analyses (~5-fold decrease). This suggests that either there is poor correlation between lbp-3 mRNA and protein levels, or there is reduced enrichment due to partial oxidation of Cys154 in daf-16;daf-2 mutants. The higher levels of ROS observed in daf-16;daf-2 double mutants supports the possibility of increased protein oxidation in this strain compared to the single daf-2 mutants (Honda and Honda, 1999; Zarse et al., 2012).
K02D7.1 is an uncharacterized protein in C. elegans but homologous to human purine nucleoside phosphorylase (PNP) with 47% sequence identity. The IA-modified cysteines have no known functional annotation, but one of these cysteines are located in a highly conserved region (IIC*GSGLG), with conservation seen throughout humans, mice, flies, and yeast (Figure S5B). PNP catalyzes cleavage of the glycosidic bond of (deoxy)ribonucleosides, forming the corresponding free purine base and pentose-1-phosphate in the purine salvage pathway (Bzowska et al., 2000). Although PNP has not been directly implicated in IIS and lifespan regulation, the purine nucleotide synthesis pathway has been shown to be regulated by PI3K/AKT signaling, suggesting that enzymes within this pathway are under IIS control (Wang et al., 2009). Furthermore, a downstream enzyme involved in purine metabolism, xanthine dehydrogenase (XDH), was identified in a systematic screen for longevity genes in C. elegans. RNAi knockdown of XDH in C. elegans caused a ~12% increase in median lifespan (Hamilton et al., 2005), suggesting that perturbations in purine metabolism can modulate longevity. These previous studies indicate that K02D7.1, similar to XDH, is a regulator of C. elegans lifespan and is a promising target for further characterization to determine the exact biochemical function and role of this protein in IIS.
Significance
Collectively, these studies constitute the first reported application of ABPP in C. elegans. Specifically, a cysteine-reactive chemical probe was applied to identify hyperreactive cysteines in C. elegans, revealing that these hyperreactive cysteines are enriched in functional residues critical to catalysis and regulation. Amongst the subset of hyperreactive cysteines were several unannotated cysteines for future functional characterization in C. elegans and other organisms in which these cysteines are conserved. Given the wide utility of C. elegans as a model organism for aging, and the well-characterized role of impaired IIS in regulating longevity in this organism, we applied chemical proteomics to identify dysregulated protein activities with potential implications in IIS-mediated longevity regulation. Importantly, chemical-proteomic approaches such as ABPP have the advantage of identifying changes in posttranslational modifications as well as low-abundance proteins, which are intractable to abundance-based transcriptomic and proteomic approaches. Comparison of cysteine reactivity across daf-2 and daf-16;daf-2 mutants, identified 40 proteins with >2-fold change across these proteomes. The majority of these changes were previously identified in transcriptomic studies and validated to regulate lifespan, serving to substantiate our chemical-proteomic data. Previously uncharacterized proteins were also identified, underscoring the complementarity of chemical-proteomic techniques to existing global transcriptomic and proteomic studies. Coupling chemical-proteomic tools with RNAi-mediated knockdown and phenotypic assays resulted in the identification of two proteins, LBP-3 and K02D7.1, as novel mediators of C. elegans lifespan and dauer formation.
Experimental Procedures
C. elegans culture and maintenance
Worm strains were grown on OP50 E. coli-seeded nematode growth medium (NGM) stock plates using standard C. elegans culturing techniques (Girard et al., 2007). C. elegans strains were provided by the Caenorhabditis Genetics Center (CGC) and details on the specific C. elegans strains are provided in the Supplemental Information.
Preparation of 4-day daf-2 and daf-16;daf-2 worm lysates for MS analysis
Synchronized worm populations for MS analysis were obtained by shaking ~3000 gravid adult worms in a solution of sterile water (5.0 mL), KOH (1.0 mL, 5 M), and bleach (4.0 mL) for 5 minutes until only the eggs remained. The eggs were pelleted, washed with S Medium (5 × 10 mL), resuspended in S Medium (8 mL) and allowed to hatch overnight in a 15 °C incubator. The hatched L1 worms (~100,000) were aliquoted onto 10 NGM plates for synchronized growth with the addition of OP50 E. coli (100 L, 100 mg/mL) at 15 °C. At the L4 larval stage, the worms were trans ferred to 25 floxuridine-containing plates (FUDR, 0.05 mg/mL) to prevent reproduction and transferred to a 25 °C incubator. After 4 days of growth, worms were harvested and sonicated to provide lysates for MS analysis. Detailed protocols for growth, synchronization, FUDR-treatment, harvesting and lysis are provided in the Supplemental Information.
Quantitative mass-spectrometry analysis
For cysteine-reactivity studies, daf-16;daf-2 C. elegans lysates (2 × 500 L, 2 mg/mL) in PBS were treated with either 10 M or 100 M IA for 1 hour at room temperature. For daf-2 and daf-16;daf-2 comparative studies, both lysates were treated with 100 M IA for 1 hour at room temperature. The IA-labeled samples were conjugated to either the heavy or light azobenzene tags (100 μM) (Qian et al., 2013) using CuAAC by addition of TCEP (1.0 mM from fresh 50X stock in water), TBTA ligand (100 μM from 17X stock in DMSO:t-Butanol 1:4) and CuSO4 (1.0 mM from 50X stock in water), and allowed to react at room temperature for 1 hour. For reactivity studies, 10 M IA-treated samples were conjugated to the heavy tag, and 100 M IA-treated samples were conjugated to the light tag. For daf-2 and daf-16;daf-2 comparative studies, daf-2 lysates were conjugated to the light tag, and daf-16;daf-2 lysates were conjugated to the heavy tag. After click chemistry, the IA-labeled proteins were enriched on streptavidin beads and subjected to on-bead trypsin digestion and sodium dithionite treatment, and LC-MS/MS analysis was performed on an LTQ-Orbitrap Discovery mass spectrometer (ThermoFisher) as detailed in the Supplemental Information. The generated tandem MS data were searched using the SEQUEST algorithm against the Uniprot C. elegans database. A static modification of +57.02146 on cysteine was specified to account for alkylation by iodoacetamide and differential modifications of +443.2897 (Azo-L tag) and +449.3035 (Azo-H tag) were specified on cysteine to account for probe modifications. SEQUEST output files were filtered using DTASelect2.0.5 and quantification of light:heavy ratios was performed using the CIMAGE quantification package as previously described (Weerapana et al., 2010).
RNAi-mediated knockdown
RNAi bacterial clones were obtained from the Ahringer Lab RNAi feeding library (Timmons et al., 2001) and feeding plates were prepared as described in Supplemental Information.
Lifespan and dauer assays
Lifespan assays were performed using L4 animals at 20 °C (30 animals per plate × 4), and scored every 2-3 days. Dauer asays were performed at 22.5 °C and scored for dauer larva after 4 days. Detailed protocols for the assays, as well RT-PCR experiments to verify knockdown are provided in the Supplemental Information.
Supplementary Material
Highlights.
Hyperreactive and functional cysteines were identified in C. elegans lysates
Comparing daf-2 and daf-16;daf-2 mutants revealed 40 proteins with >2 fold change
RNAi-mediated knockdown characterized lbp-3 and K02D7.1 as regulators of lifespan
These lifespan regulators are involved in fatty-acid transport and purine metabolism
ACKNOWLEDGEMENTS
We thank members of the Weerapana and Tissenbaum Lab for assistance with experiments and manuscript preparation. This work was funded by Boston College (E.W.), The Smith Family Foundation (E.W.) and NIH grant 1R01GM118431-01A1 (E.W.). C. elegans strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information includes Supplemental Experimental Procedures, eight Supplemental Figures (Figures S1-S5) and five data tables (Tables S1-S5). This information can be found with this article online.
Author Contributions
Conceptualization, J.M. and E.W.; Formal Analysis, J.M., D.W.B., Y.H.S., H.A.T. and E.W.; Investigation, Y J.M., D.W.B., and Y.H.S.; Resources, H.A.T. and E.W.; Writing – Original Draft, J.M. and E.W.; Writing – Review & Editing, J.M., D.W.B., H.A.T. and E.W.; Visualization, J.M. and E.W.; Supervision, H.A.T. and E.W.; Project Administration, H.A.T. and E.W.; Funding Acquisition, H.A.T. and E.W.
References
- Adam GC, Sorensen EJ, Cravatt BF. Chemical strategies for functional proteomics. Molecular & Cellular Proteomics. 2002;1:781–790. doi: 10.1074/mcp.r200006-mcp200. [DOI] [PubMed] [Google Scholar]
- Anselmi CV, Malovini A, Roncarati R, Novelli V, Villa F, Condorelli G, Bellazzi R, Puca AA. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12:95–104. doi: 10.1089/rej.2008.0827. [DOI] [PubMed] [Google Scholar]
- Ayyadevara S, Dandapat A, Singh SP, Siegel ER, Shmookler Reis RJ, Zimniak L, Zimniak P. Life span and stress resistance of Caenorhabditis elegans are differentially affected by glutathione transferases metabolizing 4-hydroxynon-2-enal. Mech Ageing Dev. 2007;128:196–205. doi: 10.1016/j.mad.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsyte D, Lovejoy DA, Lithgow GJ. Longevity and heavy metal resistance in daf-2 and age-1 long-lived mutants of Caenorhabditis elegans. Faseb J. 2001;15:627–634. doi: 10.1096/fj.99-0966com. [DOI] [PubMed] [Google Scholar]
- Bennaars-Eiden A, Higgins L, Hertzel AV, Kapphahn RJ, Ferrington DA, Bernlohr DA. Covalent modification of epithelial fatty acid-binding protein by 4-hydroxynonenal in vitro and in vivo. Evidence for a role in antioxidant biology. J Biol Chem. 2002;277:50693–50702. doi: 10.1074/jbc.M209493200. [DOI] [PubMed] [Google Scholar]
- Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- Brejning J, Norgaard S, Scholer L, Morthorst TH, Jakobsen H, Lithgow GJ, Jensen LT, Olsen A. Loss of NDG-4 extends lifespan and stress resistance in Caenorhabditis elegans. Aging Cell. 2014;13:156–164. doi: 10.1111/acel.12165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzowska A, Kulikowska E, Shugar D. Purine nucleoside phosphorylases: properties, functions, and clinical aspects. Pharmacol Ther. 2000;88:349–425. doi: 10.1016/s0163-7258(00)00097-8. [DOI] [PubMed] [Google Scholar]
- Cornejo Castro EM, Waak J, Weber SS, Fiesel FC, Oberhettinger P, Schutz M, Autenrieth IB, Springer W, Kahle PJ. Parkinson's disease-associated DJ-1 modulates innate immunity signaling in Caenorhabditis elegans. J Neural Transm. 2010;117:599–604. doi: 10.1007/s00702-010-0397-4. [DOI] [PubMed] [Google Scholar]
- Couvertier SM, Zhou Y, Weerapana E. Chemical-proteomic strategies to investigate cysteine posttranslational modifications. Biochim Biophys Acta. 2014;1844:2315–2330. doi: 10.1016/j.bbapap.2014.09.024. [DOI] [PubMed] [Google Scholar]
- Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Weerapana E, Ulanovskaya O, Sun F, Liang H, Ji Q, Ye Y, Fu Y, Zhou L, Li J, et al. Proteome-wide quantification and characterization of oxidation-sensitive cysteines in pathogenic bacteria. Cell Host Microbe. 2013;13:358–370. doi: 10.1016/j.chom.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePina AS, Iser WB, Park SS, Maudsley S, Wilson MA, Wolkow CA. Regulation of Caenorhabditis elegans vitellogenesis by DAF-2/IIS through separable transcriptional and posttranscriptional mechanisms. BMC physiology. 2011;11:11. doi: 10.1186/1472-6793-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt G, Xie F, Petyuk VA, Shanmugam N, Smolders A, Dhondt I, Brewer HM, Camp DG, Smith RD, Braeckman BP. Reduced insulin/IGF-1 signaling and dietary restriction inhibit translation but preserve muscle mass in Caenorhabditis elegans. Mol Cell Proteomics. 2013 doi: 10.1074/mcp.M113.027383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong MQ, Venable JD, Au N, Xu T, Park SK, Cociorva D, Johnson JR, Dillin A, Yates JR., 3rd Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science. 2007;317:660–663. doi: 10.1126/science.1139952. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Cravatt BF. Mechanism-based profiling of enzyme families. Chem Rev. 2006;106:3279–3301. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- Fischer M, Regitz C, Kull R, Boll M, Wenzel U. Vitellogenins increase stress resistance of Caenorhabditis elegans after Photorhabdus luminescens infection depending on the steroid-signaling pathway. Microbes and infection / Institut Pasteur. 2013;15:569–578. doi: 10.1016/j.micinf.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Flachsbart F, Caliebe A, Kleindorp R, Blanche H, von Eller-Eberstein H, Nikolaus S, Schreiber S, Nebel A. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A. 2009;106:2700–2705. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi M, Hotamisligil GS. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov. 2008;7:489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles NM, Giles GI, Jacob C. Multiple roles of cysteine in biocatalysis. Biochem Biophys Res Commun. 2003;300:1–4. doi: 10.1016/s0006-291x(02)02770-5. [DOI] [PubMed] [Google Scholar]
- Girard LR, Fiedler TJ, Harris TW, Carvalho F, Antoshechkin I, Han M, Sternberg PW, Stein LD, Chalfie M. WormBook: the online review of Caenorhabditis elegans biology. Nucleic Acids Res. 2007;35:D472–475. doi: 10.1093/nar/gkl894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohoff C, Borchers T, Rustow B, Spener F, van Tilbeurgh H. Expression, purification, and crystal structure determination of recombinant human epidermal-type fatty acid binding protein. Biochemistry. 1999;38:12229–12239. doi: 10.1021/bi990305u. [DOI] [PubMed] [Google Scholar]
- Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. Faseb J. 1999;13:1385–1393. [PubMed] [Google Scholar]
- Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol. 2008;9:9. doi: 10.1186/1471-2199-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Jensen VL, Gallo M, Riddle DL. Targets of DAF-16 involved in Caenorhabditis elegans adult longevity and dauer formation. Exp Gerontol. 2006;41:922–927. doi: 10.1016/j.exger.2006.06.058. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kumsta C, Thamsen M, Jakob U. Effects of oxidative stress on behavior, physiology, and the redox thiol proteome of Caenorhabditis elegans. Antioxid Redox Signal. 2011;14:1023–1037. doi: 10.1089/ars.2010.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamitina ST, Strange K. Transcriptional targets of DAF-16 insulin signaling pathway protect C. elegans from extreme hypertonic stress. Am J Physiol Cell Physiol. 2005;288:C467–474. doi: 10.1152/ajpcell.00451.2004. [DOI] [PubMed] [Google Scholar]
- Lee JY, Kim C, Kim J, Park C. DJR-1.2 of Caenorhabditis elegans is induced by DAF-16 in the dauer state. Gene. 2013;524:373–376. doi: 10.1016/j.gene.2013.04.032. [DOI] [PubMed] [Google Scholar]
- Li X, Matilainen O, Jin C, Glover-Cutter KM, Holmberg CI, Blackwell TK. Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet. 2011;7:e1002119. doi: 10.1371/journal.pgen.1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang WJ, Cao H, Lu J, Wu C, Hu FY, Guo J, Zhao L, Yang F, Zhang YX, et al. Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum Mol Genet. 2009;18:4897–4904. doi: 10.1093/hmg/ddp459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A, Oh SW, Tissenbaum HA. Worming pathways to and from DAF-16/FOXO. Exp Gerontol. 2006;41:928–934. doi: 10.1016/j.exger.2006.05.020. [DOI] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet. 2006;38:251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- Olsen A, Vantipalli MC, Lithgow GJ. Using Caenorhabditis elegans as a model for aging and age-related diseases. Ann N Y Acad Sci. 2006;1067:120–128. doi: 10.1196/annals.1354.015. [DOI] [PubMed] [Google Scholar]
- Ookuma S, Fukuda M, Nishida E. Identification of a DAF-16 transcriptional target gene, scl-1, that regulates longevity and stress resistance in Caenorhabditis elegans. Curr Biol. 2003;13:427–431. doi: 10.1016/s0960-9822(03)00108-8. [DOI] [PubMed] [Google Scholar]
- Pace NJ, Weerapana E. Diverse functional roles of reactive cysteines. ACS Chem Biol. 2013;8:283–296. doi: 10.1021/cb3005269. [DOI] [PubMed] [Google Scholar]
- Pace NJ, Weerapana E. A competitive chemical-proteomic platform to identify zinc-binding cysteines. ACS Chem Biol. 2014;9:258–265. doi: 10.1021/cb400622q. [DOI] [PubMed] [Google Scholar]
- Plenefisch J, Xiao H, Mei B, Geng J, Komuniecki PR, Komuniecki R. Secretion of a novel class of iFABPs in nematodes: coordinate use of the Ascaris/Caenorhabditis model systems. Mol Biochem Parasitol. 2000;105:223–236. doi: 10.1016/s0166-6851(99)00179-6. [DOI] [PubMed] [Google Scholar]
- Qian Y, Martell J, Pace NJ, Ballard TE, Johnson DS, Weerapana E. An isotopically tagged azobenzene-based cleavable linker for quantitative proteomics. Chembiochem. 2013;14:1410–1414. doi: 10.1002/cbic.201300396. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Blumenthal T, Meyer BJ, Priess JR. Introduction to C. elegans. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C elegans II. Cold Spring Harbor; NY: 1997. [Google Scholar]
- Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective "ligation" of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Smathers RL, Fritz KS, Galligan JJ, Shearn CT, Reigan P, Marks MJ, Petersen DR. Characterization of 4-HNE modified L-FABP reveals alterations in structural and functional dynamics. PLoS One. 2012;7:e38459. doi: 10.1371/journal.pone.0038459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smathers RL, Galligan JJ, Shearn CT, Fritz KS, Mercer K, Ronis M, Orlicky DJ, Davidson NO, Petersen DR. Susceptibility of L-FABP−/− mice to oxidative stress in early-stage alcoholic liver. J Lipid Res. 2013;54:1335–1345. doi: 10.1194/jlr.M034892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smathers RL, Petersen DR. The human fatty acid-binding protein family: evolutionary divergences and functions. Hum Genomics. 2011;5:170–191. doi: 10.1186/1479-7364-5-3-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch J, Corsico B. The emerging functions and mechanisms of mammalian fatty acid-binding proteins. Annu Rev Nutr. 2008;28:73–95. doi: 10.1146/annurev.nutr.27.061406.093710. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Tullet JM. DAF-16 target identification in C. elegans: past, present and future. Biogerontology. 2015;16:221–234. doi: 10.1007/s10522-014-9527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GA, White TM, McColl G, Jenkins NL, Babich S, Candido EPM, Johnson TE, Lithgow GJ. Heat shock protein accumulation is upregulated in a long-lived mutant of Caenorhabditis elegans. J Gerontol a-Biol. 2001;56:B281–B287. doi: 10.1093/gerona/56.7.b281. [DOI] [PubMed] [Google Scholar]
- Walsh CT, Garneau-Tsodikova S, Gatto GJ., Jr. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- Wang C, Weerapana E, Blewett MM, Cravatt BF. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat Methods. 2014;11:79–85. doi: 10.1038/nmeth.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Fridman A, Blackledge W, Connelly S, Wilson IA, Pilz RB, Boss GR. The phosphatidylinositol 3-kinase/akt cassette regulates purine nucleotide synthesis. J Biol Chem. 2009;284:3521–3528. doi: 10.1074/jbc.M806707200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Joo HJ, Paik YK. Novel functions of lipid-binding protein 5 in Caenorhabditis elegans fat metabolism. J Biol Chem. 2011;286:28111–28118. doi: 10.1074/jbc.M111.227165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Gupta V, Carroll KS, Liebler DC. Site-specific mapping and quantification of protein S-sulphenylation in cells. Nat Commun. 2014;5:4776. doi: 10.1038/ncomms5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu RX, Liu J, True N, Wang W. Identification of direct target genes using joint sequence and expression likelihood with application to DAF-16. PLoS One. 2008;3:e1821. doi: 10.1371/journal.pone.0001821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Kadiyala CS, Ching TT, Hakimi P, Saha S, Xu H, Yuan C, Mullangi V, Wang L, Fivenson E, et al. Enhanced energy metabolism contributes to the extended life span of calorie-restricted Caenorhabditis elegans. J Biol Chem. 2012;287:31414–31426. doi: 10.1074/jbc.M112.377275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarse K, Schmeisser S, Groth M, Priebe S, Beuster G, Kuhlow D, Guthke R, Platzer M, Kahn CR, Ristow M. Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab. 2012;15:451–465. doi: 10.1016/j.cmet.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.