Abstract
Information on multiple synteny between plants and/or within a plant is key information to understand genome evolution. In addition, visualization of multiple synteny is helpful in interpreting evolution. So far, some web applications have been developed to determine and visualize multiple homology regions at once. However, the applications are not fully convenient for biologists because some of them do not include the function of synteny determination but visualize the multiple synteny plots by allowing users to upload their synteny data by determining the synteny based only on BLAST similarity information, with some algorithms not designed for synteny determination. Here, we introduce a web application that determines and visualizes multiple synteny from two types of files, simplified browser extensible data and protein sequence file by MCScanX algorithm, which have been used in many synteny studies.
Keywords: multiple synteny, webtool, detection, visualization, plant
Introduction
Recent technological advances in sequencing make it possible to sequence genomes of various species rapidly. Given the genome sequences, understanding of a genome as well as the relationship between genomes has expanded enormously. Specifically, traces of special evolutionary events in plants such as genome duplication and speciation have been found. Synteny, a homologous region between or within chromosomes, is the most representative trace and has been widely studied to answer questions in evolution and composition of the plant genome. Thus, identifying the synteny in genomes at the gene level is important; hence, many tools have been developed to identify the synteny, such as DAGchainer1 i-ADHoRe,2 MCScanX,3 and OrthoCluster.4
To interpret synteny precisely, visualization of synteny is vital for comparative genome analysis because of the complex genome conservation and rearrangement.5 Thus, many programs and databases provide synteny information as an image in various forms such as OMA6 and PGDD.7 In addition, visualization of the multiple synteny that shows the relationship of various genes present in multiple chromosomes in a linear manner is helpful in interpreting many evolutionary events such as duplication and speciation. Especially, multiple synteny visualization at gene level displays the relationship among several species importantly to identify the evolutionary events and origin of the features. To date, there are few multiple homology visualization tools (eg, GEvo,8 mGSV,9 mVISTA10) that have helped in various genome studies. However, several demands by users are not covered by these tools. For example, the GEvo and the mVISTA determine and show homologous regions between sequences uploaded and/or selected by a user. However, the tools determine the regions just based on the similarity of sequences, so the reliability of the result regarding synteny is likely to be insufficient. The mGSV provides various options for the visualization; hence, the tool makes it possible to generate a high-quality image to help in the result interpretation. However, the tool does not include any synteny identification tools; hence, users need to make synteny information files. This can be a limitation for biologists who are not familiar with dealing large genomic data such as blasting thousands of protein sequences. For convenient and efficient synteny analysis, a fine plot for accurate multiple synteny should be generated from familiar raw data such as protein sequence file in FASTA format.
We have developed the MultiSyn (multiple synteny determination and visualization), a web-based tool for multiple synteny determination and visualization between user’s genomic data and/or published plant genome data. For identification of syntenic regions, we adopted the MCScanX as an algorithm to determine syntenies, which have been used to detect the synteny blocks for many plants (eg, Beta vulgaris,11 Brassica rapa12) and the MCScanX algorithm that can be used by statistical evaluation and correction repeatedly to search for a specific type of synteny.3 At present, 18 angiosperm species including Oryza sativa and Arabidopsis thaliana are provided in the tool. After detection of synteny, the information of synteny is linked with various visualization options and a multiple synteny map is drawn.
Materials and Methods
Overview of the MultiSyn method and procedure
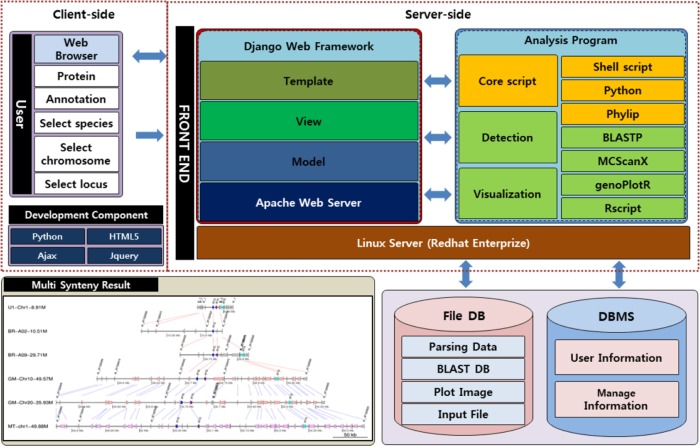
The MultiSyn was implemented on the server-side as well as on the client-side. The server-side program consists of BLASTP, MCScanX, and core script of programming using Python and Linux shell scripts in Django web framework.13 On the client-side, we implement the user interface to the Web-based program using jQuery14 and HTML5.15
There are two main components: detection of synteny and visualization of multiple synteny, which were developed in Python and R package, respectively, in order to generate an image of multiple synteny (Fig. 1). The webtool proceeds in four steps: (i) upload and/or set input data files; (ii) species selection for comparison and determination of the synteny by MCScanX; (iii) set options for drawing multiple synteny plot; and (iv) visualization of multiple synteny and adjusting the plot using core script. Users can progress onto the next step just by clicking the “next” button or the “before” button to go back.
Figure 1.
Architecture of MultiSyn webtool.
Notes: Architecture shows two configurations, the client-side and server-side. The client-side provides the interface for input and output for the MultiSyn user using HTML5 and jQuery. The user performs an analysis to enter the proteins (pep) and annotation (simplified BED) file or select species, chromosome, locus in a Web browser. The server-side consists of the core script for processing in connection with the webtool and for the determination and visualization of synteny. In the core script using python, phylip, shell script to connect the user to enter data and other utilities to detect and visualize synteny treated as a result. Input information is stored in the user information for the file DB and the analysis conducted provides the final output product.
The webtool provides a progress bar that is located at the bottom of the web page and shows the processing step of a job in real time. Table 1 was added as shown below; it shows the computation time to handle the data in MultiSyn and file size of the result plot is about 1 Mb.
Table 1.
Data computation time.
| NUMBER OF SELECT SPECIES | COMPUTATION TIME |
|---|---|
| 1 | <1 min |
| 2 | <3 min |
| 3 | ~30 mins |
| 5 | ~2 hours |
| 10 | ~10 hours |
| 15 | Over 12 hours |
Note: Computation time according to the number of selected species in the MultiSyn.
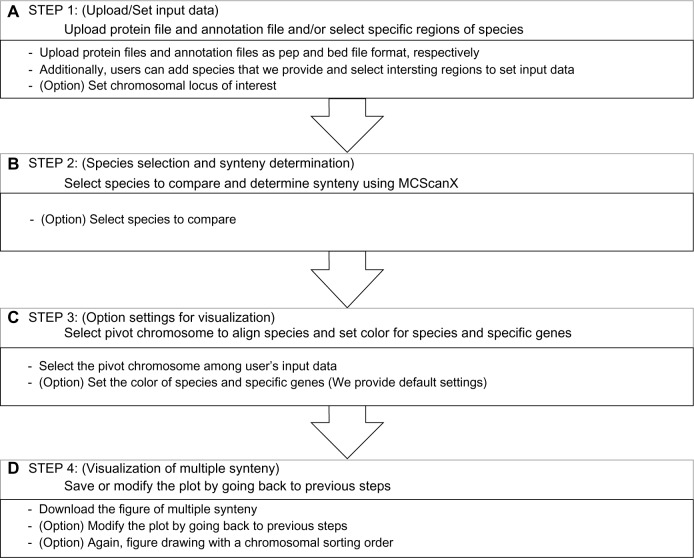
Step 1: upload and/or set input data as a pivot for a multiple synteny
In order to be used as a first-order species, MultiSyn allows users to set regions of interest of species against the 18 plants provided in the webtool (Table 2) by choosing a species, chromosome, and input locus information at the start and end positions (Fig. 2A). If the user enters the data, the webtool automatically classifies the chromosome. And it allows the user to select input in step 3. In addition to species the webtool also provides two files, a protein sequences file in FASTA format and a genome annotation file corresponding to the protein sequences; these can be used for input data. The format of the genome annotation file should be in a simplified browser extensible data (BED) format which has four columns; the name of the chromosome, the starting and ending positions of the protein in the chromosome, and the gene locus identifier corresponding to the protein respectively (Supplementary Table 1). The webtool provides a check box, “Automatic changing long name”, to decrease the length of the long gene name in input data to prevent the name overlapped with others in the plot. Concretely, the common characters in the names are determined by the longest common subsequence algorithm18 and removed.
Table 2.
Public plant genome sources.
| SPECIES NAME | COMMON NAME | ACCESS | VERSION |
|---|---|---|---|
| Arabidopsis thaliana | Arabidopsis | TAIR | TAIR10 |
| Beta vulgaris | Sugar beet | BVR | RefBeet−1.1 |
| Brassica rapa | Chinese cabbage | BRAD | Version 1.3 |
| Cajanus cajan | Pigeonpea | IIPG | Nov-11 |
| Capsicum annuum | Hot pepper | PapperGenomeDB | Version 1.55 |
| Fragaria vesca | Strawberry | PFR | Version 1.1 |
| Glycine max | Soybean | JGI | Wm82.a2.v1 |
| Lotus japonicus | Lotus | Kazusa | Version 2.5 |
| Medicago truncatula | Barrel medic | JCVI | Mt4.0v1 |
| Oryza sativa | Rice | RAP | Version 7.0 |
| Populus trichocarpa | Western poplar | JGI | Version 3.0 |
| Prunus persica | Peach | JGI | Version 1.0 |
| Pyrus bretschneideri | Pear | PGP | Version 1.0 |
| Solanum lycopersicum | Tomato | SGN | Version 2.4 |
| Solanum tuberosum | Potato | PGSC | Version 3.4 |
| Sorghum bicolor | Sorghum | JGI | Version 2.1 |
| Vitis vinifera | Grape vine | Genoscope | Genoscope (Aug 2007) |
| Zea mays | Common bean | AGI | Version 6a |
Note: The public data that include eudicots and monocots used to compare user’s sequence of interest.
Figure 2.
Progress of MultiSyn in webtool.
Notes: The yellow progress status in (A) is also presented in steps 2 and step 3 of the homepage. A: Step 1: users can upload their protein file (pep format) and annotation file (simplified bed format) or select species and set the range of chromosomal region of interest. B: Step 2: select species to compare. After species selection, synteny among user’s data and species are determined by MCScanX. C: Step 3: select pivot chromosome to align species and set colors for species and specific genes. D: Step 4: download the plot or modify the plot by going back to previous.
Step 2: select species to compare and determine synteny by MCScanX
In this step, the user chooses plants as target species that the webtool provides to determine multiple synteny (Fig. 2B). The first step in determining synteny by MCScanX is blasting protein sequences. In order to prevent superfluous modes, all the protein sequences in the provided species were blasted and the results were stored in the webtool. This method is more efficient in terms of time, similar to the results of performing BLASTP, which allows the users to shorten the time for obtaining final results of multiple synteny. Thus, the webtool only needs to blast the uploaded protein sequences set by the user against the protein sequences of the selected species. The default cutoff E-value for BLASTP is 1E-5 and the top five matches are used. Second, for the detection of synteny and collinearity in order to determine synteny for multiple species, the input results obtained from BLASTP are selected for MCScanX. Determined synteny carries out the operation visualized in the form desired by the researcher in the next step. Subsequently, synteny information is determined by MCScanX from two files: the blast result and simplified BED file.
Step 3: set options for drawing multiple synteny plot
To visualize a multiple synteny plot, the webtool requires a genomic region, pivot, used as a standard of the plot and located at the top in the plot. In step 3 of webtool, there is information about the pivot chromosome. MultiSyn provides three options for visualizing multiple synteny plots using the synteny determined in the previous step: pivot selection, color settings for species, and specific proteins (Fig. 2C). (i) The pivot has to be selected by the user during input data to align the species in order to compare by linking multiple synteny sequentially. (ii) For sophisticated visualization of multiple synteny, users can change the species colors via “color setting for species” in the webtool (MultiSyn provides differential colors for each species to default value). Moreover, (iii) proteins among input data and their synteny can be colored by putting the locus identifier that is appropriate for information on annotation file and color code in “color setting for protein”. In order to use a different color, click on “Color option” to toggle. Then, synteny is determined and the user enters option values to regenerate data sets for visualization through the core script. The core script is composed of three modules for data connected with other programs. Shell script-based program is for managing the processing and visualization of synteny with the determined information and value received from the user. And python-based programs are used to regenerate synteny, determined by the data set and R script program with genoPlotR16 to draw a plot. Through these core scripts, a plot is generated that visualizes the information for the multi synteny users.
Step 4: save or modify the multiple synteny plot
The optimally resized multiple synteny plot is displayed in the webtool, and the downloadable plot is provided in png format (Fig. 2D). The plot shows the pivot placed at the top of the plot and the colored species and proteins both in the pivot and synteny in other species as set by the user. Matching protein in synteny is linked by one of the two colored lines; red and blue lines mean same and opposite directional alignment, respectively. Each of the chromosomal aligned plots drawn can be rearranged in the order desired by the user. If necessary, the user can move back to the previous steps to modify the contents of each step for redrawing the plot.
The MultiSyn web server
MultiSyn is powered by the Apache Web server on a Red Hat Enterprise Linux operating system. The Web Framework application used for development was Django version 1.4. The server was an Intel Xeon 4core CPU at 2.2 GHz, 16 GB of RAM, 2 TB (SATA) of hard disk, and 2 ea of 1GB network interface. MultiSyn shows the multiple synteny in a plot and helps biologists to understand the pattern of evolution of the synteny regions. The webtool and example data sets are freely available at http://202.31.147.159:62001/.
Results and Discussion
Various genome sequences of plants are being rapidly identified with the development of DNA sequencing technologies that allow the evolutionary study to advance in order to understand genomes and the relationship between genomes. To compare multiple species, determination of the synteny and visualization of multiple synteny are required. Even though various types of multiple homology visualization tools are present, the absence of proper tools, which allow convenient use by biologists, is yet to be developed. Therefore, the development of the MultiSyn allows biologists to determine synteny and draw multiple synteny plots with ease.
Features of MultiSyn
MultiSyn includes features that allow biologists to upload their raw protein sequences (PEP format) with annotation files (simplified BED format) that include the chromosomal location. In addition, MultiSyn provides species to select from and settings to specify the range for data input. Other species also can be selected to compare against the 18 public species that are already run by BLASTP in order to reduce computation time. These 18 species include eudicots (asteroids and rosids) and monocots, which allow biologists to compare their sequences with. After comparison, MultiSyn determines the synteny using MCScanX and provides chromosomal pivot selection and color options to draw multiple synteny plots. Compared to the existing multiple homology visualization, we determine synteny using MCScanX. MCScanX is using the same algorithm as a program that extends the function of MCScan. MCScan uses a four-step algorithm of the top–down approach to find a synteny conservation pattern (all-against-all comparisons, a pool of syntenic chains, multiway synteny view, and interpretation of synteny). This, with respect to divergence and WGD events, allows seeing the combined result among multiple chromosomes.
Even the recent multiple synteny visualization tool, mGSV, draws the multiple synteny plot; it does not include the synteny detection due to heavy computation. However, we tried to reduce the computational time by pre-running BLASTP of the public genome data and contain the synteny determination, making it convenient for biologist by allowing them to use raw protein sequence data.
For biologists who want to determine multiple synteny among public plant genomes with their sequences of interest, MultiSyn effectively provides a suitable multiple synteny plot. Consequentially, this approach leads to greater visualization and rapid further analysis. It may be an efficient and effective tool for non-programming skilled biologists to perform comparative analysis.
Example of MultiSyn: PSY1
This example demonstrates how MultiSyn visualizes the multiple synteny plot using example gene region of phytoene synthase 1 (PSY1) with various public species. PSY1 that encodes the first dedicated step in lycopene biosynthesis was synteny analyzed in Solanum lycopersicum, which shows the neofunctionalization by Solanum triplication.17 Figure 3 shows a multiple synteny plot of PSY1 regions with B. rapa, Glycine max, Sorghum bicolor, Solanum tuberosum, and Vitis vinifera. The colors of the species are displayed as default values, whereas the colors of specific genes are visualized as our setting: AEC (Auxin efflux carrier family protein, Solyc03g031990.2) – purple, ARF8 (Auxin response factor 8, Solyc03g031970.2) – blue, HMGCR (3-hydroxy-3-methylglutaryl-coenzyme A, Solyc03g032010.2) – cyan, PSY1 (phytoene synthase, Solyc03g031860.2) – magenta, and UBQLN (Ubiquilin-1, 4, Solyc03g032160.2) – green, respectively. The syntenic matching genes in other species are also colored as the setting. The synteny blocks are linked by lines: red lines represent the same direction alignment and the blue lines mean opposite directional alignment. The steps to make the example plot are below (Supplementary Fig. 2):
Step 1: first enter the ID and click the button “Add a species”. This information is the longer gene name. Please check “Automatic changing long name”. And select the S. lycopersicum species and Sl2.40ch03 chromosome. After entering the respective 8600000 and 9000000 in the Start and End, press the “Next button”.
Step 2: select the five species (B. rapa, G. max, S. bicolor, S. tuberosum, and V. vinifera). This step takes longer than 30 minutes. The running time varies, depending on the number of selected species in MultiSyn.
Step 3: select the first start chromosome to draw the synteny among chromosomes (pivot chromosome). And you can specify the color options. Click the color options, modified in PSY1, the color setting for protein. We enter information on the locus identifier, protein name, and color code: PSY1(Tr1860.2, PSY1 and magenta), AEC(Tr1990.2, AEC and purple), ARF8(Tr1970.2, ARF8 and blue), HMGCR(Tr2010.2, HMGCR and cyan), and UBQLN(Tr2160.2, UBQLN and green). Tr character is reduced automatically by the system because of the long gene name at step 1. This information is shown on the top of the setting for color species.
Step 4: result plot shown on the screen is a preview. Click to download the results that can be found in the original plot. Each PSY1, AEC, ARF8, HMGCR, and UBQLN information entered in step 3 option is displayed on the first chromosome, depending on the color. This indicates the relationship with other species. To explain plot for result, Solyc03 g031860.2 (Tr1860.2) gene in the SL2.40ch03 chromosome of S. lycopersicum (u1: number of user input data) species is shown in magenta color. And it is shown on the synteny of PSY1 on Chr14 chromosome of G. max species and Chr10 chromosome of S. bicolor species. Thus, starting with the first chromosome (chromosome pivot) from other species, it is able to confirm the synteny of AEC, ARF8, HMGCR, and UBQLN.
Figure 3.
Multiple synteny plot of PSY1.
Notes: PSY1 encodes first dedicated step in lycopene biosynthesis, showing an expansion of genes by whole-genome triplication, neofunctionalization. The synteny of PSY1 regions (chromosome 3, 8.6 Mb ~ 9.0 Mb) are aligned with B. rapa (BR), G. max (GM), S. bicolor (SB), S. tuberosum (ST), and V. vinifera (VV). Red and blue lines mean same and opposite directional alignment, respectively. Magenta, blue, purple, cyan, and green boxes represent PSY1 (phytoene synthase, Solyc03g031860.2), ARF8 (Auxin response factor 8, Solyc03g031970.2), AEC (Auxin efflux carrier family protein, Solyc03g031990.2), HMGCR (3-hydroxy-3-methylglutaryl-coenzyme A, Solyc03g032010.2), and UBQLN (Ubiquilin-1, 4, Solyc03g032160.2), respectively.
In the same way (Add a species in Step 1 not click the Add a file button and enter a protein (.pep) and Annotation (.bed) file.), you can draw in extra sample provided by Supplementary Figure 1.
Conclusions
As the analysis of genome sequences or sequence of interest became crucial for identifying evolutionary significance, the demand for suitable tools have increased. Therefore, we developed the MultiSyn in order to aid evolutionary analysis by displaying multiple synteny. MultiSyn allows biologists to upload their protein sequence of interest, determine the synteny between their sequences, and pre-run public genome data using MCScanX and significantly present multiple synteny plots. MultiSyn provides a convenient means for biologists to analyze their sequence of interest, comparing them with public genome sequences through identification and visualization of evolutionary conserved regions.
Supplementary Material
Supplementary Figure 1. Extra sample of MultiSyn result plot.
Supplementary Figure 2. Progress of MultiSyn in webtool.
Supplementary Table 1. Data format; Protein(.pep) data, simplified annotation(.bed) data, color code.
Acknowledgments
We thank Professor Lee from Department of Computer Information Engineering, Kunsan National University for providing the support server settings.
Footnotes
ACADEMIC EDITOR: Liuyang Wang, Associate Editor
PEER REVIEW: Three peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1034 words, excluding any confidential comments to the academic editor.
FUNDING: This work was carried out with the support of “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ0108772016)”, Rural Development Administration, Republic of Korea. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived the project: D-WK, T-HL. Contributed mostly to the design: T-HL, J-HB, JK, D-WK. Developed the algorithms of optimized settings for visualization of multiple synteny files: T-HL, C-KK, S-HS, DC, MBR. Developed the web service: J-HB. Tested and revised the web application: JK. Wrote the manuscript: T-HL, J-HB, JK. Contributed equally to this work: J-HB, JK. Co-corresponding authors: D-WK and T-HL. All the authors read and approved the final manuscript.
REFERENCES
- 1.Haas BJ, Delcher AL, Wortman JR, Salzberg SL. DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics. 2004;20(18):3643–6. doi: 10.1093/bioinformatics/bth397. [DOI] [PubMed] [Google Scholar]
- 2.Simillion C, Janssens K, Sterck L, Van de Peer Y. i-ADHoRe 2.0: an improved tool to detect degenerated genomic homology using genomic profiles. Bioinformatics. 2008;24:127–8. doi: 10.1093/bioinformatics/btm449. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Tang H, Debarry JD, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng X, Pei J, Vergara IA, Nesbitt MJ, Wang K, Chen N. EDBT. Nantes, France: 2008. OrthoCluster: a new tool for mining synteny blocks and applications in comparative genomics. [Google Scholar]
- 5.Frazer KA, Elnitski L, Church DM, Dubchak I, Hardison RC. Cross-species sequence comparisons: a review of methods and available resources. Genome Res. 2003;13(1):1–12. doi: 10.1101/gr.222003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altenhoff AM, Škunca N, Glover N, et al. The OMA orthology database in 2015: function predictions, better plant support, synteny view and other improvements. Nucleic Acids Res. 2015;43:D240–9. doi: 10.1093/nar/gku1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee TH, Tang H, Wang X, Paterson AH. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 2012;41:D1152–8. doi: 10.1093/nar/gks1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyons E, Freeling M. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008;53(4):661–73. doi: 10.1111/j.1365-313X.2007.03326.x. [DOI] [PubMed] [Google Scholar]
- 9.Revanna KV, Munro D, Gao A, Chiu CC, Pathak A, Dong Q. A web-based multi-genome synteny viewer for customized data. BMC Bioinformatics. 2012;13:190. doi: 10.1186/1471-2105-13-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–9. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dohm JC, Minoche AE, Holtgräwe D, et al. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris) Nature. 2014;505(7484):546–9. doi: 10.1038/nature12817. [DOI] [PubMed] [Google Scholar]
- 12.Song X, Duan W, Huang Z, et al. Comprehensive analysis of the flowering genes in Chinese cabbage and examination of evolutionary pattern of CO-like genes in plant kingdom. Sci Rep. 2015;5:14631. doi: 10.1038/srep14631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Django. Available at: https://www.djangoproject.com/
- 14.jQuery. Available at: https://jquery.com/
- 15.HTML5. Available at: https://www.w3.org/TR/html5/
- 16.Guy L, Kultima JR, Andersson SG. genoPlotR: comparative gene and genome visualization in R. Bioinformatics. 2010;26(18):2334–5. doi: 10.1093/bioinformatics/btq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Tomato Genome Consortium The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485(7400):635–41. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergroth L, Hakonen H, Raita T. A survey of longest common subsequence algorithms. SPIRE (IEEE Computer Society) 2000:39–48. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Extra sample of MultiSyn result plot.
Supplementary Figure 2. Progress of MultiSyn in webtool.
Supplementary Table 1. Data format; Protein(.pep) data, simplified annotation(.bed) data, color code.