Abstract
Sphingosine-1-phosphate (S1P) and lysophosphatidic acid (LPA) are bioactive signaling lysophospholipids that activate specific G protein-coupled receptors on the cell surface triggering numerous biological events. In circulation, S1P and LPA associate with specific carrier proteins or chaperones; serum albumin binds both S1P and LPA while HDL shuttles S1P via interactions with apoM. We used a series of kinetic exclusion assays in which monoclonal anti-S1P and anti-LPA antibodies competed with carrier protein for the lysophospholipid to measure the equilibrium dissociation constants (Kd) for these carrier proteins binding S1P and the major LPA species. Fatty acid-free (FAF)-BSA binds these lysophospholipids with the following Kd values: LPA(16:0), 68 nM; LPA(18:1), 130 nM; LPA(18:2), 350 nM; LPA(20:4), 2.2 μM; and S1P, 41 μM. FAF human serum albumin binds each lysophospholipid with comparable affinities. By measuring the apoM concentration and expanding the model to include endogenous ligand, we were able to resolve the Kd values for S1P binding apoM in the context of human HDL and LDL particles (21 nM and 2.4 nM, respectively). The novel competitive assay and analysis described herein enables measurement of Kd values of completely unmodified lysophospholipids binding unmodified carrier proteins in solution, and thus provide insights into S1P and LPA storage in the circulation system and may be useful in understanding chaperone-dependent receptor activation and signaling.
Keywords: anti-lipid antibody, apolipoproteins, Kinetic Exclusion Assay, lipids, competitive affinity analysis, lipoproteins, lysophosphatidic acid, physical biochemistry, serum albumin, sphingosine-1-phosphate, human serum albumin
Sphingosine-1-phosphate (S1P) and lysophosphatidic acid (LPA) are bioactive lysophospholipids that bind and signal through multiple G protein-coupled receptors (GPCRs) (1–3). Many physiological processes, such as cell growth, differentiation, survival, motility, and angiogenesis (3), and pathophysiological processes, such as cancer, cardiovascular disease, multiple sclerosis, neuropathic pain, and fibrosis (4, 5), involve S1P or LPA signaling. The S1P and LPA pathways are validated therapeutic targets; many drugs and pharmacological agents have been developed to modulate the activity of receptors and enzymes in these pathways (1, 4, 6). Many of these compounds block circulating S1P and LPA from binding and activating cognate membrane-bound receptors.
Circulating S1P exists primarily bound to carrier molecules, including HDL, LDL, and serum albumin. HDL is a protein-rich lipoprotein containing multiple protein constituents (7) and reportedly binds ∼50–70% of plasma S1P, whereas serum albumin reportedly binds ∼30% or more (8–10). apoM represents the main protein component in HDL responsible for binding S1P, and the X-ray cocrystal structure of recombinant human apoM in complex with S1P has been solved (11). Human plasma contains approximately 0.9 μM apoM (11, 12), where >95% of the total apoM occupies ∼5% of the HDL (apoM-HDL) and <2% of the LDL (apoM-LDL) in plasma (13, 14); this stoichiometry results in less than 1 mol of S1P per mole of HDL in human plasma (15). S1P-associated HDL stimulates cellular pathways that promote endothelial barrier function, suggesting that S1P mediates the protective effects of HDL against atherosclerosis (16). While S1P bound apoM-HDL suppresses vascular inflammation, S1P delivered using albumin did not show this effect in vitro, suggesting divergent roles for S1P chaperones in maintaining the vasculature and other physiological processes (17, 18).
In blood, LPA also exists bound to carrier proteins, primarily serum albumin (19, 20). Total LPA in plasma comprises several distinct species, which contain esterified fatty acids with varying numbers of carbon atoms and cis double bonds (Fig. 1A) capable of activating cognate GPCRs with varying potencies (21, 22). Although albumin is the most abundant protein in human plasma and LPA is one of the first bioactive lipids identified, the stoichiometry and mechanism of interaction between these two molecules is poorly understood. As with fatty acids, serum albumin has the capacity to bind multiple LPA molecules per protein molecule (23–25). Studies suggest that albumin contains three strong affinity long-chain fatty acid binding sites, and these are the same sites occupied by LPA (26, 27).
Fig. 1.
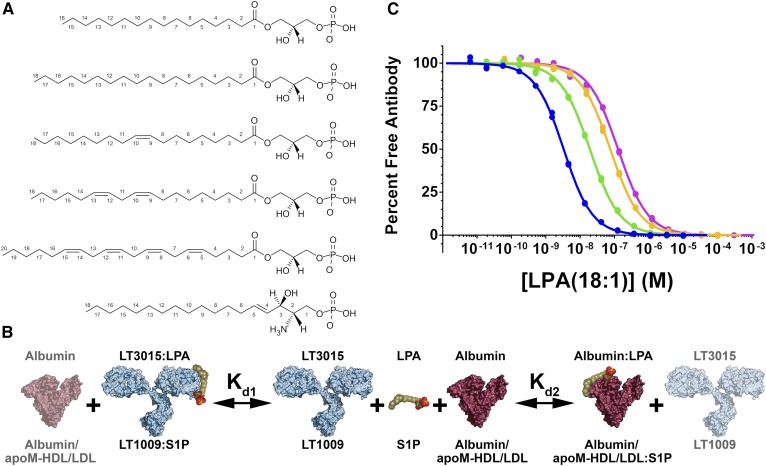
Equilibrium competition binding with native lysophospholipids in solution. A: Chemical structures of the lysophospholipids used in this study. From top to bottom: LPA(16:0), LPA(18:0), LPA(18:1), LPA(18:2), LPA(20:4), and S1P. B: Illustration showing the components in the equilibrium competition binding experiments: the lysophospholipid (olive green, orange, and red; LPA or S1P), mAb (light blue; anti-LPA, LT3015; anti-S1P, LT1009), and chaperone protein (purple; LPA, albumin; S1P, albumin and apoM-HDL/LDL). The labels printed above the picture denote components that compete for LPA binding, while the labels below the picture represent components that compete for S1P binding. Kd1 and Kd2 are the equilibrium dissociation constants for the antibody-lipid and chaperone-lipid interactions, respectively. C: Albumin competes with LT3015 for binding LPA(18:1) in vitro. Overlay of four KinExA equilibrium affinity experiments with constant antibody (1 nM) and varying concentrations of FAF-BSA: blue 3 μM, green 30 μM, orange 150 μM, and pink 300 μM show the response curves shifting toward higher lipid concentrations in response to the BSA sequestering LPA(18:1). Naively fitting any of these curves would result in an incorrect estimation of the LT3015-LPA(18:1) Kd value.
The method described in this report uses monoclonal antibodies (mAbs) to compete with purified serum albumin or isolated lipoprotein particles for binding S1P and LPA in solution (see cartoon schematic in Fig. 1B). The production and characterization of the two humanized IgG1k mAbs, LT1009 and LT3015,3 which specifically recognize S1P and LPA, respectively, and the structural basis for lipid recognition are described elsewhere (28, 29). These antibodies directly compete with carrier proteins for binding target lipids in vitro; the equilibrium binding curve for LT3015 binding LPA shifts toward weaker apparent affinity as the concentration of fatty acid-free (FAF)-BSA is increased (Fig. 1C). During competition binding, the equilibrium dissociation constants (Kd) for the antibody-lipid and protein-lipid interactions govern the concentration of antigen-free binding site on the antibody. Therefore, measuring the free antibody enables the Kd for both the antibody (Kd1) and the serum protein (Kd2) binding S1P or LPA to be determined.
The Kinetic Exclusion Assay (KinExA®) is a technique for measuring the Kd of protein-ligand interactions through direct detection of the ligand-free binding sites in a sample. This technique is compatible with a variety of biological systems and has several advantages over other methods (30–33). KinExA is particularly attractive to study protein-lipid interactions because native untagged molecules binding entirely in solution can be investigated. Using modified (non-native) lipid molecules or covalently attaching bulky tags or fluorophores may significantly alter the solubility properties of the lipids or the mechanisms of protein recognition. To overcome these issues, we developed a label-free method for determining the Kd for S1P and LPA binding carrier proteins in solution.
Using KinExA and simultaneously fitting several competitive curves (competition n-curve analysis), we measured the Kd values for FAF-BSA and FAF-human serum albumin (HSA) binding S1P and five predominant individual LPA species: palmitic (16:0), stearic (18:0), oleic (18:1), linoleic (18:2), and arachidonic (20:4). Because only the Kd value for LPA(18:1) binding albumin has been reported (34, 35), we were encouraged to measure the Kd values for other biologically active LPA species to investigate whether LPA species with different numbers of carbon atoms or unsaturated bonds in the acyl chain show different affinities for serum albumin. Indeed, we observed an ∼70-fold difference in Kd values between LPA(16:0) and LPA(20:4) binding FAF-HSA. We also used this method, along with a published apoM ELISA (36), to investigate S1P binding apoM in the context of isolated human HDL and LDL particles. Sevvana et al. (37) showed that S1P quenched the intrinsic fluorescence of purified recombinant human apoM with an IC50 value of 0.9 μM. This value is ∼45-fold and ∼375-fold higher (weaker binding) than the Kd values reported here for S1P binding to apoM-HDL and apoM-LDL, respectively. In addition, S1P appears to bind apoM-LDL with significantly stronger affinity compared with apoM-HDL, suggesting mechanistic differences in the apoM-S1P interaction between these lipoprotein particles. Finally, the Kd values for LT1009 and LT3015 binding their cognate lipid antigens absent the effects of the carrier proteins used to deliver the lipids have been determined. The use of carrier proteins to deliver lysophospholipids is ubiquitous in lipid research. Here, we demonstrate that the binding affinities of certain carrier proteins (or chaperones) for S1P or LPA are significant, and these interactions likely influence the bioavailability of the lipid and activation of their cognate receptors.
MATERIALS AND METHODS
Reagents and lipid preparations
The antigen-free binding sites on LT1009 and LT3015 were measured using a KinExA 3200 equipped with an autosampler. The antibody was captured using modified S1P and LPA, which contain a mercapto group covalently attached to the omega carbon atom and cross-linked to maleimide-activated BSA (Thermo Scientific) (3). The purified S1P-BSA and LPA-BSA conjugates were diluted (30 μg/ml) with PBS without calcium and magnesium (PBS, Cellgro), and 1 ml of the solution was added to 0.2 g of PMMA beads (Sapidyne Instruments Inc.). The slurry was rocked for 1 h at 37°C to adsorb the conjugate onto the beads. After coating, the beads were blocked with 150 μM Fraction V FAF-BSA (Calbiochem) in PBS. The concentrations of the protein stock solutions were determined by measuring the absorbance at 280 nm and using an extinction coefficient of 1.4 ml/mg for LT1009 and LT3015 and 0.66 ml/mg for FAF-BSA and FAF-HSA (Sigma-Aldrich) (38–41).
S1P, LPA(16:0), LPA(18:0), LPA(18:1), and LPA(20:4) (Avanti Polar Lipids) and LPA(18:2) (Echelon Biosciences) were resuspended in methanol by repeated sonication and vortex mixing. The lysophospholipid concentration in each stock solution was determined using a colorimetric total phosphorus assay using a protocol from Avanti Polar Lipids, which was based on work by Chen, Toribara, and Huber (42) and Fiske and Subbarow (43). Aliquots of the resuspended lipid stocks were transferred to glass vials via a glass syringe and the solvent was evaporated under a dry argon stream.
Kinetic exclusion determination
In order to demonstrate that the experiments described below were conducted in the so-called “KinExA mode,” i.e., dissociation of antibody-lipid complexes in solution does not significantly contribute to the antibody captured on the solid phase, we monitored the percentage of free antibody while systematically increasing the flow rate (32). The percentage of free antibody is calculated by dividing the signal (voltage) of the antibody in the presence of the target lipid by the signal of the antibody in the absence of the lipid (signal at 100% free antibody) after correcting for nonspecific binding. At equilibrium, samples containing the following antibody/lipid/albumin concentrations yielded approximately 50% free antibody signal at the slowest flow rate (0.25 ml/min): 1) 10 nM LT3015, 13 nM LPA(16:0), and 13 nM FAF-BSA; 2) 10 nM LT3015, 65 nM LPA(20:4), and 13 nM FAF-BSA; and 3) 10 nM LT1009, 14 nM S1P, and 1 μM FAF-BSA. As the flow rate increased, the percentage of free antibody in these solutions does not change significantly (supplemental Fig. S1), demonstrating that the fraction of antibody in complex with the lipid does not contribute to the free antibody measurements in the equilibrium affinity experiments.
Serum albumin affinity experiments
Dried aliquots of S1P were resuspended in S1P running buffer [10 mM HEPES, 150 mM NaCl, 2.5 mM CaCl2, 0.005% polysorbate 20, 0.02 NaN3 (pH 7.4)] containing 100 μM FAF-BSA by sonication and vortex mixing to yield a 0.1 mM S1P stock solution. Calcium was included in the running buffer because divalent metal ions bridge the antibody-S1P interface and are required for strong affinity binding (28). A series of 2-fold S1P dilutions were prepared in glass vials using a glass syringe using the running buffer above with 100 μM FAF-BSA. For LPA experiments, dried LPA aliquots were resuspended in PBS containing 15 μM FAF-BSA by sonication and vortex mixing to yield LPA stock solutions of 0.5 mM [LPA(16:0), LPA(18:0), LPA(18:1), LPA(18:2)] or 5 mM [LPA(20:4)], and a series of 2-fold dilutions were prepared using a glass syringe with PBS plus 15 μM FAF-BSA. During all these preparations, extra precautions were taken to minimize lipid material loss, and the syringes were washed several times between titrations to minimize lipid carry-over.
For each equilibrium affinity experiment, the antibody and serum albumin concentrations were constant. For S1P experiments, samples containing either 1 or 10 nM LT1009 and 1 or 500 μM FAF-BSA (dependent on experiment) in S1P running buffer were prepared in silanized glass tubes (Thermo Scientific). For each of the LPA species, samples containing either 0.5 or 10 nM LT3015 IgG and 13 nM or 10 μM FAF-BSA (dependent on experiment) in PBS were prepared in silanized glass tubes. S1P or LPA was added to the antibody and FAF-BSA-containing tubes from the titrated lipid stocks described above using a glass syringe from low to high lipid concentration to minimize lipid carry-over. FAF-HSA was used in place of FAF-BSA for the 10 nM LT1009, 500 μM FAF-HSA and the 10 nM LT3015, 10 μM FAF-HSA experiments.
Sample sets containing antibody, serum albumin, and titrated lipid were allowed to equilibrate (6–24 h depending on experiment) prior to data collection. Flow rates of 2.25 ml/min and 0.25 ml/min for LPA and S1P experiments, respectively, were used and shown to kinetically exclude dissociation of antibody-lipid complexes during capture of the free antibody. The captured antibody was detected using a goat anti-human Alexa Fluor or DyLight secondary (Jackson ImmunoResearch). The free antibody in each sample was measured in duplicate. Data were analyzed using drift correction in the competition n-curve software (Sapidyne Instruments); see details in the Data analysis procedures section.
HDL/LDL affinity experiments
HDL (d 1.063–1.21 g/ml) and LDL (d 1.019–1.063 g/ml) isolated from a single normal human male donor using KBr ultracentrifugation and gel filtration chromatography (HDL only) were purchased from EMD Millipore and used as lipoprotein stock solutions without further purification. Two equilibrium affinity experiments were set up where endogenous S1P in the HDL and LDL stock solutions served as the sole ligand/antigen source. Here, 2-fold serial dilutions of the HDL or LDL stocks (starting at 1:10 HDL or 1:6.1 LDL) containing either 1 or 10 nM LT1009 were prepared in S1P running buffer supplemented with 1 μM FAF-BSA.
Three additional sample sets containing a constant amount of HDL or LDL and a 2-fold serial titration of exogenous S1P were prepared. Again, S1P was delivered via glass syringe from low to high lipid concentration to samples containing either 1 or 10 nM LT1009 and a fixed dilution of the lipoprotein stock (HDL, 1:80, 1:450, and 1:1,200; LDL, 1:40, 1:225, and 1:600). The free LT1009 in each fraction was measured in duplicate, and the data were analyzed using drift correction in the competition n-curve software as detailed in Data analysis procedures below.
Data analysis procedures
The binding experiments described here involved a carrier protein (FAF-BSA, FAF-HSA, apoM-HDL, or apoM-LDL) in the reaction mixture. Generally, the Kd for the binding of the lipid to the carrier protein is more than an order of magnitude weaker than the antibody binding, however experimental conditions often dictate the carrier protein be present at concentrations more than an order of magnitude higher than the antibody. This results in a situation where the antibody and carrier protein compete for the lipid, an effect which must be taken into account when measuring the Kd values of the antibody-lipid and carrier protein-lipid interactions.
The mathematics of competitive binding is well-known and both exact and implicit solutions to the fundamental equations of binding and conservation of mass have been described (44, 45).
In the present case, we implemented an implicit solution of the following set of equations: Kd1 = [Ab][L]/[AbL], Kd2 = [P][L]/[PL], [Ab]T = [Ab] + [AbL], [P]T = [P] + [PL], and [L]T = [L] + [AbL] + [PL]; where Kd1 and Kd2 are the equilibrium binding constants of lipid (L) for antibody (Ab) and carrier protein (P), respectively. The final three equations express conservation of mass requirements for binding.
In the present analyses, the total concentrations of the antibody, [Ab]T, and the carrier protein, [P]T, were treated as independent variables. The concentration of free antibody, [Ab], was measured using the KinExA instrument. The Kd values and total lipid concentration, [L]T, were varied to minimize the least squared error to the measured data.
Uniqueness of fit was assessed by construction of error curves for the fitted parameters. After optimum values of the parameters had been determined, one parameter at a time was moved away from its optimum value and the remaining parameters were reoptimized; the residual error was plotted versus the varying parameter. Presence of a distinct minima in the resulting error graph is treated as evidence that the optimized value was unique (i.e., all other values of this parameter resulted in a worse fit to the measured data). Achieving unique fits for the parameters required that measurements be made at multiple values of the independent variables, [Ab] and [P].
The most complicated cases analyzed involved LT1009 binding S1P in the presence of apoM-HDL or apoM-LDL. The complication here is that neither lipoprotein particle was available in a form that was both active as a chaperone protein and free of S1P. Consequently, adding apoM-HDL or apoM-LDL to an experimental mixture introduces an unknown quantity of S1P. For analysis, the presence of endogenous ligand was accommodated by adding a parameter to the model to express the fraction of the apoM carrying an S1P molecule. This parameter was varied to fit the data and evaluated for uniqueness by construction of an error graph.
Confidence intervals (CIs) for the various parameters were constructed using a Monte Carlo method, as described by Straume and Johnson (46). Briefly, the optimal fitted parameters were used to construct a noise-free “data” set. Pseudo random noise was added to the simulated data and then analyzed for best fit parameters using the same algorithms used for analysis of the measured data. The magnitude of noise added was chosen such that the residual error at best fit was close to the residual for the corresponding measured data set. The addition of pseudo random noise and subsequent analysis was repeated one thousand times and the resultant one thousand best fit values of the various parameters were recorded. The 95% CI was then taken as the range from the 25th smallest parameter value to the 25th largest parameter value.
A molecular mass of 66 kDa for FAF-BSA and FAF-HSA was used for molarity calculations and was based on the manufacturer-provided molecular masses. The total HDL and LDL protein concentrations were determined using the bicinchoninic acid assay (HDL, 14.9 mg/ml; LDL, 6.56 mg/ml) and are in good agreement with concentrations provided by the manufacturer (HDL, 13.7 mg/ml; LDL, 6.39 mg/ml). apoM concentration measurements were carried out in the laboratory of C. Christoffersen by C. Wandel, as previously described (36). All data were exported to GraphPad Prism software for visualization (GraphPad Software, Inc.).
RESULTS
The competition binding between carrier proteins and mAbs that specifically bind S1P and LPA has been investigated using KinExA. The equilibrium dissociation constants for BSA and HSA binding the predominant LPA species [palmitic (16:0), stearic (18:0), oleic (18:1), linoleic (18:2), and arachidonic (20:4)] and S1P are reported, along with the Kd values for S1P binding apoM within isolated human HDL and LDL particles. These values represent interactions between unmodified protein and lipid components in solution, without using any separation procedures or molecular tags.
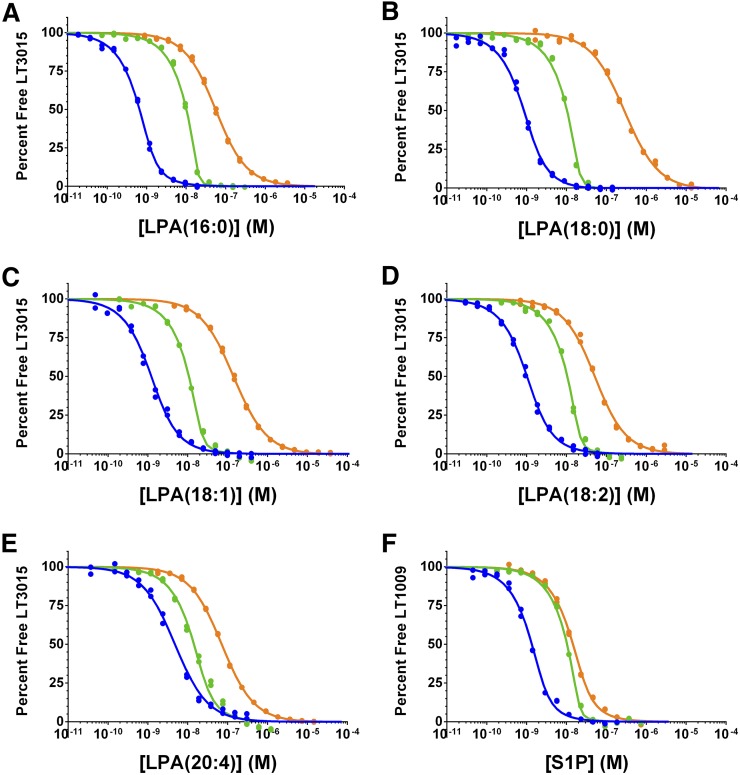
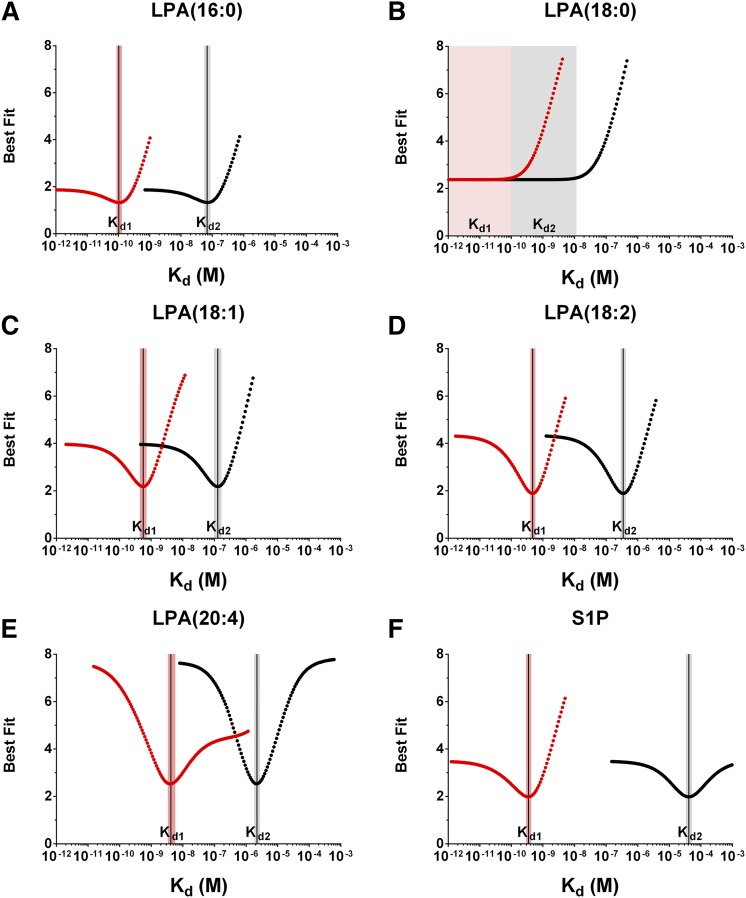
The simultaneous determination of the equilibrium dissociation constants for S1P or LPA binding the antibody (Kd1) or serum albumin (Kd2) is calculated by fitting three equilibrium affinity experiments, where the antibody or the albumin concentration is systematically altered. The first experiment (Fig. 2, blue curves) uses relatively low antibody and albumin concentrations, which should be near or below the Kd for each interaction. In the second experiment (Fig. 2, green curves), the albumin concentration remains unchanged, while the antibody concentration is increased such that the ratio of the antibody concentration to the Kd is sufficiently high to yield information about the titrant concentration. In these experiments, the nominal antibody concentration was 10 nM (20 nM antigen binding sites). The antibody concentration in the third experiment (Fig. 2, orange curves) remains unchanged at 10 nM, while the albumin concentration is increased to reshape the equilibrium binding curve, where the Kd2 parameter governs the change in curve shape and position observed between the second (green) and third (orange) curves in Fig. 2. Figure 3 shows the best fit Kd values for the albumin experiments, and the calculated Kd1 and Kd2 values, ligand activities, and curve fitting errors are reported in Tables 1, 2. An overlay of the individual 10 μM FAF-BSA affinity curves for the five LPA species in Fig. 2 shows the same relative LT3015 binding affinities as isotherms generated using a competition ELISA assay (supplemental Fig. S4): LPA(16:0) = LPA(18:2) > LPA(18:1) > LPA(20:4) > LPA(18:0). The difference between this rank order of apparent LPA binding affinities and the actual Kd1 values reported in Table 1 illustrates the effect of using FAF-BSA to deliver LPA in these types of equilibrium binding experiments.
Fig. 2.
Competition affinity experiments with FAF-BSA. Global curve fitting of the three affinity experiments used to determine the equilibrium dissociation constants for the individual LPA species (A–E) or S1P (F) binding the antibody or FAF-BSA in solution. The percentage of antigen-free binding sites on the antibody (in duplicate) is plotted as a function of the lysophospholipid concentration in each sample. The LT3015 and FAF-BSA concentrations used in each LPA experiment are as follows: blue curve, 0.5 nM LT3015 and 13 nM FAF-BSA; green, 10 nM LT3015 and 13 nM FAF-BSA; orange, 10 nM LT3015 and 10 μM FAF-BSA. For the S1P, the LT1009 and FAF-BSA concentrations are as follows: blue, 1 nM LT1009 and 1 μM FAF-BSA; green 10 nM LT1009 and 1 μM FAF-BSA; orange, 10 nM LT1009 and 500 μM FAF-BSA.
Fig. 3.
Error curves for FAF-BSA experiments. Competition n-curve error curves for the experiments in Fig. 2. The Kd values for LPA (A–E) and S1P (F) binding either the antibody (Kd1, red dotted curve) or FAF-BSA (Kd2, black dotted curve) correspond to the position the solid black vertical line intersects the x axis, and the shaded red and black bars span the 95% CI for the Kd1 and Kd2 value, respectively, except for LPA(18:0) (B), where the lower bound did not resolve. All values are reported in Table 1.
TABLE 1.
Results of competition affinity analysis for LPA and S1P binding mAbs and BSA in solution
| Lysophospholipid | Kd1 Antibodya | Kd2 FAF-BSAb | Active Ligandc (%) | Errord (%) |
| LPA(16:0) | 100 pM (86–130 pM) | 68 nM (56–87 nM) | 73 (71–75) | 1.3 |
| LPA(18:0) | UD (UD–93 pM) | UD (UD–11 nM) | 34 (32–36) | 2.4 |
| LPA(18:1) | 560 pM (440–710 pM) | 130 nM (98–170 nM) | 94 (88–100) | 2.2 |
| LPA(18:2) | 470 pM (400–580 pM) | 350 nM (290–430 nM) | 57 (55–60) | 1.9 |
| LPA(20:4) | 4.2 nM (3.3–5.8 nM) | 2.2 μM (1.8–2.8 μM) | 29 (25–35) | 2.5 |
| S1P | 350 pM (290–430 pM) | 41 μM (34–52 μM) | 83 (70–98) | 2.0 |
UD, undefined.
Anti-LPA, LT3015; anti-S1P, LT1009.
3:1 LPA:BSA and 1:1 S1P:BSA stoichiometry (see text for discussion of alternate stoichiometries).
Fit parameter describing the amount of active ligand, expressed as a percent of the nominal LPA or S1P concentration, determined for each set of experiments in Fig. 2.
Residual error between the data points and the theoretical curves.
TABLE 2.
Results of competition affinity analysis for LPA and S1P binding mAbs and HSA in solution
| Lysophospholipid | Kd1 Antibodya | Kd2 FAF-HSAb | Active Ligandc (%) | Errord (%) |
| LPA(16:0) | 74 pM (56–100 pM) | 31 nM (23–42 nM) | 73 (71–75) | 1.4 |
| LPA(18:0) | UD (UD–88 pM) | UD (UD–8.7 nM) | 34 (33–37) | 2.4 |
| LPA(18:1) | 560 pM (430–710 pM) | 130 nM (99–170 nM) | 94 (88–100) | 2.2 |
| LPA(18:2) | 470 pM (410–580 pM) | 320 nM (270–400 nM) | 57 (55–60) | 1.8 |
| LPA(20:4) | 4.1 nM (3.2–5.9 nM) | 1.7 μM (1.3–2.1 μM) | 29 (25–35) | 2.6 |
| S1P | 330 pM (280–410 pM) | 22 μM (18–27 μM) | 82 (69–97) | 1.9 |
UD, undefined.
Anti-LPA, LT3015; anti-S1P, LT1009.
3:1 LPA:HSA and 1:1 S1P:HSA stoichiometry (see text for discussion of alternate stoichiometries).
Fit parameter describing the amount of active ligand, expressed as a percent of the nominal LPA or S1P concentration, determined for each set of experiments in supplemental Fig. S2.
Residual error between the data points and the theoretical curves.
Serum albumin and LT3015 binding the major LPA species
Serum albumin contains three high-affinity long-chain fatty acid binding sites (27, 47, 48), which can be occupied by LPA (26). Using this stoichiometry [3:1 LPA:albumin (49)], the Kd1 values for LT3015 binding LPA(16:0), LPA(18:1), LPA(18:2), and LPA(20:4) are 100 pM, 560 pM, 470 pM, and 4.2 nM, respectively, and the Kd2 values for FAF-BSA binding LPA(16:0), LPA(18:1), LPA(18:2), and LPA(20:4) are 68 nM, 130 nM, 350 nM, and 2.2 μM, respectively (Table 1; Figs. 2A, C–E; 3A, C–E). For LPA(18:0), the upper 95% CI is 93 pM for Kd1 and 11 nM for Kd2, and the lower 95% CI is unresolved (Figs. 2B, 3B). We also analyzed these data using 2:1 and 1:1 LPA:albumin ratios (supplemental Table S1). For 1:1 LPA:albumin, the Kd2 value is precisely one-third the value reported in Table 1, while the other parameters do not significantly change. This direct proportional relationship between the concentration of ligand binding sites on albumin and the Kd2 value was observed for all LPA:albumin ratios analyzed.
In the competition n-curve analysis, Kd2 is overwhelmingly based on the 10 μM albumin experiment (orange curve, Fig. 2). Separate competition n-curve analyses using 10 μM FAF-HSA instead of FAF-BSA provides Kd2 values for FAF-HSA binding each of the individual LPA species (Table 2; supplemental Figs. S2A–E, S3A–E). A comparison of the Kd2 values between FAF-BSA (Table 1) and FAF-HSA (Table 2) suggests that: 1) FAF-BSA and FAF-HSA bind each LPA species with similar affinities; 2) both FAF-BSA and FAF-HSA bind stronger (lower Kd2 value) to fully saturated LPA species; and 3) as the number of double bonds in the LPA species increase, the affinity for serum albumin weakens (higher Kd2 value).
In this analysis, the antibody concentration is used as a reference, which assumes the antibody preparation is fully active. The advantage of fixing the antibody concentration as the reference is that we could float a parameter that represents the amount of ligand (S1P or LPA) present in the sample sets as a percent of the nominal lipid concentration. Including this parameter is informative when hydrophobic lipids are titrated in aqueous solution and the loss or carry-over of lipid material due to pipetting is presumed. The calculated lipid concentration is the highest for LPA(18:1) across the three affinity experiments at 94% nominal and lowest for LPA(18:0) and LPA(20:4) at 34 and 29% nominal, respectively. We believe these values reflect the solubility of these species in their stock solutions; LPA(18:0) has a relatively low critical micelle concentration (CMC) value (50), and the LPA(20:4) stock solution was prepared at a 10-fold higher concentration (see Materials and Methods). The apparent loss of LPA(18:0) and LPA(20:4) likely occurred during sample preparations even though extensive precautions and measures were taken to minimize this effect. One strength of this analysis is that all Kd values reported depend on the antibody and competitive protein (FAF-BSA, FAF-HSA, apoM-HDL, or apoM-LDL) concentrations rather than the lipid concentration, which is difficult to accurately control. The nearly 100% activity of the LPA(18:1) supports the assumption that the LT3015 antibody is 100% active.
Serum albumin and LT1009 binding S1P
We also investigated competition between the anti-S1P antibody, LT1009, and FAF-BSA for S1P binding using the same approach described for LPA above. A competition n-curve analysis of two 1 μM FAF-BSA experiments (one with 1 nM and the other with 10 nM LT1009) along with a 10 nM LT1009, 500 μM FAF-BSA experiment results in Kd1 and Kd2 values of 350 pM (95% CI 290–430 pM) and 41 μM (95% CI 34–52 μM) for S1P binding LT1009 and FAF-BSA, respectively (Table 1; Figs. 2F, 3F). Substituting the 500 μM FAF-HSA experiment yielded a Kd2 value of 22 μM (95% CI 18–27 μM) (Table 2; supplemental Figs. S2F, S3F) for the S1P-HSA interaction.
apoM-HDL and apoM-LDL binding S1P
As mentioned above, the total concentration of ligand binding sites for the chaperone protein must be defined to resolve the Kd2 value using the competition n-curve analysis. To gain this information, the concentration of apoM in pooled human HDL and LDL preparations was measured using a published ELISA method (36). The concentration of apoM in our human HDL and LDL preparations measured 21.0 μM and 0.83 μM, respectively, which equates to 1.41 μmol apoM per gram total protein and 0.13 μmol apoM per gram total protein for HDL and LDL, respectively.
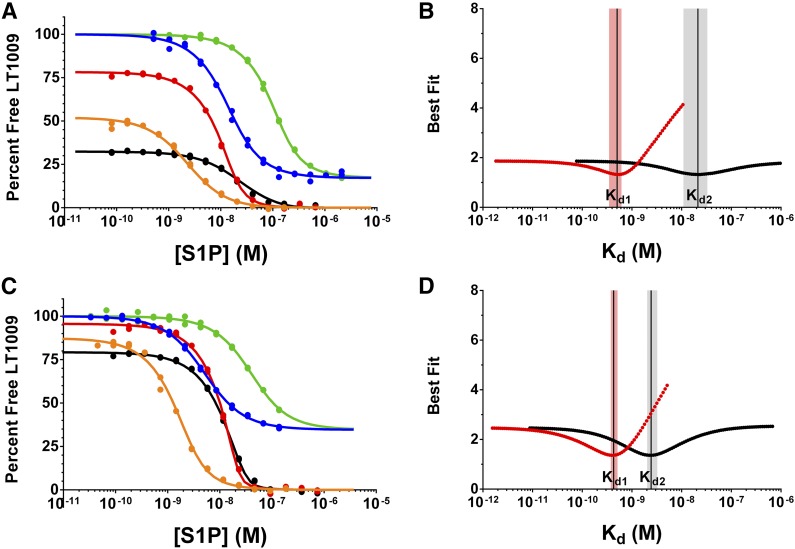
Using these concentrations and 1:1 S1P:apoM stoichiometry, the Kd values for S1P binding apoM-HDL and apoM-LDL, the ligand activity (percent nominal S1P concentration), and the concentrations of S1P in the HDL and LDL stock solutions were determined. Five equilibrium affinity experiments were used to resolve these parameters; three experiments maintained a fixed concentration of apoM-HDL or apoM-LDL while titrating exogenous S1P (Fig. 4; red, black, and orange curves) and two experiments titrated the HDL/LDL component without adding exogenous S1P (Fig. 4; blue and green curves). Competition n-curve analysis yielded Kd2 values of 21 nM (95% CI 11–33 nM) and 2.4 nM (95% CI 2.0–3.2 nM) for S1P binding apoM-HDL and apoM-LDL, respectively, and Kd1 values of 500 pM (95% CI 350–610 pM) and 430 pM (95% CI 370–510 pM), respectively. These Kd1 values for S1P-LT1009 interactions determined from the apoM-HDL/LDL analysis are in good agreement with the values determined when S1P is delivered using FAF-BSA/HSA (Tables 1, 2). The activity of the titrated S1P was 74% (95% CI 57–103%) and 83% (95% CI 69–99%) for apoM-HDL and apoM-LDL experiments, respectively, which is also similar to the percent S1P activity in the FAF-BSA/HSA experiments.
Fig. 4.
Competition affinity experiments and error curves for S1P binding apoM-HDL and apoM-LDL. Competition n-curve global fitting and error curves for the five equilibrium affinity experiments used to determine the Kd2 value for S1P binding isolated HDL (A, B) and LDL (C, D) particles in solution. A, C: Two affinity experiments (blue and green curves) used a constant concentration of the anti-S1P antibody (blue, 1 nM LT1009; green, 10 nM LT1009), and the lipoprotein particles were titrated starting at 1:10 and 1:6.1 dilutions of the neat HDL and LDL preparations, respectively. The decrease in the percent of antigen-free binding sites on LT1009 is due to binding the endogenous S1P in the lipoprotein preparations. The three additional affinity experiments (black, red, and orange curves) used constant antibody concentrations (orange, 1 nM LT1009; red, 10 nM LT1009; black, 10 nM LT1009) and a constant dilution of neat HDL (orange, 1:1,200; red 1:450; black, 1:80) or LDL (orange, 1:600; red, 1:225; black 1:40) preparations. In these three experiments, exogenous S1P was titrated using 1 μM FAF-BSA to deliver the ligand. Error curves showing the best fit Kd parameters for S1P binding to LT1009 [Kd1 (B, D)], apoM-HDL [Kd2 (B)], and apoM-LDL [Kd2 (D)]. The shaded red and black bars span the 95% CIs for the Kd1 and Kd2 values, respectively, and the reported value is represented as a vertical black line.
The concentration of endogenous S1P in the HDL and LDL preparations was calculated using a parameter that represents the S1P concentration in the stock solutions as a fraction of the total S1P binding sites on the chaperone, or the total apoM concentration in this analysis. The multiplier values were determined to be 0.10 (95% CI 0.096–0.11) and 0.25 (95% CI 0.22–0.28) for apoM-HDL and apoM-LDL, respectively, which calculates to 2.1 μM and 0.21 μM S1P in the HDL and LDL preparations, respectively. By comparison, the total S1P (S1P + dihydroS1P) in these preparations measured 1.7 μM and 0.11 μM, respectively, using HPLC-MS/MS. The S1P content in these HDL and LDL preparations (normalized for total protein) is 140 pmol/mg (95% CI 134–154 pmol/mg) and 32 pmol/mg (95% CI 27–35 pmol/mg), respectively. These levels are consistent with previous reports describing the S1P content in analogous lipoprotein preparations (51, 52).
Lipids other than S1P, such as myristic acid and oxidized phospholipids, have been reported to associate with apoM (13). In order to address the effect of additional lipids competing with S1P for binding apoM, we carried out a series of simulations using our data and introducing an additional lipid and Kd. For this work, we expanded the model to include a second lipid that competed with S1P for apoM binding sites, but did not bind to the antibody. Here, again, we looked for uniqueness of fit by calculating error curves as described above. We were not able to determine from this work whether there was additional lipid present on the apoM or not; however, our fitting of the model to the data suggests that any lipid present is not significantly affecting the binding. We were able to find fits equivalent to the simpler model with either vanishingly small quantities of additional lipid with relatively strong/low Kd binding or large quantities of a very weak binding lipid. Forcing the fit to use a significant quantity of additional lipid with a Kd in the range of the S1P-apoM Kd resulted in large residual errors and clear systematic deviations in fit from the measured data.
DISCUSSION
The KinExA, along with mAbs and the competition n-curve analysis package, enables researchers to measure the equilibrium binding constants for plasma proteins binding native LPA and S1P in solution. In our experience, chemically modifying LPA or S1P with molecular tags or protein conjugation significantly changes their biochemical properties (hydrophobicity, solubility, conformational dynamics) and, therefore, the Kd values determined using these derivatives are not necessarily representative of native interactions. However, measuring label-free S1P and LPA binding is challenging because these lysophospholipids are not traceable by UV/Vis absorption or fluorescence spectroscopy, difficult to deliver in aqueous media, and not particularly amenable to traditional separation processes (i.e., chromatography, dialysis membranes, etc.). Competition binding with KinExA, which allows for native protein-ligand interactions to equilibrate before capturing and detecting the free antibody at kinetically excluded flow rates, overcomes these challenges associated with measuring the equilibrium binding constants of protein-lipid interactions.
In order to determine Kd2 using this method, the number of ligand binding sites on the carrier protein must be assigned. Thumser, Voysey, and Wilton (26) reported that three moles of LPA could bind to one mole of serum albumin, and likely occupy the same three high-affinity sites as long-chain fatty acids (27, 48). Based on these reports, we performed the competition n-curve analysis using three LPA binding sites per albumin molecule (3:1 LPA:albumin). We also analyzed the data using stoichiometries ranging from one to six, and in every case the number of LPA binding sites on serum albumin directly and proportionally modulated the Kd2 value for the interaction. For example, the Kd2 for FAF-BSA binding LPA(16:0) reported in Table 1 is 68 nM using three binding sites per albumin molecule. The same analysis using one binding site per albumin molecule results in a 3-fold lower Kd2 value (23 nM), and the other parameters (Kd1, percent active lipid, percent error) remain unchanged. Supplemental Table S1 shows the Kd1 and Kd2 values calculated for the LPA species assuming one, two, or three equivalent LPA-binding sites on FAF-BSA/HSA. Due to the lack of published data on the S1P:albumin stoichiometry, one S1P-binding site per serum albumin molecule was used for this analysis.
The equilibrium binding of serum albumin to LPA has been studied previously. Goetzl et al. (34) reported a Kd of 357 ± 64 nM for FAF-BSA binding LPA using a radiometric equilibrium binding assay. Although the authors did not specify which LPA species was used, we speculate that it was LPA(18:1), the most commonly used LPA reagent in the literature. Ojala et al. (35) reported an apparent dissociation constant of 606 nM for HSA binding LPA(18:1) using a 6-anilinonaphthalene-2-sulfonic acid (ANS) displacement assay. The displacement of ANS from albumin generated sigmoidal binding curves, which the authors suggest could be due to either positive cooperativity or ANS not occupying the highest affinity fatty acid binding sites. Neglecting cooperativity, these values and the Kd2 values for LPA(18:1) reported in Tables 1, 2 are all in the hundreds of nanomolar (10−7 M) range. In addition, Ojala et al. (35) report an apparent Kd for HSA binding S1P of 1.3 μM, which is somewhat stronger than the Kd2 value of 22 μM that we report in Table 2.
The competition binding analysis performed here suggests that the affinity of LT3015 is 102–103 times stronger than the affinity of serum albumin for these individual LPA species. Interestingly, both the Kd1 and Kd2 values for LT3015 and serum albumin binding LPA(20:4) are significantly weaker compared with the other LPA species tested (Tables 1, 2). The weaker affinity of LT3015 for LPA(20:4) may be due to the C5-C6 cis double bond in the acyl chain of LPA(20:4), which is not present in the other LPA species (Fig. 1A). Based on the LT3015 fragment antigen-binding LPA(14:0) and fragment antigen-binding LPA(18:2) crystal structures, the LPA epitope consists of the glycerolphosphate group plus the juxtaposed C1-C8 carbon atoms of the acyl chain (29). As visualized in both cocrystal structures, this portion of the bound LPA hydrocarbon chain is fully extended and forms extensive intermolecular hydrophobic contacts with the antibody. The C5-C6 cis double bond in LPA(20:4) may introduce a kink in this region of the hydrocarbon chain, precluding the extended conformation and the hydrophobic interactions that may promote stronger binding of the other LPA species.
In human plasma, the serum albumin concentration ranges from 0.5 to 0.8 mM, and the total LPA concentration is on the order of 10−7 M (21, 53, 54). Gelsolin, a protein that regulates actin filament structure, reportedly binds LPA [likely LPA(18:1)] with a Kd of 6 nM (34) and is present in human plasma at a concentration of ∼1–3 μM (55). Although the affinity of LPA(18:1) for serum albumin is ∼20-fold weaker than the reported affinity for gelsolin, the concentration of LPA-binding sites offered on serum albumin is approximately 600-fold higher, assuming three binding sites per molecule. These concentrations and Kd values support early observations that serum LPA is mostly associated with albumin (20) and FAF-BSA demonstrates inhibitory effects on LPA-induced platelet activation and receptor activation in vitro (49, 56, 57), possibly through direct competition with the receptors for binding LPA.
The distribution of S1P among serum proteins (HDL, LDL, and HSA) in human plasma has been investigated. Most reports cite ∼60–70% of plasma S1P associates with HDL and ∼30% associates with HSA. By solving simultaneous equations using the Kd2 values reported here and previously reported normal plasma concentrations [0.1–0.4 μM S1P (58), 0.53–0.76 mM HSA (25), 0.6–1.3 μM apoM-HDL (11, 12), and 0.01–0.04 μM apoM-LDL, which is 2% of total serum LDL particle concentration (59, 60)], the calculated S1P distribution ranges from 25 to 59% HSA bound, 35 to 68% apoM-HDL bound, and 1.6 to 16% bound to apoM-LDL. If the S1P concentration is increased to 1.2 μM, the percent of S1P associated with apoM-HDL and apoM-LDL decreases, while the percent of S1P bound to albumin increases to 70% (supplemental Table S2). This demonstrates how normal variability in plasma S1P and chaperone concentrations profoundly effects the S1P distribution among these carrier proteins. In addition, the fraction of each chaperone protein (as percent of total) associated with S1P at these concentrations is shown in supplemental Table S3. These data suggest that normal plasma levels of apoM-LDL are nearly completely bound at 1.2 μM S1P. The lipoprotein preparations used in this study were collected from a single normal male donor. Therefore, the inter-individual variation in Kd2 values, which also influences the percent distribution of S1P, cannot be addressed, but will be the focus of future investigations.
Recent reports suggest that S1P exhibits distinct biological effects depending on its chaperone protein (17, 18). Wilkerson et al. (61) demonstrated that S1P bound to apoM-HDL sustains the barrier function of human endothelial cells longer than S1P delivered using albumin. In addition, the rate of S1P1 receptor internalization and degradation in these cells is slower when S1P is complexed with HDL than with FAF-BSA. These observations are consistent with S1P having a relatively weak affinity for serum albumin, which likely poorly buffers S1P from receptor binding and internalization. Unlike albumin, our results suggest that S1P has a relatively strong affinity for apoM-LDL. One proposed mechanism for S1P and apoM clearance from circulation involves the LDL receptor (52, 62). Kurano et al. (52) demonstrated that overexpressing the LDL receptor results in lower plasma S1P and apoM concentrations in mice, and exogenous C17S1P is cleared more quickly in vivo and in vitro when the LDL receptor is overexpressed. Here, we measured ∼10-fold stronger affinity of S1P for apoM-LDL than apoM-HDL, which would be consistent with LDL-associated S1P targeted for clearance, while HDL-bound S1P is bioavailable for receptor binding and activation. However, the role HDL plays in S1P-mediated biological processes is not necessarily consistent with that of “free S1P.” In fact, sometimes it is contrary (63), suggesting a more intricate role for HDL in S1P-mediated receptor activation and signaling than solely direct competition with the S1P receptors.
Despite their relatively uncomplicated chemical structure, lipid recognition is notoriously difficult to study because the mechanisms of protein-lipid binding are often complex and unique physical properties of lipids bear particular experimental challenges. In recent years, modifying lipids with molecular tags and/or covalently attaching lipids to solid surfaces to study protein-lipid interactions has gained popularity (64, 65). While these approaches may be acceptable for identifying novel lipid-binding proteins, qualitatively evaluating proteins binding to vesicles, or investigating membrane fluidity, these modifications may significantly alter the natural mode of lipid recognition. Therefore, we developed a method using anti-lipid mAbs and KinExA to measure the equilibrium dissociation constants for two native lysophospholipids, S1P and LPA, binding several serum proteins in solution.
In conclusion, individual LPA species bind albumin with a range of affinities, where shorter fully saturated fatty acyl species demonstrate stronger affinity for albumin than polyunsaturated species. S1P exhibits a relatively weak affinity for serum albumin compared with apoM-HDL and apoM-LDL, where apoM-LDL represents the stronger-affinity lower-capacity S1P carrier and HDL represents the moderate-affinity higher-capacity S1P chaperone by comparison. These affinity measurements provide insights into the role of these serum proteins in S1P and LPA transport and storage in circulation.
Supplementary Material
Acknowledgments
The authors thank Dr. Christina Christoffersen and Charlotte Wandel for measuring the concentration of apoM in our HDL and LDL samples, Jacek Bielawski at the Medical University of South Carolina for MS analysis, and Ashlee King for performing the competition ELISA.
Footnotes
Abbreviations:
- ANS
- 6-anilinonaphthalene-2-sulfonic acid
- apoM-HDL
- apoM-associated HDL
- apoM-LDL
- apoM-associated LDL
- CI
- confidence interval
- FAF
- fatty acid-free
- GPCR
- G protein-coupled receptor
- HSA
- human serum albumin
- Kd
- equilibrium dissociation constant
- Kd1
- equilibrium dissociation constant of antibody for lipid
- Kd2
- equilibrium dissociation constant of carrier protein for lipid
- KinExA®
- 2 Kinetic Exclusion Assay
- LPA
- lysophosphatidic acid
- LT1009
- anti-S1P monoclonal antibody
- LT3015
- anti-LPA monoclonal antibody
- mAb
- monoclonal antibody
- S1P
- sphingosine-1-phosphate
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Kihara Y., Maceyka M., Spiegel S., and Chun J.. 2014. Lysophospholipid receptor nomenclature review: IUPHAR Review 8. Br. J. Pharmacol. 171: 3575–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mutoh T., Rivera R., and Chun J.. 2012. Insights into the pharmacological relevance of lysophospholipid receptors. Br. J. Pharmacol. 165: 829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chun J. 2013. Lysophospholipid Receptors: Signaling and Biochemistry. Wiley, Hoboken, NJ. [Google Scholar]
- 4.Kihara Y., Mizuno H., and Chun J.. 2015. Lysophospholipid receptors in drug discovery. Exp. Cell Res. 333: 171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tigyi G. 2010. Aiming drug discovery at lysophosphatidic acid targets. Br. J. Pharmacol. 161: 241–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishii I., Fukushima N., Ye X., and Chun J.. 2004. Lysophospholipid receptors: signaling and biology. Annu. Rev. Biochem. 73: 321–354. [DOI] [PubMed] [Google Scholar]
- 7.Xu N., and Dahlback B.. 1999. A novel human apolipoprotein (apoM). J. Biol. Chem. 274: 31286–31290. [DOI] [PubMed] [Google Scholar]
- 8.Murata N., Sato K., Kon J., Tomura H., Yanagita M., Kuwabara A., Ui M., and Okajima F.. 2000. Interaction of sphingosine 1-phosphate with plasma components, including lipoproteins, regulates the lipid receptor-mediated actions. Biochem. J. 352: 809–815. [PMC free article] [PubMed] [Google Scholar]
- 9.Schuchardt M., Tolle M., Prufer J., and van der Giet M.. 2011. Pharmacological relevance and potential of sphingosine 1-phosphate in the vascular system. Br. J. Pharmacol. 163: 1140–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammad S. M., Al Gadban M. M., Semler A. J., and Klein R. L.. 2012. Sphingosine 1-phosphate distribution in human plasma: associations with lipid profiles. J. Lipids. 2012: 180705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christoffersen C., Obinata H., Kumaraswamy S. B., Galvani S., Ahnstrom J., Sevvana M., Egerer-Sieber C., Muller Y. A., Hla T., Nielsen L. B., et al. 2011. Endothelium-protective sphingosine-1-phosphate provided by HDL-associated apolipoprotein M. Proc. Natl. Acad. Sci. USA. 108: 9613–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Axler O., Ahnstrom J., and Dahlback B.. 2007. An ELISA for apolipoprotein M reveals a strong correlation to total cholesterol in human plasma. J. Lipid Res. 48: 1772–1780. [DOI] [PubMed] [Google Scholar]
- 13.Arkensteijn B. W., Berbee J. F., Rensen P. C., Nielsen L. B., and Christoffersen C.. 2013. The apolipoprotein m-sphingosine-1-phosphate axis: biological relevance in lipoprotein metabolism, lipid disorders and atherosclerosis. Int. J. Mol. Sci. 14: 4419–4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christoffersen C., Nielsen L. B., Axler O., Andersson A., Johnsen A. H., and Dahlback B.. 2006. Isolation and characterization of human apolipoprotein M-containing lipoproteins. J. Lipid Res. 47: 1833–1843. [DOI] [PubMed] [Google Scholar]
- 15.Kontush A., Lhomme M., and Chapman M. J.. 2013. Unraveling the complexities of the HDL lipidome. J. Lipid Res. 54: 2950–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Argraves K. M., Gazzolo P. J., Groh E. M., Wilkerson B. A., Matsuura B. S., Twal W. O., Hammad S. M., and Argraves W. S.. 2008. High density lipoprotein-associated sphingosine 1-phosphate promotes endothelial barrier function. J. Biol. Chem. 283: 25074–25081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galvani S., Sanson M., Blaho V. A., Swendeman S. L., Obinata H., Conger H., Dahlback B., Kono M., Proia R. L., Smith J. D., et al. 2015. HDL-bound sphingosine 1-phosphate acts as a biased agonist for the endothelial cell receptor S1P1 to limit vascular inflammation. Sci. Signal. 8: ra79 [Erratum. 2015. Sci. Signal 8: er8.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blaho V. A., Galvani S., Engelbrecht E., Liu C., Swendeman S. L., Kono M., Proia R. L., Steinman L., Han M. H., and Hla T.. 2015. HDL-bound sphingosine-1-phosphate restrains lymphopoiesis and neuroinflammation. Nature. 523: 342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tigyi G., Henschen A., and Miledi R.. 1991. A factor that activates oscillatory chloride currents in Xenopus oocytes copurifies with a subfraction of serum albumin. J. Biol. Chem. 266: 20602–20609. [PubMed] [Google Scholar]
- 20.Tigyi G., and Miledi R.. 1992. Lysophosphatidates bound to serum albumin activate membrane currents in Xenopus oocytes and neurite retraction in PC12 pheochromocytoma cells. J. Biol. Chem. 267: 21360–21367. [PubMed] [Google Scholar]
- 21.Yung Y. C., Stoddard N. C., and Chun J.. 2014. LPA receptor signaling: pharmacology, physiology, and pathophysiology. J. Lipid Res. 55: 1192–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bandoh K., Aoki J., Taira A., Tsujimoto M., Arai H., and Inoue K.. 2000. Lysophosphatidic acid (LPA) receptors of the EDG family are differentially activated by LPA species. Structure-activity relationship of cloned LPA receptors. FEBS Lett. 478: 159–165. [DOI] [PubMed] [Google Scholar]
- 23.Kragh-Hansen U., Watanabe H., Nakajou K., Iwao Y., and Otagiri M.. 2006. Chain length-dependent binding of fatty acid anions to human serum albumin studied by site-directed mutagenesis. J. Mol. Biol. 363: 702–712. [DOI] [PubMed] [Google Scholar]
- 24.Richieri G. V., Anel A., and Kleinfeld A. M.. 1993. Interactions of long-chain fatty acids and albumin: determination of free fatty acid levels using the fluorescent probe ADIFAB. Biochemistry. 32: 7574–7580. [DOI] [PubMed] [Google Scholar]
- 25.Bhattacharya A. A., Grune T., and Curry S.. 2000. Crystallographic analysis reveals common modes of binding of medium and long-chain fatty acids to human serum albumin. J. Mol. Biol. 303: 721–732. [DOI] [PubMed] [Google Scholar]
- 26.Thumser A. E., Voysey J. E., and Wilton D. C.. 1994. The binding of lysophospholipids to rat liver fatty acid-binding protein and albumin. Biochem. J. 301: 801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bojesen I. N., and Bojesen E.. 1996. Albumin binding of long-chain fatty acids: thermodynamics and kinetics. J. Phys. Chem. 100: 17981–17985. [Google Scholar]
- 28.Wojciak J. M., Zhu N., Schuerenberg K. T., Moreno K., Shestowsky W. S., Hiraiwa M., Sabbadini R., and Huxford T.. 2009. The crystal structure of sphingosine-1-phosphate in complex with a Fab fragment reveals metal bridging of an antibody and its antigen. Proc. Natl. Acad. Sci. USA. 106: 17717–17722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fleming J. K., Wojciak J. M., Campbell M. A., and Huxford T.. 2011. Biochemical and structural characterization of lysophosphatidic acid binding by a humanized monoclonal antibody. J. Mol. Biol. 408: 462–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darling R. J., and Brault P. A.. 2004. Kinetic exclusion assay technology: characterization of molecular interactions. Assay Drug Dev. Technol. 2: 647–657. [DOI] [PubMed] [Google Scholar]
- 31.Blake R. C. 2nd, Pavlov A. R., and Blake D. A.. 1999. Automated kinetic exclusion assays to quantify protein binding interactions in homogeneous solution. Anal. Biochem. 272: 123–134. [DOI] [PubMed] [Google Scholar]
- 32.Bee C., Abdiche Y. N., Stone D. M., Collier S., Lindquist K. C., Pinkerton A. C., Pons J., and Rajpal A.. 2012. Exploring the dynamic range of the kinetic exclusion assay in characterizing antigen-antibody interactions. PLoS One. 7: e36261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drake A. W., Tang M. L., Papalia G. A., Landes G., Haak-Frendscho M., and Klakamp S. L.. 2012. Biacore surface matrix effects on the binding kinetics and affinity of an antigen/antibody complex. Anal. Biochem. 429: 58–69. [DOI] [PubMed] [Google Scholar]
- 34.Goetzl E. J., Lee H., Azuma T., Stossel T. P., Turck C. W., and Karliner J. S.. 2000. Gelsolin binding and cellular presentation of lysophosphatidic acid. J. Biol. Chem. 275: 14573–14578. [DOI] [PubMed] [Google Scholar]
- 35.Ojala P. J., Hermansson M., Tolvanen M., Polvinen K., Hirvonen T., Impola U., Jauhiainen M., Somerharju P., and Parkkinen J.. 2006. Identification of alpha-1 acid glycoprotein as a lysophospholipid binding protein: a complementary role to albumin in the scavenging of lysophosphatidylcholine. Biochemistry. 45: 14021–14031. [DOI] [PubMed] [Google Scholar]
- 36.Bosteen M. H., Dahlback B., Nielsen L. B., and Christoffersen C.. 2015. Protein unfolding allows use of commercial antibodies in an apolipoprotein M sandwich ELISA. J. Lipid Res. 56: 754–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sevvana M., Ahnstrom J., Egerer-Sieber C., Lange H. A., Dahlback B., and Muller Y. A.. 2009. Serendipitous fatty acid binding reveals the structural determinants for ligand recognition in apolipoprotein M. J. Mol. Biol. 393: 920–936. [DOI] [PubMed] [Google Scholar]
- 38.Yarmush D. M., Morel G., and Yarmush M. L.. 1987. A new technique for mapping epitope specificities of monoclonal antibodies using quasi-elastic light scattering spectroscopy. J. Biochem. Biophys. Methods. 14: 279–289. [DOI] [PubMed] [Google Scholar]
- 39.Pace C. N., Vajdos F., Fee L., Grimsley G., and Gray T.. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4: 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esposito B. P., Faljoni-Alario A., de Menezes J. F., de Brito H. F., and Najjar R.. 1999. A circular dichroism and fluorescence quenching study of the interactions between rhodium(II) complexes and human serum albumin. J. Inorg. Biochem. 75: 55–61. [DOI] [PubMed] [Google Scholar]
- 41.Shpak A. P., and Gorbik P. P.. 2009. Nanomaterials and Supramolecular Structures: Physics, Chemistry, and Applications. Springer, Dordrecht, London, New York. [Google Scholar]
- 42.Chen P. S., Toribara T. Y., and Huber W.. 1956. Microdetermination of phosphorus. Anal. Chem. 28: 1756–1758. [Google Scholar]
- 43.Fiske C. H., and Subbarow Y.. 1925. The colorimetric determination of phosphorus. J. Biol. Chem. 66: 375–400. [Google Scholar]
- 44.Wang Z. X. 1995. An exact mathematical expression for describing competitive binding of two different ligands to a protein molecule. FEBS Lett. 360: 111–114. [DOI] [PubMed] [Google Scholar]
- 45.Thomä N., and Goody R. S.. 2003. What to do if there is no signal: using competition experiments to determine binding parameters. In Kinetic Analysis of Macromolecules: A Practical Approach. K. A. Johnson, editor. Oxford University Press, Oxford, UK. 153–170. [Google Scholar]
- 46.Straume M., and Johnson M. L.. 1992. Monte Carlo method for determining complete confidence probability distributions of estimated model parameters. Methods Enzymol. 210: 117–129. [DOI] [PubMed] [Google Scholar]
- 47.Simard J. R., Zunszain P. A., Ha C. E., Yang J. S., Bhagavan N. V., Petitpas I., Curry S., and Hamilton J. A.. 2005. Locating high-affinity fatty acid-binding sites on albumin by x-ray crystallography and NMR spectroscopy. Proc. Natl. Acad. Sci. USA. 102: 17958–17963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bojesen I. N., and Bojesen E.. 1994. Binding of arachidonate and oleate to bovine serum albumin. J. Lipid Res. 35: 770–778. [PubMed] [Google Scholar]
- 49.Khandoga A. L., Fujiwara Y., Goyal P., Pandey D., Tsukahara R., Bolen A., Guo H., Wilke N., Liu J., Valentine W. J., et al. 2008. Lysophosphatidic acid-induced platelet shape change revealed through LPA(1–5) receptor-selective probes and albumin. Platelets. 19: 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Z., Mintzer E., and Bittman R.. 2004. The critical micelle concentrations of lysophosphatidic acid and sphingosylphosphorylcholine. Chem. Phys. Lipids. 130: 197–201. [DOI] [PubMed] [Google Scholar]
- 51.Kimura T., Sato K., Kuwabara A., Tomura H., Ishiwara M., Kobayashi I., Ui M., and Okajima F.. 2001. Sphingosine 1-phosphate may be a major component of plasma lipoproteins responsible for the cytoprotective actions in human umbilical vein endothelial cells. J. Biol. Chem. 276: 31780–31785. [DOI] [PubMed] [Google Scholar]
- 52.Kurano M., Tsukamoto K., Hara M., Ohkawa R., Ikeda H., and Yatomi Y.. 2015. LDL receptor and ApoE are involved in the clearance of ApoM-associated sphingosine 1-phosphate. J. Biol. Chem. 290: 2477–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baker D. L., Desiderio D. M., Miller D. D., Tolley B., and Tigyi G. J.. 2001. Direct quantitative analysis of lysophosphatidic acid molecular species by stable isotope dilution electrospray ionization liquid chromatography-mass spectrometry. Anal. Biochem. 292: 287–295. [DOI] [PubMed] [Google Scholar]
- 54.Scherer M., Schmitz G., and Liebisch G.. 2009. High-throughput analysis of sphingosine 1-phosphate, sphinganine 1-phosphate, and lysophosphatidic acid in plasma samples by liquid chromatography-tandem mass spectrometry. Clin. Chem. 55: 1218–1222. [DOI] [PubMed] [Google Scholar]
- 55.Tigyi G., and Parrill A. L.. 2003. Molecular mechanisms of lysophosphatidic acid action. Prog. Lipid Res. 42: 498–526. [DOI] [PubMed] [Google Scholar]
- 56.Hama K., Bandoh K., Kakehi Y., Aoki J., and Arai H.. 2002. Lysophosphatidic acid (LPA) receptors are activated differentially by biological fluids: possible role of LPA-binding proteins in activation of LPA receptors. FEBS Lett. 523: 187–192. [DOI] [PubMed] [Google Scholar]
- 57.Tokumura A., Sinomiya J., Kishimoto S., Tanaka T., Kogure K., Sugiura T., Satouchi K., Waku K., and Fukuzawa K.. 2002. Human platelets respond differentially to lysophosphatidic acids having a highly unsaturated fatty acyl group and alkyl ether-linked lysophosphatidic acids. Biochem. J. 365: 617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ksiązėk M., Chacińska M., Chabowski A., and Baranowski M.. 2015. Sources, metabolism, and regulation of circulating sphingosine-1-phosphate. J. Lipid Res. 56: 1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Freedman D. S., Otvos J. D., Jeyarajah E. J., Shalaurova I., Cupples L. A., Parise H., D’Agostino R. B., Wilson P. W., and Schaefer E. J.. 2004. Sex and age differences in lipoprotein subclasses measured by nuclear magnetic resonance spectroscopy: the Framingham Study. Clin. Chem. 50: 1189–1200. [DOI] [PubMed] [Google Scholar]
- 60.Matyus S. P., Braun P. J., Wolak-Dinsmore J., Jeyarajah E. J., Shalaurova I., Xu Y., Warner S. M., Clement T. S., Connelly M. A., and Fischer T. J.. 2014. NMR measurement of LDL particle number using the Vantera Clinical Analyzer. Clin. Biochem. 47: 203–210. [DOI] [PubMed] [Google Scholar]
- 61.Wilkerson B. A., Grass G. D., Wing S. B., Argraves W. S., and Argraves K. M.. 2012. Sphingosine 1-phosphate (S1P) carrier-dependent regulation of endothelial barrier: high density lipoprotein (HDL)-S1P prolongs endothelial barrier enhancement as compared with albumin-S1P via effects on levels, trafficking, and signaling of S1P1. J. Biol. Chem. 287: 44645–44653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Christoffersen C., Benn M., Christensen P. M., Gordts P. L., Roebroek A. J., Frikke-Schmidt R., Tybjaerg-Hansen A., Dahlback B., and Nielsen L. B.. 2012. The plasma concentration of HDL-associated apoM is influenced by LDL receptor-mediated clearance of apoB-containing particles. J. Lipid Res. 53: 2198–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sattler K., and Levkau B.. 2009. Sphingosine-1-phosphate as a mediator of high-density lipoprotein effects in cardiovascular protection. Cardiovasc. Res. 82: 201–211. [DOI] [PubMed] [Google Scholar]
- 64.Saliba A. E., Vonkova I., and Gavin A. C.. 2015. The systematic analysis of protein-lipid interactions comes of age. Nat. Rev. Mol. Cell Biol. 16: 753–761. [DOI] [PubMed] [Google Scholar]
- 65.Zhao H., and Lappalainen P.. 2012. A simple guide to biochemical approaches for analyzing protein-lipid interactions. Mol. Biol. Cell. 23: 2823–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.