Abstract
In this study, we investigated the role and mechanism of Niemann-Pick type C (NPC)2 in regulating lysosomal activity, mitophagy, and mitochondrial function in adipocytes. We found that knocking down NPC2 impaired lysosomal activity, as evidenced by the reduced mature cathepsin B, the increased accumulation of light chain 3 (LC3) and p62, and the decreased autophagic flux. In NPC2-knockdown (kd) adipocytes, the starvation-induced conversion of LC3-I to LC3-II was abolished. More interestingly, the majority of NPC2 was found in the mitochondrial fraction, and NPC2 deficiency led to impaired autophagic flux and decreased induction of LC3-II in the mitochondrial fraction during mitochondrial stress. Moreover, cellular respiration profiling revealed that NPC2-kd adipocytes had significantly decreased basal/maximal respiration and mitochondrial gene expression compared with scrambled cells, suggesting mitochondrial dysfunction. Additionally, we found that the mitochondrial recruitment of LC3-II induced by lipopolysaccharide (LPS), but not TNFα, was blunted in NPC2-kd adipocytes. Most intriguingly, NPC2-kd selectively diminished LPS-induced NFκB and ERK1/2 phosphorylation and the expression of pro-inflammatory genes, indicating that toll-like receptor signaling activation is impaired in the absence of NPC2. Our results suggest that NPC2 is in a mitochondrially associated autophagosome and plays an important role in regulating mitophagy, mitochondrial quality control, and mitochondrial function.
Keywords: toll-like receptor, inflammation, mitochondria, Niemann-Pick disease, obesity • lysosome
Aging is a risk factor for the development of chronic metabolic diseases such as type 2 diabetes, cardiovascular disease, neurodegenerative disease, and cancer. These aging-related diseases are known to be clustered together; aging-associated neurodegenerative disorders frequently coincide with type 2 diabetes, insulin resistance, obesity, and metabolic syndrome. This suggests that neurodegenerative disorders and type 2 diabetes share common genetic and metabolic features. Therefore, studying the role of the gene that contributes to neurodegenerative diseases in obesity and diabetes is of particular importance, as it may provide a clue for the pathological mechanism of the development of aging-related diseases. Intensive investigation on the underlying mechanisms of cellular aging has suggested that autophagy-lysosome and mitochondrial function plays an essential role in the regulation of cellular homeostasis and health (1). The autophagy-lysosome system is one of the main intracellular proteolytic systems that is responsible for the clearance of unwanted intracellular macromolecules and organelles (2). Cellular aging is associated with the disruption of this homeostatic mechanism, leading to the accumulation of oxidized/misfolded proteins, lipids, and damaged organelles (3). However, the genes and proteins that regulate autophagy-lysosomal activation and contribute to cellular aging remain largely unidentified.
Autophagy is a highly regulated process that is inducible by nutrient deprivation and cellular stress. During nutrient deprivation, multiple signaling pathways cooperate to activate autophagy. In the absence of insulin signaling and amino acids, mammalian target of rapamycin complex 1 (mTORC1) is inactivated and releases inhibition on Unc-51-like kinase 1 (ULK1) (4). Additionally, 5′AMP-activated protein kinase (AMPK) inhibits mTORC1 and may also directly interact with ULK1 to activate autophagy (4). These signaling events promote the formation of the ULK1 complex and initiate formation of the autophagosome. Induction of autophagy also requires the activity of sirtuin-1, which is activated by elevated levels of nicotinamide adenine dinucleotide (NAD) and deacetylates several key autophagy-related proteins (5). Therefore, there are redundant systems in place and multiple signaling events may be required to fully activate autophagy in response to nutrient deprivation. In addition to starvation, autophagy can be activated in response to inflammation. Activation of toll-like receptors (TLRs) by lipopolysaccharide (LPS) is known to stimulate autophagy via both the MyD88- and TRIF-dependent pathways (6–8). NFκB, a key downstream component of the TLR signaling pathway, has long been known to be essential for the activation of autophagy (9, 10). While autophagy can be activated by inflammation, it has also been shown to attenuate the inflammatory response. Autophagy activation promotes p62-mediated inflammasome degradation and prevents the conversion of pro-IL-1β to mature IL-1β (11). Hence, autophagy may serve to mitigate cellular stress to maintain homeostasis.
Niemann-Pick type C (NPC) disease is a childhood onset neurodegenerative disorder caused by mutations in the NPC1 and NPC2 genes that encode endosomal/lysosomal proteins. The major NPC phenotype is the cholesterol accumulation in endosomes and lysosomes. Most of the previous studies were focused on the role of NPC in cholesterol and other lipid metabolism. It has been documented that membrane-bound NPC1 and soluble NPC2 function in a common lysosomal pathway (12) to facilitate the transfer of unesterified cholesterol from endosomes and lysosomes to endoplasmic reticulum and plasma membrane (13, 14). However, the defective cellular functions that are attributed to NPC deficiency have not been completely characterized. In particular, it is unclear whether NPC2 deficiency contributes to the development of cellular aging. In keeping with the common features of degenerative diseases, NPC1 deficiency impairs the clearance of autophagosomes leading to dysfunctional protein degradation, suggesting that it plays a role in the regulation of autophagy-lysosomal activity (15).
While most of previous studies on NPC proteins focus on their roles in neurodegenerative, liver and spleen disorders, few studies have investigated the association of NPC proteins with obesity, diabetes, and insulin resistance. In a recent study, a SNP in NPC1 has been associated with obesity and type 2 diabetes, and NPC1-deficient mice developed insulin resistance (16). Interestingly, recent studies unveiled that NPC2 has some NPC1-independent effects on metabolism. For instance, NPC2 has been demonstrated to transport endosomal cholesterol to mitochondria, which is important for maintaining mitochondrial membrane structure and function (17). In another study, increased accumulation of mitochondrial cholesterol and mitochondrial dysfunction was observed in NPC1-deficient Chinese hamster ovary cells (18). Although the accumulation of cholesterol in the late endosomes and lysosomes is believed to be the cause of NPC disease, its underlying mechanism for the NPC2 deficiency-caused pathologies, especially defective autophagy in adipocytes, has not been fully understood.
In our previous studies, we found that the mRNA and protein expression of NPC2 was upregulated in epididymal adipose tissue, but downregulated in inguinal adipose tissue in high-fat diet-induced obese mice (19). In this study, we investigated the role and underlying mechanisms of NPC2 in regulating autophagy-lysosomal activity and maintaining mitochondrial health and function in adipocytes. We found that NPC2 plays a critical role in regulating lysosomal function and mitophagy. NPC2 deficiency reduces autophagy-lysosomal activity and impairs mitochondrial function in adipocytes. Most importantly, we discovered that NPC2 is a critical regulator of TLR signaling pathway activation and its mediated autophagy activation.
MATERIALS AND METHODS
Animals
C57BL/6J mice (Jackson Laboratory, Bar Harbor, ME) were housed in a specific pathogen-free facility and were given free access to water and food. All experimental procedures were approved by the University of Minnesota animal care and use committee. Animal handling was performed according to National Institutes of Health guidelines. The tissues were collected from 12-week-old mice fed a regular chow diet.
Generation of NPC2-kd adipocytes
To establish NPC2-kd and scrambled cell lines, the shRNA sequence variants for Npc2 gene or nonspecific scrambled were synthesized and cloned into a lentiviral-based RNAi vector, pLKO.1, using ViraPower Lentiviral Expression Systems (Invitrogen, Carlsbad, CA). The selected oligomers targeting Npc2 sequence were 5′-GCTCTCGTTCTTTGGTAGTTT-3′ and 5′-CGGTTGTAAGAGTGGAATCAA-3′. Transduction was then performed using lentivirus-carrying shRNA in the 3T3-L1 fibroblasts, as described previously (20). In brief, when 3T3-L1 fibroblasts reached 70–80% confluence, the cells were transduced with lentivirus supplemented with 6 μg/ml Polybrene (Sigma-Aldrich, St. Louis, MO) for 12 h. After washing with PBS and culture for 6 h, the infected cells were selected by 2 μg/ml puromycin (Sigma-Aldrich) for 2 days. The infected 3T3-L1 fibroblasts were then induced to undergo differentiation.
The differentiation of 3T3-L1 fibroblasts was induced by a differentiation cocktail (100 IU/ml penicillin-streptomycin, 10% FBS, 115 μg/ml methylisobutylxanthine, 390 ng/ml dexamethasone, and 1 μg/ml insulin) for 2 days. The cells were subsequently maintained in culture medium supplemented with 10% FBS and 1 μg/ml insulin for 5 days. Fully differentiated 3T3-L1 adipocytes with massive accumulation of fat droplets were used for the experiments.
Relative quantitative real-time RT-PCR
Total RNA was extracted from the cells with TRIZOL reagent (Invitrogen). First-strand cDNA was synthesized from DNase-treated total RNA using a Superscript II reverse transcriptase kit (Invitrogen). Quantitative amplification by PCR was carried out using SYBR Green qPCR Master Mix (SABiosciences, Frederick, MD) by a StepOne Real-Time PCR System (Applied Biosystem, Foster City, CA). The ΔΔCt method was used to calculate the results. For quantification, results were normalized to TATA box binding protein (TBP) and presented as the levels of expression relative to those of endogenous controls. The primer sequences for amplifying the target genes and the GenBank accession number are summarized in supplemental Table S1.
Western blot analysis
Proteins were extracted using RIPA buffer (Sigma-Aldrich) containing protease inhibitors and phosphatase inhibitor cocktail (Roche Diagnostics Corporation, Indianapolis, IN). Proteins were separated on 8–10% SDS-PAGE gel and then transferred to nitrocellulose membranes. The primary antibodies probed to the target proteins included mouse monoclonal antibodies to NFκB and rabbit polyclonal antibodies to Tom20 (Santa Cruz Biotechnology, Dallas, TX), goat polyclonal antibodies to cathepsin B (CTSB) and cathepsin L (CTSL) (R&D Systems, Minneapolis, MN), mouse monoclonal antibody to Parkin and rabbit monoclonal antibodies to NPC2, autophagy marker light chain 3 (LC3), SQSTM1/p62, autophagy-related (Atg)5, Atg7, phospho-p70 S6 kinase (Thr 389), p70 S6 kinase, phospho-NFκB p65, phospho-Akt (ser473), Akt, tubulin, and β-actin (Cell Signaling Technology, Danvers, MA); and rabbit polyclonal antibody to lysosome-associated membrane protein 1 (LAMP1; Bioss Antibodies, Woburn, MA). ECL Western Blotting Detection System (GE Healthcare BioSciences, Piscataway, NJ) was used to detect antibody reactivity. Densitometric quantification was determined using an image analysis program (Alpha Innotech, San Leandro, CA) and reported as a ratio to total protein or β-actin as indicated in the result sections.
Glucose uptake assay
Uptake of 2-deoxy-d-[3H] glucose (Amersham Biosciences, Piscataway, NJ) in 3T3-L1 adipocytes was measured as previously described (21). Briefly, the adipocytes were starved in Krebs-Ringer HEPES (KRH) buffer containing 0.5% BSA (pH 7.4) for 2 h before incubation either with or without 200 nmol/l insulin for 30 min at 37°C. Glucose uptake was initiated by adding [3H]2-deoxy-d-glucose to a final assay concentration of 100 μmol/l. After 5 min, the cells were washed with ice-cold PBS three times and then solubilized with KRH buffer containing 1% Triton X-100. Incorporated radioactivity was determined by scintillation counting. Nonspecific 2-deoxyglucose uptake was measured in the presence of 20 μmol/l cytocholasin B and subtracted from the total glucose uptake assayed to obtain specific uptake.
Cellular respiratory assay
Cellular respiration was analyzed in situ with the XF24 Extracellular Flux Analyzer (Seahorse Biosciences, Billerica, MA). The 3T3-L1 fibroblasts were plated on V7 microplates coated with 0.01% collagen solution type I (Sigma-Aldrich) and differentiated into adipocytes. Eight days postdifferentiation, cells were washed and incubated at 37°C without CO2 in bicarbonate-free DMEM containing 1.0 mM sodium pyruvate, 2.0 mM glutamax, and 25 mM d-glucose. During the assay, cell monolayers were exposed to 2.0 μM oligomycin, 0.5 μM carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP), and 4 μm antimycin A. Respiration rates were calculated as previously described (22).
Mitochondria isolation
The differentiated adipocytes with different treatments were scraped and collected with ice-cold isolation buffer [20 mmol/l Tris and 1 mmol/l EDTA (pH 7.4)] supplemented with protease inhibitors. The cells were then lysed with 20 strokes of a glass-Teflon homogenizer at a speed of 1,600 rpm. After centrifugation at 700 g for 10 min, the supernatant (cytosolic fraction) was collected and lipids were removed. The pellet (mitochondrial fraction) was washed and recovered with isolation buffer by centrifugation at 9,000 g at 4°C for 10 min.
Filipin staining
Filipin staining was performed in adipocytes treated with U18666a, as described previously (23, 24). U18666a is a widely used inhibitor of cholesterol trafficking and synthesis. Briefly, scrambled and NPC2-kd cells were seeded into 6-well plates and induced to differentiate into adipocytes. Prior to full differentiation, cells were treated with 10 μg/ml U18666a (Sigma-Aldrich) for 48 h (from day 5 to day 7 of differentiation) and 72 h (from day 4 to day 7 of differentiation), respectively; PBS was used as a vehicle control. Cells were then fixed with 4% paraformaldehyde for 1 h at room temperature. After rinsing with PBS and incubation with 1.5 mg/ml glycine in PBS for 10 min to quench paraformaldehyde, cells were stained with 50 μg/ml Filipin III (Sigma-Aldrich) in PBS for 2 h. The filipin signal was imaged using an Olympus IX70 inverted fluorescence microscope with UV filter package.
Statistical analysis
Results of gene expression are expressed as mean ± SEM. Results of protein expression by Western blot analysis are expressed as mean ± SD. Differences in the parameters between two groups were evaluated using Student’s t-test with a 0.05 two-sided significance level. P < 0.05 was considered significant.
RESULTS
Establishment of NPC2-kd cell line
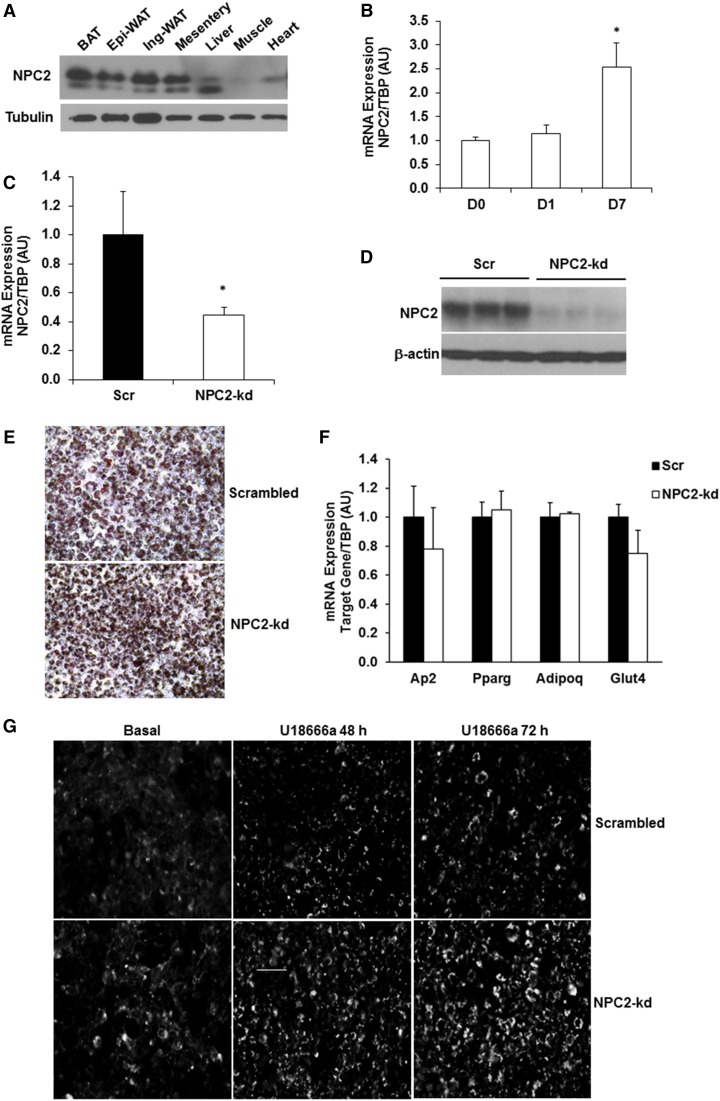
We have previously reported that NPC2 expression in adipose tissue is dysregulated in mice when challenged with high-fat diet or in macrophages in response to inflammatory stimulation (19). In this study, we showed that NPC2 was also expressed in brown adipose tissue and mesentery white adipose tissue (Fig. 1A). The gene expression of Npc2 was increased during adipocyte differentiation in 3T3-L1 cells (Fig. 1B).
Fig. 1.
NPC2 kd in 3T3-L1 adipocytes. A: NPC2 protein expression in different tissues. B: The gene expression of Npc2 in 3T3-L1 cells during differentiation. C: Npc2 mRNA expression in 3T3-L1 adipocytes was quantified by quantitative PCR. D: Protein expression of NPC2 in 3T3-L1 adipocytes by Western blot. E: Oil Red O staining of 3T3-L1 cells on day 8 of differentiation. F: Adipogenic gene expression in 3T3-L1 adipocytes on day 8 of differentiation (n = 4 in each group). G: Filipin III fluorescence image of scrambled and NPC2-kd adipocytes treated with 10 μg/ml U18666a for 48 and 72 h, respectively. Images were acquired using the automatic fluorescence microscope at 20× magnification. Scale bar = 50 μm. Data are expressed relative to the value for scrambled (Scr) cells. The values are mean ± SEM. *P < 0.001 versus scrambled cells. Ap2, adipocyte P2; PPARg, peroxisome proliferator-activated receptor gamma; Adipoq, adiponectin; Glut4, glucose transporter type 4.
To investigate the role and underlying mechanism of NPC2 in the pathogenesis of obesity and adipocyte dysfunction, we established NPC2-kd and scrambled cell lines using lentivirus delivery of shRNA directed against Npc2 mRNA or nonspecific scrambled control sequence. The selected oligomers targeting Npc2 sequence were 5′-GCTCTCGTTCTTTGGTAGTTT-3′ and 5′-CGGTTGTAAGAGTGGAATCAA-3′. As shown in Fig. 1C, lentiviral shRNA was able to knock down the gene expression of Npc2 by 60% in 3T3-L1 adipocytes. Moreover, the protein level of NPC2 was markedly reduced by more than 90% in NPC2-kd adipocytes compared with scrambled controls (Fig. 1D). Oil Red O staining of adipocytes on day 8 of differentiation showed that both scrambled and NPC2-kd 3T3-L1 cells were able to differentiate into adipocytes to a similar extent (Fig. 1E), which is confirmed by no significant difference in the expression of adipogenic genes between scrambled and NPC2-kd adipocytes (Fig. 1F).
To determine whether NPC2 kd has an impact on cholesterol metabolism, we preformed filipin staining. Adipocytes were treated without or with U18666a, an inhibitor of cholesterol transport and synthesis, for 24 and 72 h. U18666a treatment led to a time-dependent increase in intracellular accumulation of cholesterol in both scrambled and NPC2-kd adipocytes (Fig. 1G). Interestingly, we found that NPC2-kd adipocytes displayed increased levels of intracellular cholesterol accumulation compared with scrambled cells in the basal and U18666a-treated conditions (Fig. 1G). This result suggests that NPC2 is involved in cholesterol metabolism, confirming the phenotype of NPC2 kd.
NPC2 kd reduces lysosomal activity
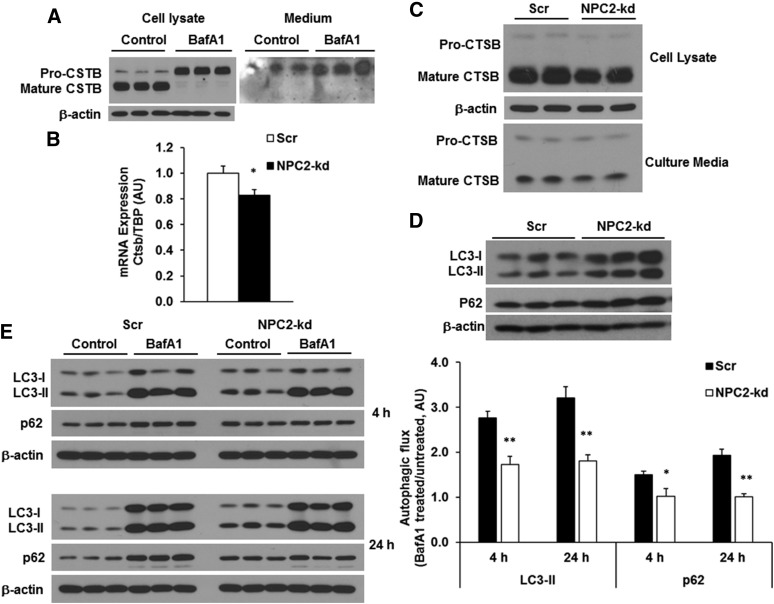
NPC2 is an endosomal/lysosomal protein and has been identified as an intracellular cholesterol transporter that acts in concert with NPC1 to remove cholesterol from the endosomal/lysosomal compartment (13, 14, 25, 26). Cathepsins, the lysosomal proteases, are the markers of lysosomal activity and have been connected to NPC2 in inflammation and obesity in our previous studies (19). To evaluate how lysosomal activity affects lysosomal cathepsins in adipocytes, we detected the regulation of protein expression and secretion of CTSB by bafilomycin A1 (BafA1), an inhibitor of lysosomal activity, in 3T3-L1 adipocytes. Treatment with BafA1 for 24 h blocked the maturation of pro-CTSB, as evidenced by inhibiting the pro-CTSB conversion to mature CTSB in the cells (Fig. 2A). Interestingly, pro-CSTB, but not mature CSTB, was secreted into culture media; BafA1 significantly increased pro-CSTB secretion (Fig. 2A). Thus, the reduced expression of mature CTSB represents the suppressed lysosomal activity in adipocytes.
Fig. 2.
Effect of NPC2 kd on lysosomal activity and autophagic flux. A: The protein expression of CTSB in the cell lysate and culture media from the differentiated 3T3-L1 adipocytes treated with 100 nM BafA1 for 24 h. B: The gene expression of CTSB in scrambled and NPC2-kd adipocytes. C: The protein expression and secretion of CTSB from the cells incubated with 10% FBS-culture media for 24 h. D: Autophagic markers LC3 and p62 in scrambled and NPC2-kd adipocytes. E: Autophagic flux measured in the cells incubated with 10% FBS-culture media for 4 and 24 h, respectively. The autophagic flux was determined by calculating the ratio of the densitometry of LC3-II and p62 levels in BafA1-treated cells after normalization by β-actin to that in untreated cells, respectively. The values of gene expression are mean ± SEM; the values of protein expression are mean ± SD. *P < 0.05; **P < 0.01; versus scrambled (Scr) cells. AU, arbitrary units.
To investigate how NPC2 deficiency affects lysosomal activity in adipocytes, we first evaluated the lysomosal activity in NPC2-kd 3T3-L1 adipocytes by examining the expression of CSTB. As shown in Fig. 2B, C, the gene expression of Ctsb was downregulated (Fig. 2B) and the mature form of CSTB was significantly reduced (Fig. 2C) in NPC2-kd adipocytes compared with scrambled cells. The secreted mature CTSB was slightly deceased in the culture media of NPC2-kd adipocytes (Fig. 2C). These results indicate that lysosomal activity is impaired in NPC2-kd adipocytes as reflected by reduced expression of Ctsb gene and mature CTSB. Because the reduced hydrolytic activity of lysosomes is expected to attenuate the degradation of autophagosomes, causing the accumulation of autophagic flux protein LC3 (27), we then determined the levels of LC3 in NPC2-kd adipocytes in the basal state. Interestingly, we found that both LC3-I and LC3-II levels were significantly higher in NPC2-kd adipocytes than those in scrambled cells, indicating a reduction in lysosomal degradation of autophagosomes (Fig. 2D). Consistently, the level of p62, a specific autophagic cargo (28), was also elevated in NPC2-kd adipocytes (Fig. 2D), suggesting the reduced clearance of autophagic proteins by NPC2 deficiency.
To further assess the impact of NPC2 kd on autophagy, we examined autophagic flux in scrambled and NPC2-kd adipocytes. Autophagic flux was measured by comparing the difference in LC3-II and p62 levels in the presence and absence of lysosomal degradation inhibitor, BafA1. Autophagic flux was determined by calculating the ratio of protein levels of LC3-II and p62 in BafA1-treated adipocytes to those in untreated cells, as previously described (29). This method determines how much LC3-II and p62 can be accumulated by blocking lysosomal degradation, which reflects the autophagic ability. As shown in Fig. 2E, both LC3-II and p62 levels were higher in NPC2-kd adipocytes compared with scrambled cells in the basal condition. BafA1 treatment for 4 or 24 h significantly increased LC3-II and p62 levels in scrambled adipocytes. However, this BafA1-induced increase in LC3-II and p62 levels was significantly reduced in NPC2-kd adipocytes (Fig. 2E). This data suggests that NPC2 kd impairs autophagic flux; the accumulation of LC3-II in NPC2-kd cells in the basal state could result from the impairment of autophagic flux.
NPC2 kd reduces starvation-induced, but not rapamycin-induced, autophagy
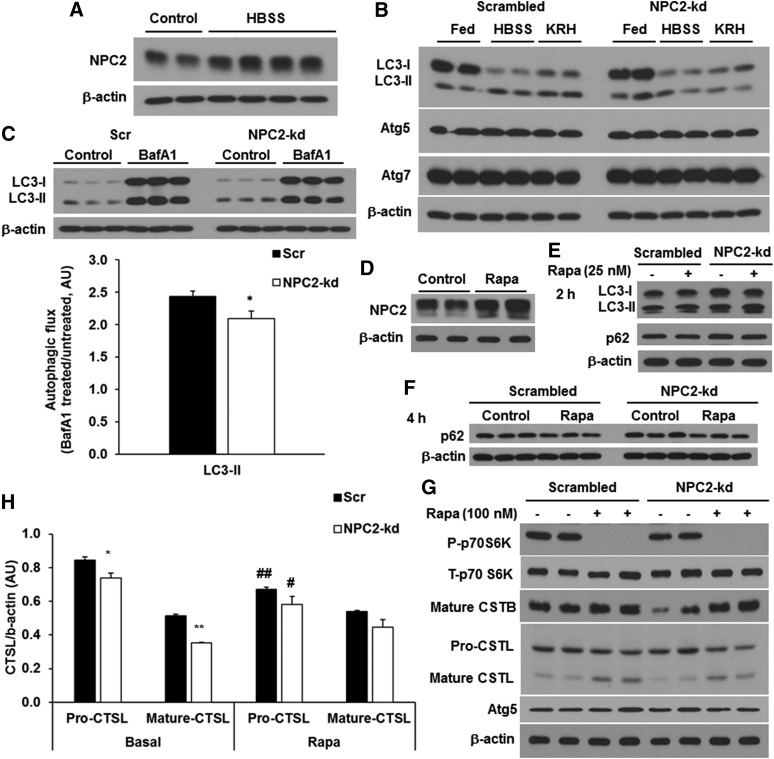
Next, we determined the effect of NPC2 deficiency on the autophagic process in adipocytes. Because nutrient deprivation is a common physiological inducer of autophagy, we examined the induction of autophagy by starvation in NPC2-kd adipocytes. First, we determined how NPC2 expression is regulated by starvation. We found that the NPC2 protein expression was significantly induced after 4 h treatment with HBSS, a medium containing 5 mM glucose and devoid of amino acids and other growth factors (Fig. 3A). Next, scrambled and NPC2-kd adipocytes were treated for 2 h with HBSS for the examination of autophagy activation as indicated by changes in LC3-II levels. As shown in Fig. 3B, there was an elevation of LC3-II in scrambled adipocytes when exposed to either HBSS or KRH buffer (containing 2.5 mM glucose) compared with those in the fed condition (Fig. 3B). However, in NPC2-kd adipocytes, neither HBSS nor KRH induced the conversion of LC3-I to LC3-II (Fig. 3B).
Fig. 3.
Effect of NPC2 kd on autophagy induction. A: The expression of NPC2 in the differentiated 3T3-L1 adipocytes incubated with HBSS (containing 5 mM glucose) for 4 h. B: The expression of autophagic markers, LC3 and Atg proteins, in NPC2-kd adipocytes incubated with culture media, HBSS (containing 5 mM glucose), and KRH buffer (containing 2.5 mM glucose) for 4 h. C: Autophagic flux measured in the cells incubated in serum-free culture media for 24 h. The autophagic flux was determined by calculating the ratio of the densitometry of LC3-II and p62 levels in BafA1-treated cells after normalization by β-actin to that in untreated cells, respectively. D: The expression of NPC2 in the differentiated 3T3-L1 adipocytes incubated with 100 nM of rapamycin for 48 h. E: LC3 and p62 expression by rapamycin treatment for 2 h. F: The expression of p62 by rapamycin treatment for 4 h. G: Scrambled or NPC2-kd cells were incubated with rapamycin in DMEM containing 25 mM of glucose, 10% of FBS, and 1 μg/ml of insulin for 48 h. H: Western blot analysis of CTSL in scrambled or NPC2-kd cells incubated with rapamycin in DMEM containing 25 mM of glucose, 10% of FBS, and 1 μg/ml of insulin for 48 h. The values are mean ± SD. *P < 0.05, **P < 0.01 versus scrambled cells; #P < 0.05, ##P < 0.01 versus basal. Rapa, rapamycin; AU, arbitrary units.
To determine whether the reduced LC3-II by starvation in NPC2-kd adipocytes was due to rapid autophagic flux or impaired induction, we treated adipocytes with or without BafA1 in a starvation milieu. Because the cells died within a very short period of time when incubated with HBSS plus BafA1, we assessed autophagic flux under the serum starvation condition. Scrambled and NPC2-kd adipocytes were treated with or without BafA1 in serum-free culture media for 24 h. Consistently, the basal level of LC3-II was higher in NPC2-kd compared with scrambled adipocytes after 24 h incubation with serum-free culture media (Fig. 3C). BafA1 treatment for 24 h led to accumulation of LC3-II in scrambled adipocytes, but this BafA1-induced LC3-II accumulation was attenuated in NPC2-kd adipocytes, as demonstrated by the ratio of LC3-II with BafA1 treatment to that without BafA1 (Fig. 3C), suggesting that the autophagic flux is defective in NPC2-kd adipocytes.
Rapamycin, an inhibitor of mTOR kinase, is another common inducer of autophagy by promoting autophagosome formation and autophagosome-lysosome fusion (30, 31). Similar to starvation, rapamycin treatment for 48 h also significantly induced NPC2 protein expression in 3T3-L1 adipocytes (Fig. 3D). Next, we assessed the effect of NPC2 kd on rapamycin-induced autophagy by detecting the protein levels of LC3 and p62. In response to rapamycin treatment for 2 h, both scrambled and NPC2-kd adipocytes had elevated levels of LC3-II, but no change in p62 levels compared with untreated controls (Fig. 3E). In order to better assess the changes in p62 proteins, cells were treated with rapamycin for a longer time (4 h). After 4 h treatment with rapamycin, p62 protein levels were decreased in both scrambled and NPC2-kd adipocytes (Fig. 3F), indicating that NPC2 kd does not affect rapamycin-induced degradation of p62 protein.
To further test to determine whether NPC2 kd impacts the mTOR signaling pathway, which is involved in the regulation of autophagy, we examined the effect of rapamycin on the mTOR signaling pathway, lysosomal proteases (CTSB and CTSL), and autophagy-related protein (Atg5). Consistent with the increased autophagy by rapamycin, we found that 48 h treatment with rapamycin significantly inhibited the phosphorylation of p70S6K in both scrambled and NPC2-kd adipocytes (Fig. 3G). Although the basal levels of CTSB and CTSL were lower in NPC2-kd adipocytes than in the scrambled controls, the expression of CTSB and CTSL was similarly upregulated by rapamycin in both types of cells (Fig. 3G, H). The expression of autophagic protein, Atg5, remained unchanged regardless of rapamycin treatment in the scrambled and NPC2-kd adipocytes (Fig. 3G). The above data suggests that NPC2 kd does not seem to affect rapamycin-induced autophagy and lysosomal activity, suggesting that NPC2 is involved in mTOR-independent signaling transduction rather than as a part of the autophagy-lysosomal degradation machinery.
NPC2 kd blunts LPS-induced, but not TNFα-induced, autophagy
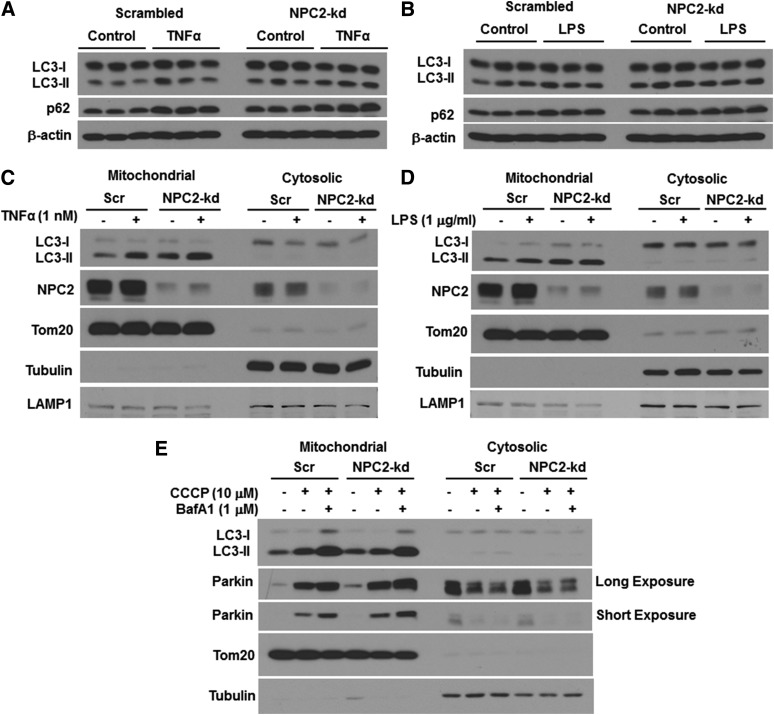
In addition to starvation, autophagy can be activated under the pathological condition (for instance, in response to inflammation/stress) and plays a role as a defensive mechanism in protecting against sustained inflammation and restoring cellular homeostasis. Numerous studies have shown that autophagy can be activated by LPS and a number of cytokines, including TNFα and IL-1β, during infection and inflammation (32). Activation of TLRs by LPS has been reported to stimulate autophagy in bone marrow-derived macrophages (33). To understand the role of NPC2 in the regulation of autophagy in response to cellular stress, we then investigated the effect of NPC2 kd on inflammation-induced autophagy in adipocytes. First, we studied the impact of NPC2 deficiency on TNFα-induced autophagy. Consistent with the results shown in Fig. 2C, the basal levels of LC3 in NPC2-kd adipocytes were higher than those in scrambled cells, indicating a reduction in lysosomal degradation (Fig. 4A). Despite an increase in basal levels of LC3 in NPC2-kd adipocytes, TNFα treatment for 4 h led to an increase in LC3-II and p62 protein in both scrambled and NPC2-kd adipocytes (Fig. 4A). This data suggests that NPC2-kd adipocytes have a normal response to TNFα-induced autophagy. Second, we tested to determine whether NPC2 deficiency influences the effect of LPS on autophagy. Interestingly, after 4 h treatment with LPS, the levels of LC3-II and p62 protein were increased in scrambled adipocytes. However, this stimulatory effect of LPS was not observed in NPC2-kd adipocytes (Fig. 4B). These results suggest that NPC2 kd selectively blunts LPS effect on autophagy activation.
Fig. 4.
Effect of NPC2 kd on inflammation-induced autophagy and mitophagy. LC3 and p62 protein levels in adipocytes treated with 1 nM of TNFα for 4 h (A) and 1 μg/ml of LPS for 4 h (B). LC3 and NPC2 protein levels in the mitochondrial fraction of adipocytes treated with 1 nM of TNFα for 4 h (C) and 1 μg/ml of LPS for 4 h (D). E: Parkin and LC3 protein levels in the mitochondrial fraction of adipocytes treated with 10 μM of CCCP and/or 1 μM of BafA1 for 4 h. Tom 20, tubulin, and LAMP1 serve as the markers of mitochondrial, cytosolic, and lysosomal proteins, respectively. Scr, scrambled.
NPC2 kd impairs mitophagy, mitochondrial function, and anti-oxidant response
One of the important purposes of inflammation-induced autophagy is to degrade mitochondria damaged by inflammation in a process known as mitophagy. To better understand the role of NPC2 in the regulation of cellular homeostasis, specifically mitochondrial quality control, we determined the cellular location of NPC2 and the effect of NPC2 kd on mitophagy and mitochondrial function. We found that NPC2 was highly abundant in the crude mitochondrial fraction and the mitochondrial abundance of NPC2 was increased in response to TNFα (Fig. 4C) and LPS treatment (Fig. 4D). While LC3-I was mostly present in the cytosolic fraction, LC3-II was more abundant in the mitochondrial fraction (Fig. 4C, D). In accordance with the results obtained from the whole cell lysate of NPC2-kd adipocytes, there was an increased accumulation of LC3-II in the mitochondrial fraction of NPC2-kd adipocytes when compared with that of scrambled cells in the basal condition (Fig. 4C, D). Both the scrambled and NPC2-kd adipocytes responded to TNFα treatment, resulting in an increase in LC3-II in the mitochondrial fraction (Fig. 4C). However, LPS treatment led to an increase in LC3-II in the mitochondrial fraction only in scrambled adipocytes, but failed to stimulate LC3-II in NPC2-kd adipocytes (Fig. 4D).
We performed an experiment to provide additional evidence for the role of NPC2 in mitophagy in adipocytes. Because mitophagy does not occur frequently in the steady state, we treated adipocytes with carbonyl cyanide m-chlorophenylhydrazone (CCCP), a mitochondrial membrane depolarizing agent, to induce mitochondrial stress and to trigger mitochondrial damage-induced mitophagy (34). The translocation of Parkin and recruitment of LC3-II to the mitochondria is one of the most important mechanisms for promoting mitophagy. Parkin is known to play a key role in initiating mitophagy by specifically targeting damaged mitochondria and promoting the removal of damaged mitochondria by autophagic clearance (35, 36). We observed that NPC2-kd adipocytes had higher levels of LC3-II and Parkin in the mitochondrial fraction than scrambled cells in the basal condition (Fig. 4E). When treated with CCCP or CCCP plus BafA1, NPC2-kd adipocytes displayed blunted induction of LC3-II with increased accumulation of Parkin in the mitochondrial fraction compared with scrambled cells (Fig. 4E).
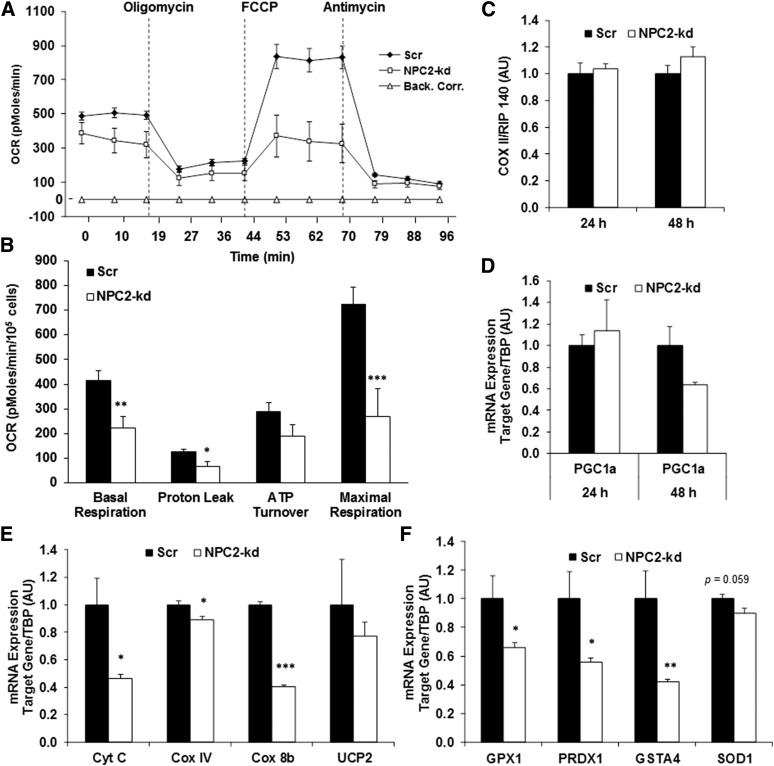
Next, cellular respiration was evaluated by the Seahorse XF24 extracellular flux analyzer to assess mitochondrial function. We observed that NPC2-kd adipocytes had significantly decreased basal respiration, reduced proton leak, and lowered maximal respiration when compared with the scrambled cells (Fig. 5A, B). This data strongly suggests that NPC2-kd adipocytes have decreased mitophagy activation and impaired mitochondrial function. To determine whether mitochondrial dysfunction results from a decreased number of mitochondria or reduced mitochondrial activity, we evaluated mitochondrial DNA (mtDNA) copy number and the expression of key genes regulating mitochondrial biogenesis and function. There was no difference in the ratio of mtDNA number cyclooxygenase (COX) II and nuclear DNA (RIP 140) between scrambled and NPC2-kd adipocytes (Fig. 5C). The gene expression of Nrf1 and Nrf2 was also not different between the scrambled and NPC2-kd adipocytes (data not shown). However, the expression of peroxisome proliferator-activated γ coactivator-1α (PGC1α) was reduced in NPC2-kd adipocytes, as was the expression of mitochondrial genes, including COX IV, cytochrome c, and cyclooxygenase 8b (COX 8b) (Fig. 5D, E). This data suggests that impaired mitochondrial function, rather than changes in mitochondrial number, is the major contributor to the reduced mitochondrial activity in NPC2-kd adipocytes.
Fig. 5.
Effect of NPC2 kd on mitochondrial function. A, B: Cellular oxygen consumption in scrambled (Scr) and NPC2-kd adipocytes. Oxygen consumption rates (OCRs) were determined in Scr and NPC2-kd adipocytes [oligomycin, 2 μM; carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP), 0.5 μM; antimycin, 4 μM]. C: mtDNA content in Scr and NPC2-kd adipocytes. D, E: The expression of genes involved in mitochondrial biogenesis and mitochondrial function in Scr and NPC2-kd adipocytes (n = 3). F: Antioxidant gene expression in Scr and NPC2-kd adipocytes (n = 3). The values are mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001 versus Scr cells. Back. Corr., background correction.
Mitochondria are an important source of superoxide production, which can be degraded to reactive oxygen species (ROS) (37). Mitochondrial dysfunction can cause increased ROS production and oxidative stress. To determine whether mitochondrial dysfunction-associated oxidative stress occurs in NPC2-kd adipocytes, we evaluated the gene expression of ROS detoxification enzymes. As shown in Fig. 5F, the gene expression of antioxidant enzymes, such as glutathione peroxidase 1 (Gpx1), peroxiredoxin 3 (Prdx3), glutathione S-transferase A4 (GstaA4), and Cu/Zn superoxide dismutase (SOD1), was significantly downregulated in NPC2-kd adipocytes compared with scrambled cells. These results support that NPC2-kd reduces the anti-oxidant response.
NPC2 kd affects insulin sensitivity in adipocytes
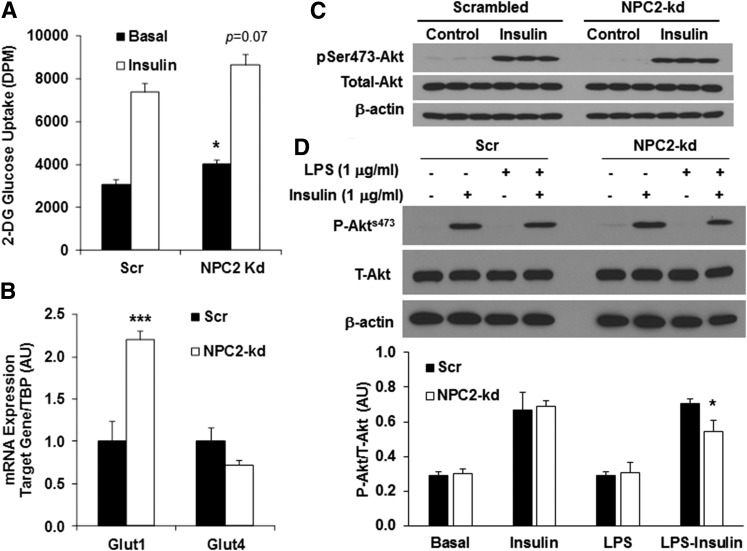
Because mitochondrial dysfunction and oxidative stress are closely associated with insulin resistance, we next evaluated the impact of NPC2 kd on insulin sensitivity by measuring insulin-stimulated glucose uptake and Akt phosphorylation. We observed that the basal glucose uptake was higher in NPC2-kd adipocytes than that in the scrambled cells (Fig. 6A). Consistently, the gene expression of Glut1, which is responsible for basal glucose uptake and highly sensitive to hypoxia, was significantly elevated in NPC2-kd adipocytes compared with scrambled cells (Fig. 6B), suggesting high activity of anaerobic metabolism of glucose in NPC2-kd adipocytes. In response to insulin stimulation, both scrambled and NPC2-kd adipocytes had a similar increase in glucose uptake (Fig. 6A). There was also no difference in the gene expression of Glut4 between the scrambled and NPC2-kd adipocytes (Fig. 6B). Additionally, insulin was able to stimulate Akt phosphorylation to a similar extent in both scrambled and NPC2-kd adipocytes (Fig. 6C). However, pretreatment with LPS for 24 h induced a greater reduction of insulin-stimulated Akt phosphorylation in NPC2-kd adipocytes than the scrambled cells (Fig. 6D), suggesting that NPC2-kd adipocytes become more insulin resistant under the condition of LPS-induced metabolic stress.
Fig. 6.
Effect of NPC2 kd on insulin sensitivity. A: The 2-deoxyglucose uptake in scrambled (Scr) and NPC2-kd adipocytes in the basal and insulin-stimulated condition. B: Gene expression of glucose transporters, Glut1 and Glut4, in Scr and NPC2-kd adipocytes. C, D: Representative Western blots for phosphorylated Akt by insulin stimulation for 30 min in Scr and NPC2-kd adipocytes with and without LPS (1 μg/ml) preincubation for 24 h. The values of gene expression are mean ± SEM. The values of protein expression are mean ± SD. *P < 0.05; ***P < 0.001 versus Scr cells.
NPC2 kd impairs TLR signaling activation
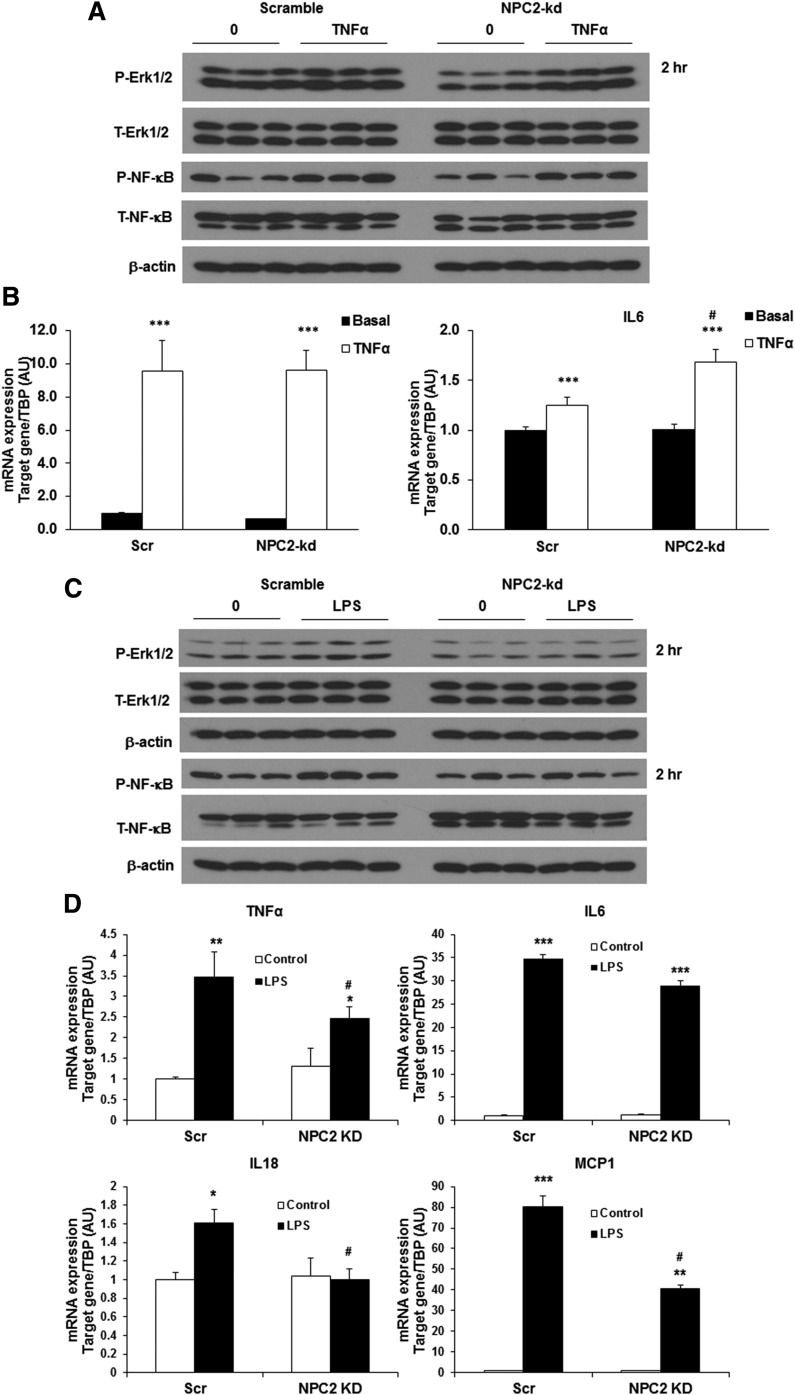
The above results that NPC2 kd selectively impairs the LPS effect on autophagy/mitophagy activation suggest that NPC2 may play a role in regulating TLR signaling pathway activation. To prove this hypothesis, we further examined how NPC2 kd impacts the effect of TNFα and LPS on the activation of TLR4 (inflammatory) signaling pathways, as well as the expression inflammatory cytokine genes by TNFα and LPS in NPC2-kd adipocytes. As shown in Fig. 7A, both scrambled and NPC2-kd adipocytes had a similar response to TNFα stimulation in NFκB and ERK1/2 phosphorylation. Consistently, the gene expression of MCP1 and IL-6 was upregulated by TNFα in both NPC2-kd and scrambled adipocytes (Fig. 7B). Strikingly, LPS treatment significantly stimulated NFκB and ERK1/2 phosphorylation in scrambled adipocytes, but this stimulatory effect was almost completely diminished in NPC2-kd adipocytes (Fig. 7C). The LPS-induced expression of inflammatory genes, including TNFα, IL-6, IL-18, and MCP1, was also significantly attenuated in NPC2-kd adipocytes (Fig. 7D). All these results suggest that NPC2 is essential for TLR signaling activation.
Fig. 7.
Effect of NPC2 kd on inflammation. A: Representative Western blots for phosphorylated p44/42 MAPK (Erk1/2) and NFκB p65 in scrambled (Scr) and NPC2-kd adipocytes treated with TNFα (1 nM) for 2 h. B: Gene expression of inflammatory cytokines in Scr and NPC2-kd adipocytes treated with TNFα (1 nM) for 48 h. C: Representative Western blots for phosphorylated p44/42 MAPK (Erk1/2) and NFκB p65 in Scr and NPC2-kd adipocytes treated with LPS (1 μg/ml) for 2 h. D: Gene expression of inflammatory cytokines in Scr and NPC2-kd adipocytes treated with LPS (1 μg/ml) for 48 h. The values are mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 versus basal levels; #P < 0.05, versus Scr cells.
DISCUSSION
NPC2 is a protein in the late endosome/lysosome that, in concert with NPC1, modulates intracellular lipid metabolism. While most of the previous studies on NPC2 focused on its role in regulating cholesterol trafficking, the pathophysiological function of this protein in cellular homeostasis remains elusive. Our previous studies have connected dysregulation of adipose NPC2 expression to diet-induced obesity (19). Studies from others have demonstrated that NPC2 has a role in adipocyte differentiation (38). In this study, we show that NPC2 plays a critical role in the regulation of mitophagy and mitochondrial function in adipocytes. We also demonstrate that the impaired mitophagy activation may be partly attributed to the failure of TLR signaling pathway activation in NPC2-kd adipocytes.
Autophagy plays an essential role in maintaining cellular homeostasis and function in many cell types, thereby regulating multiple physiological processes. In obesity, autophagy activation is necessary in order to combat metabolic stress and inflammation. Studies have indicated that this regulation of autophagy activation is defective in adipose tissue during obesity (39, 40). However, the pathological mechanism underlying this defect remains largely unknown. Autophagy activation is initiated by the formation of the autophagosome, a double-membraned vesicle, which is controlled by multiple autophagy-related proteins (ATG proteins). The mammalian homolog of yeast ATG8, LC3, is commonly used as a marker of autophagosome formation (41). The following and also most important step is the fusion of autophagosomes with late endosomes and lysosomes for the degradation of autophagic cargoes by various hydrolytic enzymes in the lysosome (42). Among the lysosomal enzymes, CTSB is one of the most abundant proteases (43) and is required for the activation of the autophagy-lysosome process (44, 45). In our study, we found that inhibiting lysosomal activity by BafA1 blocks the conversion of pro-CTSB to mature CTSB, leading to an accumulation of pro-CTSB but a decrease in mature CTSB in adipocytes. Thus, the reduction in the mature form of CTSB could indicate the impairment of lysosomal activity. In NPC2-kd adipocytes, we found that the mature form of CTSB was significantly reduced compared with scrambled cells. Consistently, the accumulation of p62 and LC3 proteins was increased in NPC2-kd adipocytes under the basal condition. During autophagy, LC3-II localizes to the inner autophagosome for degradation by lysosomal enzymes. Although there was an increase in LC3-II and p62 in the steady state, reduced induction of LC3-II and p62 was observed in NPC2-kd adipocytes treated with BafA1, indicating the impaired autophagic flux by NPC2 deficiency. All of these results indicate that NPC2 kd impairs lysosomal proteolytic activity and function, thereby reducing autophagy activity. Our results are in line with previous studies demonstrating that autophagic activity is decreased in NPC1-deficient cells (15). In the previous studies, both NPC1 deficiency and inhibition of cholesterol efflux from the lysosome could lead to the enlargement of autophagosomes, the impairment of autophagy flux, and the accumulation of ubiquitinated proteins in the endosomal/lysosomal compartment (27). Therefore, our studies have provided a new line of evidence that NPC2 has an important role in regulating autophagy-lysosomal activity.
Autophagy is involved in a wide variety of physiological and pathological processes and can be activated by various stimuli (46). Nutrient deprivation-induced autophagy is a physiological response to maintain cell survival. We found that starvation is able to induce the expression of NPC2 in adipocytes, suggesting a possible role of NPC2 in physiological autophagy. Our results showed that starvation with nutrient-deprived culture media failed to induce the recruitment of LC3-II in NPC2-kd adipocytes. Interestingly, rapamycin, a direct inhibitor of the mTORC1 and a strong inducer of autophagy, was also able to induce NPC2 expression in adipocytes. However, NPC2 deficiency does not influence the effect of rapamycin on the recruitment of LC3-II and the degradation of p62 cargo, as well as the induction of CTSB and CTSL in NPC2-kd adipocytes. Because rapamycin and nutrient deprivation have been reported to induce autophagy through two different mTOR signaling pathways, i.e., rapamycin suppresses the mTORC1 signaling pathway, while starvation inactivates the phosphoinositide-3-kinase (PI3K)-Akt and mTORC2 signaling pathway (47, 48), it is likely that NPC2 may regulate autophagy-lysosomal activation via the mTORC1-independent signaling pathway in adipocytes.
In addition to starvation, autophagy can be activated as a protective mechanism under pathological conditions, such as inflammation, oxidative and endoplasmic reticulum stress, and hypoxia (49). In obesity, autophagy is activated to alleviate stress and inflammation, thereby restoring intracellular homeostasis and limiting cellular dysfunction (50). Mitophagy, a type of selective autophagy, plays an important role in mitochondrial quality control by eliminating damaged mitochondria (51, 52). In the present study, we found that the majority of NPC2 is located in the mitochondrial fraction in adipocytes, and both TNFα and LPS increase the abundance of NPC2 protein in the mitochondrial fraction. Because the isolated crude mitochondrial fraction is potentially contaminated with other microsomes such as lysosome, we detected the lysosome marker, i.e., LAMP1, to indicate the degree of lysosomal contamination in the mitochondrial fraction. We found that LAMP1 levels are much higher in the cytosolic fraction than in the mitochondrial fraction, which is opposite to that of NPC2 levels in both LPS- and TNFα-treated conditions. Based on the distribution of TOM20 and LAMP1 between the mitochondrial and cytosolic fraction, we believe that mitochondria still account for the majority of the content in the crude mitochondrial fraction. To determine whether NPC2 is a mitochondrial targeting protein, we performed a computational prediction of mitochondrial targeting peptide on NPC2 sequence using the TargetP server. The likehood of NPC2 having a mitochondrial targeting peptide is low, suggesting that NPC2 may not be a mitochondrial protein or a mitochondrial targeting protein, but associates with mitochondria. Autophagosomes are known to have a close contact with mitochondria during mitophagy. In a previous study with electron microscopy, autophagosomes have been shown to contain mitochondria in the autophagic vacuoles (53). It is likely that the isolated mitochondrial fraction contains intact mitochondria as well as impaired mitochondria that are associated with autophagosomes, and NPC2 is located in autophagosomes/lysosomes that have been targeted to the damaged mitochondria for mitophagy.
Consistent with the marked accumulation of autophagosomes in NPC1-deficient cells in the previous report (54), we found that NPC2-kd adipocytes had significantly increased accumulation of LC3-II in the mitochondrial fraction in the steady state, indicating decreased mitochondrial degradation. Furthermore, we specifically tested the impact of NPC2 deficiency on mitophagy. Our results demonstrate that NPC2-kd adipocytes exhibit decreased induction of LC3-II and increased accumulation of Parkin in mitochondria during mitochondrial stress with CCCP. This strongly supports that NPC2 deficiency impairs mitophagy and reduces the removal of damaged mitochondria, leading to mitochondrial dysfunction. Moreover, the direct assessment of cellular respiration profiling demonstrated that NPC2-kd adipocytes have significantly impaired mitochondrial function, as evidenced by decreased basal and maximal respiration as well as reduced proton leak. This conclusion is further supported by our findings of downregulated expression of genes involved in the regulation of mitochondrial activity, including PGC1α, cytochrome c, COX IV, and COX 8b, as well as the decreased expression of anti-oxidant genes such as GPX1, PRDX1, GSTA4, and SOD1 in NPC2-kd adipocytes. In accordance with the results from a recent study (18), our data clearly support the fact that NPC2 has an essential role in the regulation of mitophagy and mitochondrial function. Because mitochondrial function is important for glucose metabolism and insulin sensitivity, we further investigated the impact of NPC2 kd on glucose uptake and insulin signaling activity. We observed that the basal level of glucose uptake was increased in NPC2-kd adipocytes compared with scrambled adipocytes. This was supported by the upregulation of Glut1, a primary transporter that is responsible for basal glucose uptake (55) and highly sensitive to hypoxia (56). The upregulation of Glut1 indicates a hypoxic condition reflecting mitochondrial dysfunction of NPC2-kd adipocytes. While NPC2 kd does not affect insulin-stimulated glucose uptake and Akt phosphorylation, it does worsen LPS-induced downregulation of insulin-stimulated Akt phosphorylation.
Inflammatory cytokines have been well-known to link inflammation to mitochondrial dysfunction and insulin resistance in obesity. TNFα has been known to alter mitochondrial membrane permeability and inhibit electron chain respiration, thereby stimulating ROS production and causing mitochondrial damage (57–59). LPS, a major component of the outer membrane of Gram-negative bacteria, has been recently considered as a critical stimulus of adipose tissue inflammation and a regulator of autophagy. LPS has been known to bind TLR2 and TLR4 on the plasma membrane with the involvement of MD-2 to activate an immune response and induce inflammation (60). The data from human studies has shown that obese subjects have altered gut microflora, leading to the elevation of circulating LPS (61). Circulating LPS levels were also found to be increased in aged humans (62). In animal studies, high-fat diet feeding has been shown to increase the proportion of LPS-containing microbiota in the gut leading to the elevation of circulating LPS levels in mice (63). In this sense, understanding the regulation of TLR signaling pathway activation and its related autophagy and metabolism in adipocytes is of importance.
Most intriguingly, we discovered that NPC2 kd selectively diminishes LPS-induced autophagy/mitophagy. NPC2-kd adipocytes have a normal response to TNFα stimulation in autophagy/mitophagy activation. LPS is known to trigger the inflammatory cascade (NFκB and Erk1/2) via activating TLR2 and TLR4 signaling (64–67), whereas TNFα stimulates NFκB and Erk1/2 activation through a TLR-independent signaling pathway. We therefore speculate that NPC2 may be critical for TLR signaling activation. Our results showed that while NPC2 kd does not affect TNFα-induced NFκB and Erk phosphorylation, LPS-stimulated phosphorylation of NFκB and Erk was almost completely diminished in NPC2-kd adipocytes. Furthermore, LPS failed to induce the expression of pro-inflammatory cytokines in NPC2-kd adipocytes as efficiently as it did in scrambled cells. This data strongly suggests that TLR signaling activation is impaired in NPC2-kd adipocytes. NFκB activation is known as an important regulator of the autophagy activation process (9, 10). It is likely that lack of NPC2 impairs the activation of TLR4 by LPS in adipocytes, leading to the failure of the NFκB signaling pathway and autophagy activation. Further studies are needed to investigate the precise molecular mechanism for the role of NPC2 in the regulation of TLR signaling pathway activation.
In summary, we have demonstrated that NPC2 kd impairs autophagy-lysosomal activity in adipocytes. We found that NPC2 kd selectively blunts the effect of LPS on TLR signaling activation, thereby leading to the defect in mitophagy activation and mitochondrial dysfunction in adipocytes. Our results support that NPC2 is an important regulator of mitophagy, mitochondrial quality control, and mitochondrial function via controlling TLR-NFκB signaling pathway activation. Our studies also suggest that NPC2 could serve as a potential therapeutic target for treating adipose tissue dysfunction in obesity and diabetes.
Acknowledgments
The authors thank Dr. Rocio Foncea and Dr. David Bernlohr from the Department of Biochemistry, Molecular Biology, and Biophysics at the University of Minnesota for technical assistance in measuring cellular respiration. They also thank Dr. Peter Lobel from the Department of Biochemistry and Molecular Biology, Rutgers University for kindly providing anti-NPC2 antibody.
Footnotes
Abbreviations:
- Atg
- autophagy-related
- BafA1
- bafilomycin A1
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- COX
- cyclooxygenase
- CTSB
- cathepsin B
- CTSL
- cathepsin L
- Gpx1
- glutathione peroxidase 1
- GstaA4
- glutathione S-transferase A4
- kd
- knockdown
- KRH
- Krebs-Ringer HEPES
- LAMP1
- lysosome-associated membrane protein 1
- LC3
- light chain 3
- LPS
- lipopolysaccharide
- mtDNA
- mitochondrial DNA
- mTORC1
- mammalian target of rapamycin complex 1
- NPC
- Niemann-Pick type C
- Prdx3
- peroxiredoxin 3
- ROS
- reactive oxygen species
- SOD1
- superoxide dismutase
- TLR
- toll-like receptor
- ULK1
- Unc-51-like kinase 1
This work was supported in part by National Institute of Diabetes and Digestive and Kidney Diseases-funded Minnesota Obesity Center (P30DK050456) and National Natural Science Foundation of China Grant 31328015. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors report no potential conflicts of interest relevant to this article.
REFERENCES
- 1.Lionaki E., Markaki M., Palikaras K., and Tavernarakis N.. 2015. Mitochondria, autophagy and age-associated neurodegenerative diseases: new insights into a complex interplay. Biochim. Biophys. Acta. 1847: 1412–1423. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N., and Komatsu M.. 2011. Autophagy: renovation of cells and tissues. Cell. 147: 728–741. [DOI] [PubMed] [Google Scholar]
- 3.Koga H., Kaushik S., and Cuervo A. M.. 2011. Protein homeostasis and aging: the importance of exquisite quality control. Ageing Res. Rev. 10: 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sengupta S., Peterson T. R., and Sabatini D. M.. 2010. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell. 40: 310–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee I. H., Cao L., Mostoslavsky R., Lombard D. B., Liu J., Bruns N. E., Tsokos M., Alt F. W., and Finkel T.. 2008. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl. Acad. Sci. USA. 105: 3374–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Travassos L. H., Carneiro L. A., Ramjeet M., Hussey S., Kim Y. G., Magalhaes J. G., Yuan L., Soares F., Chea E., Le Bourhis L., et al. . 2010. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 11: 55–62. [DOI] [PubMed] [Google Scholar]
- 7.Shi C. S., and Kehrl J. H.. 2010. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci. Signal. 3: ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi C. S., and Kehrl J. H.. 2008. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J. Biol. Chem. 283: 33175–33182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nivon M., Richet E., Codogno P., Arrigo A. P., and Kretz-Remy C.. 2009. Autophagy activation by NFkappaB is essential for cell survival after heat shock. Autophagy. 5: 766–783. [DOI] [PubMed] [Google Scholar]
- 10.Criollo A., Senovilla L., Authier H., Maiuri M. C., Morselli E., Vitale I., Kepp O., Tasdemir E., Galluzzi L., Shen S., et al. . 2010. The IKK complex contributes to the induction of autophagy. EMBO J. 29: 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi C. S., Shenderov K., Huang N. N., Kabat J., Abu-Asab M., Fitzgerald K. A., Sher A., and Kehrl J. H.. 2012. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat. Immunol. 13: 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixit S. S., Jadot M., Sohar I., Sleat D. E., Stock A. M., and Lobel P.. 2011. Loss of Niemann-Pick C1 or C2 protein results in similar biochemical changes suggesting that these proteins function in a common lysosomal pathway. PLoS One. 6: e23677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Infante R. E., Wang M. L., Radhakrishnan A., Kwon H. J., Brown M. S., and Goldstein J. L.. 2008. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc. Natl. Acad. Sci. USA. 105: 15287–15292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang M. L., Motamed M., Infante R. E., Abi-Mosleh L., Kwon H. J., Brown M. S., and Goldstein J. L.. 2010. Identification of surface residues on Niemann-Pick C2 essential for hydrophobic handoff of cholesterol to NPC1 in lysosomes. Cell Metab. 12: 166–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elrick M. J., Yu T., Chung C., and Lieberman A. P.. 2012. Impaired proteolysis underlies autophagic dysfunction in Niemann-Pick type C disease. Hum. Mol. Genet. 21: 4876–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fletcher R., Gribben C., Ma X., Burchfield J. G., Thomas K. C., Krycer J. R., James D. E., and Fazakerley D. J.. 2014. The role of the Niemann-Pick disease, type C1 protein in adipocyte insulin action. PLoS One. 9: e95598 [Erratum. 2014. PLoS One 9: e116042.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennedy B. E., Charman M., and Karten B.. 2012. Niemann-Pick type C2 protein contributes to the transport of endosomal cholesterol to mitochondria without interacting with NPC1. J. Lipid Res. 53: 2632–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy B. E., Madreiter C. T., Vishnu N., Malli R., Graier W. F., and Karten B.. 2014. Adaptations of energy metabolism associated with increased levels of mitochondrial cholesterol in Niemann-Pick type C1-deficient cells. J. Biol. Chem. 289: 16278–16289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hannaford J., Guo H., and Chen X.. 2013. Involvement of cathepsins B and L in inflammation and cholesterol trafficking protein NPC2 secretion in macrophages. Obesity (Silver Spring). 21: 1586–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., and Chen X.. 2011. Reducing selenoprotein P expression suppresses adipocyte differentiation as a result of increased preadipocyte inflammation. Am. J. Physiol. Endocrinol. Metab. 300: E77–E85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Guo H., Deis J. A., Mashek M. G., Zhao M., Ariyakumar D., Armien A. G., Bernlohr D. A., Mashek D. G., and Chen X.. 2014. Lipocalin 2 regulates brown fat activation via a nonadrenergic activation mechanism. J. Biol. Chem. 289: 22063–22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jekabsons M. B., and Nicholls D. G.. 2004. In situ respiration and bioenergetic status of mitochondria in primary cerebellar granule neuronal cultures exposed continuously to glutamate. J. Biol. Chem. 279: 32989–33000. [DOI] [PubMed] [Google Scholar]
- 23.Vanier M. T., Rodriguez-Lafrasse C., Rousson R., Gazzah N., Juge M. C., Pentchev P. G., Revol A., and Louisot P.. 1991. Type C Niemann-Pick disease: spectrum of phenotypic variation in disruption of intracellular LDL-derived cholesterol processing. Biochim. Biophys. Acta. 1096: 328–337. [DOI] [PubMed] [Google Scholar]
- 24.Tängemo C., Weber D., Theiss S., Mengel E., and Runz H.. 2011. Niemann-Pick type C disease characterizing lipid levels in patients with variant lysosomal cholesterol storage. J. Lipid Res. 52: 813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storch J., and Xu Z.. 2009. Niemann-Pick C2 (NPC2) and intracellular cholesterol trafficking. Biochim. Biophys. Acta. 1791: 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deffieu M. S., and Pfeffer S. R.. 2011. Niemann-Pick type C1 function requires lumenal domain residues that mediate cholesterol-dependent NPC2 binding. Proc. Natl. Acad. Sci. USA. 108: 18932–18936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarkar S., Carroll B., Buganim Y., Maetzel D., Ng A. H., Cassady J. P., Cohen M. A., Chakraborty S., Wang H., Spooner E., et al. . 2013. Impaired autophagy in the lipid-storage disorder Niemann-Pick type C1 disease. Cell Reports. 5: 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Overvatn A., Bjorkoy G., and Johansen T.. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282: 24131–24145. [DOI] [PubMed] [Google Scholar]
- 29.Hiebel C., Kromm T., Stark M., and Behl C.. 2014. Cannabinoid receptor 1 modulates the autophagic flux independent of mTOR- and BECLIN1-complex. J. Neurochem. 131: 484–497. [DOI] [PubMed] [Google Scholar]
- 30.Wullschleger S., Loewith R., and Hall M. N.. 2006. TOR signaling in growth and metabolism. Cell. 124: 471–484. [DOI] [PubMed] [Google Scholar]
- 31.Nyfeler B., Bergman P., Triantafellow E., Wilson C. J., Zhu Y., Radetich B., Finan P. M., Klionsky D. J., and Murphy L. O.. 2011. Relieving autophagy and 4EBP1 from rapamycin resistance. Mol. Cell. Biol. 31: 2867–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris J. 2011. Autophagy and cytokines. Cytokine. 56: 140–144. [DOI] [PubMed] [Google Scholar]
- 33.Fujita K., and Srinivasula S. M.. 2011. TLR4-mediated autophagy in macrophages is a p62-dependent type of selective autophagy of aggresome-like induced structures (ALIS). Autophagy. 7: 552–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dagda R. K., Cherra S. J. 3rd, Kulich S. M., Tandon A., Park D., and Chu C. T.. 2009. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 284: 13843–13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suen D. F., Narendra D. P., Tanaka A., Manfredi G., and Youle R. J.. 2010. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc. Natl. Acad. Sci. USA. 107: 11835–11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narendra D., Tanaka A., Suen D. F., and Youle R. J.. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183: 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim J. A., Wei Y., and Sowers J. R.. 2008. Role of mitochondrial dysfunction in insulin resistance. Circ. Res. 102: 401–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Csepeggi C., Jiang M., and Frolov A.. 2010. Somatic cell plasticity and Niemann-Pick type C2 protein: adipocyte differentiation and function. J. Biol. Chem. 285: 30347–30354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kovsan J., Bluher M., Tarnovscki T., Kloting N., Kirshtein B., Madar L., Shai I., Golan R., Harman-Boehm I., Schon M. R., et al. . 2011. Altered autophagy in human adipose tissues in obesity. J. Clin. Endocrinol. Metab. 96: E268–E277. [DOI] [PubMed] [Google Scholar]
- 40.Nuñez C. E., Rodrigues V. S., Gomes F. S., Moura R. F., Victorio S. C., Bombassaro B., Chaim E. A., Pareja J. C., Geloneze B., Velloso L. A., et al. . 2013. Defective regulation of adipose tissue autophagy in obesity. Int. J. Obes. (Lond). 37: 1473–1480. [DOI] [PubMed] [Google Scholar]
- 41.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., and Yoshimori T.. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19: 5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie Z., and Klionsky D. J.. 2007. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9: 1102–1109. [DOI] [PubMed] [Google Scholar]
- 43.Turk B., Stoka V., Rozman-Pungercar J., Cirman T., Droga-Mazovec G., Oresic K., and Turk V.. 2002. Apoptotic pathways: involvement of lysosomal proteases. Biol. Chem. 383: 1035–1044. [DOI] [PubMed] [Google Scholar]
- 44.Tatti M., Motta M., Di Bartolomeo S., Scarpa S., Cianfanelli V., Cecconi F., and Salvioli R.. 2012. Reduced cathepsins B and D cause impaired autophagic degradation that can be almost completely restored by overexpression of these two proteases in Sap C-deficient fibroblasts. Hum. Mol. Genet. 21: 5159–5173. [DOI] [PubMed] [Google Scholar]
- 45.Tatti M., Motta M., Di Bartolomeo S., Cianfanelli V., and Salvioli R.. 2013. Cathepsin-mediated regulation of autophagy in saposin C deficiency. Autophagy. 9: 241–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mizushima N., Levine B., Cuervo A. M., and Klionsky D. J.. 2008. Autophagy fights disease through cellular self-digestion. Nature. 451: 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vander Haar E., Lee S. I., Bandhakavi S., Griffin T. J., and Kim D. H.. 2007. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 9: 316–323. [DOI] [PubMed] [Google Scholar]
- 48.Soulard A., and Hall M. N.. 2007. SnapShot: mTOR signaling. Cell. 129: 434. [DOI] [PubMed] [Google Scholar]
- 49.Bachar-Wikstrom E., Wikstrom J. D., Ariav Y., Tirosh B., Kaiser N., Cerasi E., and Leibowitz G.. 2013. Stimulation of autophagy improves endoplasmic reticulum stress-induced diabetes. Diabetes. 62: 1227–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stienstra R., Haim Y., Riahi Y., Netea M., Rudich A., and Leibowitz G.. 2014. Autophagy in adipose tissue and the beta cell: implications for obesity and diabetes. Diabetologia. 57: 1505–1516. [DOI] [PubMed] [Google Scholar]
- 51.Kanki T., Furukawa K., and Yamashita S.. 2015. Mitophagy in yeast: molecular mechanisms and physiological role. Biochim. Biophys. Acta. 1853: 2756–2765. [DOI] [PubMed] [Google Scholar]
- 52.Kurihara Y., Kanki T., Aoki Y., Hirota Y., Saigusa T., Uchiumi T., and Kang D.. 2012. Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. J. Biol. Chem. 287: 3265–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamacher-Brady A., Brady N. R., Logue S. E., Sayen M. R., Jinno M., Kirshenbaum L. A., Gottlieb R. A., and Gustafsson A. B.. 2007. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 14: 146–157. [DOI] [PubMed] [Google Scholar]
- 54.Ordonez M. P., Roberts E. A., Kidwell C. U., Yuan S. H., Plaisted W. C., and Goldstein L. S.. 2012. Disruption and therapeutic rescue of autophagy in a human neuronal model of Niemann-Pick type C1. Hum. Mol. Genet. 21: 2651–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thorens B., and Mueckler M.. 2010. Glucose transporters in the 21st century. Am. J. Physiol. Endocrinol. Metab. 298: E141–E145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wood I. S., Wang B., Lorente-Cebrian S., and Trayhurn P.. 2007. Hypoxia increases expression of selective facilitative glucose transporters (GLUT) and 2-deoxy-D-glucose uptake in human adipocytes. Biochem. Biophys. Res. Commun. 361: 468–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moe G. W., Marin-Garcia J., Konig A., Goldenthal M., Lu X., and Feng Q.. 2004. In vivo TNF-alpha inhibition ameliorates cardiac mitochondrial dysfunction, oxidative stress, and apoptosis in experimental heart failure. Am. J. Physiol. Heart Circ. Physiol. 287: H1813–H1820. [DOI] [PubMed] [Google Scholar]
- 58.Mariappan N., Elks C. M., Fink B., and Francis J.. 2009. TNF-induced mitochondrial damage: a link between mitochondrial complex I activity and left ventricular dysfunction. Free Radic. Biol. Med. 46: 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mariappan N., Soorappan R. N., Haque M., Sriramula S., and Francis J.. 2007. TNF-alpha-induced mitochondrial oxidative stress and cardiac dysfunction: restoration by superoxide dismutase mimetic Tempol. Am. J. Physiol. Heart Circ. Physiol. 293: H2726–H2737. [DOI] [PubMed] [Google Scholar]
- 60.Faure M., and Lafont F.. 2013. Pathogen-induced autophagy signaling in innate immunity. J. Innate Immun. 5: 456–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., and Gordon J. I.. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 444: 1027–1031. [DOI] [PubMed] [Google Scholar]
- 62.Ghosh S., Lertwattanarak R., Garduno Jde J., Galeana J. J., Li J., Zamarripa F., Lancaster J. L., Mohan S., Hussey S., and Musi N.. 2015. Elevated muscle TLR4 expression and metabolic endotoxemia in human aging. J. Gerontol. A Biol. Sci. Med. Sci. 70: 232–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cani P. D., Bibiloni R., Knauf C., Waget A., Neyrinck A. M., Delzenne N. M., and Burcelin R.. 2008. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 57: 1470–1481. [DOI] [PubMed] [Google Scholar]
- 64.Yang R. B., Mark M. R., Gray A., Huang A., Xie M. H., Zhang M., Goddard A., Wood W. I., Gurney A. L., and Godowski P. J.. 1998. Toll-like receptor-2 mediates lipopolysaccharide-induced cellular signalling. Nature. 395: 284–288. [DOI] [PubMed] [Google Scholar]
- 65.Kirschning C. J., Wesche H., Merrill Ayres T., and Rothe M.. 1998. Human toll-like receptor 2 confers responsiveness to bacterial lipopolysaccharide. J. Exp. Med. 188: 2091–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poltorak A., He X., Smirnova I., Liu M. Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., et al. . 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 282: 2085–2088. [DOI] [PubMed] [Google Scholar]
- 67.Qureshi S. T., Lariviere L., Leveque G., Clermont S., Moore K. J., Gros P., and Malo D.. 1999. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr4). J. Exp. Med. 189: 615–625. [DOI] [PMC free article] [PubMed] [Google Scholar]