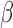Abstract
Fever is one of the most common symptoms of illness in infants and represents a clinical challenge due to the potential for serious bacterial infection. As delayed treatment for these infections has been correlated with increased morbidity and mortality, broad-spectrum 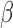 -lactam antibiotics are often prescribed while waiting for microbiological lab results (1–3 days). However, the spread of antibiotic resistance via the
-lactam antibiotics are often prescribed while waiting for microbiological lab results (1–3 days). However, the spread of antibiotic resistance via the 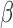 -lactamase enzyme, which can destroy
-lactamase enzyme, which can destroy 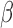 -lactam antibiotics, has confounded this paradigm; empiric antibiotic regimens are increasingly unable to cover all potential bacterial pathogens, leaving some infants effectively untreated until the pathogen is characterized. This can lead to lifelong sequela or death. Here, we introduce a fluorescent, microfluidic assay that can characterize
-lactam antibiotics, has confounded this paradigm; empiric antibiotic regimens are increasingly unable to cover all potential bacterial pathogens, leaving some infants effectively untreated until the pathogen is characterized. This can lead to lifelong sequela or death. Here, we introduce a fluorescent, microfluidic assay that can characterize 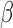 -lactamase derived antibiotic susceptibility in 20 min with a sensitivity suitable for direct human specimens. The protocol is extensible, and the antibiotic spectrum investigated can be feasibly adapted for the pathogens of regional relevance. This new assay fills an important need by providing the clinician with hitherto unavailable point of care information for treatment guidance in an inexpensive and simple diagnostic format.
-lactamase derived antibiotic susceptibility in 20 min with a sensitivity suitable for direct human specimens. The protocol is extensible, and the antibiotic spectrum investigated can be feasibly adapted for the pathogens of regional relevance. This new assay fills an important need by providing the clinician with hitherto unavailable point of care information for treatment guidance in an inexpensive and simple diagnostic format.
Keywords: Antibiotic resistance, beta-lactamase, microfluidic, point of care
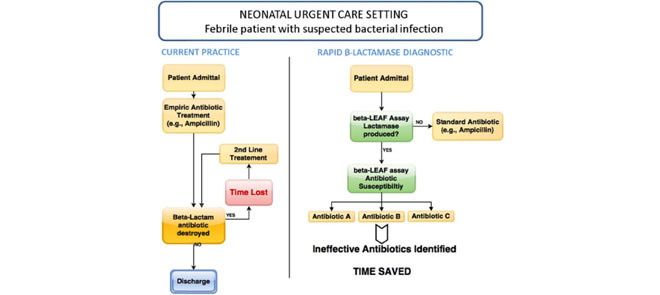
For suspected serious bacterial infection in infants, ampicillin administration is a standard treatment (left panel). However, patients with a β-lactamase positive infection may be resistant to this treatment. As delays in effective treatment are correlated with poor patient outcome, rapid identification of this situation is desirable. The assay presented here (right panel) rapidly detects β-lactamase positive infections and identifies ineffective antibiotics in less than 20 minutes, allowing clinicians to make informed treatment decisions at the point of care
I. Introduction
In the infant, fever (≥38°C) is often the only symptom of a serious bacterial infection (SBI) [1], for example, bacteremia, meningitis, urinary tract infection (UTI), etc. These patients are at serious risk of lifelong impairment or mortality if undertreated, leading to a policy of presumed SBI for febrile neonates less than 29 days old [2], [3], along with increased hospitalization, diagnostic testing and empiric antibiotic treatment. Roughly 2% of all infants less than 90 days old [4] will be taken for a medical examination due to a febrile condition. Of these patients, approximately 10% will have an SBI [5], [6], with the number approaching 20% for neonates less than 29 days old [2]. The risk of SBI decreases with age, although infants up to 3 months of age are still at risk. The majority of febrile patients have a self-limiting infection and do not require SBI treatment, but distinguishing low-risk from high-risk patients at the point of care (POC) has been an active subject of debate for more than 20 years. Of serious concern here are the severe repercussions of missed or delayed diagnoses, as delays in treatment increase the chance of serious sequela or death. Bacterial meningitis can be particularly devastating, with an approximate mortality of 10% and 20% of the survivors having a serious impairment (e.g., intellectual disability, epilepsy); another 35% will have mild to moderate disability [7]. In regards to UTI, 15% of children (0-18 years old) develop renal scarring, putting them at risk of hypertension, decreased renal function, proteinuria, and end-stage renal disease [8].
These risks notwithstanding, the general policy of presumed SBI has also led to unintended negative consequences (i.e., iatrogenic harm) to the non-SBI patients due to the additional testing, antibiotics and hospital stay as well as the emotional and financial burden placed on the family [9]–[11]. A number of studies addressing this issue suggest that appropriate criteria can identify some low-risk patients early enough to avoid unnecessary treatment [6], [12], however many patients without SBI will still receive unnecessary care and concomitant complications.
For those patients classified with a SBI, management guidelines are unclear [3], in large part due to the lack of diagnostic information available to the clinician at the point of care. For infants less than 90 days old, 63% – 92% of SBI are eventually diagnosed as UTI-related, with bacteremia/sepsis (15% – 29%) and meningitis (2% – 4%) making up the bulk of the remainder [5], [6], [13]. The standard treatment for suspected infant SBI includes ampicillin (a type of penicillin) with gentamicin or a cephalosporin. Both penicillins and cephalosporins are classed 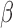 -lactam antibiotics due to their
-lactam antibiotics due to their 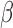 -lactam core structure (Fig. 1). Unfortunately, ampicillin resistant isolates now account for 36% to 53% of SBI in febrile infants [4], [13], [14] with 6% of Escherichia coli strains (the dominant source of SBI [5], [13], [14]) also resistant to gentamicin [4]. Despite this, a survey of 44 hospitals found that 6.3% of hospitalized febrile infants less than 29 days old still received ampicillin in combination with gentamicin as an initial treatment [6].
-lactam core structure (Fig. 1). Unfortunately, ampicillin resistant isolates now account for 36% to 53% of SBI in febrile infants [4], [13], [14] with 6% of Escherichia coli strains (the dominant source of SBI [5], [13], [14]) also resistant to gentamicin [4]. Despite this, a survey of 44 hospitals found that 6.3% of hospitalized febrile infants less than 29 days old still received ampicillin in combination with gentamicin as an initial treatment [6].
FIGURE 1.
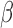 -lactam antibiotic structure.
-lactam antibiotic structure.
Thus while the need for early, effective diagnosis of SBI in the febrile infant is clear, the information required to do so is not. At present, rapid diagnostic information available to the clinician is limited. A major obstacle is the limited time available during a clinical examination. Tests requiring more than 20 minutes will not fit into a typical workflow [15]. Typical tests on the urine and cerebrospinal fluid (CSF) include glucose, protein and unspun gram stain. While these tests have been proven to help identify low risk SBI patients, they lack sensitivity and provide little information on initial antibiotic treatment guidance. Other available modalities include matrix-assisted laser desorption/ionization time of flight spectrometry and pathogen nucleic acid-based techniques. Both are powerful but can only identify pathogens that have been previously characterized [16], [17]. Cost can also be an issue for these rapid diagnostics. Definitive testing is possible with growth based assays, but requires 1-3 days of incubation time—which obviates its use for rapid testing.
Given the dearth of rapid diagnostic information, using a broad-spectrum cephalosporin antibiotic is a rational and increasingly common choice. However this is not without its own risk, as prior use of a cephalosporin has been correlated with increased risk of infection with cephalosporin-resistant pathogens [18]. While currently uncommon in the US, these pathogens are endemic in Asia [19], and pediatric infections are being increasingly reported in the West [20]–[25]. Pathogens of this sort have limited treatment options; rapid detection is essential both for effective treatment and containment of outbreaks [26], [27].
While target mutation and cell wall permeability can both cause antibiotic resistance, the root of this growing 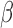 -lactam resistance stems from the spread of
-lactam resistance stems from the spread of 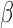 -lactamase, a plasmid-borne resistance enzyme that cleaves
-lactamase, a plasmid-borne resistance enzyme that cleaves 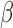 -lactam antibiotics into two fragments (Fig. 2). Hundreds of different
-lactam antibiotics into two fragments (Fig. 2). Hundreds of different 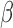 -lactamases have been characterized, each with its own spectrum of activity against different antibiotics. Thus, the antibiotic susceptibility of a pathogen to any particular
-lactamases have been characterized, each with its own spectrum of activity against different antibiotics. Thus, the antibiotic susceptibility of a pathogen to any particular 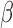 -lactam antibiotic will depend on its resistance via the
-lactam antibiotic will depend on its resistance via the 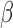 -lactamase it harbors. Identifying this rapidly is possible in principle with PCR techniques; however the genotypic detection of a resistance mechanism does not ensure its phenotypic expression. This variability can generate unwanted false positive detections. Furthermore, new types of
-lactamase it harbors. Identifying this rapidly is possible in principle with PCR techniques; however the genotypic detection of a resistance mechanism does not ensure its phenotypic expression. This variability can generate unwanted false positive detections. Furthermore, new types of 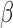 -lactamase are constantly being discovered, and, without prior knowledge of the genetic information, PCR techniques cannot be used to detect these pathogens (i.e., biased diagnostic testing). Thus, as it stands, there is no generally applicable rapid test for characterizing antibiotic susceptibility to
-lactamase are constantly being discovered, and, without prior knowledge of the genetic information, PCR techniques cannot be used to detect these pathogens (i.e., biased diagnostic testing). Thus, as it stands, there is no generally applicable rapid test for characterizing antibiotic susceptibility to 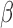 -lactam antibiotics.
-lactam antibiotics.
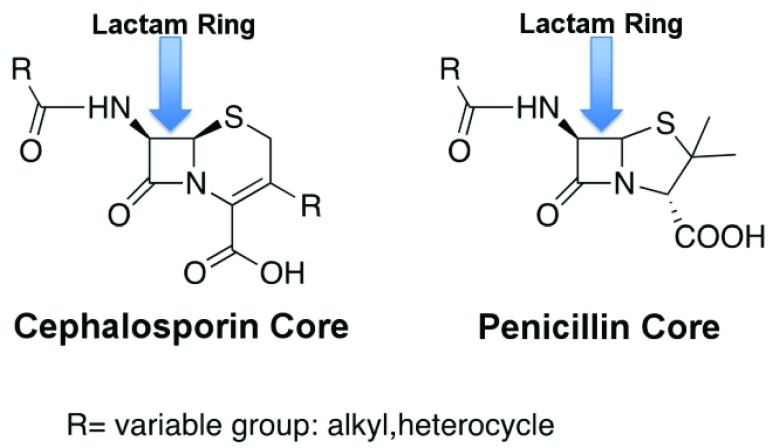
FIGURE 2.
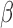 -LEAF assay principle. a) The
-LEAF assay principle. a) The 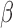 -LEAF probe comprises a
-LEAF probe comprises a 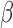 -lactam core structure (green) including the cleavable lactam ring, flanked by two fluorophores (encircled), which undergo static quenching when the probe is intact. Following cleavage by
-lactam core structure (green) including the cleavable lactam ring, flanked by two fluorophores (encircled), which undergo static quenching when the probe is intact. Following cleavage by 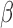 -lactamase, the fluorophores move apart and show fluorescence. b) Assay profile for
-lactamase, the fluorophores move apart and show fluorescence. b) Assay profile for 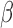 -lactamase producing bacteria. c) Assay profile for lactamase non-producing bacteria.
-lactamase producing bacteria. c) Assay profile for lactamase non-producing bacteria.
To address this unmet need, our team has been developing an innovative fluorescent assay (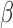 -lactamase enzyme activated fluorophore or
-lactamase enzyme activated fluorophore or 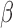 -LEAF) designed to phenotypically detect
-LEAF) designed to phenotypically detect 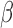 -lactamase derived antibiotic susceptibility in less than 20 minutes [28]–[32]. To achieve this,
-lactamase derived antibiotic susceptibility in less than 20 minutes [28]–[32]. To achieve this, 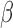 -LEAF takes advantage of the static quenching phenomenon that occurs when two fluorophores are in close proximity. To create this scenario, a cephalosporin
-LEAF takes advantage of the static quenching phenomenon that occurs when two fluorophores are in close proximity. To create this scenario, a cephalosporin 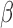 -lactam core is modified such that two fluorophore molecules are anchored to opposite ends (Fig. 2a). Consequently, the fluorophores are unable to emit fluorescence due to ground state interactions between each other. However, after
-lactam core is modified such that two fluorophore molecules are anchored to opposite ends (Fig. 2a). Consequently, the fluorophores are unable to emit fluorescence due to ground state interactions between each other. However, after 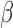 -lactamase cleaves the probe, the fluorophores diffuse away from each other and regain their fluorescent properties. This results in a time-increasing fluorescent signal, the slope of which is the readout for this assay (Fig. 3). If the assay is repeated in the presence of a cleavable antibiotic that competes with the
-lactamase cleaves the probe, the fluorophores diffuse away from each other and regain their fluorescent properties. This results in a time-increasing fluorescent signal, the slope of which is the readout for this assay (Fig. 3). If the assay is repeated in the presence of a cleavable antibiotic that competes with the 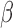 -LEAF for
-LEAF for 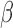 -lactamase cleavage, the rate of
-lactamase cleavage, the rate of 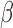 -LEAF cleavage will decrease, resulting in no fluorescence change. For example, in Fig. 3, we see cefazolin (a cephalosporin antibiotic) inhibiting the cleavage of
-LEAF cleavage will decrease, resulting in no fluorescence change. For example, in Fig. 3, we see cefazolin (a cephalosporin antibiotic) inhibiting the cleavage of  -LEAF, indicating cefazolin cleavage by
-LEAF, indicating cefazolin cleavage by 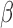 -lactamase. Thus cefazolin would be a poor treatment choice for this pathogen. We have validated this approach with multiple pathogens [29], [30] demonstrating both detection of
-lactamase. Thus cefazolin would be a poor treatment choice for this pathogen. We have validated this approach with multiple pathogens [29], [30] demonstrating both detection of 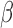 -lactamase as well as detection of several classes of
-lactamase as well as detection of several classes of 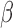 -lactamases, including the extended spectrum
-lactamases, including the extended spectrum 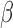 -lactamases (defined by their ability to neutralize
-lactamases (defined by their ability to neutralize 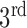 generation cephalosporin antibiotics). Thus, this test can determine which antibiotics are resistant to
generation cephalosporin antibiotics). Thus, this test can determine which antibiotics are resistant to 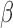 -lactamase destruction.
-lactamase destruction.
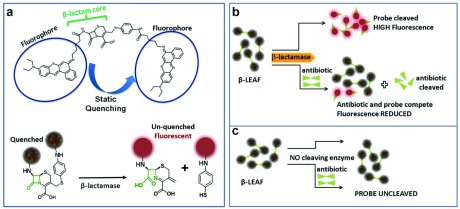
FIGURE 3.
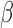 -LEAF Assay on 96 well plate. Increasing fluorescence indicates a
-LEAF Assay on 96 well plate. Increasing fluorescence indicates a 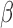 -lactamase positive organism (green). The addition of cefazolin to the lactamase-positive assay conditions leads to negligible fluorescence change (blue) due to competitive inhibition and indicates the destruction of cefazolin by
-lactamase positive organism (green). The addition of cefazolin to the lactamase-positive assay conditions leads to negligible fluorescence change (blue) due to competitive inhibition and indicates the destruction of cefazolin by 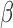 -lactamase. 1e8 CFU per well was used.
-lactamase. 1e8 CFU per well was used.
This existing 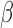 -LEAF assay has been validated [28]–[32] from cultured clinical isolates and is suitable as a laboratory tool, but rapidly assessing direct patient samples has been a challenging obstacle due to the lower pathogen concentration in the specimens of interest i.e., urine and cerebrospinal fluid (CSF). To achieve this aim, we have developed a matched diagnostic platform using disposable microfluidic cartridges to trap bacteria directly from unprocessed patient samples while simultaneously allowing antibiotic susceptibility characterization with the
-LEAF assay has been validated [28]–[32] from cultured clinical isolates and is suitable as a laboratory tool, but rapidly assessing direct patient samples has been a challenging obstacle due to the lower pathogen concentration in the specimens of interest i.e., urine and cerebrospinal fluid (CSF). To achieve this aim, we have developed a matched diagnostic platform using disposable microfluidic cartridges to trap bacteria directly from unprocessed patient samples while simultaneously allowing antibiotic susceptibility characterization with the 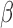 -LEAF assay. Here we compare the limits of detection of the newly established microfluidic assay with the previous well-plate assay and establish its utility as a point of care diagnostic.
-LEAF assay. Here we compare the limits of detection of the newly established microfluidic assay with the previous well-plate assay and establish its utility as a point of care diagnostic.
II. Methods
A. Reagents for 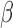 -LEAF Synthesis, Bacterial Strains and Culture Conditions
-LEAF Synthesis, Bacterial Strains and Culture Conditions
ACLE hydrochloride was a generous gift from Otsuka Chemical Co., Ltd., Tokyo, Japan. Other chemicals and solvents for the 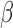 -LEAF synthesis, bacterial growth/characterization and antibiotic treatment were purchased from Sigma-Aldrich (St. Louis, Missouri) and used without further purification. Staphylococcus aureus quality control strains (used as microbiology laboratory standards) known to express
-LEAF synthesis, bacterial growth/characterization and antibiotic treatment were purchased from Sigma-Aldrich (St. Louis, Missouri) and used without further purification. Staphylococcus aureus quality control strains (used as microbiology laboratory standards) known to express 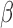 -lactamase (ATCC 29213) and not express
-lactamase (ATCC 29213) and not express 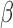 -lactamase (ATCC 25923) were purchased from ATCC (Manassas, Virginia). Brain heart infusion (BHI) broth and BHI agar were obtained from BD Difco (BD: Becton, Dickinson and Company, Franklin Lakes, New Jersey). Penicillin disks (10 U) were purchased from BD BBL. All strains were routinely cultured in BHI agar or broth at 37°C. The isolates were grown in the presence of penicillin disks to induce and enhance
-lactamase (ATCC 25923) were purchased from ATCC (Manassas, Virginia). Brain heart infusion (BHI) broth and BHI agar were obtained from BD Difco (BD: Becton, Dickinson and Company, Franklin Lakes, New Jersey). Penicillin disks (10 U) were purchased from BD BBL. All strains were routinely cultured in BHI agar or broth at 37°C. The isolates were grown in the presence of penicillin disks to induce and enhance 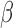 -lactamase production as required.
-lactamase production as required.
B. Synthesis of 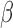 -LEAF
-LEAF
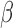 -LEAF was synthesized as previously described [32]. Briefly, the chloro- group on 7-amino-3-chloromethyl-3-cephem-4-carboxylic acid p-methoxybenzyl ester was substituted with 4-aminothiophenol with the help of 4-methylmorpholine. A mixture of carboxylic acid-modified Bodipy-FL and O-(7-azabenzotriazole-1-yl)-N,N, N,
-LEAF was synthesized as previously described [32]. Briefly, the chloro- group on 7-amino-3-chloromethyl-3-cephem-4-carboxylic acid p-methoxybenzyl ester was substituted with 4-aminothiophenol with the help of 4-methylmorpholine. A mixture of carboxylic acid-modified Bodipy-FL and O-(7-azabenzotriazole-1-yl)-N,N, N,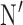 -tetramethyluroniumhexafluorophosphate in dry N,N-dimethylformamide was stirred for 30 min. Diisopropylethylamine was added to the stirring solution. The resulting reaction mixture was protected from light and stirred overnight. The solvent was removed under vacuum, and the residue was reconstituted in dichloromethane (DCM). The organic layer was washed with brine. After removing the solvent under vacuum, the crude product was purified by HPLC. This was dissolved in a solvent mixture of trifluoroacetic acid:anisole: DCM and stirred at 0°C for 1 h. The solvent was removed under vacuum, and the residue was purified by HPLC. Concentrated stocks were prepared in 100% DMSO and stored at −20°C.
-tetramethyluroniumhexafluorophosphate in dry N,N-dimethylformamide was stirred for 30 min. Diisopropylethylamine was added to the stirring solution. The resulting reaction mixture was protected from light and stirred overnight. The solvent was removed under vacuum, and the residue was reconstituted in dichloromethane (DCM). The organic layer was washed with brine. After removing the solvent under vacuum, the crude product was purified by HPLC. This was dissolved in a solvent mixture of trifluoroacetic acid:anisole: DCM and stirred at 0°C for 1 h. The solvent was removed under vacuum, and the residue was purified by HPLC. Concentrated stocks were prepared in 100% DMSO and stored at −20°C.
C.  -LEAF—Antibiotic Bacteria Well Plate Fluorescence Assays
-LEAF—Antibiotic Bacteria Well Plate Fluorescence Assays
Bacterial strains were cultured overnight on BHI agar plates in the presence of a penicillin disk (10 U). For each bacterial isolate, colonies closest to the penicillin disk were transferred to PBS to make a homogenous suspension [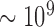 colony forming units (CFU)/ml]. Bacterial OD was measured at 600 nm. Serial dilutions were also prepared with tenfold lower bacterial concentrations in PBS. A 20
colony forming units (CFU)/ml]. Bacterial OD was measured at 600 nm. Serial dilutions were also prepared with tenfold lower bacterial concentrations in PBS. A 20 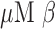 -LEAF Bodipy-FL probe solution (
-LEAF Bodipy-FL probe solution (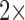 stock) was prepared in 40% DMSO in PBS, and a 100 mM cefazolin solution (
stock) was prepared in 40% DMSO in PBS, and a 100 mM cefazolin solution (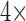 stock) was prepared by dissolving the antibiotic powder in PBS. The assays were performed in 96-well white clear-bottom plates in a total volume of
stock) was prepared by dissolving the antibiotic powder in PBS. The assays were performed in 96-well white clear-bottom plates in a total volume of 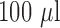 , respectively, to include bacteria and the
, respectively, to include bacteria and the 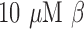 -LEAF probe, with or without 25 mM cefazolin. Each reaction was set up as follows:
-LEAF probe, with or without 25 mM cefazolin. Each reaction was set up as follows: 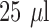 bacterial suspension,
bacterial suspension, 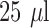 antibiotic
antibiotic 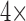 stock solution or PBS only, and
stock solution or PBS only, and 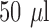 probe
probe 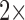 stock solution, with the resultant buffer concentration as 20% DMSO in PBS in each
stock solution, with the resultant buffer concentration as 20% DMSO in PBS in each 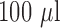 reaction. For each isolate, reactions were performed in triplicate in the absence and presence of the test antibiotic, respectively. Time course assays were carried out, monitoring
reaction. For each isolate, reactions were performed in triplicate in the absence and presence of the test antibiotic, respectively. Time course assays were carried out, monitoring  -LEAF cleavage by measuring fluorescence for 60 min, at 1 min intervals (Spectramax M5 Plate Reader, Molecular Devices). Instrument settings were kept as excitation at 450 nm and emission at 510 nm. The temperature was maintained at 37°C throughout. Fluorescence was measured in a machine specific standard unit (AU). The
-LEAF cleavage by measuring fluorescence for 60 min, at 1 min intervals (Spectramax M5 Plate Reader, Molecular Devices). Instrument settings were kept as excitation at 450 nm and emission at 510 nm. The temperature was maintained at 37°C throughout. Fluorescence was measured in a machine specific standard unit (AU). The  -LEAF cleavage rate in each case was determined as the slope, i.e., fluorescence change as a function of time (obtained from instrument software—SoftMax Pro5), normalized by bacterial OD. Antibiotic susceptibility was confirmed using standard procedures recommended by the Clinical & Laboratory Standards Institute, e.g., minimum inhibitory concentration (MIC) (E-test strip) and disk diffusion. Bovine CSF was obtained commercially from BioreclamationIVT (New York, USA).
-LEAF cleavage rate in each case was determined as the slope, i.e., fluorescence change as a function of time (obtained from instrument software—SoftMax Pro5), normalized by bacterial OD. Antibiotic susceptibility was confirmed using standard procedures recommended by the Clinical & Laboratory Standards Institute, e.g., minimum inhibitory concentration (MIC) (E-test strip) and disk diffusion. Bovine CSF was obtained commercially from BioreclamationIVT (New York, USA).
D. Microfluidic Construction and Testing
50 micron thick double sided film adhesive 3M 8212 (3M; St. Paul, MN) and 1/ clear polymethyl methacrylate ([PMMA] McMaster-Carr, Robbinsville NJ) are laser cut (Danger!Awesome, Cambridge MA) and assembled by hand with 600 nm pore size nuclear track etched polycarbonate filters (Sterlitech; Kent, WA). The viewing chambers are sized to be similar to 384 well plate dimensions. Liquid was driven through the device with a vacuum. The vacuum seal was made using a
clear polymethyl methacrylate ([PMMA] McMaster-Carr, Robbinsville NJ) are laser cut (Danger!Awesome, Cambridge MA) and assembled by hand with 600 nm pore size nuclear track etched polycarbonate filters (Sterlitech; Kent, WA). The viewing chambers are sized to be similar to 384 well plate dimensions. Liquid was driven through the device with a vacuum. The vacuum seal was made using a 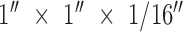 piece of plastic attached to a soft rubber gasket. A small hole was drilled through both and a small gauge needle passed through the orifice and sealed with epoxy. Typical flow rates were 1 ml/min.
piece of plastic attached to a soft rubber gasket. A small hole was drilled through both and a small gauge needle passed through the orifice and sealed with epoxy. Typical flow rates were 1 ml/min.
E.  -LEAF—Antibiotic Bacteria Microfluidic Fluorescence Assays
-LEAF—Antibiotic Bacteria Microfluidic Fluorescence Assays
Bacteria were grown and characterized as previously described. A 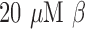 -LEAF probe solution was prepared in 40% DMSO in PBS, and 100-mM solutions of cefazoline, cefepime, and cefoxitin (
-LEAF probe solution was prepared in 40% DMSO in PBS, and 100-mM solutions of cefazoline, cefepime, and cefoxitin ( stock) was prepared by dissolving the antibiotic powder in PBS. A final working solution of 10
stock) was prepared by dissolving the antibiotic powder in PBS. A final working solution of 10 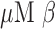 -LEAF probe solution with 25 mM antibiotic (if added) was then prepared with 20% DMSO in PBS. To perform the assay, the bacterial solution was first flushed through the device. This was immediately followed by 100
-LEAF probe solution with 25 mM antibiotic (if added) was then prepared with 20% DMSO in PBS. To perform the assay, the bacterial solution was first flushed through the device. This was immediately followed by 100 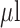 of the aforementioned probe solution. The plate was then immediately placed in the plate reader and the measurement taken as described for the well-plate assay.
of the aforementioned probe solution. The plate was then immediately placed in the plate reader and the measurement taken as described for the well-plate assay.
III. Results
The lower pathogen concentrations within human specimens is a problem for the 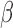 -LEAF and many other conventional assays. To circumvent this obstacle, we designed a microfluidic chamber with a porous wall to trap pathogens from a liquid specimen matrix (Fig. 4a). To run multiple conditions simultaneously, 24 chambers were created in a standard microplate reader form factor (Fig. 4b, c). With this configuration, immobilized pathogens could be easily interrogated with the
-LEAF and many other conventional assays. To circumvent this obstacle, we designed a microfluidic chamber with a porous wall to trap pathogens from a liquid specimen matrix (Fig. 4a). To run multiple conditions simultaneously, 24 chambers were created in a standard microplate reader form factor (Fig. 4b, c). With this configuration, immobilized pathogens could be easily interrogated with the 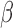 -LEAF assay and the fluorescent output monitored using a commercial plate reader (Fig. 5). A vacuum aspirator was used to drive the sample through the microfluidic system in less than 90 seconds.
-LEAF assay and the fluorescent output monitored using a commercial plate reader (Fig. 5). A vacuum aspirator was used to drive the sample through the microfluidic system in less than 90 seconds.
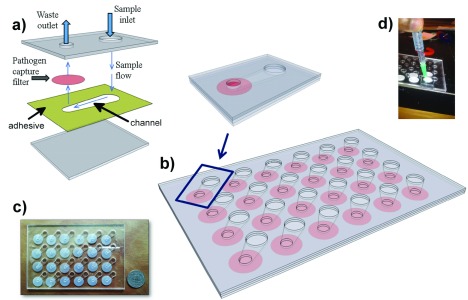
FIGURE 4.
Microfluidic Assembly. a) Schematic of single chamber. Polycarbonate filter (pink) is compressed between two sheets of clear polymethyl methacrylate (grey). A 50 micron deep channel is defined in a layer of double sided adhesive (yellow). Another layer of double sided adhesive (not shown) is placed above the filter to seal it against the outlet. b) Array of chambers is designed into a standard well-plate format. c) Photograph of microfluidic plate. d) Vacuum fitting used to drive fluids through the microfluidic.
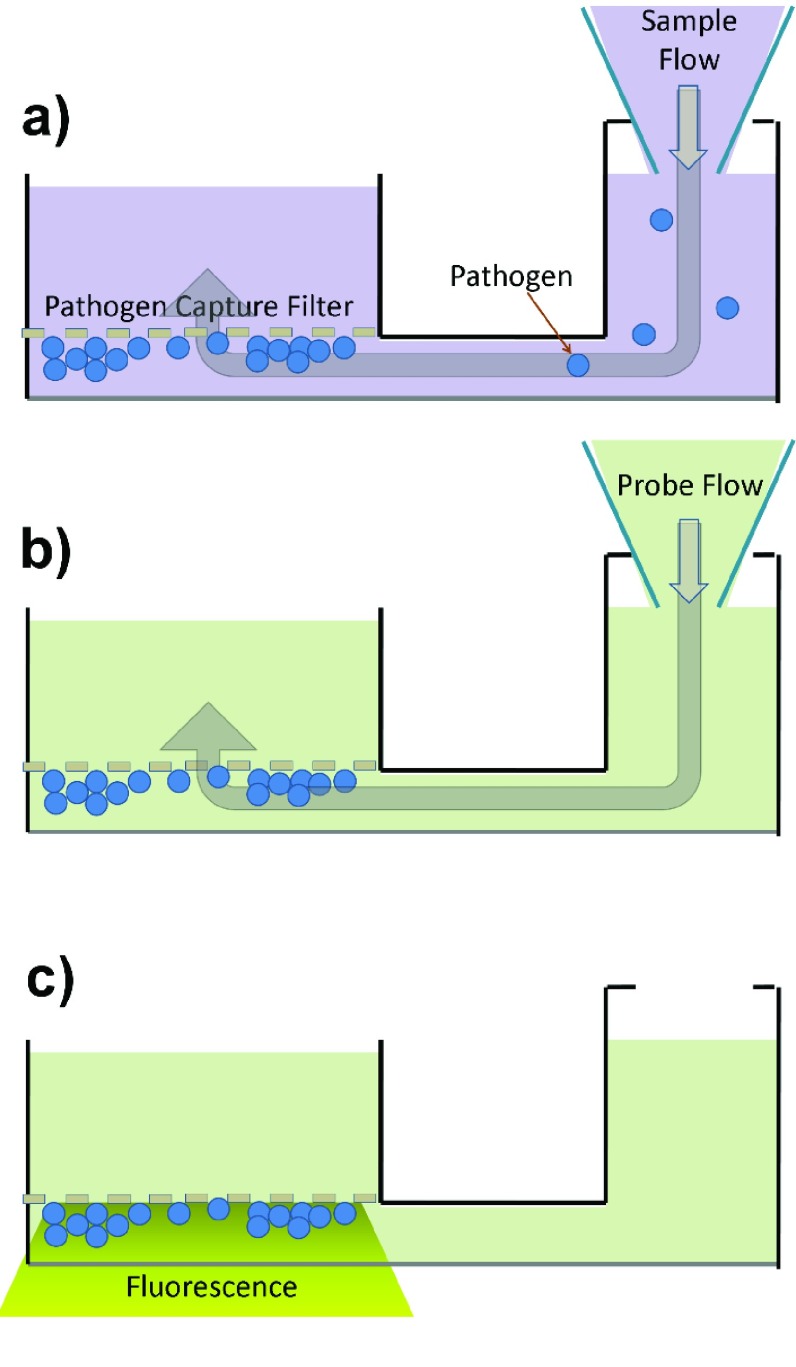
FIGURE 5.
Microfluidic principle of operation. a) sample insertion. b) 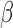 -LEAF probe insertion, c) fluorescent detection.
-LEAF probe insertion, c) fluorescent detection.
After charging the microfluidic device (as described in Fig. 5), measurements were taken on a fluorescence plate reader (representative data in Fig. 6); these displayed the expected increase in fluorescence due to the cleaving and consequent de-quenching of the 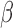 -LEAF probe. These curves are practically identical with the data acquired from the well-plate assay (Fig. 3) indicating compatibility between the two formats.
-LEAF probe. These curves are practically identical with the data acquired from the well-plate assay (Fig. 3) indicating compatibility between the two formats.
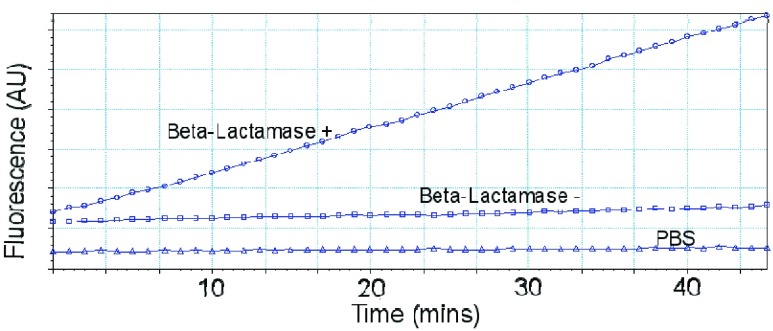
FIGURE 6.
Representative fluorescence time course on microfluidic device. 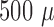 l of 5e8 CFU/ml S. aureus was inserted into the device followed by the
l of 5e8 CFU/ml S. aureus was inserted into the device followed by the 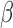 -LEAF probe. Fluorescence was measured with a commercial plate reader.
-LEAF probe. Fluorescence was measured with a commercial plate reader.
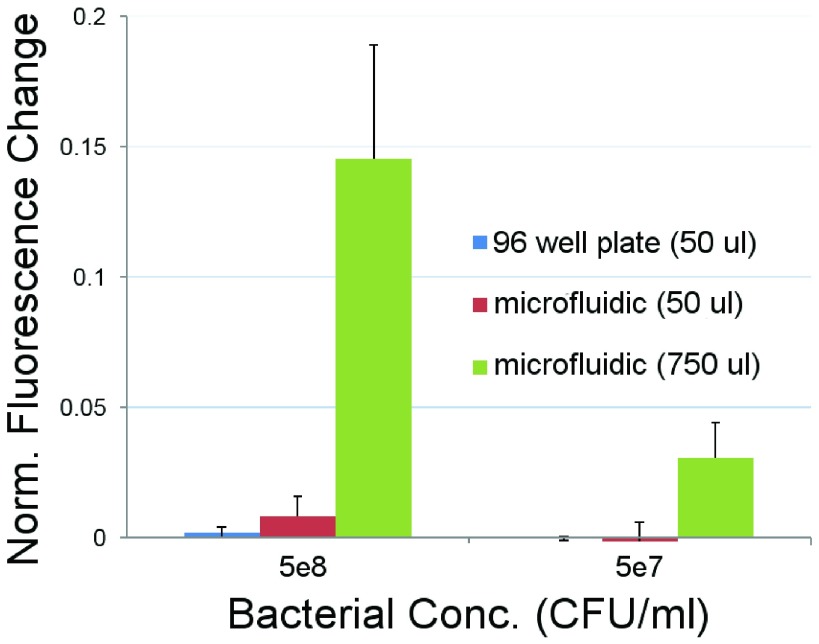
To compare the limit of detection (LOD) between the systems, different bacterial concentrations and volumes were studied (Fig. 7). A volume of 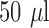 with a concentration of 5e8 CFU/ml results is just below the practical LOD for the 96 well plate protocol. Using these same conditions with the microfluidic system gives
with a concentration of 5e8 CFU/ml results is just below the practical LOD for the 96 well plate protocol. Using these same conditions with the microfluidic system gives 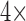 higher fluorescence emission (mostly due to the improved effective numerical aperture from confining the fluorescent signal at the bottom of the plate). Additionally, the ability to process larger quantities of sample with the microfluidic (in this case
higher fluorescence emission (mostly due to the improved effective numerical aperture from confining the fluorescent signal at the bottom of the plate). Additionally, the ability to process larger quantities of sample with the microfluidic (in this case 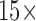 more) enables capturing larger numbers of bacteria, which gives a greater than
more) enables capturing larger numbers of bacteria, which gives a greater than 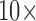 increase in the concentration LOD vs. the well plate assay (Fig. 7). This is consistent with the fact that it is the total number of pathogens in the assay which determines the fluorescence generated—and hence the ultimate LOD. We note the true concentration LOD for the microfluidic is actually better than 5e7 CFU/ml, as the fluorescence change measured is roughly
increase in the concentration LOD vs. the well plate assay (Fig. 7). This is consistent with the fact that it is the total number of pathogens in the assay which determines the fluorescence generated—and hence the ultimate LOD. We note the true concentration LOD for the microfluidic is actually better than 5e7 CFU/ml, as the fluorescence change measured is roughly 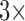 larger than the fluorescence change measured with
larger than the fluorescence change measured with 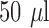 at 5e8 CFU/ml (which was easily resolved).
at 5e8 CFU/ml (which was easily resolved).
FIGURE 7.
Comparison of 96 well plate assay and new microfluidic design. Average and std. dev. from 2 separate experiments. Two different concentrations of  -lactamase positive S. aureus were studied, and the change in fluorescent emission was determined after a 20 minute incubation. Two different bacterial volume conditions were investigated with the microfluidic assay to probe its enhanced sensitivity as a function of volume. Results were normalized for instrument drift using a
-lactamase positive S. aureus were studied, and the change in fluorescent emission was determined after a 20 minute incubation. Two different bacterial volume conditions were investigated with the microfluidic assay to probe its enhanced sensitivity as a function of volume. Results were normalized for instrument drift using a 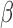 -LEAF probe only condition.
-LEAF probe only condition.
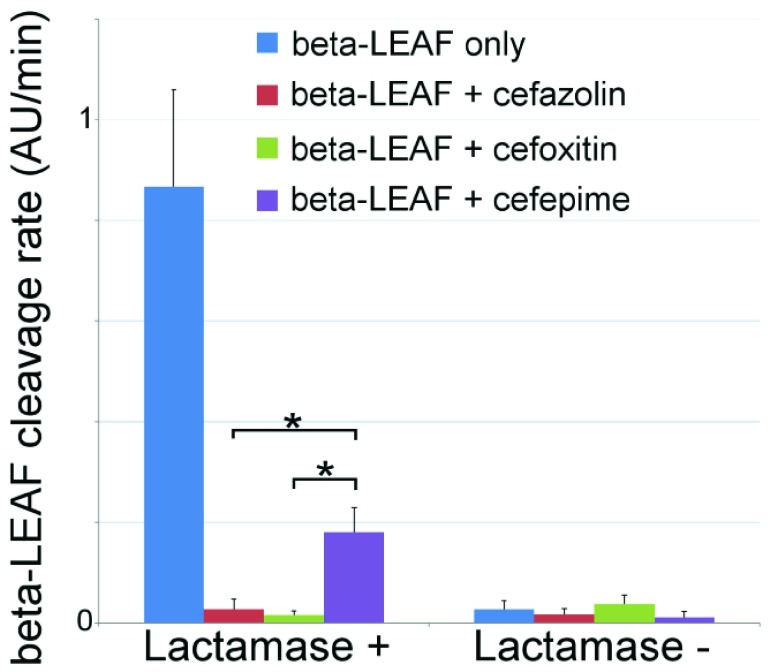
To validate the ability of the microfluidic to characterize 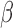 -lactamase derived antibiotic susceptibility, pathogen-spiked PBS samples were tested against a range of representative 1st, 2nd and 4th generation cephalosporin antibiotics (cefazolin, cefoxitin and cefepime respectively; Fig. 8). The rate of
-lactamase derived antibiotic susceptibility, pathogen-spiked PBS samples were tested against a range of representative 1st, 2nd and 4th generation cephalosporin antibiotics (cefazolin, cefoxitin and cefepime respectively; Fig. 8). The rate of 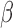 -LEAF cleavage was found to decrease dramatically with the addition of a cephalosporin, indicating the
-LEAF cleavage was found to decrease dramatically with the addition of a cephalosporin, indicating the 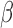 -lactamase was preferentially cleaving the antibiotics over the
-lactamase was preferentially cleaving the antibiotics over the 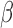 -LEAF probe. However, the rate of cleavage was not as pronounced with cefepime, indicating this antibiotic as being more robust against cleavage for this particular
-LEAF probe. However, the rate of cleavage was not as pronounced with cefepime, indicating this antibiotic as being more robust against cleavage for this particular 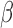 -lactamase (as compared to the other cephalosporins). Gold standard assays of antibiotic susceptibility have confirmed cefepime to be the most effective antibiotic out of the 3 for this pathogen strain [30], supporting the use of the microfluidic as a rapid platform for the
-lactamase (as compared to the other cephalosporins). Gold standard assays of antibiotic susceptibility have confirmed cefepime to be the most effective antibiotic out of the 3 for this pathogen strain [30], supporting the use of the microfluidic as a rapid platform for the 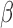 -LEAF antibiotic susceptibility assay.
-LEAF antibiotic susceptibility assay.
FIGURE 8.
Microfluidic 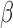 -LEAF antibiotic susceptibility assay. 1e8 CFU S. aureus in PBS was injected into the device. Cleavage rate was measured over a 45 minute incubation. Average and std. error from 5 independent experiments on
-LEAF antibiotic susceptibility assay. 1e8 CFU S. aureus in PBS was injected into the device. Cleavage rate was measured over a 45 minute incubation. Average and std. error from 5 independent experiments on 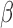 -lactamase positive and negative strains of S. aureus. * indicates
-lactamase positive and negative strains of S. aureus. * indicates 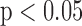 (ANOVA with 2-tailed t-test).
(ANOVA with 2-tailed t-test).
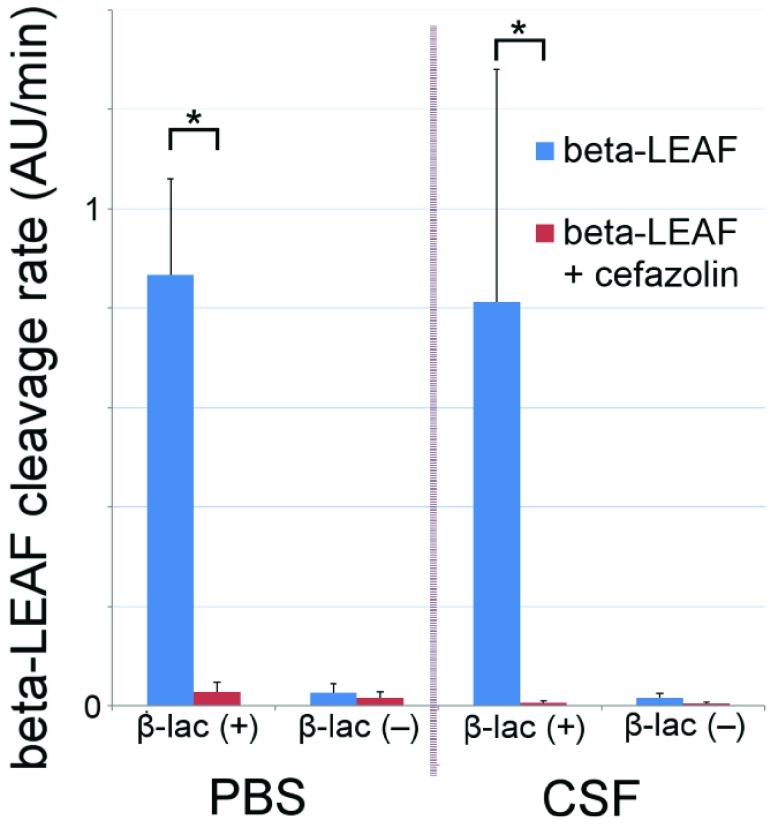
The ability of the 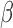 -LEAF assay to be used on direct patient samples was tested on simulated meningitis samples. S. aureus was spiked into bovine CSF and tested with the microfluidic protocol as before. These results were compared with pathogen spiked PBS samples to investigate whether the use of a human specimen could influence the results of the assay. As shown in Fig. 9, no significant difference between the two samples was found, indicating this protocol is suitable for point of care use with suspected meningitis cases.
-LEAF assay to be used on direct patient samples was tested on simulated meningitis samples. S. aureus was spiked into bovine CSF and tested with the microfluidic protocol as before. These results were compared with pathogen spiked PBS samples to investigate whether the use of a human specimen could influence the results of the assay. As shown in Fig. 9, no significant difference between the two samples was found, indicating this protocol is suitable for point of care use with suspected meningitis cases.
FIGURE 9.
Validation of microfluidic 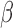 -LEAF assay with pathogen-spiked CSF. 1e8 CFU S. aureus in either CSF or PBS were injected into the microfluidic device. PBS data was taken with 5 independent experiments; CSF data was taken with 3 experiments. Cleavage rate was measured over 20 minutes. Error bars are standard error. * indicates
-LEAF assay with pathogen-spiked CSF. 1e8 CFU S. aureus in either CSF or PBS were injected into the microfluidic device. PBS data was taken with 5 independent experiments; CSF data was taken with 3 experiments. Cleavage rate was measured over 20 minutes. Error bars are standard error. * indicates 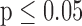 (Mann-Whitney).
(Mann-Whitney).
IV. Discussion
Treatment guidance for the febrile infant lacking any other distinguishing symptoms remains a difficult situation. Due to the serious consequences of a missed diagnosis, treatment tends to be very conservative. However, this push is tempered by the need of the medical community to conserve the use of its broad-spectrum antibiotics, which are gradually being rendered ineffective by growing antibiotic resistance. Thus the most widely effective antibiotics cannot be used as an empiric treatment, as doing so will jeopardize our ability to treat resistant organisms in the future. Indeed, a number of pathogens with no effective treatment have been documented in medical facilities around the world [33] due to the growing use of our remaining, broadly-effective antibiotics. The problem is most SBI can be treated with common antibiotics (e.g., ampicillin, cefazolin); however a small fraction will be resistant. Because of this, neonates with suspected SBI are often kept at the hospital for 24 hours after empiric treatment to ensure their condition has stabilized. This policy, while necessary, has been shown to induce psychological trauma and has been correlated with long-term behavioral disturbance [10]. These factors put increasing pressure on the clinicians to use a broad-spectrum antibiotic more widely, hastening the development of resistance.
Adding to the confusion is the poor tracking of antibiotic resistance generally. 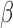 -lactamase is often not tested for in microbiological workups, as the information comes too late (2-3 days or longer) to be of practical use (though this conclusion is not unanimous [34]). This general lack of testing has contributed to the problem—it is difficult to allocate resources to a problem when the scope of the problem itself is poorly understood. This is an even greater quandary for low-resource areas, where standard microbiological laboratories are not available.
-lactamase is often not tested for in microbiological workups, as the information comes too late (2-3 days or longer) to be of practical use (though this conclusion is not unanimous [34]). This general lack of testing has contributed to the problem—it is difficult to allocate resources to a problem when the scope of the problem itself is poorly understood. This is an even greater quandary for low-resource areas, where standard microbiological laboratories are not available.
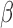 -LEAF was developed to help address these issues. Key factors necessary for its translation in these areas include sensitivity, cost and simplicity (i.e., ease of clinical adoption). Starting with sensitivity, prior studies of meningitis have found a large range of pathogen concentrations in the CSF of patients taken at the first clinical examination. However, the cases at most risk of treatment failure have CSF pathogen concentrations [35] greater than 1e7 CFU/ml, with the risk increasing with pathogen concentration [36]. Another study [37] has found 21% of CSF culture positive samples to have concentrations >5e7 CFU/ml. For positive urine cultures, the pathogen concentrations are even higher [38], with
-LEAF was developed to help address these issues. Key factors necessary for its translation in these areas include sensitivity, cost and simplicity (i.e., ease of clinical adoption). Starting with sensitivity, prior studies of meningitis have found a large range of pathogen concentrations in the CSF of patients taken at the first clinical examination. However, the cases at most risk of treatment failure have CSF pathogen concentrations [35] greater than 1e7 CFU/ml, with the risk increasing with pathogen concentration [36]. Another study [37] has found 21% of CSF culture positive samples to have concentrations >5e7 CFU/ml. For positive urine cultures, the pathogen concentrations are even higher [38], with 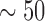 % of specimens having a mean colony count of greater than 5e7 CFU/ml. Thus this newly developed
% of specimens having a mean colony count of greater than 5e7 CFU/ml. Thus this newly developed 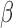 -LEAF microfluidic assay, with its LOD of 5e7 CFU/ml, is well positioned to rapidly detect urinary tract infections and meningitis for those patients most in need in of rapid diagnosis. We note the actual LOD of the microfluidic is better than 5e7 CFU/ml, and the sensitivity can be brought down to below 1e7 CFU/ml by extending the incubation time beyond 20 minutes. The practical resolution of this assay will ultimately depend on the background drift of the measurement, particularly in the context of the many different
-LEAF microfluidic assay, with its LOD of 5e7 CFU/ml, is well positioned to rapidly detect urinary tract infections and meningitis for those patients most in need in of rapid diagnosis. We note the actual LOD of the microfluidic is better than 5e7 CFU/ml, and the sensitivity can be brought down to below 1e7 CFU/ml by extending the incubation time beyond 20 minutes. The practical resolution of this assay will ultimately depend on the background drift of the measurement, particularly in the context of the many different 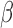 -lactamase enzymes, which may have different catalytic rates against the
-lactamase enzymes, which may have different catalytic rates against the 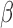 -LEAF probe. Beyond the LOD, the ability of this protocol to assess resistance at high bacterial concentrations may be of use in mitigating the “inoculum effect” which can confound conventional tests of antibiotic susceptibility [30].
-LEAF probe. Beyond the LOD, the ability of this protocol to assess resistance at high bacterial concentrations may be of use in mitigating the “inoculum effect” which can confound conventional tests of antibiotic susceptibility [30].
The material cost of the microfluidic assay was considered from the earliest stages of design. The polymethyl methacrylate was chosen due to its widespread availability, permissive fabrication characteristics and price (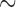 $0.02/test). For the pathogen trapping, different materials were investigated, and nuclear track etched polycarbonate filters were found to have the best combination of autofluorescence, consistent performance, flow rates and cost ($0.04/test). The price of these filters would drop if purchased in bulk and could be reduced even further by shrinking the size of the pathogen capture chamber, which is the subject of ongoing investigation. The adhesive used to assemble the system was a trivial cost in terms of the overall system (less than $0.01/test). Though disposable use seems the most practical implementation for most environments, for extremely low-resource settings, it is possible to clean and reuse the microfluidic device, thereby further reducing the expense. In this spirit, fluorescence measurements itself can be made with low cost components (LED, optical filters, silicon photosensor) for less than several dollars, while the vacuum necessary for fluid flow can be generated using a simple syringe. A secondary factor is the cost-savings which could be incurred by the use of this diagnostic for cephalosporin-resistant pathogens in hospital care facilities. The increasing prevalence of these pathogens has led to an increase in the use of carbapenem antibiotics (e.g., imipenem) as a precautionary measure, but this practice has led to outbreaks of multidrug resistant disease [39], with enormous cost to patient health as well as financial cost. Hospital infection control committees have been increasingly strict with prescription practices in an attempt to avoid this scenario, but the physicians are caught in the middle, forced to choose between providing best care to the current patient versus trying to prevent a resistant infection in the future without diagnostic guidance [40]. The microfluidic
$0.02/test). For the pathogen trapping, different materials were investigated, and nuclear track etched polycarbonate filters were found to have the best combination of autofluorescence, consistent performance, flow rates and cost ($0.04/test). The price of these filters would drop if purchased in bulk and could be reduced even further by shrinking the size of the pathogen capture chamber, which is the subject of ongoing investigation. The adhesive used to assemble the system was a trivial cost in terms of the overall system (less than $0.01/test). Though disposable use seems the most practical implementation for most environments, for extremely low-resource settings, it is possible to clean and reuse the microfluidic device, thereby further reducing the expense. In this spirit, fluorescence measurements itself can be made with low cost components (LED, optical filters, silicon photosensor) for less than several dollars, while the vacuum necessary for fluid flow can be generated using a simple syringe. A secondary factor is the cost-savings which could be incurred by the use of this diagnostic for cephalosporin-resistant pathogens in hospital care facilities. The increasing prevalence of these pathogens has led to an increase in the use of carbapenem antibiotics (e.g., imipenem) as a precautionary measure, but this practice has led to outbreaks of multidrug resistant disease [39], with enormous cost to patient health as well as financial cost. Hospital infection control committees have been increasingly strict with prescription practices in an attempt to avoid this scenario, but the physicians are caught in the middle, forced to choose between providing best care to the current patient versus trying to prevent a resistant infection in the future without diagnostic guidance [40]. The microfluidic 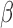 -LEAF assay is in a position to provide more clarity to this situation by providing individualized treatment information at the point of care.
-LEAF assay is in a position to provide more clarity to this situation by providing individualized treatment information at the point of care.
The adoption of a new diagnostic depends critically on the simplicity and robustness of its operation. An important aspect of the assay is its simple workflow, which provides several binary outputs for facile clinical interpretation (Fig. 10). These outputs can be programmed into the fluorimeter, reducing clinician workload and the potential for human error. Furthermore, the assay can be adapted for portable handheld readers, allowing use in smaller clinics and low-income settings. The 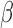 -LEAF probe itself is exceptionally stable, with only bacterial
-LEAF probe itself is exceptionally stable, with only bacterial 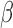 -lactamase enzymes able to degrade the probe with any significant efficiency. This ensures the assay is free from biological interference. In addition, the room temperature shelf life of the
-lactamase enzymes able to degrade the probe with any significant efficiency. This ensures the assay is free from biological interference. In addition, the room temperature shelf life of the 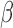 -LEAF probe is more than 1 year. One potential application for this assay is depicted in schematic form in Fig. 11, though numerous other adult applications exist as well (e.g., suspected sepsis, meningitis, UTI, infected pleural cavity).
-LEAF probe is more than 1 year. One potential application for this assay is depicted in schematic form in Fig. 11, though numerous other adult applications exist as well (e.g., suspected sepsis, meningitis, UTI, infected pleural cavity).
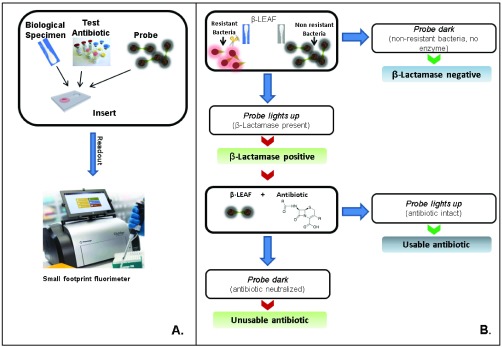
FIGURE 10.
Clinical interpretation flowchart. (a) A simple assay setup is depicted. Multiple reaction can be setup simultaneously allowing several antibiotics to be tested concurrently. A representative small footprint fluorimeter from Promega France (GloMax Discover) is shown as an example, though smaller handheld fluorimeters could also be used in principle. (b) Assay provides simple yes/no outputs as to whether 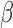 -lactamase is produced and whether an antibiotic would be stable against cleavage. These outputs can be programmed into commercially available portable fluorimeters.
-lactamase is produced and whether an antibiotic would be stable against cleavage. These outputs can be programmed into commercially available portable fluorimeters.
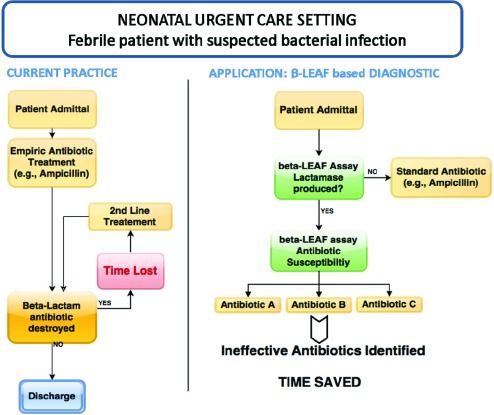
FIGURE 11.
Application of microfluidic 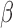 -LEAF assay. One scenario for use is presented, comparing the current practice with potential future application.
-LEAF assay. One scenario for use is presented, comparing the current practice with potential future application.
V. Conclusion
Before the advent of 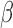 -lactam antibiotics, the prognosis for SBI was particularly dismal, as the window for effective treatment was brief. With the success of penicillins and cephalosporins as first-line empiric treatments, more sophisticated analyses of antibiotic susceptibility could be performed during the critical initial disease stages, allowing more effective narrow-spectrum antibiotics to be identified and prescribed. The growth of
-lactam antibiotics, the prognosis for SBI was particularly dismal, as the window for effective treatment was brief. With the success of penicillins and cephalosporins as first-line empiric treatments, more sophisticated analyses of antibiotic susceptibility could be performed during the critical initial disease stages, allowing more effective narrow-spectrum antibiotics to be identified and prescribed. The growth of 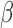 -lactamase resistance has threatened this paradigm, requiring the advent of new technologies. Characterization of pathogens at the point of care has the potential to both contain the growth of antibiotic resistance and improve patient outcomes by providing immediate treatment guidance. The assay described here is unique in that it provides phenotypic information and treatment guidance for an enzymatic resistance mechanism in the time frame of a clinical examination. Furthermore, the low cost and facile interpretation of this assay overcome two of the main obstacles for clinical adoption. By optimizing the fluorescent detection with a custom built optical train (instead of a commercial plate reader), even greater improvements in the LOD, cost-effectiveness and overall footprint are possible. In the future, the microfluidic technology can be easily adapted to other fluorescent pathogen assays (e.g., gram status). This ability to provide personalized treatment guidance is promising and deserves further development as well as validation with direct clinical specimens.
-lactamase resistance has threatened this paradigm, requiring the advent of new technologies. Characterization of pathogens at the point of care has the potential to both contain the growth of antibiotic resistance and improve patient outcomes by providing immediate treatment guidance. The assay described here is unique in that it provides phenotypic information and treatment guidance for an enzymatic resistance mechanism in the time frame of a clinical examination. Furthermore, the low cost and facile interpretation of this assay overcome two of the main obstacles for clinical adoption. By optimizing the fluorescent detection with a custom built optical train (instead of a commercial plate reader), even greater improvements in the LOD, cost-effectiveness and overall footprint are possible. In the future, the microfluidic technology can be easily adapted to other fluorescent pathogen assays (e.g., gram status). This ability to provide personalized treatment guidance is promising and deserves further development as well as validation with direct clinical specimens.
Acknowledgment
A. Palanisami thanks Emma Briars and Dr. Girgis Obaid for helpful discussions.
Biographies

Akilan Palanisami received the B.Eng. degree in engineering physics and the M.Eng. degree in applied physics from Cornell University in 1997 and 1998, respectively, and the Ph.D. degree in physics from the University of Illinois at Urbana–Champaign in 2005. He is currently a Research Scientist with the Harvard Medical School, Massachusetts General Hospital. His current projects include hyperspectral microendoscopy of peritoneal cancers and point of care diagnostic testing.

Shazia Khan received the bachelor’s degree (Hons.) in zoology and the master’s degree in biomedical sciences from the University of Delhi, New Delhi, India, in 2003 and 2005, respectively, and the Ph.D. degree in biochemistry from the National Institute of Immunology, New Delhi, in 2011, with a focus on signal transduction in mycobacterium. She was a Post-Doctoral Research Fellow with Massachusetts General Hospital and Harvard Medical School, Boston, where she was involved in development of diagnostic for rapid determination of bacterial resistance and antibiotic susceptibility and on 3-D models of cancer biology. She also received internships to gain understanding in the areas of intellectual property and biotech start-ups/innovation. She is currently a Scientific Editor for Life Sciences Journals, Elsevier (Cambridge, MA, USA, Elsevier).

Sultan Sibel Erdem received the B.S. degree in chemical education from Marmara University, Istanbul, Turkey, in 2001 and the Ph.D. degree in biorganic chemistry from Louisiana State University, in 2009. She was a Post-Doctoral Research Fellow with Massachusetts General Hospital and the Harvard Medical School, Center for Systems Biology, and the Wellman Center for Photomedicine. She is currently an Assistant Professor with the International School of Medicine, Istanbul Medipol University. Her research interests are combination therapies, synthesis and targeted delivery of photosensitizers for photodynamic therapy of cancer, and other diseases.

Tayyaba Hasan received the Ph.D. degree in organic chemistry from the University of Arkansas. She is currently a Professor of Dermatology with the Harvard Medical School and a Professor of Health Sciences and Technology. She is currently with the Wellman Center for Photomedicine. She was involved in the post-doctoral training in biochemistry with the University of Pennsylvania. She joined the Harvard Medical School, Massachusetts General Hospital in 1982 as a Research Scientist. Her research is directed to basic and translational studies in photochemistry, photobiology, and photodynamic therapy. More broadly, her research program includes investigations in cancer, microbiology, infectious diseases, arthritis, and cardiovascular pathologies.
Funding Statement
This work was supported in part by the National Institute of Health/National Institute of Biomedical Imaging and Bioengineering and Point of Care Technology in Primary Care through the Center for Integration of Medicine and Innovative Technology under Grant U54 EB015408, and the U.S. Department of Defense/Air Force Office of Research under Grant FA9550-11-1-0331.
References
- [1].Bachur R. G. and Harper M. B., “Predictive model for serious bacterial infections among infants younger than 3 months of age,” Pediatrics, vol. 108, no. , pp. 311–316, Aug. 2001. [DOI] [PubMed] [Google Scholar]
- [2].Schwartz S., Raveh D., Toker O., Segal G., Godovitch N., and Schlesinger Y., “A week-by-week analysis of the low-risk criteria for serious bacterial infection in febrile neonates,” Arch. Disease Childhood, vol. 94, pp. 287–292, Apr. 2009. [DOI] [PubMed] [Google Scholar]
- [3].ACEP Clinical Policies Committee, “Clinical policy for children younger than three years presenting to the emergency department with fever,” Ann. Emerg. Med., vol. 42, no. 3, pp. 530–545, 2003. [DOI] [PubMed] [Google Scholar]
- [4].Greenhow T. L., Hung Y. Y., and Herz A. M., “Changing epidemiology of bacteremia in infants aged 1 week to 3 months,” Pediatrics, vol. 129, no. 3, pp. e590–e596, Mar. 2012. [DOI] [PubMed] [Google Scholar]
- [5].Greenhow T. L., Hung Y.-Y., Herz A. M., Losada E., and Pantell R. H., “The changing epidemiology of serious bacterial infections in young infants,” Pediatric Infectious Disease J., vol. 33, no. 6, pp. 595–599, Jun. 2014. [DOI] [PubMed] [Google Scholar]
- [6].Aronson P. L., et al. , “Variation in care of the febrile young infant <90 days in US pediatric emergency departments,” Pediatrics, vol. 134, no. 4, pp. 667–677, Oct. 2014. [DOI] [PubMed] [Google Scholar]
- [7].de Louvois J., Halket S., and Harvey D., “Neonatal meningitis in England and Wales: Sequelae at 5 years of age,” Eur. J. Pediatrics, vol. 164, no. 12, pp. 730–734, Dec. 2005. [DOI] [PubMed] [Google Scholar]
- [8].Shaikh N., et al. , “Identification of children and adolescents at risk for renal scarring after a first urinary tract infection: A meta-analysis with individual patient data,” JAMA Pediatrics, vol. 168, no. 10, pp. 893–900, 2014. [DOI] [PubMed] [Google Scholar]
- [9].DeAngelis C., Joffe A., Wilson M., and Willis E., “Iatrogenic risks and financial costs of hospitalizing febrile infants,” Amer. J. Diseases Children, vol. 137, no. 12, pp. 1146–1149, 1983. [DOI] [PubMed] [Google Scholar]
- [10].Douglas W. B., “Early hospital admissions and later disturbances of behaviour and learning,” Develop. Med. Child Neurol., vol. 17, no. 4, pp. 456–480, Aug. 1975. [DOI] [PubMed] [Google Scholar]
- [11].Quinton D. and Rutter M., “Early hospital admissions and later disturbances of behaviour: An attempted replication of Douglas’ findings,” Develop. Med. Child Neurol., vol. 18, no. 4, pp. 447–459, Aug. 1976. [DOI] [PubMed] [Google Scholar]
- [12].Harper M. B., “Update on the management of the febrile infant,” Clin. Pediatric Emergency Med., vol. 5, no. 1, pp. 5–12, Mar. 2004. [Google Scholar]
- [13].Watt K., Waddle E., and Jhaveri R., “Changing epidemiology of serious bacterial infections in febrile infants without localizing signs,” PLoS One, vol. 5, no. 8, p. e12448, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Byington C. L., et al. , “Serious bacterial infections in febrile infants younger than 90 days of age: The importance of ampicillin-resistant pathogens,” Pediatrics, vol. 111, no. 5, pp. 964–968, 2003. [DOI] [PubMed] [Google Scholar]
- [15].Yager P., Domingo G. J., and Gerdes J., “Point-of-care diagnostics for global health,” Annu. Rev. Biomed. Eng., vol. 10, pp. 107–144, Aug. 2008. [DOI] [PubMed] [Google Scholar]
-
[16].Sparbier K., Schubert S., Weller U., Boogen C., and Kostrzewa M., “Matrix-assisted laser desorption ionization–time of flight mass spectrometry-based functional assay for rapid detection of resistance against
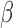 -lactam antibiotics,” J. Clin. Microbiol., vol. 50, no. 3, pp. 927–937, Mar.
2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
-lactam antibiotics,” J. Clin. Microbiol., vol. 50, no. 3, pp. 927–937, Mar.
2012. [DOI] [PMC free article] [PubMed] [Google Scholar] -
[17].Woodford N., “Rapid characterization of
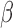 -lactamases by multiplex PCR,” in Antibiotic Resistance Protocols. London, U.K.: Springer, 2010, pp. 181–192. [Google Scholar]
-lactamases by multiplex PCR,” in Antibiotic Resistance Protocols. London, U.K.: Springer, 2010, pp. 181–192. [Google Scholar] - [18].Hague R., “What is the threat from extended spectrum beta-lactamase-producing organisms in children?” Arch. Disease Childhood, vol. 96, pp. 325–327, Apr. 2011. [DOI] [PubMed] [Google Scholar]
- [19].Al-Taiar A., et al. , “Neonatal infections in China, Malaysia, Hong Kong and Thailand,” Arch. Disease Childhood-Fetal Neonatal Ed., vol. 98, no. 3, pp. F249–F255, May 2013. [DOI] [PubMed] [Google Scholar]
-
[20].Blaschke A. J., Korgenski E. K., Daly J. A., LaFleur B., Pavia A. T., and Byington C. L., “Extended-spectrum
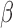 -lactamase-producing pathogens in a children’s hospital: A 5-year experience,” Amer. J. Infectious Control, vol. 37, no. 6, pp. 435–441, Aug.
2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
-lactamase-producing pathogens in a children’s hospital: A 5-year experience,” Amer. J. Infectious Control, vol. 37, no. 6, pp. 435–441, Aug.
2009. [DOI] [PMC free article] [PubMed] [Google Scholar] - [21].Birgy A., et al. , “Characterization of extended-spectrum-beta-lactamase-producing Escherichia coli strains involved in maternal-fetal colonization: Prevalence of E. coli ST131,” J. Clin. Microbiol., vol. 51, no. 6, pp. 1727–1732, Jun. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
[22].Logan L. K., Braykov N. P., Weinstein R. A., and Laxminarayan R., “Extended-spectrum
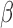 -lactamase–producing and third-generation cephalosporin-resistant enterobacteriaceae in children: Trends in the United States, 1999–2011,” J. Pediatric Infectious Diseases Soc., vol. 3, no. 4, pp. 320–328, 2014. [DOI] [PubMed] [Google Scholar]
-lactamase–producing and third-generation cephalosporin-resistant enterobacteriaceae in children: Trends in the United States, 1999–2011,” J. Pediatric Infectious Diseases Soc., vol. 3, no. 4, pp. 320–328, 2014. [DOI] [PubMed] [Google Scholar] -
[23].Woods K. L., et al. , “Community-associated Escherichia coli harboring CTX-M
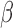 -lactamases from urine cultures from pediatric patients,” Antimicrobial Agents Chemotherapy, vol. 56, no. 4, pp. 2209–2210, Apr.
2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
-lactamases from urine cultures from pediatric patients,” Antimicrobial Agents Chemotherapy, vol. 56, no. 4, pp. 2209–2210, Apr.
2012. [DOI] [PMC free article] [PubMed] [Google Scholar] -
[24].Moissenet D., et al. , “Meningitis caused by Escherichia coli producing TEM-52 extended-spectrum
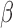 -lactamase within an extensive outbreak in a neonatal ward: Epidemiological investigation and characterization of the strain,” J. Clin. Microbiol., vol. 48, no. 7, pp. 2459–2463, Jul.
2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
-lactamase within an extensive outbreak in a neonatal ward: Epidemiological investigation and characterization of the strain,” J. Clin. Microbiol., vol. 48, no. 7, pp. 2459–2463, Jul.
2010. [DOI] [PMC free article] [PubMed] [Google Scholar] - [25].Boyer-Mariotte S., Duboc P., Bonacorsi S., Lemeland J. F., Bingen E., and Pinquier D., “CTX-M-15-producing Escherichia coli in fatal neonatal meningitis: Failure of empirical chemotherapy,” J. Antimicrobial Chemotherapy, vol. 62, no. 6, pp. 1472–1474, Dec. 2008. [DOI] [PubMed] [Google Scholar]
- [26].Woodford N., et al. , “Arrival of Klebsiella pneumoniae producing KPC carbapenemase in the United Kingdom,” J. Antimicrobial Chemotherapy, vol. 62, no. 6, pp. 1261–1264, Dec. 2008. [DOI] [PubMed] [Google Scholar]
- [27].Livermore D. M., “Current epidemiology and growing resistance of gram-negative pathogens,” Korean J. Internal Med., vol. 27, no. 2, pp. 128–142, May 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zheng X., Sallum U. W., Verma S., Athar H., Evans C. L., and Hasan T., “Exploiting a bacterial drug-resistance mechanism: A light-activated construct for the destruction of MRSA,” Angew. Chem., vol. 48, no. 12, pp. 2148–2151, 2009. [DOI] [PubMed] [Google Scholar]
-
[29].Sallum U. W., Zheng X., Verma S., and Hasan T., “Rapid functional definition of extended spectrum
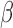 -lactamase activity in bacterial cultures via competitive inhibition of fluorescent substrate cleavage,” Photochem. Photobiol., vol. 86, no. 6, pp. 1267–1271, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
-lactamase activity in bacterial cultures via competitive inhibition of fluorescent substrate cleavage,” Photochem. Photobiol., vol. 86, no. 6, pp. 1267–1271, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar] -
[30].Khan S., Sallum U. W., Zheng X., Nau G. J., and Hasan T., “Rapid optical determination of
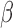 -lactamase and antibiotic activity,” BMC Microbiol., vol. 14, p. 84, Apr.
2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
-lactamase and antibiotic activity,” BMC Microbiol., vol. 14, p. 84, Apr.
2014. [DOI] [PMC free article] [PubMed] [Google Scholar] - [31].Khan S. and Hasan T., “Functional targeting of bacteria: A multimodal construct for PDT and diagnostics of drug-resistant bacteria,” in Photodynamic Therapy. Berlin, Germany: Springer, 2014, pp. 237–253. [Google Scholar]
-
[32].Erdem S. S., Khan S., Palanisami A., and Hasan T., “Rapid, low-cost fluorescent assay of
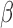 -lactamase-derived antibiotic resistance and related antibiotic susceptibility,” J. Biomed. Opt., vol. 19, no. 10, p. 105007, Oct.
2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
-lactamase-derived antibiotic resistance and related antibiotic susceptibility,” J. Biomed. Opt., vol. 19, no. 10, p. 105007, Oct.
2014. [DOI] [PMC free article] [PubMed] [Google Scholar] - [33].Laxminarayan R., et al. , “Antibiotic resistance—The need for global solutions,” Lancet Infectious Diseases, vol. 13, no. 12, pp. 1057–1098, Dec. 2013. [DOI] [PubMed] [Google Scholar]
- [34].Livermore D. M., et al. , “Are susceptibility tests enough, or should laboratories still seek ESBLs and carbapenemases directly?” J. Antimicrobial Chemotherapy, vol. 67, pp. 1569–1577, Jul. 2012. [DOI] [PubMed] [Google Scholar]
- [35].Bingen E., et al. , “Bacterial counts in cerebrospinal fluid of children with meningitis,” Eur. J. Clin. Microbiol. Infectious Diseases, vol. 9, no. 4, pp. 278–281, Apr. 1990. [DOI] [PubMed] [Google Scholar]
- [36].Feldman W. E., “Relation of concentrations of bacteria and bacterial antigen in cerebrospinal fluid to prognosis in patients with bacterial meningitis,” New England J. Med., vol. 296, pp. 433–435, Feb. 1977. [DOI] [PubMed] [Google Scholar]
- [37].Feldman W. E., “Concentrations of bacteria in cerebrospinal fluid of patients with bacterial meningitis,” J. Pediatrics, vol. 88, no. 4, pp. 549–552, Apr. 1976. [DOI] [PubMed] [Google Scholar]
- [38].Coulthard M. G., Kalra M., Lambert H. J., Nelson A., Smith T., and Perry J. D., “Redefining urinary tract infections by bacterial colony counts,” Pediatrics, vol. 125, no. 2, pp. 335–341, Feb. 2010. [DOI] [PubMed] [Google Scholar]
- [39].Diekema D. J. and Pfaller M. A., “Rapid detection of antibiotic resistant organism carriage for infection prevention,” Clin. Infectious Diseases, vol. 56, pp. 1614–1620, Jun. 2013. [DOI] [PubMed] [Google Scholar]
- [40].Manikal V. M., Landman D., Saurina G., Oydna E., Lal H., and Quale J., “Endemic carbapenem-resistant acinetobacter species in Brooklyn, New York: Citywide prevalence, interinstitutional spread, and relation to antibiotic usage,” Clin. Infectious Diseases, vol. 31, no. 1, pp. 101–106, Jul. 2000. [DOI] [PubMed] [Google Scholar]