Abstract
Eukaryotic mRNAs contain a 5' cap structure critical for recruitment of the translation machinery and initiation of protein synthesis. mRNA recognition is thought to require direct interactions between eukaryotic initiation factor 4E (eIF4E) and the mRNA cap. However, translation of numerous capped mRNAs remains robust during cellular stress, early development, and cell cycle progression1 despite eIF4E inactivation. Here we describe a new cellular cap-dependent pathway of translation initiation that relies on a previously unknown cap-binding activity of eIF3d, a subunit of the 800-kilodalton eukaryotic initiation factor 3 (eIF3) complex. A 1.4 Å crystal structure of the eIF3d cap-binding domain reveals unexpected homology to endonucleases involved in RNA turnover, and allows modeling of cap recognition by eIF3d. eIF3d makes specific contacts to the cap, as exemplified by cap analog competition, and these interactions are essential for assembly of translation initiation complexes on eIF3-specialized mRNAs2 such as the cell proliferation regulator c-Jun. The c-Jun mRNA further encodes an inhibitory RNA element that blocks eIF4E recruitment, thus enforcing alternative cap recognition by eIF3d. Our results reveal a new mechanism of cap-dependent translation independent of eIF4E, and illustrate how modular RNA elements work in concert to direct specialized forms of translation initiation.
The rate-limiting step of translation initiation is the recognition of the 5' cap structure by eIF4E3,4. eIF4E activity is highly regulated by extracellular stimuli, predominantly through steric hindrance by the eIF4E-binding proteins (4E-BPs)5,6. The translational efficiencies of mRNAs range in sensitivity to 4E-BP inhibition7-9, and these differences have traditionally been addressed by categorizing translation into cap-dependent versus cap-independent pathways10. However, the mechanisms underlying mRNA sensitivity to active eIF4E levels remain enigmatic as all cellular mRNAs maintain the same 5' cap structure11.
Recently we discovered a new translation pathway driven by RNA interactions with eIF3 that is utilized by a subset of cell proliferation mRNAs, with the prototype member being the mRNA encoding the early response transcription factor c-Jun2. eIF3-specialized translation is cap-dependent and requires recruitment of eIF3 to an internal stem loop structure in the 5' untranslated region (UTR). However, the translational efficiency of a subset of these mRNAs is unaffected by eIF4E inactivation7-9, suggesting that cap recognition may proceed by a non-canonical mechanism (Supplementary Table 1).
To understand how cap recognition occurs during eIF3-specialized translation, we examined whether c-Jun mRNA utilizes the canonical eIF4F cap-binding complex during initiation. We programmed in vitro translation extracts from human 293T cells with capped and polyadenylated c-Jun mRNA, and isolated the 48S complex to assess the presence of the eIF4F factors (eIF4G1, eIF4A1, eIF4E) (Fig. 1a,b). Unexpectedly, although c-Jun mRNA translation initiation complexes contain eIF3 and the small ribosomal subunit, they are depleted of all eIF4F components. In contrast, eIF4F is readily detectable in 48S initiation complexes formed on a canonical eIF4E-dependent mRNA, ACTB (Fig. 1b)12. In agreement with the absence of eIF4F, c-Jun levels are unaffected by cell treatment with the mTOR inhibitor INK1287, which inactivates eIF4E, and eIF4A inhibitors13 (Extended Data Fig. 1). These results indicate that c-Jun mRNA translation occurs independently of eIF4F and that the process of eIF3-specialized translation is fundamentally distinct at the initial stage of 5' cap recognition.
Figure 1. 5' end recognition of c-Jun mRNA is eIF4F-independent.
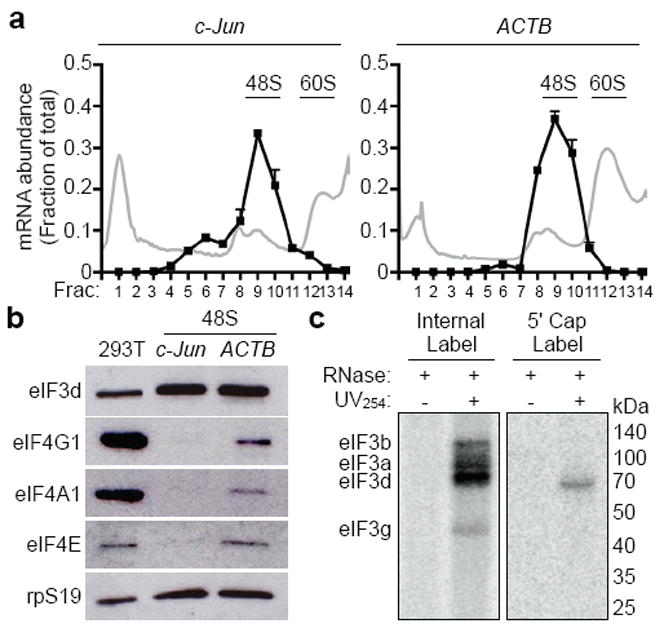
a, Distribution of c-Jun or ACTB mRNA-containing initiation complexes in programmed 293T cell in vitro translation extracts. The mRNA abundance (black line) is expressed as the fraction of total recovered transcripts. The results are given as the mean ± standard deviation (s.d.) of a representative quantitative RT-PCR experiment performed in duplicate. The polysome profile (gray line) is plotted as relative absorbance at 254 nm versus elution fractions. b, Western blot analysis of initiation factors in 48S translation complexes formed on c-Jun and ACTB mRNAs. 293T, total protein from 293T in vitro translation extracts. For gel source data, see Supplementary Figure 1. c, Phosphorimage of SDS gel resolving RNase-protected 32P-internal or 32P-cap-labeled c-Jun 5' UTR RNA crosslinked to eIF3 subunits. Recombinant eIF3a migrates at ~100 kDa due to a C-terminal truncation26. The results of a-c are representative of three independent experiments.
eIF3-specialized translation requires recognition of an internal RNA stem loop for efficient translation2. Therefore, we asked if eIF3 might also be involved in 5' cap recognition. In agreement with the previously demonstrated RNA-binding capability of eIF3, the four eIF3 RNA-binding subunits, eIF3a, b, d, and g, provide RNase protection to internally 32P-labeled c-Jun 5' UTR RNA upon UV254-induced crosslinking (Fig. 1c)2. In contrast, when the 32P label is placed in the 5' cap of c-Jun mRNA, RNase protection is observed with a single subunit of eIF3, corresponding to eIF3d (Fig. 1c, Extended Data Fig. 2a). We confirmed subunit identity by limited proteolysis and mass spectrometry, and defined a C-terminal region of eIF3d that is responsible for protection of the 5' mRNA terminus (Extended Data Fig. 2). The mapped C-terminal region of eIF3d is broadly conserved throughout plant, fungal and animal phylogeny (Fig. 2a, Extended Data Fig. 3), suggesting the apparent 5' end recognition activity of eIF3d is an evolutionarily preserved function of the eIF3 complex.
Figure 2. Structure of eIF3d reveals a conserved cap-binding domain.
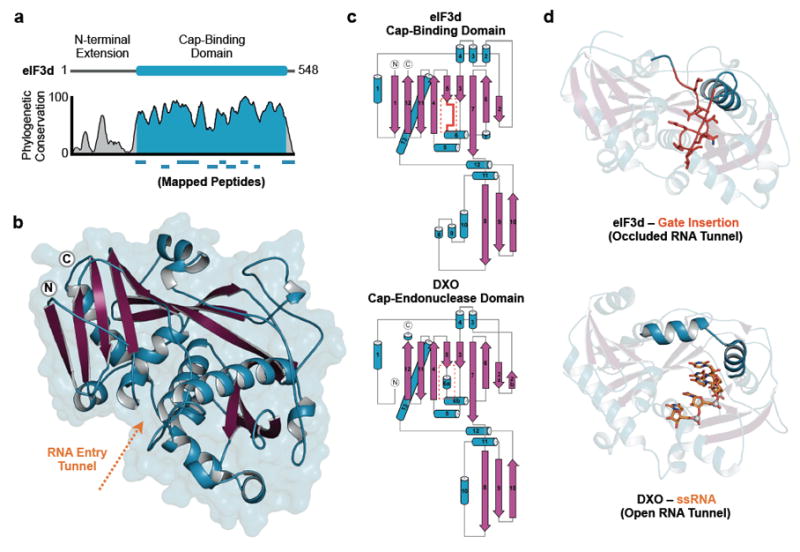
a, Cartoon schematic and phylogenetic conservation of eIF3d amino acid sequence according to physiochemical property similarity. Peptides in the cap-binding domain as identified by limited proteolysis are mapped below. b, Structure of the eIF3d cap-binding domain. α helices are colored in blue and β strands in magenta. c, Topological maps of the eIF3d cap-binding domain and the DXO cap-endonuclease domain15. d, Structures comparing the eIF3d cap-binding domain with its gate insertion to DXO bound to RNA15 (PDB 4J7L).
To understand how eIF3d recognizes the 5' RNA terminus, we determined a 1.4 Å crystal structure of the conserved C-terminal domain of eIF3d from Nasonia vitripennis (65% identical, 84% similar to human eIF3d) using sulfur anomalous dispersion for phase determination (Extended Data Table 1, Extended Data Fig. 3). The structure of eIF3d reveals a complex fold that forms a cup-shaped architecture with a positively charged central tunnel that is negatively charged at its base (Fig. 2b). Remarkably, in spite of no significant sequence homology, the structural topology of eIF3d is nearly identical to the DXO proteins, a recently described family of 5' cap-endonucleases involved in RNA quality control14-16 (Fig. 2c, Extended Data Fig. 4). In contrast to DXO, eIF3d contains a unique insertion of ~15 highly conserved amino acids between strand β5 and helix α6. The eIF3d-specific insertion folds down along the front face of the domain, making loosely packed charged interactions that close off the RNA binding tunnel (Extended Data Fig. 5). We term this insertion an “RNA gate,” as the sequence clashes with the path of single-stranded RNA (ssRNA) bound to DXO15 and must undergo a conformational change for eIF3d to become competent for RNA recognition (Fig. 2d). We determined the structure of eIF3d in two additional crystal forms, and confirmed the RNA gate exhibits a closed conformation regardless of crystal packing (Extended Data Fig. 6). As eIF3d does not bind all capped RNAs17,18 we postulate that the RNA gate regulates cap recognition to prevent promiscuous mRNA binding prior to assembly of eIF3d into the full eIF3 complex. We tested this model using c-Jun mRNA and verified eIF3d cap-recognition only occurs in the context of a full eIF3 complex and requires prior eIF3-sequence specific RNA interactions with the eIF3-recruitment stem loop (Extended Data Fig. 7). Allosteric communication between eIF3 subunits during initial RNA recruitment likely facilitates eIF3d RNA gate opening to allow 5' end recognition. The structure of eIF3d therefore reveals a new cap-binding protein and explains the ability of the eIF3 complex to protect the 5' end of mRNA (Fig. 1c).
To validate the structural finding that eIF3d is a cap-binding protein, we examined the ability of eIF3 to bind the c-Jun mRNA 5' cap in the presence of competitor ligands. eIF3d-cap recognition is sensitive to m7GDP competition but resistant to GDP, indicating that, analogous to eIF4E-cap cap binding4, eIF3d specifically interacts with the 5' cap and requires a mature methylated cap structure for recognition (Fig. 3a). Using the DXO–RNA structure as a template15, we modeled a capped ssRNA along the basic binding groove shared between eIF3d and DXO and identified two conserved helices (α5 and α11) likely to be involved in cap recognition (Fig. 3b). We purified recombinant eIF3 containing helix α5 or α11-mutated eIF3d and demonstrated that both mutants have drastically reduced ability to crosslink to the c-Jun mRNA cap (Fig. 3c). eIF3d-mutated complexes retain wild type-levels of RNA-binding, indicating that these residues specifically coordinate 5' mRNA cap recognition (Extended Data Fig. 8). We next introduced HA epitope-tagged wild type or mutant eIF3d into 293T cells, and measured assembly of 48S initiation complexes on c-Jun mRNA by quantitative RT-PCR19,20. Mutations to the predicted eIF3d cap-binding surface dramatically reduce c-Jun mRNA incorporation into translation complexes, while the control ACTB mRNA is unaffected (Fig. 3d; Extended Data Fig. 8). These results demonstrate that cap binding by eIF3d is required for efficient initiation complex formation during eIF3-specialized translation.
Figure 3. eIF3d cap-binding activity is required for efficient 48S initiation complex formation on specific mRNAs.
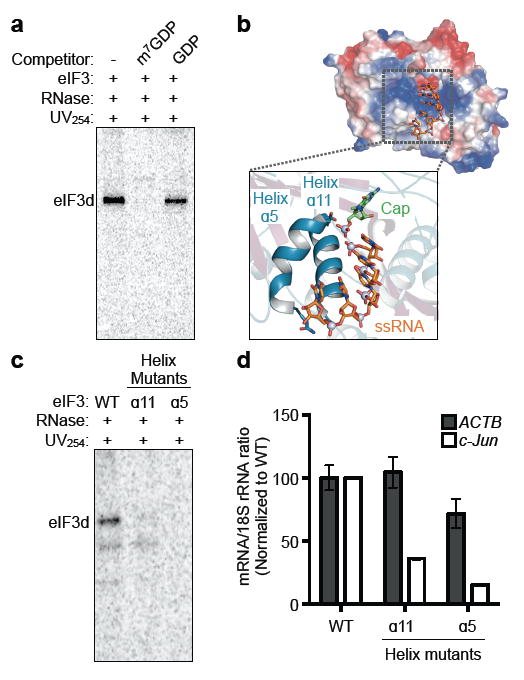
a, Phosphorimage of SDS gel resolving RNase-protected 32P-cap-labeled c-Jun 5′ UTR RNA crosslinked to eIF3 in the presence of competitor ligands. b, Electrostatic surface view of the eIF3d cap-binding domain colored by charge, with a zoomed view of single stranded RNA (ssRNA) and cap analog modeled according to their positions bound to DXO15. Positive charge is colored blue and negative charge is in red, and the RNA gate is removed for clarity. c, Phosphorimage of SDS gel resolving RNase-protected 32P-cap-labeled c-Jun 5′ UTR RNA crosslinked to wild type or helix α5 or helix α11-mutant eIF3. eIF3d-helix α5 mutant (D249Q/V262I/Y263A), helix α11 mutant (T317E/N320E/H321A). WT, wild type. d, Incorporation of c-Jun and ACTB mRNA into initiation complexes by wild type, helix α5, or helix α11-mutant eIF3d as measured by quantitative RT-PCR. mRNA-ribosome association is expressed as the ratio between the quantity of mRNA transcripts to 18S rRNA and normalized to the wild type sample. The results are representative of three independent experiments and given as the mean ± s.d. from a representative quantitative RT-PCR experiment performed in duplicate.
eIF3d recognition of the 5' cap structure provides an alternative cap-dependent translation mechanism from canonical eIF4F cap recognition. Perplexingly, when the RNA stem loop element that recruits eIF3 to the c-Jun mRNA is deleted, translation is inhibited even though the mRNA contains a 5' cap2. We hypothesized that an RNA element within the c-Jun mRNA blocks recruitment of the eIF4F complex. In support, the 5' cap of c-Jun mRNA crosslinks less efficiently to purified eIF4E than that of the ACTB mRNA (Extended Data Fig. 9). To identify the eIF4F inhibitory element, we constructed luciferase reporters to test deletions in the c-Jun 5' UTR (Fig. 4a). Deletion of the 5' 153 nucleotides, but not the initial 67 nucleotides, was sufficient to allow c-Jun mRNA translation to occur independently of the eIF3-recruitment stem loop, suggesting that canonical cap dependent translation is no longer blocked (Fig. 4b). We confirmed by western blot analysis of the 48S initiation complex formed on c-Jun mRNA lacking the 5' 153 nucleotides that the eIF4F components are now present (Fig. 4c).
Figure 4. An RNA element inhibits eIF4F recruitment and directs mRNAs to use an eIF3-specialized translation pathway.
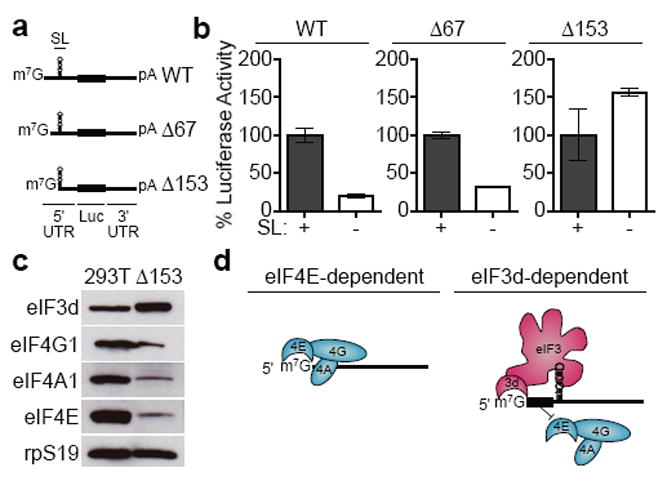
a, Schematic of c-Jun 5′ UTR truncation-luciferase reporter mRNAs. SL, stem loop; Luc, luciferase. b, Luciferase activity from in vitro translation of mRNAs containing truncations of the c-Jun 5′ UTR, with or without the internal eIF3-recruitment stem loop sequence. The results are given as the mean and standard deviation (s.d.) of three independent experiments, each performed in triplicate. c, Western blot analysis of initiation factors in 48S translation initiation complexes formed on c-Jun mRNA with a 5′ 153-nucleotide truncation. 293T, total protein from 293T in vitro translation extracts. The result is representative of three independent experiments. For gel source data, see Supplementary Figure 1. d, Model for eIF3d-directed cap-dependent mRNA translation. An eIF4F-inhibitory RNA element ensures that mRNA translation occurs through an eIF3-specialized pathway.
Together, we put forth a model of a previously undiscovered cap-dependent translation initiation pathway controlled by eIF3d recognition of the 5' mRNA cap (Fig. 4d). We postulate that encoding more than one mechanism of cap-dependent translation allows cells to specifically control protein synthesis in cellular environments where eIF4E is inactivated. In support, c-Jun mRNA translation is resistant to treatment of cells with chemicals that activate the 4E-BPs7,8,21 (Extended Data Fig. 1). As modulation of eIF4E cap-binding activity allows cells to incorporate extracellular stimuli into altered translation outputs22, it will be important to discover whether eIF3d activity is analogously regulated. Furthermore, our data indicates that the c-Jun mRNA encodes an additional cis-acting RNA element that blocks eIF4F to ensure translation can only occur through an eIF3-specialized pathway. RNA elements that block eIF4F recruitment may be a common theme to direct mRNAs into specific translation pathways to ensure controlled protein expression. For example, a subset of homeobox mRNAs contain an RNA element that blocks cap-dependent translation to ensure usage of an internal ribosome entry site and to allow for correct homeobox expression during embryonic development23. A number of eIF3-specialized mRNAs encode proteins involved in the control of cell proliferation, suggesting their translation may also require enhanced regulation2,24.
While considerable advances have been made in the structural understanding of eIF3 bound to the ribosome, direct localization of eIF3d in a 48S complex remains unclear25. Thus, understanding how eIF3d functions and assembles within the full translation initiation complex will have significant mechanistic implications in how cap recognition links to mRNA ribosomal recruitment. Our discovery of eIF3d as a cap-binding protein now reveals a new translation pathway independent of eIF4E, and adds another layer of cap-dependent translation.
Methods
Cells and transfections
Human 293T cells were maintained in DMEM (Invitrogen) supplemented with 10% FBS (Seradigm). The cells were obtained from the University of California, Berkeley, Cell Culture Facility, which authenticates cells by STR profiling and tests for mycoplasma contamination. Plasmid transfections were performed using Lipofectamine 2000 (Invitrogen), following the manufacturer’s protocol, and polysome or immunoprecipitation analyses were performed at 48 h after transfection. For INK128 (Cayman Chemical) cell treatment, 293T cells were incubated with the indicated concentration of INK128 for ~14–16 h before cell lysis.
Plasmids
To generate the eIF3d expression plasmids, eIF3d was amplified from human cDNA and inserted into pcDNA5/FRT. A 39-nucleotide linker followed by the HA epitope tag (YPYDVPDYA) was subsequently inserted prior to the eIF3d stop codon. The wild type c-Jun 5′ UTR luciferase reporter plasmid was previously described2. To generate the c-Jun in vitro transcription template, the 5′ UTR, ORF, and 3′ UTR were separately amplified from human cDNA and stitched together downstream of a T7 promoter by Gibson cloning into pcDNA4. The ACTB in vitro transcription template was constructed by addition of a T7 promoter during amplification of the full mRNA from human cDNA and inserted into pcDNA4.
Western blot
Western blot analyses were performed using the following antibodies: anti-eIF3d (Bethyl A301-758A), anti-eIF4A1 (Cell Signaling 2490), anti-eIF4G1 (Cell Signaling 2858), anti-rpS19 (Bethyl A304-002A), anti-eIF4E (Bethyl A301-154A), anti-rpLP0 (Bethyl A302-882A), anti-HA epitope tag (Pierce 26183), anti-c-Jun (Cell Signaling 9165), anti-Hsp90 (BD Biosciences 610418), and anti-4EBP1 (Cell Signaling 9644).
In vitro RNA transcription and labeling
Unlabeled RNAs were in vitro transcribed, polyadenylated, and capped as previously described2. For internal radiolabeling of RNAs, in vitro transcription was performed in the presence of 0.1 µM [α-32P]-ATP, then the RNA was subsequently capped with vaccinia virus enzymes (NEB). For radiolabeling of the 5′ cap, in vitro transcribed RNAs were capped with vaccinia virus enzymes and [α-32P]-GTP. RNAs were purified by phenol-chloroform extraction and ethanol precipitation.
In vitro translation
In vitro translation extracts were made from human 293T cells as previously described21. Lysates were nuclease-treated with 18 gel units μl-1 micrococcal nuclease (NEB) in the presence of 0.7 mM CaCl2 for 10 min at 25 °C, and the digestion was stopped by addition of 2.24 mM EGTA. Each translation reaction contained 50% in vitro translation lysate and buffer to make the final reaction with 0.84 mM ATP, 0.21 mM GTP, 21 mM creatine phosphate (Roche), 45 U ml-1 creatine phosphokinase (Roche), 10 mM HEPES-KOH pH 7.6, 2 mM DTT, 8 mM amino acids (Promega), 255 mM spermidine, 1 U ml-1 murine RNase inhibitor (NEB), and mRNA-specific concentrations of Mg(OAc)2 and KOAc. The optimal magnesium and potassium levels to add were determined to be 1.5 mM Mg(OAc)2 and 150 mM KOAc for c-Jun mRNA, and 1 mM and 150 mM KOAc for ACTB mRNA. For luciferase assays, translation reactions were incubated for 1 h at 30 °C, then luciferase activity was assayed.
48S initiation complex purification
For 48S initiation complex purification from in vitro translation reactions, 180 μl reactions were incubated for 20 min at 30 °C and centrifuged for 6 min at 12,000g at 4 °C. Lysates were purified by size exclusion chromatography through a 1 ml column packed with Sephacryl S-400 gel filtration resin (GE Healthcare) and the elutant was centrifuged through a 10-25% w/v sucrose gradient by centrifugation for 3.5 h at 38,000 rpm at 4 °C in a Beckman SW41 Ti rotor27. Fractions were collected from the top of the gradient using a peristaltic pump with a Brandel tube piercer. From the appropriate fractions, RNA was purified by phenol-chloroform extraction and ethanol precipitation and protein was precipitated with trichloroacetic acid.
For affinity purification of HA epitope-tagged eIF3d-associated 48S initiation complexes from cells, three 10 cm plates of transfected 293T cells were treated with 100 μg ml-1 cycloheximide for 5 min. Cells were washed with ice cold PBS (137 mM NaCl, 2.7 mM KCl, 100 mM Na2HPO4, 2 mM KH2PO4) with 100 μg ml-1 cycloheximide and collected in lysis buffer (20 mM HEPES-KOH pH 7.4, 150 mM KOAc, 2.5 mM Mg(OAc)2, 1 mM DTT, 100 μg ml-1 cycloheximide, 1% v/v Triton X-100). Lysates were centrifuged for 6 min at 12000g at 4 °C and purified by S-400 size exclusion chromatography. 80 μl of anti-HA antibody-conjugated agarose beads (Sigma) were added to the elutants, tumbled for 1.5 h at 4 °C, and beads were washed three times with lysis buffer without Triton X-100. Bound complexes were eluted twice with 100 μg ml-1 HA peptide and elutants were centrifuged and analyzed the same as for the in vitro purification reactions.
Quantitative real-time PCR
cDNA was reverse-transcribed from RNA using random hexamers and Superscript III (Invitrogen), following the manufacturer’s protocol. Real-time PCR was performed using DyNAmo HS Sybr Green (ThermoFisher), with a 20 μl reaction volume containing 2 μl cDNA and 0.5 μM of each primer. The following oligonucleotides were used: 18S rRNA-Forward, 5′ – GGCCCTGTAATTGGAATGAGTC–3′; 18S rRNA-Reverse, 5′ –CCAAGATCCAACTACGAGCTT–3′; ACTB-Forward, 5′ – CTCTTCCAGCCTTCCTTCCT–3′; ACTB-Reverse, 5′ -AGCACTGTGTTGGCGTACAG-3′; c-Jun-Forward, 5′–TGACTGCAAAGATGGAAACG–3′; c-Jun-Reverse; 5′–CAGGGTCATGCTCTGTTTCA–3′.
eIF3-RNA crosslinking and gel shift
Recombinant eIF3 was expressed and purified from E. coli as previously described26. For each crosslinking reaction, 1 μl water, 1 μl 125 nM labeled RNA, 1 μl 10 μg ml-1 heparan sulfate (Sigma), 1 μl 5× binding buffer (125 mM Tris-HCl pH 7.5, 25mM Mg(OAc)2, 350 mM KCl, 0.5 mM CaCl2, 0.5 mg ml-1 BSA, 10 mM TCEP), and 1 μl 1.5 μM purified eIF3 were added, in the listed order, and incubated for 30 min at 25 °C. For competition experiments, the water was substituted with 1 μl of 1 mM m7GDP/Mg2+ or GDP/Mg2+. UV254-induced crosslinking was performed using a short-wave UV lamp placed ~4 cm above the samples on ice for 10 min. After treatment with RNase for 10 min at 37 °C, proteins were separated by 12% SDS-PAGE, the gel was dried, and imaged using a phosphorimager. For digestion of internal labeled RNA, 2.5 U benzonase (Novagen) and 250 U RNase T1 (ThermoFisher) were used; for digestion of cap labeled RNA, 4 U RNase R (Epicentre) and 1 U RiboShredder (Epicentre) were used. For eIF3d subunit identification, after RNase treatment, samples were denatured and immunoprecipitation was performed as previously described2. For limited proteolysis, after RNase treatment, the reactions were treated with 2 or 20 μg ml-1 sequencing grade trypsin (Promega) for 30 min at 25 °C, prior to gel electrophoresis. Mass spectrometry samples were prepared as previously described2. Gel shift assays were performed as previously described, using 50 nM labeled c-Jun stem loop RNA and 300 nM purified eIF32.
Recombinant eIF3d protein purification
Candidate eIF3d cap-binding domain fragments were amplified by PCR and cloned into a modified pET vector to express an N-terminal 6× His (KSSHHHHHHGSS)-MBP-TEV fusion protein as previously described28. Extensive expression trials were conducted to determine optimal amino- and carboxy-terminal domain boundaries and identified a minimal stable human cap-binding domain S161–F527. Recombinant protein was expressed in BL21-RIL DE3 E. coli cells co-transformed with a pRARE2 tRNA plasmid (Agilent). E. coli was grown in 2×YT media at 37 °C to an OD600 of ~0.5, cooled at 4 °C for 15 min, induced with addition of 0.5 mM IPTG and then incubated with shaking for ~20 h at 16 °C. Pelleted cells were washed with PBS and then lysed by sonication in lysis buffer (20 mM HEPES-KOH pH 7.5, 400 mM NaCl, 10% glycerol, 30 mM imidazole, 1 mM TCEP) in the presence of EDTA-free Complete Protease Inhibitor (Roche). Following centrifugation for 30 min at 23,000g and 4 °C, clarified lysate was incubated with Ni-NTA agarose resin (QIAGEN) for 1 h at 4 °C with gentle rocking. Resin was washed with lysis buffer supplemented to 1 M NaCl and eluted by gravity-flow chromatography at 4 °C with lysis buffer supplemented to 300 mM imidazole. The eluted fraction was diluted to ~50 mM imidazole and 5% glycerol prior, concentrated to ~50 mg ml-1 and incubated with Tobacco Etch Virus protease for ~12 h at 4 °C to remove the MBP tag. Recombinant eIF3d was isolated from free MBP by diluting with gel-filtration buffer (20 mM HEPES-KOH pH 7.5, 250 mM KCl, 1 mM TCEP) and passing over a 5 ml Ni-NTA column (QIAGEN) connected in line with a 5 ml MBP-Trap column (GE Life Sciences) prior to additional purification by size-exclusion chromatography on a Superdex 75 16/60 column. Final purified eIF3d was concentrated to ~20–50 mg ml-1, used immediately for crystallography, or flash frozen in liquid nitrogen for storage at −80 °C.
Crystallization and structure determination
Initial crystals of human eIF3d were grown at 18 °C by hanging drop vapor diffusion, but diffracted poorly. Analogous eIF3d cap-binding domain sequences were cloned from a panel of highly homologous animal sequences, with the equivalent domain from the parasitic wasp Nasonia vitripennis producing the best crystals. Optimized N. vitripennis eIF3d crystals were grown in 2 μl hanging drops set at a 1:1 ratio over 300 μl of reservoir liquid: 200 mM (NH4)2SO4, 100 mM Bis-Tris 6.5, 23–27% PEG-3350 (Crystal Form 1), 1.6–1.8 M Ammonium Citrate pH 7.0 (Crystal Form 2), or 200 mM NaCl, 100 mM Tris 8.5, 25% PEG-3350 (Crystal Form 3). eIF3d crystals (Crystal Forms 1 and 2) were cryoprotected by covering the drop with a layer of saturated paratone-N or NVH oil (Hampton) and crystals were transferred into the oil emersion and cleaned using a Kozak cat whisker as previously described29, or cryoprotected by transferring to a reservoir solution supplemented with 20% ethylene glycol (Crystal Form 3). Crystals were harvested with a nylon loop and then flash-frozen in liquid nitrogen. X-ray diffraction data were collected under cryogenic conditions at the Lawrence Berkeley National Laboratory Advanced Light Source (Beamline 8.3.1).
Data were processed with XDS and AIMLESS30 using the SSRL autoxds script (A. Gonzalez, Stanford SSRL). eIF3d crystals belonged to the orthorhombic space group P 21 21 21, and contained either two copies per asymmetric unit (Crystal Form 1) or one copy (Crystal Form 2) or the space group P 21 and contained two copies per asymmetric unit (Crystal Form 3). Experimental phase information was collected from a native crystal using sulfur single-wavelength anomalous dispersion. Data was collected at a minimal accessible wavelength (~7235 eV) and iterative data sets were completed and merged from independent portions of an exceptionally large eIF3d crystal. After ~90× multiplicity, anomalous signal was detected to ~2.4 Å, and a clear phase solution was obtained at ~120× multiplicity. 35 sites were identified with HySS in PHENIX31 corresponding to 32 sulfur atoms in eIF3d and 3 potassium/chloride positions. Phases were extended to the native eIF3d data set processed to ~1.40 Å using SOLVE/RESOLVE32, and model building was completed in Coot33 prior to refinement with PHENIX. X-ray data for refinement were extended according to an I/σ resolution cutoff of ~1.5, CC* correlation and Rpim parameters, and visual inspection of the resulting map34. A completed eIF3d cap-binding domain from Crystal Form 1 was used as a search model to determine phases for Crystal Form 2 and 3 using molecular replacement. Final structures were refined to stereochemistry statistics for Ramachandran Plot (Favored/Allowed), Rotamer outliers, and MolProbity score as follows: Crystal Form 1, 96.8% / 3.2%, 0.2% and 1.40; Crystal Form 2, 97.3% / 2.7%, 0% and 1.29; Crystal Form 3 97.4% / 2.6%. 0.9% and 1.26.
Recombinant eIF4E protein purification and RNA crosslinking
Full length human eIF4E was cloned and expressed using the same protocol as for eIF3d. eIF4E–RNA crosslinking was performed as described for eIF3–RNA crosslinking, but using 25 nM RNA with normalized cpm and the indicated concentration of eIF4E. RNase treatment was performed using 4 U RNase R and 250 U RNase T1.
Extended Data
Extended Data Figure 1. c-Jun expression is unaffected by 4E-BP1 activation.
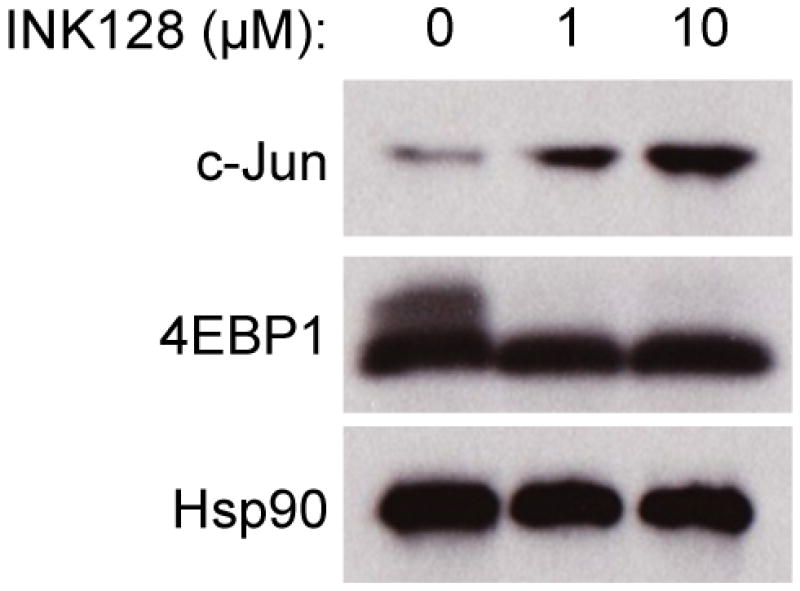
Representative western blot of 293T cells after 24 h treatment with INK128. The results are representative of three independent experiments. For gel source data, see Supplementary Figure 1.
Extended Data Figure 2. Mapping of a C-terminal region of eIF3d that protects the c-Jun 5′ cap structure.
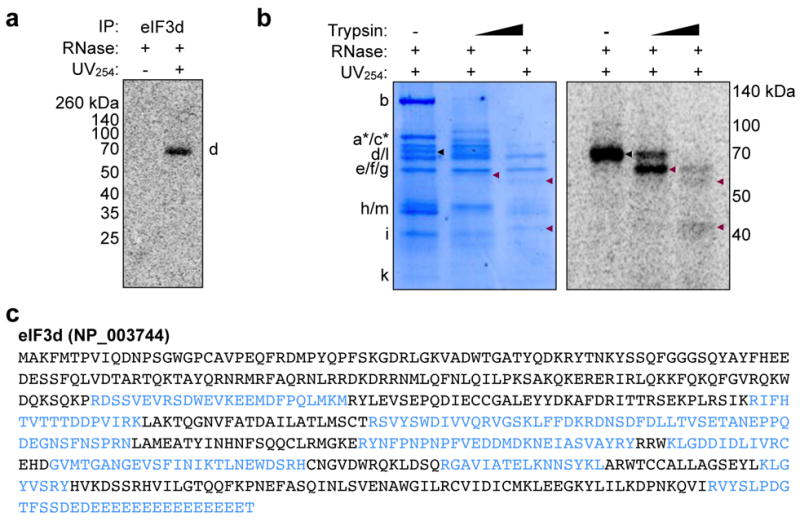
a, Validation of eIF3d subunit identification. eIF3d-cap crosslinking was validated by immunoprecipitation of eIF3d after crosslinking and denaturing the eIF3 complex by boiling in SDS. The result is representative of biological replicates. b, Limited proteolysis of eIF3 crosslinked to 32P-cap-labeled c-Jun 5′ UTR RNA. Full length and proteolysis fragments of eIF3d are indicated with black and maroon arrows, respectively, on the phosphorimage and coomassie-stained SDS gels. c, Mass spectrometry identification of trypsinized peptides from limited proteolysis of cap-crosslinked eIF3d. Identified peptides are highlighted in blue. The results in b-c are representative of three independent experiments.
Extended Data Figure 3. Purification of eIF3d cap-binding domain.
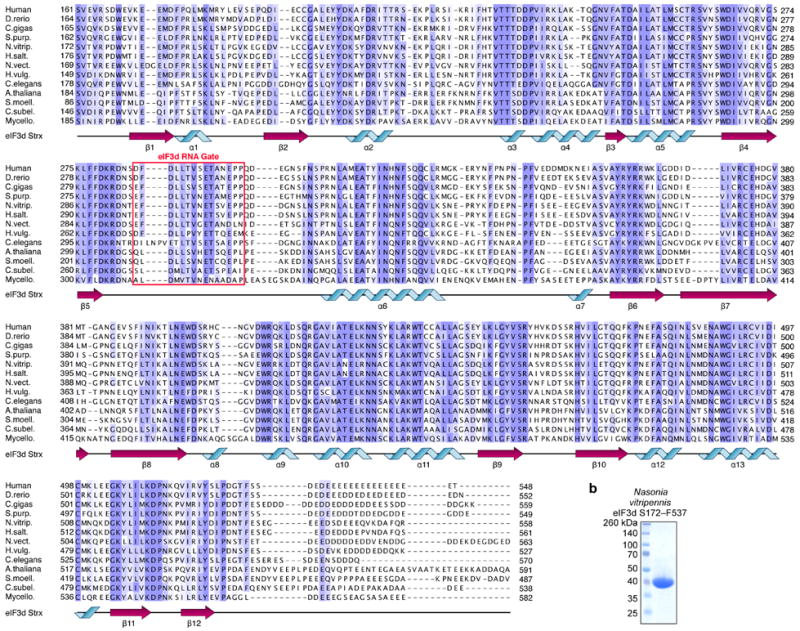
a, Alignment is colored by phylogenetic conservation of amino acid physiochemical property similarity and a cartoon schematic of the eIF3d secondary structure is depicted below the sequences. Coloring begins at 30% conservation (lightest blue). b, Coomassie blue stained SDS gel of recombinant Nasonia vitripennis eIF3d S162–F527 protein expressed in E. coli.
Extended Data Figure 4. Structure-based alignment of eIF3d cap-binding domain and DXO cap-endonuclease domain sequences.
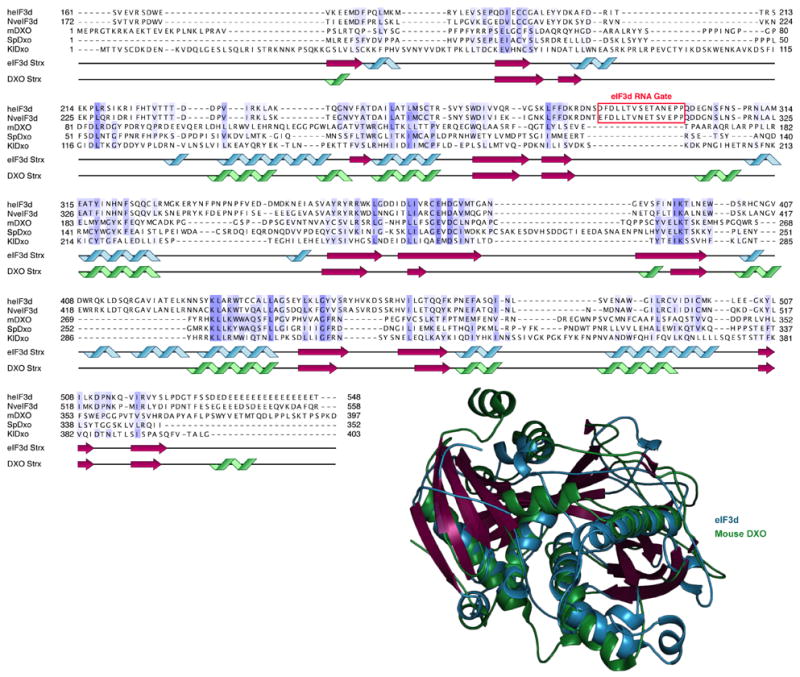
Structure-based alignment of eIF3d and DXO sequences according to superposition of eIF3d and DXO (PDB 4J7L) structures15. Alignment is colored by phylogenetic conservation as in Extended Data Fig. 2 and cartoon schematics of the secondary structures are depicted below the sequences. eIF3d is colored in blue and magenta as in Fig. 2, and DXO is colored in green and magenta.
Extended Data Figure 5. Structural details of eIF3d “RNA gate” stabilizing interactions.
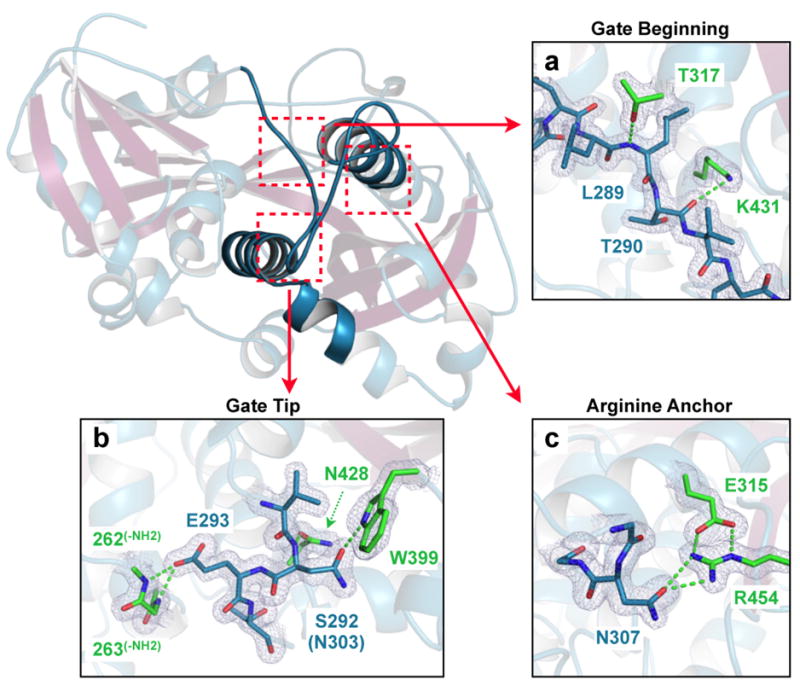
a–c. Structural overview of eIF3d with cut-away sections highlighting charged-interactions stabilizing the closed “RNA gate” conformation. No significant van der Waals interactions stabilize the closed gate conformation, supporting likely repositioning of the RNA gate prior to 5′ mRNA cap recognition. Charged interactions occur in three areas: a, at the beginning of the gate insertion sequence (Gate Beginning); b, at the tip of the unstructured loop (Gate Tip); and c, at an “Arginine Anchor” point stabilizing the return of the loop insertion sequence to the α-helix shared with DXO-family endonucleases. Residues are numbered according to the human eIF3d sequence, and all positions are conserved between human and N. vitripennis except S292N. eIF3d RNA gate residues are displayed with blue side-chains and the residues making stabilizing contacts are colored as green. 2Fo–Fc map regions are shown at 1.5σ.
Extended Data Figure 6. Packing interactions observed in alternative eIF3d crystal forms.
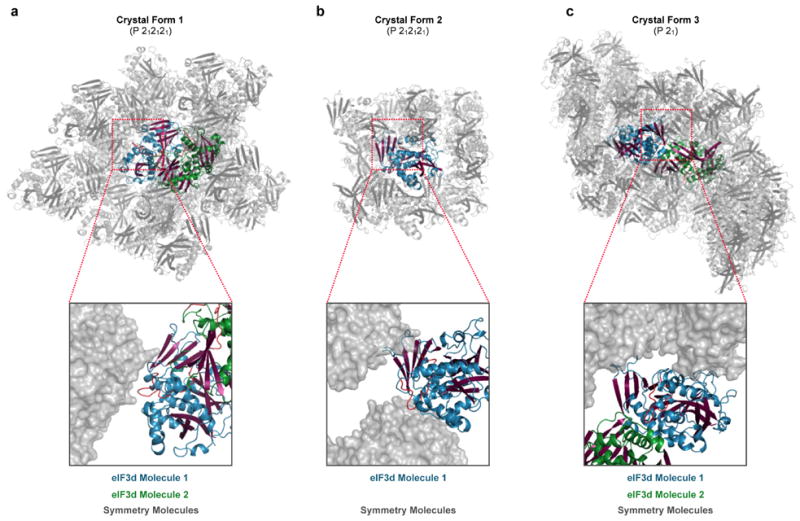
Cartoon representation of crystallographic packing in eIF3d crystal form 1, 2 and 3 (a, b, c). Crystal forms 1 and 3 have two copies of eIF3d in the asymmetric unit colored in blue/magenta and green/magenta respectively; crystal form 2 has only one copy of eIF3d per asymmetric unit. Symmetry related molecules are depicted in grey. Cut-away zoom illustrates position of the eIF3d RNA gate (red) relative to nearest symmetry related molecule. In crystal form 1 the RNA gate is packed against a neighboring symmetry molecule, but in crystal forms 2 and 3 the RNA gate is positioned towards a major solvent channel. Relative conformation of the RNA gate remains unchanged in either eIF3d crystal form.
Extended Data Figure 7. eIF3d cap-binding activity requires the eIF3-recruitment stem loop RNA.
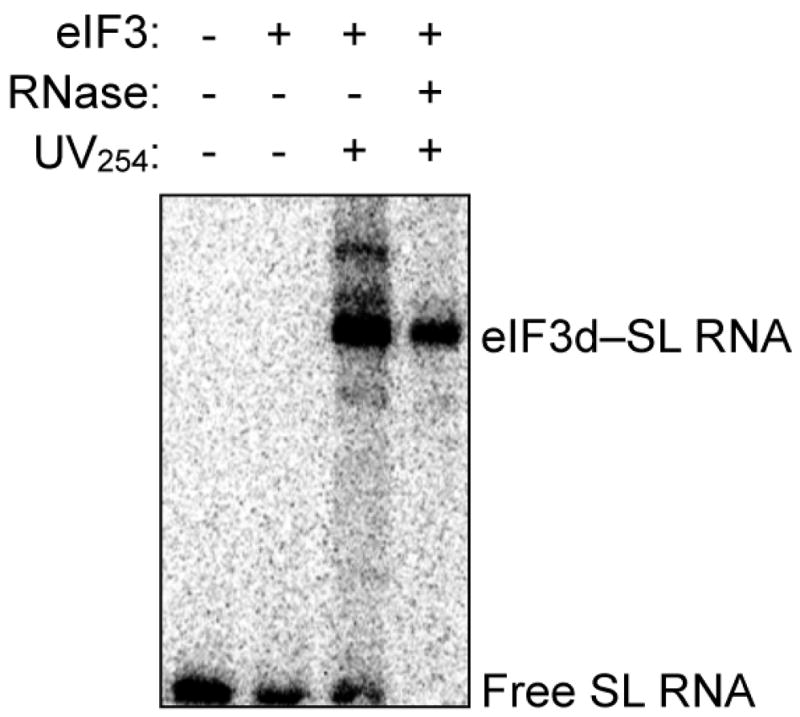
Phosphorimage of SDS gel resolving RNase-protected 32P-cap-labeled c-Jun stem loop RNA crosslinked to eIF3 subunits. The result is representative of three independent experiments.
Extended Data Figure 8. Incorporation of HA epitope-tagged eIF3d into translation initiation complexes.
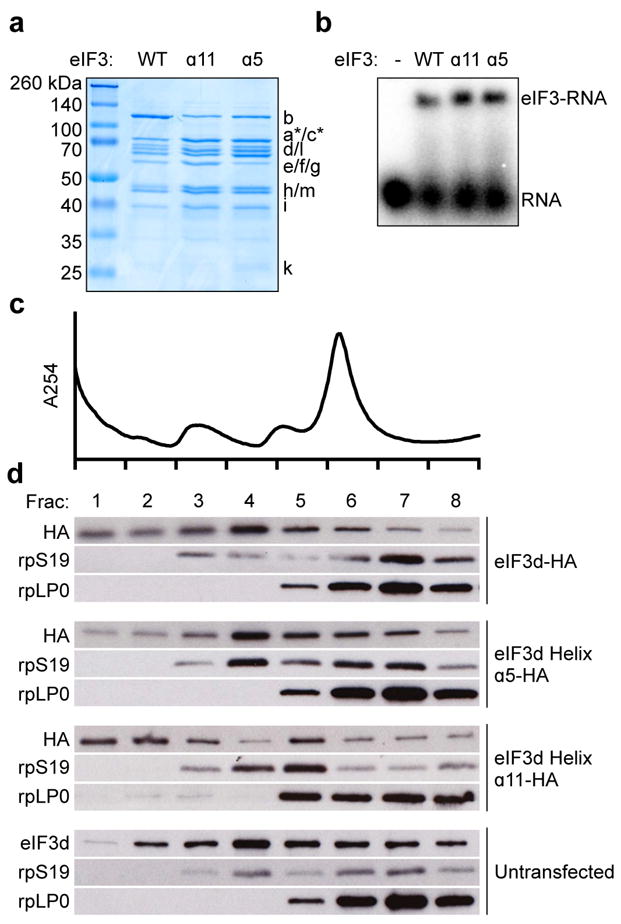
a, Coomassie blue stained SDS gel of recombinant eIF3 containing wild type (WT) or helix α5 or α11 mutated eIF3d. b, Representative native agarose gel electrophoresis of recombinant wild type and mutant eIF3 complexes bound to the c-Jun stem loop. c, Polysome profiles of untransfected 293T cells, plotted as relative absorbance at 254 nm versus elution fractions. d, Western blot analysis of eIF3d and the small (rpS19) and large (rpLP0) ribosomal subunits. The results in b-d are representative of three independent experiments. For gel source data, see Supplementary Figure 1.
Extended Data Figure 9. eIF4E recognizes the 5′ end of the c-Jun mRNA less efficiently than ACTB mRNA.
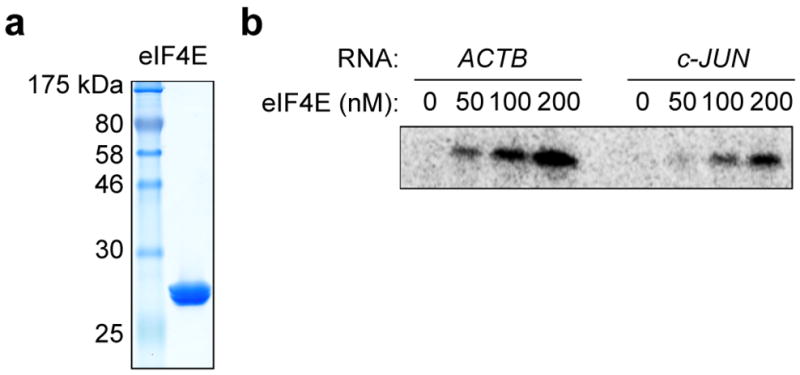
a, Coomassie blue stained SDS gel of recombinant human eIF4E expressed in E. coli. b, Phosphorimage of SDS gel resolving RNase-protected 32P-cap-labeled ACTB or c-Jun 5′ UTR RNA crosslinked to eIF4E. The result is representative of three independent experiments. For gel source data, see Supplementary Figure 1.
Extended Data Table 1. Summary of data collection, phasing and refinement statistics.
Single crystals were used to collect data for each structure.
| eIF3d Crystal Form 1 (5K4B) | eIF3d Crystal Form 2 (5K4C) | eIF3d Crystal Form 3 (5K4D) | eIF3d (S-SAD) | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P 212121 | P 212121 | P 21 | P 212121 |
| Cell dimensions | ||||
| a, b, c (Å) | 61.90, 62.93, 192.24 | 49.01, 61.84, 138.31 | 49.97, 144.32, 55.30 | 61.66, 62.72, 193.28 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 109.12, 90.0 | 90.0, 90.0, 90.0 |
| Wavelength | 1.11586 | 1.11586 | 1.11587 | 1.71371 |
| Resolution (Å)a | 48.06–1.40 (1.42–1.40) | 46.20–1.70 (1.73–1.70) | 49.13–2.00 (2.05–2.00) | 38.66–1.92 (1.96–1.92) |
| Rpim | 2.6 (45.3) | 5.0 (45.0) | 8.6 (52.3) | 0.9 (8.1) |
| I/σ (I) | 12.9 (1.5) | 9.1 (1.6) | 6.5 (1.4) | 65.2 (6.7) |
| CC1/2 | 99.9 (57.9) | 99.6 (63.9) | 98.8 (55.6) | 100 (94.7) |
| Completeness (%) | 100 (99.2) | 99.4 (89.4) | 99.8 (98.3) | 98.5 (86.1) |
| Redundancy | 15.8 (10.1) | 4.1 (3.6) | 3.0 (2.7) | 129.0 (46.6) |
| Refinement | ||||
| Resolution (Å) | 48.06–1.40 | 46.20–1.70 | 49.13–2.00 | |
| No reflections | ||||
| Total | 2,349,428 | 193,627 | 147,613 | |
| Unique | 148,455 | 47,123 | 49,757 | |
| Free (%) | 2 | 5 | 5 | |
| Rwork / Rfree | 17.5 / 19.3 | 16.2 / 19.6 | 17.7 / 20.8 | |
| No. atoms | ||||
| Protein | 5837 | 2953 | 5686 | |
| Ligand/ion | 3 (CI) | 18 (glycerol) | - | |
| Water | 910 | 385 | 741 | |
| B factors | ||||
| Protein | 21.7 | 24.2 | 26.6 | |
| Ligand/ion | 31.7 | 29.1 | - | |
| Water | 33.1 | 36.7 | 34.3 | |
| r.m.s deviations | ||||
| Bond lengths (Å) | 0.007 | 0.012 | 0.004 | |
| Bond angles (°) | 1.167 | 1.359 | 0.638 |
Values in parentheses are for highest-resolution shell.
Supplementary Material
Acknowledgments
The authors thank J. Berger and K. Chat for helpful discussions. X-ray data were collected at Beamline 8.3.1 of the Lawrence Berkeley National Lab Advanced Light Source (ALS), supported in part by the UC Office of the President, Multicampus Research Programs and Initiatives grant MR-15-328599 and the Program for Breakthrough Biomedical Research, which is partially funded by the Sandler Foundation. The authors are grateful to J. Holton, G. Meigs (ALS), and T. Doukov (SSRL) for help with S-SAD data collection. This work used the Vincent J. Proteomics/Mass Spectrometry Laboratory at UC Berkeley, supported in part by NIH S10 Instrumentation Grant S10RR025622. This work was funded by the NIGMS Center for RNA Systems Biology (P50-GM201706). J.A.D. is an HHMI Investigator. A.S.Y.L is supported as an American Cancer Society Postdoctoral Fellow (PF-14-108-01-RMC) and P.J.K. is supported as an HHMI Fellow of the Life Sciences Research Foundation.
Footnotes
Author Contributions: Project and experiments were conceived and designed by A.S.Y.L. with consultation with J.H.D.C. Protein purification, biochemistry, and cell biology experiments and analyses were performed by A.S.Y.L. Crystallography and structure determination was performed by P.J.K. The manuscript was written by A.S.Y.L, P.J.K., and J.H.D.C, with editing by J.A.D.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information: Atomic coordinates and structure factors for the reported crystal structures are deposited at the Protein Data Bank under accession codes 5K4B (Crystal Form 1), 5K4C (Crystal Form 2), and 5K4D (Crystal Form 3).
The authors declare no competing financial interests.
References
- 1.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annual Review of Biochemistry. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 2.Lee AS, Kranzusch PJ, Cate JH. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature. 2015;522:111–114. doi: 10.1038/nature14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sonenberg N. eIF4E, the mRNA cap-binding protein: from basic discovery to translational research. Biochemistry and cell biology. 2008;86:178–183. doi: 10.1139/O08-034. [DOI] [PubMed] [Google Scholar]
- 4.Sonenberg N, Morgan MA, Merrick WC, Shatkin AJ. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5'-terminal cap in mRNA. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:4843–4847. doi: 10.1073/pnas.75.10.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pause A, et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5'-cap function. Nature. 371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 6.Gingras AC, et al. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes & development. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thoreen CC, et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gandin V, et al. nanoCAGE reveals 5' UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res. 2016;26:636–648. doi: 10.1101/gr.197566.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shatkin AJ. Capping of eucaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 12.Andreev DE, et al. Differential contribution of the m7G-cap to the 5' end-dependent translation initiation of mammalian mRNAs. Nucleic Acids Res. 2009;37:6135–6147. doi: 10.1093/nar/gkp665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolfe AL, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang JH, et al. Dxo1 is a new type of eukaryotic enzyme with both decapping and 5'-3' exoribonuclease activity. Nat Struct Mol Biol. 2012;19:1011–1017. doi: 10.1038/nsmb.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiao X, Chang JH, Kilic T, Tong L, Kiledjian M. A mammalian pre-mRNA 5' end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol Cell. 2013;50:104–115. doi: 10.1016/j.molcel.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang S, et al. Structure and function of the 5'-->3' exoribonuclease Rat1 and its activating partner Rai1. Nature. 2009;458:784–788. doi: 10.1038/nature07731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sonenberg N, Morgan MA, Testa D, Colonno RJ, Shatkin AJ. Interaction of a limited set of proteins with different mRNAs and protection of 5'-caps against pyrophosphatase digestion in initiation complexes. Nucleic Acids Res. 1979;7:15–29. doi: 10.1093/nar/7.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindqvist L, Imataka H, Pelletier J. Cap-dependent eukaryotic initiation factor-mRNA interactions probed by cross-linking. RNA. 2008;14:960–969. doi: 10.1261/rna.971208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Locker N, Easton LE, Lukavsky PJ. Affinity purification of eukaryotic 48S initiation complexes. RNA. 2006;12:683–690. doi: 10.1261/rna.2227906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Pan X, Hershey JW. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. J Biol Chem. 2007;282:5790–5800. doi: 10.1074/jbc.M606284200. [DOI] [PubMed] [Google Scholar]
- 21.Blau L, et al. Aberrant expression of c-Jun in glioblastoma by internal ribosome entry site (IRES)-mediated translational activation. Proc Natl Acad Sci USA. 2012;109:E2875–2884. doi: 10.1073/pnas.1203659109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 23.Xue S, et al. RNA regulons in Hox 5' UTRs confer ribosome specificity to gene regulation. Nature. 2015;517:33–38. doi: 10.1038/nature14010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wisdom R, Johnson RS, Moore C. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 1999;18:188–197. doi: 10.1093/emboj/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.des Georges A, et al. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 2015;525:491–495. doi: 10.1038/nature14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun C, et al. Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3) Proc Natl Acad Sci USA. 2011;108:20473–20478. doi: 10.1073/pnas.1116821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee AS, Burdeinick-Kerr R, Whelan SP. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc Natl Acad Sci USA. 2013;110:324–329. doi: 10.1073/pnas.1216454109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kranzusch PJ, et al. Structure-guided reprogramming of human cGAS dinucleotide linkage specificity. Cell. 2014;158:1011–1021. doi: 10.1016/j.cell.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kranzusch PJ, et al. Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling. Mol Cell. 2015;59:891–903. doi: 10.1016/j.molcel.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabsch W. Xds. Acta Crystallographica Section D, Biological crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terwilliger TC. Reciprocal-space solvent flattening. Acta Crystallographica Section D, Biological crystallography. 1999;55:1863–1871. doi: 10.1107/S0907444999010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D, Biological crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


