Abstract
Electrically conductive materials provide a suitable platform for the in vitro study of excitable cells, such as skeletal muscle cells, due to their inherent conductivity and electro-activity. Here we demonstrate that bioinspired electroconductive nanopatterned substrates enhanced myogenic differentiation and maturation. The topographical cues from the highly-aligned collagen bundles that form the extracellular matrix (ECM) of skeletal muscle tissue were mimicked using nanopatterns created with capillary force lithography. Electron beam deposition was then utilized to conformally coat nanopatterned substrates with a thin layer of either gold or titanium to create electroconductive substrates with well-defined, large-area nanotopographical features. C2C12 cells, a myoblast cell line, were cultured for 7 days on substrates, and the effects of topography and electrical conductivity on cellular morphology and myogenic differentiation were assessed. We found that biomimetic nanotopography enhanced the formation of aligned myotubes, and the addition of an electroconductive coating promoted myogenic differentiation and maturation, as indicated by the upregulation of myogenic regulatory factors Myf5, MyoD and myogenin (MyoG). These results suggest the suitability of electroconductive nanopatterned substrates as a biomimetic platform for the in vitro engineering of skeletal muscle tissue.
Keywords: electroconductive substrate, nanotopography, myogenic differentiation, maturation, tissue engineering
1. Introduction
Skeletal muscle possesses a remarkable ability for self-repair and regeneration following injury. [1, 2] However, loss of muscle tissue and function due to chronic disease and traumatic injury is difficult to restore.[3–7] Much of this is due to the morphology of mature skeletal muscle tissue, which is comprised of many large multinucleated fibers whose nuclei cannot divide.[3] Additionally, the incidence of satellite cells, a sub-population of myoblasts capable of regeneration, in mature tissues is very low (typically less than 5%).[8] Engineered muscle constructs could thus potentially provide the means to replace permanently damaged tissue by mimicking the structure and function of healthy skeletal muscle.[6, 9–12] Successful engineering of skeletal muscle depends on the ability to stimulate myogenic differentiation in vitro in order to generate large numbers of proliferative cells and/or mature tissue constructs.[13]
Previous studies have demonstrated that the regenerative performance of engineered scaffolds are influenced by various factors, including the substrate surface functionality[14–16] and stiffness[17–19]. Also important is the influence of surface topography or geometry, as muscle tissue in vivo is characterized by the parallel alignment of the multinucleated fibers, as well as the highly-aligned nature of the collagen bundles that make up the underlying extracellular matrix (ECM).[20, 21] Studies have shown the importance of ECM topography as a potent cue for defining cell shape, orientation, alignment, and regulation of tissue-level functions.[22–24] Thus, when designing scaffolds for skeletal muscle regeneration, mimicking the nanotopographic features of native extracellular matrix of skeletal muscle may prove advantageous for tissue development and regeneration by promoting cell adhesion, mobility and differentiation. Recent advances in the micro- and nanofabrication of biomaterials have enabled more precise control over cell patterning and morphology, and such techniques have been applied to the design of scaffolds for engineering tissues such as skeletal muscle.[25–29]
In addition to topographical cues, electrical stimuli can play an important role in regulating the differentiation and behavior of certain electroactive cell and tissue types. Biocompatible metals such as gold,[12, 30] and titanium [15, 25], or electrically conducting polymers such as polyaniline,[9, 11, 14] and poly(3,4-ethylenedioxythiophene) (PEDOT)[31, 32] have been used as substrates for a range of cell types, including cardiomyocytes,[33] PC12 cells,[9, 14] endothelial cells,[34] fibroblasts,[16] and keratinocytes.[35] There have been examples of skeletal muscle tissue engineering studies which have used electrically conductive polymers and/or metals, and the early indications are that such biomaterials are indeed beneficial for the differentiation of myoblasts towards a functional phenotype.[36–38] However, the augmentation of myoblast differentiation to form multinucleated myofibers on electrically conductive scaffolds that also present topographical cues has not yet been extensively studied.
In this study, we developed a method of generating electrically conductive nanopatterned substrates for enhanced myogenic differentiation and maturation. Inspired by the structural organization of mature skeletal muscle ECM, nanopatterned poly urethane acrylate (PUA) substrates were fabricated using capillary force lithography (CFL) and subsequently coated with a thin metal layer of gold or titanium by an electron-beam evaporator. In this instance, gold and titanium were selected as the conductive materials of choice as they are two of most commonly used metals that are used in medical applications, and thus would generate limited complications in regards to biocompatibility. Metal deposition and its limited impact on the fidelity of the nanopatterning were validated through the use of electron microscopy, and the capability of these substrates at simultaneously providing topographical and electrical cues on the growth, morphology, and maturation of cultured myoblasts were then investigated.
2. Results and Discussion
Skeletal muscle tissue is structurally complex, comprised of highly-aligned multinucleated myofiber bundles that allow for the unidirectional and synchronized exertion of force. Additionally, skeletal muscle cells are frequently exposed to electrical stimuli in the body, as activation of voltage-gated calcium channels in the cell membranes are closely linked to the contraction mechanism of muscle tissue.[39] Therefore, engineering functional muscle tissue requires the parallel alignment of myoblasts in culture while exposed to chronic electrical stimulation such that they will differentiate towards mature tissue phenotypes. In this study, we demonstrated that electrically conductive materials, in combination with geometrically controlled nanopatterned substrates, enhanced both myoblast differentiation and ultrastructural organization without the need for external mechanical stimuli.
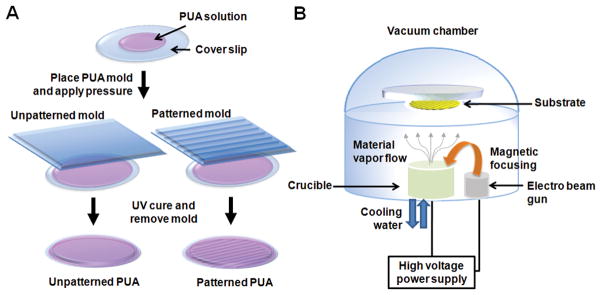
The capability for the reproducible production of well-defined nanotopographies in a scalable manner has been a significant challenge for the application of nanopatterned substrates on the study of cell biology and for applications such as tissue engineering. For example, while electrospinning is capable of producing substrates with relatively aligned nanotopographies, the controllability of not only fiber dimensions but also fiber-to-fiber spacing is poor and fabrication parameters often require frequent fine-tuning due to changing environmental conditions.[40] Microcontact printing techniques offer advantages in reproducibility of intricate patterns; however, feature sizes are limited in that they cannot produce nanoscale topographies.[41,42] Bench-top lithographic techniques, such as CFL, can address these issues, as standard photolithography methods can create initial molds with nanoscale features, which then can be mass replicated at low cost with curable polymers. Furthermore, CFL allows for the fabrication of large area substrates that would be well-suited for engineering tissue constructs at physiologically-relevant size scales, while maintaining the nanoscale definition of the designed features. When electron beam evaporation of metallic species is coupled with this ability to reproducibly fabricate substrates with defined nanotopographies, electroconductive properties can easily be imparted without the need for complex chemistries, leading to the development of a unique platform (Fig. 1).
Figure 1.
A schematic illustration of the fabrication of (A) unpatterned and patterned PUA substrates and (B) of electron beam evaporation deposition of Au and Ti onto PUA substrates.
For this study, we utilized nanopatterning featuring parallel grooves and ridges 800 nm in width and 600 nm in height, as not only does this mimic the spacing and organization of the collagen fiber bundles that make up skeletal muscle ECM,[20] but this has also been shown in our previous studies to produce an enhancement in primary myoblast maturation.[43] The nanopatterned PUA substrates demonstrated well-defined ridge-and-groove structures (Fig. 2A), and the approximately 30 nm-thick metal layers deposited via electron beam evaporation did not significantly alter the initial fidelity of these patterned topographies (Fig. 2B, 2C). Not only were these thin layers of deposited metals able preserve the topographical cues presented by the substrates, but they also were able to impart electroconductive properties as well. We observed changes in surface electrical conductivity with respect to the metal coating, or lack thereof (Fig. 2D). Bare (un-coated) PUA substrates had no detectable electrical conductivity, while PUA substrates coated with a metal layer demonstrated significantly greater electrical conductivity, with values of 2.0 × 105 S/m and 1.2 × 103 S/m obtained from the gold and titanium coated PUA surfaces, respectively. Measured conductivity values were consistent regardless of nanofeature orientation relative to four-point probe alignment, an indication that topographical anisotropy does not have a significant effect on global surface conductivity.
Figure 2.
SEM images of nanopatterned (A) PUA, (B) Au-PUA, (C) Ti-PUA, respectively. Scale bars in top-down images indicate 5 μm, while scale bars in inset cross-section images indicate 1 μm. Thickness of Ti and Au coating on PUA substrates was measured to be approximately 30 nm. (D) The electrical conductivity of various unpatterned and patterned PUA substrates. (E) Representative images of an approximately 5 μL drop of water on unpatterned and patterned PUA substrates with and without Ti or Au coating. Presented water contact angle values are means ± SD, n = 10, and there were no significant differences found
It is well known that cell reorganization, motility, and adhesion is greatly affected by substrate surface wettability.[44–47] Therefore, to ensure that changes in myoblast morphology and maturation would be due to differences in conductivity and topographies, rather than surface chemistries, surface wettability measurements on unpatterned and patterned substrates with and without metallic coatings were taken. No significant differences were found between all sample groups (Fig. 2E), indicating that cell adhesion was not affected significantly by the different metallic coatings, and that the effects on cellular differentiation was likely due to surface conductivity.
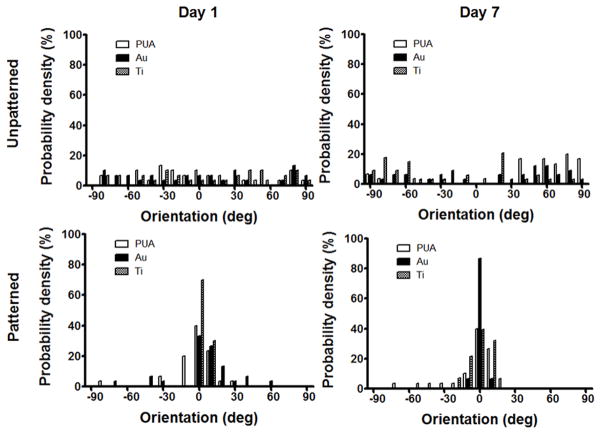
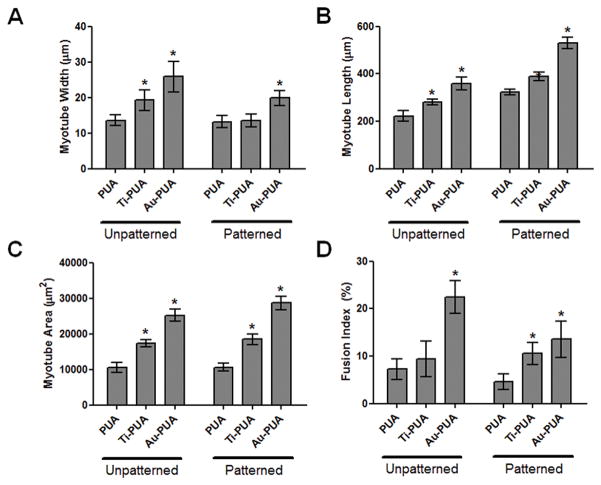
Myotube formation is a highly orchestrated terminal differentiation process in which the proliferating mononucleated myoblasts differentiated and fused to multinucleated myotubes. In previous studies, we had observed that nanopatterning induced the anisotropic and parallel alignment of cultured myocytes,[43, 48, 49] and the results of this study confirm the beneficial effects of such nanotopographies. Patterned substrates with and without metal coatings exhibited enhanced myotube formation over unpatterned counterparts. However, myotubes formed on both electrically conductive substrates after 7 days were larger and more numerous (Fig. 3A, 3B). Additionally, cell alignment on nanopatterned Au-PUA substrates was significantly greater than on nanopatterned Ti-PUA and PUA substrates, with matrix topography exerting a significant effect within 24 hours of cell introduction, regardless of electroconductivity (Fig. 4). Myoblasts expressed myosin heavy chain, a marker for more mature skeletal muscle cells, and fused into multinucleated myotubes on the electrically conductive substrates, and greater numbers of multinucleated myotubes were observed on the electrically conductive unpatterned and patterned PUA substrates compared to bare PUA substrates (Fig. 5). The effect of electrically conductive substrates on the differentiation of myoblasts (the width, length, area, and fusion index of multinucleated myotubes) were also measured (Fig. 6) by ImageJ software. The width and length of myotubes on the electrically conductive substrates were significantly greater than those cultured on bare non-conductive PUA substrates. Similarly, myotube area was significantly increased on electrically conductive unpatterned and patterned PUA substrates compared to bare PUA substrates. Myotube area on Au and Ti coated PUA substrates were approximately 2.5 and 1.8 times greater than that on the bare PUA substrates. Furthermore, the formations of myotubes on electrically conductive substrates also exhibited increased multinuclear fusion compared to bare PUA substrates.
Figure 3.
Bright-field microscopy showing attachment, growth, and alignment of C2C12 cells on (A) unpatterned and (B) patterned PUA substrates after 1, 3, and 7 days (from top to bottom). After 24 hours of culture, media was changed to one containing horse serum for inducing differentiation, and cells were cultured for an additional 6 days. Double-headed yellow arrows indicate direction of nanopatterning. Scale bars indicate 100 μm.
Figure 4.
Quantitative analysis of overall orientation of cells cultured on unpatterned and patterned bare and electroconductive PUA substrates after 1 and 7 days as performed by using a MATLAB script in conjunction with ImageJ software.
Figure 5.

Representative fluorescent images of C2C12 cells cultured on (A) unpatterned and (B) patterned PUA substrates at 7 days. Cells were stained for sarcomeric-myosin heavy chain (green), F-actin (red), and nuclei (blue). Scale bars indicate 50 μm.
Figure 6.
Morphometric analysis of the C2C12 cells cultured on various unpatterned and patterned PUA substrates at 7 days. (A) myotube width, (B) myotube length, (C) myotube area, and (D) fusion index. The fusion index was calculated as the ratio of the nuclei number in myocytes with two or more nuclei versus the total number of nuclei. Measurements are represented as means ± SD. Student’s t-test *P < 0.05.
To confirm the differentiation of cultured cells towards skeletal muscle, quantitative RT-PCR analysis was performed on three genes recognized as important for skeletal muscle development (Fig. 7). Myf5 and MyoD expression is initiated upon cell activation, and MyoD expression is sustained in proliferating and differentiating progeny, while MyoG expression is associated specifically with late-stage myogenic differentiation.[50–52] There was a higher expression of myogenic regulatory genes Myf5, MyoD, and MyoG on the electrically conductive substrates compared to non-conductive bare PUA substrates. Most importantly, cells cultured on the unpatterned and patterned Au-PUA substrates showed a significantly enhanced expression of these genes compared to bare PUA substrates. The expression of almost all the tested genes for cells cultured on Au-PUA substrates was 2 to 4 times higher expression than cells from bare PUA substrates.
Figure 7.

Quantitative RT-PCR analysis of C2C12 cells cultured on substrates after 7 days. Genes assayed are markers for myogenic differentiation and maturation. Myf5 and MyoD expression is initiated upon cell activation, and MyoD expression is sustained in proliferating and differentiating progeny, while MyoG expression is associated specifically with late-stage myogenic differentiation. Relative expression levels are represented as means ± SD. Student’s t-test *P< 0.05.
Interestingly, in these results from the genetic analyses of C2C12 cells cultured on electroconductive substrates, electroconductivity appears to be the dominant factor in play in regards to producing significant differences in myotube dimensions, fusion, and myogenic gene expression. A potential mechanism for the observed phenomenon involves the regulation of calcium levels within cultured myoblasts. Calcium is an intracellular messenger that is important in muscle cells for excitation-contraction coupling, as well as for modulating many aspects of cell physiology.[53, 54] Calcium-signaling pathways modulate the expression of muscle-specific genes during differentiation by altering the phosphorylation and activity of a number of transcriptional factors.[53, 55] Indeed, when L-type calcium channels in C2C12 myoblasts were blocked, thereby reducing intracellular calcium levels, it was observed that differentiation was significantly inhibited.[56] It has also been found that exposure of skeletal myoblasts to chronic electrical stimulation results in greater proliferation and maturation, and that these responses may be regulated by L-type calcium channels present in the cell membrane. When these channels are blocked, beneficial effects on proliferation are not observed even when myoblasts are subjected to the same electrical stimulation as before.[57] Therefore, by culturing skeletal myoblasts on electroconductive substrates, intracellular calcium levels are increased, leading to enhanced myogenic differentiation. At the same time, the underlying nanotopography still plays an important role in establishing the structural organization required for the generation of physiologically-relevant and mature muscle tissues. Each tissue in the body possesses distinct structural differences which play important roles in their function, and in skeletal muscle, parallel bundles of multinucleated myotubes and collagen allow for the propagation of electrical signals and force generation along a unified axis. These topographical cues are mimicked by the nanopatterned substrates and are shown to direct cells to align anisotropically through contact guidance cues, as regulated by cytoskeletal actin filament organization,[58] thereby recapitulating native tissue structure.
Thus, while there is not a true synergistic response observed, the goals of simultaneously inducing greater cell alignment and enhanced differentiation of myoblasts were still achieved with this platform. It should be noted, however, that these results were obtained using an immortalized cell line, which in many studies have been shown to behave and mature quite differently from primary myoblasts.[59] Specifically, the lack of an active progenitor or satellite cell population in these cultures could very well inhibit the degree of maturation achievable. Future work will therefore investigate this further by utilizing cultures of primary mononuclear cells isolated from muscle, and it is anticipated that the combination of ordered bio-inspired nanotopography and electroconductive coatings will indeed result in a compounding effect on differentiation and subsequent skeletal muscle tissue formation, although mechanistic studies into the underlying biology should be further examined. Additional studies will also examine the role of the modulus and thickness of electron beam evaporated metal coatings on the mechanical and electrical properties of substrates, and how these factors in turn mediate the differentiation of myoblasts and other electrically-sensitive cell types.
3. Conclusion
In this study, we have demonstrated a novel method of generating electroconductive nanopatterned substrates with lithographically-defined nanoscale features in a cost-effective, scalable and reproducible manner, and the feasibility of this platform for use in engineering electroactive tissues such as skeletal muscle. By combining CFL and electron beam evaporation, bioinspired substrates with high topographical fidelity and high electrical conductivity could be fabricated with ease. Multinucleated myotubes were formed more readily on conductive unpatterned and patterned substrates compared to non-conductive substrates, but were significantly more aligned and organized on patterned substrates, regardless of conductivity. Electroconductive substrates were shown induce the increased expression of genes that play significant roles in myogenic differentiation, indicating that substrate conductivity could play an important role in engineering functional and biomimetic skeletal muscle tissues. With the versatility afforded by this platform in regards to topographical design and metal species deposition, this functional scaffold can have a broad range of applications in engineering mature muscle (skeletal, smooth, or cardiac) or neural tissues, and can be used to generate tissue constructs for in vitro drug-screening studies or for therapeutic purposes. The findings from this study could also be valuable in the context of biomedical device or instrument design, as Au-coated nanopatterned substrates could be utilized to differentiate cultured cells while simultaneously measuring their electrophysiological responses to external stimuli.
4. Experimental Section
Fabrication of polyurethane-acrylate master molds
Polyurethane-acrylate (PUA; MINS 301 RM, Minuta Tech, Gyeonggi, Korea) polymer molds were fabricated using capillary force lithography techniques as described elsewhere.[60, 61] Briefly, polymer master molds were created by first dispensing PUA precursor solution onto a patterned silicon wafer that was generated using standard photolithography. A nanopattern comprising a series of parallel ridges and grooves with 800 nm × 800 nm × 600 nm (ridge width × groove width × ridge height) dimensions was used for these studies. A poly(ethylene terephthalate) (PET; Skyrol®, SKC Co., Ltd., Seoul, Korea) film (thickness: 75 μm) was used as a supporting backplane and pressed on top of the PUA. A polymer replica of the silicon pattern was then formed by exposing the PUA to UV light for 30 seconds before removing the silicon wafer and exposing the patterned PUA to UV light overnight to complete curing.
Fabrication of unpatterned and patterned substrates
Substrates were fabricated using capillary force lithography in a similar manner to the PUA molds (Fig. 1A). Briefly, cover glasses (ø 25 mm, Fisher Scientific, Waltham, MA, USA) were washed with isopropyl alcohol for 30 min in a bath sonicator and dried under a nitrogen stream. Unpatterned PUA substrates were prepared by drop-dispensing PUA solution (30 μL) onto the cover glass, spreading the polymer evenly over the surface, and exposing it to UV light for 20 seconds. Nanopatterned PUA substrates were fabricated by pressing a PUA mold on top of the drop-dispensed PUA solution glass and exposing the assembly to UV for 20 seconds before removing the mold. After initial UV cures, both unpatterned and patterned samples were subjected to UV light exposure overnight for complete curing.
Deposition of titanium and gold
Ti and Au thin films were prepared by using an electron-beam evaporator (A-Tech System, Co., Ltd., Incheon, Korea) (Fig. 1B). The base pressure of the deposition chamber was 7.0 × 10−6 Torr. Pure Ti and Au tablets (Thifine Co., Ltd., Incheon, Korea) were loaded into the electron beam source and film thickness was monitored in situ by a quartz crystal thickness monitor. A deposition rate of 1.0 Å/s was used, and the final thickness of the film on substrates was 30 nm.
Surface imaging with scanning electron microscopy
The topography of substrates, with and without Ti or Au deposition, was observed using scanning electron microscopy (SEM; Sirion SEM, FEI, Hillsboro, OR, USA). The specimens were mounted on an aluminum stub using double-sided carbon tape. The non-conductive specimens were coated with a 10 nm thick Au/Pd alloy layer using an ion sputter coater prior to imaging. All samples were imaged with an acceleration voltage of 18 kV.
Electrical conductivity measurements
The electrical conductivity of Au-PUA and Ti-PUA substrates were measured using a four-point probe with dual configuration method and sheet resistance meter (FPP-2400, BEGA Technologies, Deajeon, Korea). The conductivity (σ, S · cm−1) of the electroconductive substrates (n = 10) was calculated based on the equation: σ = l/(Rs·S) (where l is the distance between reference electrodes, Rs is the bulk resistance of the membrane sample derived from an impedance analyzer, and S is the cross-sectional area of the sample) as previously reported.[62]
Surface wettability analysis
The water contact angles of substrates were measured to examine surface wettability in relation to topography and surface coating. The contact angles were measured by adding a droplet of deionized water (5 μL) onto a dry sample at room temperature and allowing the droplet to equilibrate for 10 sec. The water droplet images were captured by a CCD camera, and the mean contact angles were then calculated using ImageJ software analysis (NIH, Bethesda, MD).
Culture and differentiation of C2C12 myoblasts
The fabricated substrates were rehydrated in phosphate-buffered saline (PBS; Sigma) for 2 hours at room temperature, sterilized by a series of ethyl alcohol washes from 50% to 100%, and washed with PBS three times. All the substrates were coated with fibronectin by incubating the substrates in a fibronectin solution (50 μg/mL) overnight. To remove unbound fibronectin, the surface was washed with PBS three times. C2C12 myoblasts were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco BRL, Gaithersburg, MD, USA) containing 10% (v/v) fetal bovine serum (Gibco BRL), 1% (v/v) penicillin-streptomycin (PS; Gibco BRL) under standard culture condition at 37°C in a humidified 5% CO2 atmosphere. Myoblasts were seeded onto substrates at a density of 6 × 104 cell/cm2 and cultured under growth media for 24 hours. To induce myotube formation, the media was then replaced with differentiation media (DMEM supplemented with 2% horse serum and 1% PS) and cultured for an additional 6 days.
Immunohistochemistry
Myotube formation was evaluated on cells fixed after six days of differentiation. The C2C12 cells cultured with differentiation media on bare PUA and electrically conductive PUA substrates were immunostained for sarcomeric-myosin heavy chain (MHC) and F-actin to indicate myotube formation. The cells were washed with PBS and fixed with 4% paraformaldehyde (Sigma-Aldrich, St Louis, MO, USA) at room temperature for 15 min and then gently washed with PBS and additional two times. Cell membranes were then permeabilized with 0.1% Triton X-100 (Sigma-Aldrich, St Louis, MO, USA) in PBS for five minutes. After rinsing three times with PBS, nonspecific binding sites were blocked with 5% bovine albumin serum with protein blocker solution (Sigma-Aldrich, St Louis, MO, USA) for 60 minutes at room temperature. Then, the samples were sequentially incubated with MF20 (1:50, Anti-MHC Alexa Fluor 488) and Alexa Fluor 594 conjugated anti-F-actin at 1:300 concentrations for 60 minutes at room temperature in a moist chamber. The stained samples were mounted with Vectashield with DAPI (Vector laboratories, Burlingame, CA) and images were captured with confocal microscopy (Nikon A1 confocal).
Cell Morphology Analyses
Myotube numbers were counted and averaged from 10 random fields of view per sample. The myotube area and length were measured using ImageJ software and averaged from 10 representative fluorescent fields of view per sample. The fusion index was determined by dividing the total number of nuclei in myotubes (≥2 nuclei) by the total number of counted nuclei for each image. Cell orientation was also measured using ImageJ software using a previously published MATLAB script utilizing pixel gradient analysis, and averaged from 10 representative images per sample.[63] Briefly, the images were passed through a Gaussian low-pass filter and Sobel horizontal edge-emphasized filter (predefined MATLAB Image Analysis toolbox functions) to generate a 2Dconvolution. The Sobel filter was then transposed to extract the vertical edge, and the horizontal and vertical edges were combined to calculate the gradient magnitude of each pixel in the image. The image was then thresholded to determine the borders of the areas of interest, and the orientation of the gradient was calculated via respect to the x-axis (0°). The orientation gradient data obtained from the pixel gradient analysis MATLAB script was then used to generate representative plots of overall cell alignment using a second custom MATLAB script. Each image was segmented into a grid of user-designated size. The gradient orientation angle for each pixel in a given grid square was then shifted in order to take a circular average with respect to the angle present with the highest incidence within that grid square’s orientation angle data. This circular average is representative of the mean orientation of each pixel gradient within a given gridsquare.
Quantitative RT-PCR
Gene expression of specific markers was evaluated via quantitative real time PCR and performed on C2C12 cultured on the different substrates after 6 days in differentiation medium, according to the following procedure. Total RNA were extracted from the C2C12 cultured on various substrates (n=4) using a protocol with RNeasy Minikit (Qiagen Sciences, Valencia, CA, USA). After RNA extraction, cDNA was prepared from 1 μg RNA using reverse transcription reagents. Quantitative PCR was performed using Power SYBR Green PCR Master Mix. The expression levels of relative myogenic differentiation genes (Myf5, MyoD, and MyoG) were measured by quantitative real-time-PCR (PCR system 7900HT). Quantification of target genes was performed relative to the reference GAPDH gene, using the following formula: relative expression = 2[−(Ct sample − Ct GAPDH)]. The mean minimal cycle threshold values (Ct) were calculated from quadruplicate reactions. Then, the relative gene expression in each experimental group was normalized to the relative gene expression found in the reference sample.
Statistical analysis
All quantitative data were expressed as the mean ± standard deviation. A one-way analysis of variance (ANOVA) using the Bonferroni test was performed on samples to determine significant differences. The assumptions for ANOVA were found to satisfy Levene’s test for homogeneity of variance and passed tests for normality. A value of p < 0.05 was considered statistically significant.
Acknowledgments
D. H. Kim acknowledges Department of Bioengineering at the University of Washington for the new faculty startup fund. This work was supported by a National Institutes of Health R21 Grant (R21AR064395) and a Muscular Dystrophy Association Research Grant (MDA255907). This work was also supported by the Basic Science Research Program through National Research Foundation of Korea (NRF) grants funded by the Korean Government the Ministry of Education, Science and Technology (MEST) (Grant Nos. 2010-0027771 and 2012-0003282), and by the Advanced Biomass R&D Center (ABC) of Global Frontier Project funded by MEST (ABC-2011-0031356). The authors thank Rachel Golan for her technical assistance in conducting the cellular orientation analyses.
Contributor Information
Hee Seok Yang, Department of Bioengineering, University of Washington, Seattle, WA 98195, USA. Department of Nanobiomedical Science and BK21 PLUS NBM Global Research Center, Dankook University, Cheonan, 330-714, Republic of Korea.
Bora Lee, Department of Chemical and Biomolecular Engineering and KAIST Institute for the NanoCentury, Korea Advanced Institute Science and Technology, Daejeon, 305-701, Republic of Korea.
Jonathan H. Tsui, Department of Bioengineering, University of Washington, Seattle, WA 98195, USA
Jesse Macadangdang, Department of Bioengineering, University of Washington, Seattle, WA 98195, USA.
Seok-Young Jang, Department of Chemical and Biomolecular Engineering and KAIST Institute for the NanoCentury, Korea Advanced Institute Science and Technology, Daejeon, 305-701, Republic of Korea.
Prof. Sung Gap Im, Department of Chemical and Biomolecular Engineering and KAIST Institute for the NanoCentury, Korea Advanced Institute Science and Technology, Daejeon, 305-701, Republic of Korea
Prof. Deok-Ho Kim, Department of Bioengineering, University of Washington, Seattle, WA 98195, USA. Institute for Stem Cell and Regenerative Medicine, University of Washington, Seattle, WA 98109, USA
References
- 1.Vandenburgh HH, Karlisch P, Farr L. In Vitro Cell Dev Biol. 1988;24:166. doi: 10.1007/BF02623542. [DOI] [PubMed] [Google Scholar]
- 2.Musaro A, Giacinti C, Pelosi L, Dobrowolny G, Barberi L, Nardis C, Coletti D, Scicchitano BM, Adamo S, Molinaro M. Eur J Histochem. 2007;51(Suppl 1):35. [PubMed] [Google Scholar]
- 3.Carlson BM. Am J Anat. 1973;137:119. doi: 10.1002/aja.1001370202. [DOI] [PubMed] [Google Scholar]
- 4.Braund KG, McGuire JA, Lincoln CE. Vet Pathol. 1982;19:577. doi: 10.1177/030098588201900601. [DOI] [PubMed] [Google Scholar]
- 5.Galavazi G. Z Zellforsch Mikrosk Anat. 1971;121:531. doi: 10.1007/BF00560158. [DOI] [PubMed] [Google Scholar]
- 6.Karatzaferi C, Franks-Skiba K, Cooke R. Am J Physiol Regul Integr Comp Physiol. 2008;294:R948. doi: 10.1152/ajpregu.00541.2007. [DOI] [PubMed] [Google Scholar]
- 7.Mauro A, Adams WR. J Biophys Biochem Cytol. 1961;10(4 Suppl):177. doi: 10.1083/jcb.10.4.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen RE, Temm-Grove CJ, Sheehan SM, Rice G. Methods Cell Biol. 1997;52:155. doi: 10.1016/s0091-679x(08)60378-7. [DOI] [PubMed] [Google Scholar]
- 9.Bhang SH, Jeong SI, Lee TJ, Jun I, Lee YB, Kim BS, Shin H. Macromol Biosci. 2012;12:402. doi: 10.1002/mabi.201100333. [DOI] [PubMed] [Google Scholar]
- 10.Jiang L, Sun Y, Chen X. Small. 2012;8:333. doi: 10.1002/smll.201102275. [DOI] [PubMed] [Google Scholar]
- 11.Jun I, Jeong S, Shin H. Biomaterials. 2009;30:2038. doi: 10.1016/j.biomaterials.2008.12.063. [DOI] [PubMed] [Google Scholar]
- 12.Yoon SH, Mofrad MR. Biomaterials. 2011;32:7286. doi: 10.1016/j.biomaterials.2011.05.077. [DOI] [PubMed] [Google Scholar]
- 13.Florini JR, Ewton DZ, Coolican SA. Endocr Rev. 1996;17:481. doi: 10.1210/edrv-17-5-481. [DOI] [PubMed] [Google Scholar]
- 14.Bhang SH, Lee TJ, Yang HS, La WG, Han AM, Kwon YH, Kim BS. Biochem Biophys Res Commun. 2009;382:315. doi: 10.1016/j.bbrc.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 15.Campoccia D, Arciola CR, Cervellati M, Maltarello MC, Montanaro L. Biomaterials. 2003;24:587. doi: 10.1016/s0142-9612(02)00373-3. [DOI] [PubMed] [Google Scholar]
- 16.Park JY, Lee DH, Lee EJ, Lee SH. Lab Chip. 2009;9:2043. doi: 10.1039/b820955c. [DOI] [PubMed] [Google Scholar]
- 17.Ogut O, Granzier H, Jin JP. Am J Physiol. 1999;276:C1162. doi: 10.1152/ajpcell.1999.276.5.C1162. [DOI] [PubMed] [Google Scholar]
- 18.Rafolt D, Gallasch E, Fend M, Bijak M, Lanmuller H, Mayr W. Artif Organs. 2002;26:244. doi: 10.1046/j.1525-1594.2002.06943.x. [DOI] [PubMed] [Google Scholar]
- 19.Gilbert PM, Havenstrite KL, Magnusson KE, Sacco A, Leonardi NA, Kraft P, Nguyen NK, Thrun S, Lutolf MP, Blau HM. Science. 2010;329:1078. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillies AR, Lieber RL. Muscle Nerve. 2011;44:318. doi: 10.1002/mus.22094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trotter JA, Purslow PP. J Morphol. 1992;212:109. doi: 10.1002/jmor.1052120203. [DOI] [PubMed] [Google Scholar]
- 22.Calve S, Odelberg SJ, Simon HG. Dev Biol. 2010;344:259. doi: 10.1016/j.ydbio.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Science. 2004;303:1352. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 24.Kim DH, Provenzano PP, Smith CL, Levchenko A. J Cell Biol. 2012;197:351. doi: 10.1083/jcb.201108062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park J, Bauer S, Pittrof A, Killian MS, Schmuki P, von der Mark K. Small. 2012;8:98. doi: 10.1002/smll.201100790. [DOI] [PubMed] [Google Scholar]
- 26.Kim HN, Jiao A, Hwang NS, Kim MS, Kang do H, Kim DH, Suh KY. Adv Drug Deliv Rev. 2013;65:536. doi: 10.1016/j.addr.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang PY, Thissen H, Tsai WB. Biotechnol Bioeng. 2012;109:2104. doi: 10.1002/bit.24452. [DOI] [PubMed] [Google Scholar]
- 28.Altomare L, Riehle M, Gadegaard N, Tanzi MC, Fare S. Int J Artif Organs. 2010;33:535. doi: 10.1177/039139881003300804. [DOI] [PubMed] [Google Scholar]
- 29.Lee EA, Jung G, Im SG, Hwang NS. J Biomed Mater Res B Appl Biomater. 2014. [DOI] [PubMed] [Google Scholar]
- 30.Englebienne P, Van Hoonacker A. J Colloid Interface Sci. 2005;292:445. doi: 10.1016/j.jcis.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 31.Kuniaki N, Takeaki K, Soichiro S, Yuichiro I, Makoto K, Matsuhiko N. Lab Chip. 2011;11:513. [Google Scholar]
- 32.Salto C, Saindon E, Bolin M, Kanciurzewska A, Fahlman M, Jager EW, Tengvall P, Arenas E, Berggren M. Langmuir. 2008;24:14133. doi: 10.1021/la8028337. [DOI] [PubMed] [Google Scholar]
- 33.Dvir T, Timko BP, Brigham MD, Naik SR, Karajanagi SS, Levy O, Jin H, Parker KK, Langer R, Kohane DS. Nat Nanotechnol. 2011;6:720. doi: 10.1038/nnano.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang SJ, Omori N, Li F, Jin G, Hamakawa Y, Sato K, Nagano I, Shoji M, Abe K. Neurol Res. 2003;25:516. doi: 10.1179/016164103101201751. [DOI] [PubMed] [Google Scholar]
- 35.Tsutsumi M, Inoue K, Denda S, Ikeyama K, Goto M, Denda M. Cell Tissue Res. 2009;338:99. doi: 10.1007/s00441-009-0848-0. [DOI] [PubMed] [Google Scholar]
- 36.Balint R, Cassidy NJ, Cartmell SH. Acta Biomater. 2014;10:2341. doi: 10.1016/j.actbio.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 37.Thrivikraman G, Mallik PK, Basu B. Biomaterials. 2013;34:7073. doi: 10.1016/j.biomaterials.2013.05.076. [DOI] [PubMed] [Google Scholar]
- 38.Gilmore KJ, Kita M, Han Y, Gelmi A, Higgins MJ, Moulton SE, Clark GM, Kapsa R, Wallace GG. Biomaterials. 2009;30:5292. doi: 10.1016/j.biomaterials.2009.06.059. [DOI] [PubMed] [Google Scholar]
- 39.Lorenzon P, Giovannelli A, Ragozzino D, Eusebi F, Ruzzier F. Eur J Neurosci. 1997;9:800–808. doi: 10.1111/j.1460-9568.1997.tb01429.x. [DOI] [PubMed] [Google Scholar]
- 40.Szentivanyi AL, Zernetsch H, Menzel H, Glasmacher B. Int J Artif Organs. 2011;34:986. doi: 10.5301/ijao.5000062. [DOI] [PubMed] [Google Scholar]
- 41.Perl A, Reinhoudt DN, Huskens J. Adv Mater. 2009;21:2257. [Google Scholar]
- 42.Hui CY, Jagota A, Lin YY, Kramer EJ. Langmuir. 2002;18:1394. [Google Scholar]
- 43.Yang HS, Ieronimakis N, Tsui JH, Kim HN, Suh KY, Reyes M, Kim DH. Biomaterials. 2014;35:1478. doi: 10.1016/j.biomaterials.2013.10.067. [DOI] [PubMed] [Google Scholar]
- 44.Lampin M, Warocquier-Clerout R, Legris C, Degrange M, Sigot-Luizard MF. J Biomed Mater Res. 1997;36:99–108. doi: 10.1002/(sici)1097-4636(199707)36:1<99::aid-jbm12>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 45.Webb K, Hlady V, Tresco PA. J Biomed Mater Res. 1998;41:422–430. doi: 10.1002/(sici)1097-4636(19980905)41:3<422::aid-jbm12>3.0.co;2-k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Wachem PB, Beugeling T, Feijen J, Bantjes A, Detmers JP, van Aken WG. Biomaterials. 1985;5:403–408. doi: 10.1016/0142-9612(85)90101-2. [DOI] [PubMed] [Google Scholar]
- 47.Lee SJ, Khang G, Lee YM, Lee HB. J Colloid Interface Sci. 2003;259:228–235. doi: 10.1016/S0021-9797(02)00163-7. [DOI] [PubMed] [Google Scholar]
- 48.Kim DH, Lipke EA, Kim P, Cheong R, Thompson S, Delannoy M, Suh KY, Tung L, Levchenko A. Proc Natl Acad Sci U S A. 2010;107:565. doi: 10.1073/pnas.0906504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiao A, Trosper NE, Yang HS, Kim J, Tsui JH, Frankel SD, Murry CE, Kim DH. ACS Nano. 2014;8:4430. doi: 10.1021/nn4063962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ieronimakis N, Balasundaram G, Rainey S, Srirangam K, Yablonka-Reuveni Z, Reyes M. PLoS One. 2010;5:e10920. doi: 10.1371/journal.pone.0010920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kitzmann M, Carnac G, Vandromme M, Primig M, Lamb NJ, Fernandez A. J Cell Biol. 1998;142:1447. doi: 10.1083/jcb.142.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Venuti JM, Cserjesi P. Curr Top Dev Biol. 1996;34:169. doi: 10.1016/s0070-2153(08)60711-5. [DOI] [PubMed] [Google Scholar]
- 53.Frey N, McKinsey TA, Olson EN. Nat Med. 2000;6:1221–1227. doi: 10.1038/81321. [DOI] [PubMed] [Google Scholar]
- 54.McKinsey TA, Zhang CL, Olson EN. Curr Opin Genet Dev. 2001;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- 55.Balke CW, Shorofsky SR. Cardiovasc Res. 1998;37:290–299. doi: 10.1016/s0008-6363(97)00272-1. [DOI] [PubMed] [Google Scholar]
- 56.Porter GA, Makuck RF, Rivkees SA. J Biol Chem. 2002;277:28942–28947. doi: 10.1074/jbc.M203961200. [DOI] [PubMed] [Google Scholar]
- 57.Pedrotty DM, Koh J, Davis BH, Taylor DA, Wolf P, Niklason LE. Am J Physiol Heart Circ Physiol. 2005;288:H1620–H1626. doi: 10.1152/ajpheart.00610.2003. [DOI] [PubMed] [Google Scholar]
- 58.Kim DH, Han K, Gupta K, Kwon K, Suh KY, Levchenko A. Biomaterials. 2009:5433–5444. doi: 10.1016/j.biomaterials.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langelaan ML, Boonen KJ, Rosaria-Chak KY, van der Schaft DW, Post MJ, Baaijens FP. J Tissue Eng Regen Med. 2011;5:529. doi: 10.1002/term.345. [DOI] [PubMed] [Google Scholar]
- 60.Kim DH, Kshitiz, Smith RR, Kim P, Ahn EH, Kim HN, Marban E, Suh KY, Levchenko A. Integr Biol (Camb) 2012;4:1019. doi: 10.1039/c2ib20067h. [DOI] [PubMed] [Google Scholar]
- 61.Chaterji S, Kim P, Choe SH, Tsui JH, Lam CH, Ho DS, Baker AB, Kim DH. Tissue Eng Part A. 2014;20:2115. doi: 10.1089/ten.tea.2013.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li M, Guo Y, Wei Y, MacDiarmid AG, Lelkes PI. Biomaterials. 2006;27:2705. doi: 10.1016/j.biomaterials.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 63.Cho H, Jönsson H, Campbell K, Melke P, Williams JW, Jedynak B, Stevens AM, Groisman A, Levchenko A. PLoS Biol. 2007;5:e302. doi: 10.1371/journal.pbio.0050302. [DOI] [PMC free article] [PubMed] [Google Scholar]