Abstract
Background/Aims
Aberrant DNA methylation has a specific role in field cancerization. Certain molecular markers, including secreted frizzled-related protein 2 (SFRP2), tissue factor pathway inhibitor 2 (TFPI2), N-Myc downstream-regulated gene 4 (NDRG4) and bone morphogenic protein 3 (BMP3), have previously been shown to be hypermethylated in colorectal cancer (CRC). We aim to examine field cancerization in CRC based on the presence of aberrant DNA methylation in normal-appearing tissue from CRC patients.
Methods
We investigated promoter methylation in 34 CRC patients and five individuals with normal colonoscopy results. CRC patients were divided into three tissue groups: tumor tissue, adjacent and nonadjacent normal-appearing tissue. The methylation status (positive: methylation level >20%) of SFRP2, TFPI2, NDRG4, and BMP3 promoters was investigated using methylation-specific PCR.
Results
The methylation frequencies of the SFRP2, TFPI2, NDRG4 and BMP3 promoters in tumor/adjacent/nonadjacent normal-appearing tissue were 79.4%/63.0%/70.4%, 82.4%/53.6%/60.7%, 76.5%/61.5%/69.2%, 41.2%/35.7%/50.0%, respectively. The methylation levels of the SFRP,TFPI2, NDRG4 and BMP3 promoters in tumor tissues were significantly higher than those in normal-appearing tissue (SFRP2, p=0.013; TFPI2, p<0.001; NDRG4, p=0.003; BMP3, p=0.001). No significant correlation was observed between the methylation levels of the promoters and the clinicopathological variables.
Conclusions
The field effect is present in CRC and affects both the adjacent and nonadjacent normal-appearing mucosa.
Keywords: Colorectal neoplasms, DNA methylation, Field effect, Epigenomics
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer worldwide. Most CRC develops through an adenoma-carcinoma progression sequence, which suggests that the normal colorectal epithelium transforms into an adenoma, then progresses to cancer via the accumulation of progressive molecular changes, including both genetic and epigenetic alterations.1,2 Epigenetic changes, alterations in the regulation of gene expression that do not involve a change in the DNA sequence of the cell, are carried out via DNA methylation, histone modification and polycomb complex formation.3 With regard to the epigenetic alterations observed in CRC, aberrant DNA methylation has been extensively studied.4–7
In carcinogenesis, the “field effect” concept developed from the observation that survivors of certain cancers are prone to develop other malignancies of the same tissue type near the primary cancer.8 Epigenetic alteration has a specific role in the field effect and several studies have provided evidence that specific aberrant DNA methylation may be a potential marker of the CRC field effect.9,10 In the present study, we selected four previously demonstrated promoters, secreted frizzled-related protein 2 (SFRP2), tissue factor pathway inhibitor 2 (TFPI2), N-Myc downstream-regulated gene 4 (NDRG4) and bone morphogenic protein 3 (BMP3), to demonstrate the field effect in CRC.
MATERIALS AND METHODS
1. Sample collection and DNA preparation
The study was approved by the Institutional Review Board of Kangbuk Samsung Hospital. All patients provided written informed consent as required by the Institutional Review Board. None of the patients had clinically apparent polyposis syndrome or hereditary nonpolyposis colon cancer syndrome. Patients with inflammatory bowel disease, prior colorectal resection, a history of any cancer, or a major psychological illness were excluded from the study.
Tissue samples were obtained from 34 patients who underwent surgery for CRC and 5 normal subjects without CRC or adenoma who underwent colonoscopy at Kangbuk Samsung Hospital in Seoul, Korea from 2012 to 2013. We examined samples taken from the sigmoid colon of endoscopically normal subjects. We collected samples of primary CRC tissue (T), adjacent normal-appearing tissue (AN), and nonadjacent normal-appearing tissue (NN) from each patient with CRC. All samples of adjacent normal-appearing tissues and nonadjacent normal-appearing tissues were derived from tissue located 2 cm and 8 cm, respectively, from the tumor. The status of all tissue specimens was confirmed histologically. Clinical and pathologic data were obtained for all 34 patients with CRC.
2. Isolation of DNA and sodium bisulfite conversion
Formalin-fixed, paraffin-embedded tissues were mounted on glass slides and stained with hematoxylin and eosin. Microdis-section and DNA extraction were performed as previously described.11
Epithelium and tumor tissue were carefully microdissected using a microtome (RM2255; Leica, Nussloch, Germany). The dissected tissues were placed individually in 1.5-mL microcentrifuge tubes with phosphate-buffered saline and deparaffinized by heating for 5 minutes at 75°C. The mixtures obtained were then centrifuged at 13,000 rpm for 2 minutes, and the supernatants were removed. Pellets were mixed with DNA extraction buffer (Biosewoom, Seoul, Korea) and heated for 5 minutes at 56°C, and an additional 8 minutes at 100°C to destroy the cells and remaining tissues. The mixtures obtained were then centrifuged at 13,000 rpm for 2 minutes, and the supernatants, which contained DNA, were then used for further studies. Genomic DNA was chemically modified by sodium bisulfite to convert all unmethylated cytosines to uracils while leaving the methylcytosines unaltered (EZ DNA MethylationTM kit; Zymo Research, Irvine, CA, USA).
3. Methylation-specific PCR
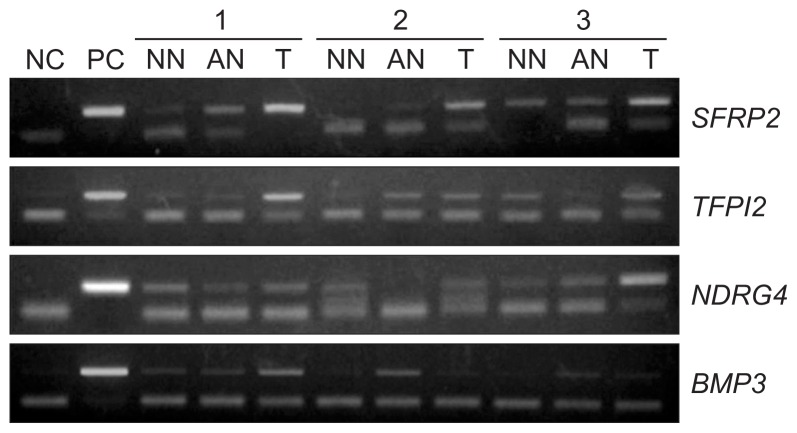
Methylation of the SFRP2, TFPI2, NDRG4 and BMP3 promoters in the bisulfite-modified DNA was investigated using methylation-specific PCR (MSP) with primer pairs designed to specifically amplify methylated or unmethylated alleles. The nucleotide sequences of the primers previously reported are listed in Table 1. Commercially available methylated human genomic DNA (CpGenomeTM Universal Methylated DNA; Chemicon International, Temecula, CA, USA) was used as a positive control for unmethylated and methylated alleles and reagents without the addition of DNA served as negative controls. The thermocycler conditions were, in general, as follows: 95°C for 15 minutes, 39 cycles of 95°C for 30 seconds, specific annealing temperature for 30 to 60 seconds, 72°C for 30 seconds, followed by a final extension at 72°C for 10 minutes (Table 1). The MSP products were then subjected to horizontal gel electrophoresis through 1.2% agarose gel, stained with ethidium bromide and visualized with UV transillumination by using the Quality One Image Analyzer system (Bio-Rad, Hercules, CA, USA) (Fig. 1). Normalization of methylation level (%) was defined based on the following calculation: (Measured-negative control/positive control-negative control).
Table 1.
Summary of the Primer Sequences, Polymerase Chain Reaction (PCR) Product Sizes and Annealing Temperatures Used for Methylation-Specific PCR Assays
| Gene | Primer sequence (5′→3′) | PCR product size, bp | Annealing temperature, °C | |
|---|---|---|---|---|
| SFRP2 | M | S: GGGTCGGAGTTTTTCGGAGTTGCGC | 138 | 62 |
| A: CCGCTCTCTTCGCTAAATACGACTCG | ||||
| U | S: TTTTGGGTTGGAGTTTTTTGGAGTTGTGT | 145 | 50 | |
| A: ACCCACTCTCTTCACTAAATACAACTCA | ||||
| BMP3 | M | S: GTTTGGAGTTTAATTTTCGGTTTC | 179 | 54 |
| A: ATAACTTCGATCTCTCTCCCTACG | ||||
| U | S: GGTTTGGAGTTTAATTTTTGGTTTT | 178 | 54 | |
| A: AACTTCAATCTCTCTCCCTACACC | ||||
| NDRG4 | M | S: TTTAGGTTCGGTATCGTTTCGC | 110 | 61 |
| A: CGAACTAAAAACGATACGCCG | ||||
| U | S: GATTAGTTTTAGGTTTGGTATTGTTTTGT | 105 | 61 | |
| A: AAAACCAAACTAAAAACAATACACCA | ||||
| TFPI2 | M | S: ATTTTTTAGGTTTCGTTTCGGC | 118 | 57 |
| A: GCCTAACGAAAAAAAATACGCG | ||||
| U | S: TTAGTTATTTTTTAGGTTTTGTTTTGGT | 105 | 57 | |
| A: AAAACACCTAACAAAAAAAAATACACA |
bp, base pair; SFRP2, secreted frizzled-related protein 2; M, methylated; S, sense; A, antisense; U, unmethylated; BMP3, bone morphogenic protein 3; NDRG4, N-Myc downstream-regulated gene 4; TFPI2, tissue factor pathway inhibitor 2.
Fig. 1.
Representative methylation-specific polymerase chain reaction of promoters in tissues. (a) SFRP2; (b) TFPI2; (c) NDRG4; and (d) BMP3.
NC, negative control; PC, positive control; NN, nonadjacent normal-appearing tissue; AN, adjacent normal-appearing tissue; T, primary colorectal tumor tissue; SFRP2, secreted frizzled-related protein 2; TFPI2, tissue factor pathway inhibitor 2; NDRG4, N-Myc downstream-regulated gene 4; BMP3, bone morphogenic protein 3.
4. Statistics
Presence of methylated promoters was analyzed initially as a categorical variable (negative, methylation level <20%; positive, methylation level >20%). The cutoff value was selected because lower marginal values could not be distinguished from background staining of the gels, as described in the previous study.12 We analyzed the levels of methylation as a continuous variable. We computed means, standard deviations, medians, and ranges with levels of methylation and analyzed data with one-way analysis of variance followed by Tukey’s posttest. The differences in the methylation frequency of each promoter between patients with CRC and normal subjects were analyzed using the Fisher exact test. The association between levels of methylated promoter and clinicopathological variables was analyzed using the Mann-Whitney test or Kruskal-Wallis test. All reported p-values were two-sided, and p-values <0.05 indicated statistical significance. Statistical analysis was performed using PASW 18.0 version software (SPSS Inc., Chicago, IL, USA).
RESULTS
A total of 39 samples from 34 patients with CRC (mean age, 57.0 years old; range, 36 to 80 years; 22 male and 12 female) and five normal subjects (mean age, 68.2 years old; range, 56 to 86 years; three male and two female) were analyzed. Clinicopathologic features of the patients with CRC are shown in Table 2. Nodal spread and distant metastasis were detected in 17.6% and 32.4%, respectively.
Table 2.
Clinicopathological Features of the Patients
| Clinicopathologic feature | No. of patients (%) |
|---|---|
| Total no. | 34 |
| Age, yr | |
| ≤60 | 22 (64.7) |
| >60 | 12 (35.3) |
| Sex | |
| Male | 22 (64.7) |
| Female | 12 (35.3) |
| Smoking | |
| Nonsmoker | 26 (76.5) |
| Ex-smoker | 5 (14.7) |
| Current smoker | 3 (3.0) |
| TNM stage* | |
| T1/T2/T3/T4 | 2 (5.9)/4 (11.8)/24 (70.6)/4 (11.8) |
| N0/N1/N2 | 16 (47.1)/12 (35.3)/6 (17.6) |
| M0/M1 | 23 (37.6)/11 (32.4) |
| Stage I/II/III/IV | 5 (14.7)/8 (23.5)/11 (32.4)/10 (29.4) |
T, tumor; N, node; M, metastasis.
Tumor stage was determined with the use of the American Joint Committee on Cancer (AJCC) staging system (2009).
1. Methylation frequency and levels of methylation of the SFRP2 promoter
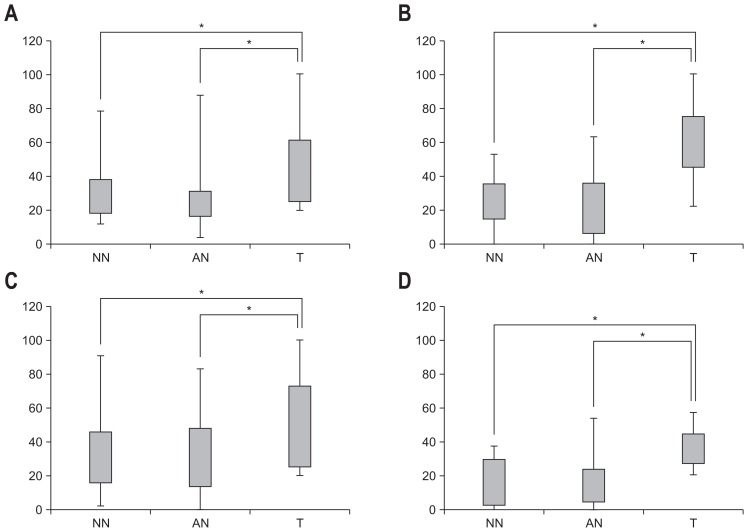
When analyzed as a categorical variable, promoter hypermethylation of SFRP2 in tumor tissues was observed in 27 of the 34 patients with CRC (79.4%). Among these 27 patients with methylation-positive tumor tissue, promoter hypermethylation of SFRP2 was observed in adjacent normal-appearing tissue in 17 (63.0%) patients and in 19 nonadjacent normal-appearing tissue samples (70.4%). Levels of methylation in tumor tissue, adjacent normal-appearing tissue, and nonadjacent normal-appearing tissue are shown in Table 3 and Fig. 2. Levels of methylated SFRP2 promoter in tumor tissue were significantly higher than in adjacent normal-appearing tissue or nonadjacent normal-appearing tissue (p=0.013).
Table 3.
Methylation Levels of SFRP2, TFPI2, NDRG4, and BMP3
| Mean±SD | p-value | ||
|---|---|---|---|
|
| |||
| Tukey’s posttest | One-way ANOVA | ||
| SFRP2, % | |||
| NN | 29.9±16.5 | 0.039 | 0.013 |
| AN | 29.4±21.3 | 0.021 | |
| T | 49.8±26.1 | ||
| TFPI2, % | |||
| NN | 24.1±14.6 | 0.000 | 0.000 |
| AN | 23.6±18.4 | 0.000 | |
| T | 59.5±21.4 | ||
| NDRG4, % | |||
| NN | 33.8±21.8 | 0.024 | 0.003 |
| AN | 30.0±21.9 | 0.004 | |
| T | 51.8±27.5 | ||
| BMP3, % | |||
| NN | 16.9±13.6 | 0.003 | 0.001 |
| AN | 17.9±16.3 | 0.004 | |
| T | 35.7±10.8 | ||
SFRP2, secreted frizzled-related protein 2; TFPI2, tissue factor pathway inhibitor 2; NDRG4, N-Myc downstream-regulated gene 4; BMP3, bone morphogenic protein 3; ANOVA, analysis of variance; NN, non-adjacent normal appearing tissue; AN, adjacent normal appearing tissue; T, primary tumor tissue.
Fig. 2.
Distribution of methylation level (%) for nonadjacent normal-appearing tissue (NN), adjacent normal-appearing tissue (AN), and primary colorectal tumor tissue (T) in (A) SFRP2, (B) TFPI2, (C) NDRG4, and (D) BMP3, respectively.
SFRP2, secreted frizzled-related protein 2; TFPI2, tissue factor pathway inhibitor 2; NDRG4, N-Myc downstream-regulated gene 4; BMP3, bone morphogenic protein 3. *p<0.05.
2. Methylation frequency and levels of methylation of the TFPI2 promoter
Promoter hypermethylation of TFPI2 in tumor tissue was observed in 28 of the 34 patients with CRC (82.4%). Of these 28 patients, promoter hypermethylation of TFPI2 was also observed in adjacent normal-appearing tissue in 15 (53.6%) patients and in 17 nonadjacent normal-appearing tissue samples (60.7%). Levels of methylated TFPI2 promoter in tumor tissue appeared to be significantly higher than in adjacent normal-appearing tissue or nonadjacent normal-appearing tissue (p<0.001).
3. Methylation frequency and levels of methylation of the NDRG4 promoter
For the NDRG4 promoter, promoter hypermethylation in tumor tissue was observed in 26 of the 34 patients with CRC (76.5%). Of these 26 patients, promoter hypermethylation of NDRG4 was observed in adjacent normal-appearing tissue in 16 patients (61.5%) and in 18 nonadjacent normal-appearing tissue samples (69.2%). Levels of methylated NDRG4 in tumor tissue were significantly higher than in adjacent normal-appearing tissue or nonadjacent normal-appearing tissue (p=0.003).
4. Methylation frequency and levels of methylation of the BMP3 promoter
Promoter hypermethylation of BMP3 in tumor tissue was observed in 14 out of the 34 patients with CRC (41.2%). Of these 14 patients, methylation frequency of the BMP3 promoter was observed in five adjacent normal-appearing tissue samples (35.7%) and in seven nonadjacent normal-appearing tissue samples (50.0%). Levels of methylated BMP3 in tumor tissue were significantly higher than both normal-appearing tissues (p=0.001).
5. Promoter methylation frequency of individual genes in normal subjects and methylation-negative CRC patients
Normal subjects did not exhibit methylation of each tested promoter. Mean levels of methylated promoters in normal subjects were observed to be low (SFRP2, 1.6%; TFPI2, 0.23%; NDRG4, 1.72%; BMP3, 0.86%). Promoter hypermethylation of SFRP2, TFPI2, NDRG4 and BMP3 in tumor tissues was not observed in seven (20.5%), six (17.6%), eight (23.5%), and 20 (58.8%) patients. Of these patients, the methylation frequency of SFRP2, TFPI2, NDRG4 and BMP3 was 0%/42.8% (3/7), 0%/50% (3/6), 37.5% (3/8) and 0%/10% (2/20) in adjacent/nonadjacent normal-appearing tissues respectively.
6. Levels of methylated promoters in tumor tissue and clinicopathologic features
The association between methylated promoters in tumor tissue and clinicopathologic features is shown in Table 4. There was no statistically significant association observed between the methylated promoters in tumor tissue and patient age, sex, smoking status, tumor size, nodal status, metastasis or TNM stage. In correlation analysis of methylation level and age, there was a trend toward a higher level of SFRP2 (58.2 vs 43.9, p=0.08) and NDRG4 (59.7 vs 37.3, p=0.07) in age ≥60 compared to age <60 patients. In correlation analysis of methylation frequency and level with tumor location, there was a trend toward a higher frequency and level of NDRG4 (frequency, 100% vs 71.4%, p=0.29; level, 54.6 vs 43.5, p=0.46) and BMP3 (frequency, 66.7% vs 35.7%, p=0.20; level, 34.5 vs 30.0, p=0.57) promoters in the right colon than the left colon. There was no difference in methylation frequency or the levels of the four promoters between the colon and rectum (data not shown).
Table 4.
Clinicopathological Features and Methylation of SFRP2, TFPI2, NDRG4 and BMP
| Clinicopathological feature | SFRP2 | TFPI2 | NDRG4 | BMP3 | ||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||
| No. (%) | p-value | No. (%) | p-value | No. (%) | p-value | No. (%) | p-value | |
| Total positive | 27 | 28 | 26 | 14 | ||||
| Age, yr | 0.681 | 0.578 | 0.077 | 0.225 | ||||
| ≤60 | 17 (63.0) | 17 (60.7) | 16 (61.5) | 11 (78.6) | ||||
| >60 | 10 (37.0) | 11 (39.3) | 10 (38.5) | 3 (21.4) | ||||
| Sex | 0.624 | 0.498 | 0.070 | 0.662 | ||||
| Male | 19 (70.4) | 19 (67.9) | 18 (69.2) | 8 (57.1) | ||||
| Female | 8 (29.6) | 9 (32.1) | 8 (30.8) | 6 (32.9) | ||||
| Smoking | 0.490 | 0.902 | 0.811 | 0.468 | ||||
| Nonsmoker | 19 (70.4) | 22 (78.6) | 19 (73.1) | 11 (78.6) | ||||
| Ex-smoker | 5 (18.5) | 4 (14.3) | 4 (15.4) | 2 (14.3) | ||||
| Current smoker | 3 (11.1) | 2 (7.1) | 3 (11.5) | 1 (7.1) | ||||
| Tumor depth | 0.344 | 0.416 | 0.364 | 0.498 | ||||
| T1 | 2 (7.4) | 2 (7.1) | 1 (3.8) | 0 | ||||
| T2 | 4 (14.8) | 3 (10.7) | 4 (15.4) | 3 (21.4) | ||||
| T3 | 17 (63.0) | 19 (67.9) | 17 (65.4) | 10 (71.5) | ||||
| T4 | 4 (14.8) | 4 (14.3) | 4 (15.4) | 1 (7.1) | ||||
| Nodal status | 0.180 | 0.853 | 0.150 | 0.205 | ||||
| N0 | 14 (51.9) | 13 (46.4) | 11 (42.3) | 9 (64.3) | ||||
| N1 | 8 (29.6) | 11 (39.3) | 9 (34.6) | 5 (35.7) | ||||
| N2 | 5 (18.5) | 4 (14.3) | 6 (23.1) | 0 | ||||
| Metastasis | 0.109 | 0.382 | 0.979 | 0.839 | ||||
| M0 | 19 (70.4) | 18 (64.3) | 15 (57.7) | 10 (71.4) | ||||
| M1 | 8 (29.6) | 10 (35.7) | 11 (42.3) | 4 (28.6) | ||||
| TNM stage* | 0.120 | 0.399 | 0.092 | 0.798 | ||||
| I | 5 (18.5) | 4 (14.3) | 4 (15.4) | 2 (14.2) | ||||
| II | 6 (22.3) | 6 (21.4) | 4 (15.4) | 4 (28.6) | ||||
| III | 8 (29.6) | 8 (28.6) | 8 (30.8) | 4 (28.6) | ||||
| IV | 8 (29.6) | 10 (35.7) | 10 (38.4) | 4 (28.6) | ||||
SFRP2, secreted frizzled-related protein 2; TFPI2, tissue factor pathway inhibitor 2; NDRG4, N-Myc downstream-regulated gene 4; BMP3, bone morphogenic protein 3; T, tumor; N, node; M, metastasis.
Tumor stage was determined with the use of the American Joint Committee on Cancer (AJCC) staging system (2009).
DISCUSSION
In this study, hypermethylation of SFRP2, TFPI2, NDRG4, and BMP3 was observed in normal-appearing tissue of patients with CRC and demonstrated the field effect in CRC. The methylation frequency in normal-appearing tissue was over 50% for each promoter. In addition, we also showed that the field effect was observed not only in adjacent normal-appearing tissue, but also in nonadjacent normal-appearing tissue.
In colorectal carcinogenesis, the possibility of field cancerization was first proposed due to the increased occurrence of flat dysplasia and CRC in patients with inflammatory bowel disease.13 In cases of sporadic CRC, individuals who had a personal history of colon adenoma or adenocarcinoma were at increased risk of developing metachronous adenoma or adenocarcinoma14 and these results supported that the field effect may occur in the colon and could consequently increase the risk of CRC. Recent studies showed that there were several changes such as increased occurrence of chromosomal aberrations or aberrant DNA methylation in the normal colon mucosa adjacent to colon cancer.15
Aberrant DNA methylation is a key mechanism of tumor suppressor gene inactivation in certain malignancies including CRC, and many genes that are targets of aberrant methylation have been identified.4 However, only a few studies have demonstrated the field effect and assessed the methylation status of specific loci in normal colon mucosa. In previous studies, methylation of five genes (RUNX3, SOCS1, NEUROG1, CACNA1G, and IGF2) was found to be increased in the morphologically normal colon mucosa of patients with advanced proximal sessile polyps, a precursor lesion to CpG island methylator phenotype (CIMP) cancers.16 In addition to CIMP genes, methylation of the O6-methylguanine-DNA methyltransferase (MGMT) gene promoter was detected in normal-appearing mucosa adjacent to CRC, which was methylated in 46% of colorectal tumors and in 26% of corresponding adjacent normal-appearing mucosa.12 Grady et al.17 suggested another locus, EVL/miR-342, to be a marker of field cancerization in the colon, and found methylation in 86% of colorectal adenocarcinomas, 56% of histologically normal colorectal mucosa 10 cm away from CRC, and in 12% of normal colorectal mucosa from individuals without CRC.
We selected four promoters, SFRP2,18–20 TFPI2,21 NDRG4,22 and BMP3,23 as they previously showed aberrant DNA methylation in CRC tumor tissue with high sensitivity and specificity compared with normal subjects and were included in the recent multitarget stool DNA test.24,25 Aberrant Wnt pathway signaling is observed in approximately 90% of CRC tumors; SFRPs possess a domain similar to Wnt-receptor frizzled proteins and can inhibit Wnt receptor binding to downregulate pathway signaling during development.26,27 In a previous study, methylation of the SFRP2 promoter was present in over 60% of advanced CRC cases and in less than 3.1% of normal subjects.19 TFPI2 is a Kunitz-type serine proteinase inhibitor that protects the extracellular matrix of cancer cells from degradation and inhibits in vitro colony formation and proliferation.28,29 Methylation of TFPI2 was detected with a high frequency in over 62% of CRC patients, and TFPI2 was more frequently methylated in well-differentiated colorectal carcinomas and lymph node metastases.21 NDRG4 is a member of the NDRG protein family that showed 57% to 65% amino acid sequence homology. NDRG4 was suggested to be a tumor suppressor gene in CRC whose expression is frequently inactivated by promoter methylation.30 In a previous study, methylation of the NDRG4 gene was detected in over 86% of CRC patients and in less than 4% of normal subjects.22 BMPs are members of the TGFβ growth factor superfamily and disrupted BMP signaling in tumor development has recently been studied.23,31 BMP3 inactivation was observed in early-onset tumorigenesis of colorectal cancer and approximately 55% of CRC patients showed BMP3 promoter methylation.23 In this study, the methylation frequency of CRC was over 75% for each promoter except BMP3, which was in agreement with previous studies.19,21,22 However, even though there was a trend toward a higher level of methylation in older patients, there was no significant association between methylation status and clinicopathologic features.
Our results showed that the methylation frequency of normal-appearing tissue in patients was over 50% for each promoter, thus demonstrating the field effect. To evaluate the extent of the field effect, we obtained normal-appearing mucosal specimens that were located 2 and 8 cm from the CRC, and provided evidence that the field effect was also observed in nonadjacent normal-appearing tissue in patients with CRC. These results are consistent with a previous study in which MGMT methylation was detected 10 cm away from tumor tissue in 77% of cases.12 In addition, we compared methylation frequencies and levels between adjacent and nonadjacent normal-appearing tissues, but the results were similar. These findings suggest that the field effect occurs in CRC, but the mechanism of tumorigenesis from the cancerized field is not yet clear.
There are several limitations to our study. First, this study examined a limited number of cases. A large study should be performed to examine the field effect in CRC with the genes investigated in this study. Second, the distance between adjacent and nonadjacent normal appearing tissue might have been insufficient. In our study, 8 cm from CRC tissues was the maximum distance used due to the limitations associated with resected specimens. Although there is no standard distance between adjacent and nonadjacent tissues, samples further from CRC tissues should be examined to investigate the extent of the field effect. Third, real-time quantitative MSP was not available in our study. Further studies with more sensitive MSP assays are needed.
In conclusion, this study provides evidence that the field effect is present in CRC, and that it affects both adjacent and non-adjacent normal-appearing mucosa. The levels of methylated promoters and methylation frequency in CRC tumor tissues are higher than in adjacent and nonadjacent normal-appearing mucosa. Further research is needed to validate methylated promoters as a biomarker for CRC, to clarify the biological mechanisms of the field effect in CRC and to evaluate the usefulness of DNA methylation levels as a risk marker of CRC.
Footnotes
CONFLICTS OF INTEREST
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim ER, Kim YH. Clinical application of genetics in management of colorectal cancer. Intest Res. 2014;12:184–193. doi: 10.5217/ir.2014.12.3.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer: a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 4.Goel A, Boland CR. Epigenetics of colorectal cancer. Gastroenterology. 2012;143:1442–1460.e1. doi: 10.1053/j.gastro.2012.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim MS, Lee J, Sidransky D. DNA methylation markers in colorectal cancer. Cancer Metastasis Rev. 2010;29:181–206. doi: 10.1007/s10555-010-9207-6. [DOI] [PubMed] [Google Scholar]
- 6.Kondo Y, Issa JP. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev. 2004;23:29–39. doi: 10.1023/A:1025806911782. [DOI] [PubMed] [Google Scholar]
- 7.Zou H, Harrington JJ, Shire AM, et al. Highly methylated genes in colorectal neoplasia: implications for screening. Cancer Epidemiol Biomarkers Prev. 2007;16:2686–2696. doi: 10.1158/1055-9965.EPI-07-0518. [DOI] [PubMed] [Google Scholar]
- 8.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium: clinical implications of multi-centric origin. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::AID-CNCR2820060515>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 9.Luo Y, Yu M, Grady WM. Field cancerization in the colon: a role for aberrant DNA methylation? Gastroenterol Rep (Oxf) 2014;2:16–20. doi: 10.1093/gastro/got039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein C, Bernstein H, Payne CM, Dvorak K, Garewal H. Field defects in progression to gastrointestinal tract cancers. Cancer Lett. 2008;260:1–10. doi: 10.1016/j.canlet.2007.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turenne CY, Tschetter L, Wolfe J, Kabani A. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J Clin Microbiol. 2001;39:3637–3648. doi: 10.1128/JCM.39.10.3638-3648.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen L, Kondo Y, Rosner GL, et al. MGMT promoter methylation and field defect in sporadic colorectal cancer. J Natl Cancer Inst. 2005;97:1330–1338. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- 13.Van Assche G, Dignass A, Bokemeyer B, et al. Second European evidence-based consensus on the diagnosis and management of ulcerative colitis part 3: special situations. J Crohns Colitis. 2013;7:1–33. doi: 10.1016/j.crohns.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Levi F, Randimbison L, Blanc-Moya R, et al. High constant incidence of second primary colorectal cancer. Int J Cancer. 2013;132:1679–1682. doi: 10.1002/ijc.27780. [DOI] [PubMed] [Google Scholar]
- 15.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 16.Worthley DL, Whitehall VL, Buttenshaw RL, et al. DNA methylation within the normal colorectal mucosa is associated with pathway-specific predisposition to cancer. Oncogene. 2010;29:1653–1662. doi: 10.1038/onc.2009.449. [DOI] [PubMed] [Google Scholar]
- 17.Grady WM, Parkin RK, Mitchell PS, et al. Epigenetic silencing of the intronic microRNA hsa-miR-342 and its host gene EVL in colorectal cancer. Oncogene. 2008;27:3880–3888. doi: 10.1038/onc.2008.10. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki H, Gabrielson E, Chen W, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31:141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki H, Watkins DN, Jair KW, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 20.Sui C, Wang G, Chen Q, Ma J. Variation risks of SFRP2 hypermethylation between precancerous disease and colorectal cancer. Tumour Biol. 2014;35:10457–10465. doi: 10.1007/s13277-014-2313-2. [DOI] [PubMed] [Google Scholar]
- 21.Hibi K, Goto T, Kitamura YH, et al. Methylation of TFPI2 gene is frequently detected in advanced well-differentiated colorectal cancer. Anticancer Res. 2010;30:1205–1207. [PubMed] [Google Scholar]
- 22.Melotte V, Lentjes MH, van den Bosch SM, et al. N-Myc downstream-regulated gene 4 (NDRG4): a candidate tumor suppressor gene and potential biomarker for colorectal cancer. J Natl Cancer Inst. 2009;101:916–927. doi: 10.1093/jnci/djp131. [DOI] [PubMed] [Google Scholar]
- 23.Loh K, Chia JA, Greco S, et al. Bone morphogenic protein 3 inactivation is an early and frequent event in colorectal cancer development. Genes Chromosomes Cancer. 2008;47:449–460. doi: 10.1002/gcc.20552. [DOI] [PubMed] [Google Scholar]
- 24.Ahlquist DA, Zou H, Domanico M, et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology. 2012;142:248–256. doi: 10.1053/j.gastro.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 26.Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer. 2001;1:55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- 27.Xu Q, D’Amore PA, Sokol SY. Functional and biochemical interactions of Wnts with FrzA, a secreted Wnt antagonist. Development. 1998;125:4767–4776. doi: 10.1242/dev.125.23.4767. [DOI] [PubMed] [Google Scholar]
- 28.Wong CM, Ng YL, Lee JM, et al. Tissue factor pathway inhibitor-2 as a frequently silenced tumor suppressor gene in hepatocellular carcinoma. Hepatology. 2007;45:1129–1138. doi: 10.1002/hep.21578. [DOI] [PubMed] [Google Scholar]
- 29.Sato N, Parker AR, Fukushima N, et al. Epigenetic inactivation of TFPI-2 as a common mechanism associated with growth and invasion of pancreatic ductal adenocarcinoma. Oncogene. 2005;24:850–858. doi: 10.1038/sj.onc.1208050. [DOI] [PubMed] [Google Scholar]
- 30.Zhou RH, Kokame K, Tsukamoto Y, Yutani C, Kato H, Miyata T. Characterization of the human NDRG gene family: a newly identified member, NDRG4, is specifically expressed in brain and heart. Genomics. 2001;73:86–97. doi: 10.1006/geno.2000.6496. [DOI] [PubMed] [Google Scholar]
- 31.Koinuma K, Kaneda R, Toyota M, et al. Screening for genomic fragments that are methylated specifically in colorectal carcinoma with a methylated MLH1 promoter. Carcinogenesis. 2005;26:2078–2085. doi: 10.1093/carcin/bgi184. [DOI] [PubMed] [Google Scholar]