Significance
Understanding divisome assembly and activation has become the focus of research on bacterial cytokinesis. However, very little is known about how this process is regulated. Here, we find that FtsEX (an ATP-binding cassette transporter-like complex) acts on the bacterial actin homolog FtsA to regulate divisome assembly and function in Escherichia coli. Our results suggest that FtsEX antagonizes FtsA polymerization to promote divisome assembly and continual ATP hydrolysis by FtsEX is needed for the divisome to synthesize septal peptidoglycan. Because FtsEX is also required for cell wall hydrolysis at the septum, our study indicates that FtsEX couples cell wall synthesis and hydrolysis at the septum by acting through FtsA. Our study also implies that unpolymerized FtsA is favored for division and FtsW plays a critical role in divisome activation.
Keywords: divisome, FtsZ, FtsA, FtsEX, Z ring
Abstract
Bacterial cell division is driven by the divisome, a ring-shaped protein complex organized by the bacterial tubulin homolog FtsZ. Although most of the division proteins in Escherichia coli have been identified, how they assemble into the divisome and synthesize the septum remains poorly understood. Recent studies suggest that the bacterial actin homolog FtsA plays a critical role in divisome assembly and acts synergistically with the FtsQLB complex to regulate the activity of the divisome. FtsEX, an ATP-binding cassette transporter-like complex, is also necessary for divisome assembly and inhibits division when its ATPase activity is inactivated. However, its role in division is not clear. Here, we find that FtsEX acts on FtsA to regulate both divisome assembly and activity. FtsX interacts with FtsA and this interaction is required for divisome assembly and inhibition of divisome function by ATPase mutants of FtsEX. Our results suggest that FtsEX antagonizes FtsA polymerization to promote divisome assembly and the ATPase mutants of FtsEX block divisome activity by locking FtsA in the inactive form or preventing FtsA from communicating with other divisome proteins. Because FtsEX is known to govern cell wall hydrolysis at the septum, our findings indicate that FtsEX acts on FtsA to promote divisome assembly and to coordinate cell wall synthesis and hydrolysis at the septum. Furthermore, our study provides evidence that FtsA mutants impaired for self-interaction are favored for division, and FtsW plays a critical role in divisome activation in addition to the FtsQLB complex.
Bacteria assemble a ring-shaped multiprotein complex called the divisome to drive division (1, 2). Studies of cell division in the model organism Escherichia coli suggest that the division process can be divided into at least three steps (1, 2). In the first step a Z ring is formed at the future division site by the bacterial tubulin homolog FtsZ (3). The Z ring consists of FtsZ polymers tethered to the membrane by the membrane anchors FtsA and ZipA (4–6). A number of nonessential proteins, called Zaps (FtsZ-associated proteins, ZapA, ZapB, ZapC, and ZapD), coassemble with FtsZ to promote the integrity of the Z ring (7). FtsEX, an ATP-binding cassette transporter-like complex, and its interaction partner EnvC are also recruited to the Z ring at this step (8, 9). FtsEX is required for the recruitment of downstream division proteins that occurs in the second step (8), and normally ensues after a delay equal to one-third of the cell cycle (10). During this step, numerous essential division proteins are recruited to the Z ring in a linear sequential pathway (Fig. 1A) (1, 11). These proteins include FtsK, FtsQ, FtsL, FtsB, FtsW, FtsI, and FtsN. The third step involves activation of the divisome complex by the arrival of FtsN to start peptidoglycan (PG) synthesis (12, 13). The onset of septal PG synthesis results in the recruitment of many additional proteins involved in cell wall remodeling and invagination of the envelope, including amidases (14), the denuded PG strand binding proteins (containing SPOR domains) (12, 15, 16), the transenvelope Tol–Pal complex, and a number of other proteins with unknown function (2, 17). Together, these proteins ensure that the constriction process proceeds smoothly and that daughter cells are separated without lysis.
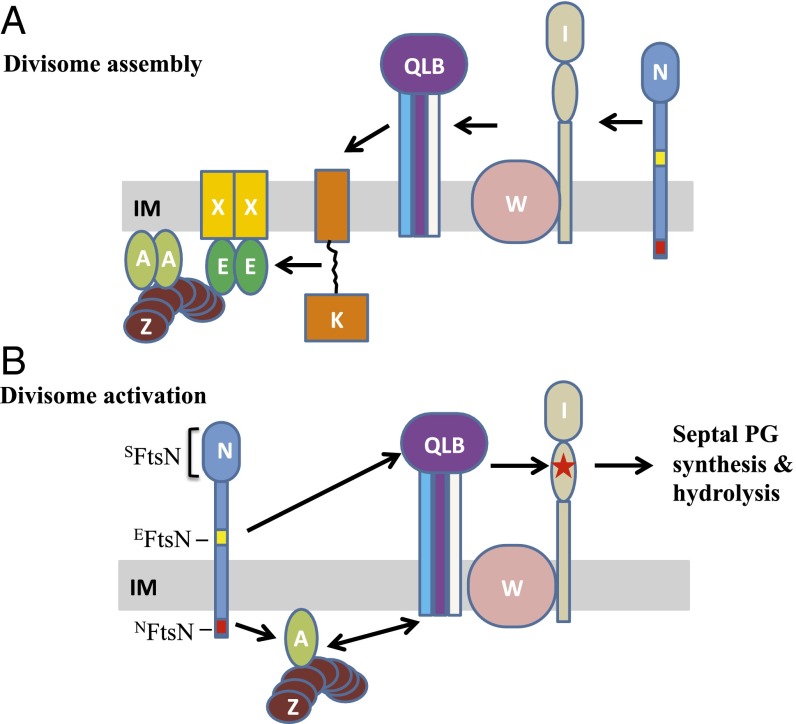
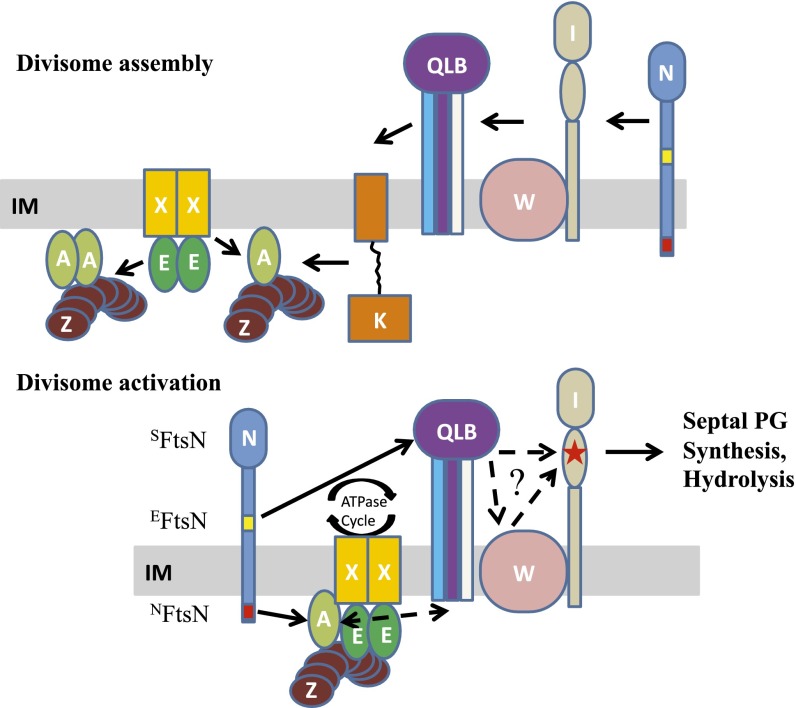
Fig. 1.
Current models for divisome assembly and activation. In these models, the division proteins (Fts) are indicated with capital letters and some proteins are not depicted for simplicity. (A) The linear sequential divisome assembly pathway based on depletion studies. FtsZ polymers anchored to the membrane by FtsA and ZipA (not shown) coalesce into a Z ring at the future division site. The ABC transporter-like complex FtsEX localizes to the Z ring immediately. After roughly one-third of the cell cycle, the downstream division proteins FtsK, FtsQ, FtsL, FtsB, FtsW, FtsI, and FtsN are recruited to the Z ring sequentially. FtsQ, FtsB, and FtsL form a tight complex and are pictured as a complex. Similarly, FtsW and FtsI are pictured as a complex. The cytoplasmic (NFtsN) and essential (EFtsN) domains of FtsN are colored red and yellow, respectively. (B) Model for FtsN-triggered divisome activation adapted from ref. 24. In this model, the divisome is kept inactive by FtsA and FtsQLB before FtsN arrives at the Z ring. Interaction between NFtsN and FtsA switches FtsA to the on state and brings EFtsN to the divisome complex to derepress the FtsQLB complex. Together, these two activation signals converge to activate FtsI (indicated with a red star) to start septal PG synthesis.
Although the first step of divisome assembly—formation of the Z ring—is relatively well understood, how the downstream division proteins (from FtsK to FtsN) are recruited to the Z ring that form the core of the divisome is poorly understood. Depletion studies indicate that they are recruited in a linear sequential pathway, although multiple interactions exist among these proteins (Fig. 1A) (11, 18). Accumulating evidence indicates that FtsA, an actin-related protein, plays a critical role. First, other than FtsA and FtsZ, the early divisome proteins (ZipA, FtsEX, EnvC, and the Zaps) can be deleted under certain conditions and the divisome can still assemble and function (7, 8, 19–21). Second, FtsA has been reported to interact with many downstream division proteins (22), including FtsN (23–25). Importantly, FtsA mutants impaired for self-interaction and overexpression of FtsN bypass ZipA, supporting a model in which FtsA monomers recruit downstream division proteins to the Z ring (20, 25). In this model the essential role of ZipA in recruitment is to antagonize FtsA polymerization. FtsEX is also necessary for recruitment of the downstream division proteins to the Z ring, but why is not clear. Many conditions, such as high osmolarity, FtsA* (FtsAR286W, a gain-of-function mutation that impairs self-interaction), or overexpression of FtsN can suppress the growth defect of a ∆ftsEX strain (21). The conditional essentiality of FtsEX indicates that it is probably not directly involved in recruiting but regulates the function of an early-division protein to promote divisome assembly. Interestingly, many conditions that bypass FtsEX also bypass ZipA, such as overexpression of FtsN or the ftsA* mutation, suggesting that FtsEX may be required for the recruitment of downstream division proteins for the same reason as ZipA.
FtsN is the last essential protein to arrive at the Z ring and is thought to be the trigger for septation (12, 13, 26), but how it does this is not clear. It is a bitopic membrane protein with its N-terminal domain in the cytoplasm (NFtsN) that interacts with FtsA and aids the localization of FtsN to the Z ring (27, 28). The extreme C-terminal periplasmic part of FtsN is a SPOR (SFtsN) domain that recognizes denuded glycan strands generated during constriction and enhances FtsN localization once constriction starts (12). The essential function of FtsN (EFtsN) is located within a helix preceding the SPOR domain (12, 24). Although EFtsN does not display a midcell localization when overexpressed and exported to the periplasm, it is sufficient to stimulate septum synthesis (12). Insight into the role of EFtsN comes from the isolation of extragenic mutations that allow cell division when EFtsN is inactivated. These mutations are located in FtsA, FtsB, and FtsL, leading to a model in which the divisome is kept inactive by FtsA and the FtsQLB complex (24). Interaction between NFtsN and FtsA brings EFtsN to the division site so that these two signals synergize to derepress the FtsQLB complex and activate septal PG synthesis (Fig. 1B) (24, 29, 30). As new PG is synthesized, amidases regulated by FtsEX generate denuded glycan chains, resulting in more recruitment of FtsN by binding of SFtsN and forming a positive feedback loop (12).
In addition to FtsA, FtsQLB, and FtsN, FtsEX may also play a role in divisome activation. FtsE mutants predicted to be defective in ATP binding (FtsEK41A and FtsED162A) or hydrolysis (FtsEE163A) support divisome assembly but not cell constriction when expressed with FtsX (31). However, this role of FtsEX has been largely overlooked because the division defect of a ∆ftsEX mutant can be suppressed by many conditions and the emphasis on the role of FtsEX in governing cell wall hydrolysis in diverse bacteria. In this study we focus on the role of FtsEX and find that FtsEX interacts with FtsA so that FtsA can recruit downstream proteins. This interaction also mediates the inhibition of divisome activity by ATPase mutants of FtsEX. Our results suggest that FtsEX, similar to ZipA, antagonizes FtsA polymerization to promote divisome assembly and ATP hydrolysis by FtsEX is necessary for FtsA to promote the start of cell constriction. In addition, our study also provides evidence that FtsA mutants impaired for self-interaction are less dependent on FtsN for division and FtsW participates in divisome activation.
Results
ATPase Activity of FtsEX but Not Its Role in Regulation of Amidase Activity Is Essential for Division.
FtsEX is an early-divisome component and required for E. coli cell division in media with low osmolarity (8, 21). Its ATPase activity is necessary for the activation of two amidases, AmiA and AmiB, which hydrolyze cell wall at the septum, via EnvC (9). Interestingly, its ATPase activity has also been implicated in initiation of cell constriction (31). These findings raise the question of whether the essential function of the ATPase activity of FtsEX is to activate the amidases for cell separation. Several lines of evidence suggest that this is probably not the case. The major phenotype of ATPase-defective mutants of FtsEX is a failure of cell division under the nonsuppressed conditions (31), whereas deletion of AmiA and AmiB or their activator EnvC results in a mild cell-chaining defect with little effect on division and growth (14, 32, 33). This discrepancy suggests that although the ATPase activity of FtsEX coordinates cell constriction and cell wall hydrolysis at the septum, its role in cell wall hydrolysis may be nonessential. To confirm this idea, we tested whether FtsEX∆152–161, which lacks the EnvC interaction domain (9), could complement an ftsEX::cat strain in LB medium without NaCl (LB0N), a condition where ftsEX::cat cells cannot grow (34). FtsEX∆152–161 rescued the growth of the ftsEX::cat strain in LB0N medium at low induction levels as well as wild-type FtsEX (SI Appendix, Fig. S1). In contrast, none of the predicted ATPase mutants of FtsEX (FtsEK41MX, FtsED162NX, and FtsEE163QX) (9, 31) could complement the deletion strain at any level of expression. Thus, we conclude that the essential role of the ATPase activity of FtsEX is for cell constriction and not regulation of the amidases.
Overexpression of FtsED162NX Blocks Cell Constriction but Not Divisome Assembly.
One ATPase mutant of FtsEX, FtsED162AX, has been reported to block cell division when expressed in wild-type cells (31). Consistent with this finding, we found that overexpression of a similar mutant (FtsED162NX) in wild-type cells, as well as ftsEX::cat cells, was toxic on either LB or LB with sucrose, a condition suppressing the growth defect of the ftsEX::cat strain (SI Appendix, Figs. S1 and S2). Overexpression of the other two ATPase mutants was also toxic, but to a lesser extent, presumably because they affect different steps of the ATPase cycle. Nonetheless, with all three mutants expression led to filamentation (SI Appendix, Fig. S3). Overexpression of wild-type FtsEX or FtsEX∆152–161 also blocked cell division, but they required a higher concentration of inducer (SI Appendix, Figs. S1 and S2). Deletion of the EnvC interacting domain from FtsX did not abolish the toxicity of FtsED162NX (FtsED162NX∆152–161), indicating that inhibition of cell division by FtsED162NX is independent of its function in governing the amidases.
Because the ATPase mutants of FtsE were reported to support divisome assembly but not cell constriction (31), we suspected that the divisome was assembled in cells overexpressing the ATPase mutants of FtsEX. We focused on FtsED162NX because it was the most potent in inhibiting cell division. We visualized Z-ring formation by using ZapA–GFP and assessed divisome assembly by monitoring the localization of two late cell-division proteins, GFP–FtsI and GFP–FtsN. Note that the gfp fusions are integrated in the chromosome with zapA-gfp under its own promoter and gfp-ftsI and gfp-ftsN under an isfopropyl-β-d-thiogalactopyranoside (IPTG) inducible promoter (14, 35, 36). The plasmids used here express a lower level of FtsEX but, they still produce enough FtsED162NX to block division (Fig. 2 and SI Appendix, Fig. S4). As shown in Fig. 2A, cells overexpressing wild-type FtsEX displayed a normal cell length and ZapA–GFP localized to midcell, whereas cells overexpressing FtsED162NX became filamentous. However, ZapA–GFP still localized to potential division sites in these filaments, indicating that Z-ring formation was not affected. Consistent with previous findings (31), we found that GFP–FtsI and GFP–FtsN also localized to potential division sites in cells overexpressing FtsED162NX (Fig. 2B). Although cells expressing GFP–FtsI formed smooth filaments, cells expressing GFP–FtsN formed filaments with some constrictions, suggesting that GFP–FtsN counteracts the division inhibitory activity of FtsED162NX. This result is consistent with a previous observation that GFP–FtsN alleviated the mild dominant-negative effect of FtsED162AX on division (31).
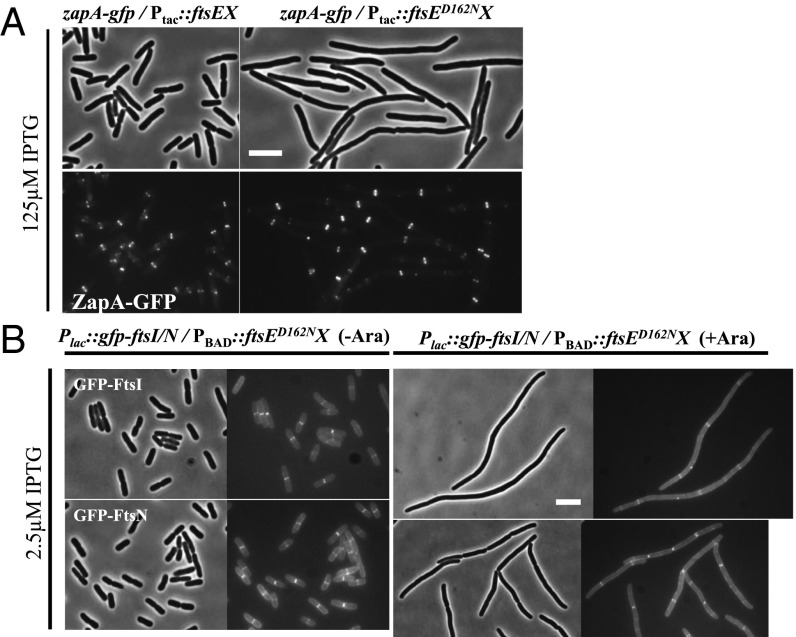
Fig. 2.
Overexpression of FtsED162NX blocks cell division but not divisome assembly. (A) Overexpression of FtsED162NX does not block Z ring formation. Cells of HC261 (TB28, zapA-gfp) harboring plasmid pSD221 (pEXT22, Ptac::ftsEX) or its variant pSD221-D162N were grown to exponential phase in LB + 0.2 M sucrose at 30 °C. The cultures were then diluted 1:10 in fresh LB medium with 0.2 M sucrose and antibiotics. IPTG was added to a final concentration of 125 µM to induce expression of FtsEX or FtsED162NX. Localization of ZapA–GFP was visualized 2 h later. (Scale bar, 3 µm.) (B) Overexpression of FtsED162NX does not block recruitment of late-division proteins. Cells of EC436 (MC4100, Plac::gfp-ftsI) or EC440 (MC4100, Plac::gfp-ftsN) harboring pDSW610-D162N (pBAD33, PBAD::ftsED162NX) were grown to exponential phase in LB medium at 30 °C. The cultures were then diluted 1:10 in fresh LB medium with antibiotics. IPTG and arabinose were added to a final concentration of 2.5 µM and 0.2% to induce expression of GFP-ftsI or GFP-ftsN and FtsED162NX, respectively. Localization of GFP-FtsI and GFP-FtsN were visualized 5 h after the induction of FtsED162NX. (Scale bar, 3 µm.)
Overexpression of FtsN Suppresses the Division Inhibitory Activity of FtsED162NX.
To further confirm that FtsN alleviates the division inhibitory effect of FtsED162NX, we tested whether overexpression of FtsN would allow cells overexpressing FtsED162NX to survive. As shown in Fig. 3, cells carrying the control plasmid pBR322 were killed by overexpression of FtsED162NX at 60 µM IPTG, whereas cells containing pKD140 (pBR322-FtsN) survived at 1 mM. Because FtsN activates the divisome for constriction, whereas FtsED162NX allows divisome assembly but not cell constriction, it suggests they have opposing activities in the divisome. Because both the cytoplasmic (NFtsN) and the essential (EFtsN) domains of FtsN play important roles in activating the divisome (24), we tested their role in counteracting FtsED162NX by using FtsN mutants with mutations that inactivate NFtsN (FtsND5N) or EFtsN (FtsNWYAA) (24, 25). A Western blot showed that these mutants were expressed at the same level as wild-type FtsN (SI Appendix, Fig. S5). As shown in Fig. 3, although the mutation in the NFtsN domain affected the efficiency of FtsN in counteracting FtsED162NX, the mutation in the EFtsN domain completely abolished the ability of FtsN to counteract FtsED162NX. The partial suppression of FtsED162NX by FtsND5N may be a result of a weak interaction between FtsND5N and FtsA because we showed previously that the D5N mutation does not completely eliminate the FtsN interaction with FtsA (25). These results demonstrate that NFtsN contributes to the ability of FtsN to suppress the division block posed by FtsED162NX and that EFtsN is critical for this suppression.
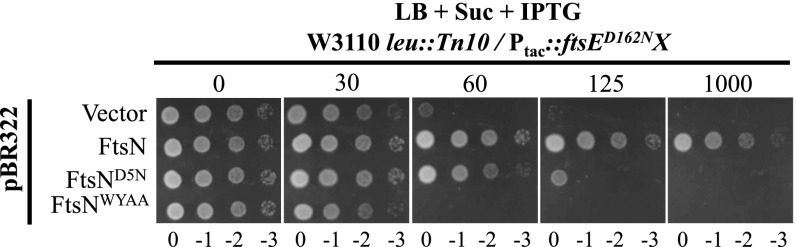
Fig. 3.
Domains of FtsN required to counteract the killing by overexpressed FtsED162NX. Plasmid pBR322 or its derivatives carrying FtsN, FtsND5N or FtsNWYAA were transformed into strain S3/pSD221-D162N (W3110, leu::Tn10/Ptac::ftsED162NX). A transformant of each strain from LB + 0.2 M sucrose plates was resuspended in 1 mL LB medium and serially diluted. Three microliters of each aliquot was then spotted on LB + 0.2 M sucrose plates with appropriate antibiotics and increasing concentrations of IPTG. The plates were then incubated at 30 °C overnight before photography.
Hyperactive ftsB and ftsL but Not ftsA Mutations Confer Resistance to the Division Inhibitory Activity of FtsED162NX.
In the current model of divisome activation, NFtsN and EFtsN act synergistically on the two sides of the cytoplasmic membrane to activate the divisome, so losing either NFtsN or EFtsN function results in failure of divisome activation at the physiological level (24, 25, 28). FtsED162NX could block cell constriction by inhibiting either NFtsN or EFtsN function, or the synergism, or even a downstream event. To get some insight into how FtsED162NX inhibits division, we took advantage of the recently isolated ftsA, ftsB, and ftsL mutations that make cells less dependent upon FtsN (hyperactive division mutations) (24, 37). These mutations act on the periplasmic side of the divisome (ftsB and ftsL mutations) or on the cytoplasmic side of the divisome (ftsA mutations), such that EFtsN is no longer required to derepress the FtsQLB complex in the periplasm or for NFtsN to bind and stimulate FtsA in the cytoplasm (24). As a consequence, cells containing one of these mutations require less FtsN for division. If FtsED162NX inhibits constriction by blocking FtsN function, these mutants may be resistant because less FtsN is required.
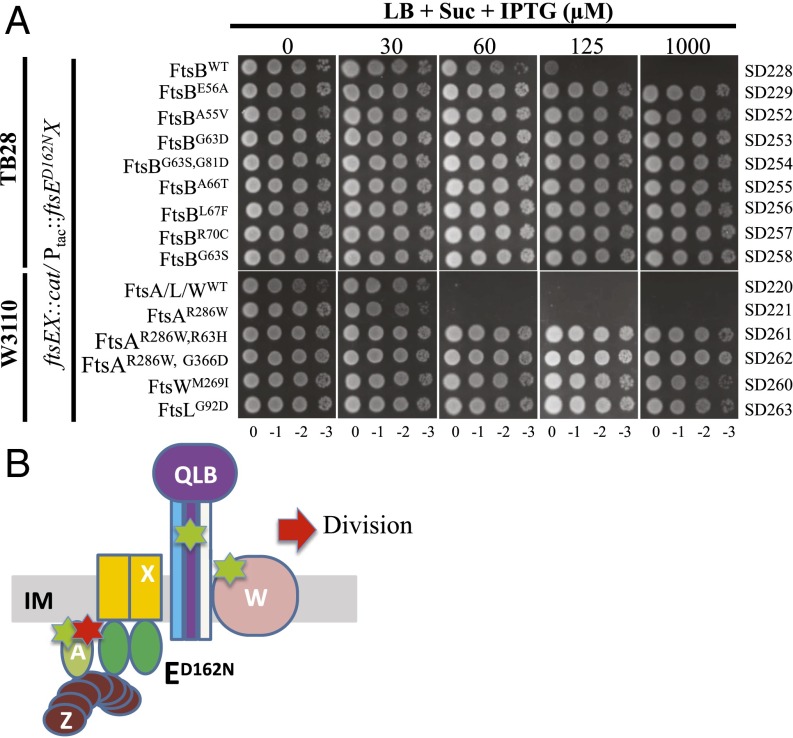
We first tested whether ftsB and ftsL mutations confer resistance to FtsED162NX. Two mutations in each gene (ftsBE56A and ftsBD59H, ftsLE88K and ftsLD93G) were chosen. FtsBE56A is stronger than FtsBD59H (requires less FtsN) and FtsLE88K is stronger than FtsLD93G (24). The plasmid pSD221-D162N (Ptac::ftsED162NX) was transformed into strains carrying ftsB or ftsL mutations and the transformants tested on plates containing IPTG to induce expression of FtsED162NX. As shown in Fig. 4, growth of the wild-type strain was blocked by expression of FtsED162NX at 60 µM IPTG, whereas the FtsB and FtsL mutant strains grew at this and higher IPTG concentrations. Importantly, the resistance of the mutant strains correlated with the strength of the mutation. For example, the strain carrying ftsBD59H grew on plates containing less than 250 µM IPTG, whereas the strain harboring the stronger ftsBE56A mutation was resistant to the killing of FtsED162NX even at 500 µM IPTG. A similar phenomenon was observed for the FtsL mutant strains. To make sure the resistance to FtsED162NX was not caused by the presence of wild-type FtsEX in the chromosome, ftsEX was deleted from these strains. Such strains were still resistant to the killing by FtsED162NX (SI Appendix, Fig. S6), indicating that a divisome rendered hyperactive (requiring less FtsN) by ftsB or ftsL mutations displayed resistance to FtsED162NX, regardless of the presence of wild-type FtsEX.
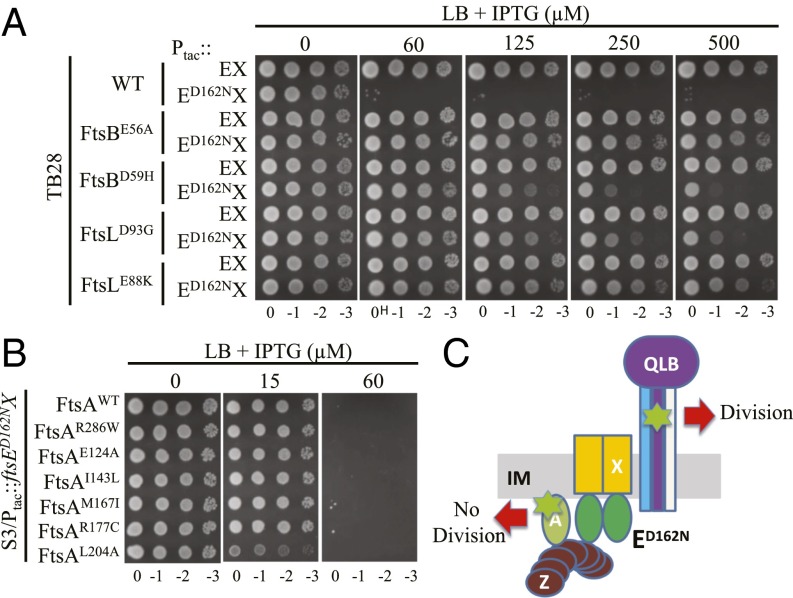
Fig. 4.
Hyperactive ftsB and ftsL mutations, but not hyperactive ftsA mutations, confer resistance to overexpressed FtsED162NX. (A) Hyperactive ftsB and ftsL mutations confer resistance to overexpressed FtsED162NX. Plasmid pSD221 and its variant pSD221-D162N were transformed into strains TB28, BL167 (TB28, ftsBE56A), BL140 (TB28, ftsBD59H), MT10 (TB28, ftsLE88K), and BL154 (TB28, ftsLD93G). A single transformant of each strain from LB + 0.2 M sucrose plates was then resuspended in 1 mL of LB medium and serially diluted. Three microliters of each aliquot was then spotted on the LB plates or the LB + 0.2 M sucrose plates with appropriate antibiotics and increasing concentrations of IPTG. The plates were incubated at 30 °C overnight before photography and only results from LB plates are shown. (B) Hyperactive ftsA mutations do not confer resistance to overexpressed FtsED162NX. Plasmid pSD221-D162N was transformed into strain S3 and its derivatives carrying hyperactive ftsA mutations. A spot test was performed as in A. (C) Diagram indicating the location of hyperactive mutations and the response to FtsED162NX. The hyperactive mutations are indicated by the green symbol and are in FtsA or FtsL/FtsB.
We next tested whether ftsA mutations could confer resistance to the killing of FtsED162NX. We chose two ftsA mutations that have been reported to require less FtsN for division (ftsAE124A and ftsAI143L, because they bypass the function of EFtsN) as well as several mutations that have been reported to bypass ZipA (ftsAM167I, ftsAR177C, ftsAL204, and ftsAR286W) (20, 24, 38). The mutations in the former class were considered unique because they affect residues in the IC domain of FtsA (SI Appendix, Fig. S7A); however, these mutations also bypass ZipA, indicating they share some common properties. Indeed, we found that the four mutations in the latter class behaved similarly to ftsAE124A and ftsAI143L, enabling cells to grow with a much lower level of FtsN1-140-D5N than cells with wild-type FtsA (SI Appendix, Fig. S7B). Thus, all ftsA mutations that bypass ZipA require less FtsN for division regardless of their location in FtsA. Importantly, and unlike ftsB or ftsL mutations, ftsA mutations were unable to provide resistance to FtsED162NX as strains with the ftsA mutations were unable to grow with more than 60 µM IPTG (Fig. 4B). To make sure this lack of resistance to FtsED162NX was not a result of the presence of wild-type FtsEX on the chromosome, ftsEX was deleted in a few of these ftsA mutant strains. Such strains became even more sensitive to the killing by FtsED162NX (SI Appendix, Fig. S6). Taken together, these results show that hyperactive mutations (requiring less FtsN) acting on the periplasmic side of the divisome (ftsB and ftsL) render cells more resistant to FtsED162NX, whereas hyperactive mutations on the cytoplasmic side (ftsA) of the divisome do not (Fig. 4C). Furthermore, these results imply that FtsED162NX blocks divisome activity by locking the cytoplasmic side of the divisome in an inactive state, or preventing communication between the cytoplasmic and the periplasmic sides of the divisome, where septal PG synthesis occurs.
Because FtsAR286W can bypass FtsEX (21), we tested whether the other hyperactive FtsA mutants also have this ability. Strains containing the ftsA mutations that bypass ZipA grew on LB or LB0N plates without FtsEX (SI Appendix, Fig. S8). In addition, the ftsB and ftsL hyperactive mutations allowed growth without FtsEX on LB or LB0N plates (SI Appendix, Fig. S8). Because the ftsLE88K mutation was also reported to bypass ZipA (37), our results imply that any mutation that bypasses ZipA would also bypass FtsEX, indicating that ZipA and FtsEX share a common function in cell division. However, it should be noted that ZipA and FtsEX cannot substitute for each other.
Isolation of Additional Division Protein Mutants That Provide Resistance to FtsED162NX.
The above results strongly suggest that FtsED162NX inhibits divisome activation, but how it does so is not clear. Because FtsED162NX localized to the Z ring, it must interact with a division protein or proteins to execute its inhibitory function; we screened for suppressor mutations that allow survival in the presence of FtsED162NX, thinking that such mutations might reveal insights into the inhibitory mechanism. We expected to obtain mutations that specifically impair or eliminate the interaction between FtsED162NX and its target protein. However, mutations in ftsB and ftsL were also expected because mutations in these genes also confer resistance to FtsED162NX, as described above. We also expected to obtain mutations that prevent the localization of FtsED162NX because FtsED162NX is unlikely to block cell division if it does not localize to the Z ring.
For this screen, we used strain SD221 (W3110, leu::Tn10 ftsAR286W ftsEX::cat). The ftsAR286W mutation is sensitive to FtsED162NX and allows the bypass of FtsEX so that mutations that prevent FtsEX from localizing to the Z ring would not have a growth disadvantage. This strain also allowed us to check the linkage between mutations resistant to FtsED162NX and the leu::Tn10 marker, which is about 10 kb away from the 2-min region where most essential cell division genes are located. SD221 was mutagenized with ethyl methanesulfonate and then transformed with plasmid pSD221-D162N. Suppressors were selected on LB plates with appropriate antibiotics, sucrose and 250 µM IPTG. Of 41 suppressors picked for analysis, 21 had mutations in the plasmid and were not studied further. Eight of the remaining 20 showed ∼60–90% linkage with leu::Tn10. Sequencing the division genes in the 2-min region of these eight suppressors revealed that five contained missense mutations in ftsL (ftsLE88K, ftsLD92G and ftsLH94Y) (SI Appendix, Table S1). Obtaining ftsLE88K, which bypasses EFtsN (24), and as we showed above, provides resistance to FtsED162NX, validated our approach. Two suppressors contained missense mutations in ftsA (ftsAR286W, R63H and ftsAR286W, G366D) and one contained a mutation in ftsW (ftsWM269I). Of the 12 suppressors that showed no linkage with leu::Tn10, we sequenced ftsB and found that 7 contained a missense mutation in ftsB [ftsBA55V, ftsBG63D, ftsBG63S,G81D (containing two mutations), ftsBA66T, ftsBL67F and ftsBR70C], none of which were isolated previously. The remaining five suppressors did not contain a mutation in ftsB and were not further investigated.
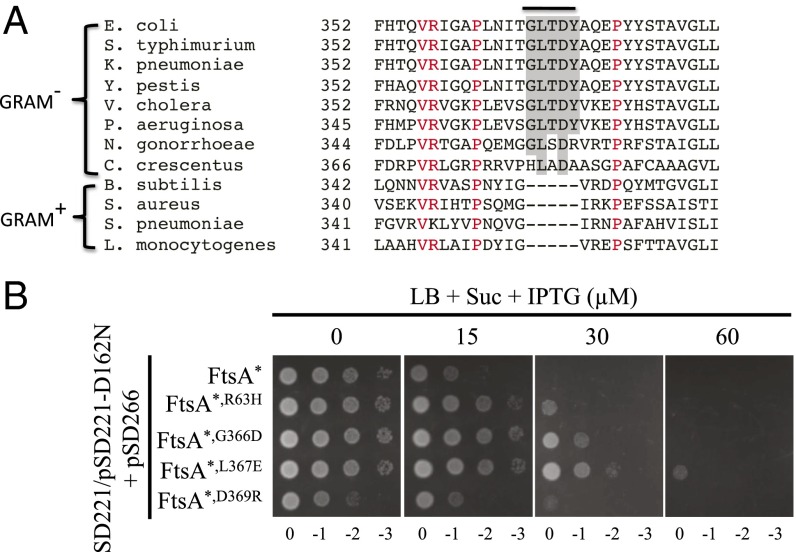
To make sure that the resistance to FtsED162NX in these suppressors was a result of the identified mutations, each mutation was introduced into a naïve strain S3 (W3110, leu::Tn10) or TB28 (MG1655, ∆lacIZYA) by allelic replacement or λ-Red–mediated recombineering. All of the mutations were successfully introduced into the chromosome except for ftsAR63H and ftsAG366D. However, these two ftsA mutations could easily be introduced into the chromosome in combination with ftsAR286W, suggesting that these two mutations impair an important function of FtsA, which is rescued by ftsAR286W (see FtsED162NX-Resistant FtsA Mutants Are Impaired for Interaction with FtsX). As shown in Fig. 5A, all of the mutations confer resistance to FtsED162NX in the absence of wild-type FtsEX, similar to the ftsBE56A mutation. Among these mutations, ftsAR286W, G366D provided greater resistance because the cells were not elongated at the highest IPTG concentration, whereas the others were elongated. The resistance of the strain containing two mutations in ftsB (ftsBG63S, G81D) was largely a result of the ftsBG63S mutation because a strain containing only this mutation showed a similar level of resistance. The resistance in the strains containing the ftsAR286W, R63H and ftsAR286W, G366D mutations must be a result of the R63H and G366D mutations because cells carrying ftsAR286W are sensitive to FtsED162NX (Fig. 5B).
Fig. 5.
Newly isolated ftsA, ftsB, ftsL, and ftsW mutations provide resistance to overexpressed FtsED162NX. (A) Plasmid pSD221-D162N was transformed into TB28 and W3110 derivatives carrying the ftsEX::cat allele and different mutations in ftsA, ftsB, ftsL, or ftsW. A spot test was performed as Fig. 4A. Strains are indicated to the right. (B) Summary of mutations that provide resistance to FtsED162NX. The symbols are as in Fig. 4B with the addition of FtsW and the red star indicating a second mutation in ftsA that provides resistance to FtsED162NX.
How do mutations in so many different division genes confer resistance to the toxicity of FtsED162NX? Examination of the mutations in FtsB and FtsL revealed that the mutations in FtsL are located in the recently defined constriction control domain (CCD), whereas the mutations in FtsB, except for ftsBA55V, are adjacent to the CCD of FtsB (24). This finding suggests that all of the ftsB and ftsL mutations are hyperactive division mutations similar to those reported by de Boer and colleagues (24), and probably confer resistance to FtsED162NX by the same mechanism. The ftsWM269I mutation is located in the periplasmic loop between transmembrane segments 7/8 of FtsW (39). Interestingly, this residue is next to residue ftsWA270, which corresponds to residue ftsWA246 of Caulobacter crescentus. Mutation of this residue in C. crescentus is hyperactive for division because it results in shorter cells (40). This finding implies that ftsWM269I provides resistance to FtsED162NX through a similar mechanism as the ftsB and ftsL mutations. Because FtsA resides in the cytoplasm and cells carrying hyperactive ftsA mutations are sensitive to FtsED162NX, the two ftsA mutations must act by a different mechanism. The ftsAR63H and ftsAG366D mutations are located in the H1 helix and a loop before the H11 helix of FtsA (SI Appendix, Fig. S7A) (41), respectively. Although far away in the linear amino acid sequence, the two residues are quite close in the structure. It is possible that they constitute a binding site for FtsED162NX and the mutations impair the interaction between FtsA and FtsED162NX, resulting in resistance to the toxicity of FtsED162NX. These hypotheses were tested as described in the following sections.
FtsED162NX-Resistant FtsB, FtsL, and FtsW Mutants Are Hyperactive for Division.
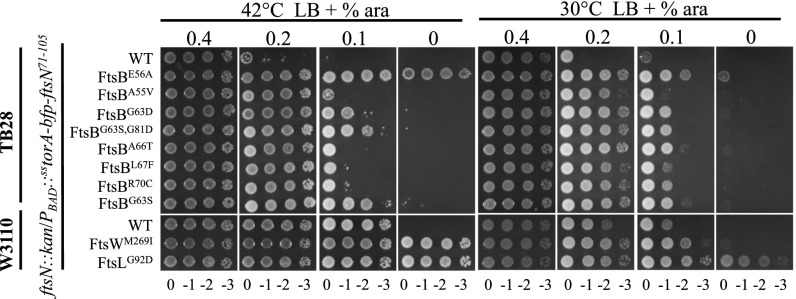
Based on the analysis above, we speculated that ftsB, ftsL, and ftsW mutations provide resistance to FtsED162NX by a similar mechanism. We thus tested whether these mutations allowed the cells to divide without or with less FtsN, an indication of hyperactivation. To do that, strains containing the ftsB, ftsL, or ftsW mutations were transformed with a plasmid harboring FtsN that is temperature sensitive for replication. Chromosomal ftsN in the resultant transformants was then deleted by P1 transduction and the transductants tested for growth at nonpermissive temperature. Although none of the newly isolated FtsB mutants supported growth of the transductants at the nonpermissive temperatures of 37 °C and 42 °C, they all improved survival (SI Appendix, Fig. S9). Strikingly, the FtsLG92D and FtsWM269I mutants could fully support the growth of the transductants at 37 °C and 42 °C, suggesting that they bypassed FtsN. To further prove that these ftsB, ftsL, and ftsW mutations required less FtsN for division, we tested whether they allowed the cells to survive with less EFtsN. An FtsN-null strain carrying the plasmid pMG20, which harbors sstor-bfp-EftsN under the arabinose inducible promoter, only grows on plates with a high level of arabinose (12). As shown in Fig. 6, strains SD274 (W3110, ftsN::kan/pMG20) and SD276 (TB28, ftsN::kan/pMG20) required 0.2% and 0.4% arabinose, respectively, whereas strains carrying the ftsB, ftsL, or ftsW mutations grew at lower arabinose concentrations. Strains carrying ftsLG92D or ftsWM269I even grew without arabinose at 42 °C, consistent with their ability to bypass FtsN reported above. However, attempts to introduce ftsN::kan into strains carrying ftsLG92D or ftsWM269I without a complementing plasmid were unsuccessful, suggesting that a residual level of FtsN is required for survival under our conditions. All of the newly isolated ftsB, ftsL, and ftsW mutations allowed cells to grow without FtsEX (SI Appendix, Fig. S10), consistent with the above results showing that known ftsB and ftsL mutations bypass FtsEX. Taken together, these results confirm that the newly isolated ftsB, ftsL, and ftsW mutations result in a hyperactive divisome (requiring less FtsN), which is sufficient to provide resistance to FtsED162NX.
Fig. 6.
Newly isolated FtsB, FtsL, and FtsW mutants are hyperactive for division as they require less FtsN. TB28 or W3110 derivatives carrying the ftsN::kan allele and different mutations in ftsB, ftsL, or ftsW were transformed with pMG20 (pBAD33, PBAD::sstor-bfp-ftsN71-105) on LB plates with 0.4% arabinose. Single colonies of each strain were then spotted on plates with increasing concentration of arabinose and incubated overnight before photography, as in Fig. 4. The strains are indicated on the right.
FtsED162NX-Resistant FtsA Mutants Are Impaired for Interaction with FtsX.
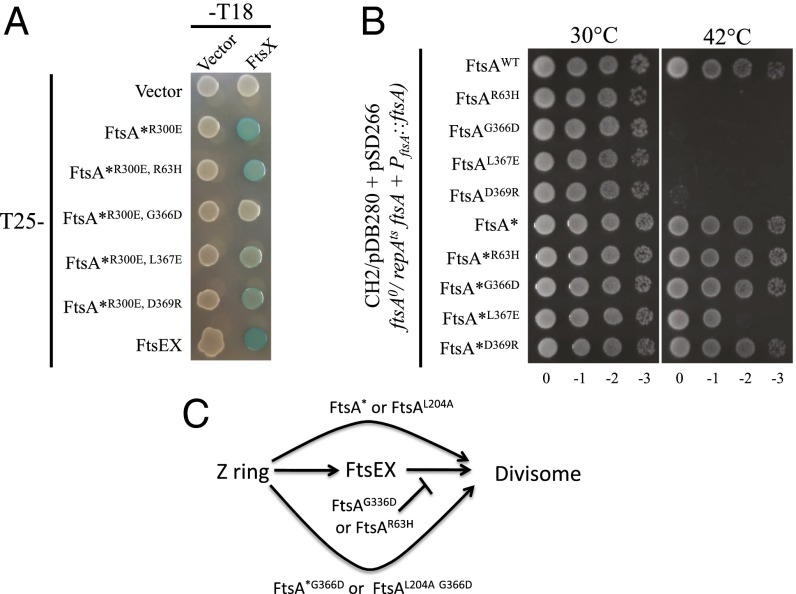
We hypothesized that the newly isolated ftsA mutations confer resistance to FtsED162NX by impairing the interaction between FtsA and FtsED162NX. This hypothesis is based on the fact that cells carrying hyperactive FtsA mutants are more sensitive to FtsED162NX and that FtsA and FtsX have been reported to interact directly (22). To test this hypothesis, we used the bacterial two-hybrid assay (BTH). Although FtsE did not interact with FtsA, FtsZ, or itself unless coexpressed with FtsX, FtsX interacted with FtsA when expressed alone or with FtsE (SI Appendix, Fig. S11). Because the BTH does not necessarily report a direct interaction between two proteins if they are part of a larger protein complex, we used a variant of FtsA, FtsAR286W, R300E, which prevents FtsA from going to the divisome, making a false-positive less likely (25). The R300E mutation reduces the interaction between FtsA and FtsZ (42), whereas the R286W mutation impairs FtsA’s self-interaction (20). Importantly, this FtsA variant still interacted strongly with FtsX (Fig. 7A and SI Appendix, Fig. S11B). Introduction of the R63H mutation into this variant resulted in only a slight reduction in interaction in the BTH assay, whereas introduction of the G366D mutation completely eliminated the interaction with FtsX (Fig. 7A). This finding supports our hypothesis that the mutations in ftsA that provide resistance to FtsED162NX impair the interaction between FtsA and FtsX.
Fig. 7.
FtsED162NX resistant ftsA mutations impair the FtsA–FtsX interaction and inactivate FtsA function. (A) BTH assay to assess the interaction between FtsA mutants and FtsX. Pairs of plasmids harboring T25-FtsA*R300E (T25-FtsAR286W, R300E) and FtsX-T18 or their variants were cotransformed into BTH101. Single transformants of each combination were then resuspended in LB medium and 3 µL of each aliquot was spotted on LB plates containing appropriate antibiotics, 25 µM IPTG and 40 µg/mL X-gal. The plates were then incubated at 30 °C overnight before photography. (B) FtsA mutants impaired for interaction with FtsX cannot complement an FtsA-depletion strain and the ftsAR286W mutation suppresses this defect. Plasmid pSD266 (pACYC184, PftsA::ftsA) or its variants harboring different ftsA mutations were transformed into strain CH2/pDB280 (ftsA0 recA56 srlD::Tn10/pSC101ts, PftsA::ftsA). Single transformants of each strain obtained at 30 °C were then resuspended in LB medium and serially diluted. Three microliters of each aliquot was spotted on LB plates with appropriate antibiotics. The plates were then incubated at 30 °C and 42 °C overnight before photography. (C) Summary of the behavior of ftsA alleles in divisome assembly. ftsAG366D and ftsAR63H block divisome assembly; however, they are rescued by some ftsA mutations that bypass FtsEX. Note, some ftsA mutations that bypass FtsEX do not rescue and are considered weak, as explained in the text.
Interaction Between FtsA and FtsX Is Important for Divisome Assembly.
The failure to introduce the ftsAR63H and ftsAG366D mutations into the chromosome suggests that these mutations affect an essential function of FtsA, likely the interaction with FtsX, which is suppressed by the ftsAR286W mutation. To further examine this idea, we tested whether the FtsAR63H or FtsAG366D mutant could complement an FtsA-depletion strain CH2/pDB280 (plasmid pDB280 carrying ftsA is temperature sensitive for replication) (43). Plasmid pSD266 or variants harboring the ftsA mutations was transformed into the CH2/pDB280 strain and the transformants tested for growth at 42 °C. As shown in Fig. 7B and summarized in Fig. 7C, wild-type FtsA complemented the depletion strain, but neither FtsAR63H nor FtsAG366D could support cell growth at 42 °C. Western blot showed that the FtsA mutants were expressed at the same level as wild-type FtsA (SI Appendix, Fig. S12). As expected, combining the ftsAR63H or ftsAG366D mutation with ftsAR286W fully supported cell growth. Together, these results suggest that in the presence of an ftsA mutation, like ftsAR286W, which bypasses FtsEX, the interaction between FtsA and FtsX becomes dispensable. To determine whether this was the case, we tested three other ftsA mutations that bypass FtsEX (ftsAE124A, ftsAI143L, and ftsAL204A). As shown in Fig. 7C (SI Appendix, Fig. S13), the ftsAL204A mutation rescued both FtsAR63H and FtsAG366D, whereas ftsAE124A and ftsAI143L failed to rescue. This difference may be because of a difference in the strength of the mutation. FtsAR286W and FtsAL204A appear to be stronger because they bypass ZipA at a lower level of expression than FtsAE124A and FtsAI143L (20). In addition, the stronger ftsBE56A mutation could fully rescue FtsAR63H or FtsAG366D at both 37 °C and 42 °C, whereas the weaker ftsBD59H mutation only fully rescued at 42 °C (SI Appendix, Fig. S14). Taken together, these results suggest that in the absence of the FtsA–FtsX interaction, only strong suppressor mutations rescue FtsAR63H or FtsAG366D.
Examination of cells in which FtsA was depleted so that only FtsAR63H or FtsAG366D was expressed revealed that the cells became filamentous and eventually lysed, suggesting that it was a failure to divide that prevented growth on plates. Because FtsA has been reported to be necessary for FtsEX to localize to the Z ring (8), we tested whether FtsEX localized to midcell in the FtsAR286W, R63H or FtsAR286W, G366D mutants. FtsEX–GFP localized efficiently to the midcell in strains SD220 (ftsEX::cat) and SD221 (ftsEX::cat ftsAR286W) as expected, but also in SD261 (ftsEX::cat ftsAR286W, R63H) and SD262 (ftsEX::cat ftsAR286W, G366D), indicating that the FtsA–FtsX interaction is not necessary for FtsEX localization to the Z ring (SI Appendix, Fig. S15). FtsX–GFP alone, however, displayed very poor midcell localization in strains lacking FtsE, although it localizes strongly to midcell in wild-type cells, suggesting that FtsX depends on FtsE for efficient localization to the Z ring. This result is in contrast to a previous finding that FtsX can localize to the Z ring without FtsE (31). One possibility is that our FtsX–GFP is not fully functional; however, FtsEX–GFP fully complemented ftsEX::cat cells, suggesting that it is functional.
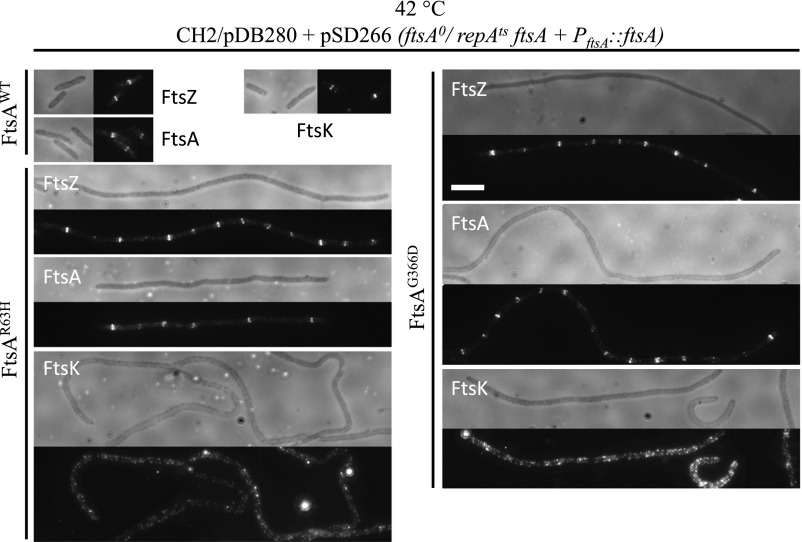
To understand the defect of the FtsAR63H or FtsAG366D mutants, we checked their localization by immunofluorescence microscopy when they were expressed in cells depleted of FtsA. As shown in Fig. 8, both FtsA mutants localized to potential division sites in the filaments, indicating that they did not have a defect in localization. We also examined the localization of FtsZ and the localization of the downstream division protein FtsK. Although FtsZ localized well to potential division sites in the filaments as expected, FtsK resided at the membrane but displayed no localization to potential division sites. Consistent with this finding, GFP–FtsW also failed to localize to potential division sites in cells expressing only FtsAR63H or FtsAG366D (SI Appendix, Fig. S16). Together, these results indicate that the interaction between FtsA and FtsX is not required for FtsEX to localize to the Z ring but is required for the recruitment of downstream division proteins to the Z ring.
Fig. 8.
FtsA mutants impaired for interaction with FtsX fail to recruit downstream division proteins. Localization of FtsZ, FtsA, and FtsK was determined by immunofluorescence microscopy in CH2/pDB280 (ftsA0/repAts, ftsA) cells expressing wild-type FtsA or FtsA mutants as the only copy of FtsA at 42 °C. Detailed information about the experiment is provided in SI Appendix. (Scale bar, 3 μm.)
Although FtsEX is a known component of the divisome in many Gram-negative bacteria, FtsEX in the Gram-positive bacterium Bacillus subtilis is involved in cell wall hydrolysis, but not at the septum (44–47). Alignment of FtsA sequences from members of both groups revealed a motif (GLTDY) in FtsA of many Gram-negative bacteria that is not present in FtsA of Gram-positive bacteria (Fig. 9A). Strikingly, the ftsAG366D mutation affects the first residue of this motif. This finding prompted us to test whether other residues of this motif were important for FtsA–FtsX interaction. As shown in Fig. 7A, the ftsAL367E and ftsAD369R mutations reduced the interaction between FtsA and FtsX in the BTH assay. Furthermore, FtsAL367E and FtsAD369R failed to complement an FtsA depletion strain when expressed as the only copy of FtsA, but they could be rescued or largely rescued by the ftsAR286W mutation (Fig. 7B). Western blot showed that both proteins were expressed at the same level as wild-type FtsA (SI Appendix, Fig. S12). Because cells did not grow very well when FtsAR286W, L367E and FtsAR286W, D369R were expressed as the only copy of FtsA, we tested their resistance to FtsED162NX in the presence of chromosomal FtsAR286W. As shown in Fig. 9B, cells expressing the FtsAR286W, L367E mutant showed similar or greater resistance to FtsED162NX than cells expressing FtsAR286W, G366D, suggesting that L367 is indeed an important residue for FtsA–FtsX interaction. Cells expressing FtsAR286W, D369R showed no resistance to FtsED162NX for unknown reasons. Immunofluorescence microscopy showed that both FtsAL367E and FtsAD369R localized to potential division sites when they were expressed as the only copy of FtsA (SI Appendix, Fig. S17). In these cells, FtsZ localization was not affected, but FtsK failed to localize to potential division sites. Taken together, these results strongly suggest that this motif is critical for the FtsA–FtsX interaction in FtsAs from many Gram-negative bacteria.
Fig. 9.
Identification of a conserved motif in FtsA from Gram-negative bacteria required for interaction with FtsX. (A) Alignment of FtsA amino acid sequences from diverse Gram-negative and Gram-positive bacteria. Residues conserved in all species are colored red. The GLTDY motif (shaded) is present in many Gram-negative bacteria, but not in Gram-positive bacteria. (B) Mutations in the GLTDY motif provide resistance to overexpressed FtsED162NX. Plasmids pSD221-D162N and pSD266* (pACYC184, PftsA::ftsAR286W) or its variants were cotransformed into strain SD221 (S3, ftsAR286W ftsEX::cat). A single transformant of each strain was then picked and spot tested for resistance to overexpressed FtsED162NX, as in Fig. 4A.
Discussion
FtsEX has been studied in diverse bacterial species for its role in regulating cell wall hydrolysis (9, 44–47), but its essential role in E. coli cell division, where it was discovered, remained enigmatic because it can be bypassed. Nevertheless, in medium with low osmolarity, FtsEX is essential for division (8, 21, 31, 34). Under these conditions, FtsEX has at least three functions in cell division: (i) recruitment of downstream divisome proteins to the Z ring (8); (ii) its ATPase activity is necessary for cell constriction to occur (31); and (iii) activation of the amidases to separate daughter cells (9). Here we show that the first two functions, which can be suppressed by increased osmolarity, are essential for cell survival, whereas the third, which cannot be suppressed, is dispensable. The conditional essentiality of FtsEX suggests that it is probably a regulator of the division apparatus instead of a core component. Indeed, in this study we find that FtsEX acts on FtsA to regulate both divisome assembly and activation. Our results suggest FtsEX interacts with a conserved region of FtsA to promote recruitment of downstream division proteins to the Z ring and also to activate the divisome to synthesize septal PG. Only the latter process requires ATP binding and hydrolysis. Because FtsEX has been proposed to use ATPase activity to govern cell wall hydrolysis at the septum in E. coli (9), our findings indicate that FtsEX acts on FtsA to couple cell wall synthesis and hydrolysis at the septum.
FtsEX’s role in divisome assembly depends on its interaction with FtsA. In the absence of FtsEX, none of the late-division proteins localize to the Z ring in medium with low osmolarity (8). Strikingly, a single mutation in FtsA that disrupts the FtsA–FtsX interaction also produces this phenotype. In such a mutant, both FtsA and FtsEX localize to the Z ring, but the downstream division proteins, such as FtsK and FtsW, are not recruited. Although it is unclear how this interaction leads to recruitment of downstream division proteins, the fact that all FtsA mutants that bypass ZipA also bypass FtsEX suggests that FtsEX, like ZipA, modulates FtsA’s ability to recruit downstream divisome proteins. Because most of the FtsA mutants that bypass ZipA are impaired for self-interaction, we proposed previously that ZipA acts by antagonizing FtsA polymerization although the mechanism is unclear (20). Here, we propose that FtsEX also does this, but by directly interacting with FtsA. In fact, ZipA may be necessary to stabilize the Z ring so that FtsEX can act. This function of FtsEX does not require its ATPase activity because the ATPase mutants support divisome assembly (31).
The mutations in FtsA that affect the interaction with FtsX in the BTH and confer resistance to FtsED162NX are located in the GLTDY motif and in the nearby H1 helix (SI Appendix, Fig. S7A). These residues are likely part of the binding site for FtsX. Consistent with this supposition, the residues are located on the predicted membrane proximal side of FtsA (opposite the FtsZ binding site) and therefore are in position to interact with the integral membrane protein FtsX (SI Appendix, Fig. S7A). Because these residues are not directly involved in FtsA polymerization, FtsEX binding to FtsA probably causes a conformational change in FtsA, resulting in unpolymerized FtsA that can recruit the downstream division proteins (Fig. 10). The GLTDY motif is present in many Gram-negative but not Gram-positive bacteria, consistent with the observation that FtsEX is used for divisome assembly, activation, and septal cell wall hydrolysis in a Gram-negative bacterium like E. coli, but only used for cell wall hydrolysis in Gram-positive bacteria like Bacillus subtilis and Streptococcus pneumoniae (8, 9, 31, 44–46).
Fig. 10.
Model for role of FtsEX in divisome assembly and function. The division proteins are indicated with capital letters as in Fig. 1. During divisome assembly, FtsEX localizes to the Z ring by an interaction between FtsE and FtsZ. FtsEX then activates FtsA by antagonizing FtsA polymerization, leading to the sequential recruitment of downstream divisome proteins. ATP binding/hydrolysis is not required for this step. Before FtsN arrives at the Z ring, FtsQLB and FtsW keep FtsI in the inactive state. When FtsN arrives at the Z ring it interacts with FtsA in the cytoplasm through NFtsN and EFtsN relieves the repression of FtsQLBW in the periplasm to start septal PG synthesis. FtsEX hydrolyzes ATP to activate the amidases to cleave the newly synthesized septal PG. After ATP hydrolysis, FtsEX (in the ADP form) locks FtsA in the inactive form or prevents FtsA from communicating with other components of the divisome so that septal PG synthesis is blocked. Repeated rounds of ATP hydrolysis by FtsEX results in cycling of synthesis and hydrolysis of cell wall at the septum. For simplicity, many division proteins, such as ZipA, are left out of the picture. FtsA is drawn as a monomer, which is active during assembly and constriction. NFtsN and EFtsN are colored red and yellow, respectively. Active FtsI is indicated by a red star. How the activation signals in FtsA and FtsBLQ are transmitted to FtsI is not clear and dashed arrows are used in the diagram.
One intriguing observation is that many conditions that bypass FtsEX cannot suppress the FtsA mutants impaired for interaction with FtsEX. For example, all ftsA mutations that bypass ZipA can suppress the growth defect of an ftsEX::cat strain, but only a few can suppress the FtsAR63H or FtsAG366D mutants. It is not clear why there is a difference, but the ability of these mutations to suppress the FtsAR63H or FtsAG366D mutants appears to correlate with their ability to bypass ZipA. For example, FtsAR286W and FtsAL204, which suppress, are stronger (require less FtsA to bypass ZipA) than FtsAE124A or FtsAI143L, which don’t suppress (20). It is also possible that these mutations affect FtsA differently, which is not revealed by our tests to date. Similarly, FtsBE56A, which is stronger (less FtsN-dependent) than FtsBD59H, suppresses, whereas FtsBD59H cannot suppress (24). One possibility is that FtsEX becomes inhibitory to the Z ring when its interaction with FtsA is lost, such that a stronger mutation is required. Another possibility is that the ftsAR63H or ftsAG366D mutation affects another function of FtsA, which would require a strong hyperactive mutation to overcome the defect. Whatever the mechanism is, this difference is intriguing and warrants further investigation.
Although the ATPase mutants of FtsEX support divisome assembly, they do not support cell constriction (31). Here we showed that the FtsED162NX mutant, which is predicted to be deficient in ATP binding, acts on FtsA to block the assembled divisome from starting constriction, indicating ATP binding or hydrolysis by FtsEX is necessary. This role of FtsEX in divisome activation is independent of its role in promoting divisome assembly because cells expressing the mutant do not have a defect in divisome assembly (31). It is also independent of its role in controlling the activity of cognate amidases because the FtsED162NX∆152–161 mutant, which lacks the domain for interaction with the amidase activator EnvC, still prevents constriction. This block to division, however, can be suppressed by some conditions that activate the divisome, such as overexpression of FtsN and hyperactive mutations in ftsB, ftsL, and ftsW but not ftsA.
Reducing the FtsA–FtsX interaction by FtsA mutations isolated here renders cells less sensitive to the division inhibitory activity of FtsED162NX, given that a suppressor mutation (like ftsAR286W) is present to suppress the defect of the FtsA mutations. These results suggest that FtsED162NX inhibits divisome activation by locking FtsA in an inactive state or by preventing FtsA from communicating with the periplasmic side of the divisome to start septal PG synthesis. Because the ATPase activity is also required for FtsEX to activate the amidases at the septum (9), our results suggest that FtsEX couples divisome activation (septal PG synthesis) with cell wall hydrolysis through acting on FtsA (Fig. 10). In this model, when FtsN arrives at the Z ring, FtsEX hydrolyzes ATP to activate the divisome in an FtsA-dependent manner to synthesize septal PG and also to activate the amidases via EnvC to hydrolyze septal PG. In the absence of ATP hydrolysis, both the divisome and amidases are inactive, indicating that a continuous ATP cycle is necessary for septal PG synthesis and hydrolysis. This coordination of septal PG synthesis and hydrolysis ensures that the division process proceeds smoothly to avoid possible catastrophic consequences if the divisome malfunctions. Consistent with this model, cell separation is delayed in cells where septal PG synthesis occurs without septal cell wall hydrolysis controlled by FtsEX (cells form chains in the FtsEX∆152–161 mutant strain, ∆envC strain, or ∆amiA ∆amiB strain) (9) and cells lyse when cell wall hydrolysis occurs without septal PG synthesis (cells expressing AmiA or AmiB mutants that are active without FtsEX lyse) (48).
Whereas FtsEX’s role in amidase activation is dispensable for cell survival, its roles in divisome assembly and activation are essential for E. coli division in medium with low osmolarity. Any condition that bypasses FtsEX has to compensate for these two functions. Overexpression of FtsN bypasses FtsEX probably because it can interact with FtsA to back-recruit the late-division proteins to the Z ring and also activate the divisome. Consistent with this finding, both NFtsN and EFtsN are necessary for this bypass (SI Appendix, Fig. S18). FtsA mutants impaired for self-interaction likely bypass FtsEX because the binding site for a late-division protein has been exposed, making them hyperactive for division. How hyperactive FtsB, FtsL, and FtsW mutants bypass FtsEX is mysterious because they are downstream of FtsEX and no report suggests they antagonize FtsA polymerization. However, such mutants have been reported to survive with depleted levels of FtsA or FtsK and bypass ZipA (37), so it is not surprising that they also bypass FtsEX. These mutants probably enable one or more of the late-division proteins to interact better with an early division protein (likely FtsA) to promote divisome assembly. Several other conditions, such as high osmolarity and overexpression of FtsP, probably bypass FtsEX by increasing the level of FtsN or enhancing protein–protein interactions between the early- and late-divisome proteins, as suggested for the FtsB, FtsL, or FtsW mutants.
The binding site for FtsA on FtsX remains to be determined. Because FtsX is a membrane protein with four transmembrane domains and both its N-terminal and C-terminal domains reside in the cytoplasm (31), there are three potential binding sites for FtsA: the N-terminal domain, the C-terminal domain, or the loop between the second and the third transmembrane segments. However, based on analogy to the ABC transporters, such as the LolCDE or LptBFG systems (49, 50), the loop is probably required for interaction with FtsE. Therefore, it is likely that either the N-terminal or the C-terminal domain of FtsX interacts with FtsA. Another issue about FtsEX is how FtsEX localizes to the Z ring. One report suggested that FtsEX localizes to the midcell through an interaction between FtsX and its target protein (31), whereas another report has argued that FtsEX localizes to the Z ring through an interaction between FtsE and FtsZ (51). We showed here the interaction between FtsA and FtsX is not necessary for FtsEX localization: FtsX–GFP localizes very weakly to midcell in the absence of FtsE and FtsEX–GFP localizes to midcell in the presence of FtsA mutations that compromise the FtsA–FtsX interaction. These results suggest that FtsEX localizes to the Z ring through interaction between FtsE and FtsZ. However, as it has also been reported that FtsX–GFP localizes to the Z ring without FtsE (31), we cannot exclude the possibility that our FtsX–GFP fusion (with FtsE) is not fully functional although it does complement ΔftsEX.
In addition to elucidating the function of FtsEX, our study also provides additional insights on divisome assembly and function. First, the bypass of FtsEX by FtsA mutants impaired for self-interaction strongly supports the model in which FtsA monomers recruit downstream division proteins to the Z ring (20, 25). In this model, both ZipA and FtsEX promote divisome assembly by antagonizing FtsA polymerization, with ZipA acting indirectly and FtsEX acting directly. FtsN overexpression may also bypass FtsEX by antagonizing FtsA polymerization with FtsN binding FtsA monomers and back recruiting other late-division proteins when the linear sequential pathway is disrupted by the loss of FtsEX. In this model unpolymerized FtsA favors divisome assembly because binding sites for the late division proteins are only available in the unpolymerized form. Second, previous studies have emphasized that the IC domain of FtsA is uniquely involved in the switch to the active form because only mutations in the IC domain of FtsA were isolated or tested in the bypass of EFtsN (24, 38). However, here we showed that all mutations that impair self-interaction of FtsA, regardless of their locations in FtsA, require less FtsN for division (SI Appendix, Fig. S7). This finding suggests that monomeric FtsA, if it is not the active form, at least potentiates the active form. Third, isolation of mutations adjacent to the CCD domain of FtsB as hyperactive division mutations indicate that the CCD domain of FtsB should be extended. These mutations and those isolated by the de Boer group are all located in the coiled-coil region of FtsB (residues 50–70) (24) and likely disrupt interaction between FtsB and another protein. Finally, isolation of ftsWM269I as a hyperactive division mutation suggests that FtsW, along with the FtsQLB complex, plays an important role in keeping the divisome inactive before FtsN arrives at the Z ring. Several mutations in C. cresentus ftsW have been isolated as hyperactive division mutations (40). Strikingly, our ftsWM269I is adjacent to the corresponding residue of the most potent one (ftsWA246T) (40). These observations suggest that the domain where these mutations are located has a critical role in FtsW function in divisome activation. It will be of interest to determine how the ftsWM269I mutation results in a hyperactive divisome. More importantly, identification of FtsW as a protein involved in divisome activation suggests that the FtsN triggered activation signals from FtsA and FtsQLB converge on the FtsW–FtsI complex to start septal PG synthesis (Fig. 10).
Materials and Methods
SI Appendix, SI Materials and Methods contains descriptions of bacterial strains, plasmids, strain and plasmid constructions, growth conditions, and procedure to screen for the FtsED162NX suppressors. This section also contains procedures for immunofluorescence microscopy, BTH assays, and visualization of GFP fusion proteins, as well as Western blot. FtsED162NX suppressor mutations, bacterial strains, plasmids, and oligonucleotide primers used in this study are listed in SI Appendix, Tables S1–S4, respectively.
Supplementary Material
Acknowledgments
We thank Dr. Thomas Bernhardt, Dr. Piet de Boer, and Dr. David Weiss for sending us strains and plasmids. This study was support by NIH Grant GM29746 (to J.L.) and a Biomedical Research Training Program fellowship from University of Kansas Medical Center (to S.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606656113/-/DCSupplemental.
References
- 1.Lutkenhaus J, Pichoff S, Du S. Bacterial cytokinesis: From Z ring to divisome. Cytoskeleton (Hoboken) 2012;69(10):778–790. doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Boer PA. Advances in understanding E. coli cell fission. Curr Opin Microbiol. 2010;13(6):730–737. doi: 10.1016/j.mib.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354(6349):161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 4.Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol. 2005;55(6):1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- 5.Hale CA, de Boer PA. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88(2):175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 6.Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21(4):685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang KH, Durand-Heredia J, Janakiraman A. FtsZ ring stability: Of bundles, tubules, crosslinks, and curves. J Bacteriol. 2013;195(9):1859–1868. doi: 10.1128/JB.02157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt KL, et al. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J Bacteriol. 2004;186(3):785–793. doi: 10.1128/JB.186.3.785-793.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang DC, et al. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc Natl Acad Sci USA. 2011;108(45):E1052–E1060. doi: 10.1073/pnas.1107780108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aarsman ME, et al. Maturation of the Escherichia coli divisome occurs in two steps. Mol Microbiol. 2005;55(6):1631–1645. doi: 10.1111/j.1365-2958.2005.04502.x. [DOI] [PubMed] [Google Scholar]
- 11.Goehring NW, Beckwith J. Diverse paths to midcell: Assembly of the bacterial cell division machinery. Curr Biol. 2005;15(13):R514–R526. doi: 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 12.Gerding MA, et al. Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J Bacteriol. 2009;191(24):7383–7401. doi: 10.1128/JB.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lutkenhaus J. FtsN—Trigger for septation. J Bacteriol. 2009;191(24):7381–7382. doi: 10.1128/JB.01100-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peters NT, Dinh T, Bernhardt TG. A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators. J Bacteriol. 2011;193(18):4973–4983. doi: 10.1128/JB.00316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arends SJ, et al. Discovery and characterization of three new Escherichia coli septal ring proteins that contain a SPOR domain: DamX, DedD, and RlpA. J Bacteriol. 2010;192(1):242–255. doi: 10.1128/JB.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yahashiri A, Jorgenson MA, Weiss DS. Bacterial SPOR domains are recruited to septal peptidoglycan by binding to glycan strands that lack stem peptides. Proc Natl Acad Sci USA. 2015;112(36):11347–11352. doi: 10.1073/pnas.1508536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PA. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol. 2007;63(4):1008–1025. doi: 10.1111/j.1365-2958.2006.05571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goehring NW, Gueiros-Filho F, Beckwith J. Premature targeting of a cell division protein to midcell allows dissection of divisome assembly in Escherichia coli. Genes Dev. 2005;19(1):127–137. doi: 10.1101/gad.1253805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geissler B, Elraheb D, Margolin W. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc Natl Acad Sci USA. 2003;100(7):4197–4202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pichoff S, Shen B, Sullivan B, Lutkenhaus J. FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA’s self-interaction competes with its ability to recruit downstream division proteins. Mol Microbiol. 2012;83(1):151–167. doi: 10.1111/j.1365-2958.2011.07923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddy M. Role of FtsEX in cell division of Escherichia coli: Viability of ftsEX mutants is dependent on functional SufI or high osmotic strength. J Bacteriol. 2007;189(1):98–108. doi: 10.1128/JB.01347-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karimova G, Dautin N, Ladant D. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol. 2005;187(7):2233–2243. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Busiek KK, Eraso JM, Wang Y, Margolin W. The early divisome protein FtsA interacts directly through its 1c subdomain with the cytoplasmic domain of the late divisome protein FtsN. J Bacteriol. 2012;194(8):1989–2000. doi: 10.1128/JB.06683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu B, Persons L, Lee L, de Boer PA. Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol Microbiol. 2015;95(6):945–970. doi: 10.1111/mmi.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pichoff S, Du S, Lutkenhaus J. The bypass of ZipA by overexpression of FtsN requires a previously unknown conserved FtsN motif essential for FtsA-FtsN interaction supporting a model in which FtsA monomers recruit late cell division proteins to the Z ring. Mol Microbiol. 2015;95(6):971–987. doi: 10.1111/mmi.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Addinall SG, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol. 1997;25(2):303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- 27.Dai K, Xu Y, Lutkenhaus J. Topological characterization of the essential Escherichia coli cell division protein FtsN. J Bacteriol. 1996;178(5):1328–1334. doi: 10.1128/jb.178.5.1328-1334.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Busiek KK, Margolin W. A role for FtsA in SPOR-independent localization of the essential Escherichia coli cell division protein FtsN. Mol Microbiol. 2014;92(6):1212–1226. doi: 10.1111/mmi.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsang MJ, Bernhardt TG. Guiding divisome assembly and controlling its activity. Curr Opin Microbiol. 2015;24:60–65. doi: 10.1016/j.mib.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiss DS. Last but not least: new insights into how FtsN triggers constriction during Escherichia coli cell division. Mol Microbiol. 2015;95(6):903–909. doi: 10.1111/mmi.12925. [DOI] [PubMed] [Google Scholar]
- 31.Arends SJ, Kustusch RJ, Weiss DS. ATP-binding site lesions in FtsE impair cell division. J Bacteriol. 2009;191(12):3772–3784. doi: 10.1128/JB.00179-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernhardt TG, de Boer PA. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol Microbiol. 2004;52(5):1255–1269. doi: 10.1111/j.1365-2958.2004.04063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heidrich C, et al. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol. 2001;41(1):167–178. doi: 10.1046/j.1365-2958.2001.02499.x. [DOI] [PubMed] [Google Scholar]
- 34.de Leeuw E, et al. Molecular characterization of Escherichia coli FtsE and FtsX. Mol Microbiol. 1999;31(3):983–993. doi: 10.1046/j.1365-2958.1999.01245.x. [DOI] [PubMed] [Google Scholar]
- 35.Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol. 1999;181(2):508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S, Arends SJ, Weiss DS, Newman EB. A deficiency in S-adenosylmethionine synthetase interrupts assembly of the septal ring in Escherichia coli K-12. Mol Microbiol. 2005;58(3):791–799. doi: 10.1111/j.1365-2958.2005.04864.x. [DOI] [PubMed] [Google Scholar]
- 37.Tsang MJ, Bernhardt TG. A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol Microbiol. 2015;95(6):925–944. doi: 10.1111/mmi.12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernard CS, Sadasivam M, Shiomi D, Margolin W. An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli. Mol Microbiol. 2007;64(5):1289–1305. doi: 10.1111/j.1365-2958.2007.05738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lara B, Ayala JA. Topological characterization of the essential Escherichia coli cell division protein FtsW. FEMS Microbiol Lett. 2002;216(1):23–32. doi: 10.1111/j.1574-6968.2002.tb11409.x. [DOI] [PubMed] [Google Scholar]
- 40.Modell JW, Kambara TK, Perchuk BS, Laub MT. A DNA damage-induced, SOS-independent checkpoint regulates cell division in Caulobacter crescentus. PLoS Biol. 2014;12(10):e1001977. doi: 10.1371/journal.pbio.1001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szwedziak P, Wang Q, Freund SM, Löwe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31(10):2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pichoff S, Lutkenhaus J. Identification of a region of FtsA required for interaction with FtsZ. Mol Microbiol. 2007;64(4):1129–1138. doi: 10.1111/j.1365-2958.2007.05735.x. [DOI] [PubMed] [Google Scholar]
- 43.Hale CA, de Boer PA. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J Bacteriol. 1999;181(1):167–176. doi: 10.1128/jb.181.1.167-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meisner J, et al. FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol Microbiol. 2013;89(6):1069–1083. doi: 10.1111/mmi.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sham LT, Barendt SM, Kopecky KE, Winkler ME. Essential PcsB putative peptidoglycan hydrolase interacts with the essential FtsXSpn cell division protein in Streptococcus pneumoniae D39. Proc Natl Acad Sci USA. 2011;108(45):E1061–E1069. doi: 10.1073/pnas.1108323108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Domínguez-Cuevas P, Porcelli I, Daniel RA, Errington J. Differentiated roles for MreB-actin isologues and autolytic enzymes in Bacillus subtilis morphogenesis. Mol Microbiol. 2013;89(6):1084–1098. doi: 10.1111/mmi.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mavrici D, et al. Mycobacterium tuberculosis FtsX extracellular domain activates the peptidoglycan hydrolase, RipC. Proc Natl Acad Sci USA. 2014;111(22):8037–8042. doi: 10.1073/pnas.1321812111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang DC, Tan K, Joachimiak A, Bernhardt TG. A conformational switch controls cell wall-remodelling enzymes required for bacterial cell division. Mol Microbiol. 2012;85(4):768–781. doi: 10.1111/j.1365-2958.2012.08138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ito Y, Matsuzawa H, Matsuyama S, Narita S, Tokuda H. Genetic analysis of the mode of interplay between an ATPase subunit and membrane subunits of the lipoprotein-releasing ATP-binding cassette transporter LolCDE. J Bacteriol. 2006;188(8):2856–2864. doi: 10.1128/JB.188.8.2856-2864.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sherman DJ, et al. Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport. Proc Natl Acad Sci USA. 2014;111(13):4982–4987. doi: 10.1073/pnas.1323516111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corbin BD, Wang Y, Beuria TK, Margolin W. Interaction between cell division proteins FtsE and FtsZ. J Bacteriol. 2007;189(8):3026–3035. doi: 10.1128/JB.01581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.