Significance
Many biological processes, including splicing, translation, and genome maintenance, require structured RNAs to fold into complex three-dimensional shapes. Our current understanding of these processes is based on distilling principles from descriptive folding studies. Moving toward predictive models will require coupling observed structural changes with kinetic and thermodynamic measurements. We have dissected P4-P6 RNA folding through distinct structural states and measured the rate and equilibrium constants for transitions between these states. Common kinetics found for RNA tertiary elements embedded in different structural contexts may help develop predictive folding models. Also, our results suggest that RNA folding may be well described by a model analogous to the diffusion-collision model for protein folding.
Keywords: RNA folding, single-molecule FRET, kinetics, folding pathways, RNA tertiary motifs
Abstract
The past decade has seen a wealth of 3D structural information about complex structured RNAs and identification of functional intermediates. Nevertheless, developing a complete and predictive understanding of the folding and function of these RNAs in biology will require connection of individual rate and equilibrium constants to structural changes that occur in individual folding steps and further relating these steps to the properties and behavior of isolated, simplified systems. To accomplish these goals we used the considerable structural knowledge of the folded, unfolded, and intermediate states of P4-P6 RNA. We enumerated structural states and possible folding transitions and determined rate and equilibrium constants for the transitions between these states using single-molecule FRET with a series of mutant P4-P6 variants. Comparisons with simplified constructs containing an isolated tertiary contact suggest that a given tertiary interaction has a stereotyped rate for breaking that may help identify structural transitions within complex RNAs and simplify the prediction of folding kinetics and thermodynamics for structured RNAs from their parts. The preferred folding pathway involves initial formation of the proximal tertiary contact. However, this preference was only ∼10 fold and could be reversed by a single point mutation, indicating that a model akin to a protein-folding contact order model will not suffice to describe RNA folding. Instead, our results suggest a strong analogy with a modified RNA diffusion-collision model in which tertiary elements within preformed secondary structures collide, with the success of these collisions dependent on whether the tertiary elements are in their rare binding-competent conformations.
Structured RNAs play central roles in biology, in the splicing and alternative splicing of eukaryotic pre-mRNAs, in the synthesis of proteins and their transport, and in chromosome maintenance (1–4). Beyond the RNAs and RNA–protein machines involved in these processes, it has been increasingly recognized that Nature has taken extensive advantage of RNA at multiple levels of gene regulation, and considerable current efforts are focused on uncovering the pathways and molecular mechanisms that underlie the functions of small RNAs and long noncoding RNAs (2, 5–8). The pervasive functions of RNA in biology underscore the importance of understanding RNA’s fundamental physical properties and, ultimately, of using this understanding to predict the kinetics and thermodynamics of folding and conformational transitions responsible for RNA function.
Decades of characterization of RNA folding and structure have led to generalizable principles and provided biological insights. The observations that RNA encodes genetic information, forms highly stable local structures, and can catalyze reactions provided support for the possibility that early life evolved using functional RNAs (9–12). This high stability was also recognized to lead to misfolding. Indeed, highly stable misfolded tRNAs were identified in the 1960s (13–15), and many subsequent studies supported a strong propensity of RNA to misfold (16–20). These observations and consideration of the physical properties of RNA (21) led to the “RNA chaperone hypothesis” (22), which predicted the existence of RNA chaperones to prevent and resolve deleterious misfolded states in vivo (20, 23–28).
Due to its complexity, most studies of RNA tertiary folding have been descriptive. Studies to date have identified misfolded intermediates (29–32), revealed multiple folding pathways (33, 34) and functional states (35), and led to the general perspective that RNA folds on a rugged energetic landscape (18). Nevertheless, these conclusions drawn from qualitative studies have been limited and, even at times, misleading. For example, evidence for new intermediates arising upon mutation and for changes in conditions that supported a rugged landscape for RNA folding instead has been shown, upon placement of results within the context of a kinetic folding framework, to represent formation of the same intermediate with altered stability (36). Additionally, an alternative base-pairing of the P3 helix was implicated in forming a long-lived misfolded state of the Tetrahymena group I intron (30) but could be isolated and probed only once the kinetic folding framework was established. Subsequent studies showed that the alternative P3 helix biased partitioning toward the long-lived misfolded state but that the final state contains the native P3 helix with its kinetic stability arising from an altered folding topology that requires near-complete unfolding to rectify (37, 38).
The above examples highlight the importance and power of quantitative analysis of individual reaction steps within complex processes. Such studies have been instrumental in many areas of biology and biochemistry—providing kinetics and frameworks to consolidate existing knowledge, aid the design of new experimental tests, anchor the interpretation of new results, and allow more precise and predictive models to be built (39–43). The power of this information is fully manifest when accompanied by structural information sufficient to allow molecular, and even biological, interpretation of kinetic and thermodynamic behavior (44, 45). However, the folding of RNAs, such as the group I ribozymes, RNase P RNA, and ribosomal RNAs, is extremely complex, with many structural elements, folding intermediates, and pathways, and there is limited structural information about these intermediates, precluding them as foundational models to connect kinetic and thermodynamic measurements with specific structural changes during folding.
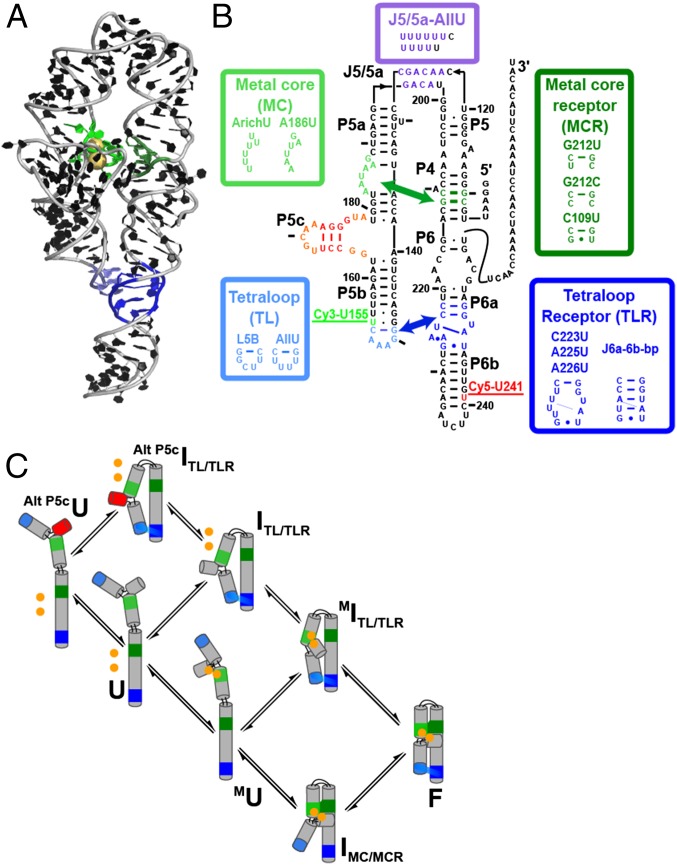
Decades of work on P4-P6 RNA (Fig. 1) have produced an X-ray structure of the folded state and rich structural information that delineate a set of potential folding intermediates and folding pathways (Fig. 1C) (47–57). Moreover, functional work has uncovered sets of mutations that allow ablation and isolation of these folding intermediates with high confidence (49, 51, 53, 56, 58–60). The control afforded by this well-behaved, established system provides an opportunity to deepen understanding and develop and test generalizable principles beyond this model system, provided that a detailed framework is in place to provide context for interpretation. Here, we use single-molecule FRET (smFRET) with a series of P4-P6 mutants to isolate and measure specific folding transitions, extending beyond measurements of overall folding rates (52–55, 61–65) and constructing a kinetic and thermodynamic framework for P4-P6 RNA. This framework defines a preferred folding pathway, allows this pathway and the partitioning of states to be interpreted in structural terms, helps to uncover the origin of the effects of mutations on folding rates and pathways, and suggests a generalized framework for considering RNA tertiary folding.
Fig. 1.
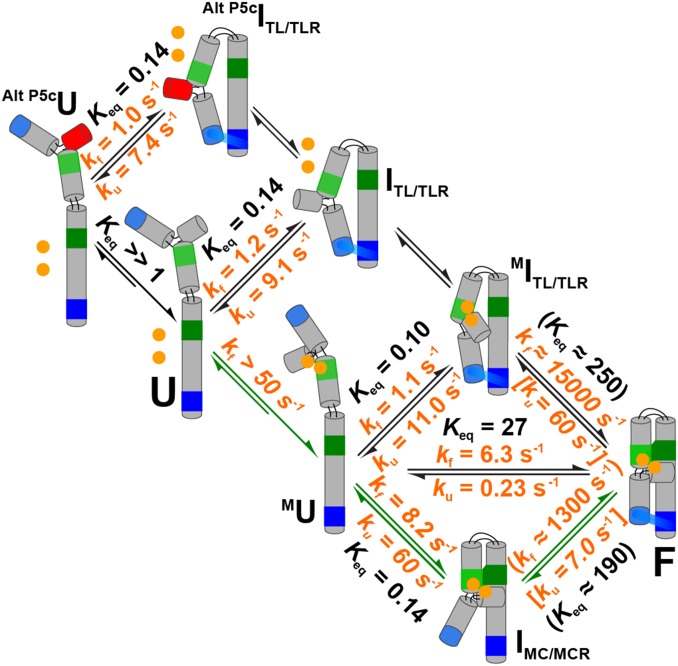
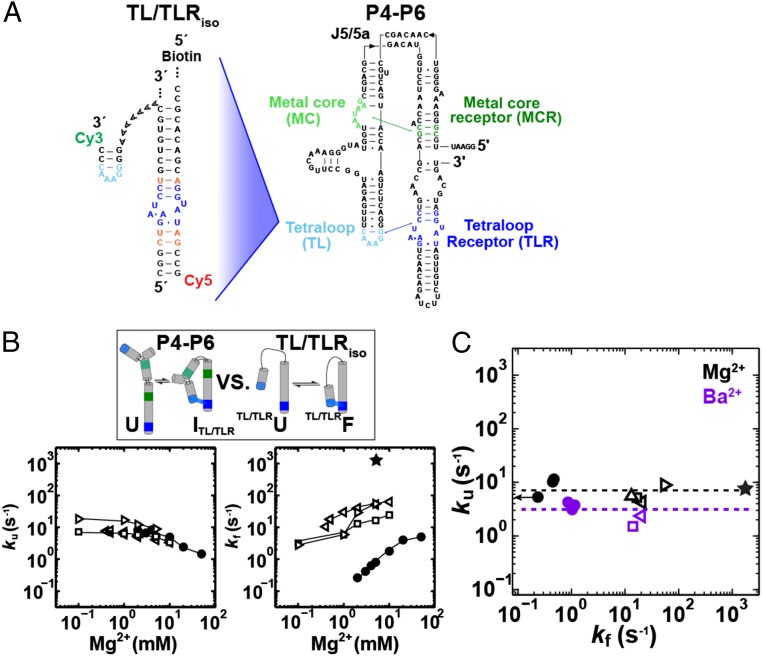
P4-P6 RNA and proposed framework. (A) P4-P6 crystal structure (PDB ID code 1GID). Tertiary contacts are colored as follows: tetraloop (TL, light blue), tetraloop receptor (TLR, dark blue), metal core (MC, light green), metal-core receptor (MCR, dark green). Crystallographically and functionally defined Mg2+ ions bound by the MC are shown as gold spheres (46, 50, 82). (B) P4-P6 secondary structure and variants used in this study to isolate specific folding intermediates (see also Fig. 2A). Tertiary contacts are colored as in A. The positions of the donor and acceptor dyes are shown. (C) Proposed thermodynamic and kinetic framework based on previously identified structural intermediates. P4-P6 is shown in a schematic depiction, color-coded as in A and B, and tertiary contacts are shown throughout as a green swatch and a blue swatch for the MC/MCR and TL/TLR, respectively. Intermediates include the following: Alt P5cU, unfolded state with an alternative P5c secondary structure; Alt P5cITL/TLR, intermediate with the TL/TLR interaction formed and the alternative P5c secondary structure; U, unfolded state with the native P5c secondary structure (states not notated “Alt P5c” have native P5c secondary structure); ITL/TLR, intermediate with only the TL/TLR interaction formed; MU, unfolded state with the MC formed through binding of two Mg2+ ions (depicted as two bound gold spheres); MITL/TLR, intermediate in which the TL/TLR interaction and the MC are formed; IMC/MCR, intermediate with the MC/MCR interaction formed; and F, the native folded state in which both tertiary contacts are formed (49, 56, 60, 77, 82, 118).
A further major goal in RNA folding, beyond a description of the kinetics and thermodynamics of individual structural transitions, is to obtain quantitative and predictive algorithms for RNA tertiary structure, akin to the predictive rules for RNA secondary structure formation. Although this problem is complex, several observations suggest that such a goal may be achievable due to a modular architecture and folding behavior of RNA (66, 67). This perspective is based on the observations that structures of tertiary contacts and junctions found in different RNA structures can be overlaid with little deviation (68–70) and that these substructures, or motifs, can be “cut and pasted” from one structural context to another with function retained (67–69, 71–75). From these observations, it has been proposed that it may be possible to predict the thermodynamics and kinetics of RNA tertiary structure formation from the energetic contributions of these motifs and other components of RNA structure (67). However, despite the structural similarity and transferability of motifs, there is as yet no quantitative assessment of whether the energetic properties of tertiary structure elements are the same in different structural contexts—a basic requirement for the viability of an energetic reconstruction of RNA tertiary structure from its components.
The P4-P6 kinetic and thermodynamic folding framework we derive provides an opportunity to assess whether components of RNA tertiary structure are energetically modular. Specifically, the P4-P6 framework isolates the formation and breaking of each of its two tertiary contacts, the tetraloop/tetraloop receptor and the metal core/metal-core receptor, allowing us to compare the rate constants for breaking these tertiary contacts in P4-P6 with those in simplified RNA constructs with each tertiary motif alone. We find kinetic properties of these motifs that are independent of structural context and thus may be transferrable, thereby providing an early step toward a predictive, energetic model for RNA tertiary structure.
Results and Discussion
We first developed the P4-P6 folding framework from structural states previously identified and measured the rate and equilibrium constants for transitions between these states with smFRET. We then used this framework to isolate the formation and dissolution of particular tertiary contacts to test whether the kinetics properties of this structural transition are transferable to other RNA contexts. Finally, we probed properties of the framework, in particular the flux through folding pathways in response to mutation, that suggest features of RNA folding that are analogous to simple protein-folding models and may be general.
Building and Determining a Kinetic and Thermodynamic Framework for P4-P6 RNA.
Building the structural states for the P4-P6 framework.
We first built on the large body of work on P4-P6 RNA (e.g., refs. 47–56, 59, 60, 65 and 76–82) to construct a framework of structural states, and we then probed transitions between these states to establish a kinetic and thermodynamic folding framework.
The P456 helical stack of P4-P6 (Fig. 1B, Right) connects to the P5abc helices (Fig. 1B, Left) via the two-way J5/5a junction, which enables a 180-degree turn that allows the P5abc helix to be roughly parallel to the P456 helices (Fig. 1A). This parallel configuration is enforced by two tertiary contacts of P4-P6, the metal core/metal-core receptor (MC/MCR) and the tetraloop/tetraloop receptor (TL/TLR). TL/TLR tertiary motifs are found in many structured RNAs (70), and, whereas the MC/MCR may be unique to this RNA (50), specific divalent binding sites are common in tertiary structures (83–87). In addition, many structured RNAs have junctions that, like J5/5a, exhibit distinct bending properties (51, 68, 88, 89).
The folding of P4-P6 has been followed in multiple studies, with folding rate constants of 2–30 s−1 observed across a range of conditions (52–55, 61–65). However, interpretation of these overall rate constants has been stymied by a lack of knowledge of which individual folding steps were followed and rate limiting. Fig. 1C diagrams potential intermediates in the folding of P4-P6 suggested by prior structural work (47–57). States differ in whether an alternative secondary structure is present (Fig. 1C, Alt P5c, red), whether Mg2+ ions are bound to the metal core (Fig. 1C, gold circles), and whether each tertiary contact is formed (Fig. 1C, MC/MCR, green and TL/TLR, blue). NMR, small angle X-ray scattering, and chemical probing data with WT and mutant P4-P6 RNAs provide evidence for each of these structural states and conditions or mutations that allow each species to predominate (49, 51, 53, 56, 58–60).
Determining the P4-P6 kinetic and thermodynamic framework.
We studied the folding of the P4-P6 constructs depicted in Fig. 1B by smFRET.
smFRET allows determination of forward and reverse reaction rate constants and highly unfavorable and favorable equilibrium constants. This ability, along with homogenous molecular behavior (ref. 90 and Materials and Methods), allows construction of a folding framework under uniform conditions, avoiding uncertainties and complications from extrapolation of RNA behavior across conditions (62, 82, 91). Example traces and data quality analyses for all constructs can be found in Dataset S1. We emphasize the importance of reporting full characterization and analysis of all traces (92), rather than just representative traces and a final model, as well as the need to have single-molecule data in a common format and publically available for future analysis (93). In the following sections, we describe how individual steps and partition points in the overall folding framework have been assessed.
Determining the predominant unfolded state.
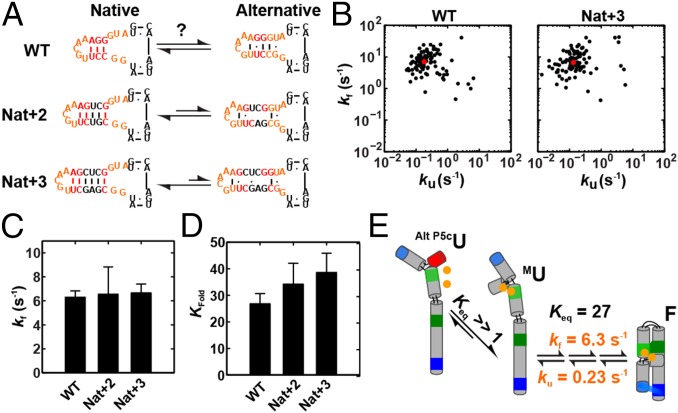
Prior studies by Wu and Tinoco (77) identified an alternative secondary structure within the P5c helix of an isolated P5abc construct in the absence of Mg2+ (Fig. 2A). Thus, it was necessary to determine, under the folding conditions used herein [5 mM Mg2+, 100 mM KCl, 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS), pH 8.0], whether the unfolded state has P5c predominantly with the native or alternative secondary structure and whether the predominant folding pathway proceeded through the alternative secondary structure state (Fig. 1C). We therefore mutated P5c to stabilize the native form of P5c over the alternatively base-paired form (Nat+2 and Nat+3) (Fig. 2A) (94). If the alternative unfolded state (Alt P5cU) were the prevalent unfolded state, then Nat+2 and Nat+3 would be predicted to increase the folding equilibrium constant and, in the simplest case of folding proceeding subsequently through U, increase the overall folding rate as well. In contrast, if P5c existed predominantly with its native base pairing in its predominant unfolded state (U or MU) (Fig. 1C), then further stabilizing the native base pairing would have little if any effect on the rate and equilibrium constants.
Fig. 2.
Overall folding of WT P4-P6 and constructs with the native P5c secondary structure enforced. (A) The local equilibrium between the native and alternative P5c secondary structure of WT P4-P6 and constructs with the native P5c secondary structure enforced by two (Nat+2) or three (Nat+3) base pairs (46, 50, 77, 82). (B) Scatter plot of folding versus unfolding rate constants for WT and Nat+3 P4-P6. Data for 100 randomly selected molecules with the median folding and unfolding rate constants denoted by red circles (Dataset S1). See SI Appendix, Fig. S1 for data for a variant with the alternative P5c secondary structure (Alt+2). (C and D) Comparison of the folding rate (C) and equilibrium (D) constants of WT, Nat+2, and Nat+3. Error bars correspond to the bootstrap-estimated 95% confidence intervals (SD = 2σbootstrap). See SI Appendix, Table S1 for data and SI Appendix, Supporting Methods for experimental details. Conditions were as follows: 50 mM MOPS, pH 8.0, 5 mM MgCl2, and 100 mM KCl at 23 °C. (E) Overall P4-P6 folding occurs from an unfolded state with native P5c secondary structure (MU).
Fig. 2B summarizes the overall folding and unfolding rate and equilibrium data for individual molecules of WT P4-P6 and Nat+3 P4-P6 (the data and analysis for all smFRET experiments are presented in Dataset S1). The rate constants for individual molecules are shown and demonstrate that the behavior of individual molecules is highly homogeneous for each construct. Nat+3 and Nat+2 P4-P6 give folding and unfolding rate and equilibrium constants that are nearly the same as WT P4-P6 (Fig. 2 C and D and SI Appendix, Table S1), suggesting that an unfolded state or states with native P5c secondary structure predominate over one with the alternative secondary structure (Alt P5cU) under the conditions of our experiments.
Consistent with these results, an Mg1/2 of ∼0.25 mM is observed for binding of Mg2+ to the metal-ion core (53), strongly suggesting that the predominant unfolded state at 5 mM Mg2+ has bound metal ions and the correct P5c secondary structure (77). This state is referred to as MU herein (Fig. 1C and Fig. 2E) and predominates over U. From the above, it seemed most likely that folding would proceed from MU to more folded states. Nevertheless, alternative pathways via the Alt P5cU or U states were possible. To rigorously assess these potential folding pathways, and others in subsequent sections, we took advantage of the fact that any step that is on the folding pathway must occur with a rate constant of 6.3 s−1 or greater because no step on a pathway can occur slower than the overall rate constant (Fig. 2E).
To test the possibility that folding could involve fast formation of Alt P5cITL/TLR from the small fraction of P4-P6 in the Alt P5cU state (Fig. 1C), we measured the Alt P5cU-to-Alt P5cITL/TLR transition, using a construct analogous to Nat+2 that stabilizes the Alt P5cU state instead (Alt+2). This construct folds at 1.0 s−1, which is substantially slower than the overall folding rate of 6.3 s−1 (SI Appendix, Fig. S1 and Table S1) and thus rules out folding via Alt P5cU as the preferred pathway (Fig. 2E).
Does P4-P6 fold via MU or ITL/TLR?
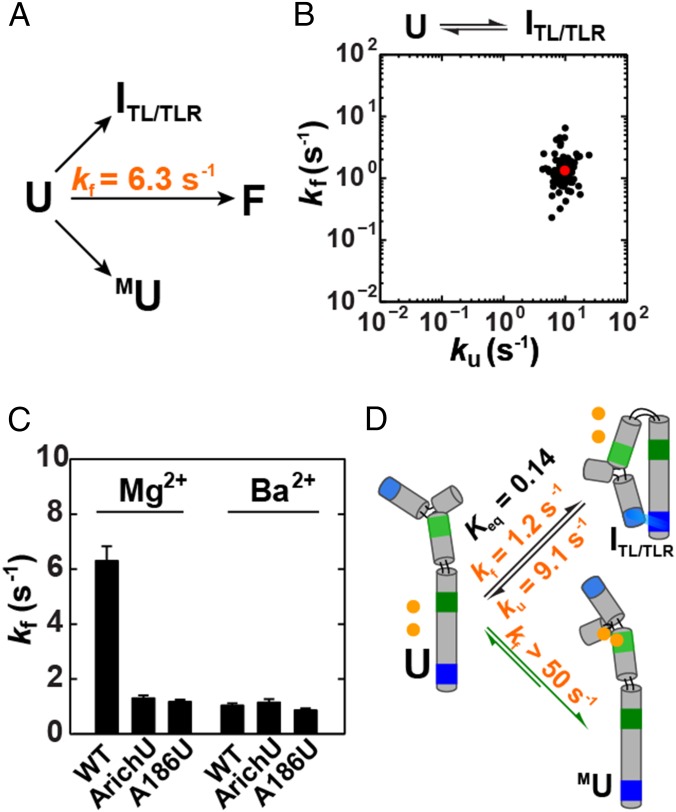
Having established MU as predominant over Alt P5cU and U, the simplest scenario would be predominant folding from it through one or both intermediates with a single tertiary interaction formed (MITL/TLR and IMC/MCR) (Fig. 1C). Nevertheless, a predominant folding from U to ITL/TLR would also have been possible. Specifically, the small amount of U present could first form the TL/TLR tertiary interaction very rapidly to give ITL/TLR, and subsequent folding could follow from that species (Fig. 3A).
Fig. 3.
Does the folding pathway proceed via formation of the tetraloop/tetraloop receptor (TL/TLR) or the metal core (MC)? (A) Folding from U could occur either through ITL/TLR or MU. (B) Scatter plot of the folding versus unfolding rate constants of the ArichU mutant (Fig. 1C) that isolates the U-to-ITL/TLR transition. (C) Comparison of the folding rate constants of WT, ArichU, and A186U P4-P6 in Mg2+ and Ba2+. Prior work has shown that Ba2+ does not bind to the metal core so the same folding behavior is predicted for WT P4-P6 and the ArichU and A186U mutants in Ba2+ (81). This expectation was met, suggesting that there are no unintended effects from the above mutations. Errors as in Fig. 2. Conditions were as follows: 50 mM MOPS, pH 8.0, 5 mM MgCl2 or BaCl2, and 100 mM KCl at 23 °C. (D) P4-P6 folds predominantly via formation of the MU intermediate (green arrows). The equilibrium and kinetic values shown for the U-to-ITL/TLR pathway are average values from A186U and ArichU variants (SI Appendix, Table S1). Estimation of the limit for the U-to-MU transition of >50 s−1 was determined from previous literature data (SI Appendix, SI Text).
We isolated the folding of U to ITL/TLR by mutating residues in the so-called A-rich bulge, an internal loop that binds Mg2+ ions; and these mutations were previously shown to prevent Mg2+ binding (Fig. 1B, ArichU) (56). Fig. 3B shows the folding and unfolding of individual ArichU RNA molecules, and Fig. 3C compares the folding rate constants for this mutant, an additional mutant that prevents Mg2+ binding (A186U) (58), and WT P4-P6. These mutants can fold only to ITL/TLR whereas WT P4-P6 folds to its final state (F) with both contacts under these conditions (56). The MC mutants fold ∼10-fold slower than WT (Fig. 3C and SI Appendix, Table S1). Because an overall process cannot occur faster than its slowest step, a pathway involving a rare state such as U would need to be correspondingly faster than the overall folding rate. Thus, these data strongly suggest that a pathway via ITL/TLR is not significantly traversed in WT P4-P6 folding—i.e., this pathway is not kinetically competent.
The observation that folding via ITL/LTR is not kinetcally competent requires that folding of U to MU be at least as fast as the overall folding rate constant of 6.3 s−1 because this step is on the pathway that must be traversed (Fig. 3D). Although our smFRET assay cannot detect the U-to-MU transition, because there is no significant FRET difference between these states, prior time-resolved hydroxyl radical footprinting results (53) suggest that Mg2+ binding is fast relative to overall P4-P6 folding, with a rate constant of >50 s−1 (Fig. 3D and SI Appendix, SI Text), further supporting the preferred folding pathway from U to MU.
Does folding proceed from MU via MITL/TLR or IMC/MCR?
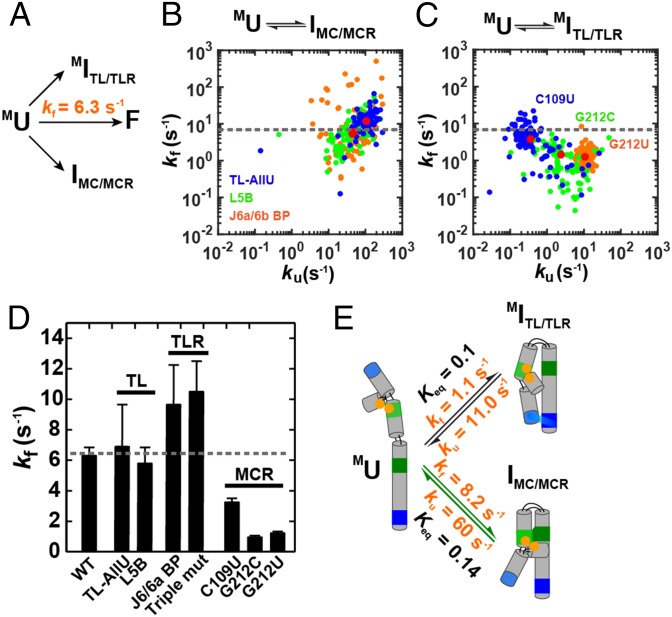
Once Mg2+ ions are bound to the metal core (MC) to give MU, two possible intermediates can form next, MITL/TLR or IMC/MCR (Fig. 1C and Fig. 4A). We distinguished between these potential folding pathways from MU using mutations to isolate the transitions to MITL/TLR and IMC/MCR (Fig. 4A). To isolate the MU-to-IMC/MCR transition, we mutated the tetraloop (TL) and tetraloop receptor (TLR) (Fig. 1B). The folding of these mutants is as fast as that for overall folding (Fig. 4B, dashed line), and the same results were obtained for multiple constructs with mutations in the TL or TLR (Fig. 4D and SI Appendix, Table S1).
Fig. 4.
Does folding proceed from MU via formation of the tetraloop/tetraloop receptor (TL/TLR) or the metal core/metal-core receptor (MC/MCR)? (A) From MU, folding can proceed through MITL/TLR or IMC/MCR. (B) Scatter plot of the folding versus unfolding rate constants for the TL-AllU (blue), L5B (green), and J6a/6b BP (orange) mutants that destabilizes the TL/TLR; and (C) MCR variants C109U (blue), G212C (green), and G212U (orange) that destabilize the formation of the MC/MCR contact (SI Appendix, Fig. S5). (D) Comparison of the folding rate constants of WT P4-P6 with those of the TL, TLR, and MCR mutants. Triple mut, the A225U/A226U/C223U variant. The dotted gray lines in C and D indicate the rate constant for WT P4-P6 folding. The values are from SI Appendix, Table S1, and the error estimates and conditions are as in Fig. 3. (E) Preferential folding via IMC/MCR. The rate constants for the MU-to-MITL/TLR transition are for the G212U mutant (SI Appendix, Table S1) because this mutant has rate constants and behavior similar to MC mutations that ablate the MC/MCR interaction (56). The rate constants for the MU-to-IMC/MCR transition are averages of the values for the TL and TLR variants in D.
These results meet a necessary criterion for folding proceeding through the IMC/MCR intermediate but alone are not sufficient to establish this pathway over pathways through the MITL/TLR intermediate (Fig. 4A): i.e., a downstream step subsequent to formation of the MITL/TLR intermediate could be slow and thereby render this path kinetically incompetent. Thus, we used MCR mutations previously shown to affect the formation of the MC/MCR (Fig. 1B) (79) to evaluate the potential for folding through the MITL/TLR intermediate (Fig. 4A). Each of the three MCR mutations led to different levels of destabilization (Fig. 4C and SI Appendix, Table S1), suggesting that the two least destabilizing mutants did not fully ablate the MC/MCR. Because the MC/MCR motif involves A-minor interactions between adenosine residues and 2′-hydroxyl groups of residues in Watson–Crick base pairs, it may be particularly refractory to full ablation (50, 79, 95, 96). Nevertheless, the most destabilizing mutant, G212U, has rate constants that are the same as variants with mutations in the MC that remove Mg2+ binding and ablate the MC/MCR (56)(SI Appendix, Table S1) (G212U vs. ArichU and A186U), suggesting that the G212U variant also ablates the MC/MCR interaction. The G212U mutant and other MCR mutants gave folding rate constants slower than overall folding (Fig. 4D, dashed line vs. MCR mutants), providing evidence that the pathway with TL/TLR formation before MC/MCR formation is not kinetically competent (Fig. 4E).
The MC and MCR tertiary contact partners that form first in folding are closer in secondary structure than the TL and TLR and are also closer to the major bend in the molecule mediated by the J5/5a junction (Fig. 1A) (49). This observation is reminiscent of the correlation over four orders of magnitude of folding rates with the closeness of tertiary contact partners in primary sequence for a subset of proteins (97). This concept has also been explored in, and suggested for, RNA folding, but only overall reaction rates were followed and effects were modest in scale (98, 99). Herein, we observed a pathway preference of only ∼10-fold (Figs. 4E and 5), despite the close positioning of the MC and MCR tertiary elements (Fig. 1), and, as shown below, this preference can be reversed by a single point mutation. These results underscore the need for more complex descriptions for the RNA folding process, and our results further suggest a general framework that may be appropriate (Analysis of Mutational Effects on Folding Pathways).
Fig. 5.
Thermodynamic and kinetic framework of P4-P6 folding (5 mM MgCl2, 100 mM KCl, 23 °C). The preferred folding pathway is depicted with the green arrows. Values for each step are taken from results in Fig. 3D, Fig. 4E, and SI Appendix, Fig. S1. The value in the square bracket for F→IMC/MCR is the average of the unfolding rate constants determined for constructs that isolate the TL/TLR (SI Appendix, Table S2). The value in the square bracket for F → MITL/TLR is the average of unfolding rate constants for constructs that isolate the MC/MCR (P4-P6, MU⇌IMC/MCR) (SI Appendix, Table S1); (MC/MCRiso, 5 mM Mg2+) (SI Appendix, Table S3). The value for Alt P5cITL/TLR→Alt P5cU was corrected by a factor of two to account for the uniform twofold slower unfolding in 5 mM Ba2+ relative to 5 mM Mg2+ (Fig. 6C and SI Appendix, Table S1) because the experiment was carried out with Ba2+ to prevent Mg2+ occupancy of the metal core (MC) (81). Values in parentheses were calculated from completion of the thermodynamic cycle (SI Appendix, Supporting Methods).
The overall kinetic and thermodynamic framework for P4-P6 folding.
Fig. 5 presents the rate and equilibrium constants for individual reaction steps in the P4-P6 folding framework, combining the results described above with additional results and inferences from the following section (Modularity of Tertiary Contact Unfolding Kinetics). Our findings can be summarized as follows: The MU state predominates over the Alt P5cU and U states, and folding does not proceed through these less populated unfolded states because folding to Alt P5cITL/TLR and ITL/TLR from Alt P5cU and U, respectively, is slower than overall folding. MU proceeds preferentially to IMC/MCR, with a rate constant similar to that for overall folding, whereas folding to MITL/TLR from MU is not kinetically competent. Thus, the final step in the preferred folding pathway is formation of F from IMC/MCR.
We could not measure the conversion of IMC/MCR to F directly because IMC/MCR does not accumulate sufficiently under conditions in which there is also significant formation of F. Nevertheless, we could calculate the equilibrium constant for the folding of IMC/MCR to F based on the values of the thermodynamic cycle in Fig. 5 involving the overall folding equilibrium and the folding equilibrium between MU and IMC/MCR (Keq = [F]/[IMC/MCR] = 190) (Fig. 5) (and see Eq. 2 in SI Appendix, Supporting Methods). In addition, commonalities in conformational steps described in the next section allow us to estimate rate constants for this step (Fig. 5, rate constants in parentheses and square brackets) and complete the P4-P6 folding framework.
Modularity of Tertiary Contact Unfolding Kinetics.
As noted in the Introduction, structured RNAs seem to be modular, made up of motifs that are seen across multiple RNAs, and are often structurally superimposable (68–70). A straightforward, but hitherto untested, extension of this perspective would be that these structural motifs are also energetically modular.
What is meant by “energetic modularity” and what would its signature be? One of course would not expect all structures that contain a given motif to have the same overall folding equilibrium. However, the rate constant for breaking of a particular tertiary motif could be independent of its surroundings: i.e., the same across different structured RNAs. Although more complex scenarios are possible, a common unfolding rate constant across different RNAs would be expected if a motif has the same structure in different RNAs and if the transition state for its dissolution entails disruption of interactions within the motif rather than distal rearrangements that induce disruption and do so differentially in different RNAs. The experiments described below use our folding framework to test this kinetic prediction of energetic modularity.
We determined the rate constants for formation and dissolution of the two tertiary contacts, the TL/TLR and the MC/MCR, in P4-P6 and in engineered constructs that isolate the folding of each tertiary contact (100–102). These engineered single-molecule FRET constructs, developed by Pardi, Nesbitt, and coworkers (100–102), contain a single tertiary contact connected by a simplified junction (Fig. 6A and SI Appendix, Fig. S3) (TL/TLRiso and MC/MCRiso) and allow us to obtain rate constants for unfolding of each contact alone.
Fig. 6.
Folding kinetics of tertiary contacts within different structural contexts. (A) Construct used to isolate the folding and unfolding of the TL/TLR (TL/TLRiso). Residues are colored as in Fig.1, and residues that are the same as in P4-P6 are colored orange. (B, Top) Constructs that isolate TL/TLR formation (see SI Appendix, Fig. S2 for TL/TLRiso sequences). (Bottom) Unfolding (Left) and folding (Right) rate constants for ArichU P4-P6 variant (●) and TL/TLRiso constructs (A7 linker, □; T14 linker, left triangle; and an extended tetraloop helix, right triangle) across a range of Mg2+ concentrations. Published data for TL/TLRiso are shown (A7 linker, □) (100). Lines are shown to guide the eye (published data for the TL/TLRiso were taken with 100 mM NaCl instead of 100 mM KCl; see SI Appendix, Tables S1 and S3 for values and condition details). (C) Unfolding versus folding rate constants of P4-P6 variants (closed circles: ArichU, A186U, and J5/5a AllU/Arichu) and TL/TLRiso constructs (open symbols, as in B, and including a P4-P6 helical context, △) in 5 mM Mg2+ (black) or Ba2+ (purple). The folding rate constant for J5/5a AllU/ArichU P4-P6 is an upper limit indicated by the left arrow. The folding rate constant for IMC/MCR→F (★) was calculated as described in the SI Appendix, Supporting Methods, Eq. 3. The average unfolding rates in 5 mM Mg2+ or Ba2+ are shown as black and purple dashed lines, respectively, and were twofold slower in Ba2+ than Mg2+. Data are from SI Appendix, Tables S1–S3.
We measured the unfolding and folding rate constants for the simplified TL/TLRiso construct previously studied (100) as well as several related TL/TLRiso constructs (SI Appendix, Fig. S2), and we compared these values with those for P4-P6 constructs that form only the TL/TLR tertiary contact (ArichU, Fig. 6B). The unfolding rate constants measured for three TL/TLRiso constructs and a mutant P4-P6 construct were nearly the same over an Mg2+ concentration range of ∼10-fold. Given the very shallow Mg2+ dependences, this correspondence likely holds over a considerably wider range (Fig. 6B, Left, open vs. closed symbols). In contrast, the folding rate constants varied considerably across this range of Mg2+ (Fig. 6B, Right), presumably reflecting the different probabilities of aligning the TL and TLR for formation (Implications).
We extended the comparisons of TL/TLR unfolding rates to include additional P4-P6 variants (Fig. 6C). For four TL/TLRiso constructs (SI Appendix, Fig. S2) and three P4-P6 variants lacking the MC/MCR in 5 mM Mg2+, the unfolding rate constants varied less than threefold (4.3–9.5 s−1) whereas the folding rate constants varied by over 100-fold (Fig. 6C, black). We also determined these rate constants with 5 mM Ba2+because Ba2+ does not bind the metal core (81) and thereby allows comparisons that include WT P4-P6. As in Mg2+, the unfolding rate constants were constant across Ba2+ concentration whereas the folding constants varied widely (Fig. 6C, purple). The above results suggest that the TL/TLR tertiary contact comes apart with the same probability in different structural contexts.
To test whether the observation of common unfolding rates for the TL/TLR also held for the MC/MCR, we compared the folding and unfolding rates of the MC/MCR in P4-P6 with that of the simplified MC/MCRiso construct (SI Appendix, Fig. S3). We found that the unfolding rate constants for a P4-P6 variant that isolates the MU-to-IMC/MCR (L5B, Fig. 1B) transition and the simpler construct were the same, within error, at each Mg2+ concentration over an ∼10-fold range (SI Appendix, Fig. S3). This observation is consistent with a common unfolding rate for the MC/MCR but does not provide as strong support for modular kinetic behavior as the TL/TLR results because the MC/MCR experiments required a construct more similar to the P4-P6 context to achieve measurable folding kinetics (SI Appendix, Fig. S3 A and D).
The close correspondence of unfolding rate constants for individual tertiary contacts in P4-P6 constructs and minimal isolated constructs allowed us to assign the two unfolding steps in the framework that we could not directly follow to these common values (Fig. 5, F→MITL/TLR and F→IMC/MCR). These values, the directly measured rate constants, and the measured overall folding equilibrium then allowed us to calculate the two remaining folding rate constants from thermodynamic cycles (MITL/TLR→F and IMC/MCR→F) (SI Appendix, Supporting Methods, Eq. 3).
Analysis of Mutational Effects on Folding Pathways.
Contact order has been, at least partially, a predictive model in protein folding (97) and has been invoked in RNA folding models (98, 99). Predictions of this model are consistent with the observation that the MC/MCR forms before the TL/TLR on the preferred folding pathway, although with a modest effect, as described above. Here, we use the WT P4-P6 RNA framework (Fig. 5) to analyze the destabilizing effects of a point mutation in the MCR. The results provide evidence that this single mutation changes the order of tertiary contact formation. These observations, additional observations from this study, and general properties of RNA tertiary folding led us to invoke an alternative formalism modified from the diffusion–collision model for protein folding (103). This model is described in Implications.
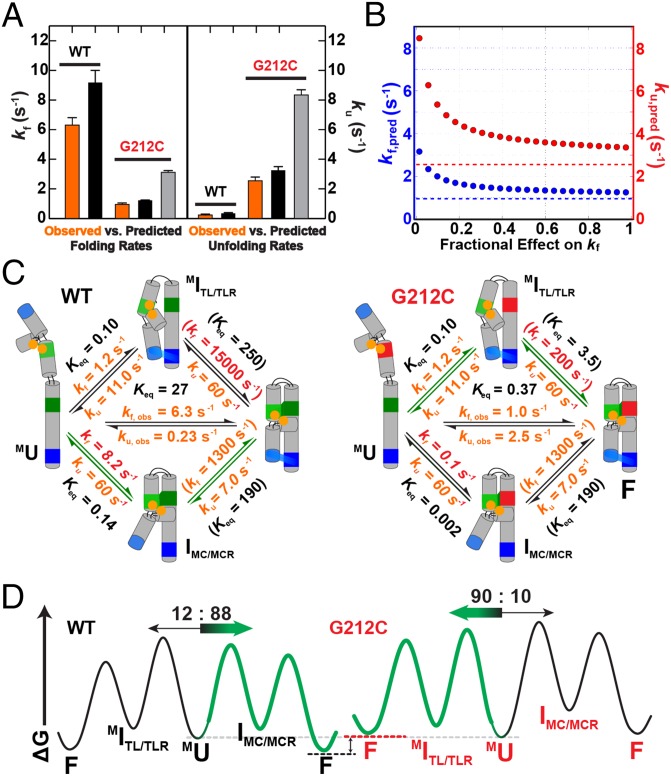
The G212C P4-P6 mutant unfolds faster than WT P4-P6 (Fig. 7A, orange). Nevertheless, it unfolds slower than variants with the MC ablated and slower than the G212U MCR mutant (SI Appendix, Table S1 and Fig. 4C), suggesting the presence of residual MC/MCR interactions in folded G212C P4-P6. Intriguingly, the G212C folding rate constant of 0.95 s−1 is slower than that for WT P4-P6 and matches that for folding to form the TL/TLR from the unfolded state (U), as determined with mutants and conditions that do not allow MC/MCR formation (Fig. 4C and SI Appendix, Table S1). This congruence in folding rate constants introduced the possibility that folding of this variant follows a pathway in which the TL/TLR forms first followed by the mutant MC/MCR interaction, an order that would be opposite to that for WT P4-P6 folding (Fig. 5).
Fig. 7.
The kinetic effects of the G212C MCR mutation. (A) Comparison of the observed (orange) versus predicted (black and gray) overall folding (Left) and unfolding (Right) rate constants of WT P4-P6 and G212C P4-P6 (SI Appendix, Supporting Methods). For WT, the predicted rate constants are calculated from the values in Fig. 5 that are reproduced in C of this figure. For G212C, the predicted folding and unfolding rate constants assume the full equilibrium effect of the mutation either on the folding (black) or unfolding (gray) rate constant for the MC/MCR interaction. (B) Predicted values for the overall folding (blue) and unfolding (red) rate constants for the G212C mutant over a range of hypothetical MC/MCR folding and unfolding rate constants, assuming that the full equilibrium effect of the mutation (72-fold) is apportioned between these rate constants, ranging from the full effect of the mutation on the unfolding rate constant [(fractional effect on kf) = 0] to the full effect on the folding rate constant [(fractional effect on kf) = 1]. The observed overall folding and unfolding rate constants for G212C are represented by the dashed lines. (C) Simplified folding framework for WT P4-P6 (Left; values from Fig. 5) and G212C P4-P6 (Right). Values for G212C P4-P6 are the same as for WT except for steps for MC/MCR formation, which are decreased by the full equilibrium effect of the mutation (72-fold). (D) Free energy diagrams for WT P4-P6 (Left) and the G212C mutant (Right) folding. The preferred folding pathway is shown in green, and the flux through each pathway is given above each pathway and is designated by the relative arrow size. Profiles are to scale.
To assess whether this alternative pathway is indeed traversed, we calculated the overall folding rate constants (MU→F) predicted based on the observed overall equilibrium effect of the G212C mutation (72-fold) (SI Appendix, Table S1), assuming that only the folding and/or unfolding steps involving MC/MCR formation are affected (Fig. 7) (i.e., that the steps involving TL/TLR formation are not affected). The predicted overall folding and unfolding rate constants vary when the overall equilibrium effect is apportioned differentially between MC/MCR folding versus unfolding (Fig. 7B): i.e., the overall equilibrium effect of 72-fold could arise with the full effect on folding [(fractional effect on kf) = 1] or on unfolding [(fractional effect on kf) = 0], or any combination of fractional effects that sum to one. The predicted values closely match the observed rate constants only when the equilibrium effect is predominantly apportioned to the MC/MCR folding rate constant (Fig. 7B, points vs. dashed lines; Fig. 7A, orange vs. black or gray).
The WT and G212C frameworks both predict overall folding and unfolding rate constants that are in good agreement with the observed rate constants for folding and unfolding (Fig. 7A, orange versus black bars), and the G212C framework has all rate constants identical to those for WT P4-P6 folding other than those involved in MC/MCR folding (Fig. 7C). Because the prediction assumes that the effects of the G212C mutation are only on steps involving its tertiary contact, this observation is also consistent with modularity in tertiary contact formation within P4-P6.
Fig. 7D shows free energy profiles for folding of WT and G212C P4-P6 RNA. For WT, folding proceeds 88% through the MC/MCR pathway whereas, for G212C P4-P6, the flux is 90% through the TL/TLR pathway. This dramatic change in the preferred folding pathway arises because the rate-limiting step for WT P4-P6 folding, formation of the MC/MCR interaction, is slowed to such an extent for the mutant that the barrier for this pathway becomes higher than the barrier for the alternative pathway through initial formation of the TL/TLR interaction. Our data suggest, most simply, that the G212C mutation renders formation of a binding-competent state of the MCR more unfavorable, and with an effect that is sufficiently large to favor formation of a more distal contact.
Implications
The folding framework for P4-P6 RNA developed herein and shown in Fig. 5 suggests kinetic properties of RNA elements that may hold across different RNAs, leads to simplifications that may aid in predicting RNA folding kinetics and thermodynamics, and provides a foundation for future fundamental studies of RNA folding. Such frameworks describing the kinetics and thermodynamics of individual steps in complex processes have been foundational in numerous areas of biology (e.g., refs. 39–45).
A Common Unfolding Rate Constant for a Tertiary Motif.
The TL/TLR tertiary motif is found in multiple RNAs and may be characteristic of at least a subset of RNA tertiary motifs. This interaction forms with rate constants spanning more than three orders of magnitude in different structural contexts and under different solution conditions but comes apart with a common unfolding rate of ∼10 s−1 (Fig. 6C and SI Appendix, Fig. S3). Uncovering this commonality required an ability to interrogate individual folding steps. The results suggest that the identity of the base pairs in the helices surrounding this tertiary contact and the junctions that bring together the tertiary contact partners do not affect the final structure of the tertiary motif or the energetics of the transition state ensemble for its disengagement. This observed modularity in tertiary contact unfolding provides a first step toward using the energetic contributions of individual tertiary motifs to predict the overall folding kinetics and thermodynamics for complex RNAs (104).
Stated another way, the data suggest a common rate for dissolution of a TL/TLR, a conclusion that, if general, could enable the prediction of the rates of conformational steps of complex RNAs that involve breaking of a particular contact. Similarly, observing a conformational step with a rate constant that is stereotypical for the dissociation of a particular tertiary interaction could suggest the breaking of that motif in the rate-limiting step, a model that could then be specifically tested by mutation of that motif. Conversely, tertiary contacts that do not follow modular behavior would be suggested to have multiple folded states and/or complex folding pathways and merit further detailed structural and dynamic study.
A Generalized Model for RNA Tertiary Folding.
A general goal in developing an RNA folding model (or any model) is to incorporate the minimal number of features sufficient for accurate quantitative prediction. As described above, contact order is a predictive, yet simple, concept in protein folding (97). Although folding of WT P4-P6 RNA follows the general prediction for a contact order model because the tertiary contact pairs that are closest to one another within the secondary structure form first [i.e., the MC/MCR forms first followed by the TL/TLR (Fig. 5)], this simple model fails to account for the reversal in the order of tertiary contact formation with the G212C mutation (Fig. 7D).
Modification of another established formalism from protein folding, the diffusion–collision model, may better describe the folding behaviors observed for P4-P6 and may prove more general in RNA folding and assembly. This model applied to proteins posits that the rate of forming a tertiary interaction between alpha helices, beta strands, or hydrophobic clusters is determined by the rate of the regions of the protein colliding, weighted by the probability that the partner elements (e.g., two alpha helices) are in a conformation competent for the tertiary contact to form when the collision occurs (103, 105). There is evidence for intermediates in protein folding that are not accounted for by this model (e.g., refs. 106–108). In contrast, given the more hierarchical nature of RNA than protein folding and the discrete nature of RNA tertiary contacts, a diffusion–collision model may be more readily and quantitatively applied to RNA, as elaborated below.
RNA secondary is preformed and stable, and RNA tertiary contact partners come together due to diffusion of the RNA helices they are embedded in (66, 104, 109). RNA tertiary contact formation also typically requires local unfavorable conformational rearrangements (104, 110–115) such that the rates of formation of these interactions are orders of magnitude slower than diffusional collision (104, 116).
In this way, RNA tertiary elements are potentially analogous to protein secondary structure elements in the diffusion–collision model. (Long-range secondary structure is also involved in RNA 3D structure formation and can be considered along with tertiary contacts in this model.) The probability that RNA tertiary contact partners associate is determined by (i) the conformational properties of the helices and the junctions that connect the tertiary elements and thus determine their collisional frequencies and (ii) the properties of the tertiary elements that determine the probability that both partners are in binding-competent conformations and the lifetimes of these states, relative to the lifetime of the collisional state. This process is akin to the need for protein secondary structure elements to be formed to allow tertiary structure formation in the original diffusion–collision model (103).
Our P4-P6 folding results can be described in terms of this RNA diffusion–collision model. Formation of one of P4-P6’s tertiary interactions increases the rate constant (i.e., the probability) of forming the other, presumably because the first contact greatly restricts the conformational space explored by the second tertiary contact elements and thereby increases their collisional probability and thus the rate of forming the second contact (Fig. 5). Cooperativity, a fundamental feature of the folding of biological macromolecules, is an emergent property of this increased collisional probability (60, 104).
Mutations in J5/5a, which forms a bend in P4-P6 (Fig. 1 A and B) (49, 51), slow formation of the TL/TLR contact, and linking the TL and TLR elements by a flexible single strand increases the folding rate; in contrast, neither alteration affects the unfolding rate (Fig. 6C and SI Appendix, Table S2), consistent with different linkages imparting different collisional probabilities but not affecting the contacts themselves. Collisional probabilities for RNA, as a polyelectrolyte, are also expected to be dependent on ionic conditions, and accordingly we observe a steeper salt dependence on the rate constant for TL/TLR formation for the more highly charged P4-P6 construct than the TL/TLRiso constructs (Fig. 6B and SI Appendix, Table S3). Finally, our results suggest that the G212C mutation decreases the probability of the MCR adopting its tertiary-competent conformation and thereby slows formation of the MC/MCR and reverses the order of tertiary contact formation in folding (Fig. 7D).
We have established a P4-P6 RNA folding framework and determined the effects of mutations within this RNA and also compared its folding to dramatically simplified constructs. The data emerging from these studies have allowed us to describe the folding of P4-P6 in terms of an RNA diffusion–collision model. Although this description is, to our knowledge, the first such description, there is no reason to think that such a description will not be generalizable because the model encompasses structural features that are common to folded RNAs. Certainly, additional complexities are expected, including parallel folding pathways from distinct secondary structure transitions, secondary structure transitions during tertiary folding, and influences from protein and other ligand binding. Thus, it will be of great interest to determine whether the RNA diffusion–collision model can be modified and adapted to describe this full range of behaviors and properties and whether it provides a foundation for building a quantitative and predictive understanding of RNA from the properties of its components.
Materials and Methods
Further details of the methods are provided in SI Appendix, Supporting Methods.
Preparation of P4-P6 smFRET (smP4-P6) Variants.
The smP4-P6 variants were prepared by a five-piece splinted ligation with protocols slightly altered from those described previously (60, 90, 117). Prior studies revealed that standard laboratory protocols introduce covalent differences in the RNA and that these modifications can affect the folding of individual molecules (90). Alterations in the protocol include substituting a synthetically derived 3′ and 5′ piece and removing intermediate gel purification and UV shadowing steps (SI Appendix, Supporting Methods). These changes increase the yield and throughput of the ligation procedure while producing RNA with the same purity, kinetics and thermodynamics as assayed by smFRET and compared with RNAs prepared by differed methods (SI Appendix, Fig. S4).
Preparation of Isolated Constructs.
Synthetic RNAs (SI Appendix, Tables S4 and S5) for each isolated construct were purchased from IDT with a 3′ primary amine, labeled with Cy3/Cy5 or Cy3b/Atto647N dyes with an NHS-ester reaction, and gel-purified.
smFRET Experiments.
smFRET measurements were performed on a custom-built prism-based total internal reflection microscope. Buffers used in smFRET measurements contained 1–50 mM MgCl2 or 5 mM BaCl2, 50 mM MOPS, pH 8.0, 100 mM KCl, 2 mg/mL glucose, 1.8 mM Trolox, 100 units/mL glucose oxidase (Sigma-Aldrich), and 1,000 units/mL catalase (Sigma-Aldrich). Depending on the folding equilibrium and change in FRET for each molecule, smFRET data were collected at an acquisition rate between 50 and 310 frames per second. Data were analyzed using the SMART data analysis package (92). See Dataset S1 for the imaging parameters and buffer conditions for each measurement reported herein. Before imaging, P4-P6 molecules were annealed to a DNA oligo containing a 5′ biotin (5′ biotin-AACCAAAATCAACCTAAAACTTACACA-3′) at 50 °C for 15 min and cooled at 0.1 °C/min to room temperature. Samples were diluted to ∼10–100 pM for imaging. Prior smFRET studies have shown that the unfolded states of P4-P6 have very low FRET (∼0.2) whereas states with the TL/TLR formed, including the fully folded state, have a high FRET value (∼0.8) and the intermediate with only the MC/MCR formed has an intermediate FRET value (∼0.5) (Fig. 1C) (60). Also, prior P4-P6 smFRET work revealed molecular heterogeneity from covalent damage caused by standard RNA treatments and showed that this damage could be avoided to yield highly homogeneous populations (90).
Data Analysis.
Thermodynamic and kinetic parameters were inferred from single-molecule FRET traces that were fit with a hidden Markov model (HMM)-based algorithm to a two-state model with a single unfolded (low FRET = 0.16) and single folded (high FRET = 0.45 or 0.8) state to extract transition probabilities, taking into account the noise observed in each intensity channel (92).
A subset of 20 traces for each construct and condition were also fit to a three-state linear model. In each case, the data were found to fit better to the two-state model than to a three-state model based on the difference in the Bayesian information criterion (BIC) (92), which includes a statistical penalty for including additional states, with the following exceptions that do not affect the conclusions of this work: WT P4-P6 folding with a Cy3b/Atto674N dye pair at 1 mM Mg, and MC/MCRiso at 5 and 10 mM Mg2+. (Example traces can be found in Dataset S1.) For WT P4-P6 folding, this behavior may be related to the dye dynamics because the apparent three-state behavior is not seen in variants with the Cy3/Cy5 dye pair. For MC/MCRiso, multistate behavior is seen only at low Mg2+ concentration, and not above 10 mM Mg2+. This construct has a lower folded FRET value than constructs that form the TL/TLR and fast transitions at lower [Mg2+], so that the signal-to-noise is relatively low; thus, the apparent three-state behavior may arise from noise in the data. We emphasize that elimination of the above data with potential three-state behavior would not change any of the conclusions of this work. We also note that, in general, best-fit models are sensitive to the penalty used for additional variables and the data quality and number of transitions (92).
Standard criteria for the identification of single-molecule traces were used: (i) single-step photobleaching, (ii) anticorrelated donor and acceptor channels, (iii) total intensity consistent with a single molecule, and (iv) stable total intensity. The folding rate constant reported for the J5/5a-AllU/ArichU P4-P6 variant is a limit because most traces for this molecule showed no folding behavior whereas traces that contained transitions had on average fewer than two transitions, preventing reliable estimation of the folding rate constant (92). All rate and equilibrium constants reported herein are the mean of the log distribution of the values for each molecule (118), and all other folding parameters are the mean of the distribution of each parameter for each molecule. Error estimates for all measurements, including the predictions of the overall folding and unfolding rate constants for WT and G212C, were determined by 95% confidence intervals of the mean from bootstrapping the parameters from individual molecules. Because the bootstrapped errors are asymmetric, the maximum of the two values is reported as the error for simplicity in the main text. Both errors are reported in SI Appendix. Signal-to-noise (SNR) cutoffs of 1 and 0.25 for molecules with high FRET states of 0.8 and 0.45, respectively, were used to minimize use of molecules for which fitting errors contributed disproportionately to the inferred rate constants. Evaluation of smFRET data for each construct is presented in Dataset S1. All smFRET data are available online for reanalysis in a standard single-molecule dataset (SMD) format (93).
Supplementary Material
Acknowledgments
We thank Julie Fiore and David Nesbitt for generously sharing their design and unpublished data on the isolated MC/MCR smFRET construct; and the D.H. laboratory and Geeta Narlikar for helpful discussions and for critical advice throughout this work. This work was supported by National Institutes of Health Grant P01GM066275. N.B. was supported by an NSF graduate fellowship. M.G. was supported in part by NIH Biotechnology Training Grant GM008412.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The single-molecule FRET data in a standardized format for reanalysis (single-molecule dataset or SMD) reported in this paper can be found at https://figshare.com/articles/Bisaria2016_PNAS_smFRETSMD_zip/3504470.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1525082113/-/DCSupplemental.
References
- 1.Warf MB, Berglund JA. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem Sci. 2010;35(3):169–178. doi: 10.1016/j.tibs.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81(1):145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20(3):300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 4.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Staley JP, Guthrie C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92(3):315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 6.Moore MJ. From birth to death: The complex lives of eukaryotic mRNAs. Science. 2005;309(5740):1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 7.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orgel LE. Evolution of the genetic apparatus. J Mol Biol. 1968;38(3):381–393. doi: 10.1016/0022-2836(68)90393-8. [DOI] [PubMed] [Google Scholar]
- 10.Woese C. The Genetic Code: The Molecular Basis for Genetic Expression. Harper & Row; New York: 1967. [Google Scholar]
- 11.Crick FH. The origin of the genetic code. J Mol Biol. 1968;38(3):367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert W. Origin of life: The RNA world. Nature. 1986;319(6055):618. [Google Scholar]
- 13.Lindahl T, Adams A, Fresco JR. Renaturation of transfer ribonucleic acids through site binding of magnesium. Proc Natl Acad Sci USA. 1966;55(4):941–948. doi: 10.1073/pnas.55.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams A, Lindahl T, Fresco JR. Conformational differences between the biologically active and inactive forms of a transfer ribonucleic acid. Proc Natl Acad Sci USA. 1967;57(6):1684–1691. doi: 10.1073/pnas.57.6.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishida T, Sueoka N. Elimination of magnesium ions as an absolute requirement for the native conformation of tryptophan transfer ribonucleic acid. J Mol Biol. 1968;37(2):313–316. doi: 10.1016/0022-2836(68)90270-2. [DOI] [PubMed] [Google Scholar]
- 16.Walstrum SA, Uhlenbeck OC. The self-splicing RNA of Tetrahymena is trapped in a less active conformation by gel purification. Biochemistry. 1990;29(46):10573–10576. doi: 10.1021/bi00498a022. [DOI] [PubMed] [Google Scholar]
- 17.Uhlenbeck OC. Keeping RNA happy. RNA. 1995;1(1):4–6. [PMC free article] [PubMed] [Google Scholar]
- 18.Treiber DK, Williamson JR. Exposing the kinetic traps in RNA folding. Curr Opin Struct Biol. 1999;9(3):339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 19.Shcherbakova I, Mitra S, Laederach A, Brenowitz M. Energy barriers, pathways, and dynamics during folding of large, multidomain RNAs. Curr Opin Chem Biol. 2008;12(6):655–666. doi: 10.1016/j.cbpa.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell R. RNA misfolding and the action of chaperones. Front Biosci. 2008;13:1–20. doi: 10.2741/2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sigler PB. An analysis of the structure of tRNA. Annu Rev Biophys Bioeng. 1975;4(00):477–527. doi: 10.1146/annurev.bb.04.060175.002401. [DOI] [PubMed] [Google Scholar]
- 22.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270(36):20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 23.Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem. 1997;272(1):196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 24.Semrad K, Green R, Schroeder R. RNA chaperone activity of large ribosomal subunit proteins from Escherichia coli. RNA. 2004;10(12):1855–1860. doi: 10.1261/rna.7121704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorsch JR. RNA chaperones exist and DEAD box proteins get a life. Cell. 2002;109(7):797–800. doi: 10.1016/s0092-8674(02)00804-8. [DOI] [PubMed] [Google Scholar]
- 26.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol. 2004;5(11):908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 27.Woodson SA. Taming free energy landscapes with RNA chaperones. RNA Biol. 2010;7(6):677–686. doi: 10.4161/rna.7.6.13615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rein A. Nucleic acid chaperone activity of retroviral Gag proteins. RNA Biol. 2010;7(6):700–705. doi: 10.4161/rna.7.6.13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodson SA, Cech TR. Alternative secondary structures in the 5′ exon affect both forward and reverse self-splicing of the Tetrahymena intervening sequence RNA. Biochemistry. 1991;30(8):2042–2050. doi: 10.1021/bi00222a006. [DOI] [PubMed] [Google Scholar]
- 30.Pan J, Woodson SA. Folding intermediates of a self-splicing RNA: Mispairing of the catalytic core. J Mol Biol. 1998;280(4):597–609. doi: 10.1006/jmbi.1998.1901. [DOI] [PubMed] [Google Scholar]
- 31.Treiber DK, Rook MS, Zarrinkar PP, Williamson JR. Kinetic intermediates trapped by native interactions in RNA folding. Science. 1998;279(5358):1943–1946. doi: 10.1126/science.279.5358.1943. [DOI] [PubMed] [Google Scholar]
- 32.Chadalavada DM, Senchak SE, Bevilacqua PC. The folding pathway of the genomic hepatitis delta virus ribozyme is dominated by slow folding of the pseudoknots. J Mol Biol. 2002;317(4):559–575. doi: 10.1006/jmbi.2002.5434. [DOI] [PubMed] [Google Scholar]
- 33.Russell R, et al. Exploring the folding landscape of a structured RNA. Proc Natl Acad Sci USA. 2002;99(1):155–160. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan J, Deras ML, Woodson SA. Fast folding of a ribozyme by stabilizing core interactions: Evidence for multiple folding pathways in RNA. J Mol Biol. 2000;296(1):133–144. doi: 10.1006/jmbi.1999.3439. [DOI] [PubMed] [Google Scholar]
- 35.Solomatin SV, Greenfeld M, Chu S, Herschlag D. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature. 2010;463(7281):681–684. doi: 10.1038/nature08717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell R, Herschlag D. Probing the folding landscape of the Tetrahymena ribozyme: Commitment to form the native conformation is late in the folding pathway. J Mol Biol. 2001;308(5):839–851. doi: 10.1006/jmbi.2001.4751. [DOI] [PubMed] [Google Scholar]
- 37.Russell R, et al. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J Mol Biol. 2006;363(2):531–544. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 38.Mitchell D, 3rd, Jarmoskaite I, Seval N, Seifert S, Russell R. The long-range P3 helix of the Tetrahymena ribozyme is disrupted during folding between the native and misfolded conformations. J Mol Biol. 2013;425(15):2670–2686. doi: 10.1016/j.jmb.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albery WJ, Knowles JR. Free-energy profile of the reaction catalyzed by triosephosphate isomerase. Biochemistry. 1976;15(25):5627–5631. doi: 10.1021/bi00670a031. [DOI] [PubMed] [Google Scholar]
- 40.Gresser MJ, Myers JA, Boyer PD. Catalytic site cooperativity of beef heart mitochondrial F1 adenosine triphosphatase: Correlations of initial velocity, bound intermediate, and oxygen exchange measurements with an alternating three-site model. J Biol Chem. 1982;257(20):12030–12038. [PubMed] [Google Scholar]
- 41.Fierke CA, Johnson KA, Benkovic SJ. Construction and evaluation of the kinetic scheme associated with dihydrofolate reductase from Escherichia coli. Biochemistry. 1987;26(13):4085–4092. doi: 10.1021/bi00387a052. [DOI] [PubMed] [Google Scholar]
- 42.Patel SS, Wong I, Johnson KA. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30(2):511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 43.Ma YZ, Taylor EW. Kinetic mechanism of a monomeric kinesin construct. J Biol Chem. 1997;272(2):717–723. doi: 10.1074/jbc.272.2.717. [DOI] [PubMed] [Google Scholar]
- 44.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95(11):6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Rashid R, Wang K, Shan S-O. Sequential checkpoints govern substrate selection during cotranslational protein targeting. Science. 2010;328(5979):757–760. doi: 10.1126/science.1186743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das R, Travers KJ, Bai Y, Herschlag D. Determining the Mg2+ stoichiometry for folding an RNA metal ion core. J Am Chem Soc. 2005;127(23):8272–8273. doi: 10.1021/ja051422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murphy FL, Cech TR. An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry. 1993;32(20):5291–5300. doi: 10.1021/bi00071a003. [DOI] [PubMed] [Google Scholar]
- 48.Murphy FL, Wang YH, Griffith JD, Cech TR. Coaxially stacked RNA helices in the catalytic center of the Tetrahymena ribozyme. Science. 1994;265(5179):1709–1712. doi: 10.1126/science.8085157. [DOI] [PubMed] [Google Scholar]
- 49.Cate JH, et al. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science. 1996;273(5282):1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 50.Cate JH, Hanna RL, Doudna JA. A magnesium ion core at the heart of a ribozyme domain. Nat Struct Biol. 1997;4(7):553–558. doi: 10.1038/nsb0797-553. [DOI] [PubMed] [Google Scholar]
- 51.Szewczak AA, Cech TR. An RNA internal loop acts as a hinge to facilitate ribozyme folding and catalysis. RNA. 1997;3(8):838–849. [PMC free article] [PubMed] [Google Scholar]
- 52.Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science. 1998;279(5358):1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 53.Deras ML, Brenowitz M, Ralston CY, Chance MR, Woodson SA. Folding mechanism of the Tetrahymena ribozyme P4-P6 domain. Biochemistry. 2000;39(36):10975–10985. doi: 10.1021/bi0010118. [DOI] [PubMed] [Google Scholar]
- 54.Silverman SK, Deras ML, Woodson SA, Scaringe SA, Cech TR. Multiple folding pathways for the P4-P6 RNA domain. Biochemistry. 2000;39(40):12465–12475. doi: 10.1021/bi000828y. [DOI] [PubMed] [Google Scholar]
- 55.Silverman SK, Cech TR. An early transition state for folding of the P4-P6 RNA domain. RNA. 2001;7(2):161–166. doi: 10.1017/s1355838201001716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takamoto K, et al. Principles of RNA compaction: Insights from the equilibrium folding pathway of the P4-P6 RNA domain in monovalent cations. J Mol Biol. 2004;343(5):1195–1206. doi: 10.1016/j.jmb.2004.08.080. [DOI] [PubMed] [Google Scholar]
- 57.Das R, Baker D. Automated de novo prediction of native-like RNA tertiary structures. Proc Natl Acad Sci USA. 2007;104(37):14664–14669. doi: 10.1073/pnas.0703836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murphy FL, Cech TR. GAAA tetraloop and conserved bulge stabilize tertiary structure of a group I intron domain. J Mol Biol. 1994;236(1):49–63. doi: 10.1006/jmbi.1994.1117. [DOI] [PubMed] [Google Scholar]
- 59.Das R, et al. Structural inference of native and partially folded RNA by high-throughput contact mapping. Proc Natl Acad Sci USA. 2008;105(11):4144–4149. doi: 10.1073/pnas.0709032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sattin BD, Zhao W, Travers K, Chu S, Herschlag D. Direct measurement of tertiary contact cooperativity in RNA folding. J Am Chem Soc. 2008;130(19):6085–6087. doi: 10.1021/ja800919q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silverman SK, Cech TR. RNA tertiary folding monitored by fluorescence of covalently attached pyrene. Biochemistry. 1999;38(43):14224–14237. doi: 10.1021/bi991333f. [DOI] [PubMed] [Google Scholar]
- 62.Young BT, Silverman SK. The GAAA tetraloop-receptor interaction contributes differentially to folding thermodynamics and kinetics for the P4-P6 RNA domain. Biochemistry. 2002;41(41):12271–12276. doi: 10.1021/bi0264869. [DOI] [PubMed] [Google Scholar]
- 63.Lee TH, et al. Measuring the folding transition time of single RNA molecules. Biophys J. 2007;92(9):3275–3283. doi: 10.1529/biophysj.106.094623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lamb J, et al. Reconstructing three-dimensional shape envelopes from time-resolved small-angle X-ray scattering data. J Appl Cryst. 2008;41(Pt 6):1046–1052. doi: 10.1107/S0021889808028264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schlatterer JC, et al. Hinge stiffness is a barrier to RNA folding. J Mol Biol. 2008;379(4):859–870. doi: 10.1016/j.jmb.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leontis NB, Westhof E. Analysis of RNA motifs. Curr Opin Struct Biol. 2003;13(3):300–308. doi: 10.1016/s0959-440x(03)00076-9. [DOI] [PubMed] [Google Scholar]
- 67.Tinoco I, Jr, Bustamante C. How RNA folds. J Mol Biol. 1999;293(2):271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 68.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: A new RNA secondary structure motif. EMBO J. 2001;20(15):4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nat Struct Biol. 2003;10(12):1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 70.Wu L, Chai D, Fraser ME, Zimmerly S. Structural variation and uniformity among tetraloop-receptor interactions and other loop-helix interactions in RNA crystal structures. PLoS One. 2012;7(11):e49225. doi: 10.1371/journal.pone.0049225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ishikawa J, Fujita Y, Maeda Y, Furuta H, Ikawa Y. GNRA/receptor interacting modules: Versatile modular units for natural and artificial RNA architectures. Methods. 2011;54(2):226–238. doi: 10.1016/j.ymeth.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 72.Costa M, Michel F. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J. 1995;14(6):1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Geary C, Baudrey S, Jaeger L. Comprehensive features of natural and in vitro selected GNRA tetraloop-binding receptors. Nucleic Acids Res. 2008;36(4):1138–1152. doi: 10.1093/nar/gkm1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc Natl Acad Sci USA. 2001;98(9):4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nagaswamy U, Fox GE. Frequent occurrence of the T-loop RNA folding motif in ribosomal RNAs. RNA. 2002;8(9):1112–1119. doi: 10.1017/s135583820202006x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Basu S, et al. A specific monovalent metal ion integral to the AA platform of the RNA tetraloop receptor. Nat Struct Biol. 1998;5(11):986–992. doi: 10.1038/2960. [DOI] [PubMed] [Google Scholar]
- 77.Wu M, Tinoco I., Jr RNA folding causes secondary structure rearrangement. Proc Natl Acad Sci USA. 1998;95(20):11555–11560. doi: 10.1073/pnas.95.20.11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Uchida T, He Q, Ralston CY, Brenowitz M, Chance MR. Linkage of monovalent and divalent ion binding in the folding of the P4-P6 domain of the Tetrahymena ribozyme. Biochemistry. 2002;41(18):5799–5806. doi: 10.1021/bi020042v. [DOI] [PubMed] [Google Scholar]
- 79.Battle DJ, Doudna JA. Specificity of RNA-RNA helix recognition. Proc Natl Acad Sci USA. 2002;99(18):11676–11681. doi: 10.1073/pnas.182221799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schwans JP, Cortez CN, Olvera JM, Piccirilli JA. 2′-mercaptonucleotide interference reveals regions of close packing within folded RNA molecules. J Am Chem Soc. 2003;125(33):10012–10018. doi: 10.1021/ja035175y. [DOI] [PubMed] [Google Scholar]
- 81.Travers KJ, Boyd N, Herschlag D. Low specificity of metal ion binding in the metal ion core of a folded RNA. RNA. 2007;13(8):1205–1213. doi: 10.1261/rna.566007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Frederiksen JK, Li N-S, Das R, Herschlag D, Piccirilli JA. Metal-ion rescue revisited: Biochemical detection of site-bound metal ions important for RNA folding. RNA. 2012;18(6):1123–1141. doi: 10.1261/rna.028738.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang S, Karbstein K, Peracchi A, Beigelman L, Herschlag D. Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of a cleavage site phosphorothioate. Biochemistry. 1999;38(43):14363–14378. doi: 10.1021/bi9913202. [DOI] [PubMed] [Google Scholar]
- 84.Shan So, Yoshida A, Sun S, Piccirilli JA, Herschlag D. Three metal ions at the active site of the Tetrahymena group I ribozyme. Proc Natl Acad Sci USA. 1999;96(22):12299–12304. doi: 10.1073/pnas.96.22.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maderia M, Hunsicker LM, DeRose VJ. Metal-phosphate interactions in the hammerhead ribozyme observed by 31P NMR and phosphorothioate substitutions. Biochemistry. 2000;39(40):12113–12120. doi: 10.1021/bi001249w. [DOI] [PubMed] [Google Scholar]
- 86.Murray JB, Dunham CM, Scott WG. A pH-dependent conformational change, rather than the chemical step, appears to be rate-limiting in the hammerhead ribozyme cleavage reaction. J Mol Biol. 2002;315(2):121–130. doi: 10.1006/jmbi.2001.5145. [DOI] [PubMed] [Google Scholar]
- 87.Pyle AM. Metal ions in the structure and function of RNA. J Biol Inorg Chem. 2002;7(7-8):679–690. doi: 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]
- 88.Thomson JB, Lilley DM. The influence of junction conformation on RNA cleavage by the hairpin ribozyme in its natural junction form. RNA. 1999;5(2):180–187. doi: 10.1017/s1355838299981670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dibrov SM, McLean J, Parsons J, Hermann T. Self-assembling RNA square. Proc Natl Acad Sci USA. 2011;108(16):6405–6408. doi: 10.1073/pnas.1017999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Greenfeld M, Solomatin SV, Herschlag D. Removal of covalent heterogeneity reveals simple folding behavior for P4-P6 RNA. J Biol Chem. 2011;286(22):19872–19879. doi: 10.1074/jbc.M111.235465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Leipply D, Draper DE. Dependence of RNA tertiary structural stability on Mg2+ concentration: Interpretation of the Hill equation and coefficient. Biochemistry. 2010;49(9):1843–1853. doi: 10.1021/bi902036j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Greenfeld M, Pavlichin DS, Mabuchi H, Herschlag D. Single Molecule Analysis Research Tool (SMART): An integrated approach for analyzing single molecule data. PLoS One. 2012;7(2):e30024. doi: 10.1371/journal.pone.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Greenfeld M, et al. Single-molecule dataset (SMD): A generalized storage format for raw and processed single-molecule data. BMC Bioinformatics. 2015;16:3. doi: 10.1186/s12859-014-0429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Silverman SK, Zheng M, Wu M, Tinoco I, Jr, Cech TR. Quantifying the energetic interplay of RNA tertiary and secondary structure interactions. RNA. 1999;5(12):1665–1674. doi: 10.1017/s1355838299991823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cate JH, Doudna JA. Metal-binding sites in the major groove of a large ribozyme domain. Structure. 1996;4(10):1221–1229. doi: 10.1016/s0969-2126(96)00129-3. [DOI] [PubMed] [Google Scholar]
- 96.Silverman SK, Cech TR. Energetics and cooperativity of tertiary hydrogen bonds in RNA structure. Biochemistry. 1999;38(27):8691–8702. doi: 10.1021/bi9906118. [DOI] [PubMed] [Google Scholar]
- 97.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J Mol Biol. 1998;277(4):985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 98.Sosnick TR, Pan T. Reduced contact order and RNA folding rates. J Mol Biol. 2004;342(5):1359–1365. doi: 10.1016/j.jmb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 99.Heilman-Miller SL, Woodson SA. Perturbed folding kinetics of circularly permuted RNAs with altered topology. J Mol Biol. 2003;328(2):385–394. doi: 10.1016/s0022-2836(03)00304-8. [DOI] [PubMed] [Google Scholar]
- 100.Hodak JH, Downey CD, Fiore JL, Pardi A, Nesbitt DJ. Docking kinetics and equilibrium of a GAAA tetraloop-receptor motif probed by single-molecule FRET. Proc Natl Acad Sci USA. 2005;102(30):10505–10510. doi: 10.1073/pnas.0408645102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Downey CD, et al. Metal ion dependence, thermodynamics, and kinetics for intramolecular docking of a GAAA tetraloop and receptor connected by a flexible linker. Biochemistry. 2006;45(11):3664–3673. doi: 10.1021/bi0520941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fiore JL, Holmstrom ED, Nesbitt DJ. Entropic origin of Mg2+-facilitated RNA folding. Proc Natl Acad Sci USA. 2012;109(8):2902–2907. doi: 10.1073/pnas.1114859109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Karplus M, Weaver DL. Protein-folding dynamics. Nature. 1976;260(5550):404–406. doi: 10.1038/260404a0. [DOI] [PubMed] [Google Scholar]
- 104.Herschlag D, Allred BE, Gowrishankar S. From static to dynamic: The need for structural ensembles and a predictive model of RNA folding and function. Curr Opin Struct Biol. 2015;30:125–133. doi: 10.1016/j.sbi.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Karplus M, Weaver DL. Protein folding dynamics: The diffusion-collision model and experimental data. Protein Sci. 1994;3(4):650–668. doi: 10.1002/pro.5560030413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ohgushi M, Wada A. ‘Molten-globule state’: A compact form of globular proteins with mobile side-chains. FEBS Lett. 1983;164(1):21–24. doi: 10.1016/0014-5793(83)80010-6. [DOI] [PubMed] [Google Scholar]
- 107.Baldwin RL, Rose GD. Molten globules, entropy-driven conformational change and protein folding. Curr Opin Struct Biol. 2013;23(1):4–10. doi: 10.1016/j.sbi.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 108.Bachmann A, Wildemann D, Praetorius F, Fischer G, Kiefhaber T. Mapping backbone and side-chain interactions in the transition state of a coupled protein folding and binding reaction. Proc Natl Acad Sci USA. 2011;108(10):3952–3957. doi: 10.1073/pnas.1012668108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chu VB, et al. Do conformational biases of simple helical junctions influence RNA folding stability and specificity? RNA. 2009;15(12):2195–2205. doi: 10.1261/rna.1747509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Butcher SE, Dieckmann T, Feigon J. Solution structure of a GAAA tetraloop receptor RNA. EMBO J. 1997;16(24):7490–7499. doi: 10.1093/emboj/16.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Al-Hashimi HM, Walter NG. RNA dynamics: It is about time. Curr Opin Struct Biol. 2008;18(3):321–329. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jiang F, Kumar RA, Jones RA, Patel DJ. Structural basis of RNA folding and recognition in an AMP-RNA aptamer complex. Nature. 1996;382(6587):183–186. doi: 10.1038/382183a0. [DOI] [PubMed] [Google Scholar]
- 113.Allain FH, et al. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formulation. Nature. 1996;380(6575):646–650. doi: 10.1038/380646a0. [DOI] [PubMed] [Google Scholar]
- 114.Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992;257(5066):76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 115.Sussman D, Nix JC, Wilson C. The structural basis for molecular recognition by the vitamin B 12 RNA aptamer. Nat Struct Biol. 2000;7(1):53–57. doi: 10.1038/71253. [DOI] [PubMed] [Google Scholar]
- 116.Karbstein K, Herschlag D. Extraordinarily slow binding of guanosine to the Tetrahymena group I ribozyme: Implications for RNA preorganization and function. Proc Natl Acad Sci USA. 2003;100(5):2300–2305. doi: 10.1073/pnas.252749799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Solomatin SV, Greenfeld M, Herschlag D. Implications of molecular heterogeneity for the cooperativity of biological macromolecules. Nat Struct Mol Biol. 2011;18(6):732–734. doi: 10.1038/nsmb.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91(5):1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.