Significance
Ferroptosis is a regulated form of cell death induced by loss of glutathione peroxidase 4 (GPX4) phospholipid peroxidase activity and lethal accumulation of reactive oxygen species. Small-molecule inhibitors of GPX4 induce ferroptosis; however, the interaction between these inhibitors and GPX4 has remained elusive, as has the identity of the reactive oxygen species that drive execution of ferroptosis. We identified here a ligand-binding site on GPX4 and determined the specific lipids oxidized during ferroptosis. We further identified two key drivers of lipid peroxidation during ferroptosis: lipoxygenases and phosphorylase kinase G2. These findings reveal a previously enigmatic mechanism of ferroptotic lipid peroxide generation and suggest new strategies for pharmacological control of ferroptosis and diseases associated with this mode of cell death.
Keywords: ferroptosis, Gpx4, PUFAs, lipoxygenase, PHKG2
Abstract
Ferroptosis is form of regulated nonapoptotic cell death that is involved in diverse disease contexts. Small molecules that inhibit glutathione peroxidase 4 (GPX4), a phospholipid peroxidase, cause lethal accumulation of lipid peroxides and induce ferroptotic cell death. Although ferroptosis has been suggested to involve accumulation of reactive oxygen species (ROS) in lipid environments, the mediators and substrates of ROS generation and the pharmacological mechanism of GPX4 inhibition that generates ROS in lipid environments are unknown. We report here the mechanism of lipid peroxidation during ferroptosis, which involves phosphorylase kinase G2 (PHKG2) regulation of iron availability to lipoxygenase enzymes, which in turn drive ferroptosis through peroxidation of polyunsaturated fatty acids (PUFAs) at the bis-allylic position; indeed, pretreating cells with PUFAs containing the heavy hydrogen isotope deuterium at the site of peroxidation (D-PUFA) prevented PUFA oxidation and blocked ferroptosis. We further found that ferroptosis inducers inhibit GPX4 by covalently targeting the active site selenocysteine, leading to accumulation of PUFA hydroperoxides. In summary, we found that PUFA oxidation by lipoxygenases via a PHKG2-dependent iron pool is necessary for ferroptosis and that the covalent inhibition of the catalytic selenocysteine in Gpx4 prevents elimination of PUFA hydroperoxides; these findings suggest new strategies for controlling ferroptosis in diverse contexts.
Ferroptosis is a regulated form of cell death characterized by loss of glutathione peroxidase 4 (GPX4) activity and subsequent accumulation of lipid peroxides of an unknown nature, which are required for destruction of cells (1, 2). Cells undergoing ferroptosis cannot be rescued by chemical or genetic inhibitors of apoptosis (such as zVAD-fmk or Bax/Bak double knockout) (3–6) or inhibitors of necroptosis (such as Nec-1 or RIPK1/3 knockdown) (1, 7), which indicates that ferroptosis is a distinct cell death modality. Inhibition of ferroptosis is protective in models of Huntington’s disease (8), periventricular leukomalacia (8–10), and kidney dysfunction (8, 11, 12). These results suggest that ferroptosis may play a detrimental role in pathological conditions. Gpx4-deleted mice do not survive past embryonic day 8, indicating that protection from ferroptosis is likely essential during normal mammalian development (13, 14).
Two ferroptosis-triggering mechanisms have been reported. The first is inhibition of system xc−, the cystine-glutamate antiporter, which limits cellular availability of cysteine and glutathione (GSH) (1, 15). GSH is a cofactor for GPX4, which is a critical antioxidant enzyme that detoxifies diverse lipid peroxides in the context of cell membranes. GPX4 uses GSH as a reducing agent in its peroxidase reaction cycle. Therefore, inhibition of system xc− indirectly inhibits GPX4 to cause accumulation of lethal lipid peroxides and to initiate execution of ferroptosis. Erastin, sulfasalazine, and sorafenib are small molecules that induce ferroptosis through this mechanism (1, 16).
A second ferroptosis-triggering mechanism is direct binding of small-molecule inhibitors to GPX4 to inhibit its phospholipid peroxidase activity. Indeed, GPX4 was identified as a target protein of (1S, 3R)-RSL3, a ferroptosis-inducing compound, through an affinity purification experiment (2), and knockdown of GPX4 expression using siRNA reagents is sufficient to induce ferroptosis (2). (1S, 3R)-RSL3 contains an electrophilic chloroacetamide: Gpx4 contains nucleophilic thiol-containing cysteine residues and a nucleophilic selenol moiety on the active site selenocysteine. The relevant nucleophilic site on GPX4 has remained elusive, making it difficult to design new Gpx4 inhibitors.
Upon inhibition of GPX4, sensitive cells cannot eliminate lipid hydroperoxides that accumulate, leading to ferroptotic cell death. Although lipid peroxides have been detected using a generic lipophilic reactive oxygen species (ROS) sensor (e.g., BODIPY-C11) (1, 2), the identity of the lipid species oxidized during ferroptosis and that drive lethality is not known. Identifying lipid oxidation events in the course of ferroptosis may provide insights into the mechanism of GPX4-regulated ferroptosis.
Here, we found that (1S, 3R)-RSL3 covalently interacts with the active site selenocysteine of GPX4 to inhibit its enzymatic activity. We used lipidomics to discover that polyunsaturated fatty acids (PUFAs) are the most susceptible lipids to peroxidation in the course of ferroptosis, and that preventing this peroxidation by supplementing cells with PUFAs deuterated at the susceptible bis-allyclic carbon suppresses ferroptosis. This led in turn to the identification of lipoxygenases and PHKG2 as regulators of PUFA peroxidation during ferroptosis.
Results
Characterization of Binding Between Small-Molecule Inducers of Ferroptosis and GPX4.
Previously, GPX4 was identified as the relevant target protein of the ferroptosis inducer (1S, 3R)-RSL3 using an affinity purification approach that made use of (1S, 3R)-RSL3-fluorescein [(1S, 3R)-RSL3-Fcn]; this compound is an analog that consists of (1S, 3R)-RSL3, a PEG linker, and isobutyryl-protected fluorescein, which is used here as an affinity tag (Fig. 1A) (2). (1S, 3R)-RSL3 contains a chloroacetamide moiety (red, Fig. 1A) that reacts with nucleophilic amino acid residues; indeed, (1S, 3R)-RSL3 acts as a covalent inhibitor of GPX4 (2).
Fig. 1.
Characterization of RSL3 binding to GPX4. (A) Structure of RSL3 affinity probe used (Upper) and schematic drawing of the affinity complex formed during the pull-down assay (Lower). (B) A pull-down analysis showing that RSL3 does not bind to a mutant GPX4 where the active-site selenocysteine and all other cysteines were replaced with either alanine or serine. The schematic structure of GFP-GPX4 protein is shown (Upper). Cells stably expressing either WT or mutant [AllCys(-)] GFP-GPX4 protein were treated with (1S, 3R)-RSL3-Fcn, then the cell lysates were subjected to affinity purification using anti–Fcn-conjugated Sepharose beads. The binding between GFP-GPX4 protein and (1S, 3R)-RSL3-Fcn was determined by detecting the presence of GFP-GPX4 protein in the eluent using Western blot with anti-GFP antibody. Arrowheads indicate GFP-GPX4 protein; asterisks indicate nonspecific bands. W.C.L., whole-cell lysate before pulldown. (C) Selenocysteine is much more reactive to RSL3 than cysteine at the active site. Cells stably expressing A46U or A46C protein (see diagram in the figure) were treated with (1S, 3R)-RSL3-Fcn and subjected to the same analysis as in B. (1S, 3R)-RSL3-Fcn interacted with A46U protein but not with A46C protein. Arrowheads indicate GFP-GPX4 protein; asterisks indicate nonspecific bands. (D) RSL3 interacts weakly with other cysteines on GPX4. Cells stably expressing indicated mutant protein (see text for detailed information) were treated with (1S, 3R)-RSL3-Fcn and subjected to the same analysis as in B. Some mutant proteins such as A10C and S37C interacted with (1S, 3R)-RSL3-Fcn whereas others did not. The interaction was the strongest when WT GPX4 was expressed (thickest band in WT sample). Arrowheads indicate GFP-GPX4 protein; asterisks indicate nonspecific bands. (E) Some ferroptosis-inducing compounds competed off RSL3 binding to GPX4. HT-1080 cells were treated with (1S, 3R)-RSL3-Fcn in the presence or absence of indicated competitors, and cell lysates were subjected to pull-down experiment with anti-Fcn antibody. The binding between (1S, 3R)-RSL3-Fcn and endogenous GPX4 protein was determined by Western blotting with anti-GPX4 antibody. The (1S, 3R)-RSL3-Fcn binding to GPX4 was suppressed in samples where free (1S, 3R)-RSL3 was used as a competitor. Different GPX4 inhibitors competed off (1S, 3R)-RSL3-Fcn binding to GPX4 protein with varying degrees. (F) The amount of endogenous GPX4 protein bound to the affinity probe in E was quantified using Odyssey software (LI-COR Biosciences). Data represent mean ± SD calculated from technical triplicates. (G) Covalent interaction between GPX4 and RSL3 was confirmed by immunoprecipitating GFP-GPX4/(1S, 3R)-RSL3-Fcn complex with anti-GPX4 antibody and performing Western blot with anti-GFP or anti-Fcn antibodies. Arrow indicates GFP-GPX4 protein band in the Western blot analysis. (H) The (1S, 3R)-RSL3 covalently interacts with purified GPX4U46C protein. Purified GPX4U46C protein was mixed with (1S, 3R)-RSL3-Fcn, then the mixture was resolved on a denaturing gel and transferred to a membrane. Western blotting with anti-Fcn antibody detected (1S, 3R)-RSL3-Fcn in a position corresponding to the size of GPX4U46C protein (∼21 kDa), suggesting that (1S, 3R)-RSL3-Fcn covalently attached to GPX4U46C protein and migrated together in this denaturing condition. The Western blot band became thinner when a lesser amount of (1S, 3R)-RSL3-Fcn was added to the mixture (Upper). The band size remained the same across the sample when anti-GPX4 antibody was used because equal amount of GPX4U46C protein was added across the samples (Lower). (I) Inhibition of GPX4 enzyme activity by (1S, 3R)-RSL3 or (1R, 3R)-RSL3. GPX4 enzyme activity was assayed by mixing cell lysates with PC hydroperoxides (PC-OOH), a GPX4-specific substrate, and by determining the amount of substrate left in the reaction mixture using an LC-MS instrument. The arrow indicates the LC-MS peak corresponds to PC-OOH. (J) Area under the curve was determined from each mass chromatogram in I (retention time from 8.2 to 8.8 min; dotted lines in the chromatogram) and used to draw a concentration-dependent curve of GPX4 inhibition by (1S, 3R)-RSL3.
GPX4 contains eight nucleophilic amino acids: one selenocysteine (Sec) at the active site and seven other cysteines (Cys), which are potentially reactive with electrophiles (Fig. 1B). We replaced the Sec and all Cys residues with Ala or Ser (C2A, C10A, C37S, Sec46A, C66A, C75S, C107A, and C148A) and expressed the mutant GPX4 protein, termed allCys(-), in G401 renal carcinoma cells as a GFP fusion (Fig. 1B).
G401 cells stably expressing GFP-allCys(-) were treated with (1S, 3R)-RSL3-Fcn. The treated cells were lysed, and fluorescein-tagged proteins were affinity-purified using Sepharose beads coupled to an antifluorescein antibody. Eluted proteins were analyzed for GFP-GPX4 abundance by Western blot using an anti-GFP antibody. A control sample was prepared by adding (1S, 3R)-RSL3-Fcn to G401 cells stably expressing WT GPX4 tagged with GFP.
The (1S, 3R)-RSL3-Fcn purified the WT GPX4-GFP fusion protein, confirming covalent binding between (1S, 3R)-RSL3 and GPX4 (Fig. 1B). However, we did not detect the allCys(-) mutant GPX4 protein in the pull-down sample, despite much higher basal expression level of allCys(-) GPX4 compared with WT GPX4-GFP (Fig. 1B). This indicates that there is no detectable binding between (1S, 3R)-RSL3-Fcn and allCys(-) GPX4-GFP in this assay, likely due to mutations in the nucleophilic Sec and Cys amino acids in this construct. Therefore, the RSL3 labeling site on GPX4 is likely to be one or more of these eight nucleophilic amino acids (Fig. 1B).
The Sec in the active site of GPX4 has a lower pKa value (5.47) than Cys (8.37), which makes Sec more amenable to electrophilic attack at neutral pH. Moreover, RSL3 binding to GPX4 completely inhibits the activity of GPX4 (2), which suggests that either the active-site Sec is covalently modified by (1S, 3R)-RSL3 or one of the other cysteine residues is modified and the resulting enzyme is fully inhibited. To test the hypothesis that (1S, 3R)-RSL3 primarily targets the active-site Sec, we reverted only Ala46 in the allCys(-) protein to Sec46 (termed A46U) (Fig. 1C). Then, we generated a G401-derived cell line stably expressing GFP-A46U-GPX4 and performed the pull-down assay. As shown in Fig. 1C, (1S, 3R)-RSL3 successfully affinity-purified GFP-A46U-GPX4, supporting the hypothesis that RSL3 covalently interacts with GPX4 via its active-site Sec.
To further evaluate the requirement of Sec in the active site of GPX4 for covalent binding with (1S, 3R)-RSL3, we generated a G401 cell line stably expressing the GFP-A46C protein in which the A46 in allCys(-) was replaced with Cys. A pull-down assay using this cell line with (1S, 3R)-RSL3-Fcn showed no GFP-A46C protein band on a Western blot, suggesting a less efficient interaction between GFP-A46C-GPX4 and (1S, 3R)-RSL3-Fcn (Fig. 1C). The result suggests a critical importance of the active-site Sec in (1S, 3R)-RSL3 for binding to GPX4. Moreover, these results are consistent with the hypothesis that the active-site Sec is advantageous for GPX4 enzyme activity and has been selected for during evolution despite the energetic expense and low expression caused by incorporating Sec into the enzyme. Indeed, there is a significant decrease in peroxidase activity when the active-site Sec is replaced with Cys (17, 18).
Although less reactive than Sec, Cys is also nucleophilic. To test whether other Cys residues on GPX4 contribute to RSL3 binding, we generated G401 cell lines stably expressing individual revertants of allCys(-) in which each mutated residue was individually reverted to the original Cys (Fig. 1D). A total of seven stable cell lines (A2C, A10C, S37C, A66C, S75C, A107C, and A148C) were generated and subjected to binding analysis through the pull-down assay. We found that the RSL3 affinity probe bound moderately with a few other Cys residues, such as A10C and S37C (Fig. 1D). However, the amount of purified protein was substantially less than with WT GPX4 or the A46U revertant, suggesting that the binding between GPX4 and the RSL3-containing affinity probe is mainly driven by the reaction with the active-site selenocysteine.
Previously, several ferroptosis-inducing compounds, referred to as DPI compounds (19), were discovered, which displayed RSL3-like inhibition in a GPX4 activity assay (2). However, the interaction of these compounds with GPX4 has not been characterized. We thus cultured HT-1080 cells stably expressing GFP-GPX4 (WT), treated cells with (1S, 3R)-RSL3-Fcn in the presence or absence of each DPI compound, and ran the pull-down assay to test whether (1S, 3R)-RSL3-Fcn binding to GFP-GPX4 could be competed off by each DPI compound. In this experiment, we used two controls, free (1S, 3R)-RSL3 as a positive control and erastin as a negative control (Fig. 1E), which does not bind to Gpx4 directly.
Free (1S, 3R)-RSL3 indeed competed for (1S, 3R)-RSL3-Fnc binding to GPX4, which resulted in a much smaller amount of GPX4 protein pulled down by antifluorescein antibody beads compared with a sample without any competitor (Fig. 1E). However, erastin, which does not bind to GPX4, did not compete with (1S, 3R)-RSL3-Fcn binding to GPX4 and allowed pull-down of a similar amount of GPX4 protein with antifluorescein antibody beads (Fig. 1E). The DPI compounds inhibited the binding between (1S, 3R)-RSL3-Fcn and GPX4 protein to varying degrees. The level of competition by DPI7, DPI12, and DPI17 was highly significant (Fig. 1F; P < 0.01 by Student’s t test), suggesting that these DPIs share the same binding site as RSL3, namely the catalytic Sec. However, DPI13 and DPI10 showed marginal or no competition activity, which suggests that these compounds inhibit GPX4 indirectly or perhaps bind to a different site (Fig. 1F).
These data thus revealed which ferroptosis inducers bind competitively with (1S, 3R)-RSL3-fluorescein. We then tested whether (1S, 3R)-RSL3 indeed covalently interacts with GPX4 by adding (1S, 3R)-RSL3-fluorescein to GFP-GPX4–expressing HT-1080 cells and immunopurifying the GFP-GPX4 protein from cell lysates. If the interaction between (1S, 3R)-RSL3-fluorescein and GFP-GPX4 were to be noncovalent, Western blotting with an antifluorescein antibody would not detect the GFP-GPX4 band, because free (1S, 3R)-RSL3 would not be transferred to the Western blot membrane under the denaturing condition of SDS/PAGE.
Immunopurification of GFP-GPX4 with an anti-GPX4 antibody and detection by Western blot with an anti-GFP antibody revealed a clear protein band migrating at the expected size of the GFP-GPX4 fusion protein (Fig. 1G, Left). When the Western blot membrane was incubated with an antifluorescein antibody, we indeed detected a protein band migrating at the size of GFP-GPX4 (Fig. 1G, Right). This demonstrates that (1S, 3R)-RSL3-Fcn covalently labels the GFP-GPX4 protein in the cells.
We then used bacterially expressed and in vitro-purified GPX4U46C protein to confirm the presence of a covalent interaction between (1S, 3R)-RSL3 and GPX4 protein; note that due to the challenge of Sec incorporation in bacteria it is extremely difficult to produce and purify WT GPX4 protein using common expression systems. The GPX4U46C protein contains Cys in place of Sec in the active site, which allows for efficient bacterial protein production; GPX4U46C protein contains reduced peroxidase activity compared with WT GPX4 (20). Despite the lack of binding of (1S, 3R)-RSL3-Fcn to U46C GPX4 in the pull-down assay, we suspected that an in vitro reaction with greater protein abundance and sensitivity might allow for detection of the less efficient covalent interaction between these two species.
The (1S, 3R)-RSL3-fluorescein probe was incubated with GPX4U46C protein in a twofold dilution series. The GPX4U46C protein was resolved on SDS/PAGE and transferred to a PVDF membrane for the Western blotting. The (1S, 3R)-RSL3-fluorescein probe remains bound to GPX4U46C in this blot only if it interacts with the protein covalently under denaturing conditions. Western blotting with a fluorescein antibody revealed fluorescein bound to GPX4U46C, confirming the covalent interaction between the (1S, 3R)-RSL3-fluorescein probe and GPX4U46C protein (Fig. 1H).
The covalent interaction between (1S, 3R)-RSL3 and GPX4 should inhibit GPX4 enzymatic activity in an irreversible manner. Covalent inhibitors display time-dependent changes in IC50 values, making it difficult to derive enzyme kinetic parameters (21). Thus, we adapted a liquid chromatography (LC)-MS–based GPX4 assay (2) to measure GPX4 inhibition by (1S, 3R)-RSL3 in a concentration series at a specific time point to determine an effective IC50 for (1S, 3R)-RSL3. We included (1R, 3R)-RSL3 in this analysis as a negative control for enzyme inhibition, because this compound contains the same structure but differs only in the stereochemical configuration at one center, which results in a loss of activity in a cellular context.
The active stereoisomer of RSL3, (1S, 3R)-RSL3, effectively inhibited cellular GPX4 activity in a concentration-dependent manner (Fig. 1 I and J, EC50 = 10 μM). However, (1R, 3R)-RSL3, an inactive diastereomer of RSL3, showed no GPX4 inhibition up to 10 μM and exhibited only a small inhibitory effect at 100 μM, the highest concentration tested (Fig. 1 I and J).
Ferroptosis Is Driven by Peroxidation of PUFAs.
Upon inhibition of GPX4, cells accumulate lipid peroxides to a lethal level, leading to ferroptotic cell death. There are many different classes of lipids in cells, including fatty acids, phospholipids, cholesterols, cardiolipins, and sphingolipids. It was not clear which of these are oxidized during ferroptosis, and whether each lipid species has any functional role in the lethal phenotype of ferroptosis. Delineation of lipid oxidation events in the context of ferroptosis may provide insights into the mechanism of GPX4 regulation, as well as the execution mechanism for ferroptosis; many oxidized lipids (oxylipins) act as signaling molecules (22).
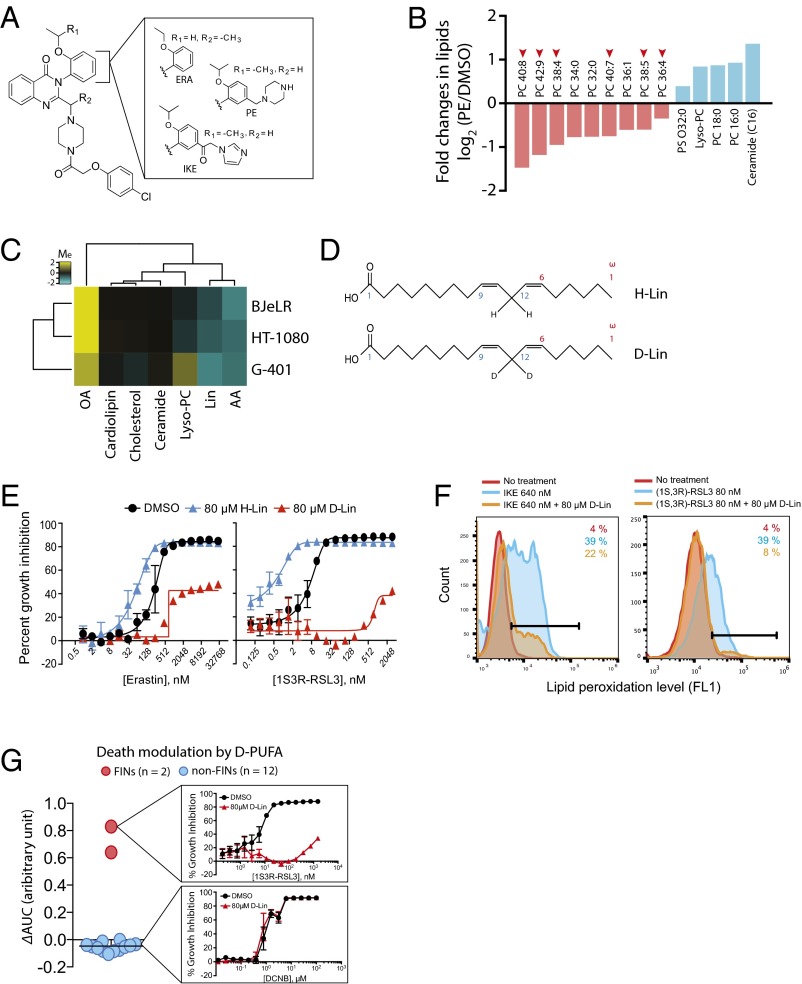
To examine global changes in lipid metabolites associated with ferroptosis, we added piperazine erastin (PE), a metabolically stable and water-soluble analog of erastin (2), to HT-1080 ferroptosis-sensitive fibrosarcoma cells and extracted cellular lipid metabolites for analysis by LC tandem mass spectrometry (LC-MS/MS) (Fig. 2A).
Fig. 2.
PUFAs play a functional role in ferroptotic cell death. (A) Structure of erastin analogs used in this study. ERA, erastin (3); IKE, imidazole ketoerastin (61); and PE, piperazine erastin (2). (B) Lipidomics analysis revealed loss of PUFAs is the most prominent change during ferroptosis. HT-1080 cells were treated with PE or vehicle (DMSO) and lipid metabolites were extracted as described in Methods. The amount of individual lipid metabolites in the sample was determined using an LC-MS instrument, and changes in the amount of lipid metabolites were calculated by dividing the amount in PE sample by that in DMSO sample. The graph shows the name of top changing lipid metabolites (nine down-regulated and five up-regulated) and the value of fold changes in log2 scale. The number of carbons and double bonds in sn-2 position fatty acids are indicated. For example, PC 40:8 indicates phosphatidyl choline (PC) that has a fatty acid of 40 carbon and 8 double bonds in sn-2 position. PS, phosphatidyl serine. (C) Modulation of ferroptosis sensitivity by different lipid species. PUFAs sensitized cells to ferroptosism, whereas OA prevented ferroptosis. Yellow indicates cell death suppression and blue represents cell death sensitization. Modulatory index (Me) was calculated as previously described (1). (D) Structures of natural LA (H-Lin) and deuterated LA (D-Lin). (E) Suppression of ferroptosis by D-Lin. G-401 cells were treated with either erastin or (1S, 3R)-RSL3 in the presence or absence of either H-Lin or D-Lin. Cells became rescued from ferroptosis upon D-Lin treatment, whereas they became more sensitive to ferroptosis upon H-Lin treatment. (F) D-Lin prevented generation of lipid peroxides. (G) D-Lin did not suppress lethality of 12 cytotoxic compounds, highlighting the specificity on ferroptosis.
The LC-MS/MS conditions were established to monitor changes in diverse lipid classes simultaneously (23). The lipid species detected under these conditions include phospholipids, lysophospholipids, sphingolipids, cholesterols, diacylglycerols, and triglycerides. We calculated the ratio of mass signal intensities between PE-treated and vehicle-treated samples to generate the differential lipidomic profiles and identified 14 lipid metabolites (nine down-regulated and five up-regulated) that displayed the most significant changes upon PE treatment (Fig. 2B and Dataset S1).
Many phosphatidylcholines (PCs) with polyunsaturated fatty acyl moieties (PUFAs) were depleted, whereas the levels of ceramide and lysophosphatidylcholine (lyso-PC) accumulated during PE-induced ferroptosis (Fig. 2B). Ceramide has been implicated in cell death following activation of stress responses (24, 25). Treatment with lyso-PC has been reported to generate ROS and destabilize plasma membranes in cultured human fibroblast cells (26). Therefore, we evaluated the functional relevance of these lipids in ferroptosis by treating three ferroptosis-sensitive cell lines, BJeLR, HT-1080, and G-401, with (1S, 3R)-RSL3 in the presence or absence of each lipid species. Arachidonic acid (AA, C20:4) and linoleic acid (Lin, C18:2) were selected as representative PUFAs, and oleic acid (OA, C18:1) was selected as a representative monounsaturated fatty acid (MUFA). We also tested cholesterol and cardiolipin, because they constitute major lipid classes and have been implicated in cell death processes in their oxidized forms (27, 28). We note that we focused on identifying lipid classes that are functionally relevant to ferroptosis, rather than identifying individual lipid species contributing to ferroptosis. Therefore, we tested representative lipid species from each lipid class in our analysis.
Cells were treated with (1S, 3R)-RSL3 in a 16-point, twofold dilution series in the presence or absence of added lipids, and the impact of the dose–response curves on cell survival was determined. Then, we computed the modulatory effect (Me) of each lipid on the normalized viability of cells treated with (1S, 3R)-RSL3 alone (Me < 0, death sensitization; Me = 0, no effect; and Me > 0, death rescue). The resulting values were clustered hierarchically in an unsupervised fashion and displayed as a heat map (Fig. 2C). Using this analysis, we observed that ferroptosis induced by (1S, 3R)-RSL3 was not consistently modulated by lyso-PC, ceramide, cholesterol, or cardiolipin. OA, however, strongly suppressed ferroptosis in all of the three cell lines; this raises the possibility that OA could be used as a cell death inhibitor in models in which ferroptosis is operative. In contrast, the PUFAs AA and Lin sensitized cells to (1S, 3R)-RSL3–induced ferroptosis, suggesting a functional role for PUFA oxidation in ferroptosis (Fig. 2C).
PUFAs are highly susceptible to oxidative damage due to the presence of bis-allylic protons that are vulnerable to hydrogen atom abstraction (Fig. 2D). Hydrogen abstraction produces alkyl radicals that react readily with molecular oxygen to produce peroxyl radicals, which subsequently react with other PUFAs to propagate a chain reaction of lipid peroxidation. PUFA peroxides, along with their end product reactive aldehydes, such as malondialdehydes and 4-hydroxynonenal, are damaging to cells.
To further test the functional role of PUFAs in ferroptosis, we replaced natural PUFAs in cells (H-PUFAs) with deuterated PUFAs (D-PUFAs) (Fig. 2D). D-PUFAs have deuterium in place of the bis-allylic hydrogens, which slows the initiation of deuterium abstraction and subsequent radical generation compared with H-PUFAs (29). It was previously found that growing yeast in D-PUFA–enriched medium allowed uptake of D-PUFAs into yeast cell membranes within 4 h, and D-PUFA uptake resulted in protection of yeasts from copper-induced toxicity (30).
We incubated G-401 cells with D-linoleate (D-Lin), H-linoleate (H-Lin), or vehicle only (0.1% ethanol) overnight and then treated with erastin or (1S, 3R)-RSL3 to examine the impact of D-PUFAs on ferroptosis. We observed a strong protective effect of D-PUFAs in both erastin-treated and (1S, 3R)-RSL3–treated conditions (Fig. 2E). Cells treated with D-PUFAs did not generate lipid peroxides, as measured by C11-BODIPY, upon imidazole ketoerastin (IKE) or (1S, 3R)-RSL3 treatment, confirming that deuterium in the bis-allylic position retards the radical chain reaction of lipid peroxidation (Fig. 2F). In contrast to cell death induced by IKE or (1S, 3R)-RSL3, cell death induced by 12 other cytotoxic compounds with nonferroptotic cell death mechanisms was not rescued by D-PUFA treatment (Fig. 2G), highlighting a unique role for PUFA oxidation in executing ferroptotic cell death.
Lipoxygenases Regulate Erastin-Induced Ferroptosis.
Abstraction of hydrogen and subsequent oxidation of PUFAs can occur either nonenzymatically or enzymatically. Lipoxygenases are a family of iron-containing enzymes that catalyze dioxygenation of PUFAs to produce fatty acid hydroperoxides in a stereospecific manner (Fig. 3A) (31). Fatty acid hydroperoxides are then converted to fatty acid alcohols, generating lipid-signaling molecules, such as eicosanoids (31). Oxidation of PUFAs by lipoxygenases has been implicated in cell death under specific conditions, such as GSH depletion, which led us to hypothesize that ferroptosis can be modulated by lipoxygenases (32–35).
Fig. 3.
Lipoxygenases are involved in ferroptosis by system xc− inhibition. (A) Dioxygenation reaction induced by lipoxygenases. (B) G-401 cell line expressed all isoforms of ALOXs as assessed by qPCR experiment. ACTB, Beta-actin. (C) A pool of siALOX targeting all ALOX genes successfully decreased mRNA levels of ALOX genes. (D) Silencing ALOXs expression conferred resistance to IKE-induced ferroptosis, whereas it did not affect RSL3 lethality in G-401 cells. (E) Silencing ALOX15B and ALOXE3 in HT-1080 cells made cells resistant to erastin. (F) Lipoxygenase inhibitors rescued cells from erastin-induced ferroptosis. A cyclooxygenase inhibitor did not affect erastin’s lethality. (G) GFP-ALOX5 translocated to the nuclear membrane only in sensitive cells upon 10 µM erastin treatment. Bar graph: mean + SD; n = 3–4; ***P < 0.001. (H) Time course of GFP-ALOX5 translocation in HT-1080 cells upon treatment with erastin or ionomycin.
There are six ALOX (arachidonate lipoxygenase) genes in humans: ALOX5, ALOX12, ALOX12B, ALOX15, ALOX15B, and ALOXE3. These genes have different expression patterns in different tissues; therefore, we examined which ALOX genes were expressed in the G-401 cells using quantitative PCR (qPCR) and observed expression of all ALOX genes in this cell line (Fig. 3B). Because we expected that each ALOX isoform might be functionally redundant in terms of PUFA oxidation, we suppressed expression of all ALOX genes using a pool of siRNAs targeting all ALOX genes (Fig. 3C) and then added IKE or (1S, 3R)-RSL3 to examine changes in ferroptosis sensitivity.
Silencing ALOX genes made cells resistant to IKE treatment; however, it did not change sensitivity to (1S, 3R)-RSL3 (Fig. 3D). Mechanistically, IKE inhibits system xc− to deplete cellular GSH and inhibits GPX4 indirectly, whereas (1S, 3R)-RSL3 binds to and inhibits GPX4 in the absence of cellular GSH depletion. It is likely that lipoxygenase-mediated PUFA oxidation exerts a lethal effect under GSH depletion conditions, whereas the effect is negligible when the cellular GSH level is normal.
We further tested the relevance of lipoxygenases to ferroptosis mediated by system xc− inhibition in two other cell lines, BJeLR and HT-1080. Of the six ALOX isoforms, ALOX15B and ALOXE3 were consistently expressed in these cell lines, whereas ALOX5, ALOX12, ALOX12B, and ALOX15 did not show consistent expression by qPCR analysis (Fig. S1A). Accordingly, we prepared pools of siRNAs targeting ALOX15B and ALOXE3 and tested the effect of each siRNA pool on the lethality of erastin. The siRNA pools targeting ALOX15B and ALOXE3 decreased their target mRNA levels by greater than 6- and 20-fold, respectively (Fig. S1B). System xc− inhibition by erastin induced cell death in control siRNA-treated cells; however, erastin-induced cell death was rescued by silencing either ALOX15B or ALOXE3, which supported the hypothesis that lipoxygenases are required for ferroptosis under GSH-depleted condition (Fig. 3E). Finally, five different pharmacological inhibitors of ALOXs prevented erastin-induced cell death, supporting the role of lipoxygenase activity in erastin lethality (Fig. 3F). Indomethacin, a cyclooxygenase inhibitor, was minimally effective in suppressing erastin lethality, which highlights the importance of lipoxygenases, but not cyclooxygenases, in erastin-mediated ferroptosis.
Fig. S1.
(A) Expression analysis of ALOX genes in BJeLR and HT-1080 cells. The figures show the amplification plot of each ALOX isoform. Triplicate samples were analyzed for each gene using mRNA preparation from BJeLR cells. The red lines in each plot indicate ACTB gene amplification that served as endogenous control in the quantification. The gene name and the Ct number, if was possible to determine, are presented. A Ct value greater than 35 is considered a weak expression level, which suggests that ALOXE3 is the major isoform expressed in these cell lines. N.D., not determined. (B) Knockdown of ALOX15B and ALOXE3 expression by the pool of siRNAs was confirmed using qPCR analysis. Data are presented as mean ± SD; n = 3.
ALOX5 is one of the six human ALOX isoforms and plays a critical role in leukotriene synthesis (31). In the basal state, the ALOX5 protein remains in the nucleus; however, upon activation, it translocates to the nuclear membrane (36). To examine whether ALOX proteins are activated upon erastin treatment, we expressed GFP-tagged ALOX5 in BJ-derived cell lines and examined whether erastin treatment had any effect on the localization of GFP-ALOX5 (Fig. 3G). A positive control for GFP-ALOX5 translocation, treatment with ionomycin, induced localization of GFP-ALOX5 to the nuclear membrane in all BJ-derived cell lines (Fig. S2). In contrast, GFP-ALOX5 was translocated to the nuclear membrane only in ferroptosis-sensitive BJeLR cells upon erastin treatment (Fig. 3G). These results suggested that activation of ALOX proteins after system xc− inhibition occurs selectively in ferroptosis-sensitive cell lines, leading to lipid peroxidation and oxidative cell death. We expressed GFP-ALOX5 in HT-1080 cells and observed the same translocation event upon erastin treatment (Fig. 3H, Upper).
Fig. S2.
GFP-ALOX5 translocated to the perinuclear membrane region upon ionomycin and erastin treatment. GPF-ALOX5 remained within the nucleus when expressed in BJeH, BJeHLT, and BJeLR cells (Upper) but translocated to the perinuclear membrane region upon ionomycin treatment (Lower). Unlike erastin-induced translocation (Fig. 3G), all three BJ cells responded equally to the ionomycin treatment. BJ cells were treated with 113 μM ionomycin for 12 h. Bar graph, n = 3–4; n.s., not significant. (Scale bars, 60 μm.)
Elevation of cellular calcium level is a well-known trigger for nuclear membrane translocation of ALOX5 (37). However, it is unlikely that erastin activates lipoxygenases through calcium up-regulation for multiple reasons. First, the kinetics of GFP-ALOX5 translocation in response to erastin differed from those seen upon ionomycin treatment, a positive control for intracellular calcium up-regulation (Fig. 3H, Lower). Second, calcium chelators were not effective in suppressing erastin-induced cell death (4).
PHKG2 Regulates Ferroptosis Through Modulation of Available Iron.
Considering the critical role of lipid oxidation on ferroptosis, we determined that an shRNA suppressor screen with erastin might identify genetic modifiers of PUFA oxidation. The identified genes might encode proteins that alter PUFAs at the intrinsic level, such as enzymes regulating abundance or saturation level of PUFAs, or encode proteins that belong to a molecular pathway modulating the rate of PUFA oxidation.
To identify shRNA suppressors of erastin, we infected U-2-OS cells with lentiviruses that harbor expression cassettes for shRNAs targeting human kinases and related genes (38, 39). Adding 10 μM erastin to the culture induced 99% lethality in cells infected with a control shRNA virus targeting no known gene, whereas there were several shRNAs that consistently rescued U-2-OS cells by greater than 50%. Because we were interested in robust shRNA suppressors with activity in multiple cell lines, we retested candidate hits from U-2-OS cells in HT-1080 cells. This two-cell-line testing was a stringent means to select only one top confident hit, PHKG2 (Fig. 4A).
Fig. 4.
Functional RNAi screening identified an iron regulatory role of PHKG2. (A) Screening outline to identify PHKG2 as a final shRNA screen hit. (B) U-2-OS cells became resistant to erastin upon PHKG2 silencing by two independent shRNAs. (C) Erastin-treated HT-1080 cells were rescued by shPHKG2 used in B. (D) Silencing of PHKG2 expression in HT-1080 cells was confirmed by qPCR experiment. (E) A known role of PHKG2 in glycogen metabolism. (F) Kinase inhibitor of PHKG2 conferred resistance to erastin in HT-1080 cells; 10 µM K252a was used. (G) Two kinase inhibitors of GP, CP-91149 (100 μM) or GP-I (5 μM), were not effective in rescuing HT-1080 cells from erastin-induced ferroptosis. (H) Silencing PHKG2 prevented accumulation of lipid peroxides upon erastin treatment. (I) Silencing PHKG2 by shRNA decreased cellular iron level. (J) A diagram showing the RNAi target site of shRNAs and siRNAs used in this study. Note that the target site is different between shRNA and siRNA. (Lower) Chart shows silencing efficiency of siRNAs assessed by qPCR. (K) Silencing PHKG2 by siRNA decreased cellular iron level. (L) A model summarizing the findings from our study.
We confirmed that two independent shRNAs targeting PHKG2 raised the EC50 of erastin by two- to fourfold in U-2-OS cells (Fig. 4B). Furthermore, both shRNAs suppressed erastin lethality in erastin-sensitive HT1080 cells, confirming that the erastin-suppressing effects of PHKG2 knockdown were not limited to one cell line (Fig. 4C). The knockdown of PHKG2 gene expression by both shRNAs in the HT1080 cell line was confirmed by qPCR (Fig. 4D).
PHKG2 encodes the catalytic subunit of the PHK (phosphorylase kinase) complex, which activates glycogen phosphorylase (GP) to release glucose-1-phosphate from glycogen (Fig. 4E) (40). To investigate whether the kinase function of PHK is important for regulating erastin sensitivity, we treated cells with both erastin and a general kinase and PHK inhibitor, K252a (41), and found that K252a suppressed erastin’s lethality (Fig. 4F).
We speculated that the metabolic cascade consisting of PHK, GP, and glycogen might modulate erastin sensitivity. Glycogen is found mainly in liver and muscle tissues, and the level of glycogen is generally low in other tissues. However, the occurrence of glycogen has been reported in cancer cells from diverse tissue origins that normally do not store glycogen (42–44). To test the functional role of this metabolic pathway on erastin-induced ferroptosis, we treated cells with either of two GP inhibitors, CP-91149 (45) or GP inhibitor (46), and monitored the effects on erastin sensitivity. These inhibitors did not rescue cells from erastin lethality (Fig. 4G); on the contrary, we observed a sensitization effect of one inhibitor, CP-91149 (Fig. 4G). We concluded that it is likely not the metabolic pathway of glycogen breakdown that is responsible for the suppression of erastin’s lethality by shPHKG2, but rather an unidentified function of PHKG2.
When we examined changes in lipid oxidation level, we observed that PHKG2-silenced cells did not have elevated lipid peroxidation upon erastin treatment (Fig. 4H). We hypothesized that PHKG2 silencing might perturb ROS generation or defenses, or affect cellular iron homeostasis, causing lack of lipid oxidation upon erastin treatment. To test this latter hypothesis, we used calcein-AM, a fluorescein-derived dye with green fluorescence that is quenched upon binding to ferrous ion (47). Calcein-AM staining of HT-1080 cells infected with a nontargeting shRNA defined a basal level of fluorescence (Fig. 4I). Cells infected with both shRNAs targeting PHKG2 shifted the fluorescence to a higher intensity, indicating that calcein AM was dequenched in these cells compared with nontargeting shRNA-infected cells (Fig. 4I). The result implies that cells infected with shPHKG2 have less iron available for chelating calcien AM. The iron-chelating effect of PHKG2 silencing was reproducible in the U-2-OS cell line when an independent siRNA pool (Fig. 4J) and a different iron sensor, PhenGreen SK, were used (Fig. 4K). Using Ingenuity pathway analysis, we found a hypothetical connection between PHKG2 and iron metabolism involving p53 (Fig. S3), which has itself been linked to ferroptosis regulation (48, 49), providing an avenue for future exploration. Because we know that decreased iron levels suppress erastin lethality (6), this iron regulatory function of PHKG2 is likely responsible for modulating sensitivity to erastin. Therefore, this small-scale shRNA suppressor screen identified an extrinsic factor for PUFA oxidation, iron depletion by PHKG2 silencing.
Fig. S3.
There is a possible link between PHKG2 and cellular iron involving tumor suppressor p53. PHKG2 gene and biomolecules associated with cellular iron were put into a single network space in Ingenuity Pathway Analysis (IPA) software. The “grow” function of IPA software was used to expand the number of edges in the network space, and then the “connect” function was used to identify possible connections among the molecules. This revealed p53 as a hypothetical link between PHKG2 and cellular iron.
In summary, we demonstrated that (1S, 3R)-RSL3 covalently interacts with the active-site selenocysteine of GPX4 to inhibit its enzymatic activity. Inhibition of GPX4 initiates uncontrolled PUFA oxidation and fatty acid radical generation, causing ferroptotic cell death. We identified intrinsic and extrinsic factors such as D-PUFAs, lipoxygenases, and PHKG2 that modulate the status of cellular PUFAs and thereby cells’ sensitivity to ferroptosis (Fig. 4L).
Discussion
Although we identified the RSL3 binding site on GPX4, the structural basis of RSL3 inhibition on GPX4 remains to be elucidated. Ideally, a cocrystal structure of RSL3-bound GPX4 would be solved to obtain structural information on the binding mechanism and to initiate a search for drug-like GPX4 inhibitors. The apo-GPX4 structure is available (Protein Data Bank ID code 2OBI) (20). However, no cocrystal structure containing GPX4 is available, implying technical difficulties in achieving this goal, consistent with our experience. Two technical hurdles are worth mentioning here. First, GPX4 is a selenoprotein requiring a complicated machinery to incorporate Sec in the active site (50). The UAG codon for selenocysteine is normally a STOP codon. Although Escherichia coli does have a native selenoprotein, formate dehydrogenase (51), the selenocysteine incorporation mechanism is sufficiently different to impose a strict species barrier in direct heterologous synthesis of recombinant selenoproteins in E.coli (52). Indeed, the apo-GPX4 crystal structure available is the Sec-to-Cys mutant GPX4 protein, not WT protein (20). Engineering of a bacterial expression system has reportedly produced rat thioredoxin reductase, another mammalian selenoprotein (53). Such an approach might be explored to produce WT GPX4 protein. Second, apo-GPX4 has a P3121 space group (Protein Data Bank ID code 2OBI), which might be most favorable for crystal formation. The C terminus of one GPX4 monomer occludes the active site of an adjacent GPX4 protein during the formation of the crystal with a P3121 space group. Because RSL3 binds to the active site, RSL3 addition to the crystal tray would inhibit formation of GPX4 crystal with the P3121 space group. Deleting C-terminal amino acids and optimizing crystal formulation to avoid formation of the P3121 space group may be considered in the cocrystallization experiments.
D-PUFAs have demonstrated efficacy in several human disease models, including Parkinson’s disease (29) and Friedreich’s ataxia (54). Strong and specific suppression of ferroptosis by D-PUFA (Fig. 2G) suggests the possible involvement of ferroptosis in such disease contexts and supports the idea of using ferroptosis inhibitors to delay progression of these diseases. Relevant to this, it is intriguing that OA, a MUFA, strongly rescued cells from ferroptosis (Fig. 2C). Treatment of OA might decrease the abundance of H-PUFAs in the cell membrane or activate a downstream signaling pathway to exert an indirect antioxidant effect. Such an indirect antioxidant effect was observed when DHA was added to a murine hippocampal cell line and up-regulated GPX4 gene expression (55). Delineation of an indirect antioxidant mechanism induced by OA should provide an alternative means to protect cells.
Inhibition of lipoxygenases suppressed erastin-induced, but not RSL3-induced, ferroptosis (Fig. 3D). Our observation is analogous to the suppression of glutamate-induced toxicity by inhibition of 12-lipoxygenase in cortical neurons where excessive glutamate inhibits system xc−, the glutamate/cysteine antiporter, and depletes cellular GSH (33). Similarly, cysteine depletion in the culture media for oligodendrocytes activated 12-lipoxygenase activity (56). Moreover, depletion of GSH by buthionine sulfoximine treatment was lethal to WT mouse embryonic fibroblasts (MEFs), whereas MEFs with 12/15-LOX gene deletion were resistant to the same treatment (57), which supports the functional role of lipoxygenases in ferroptosis induced by GSH depletion. In contrast, the lethal phenotype of Gpx4 knockout mice was independent of the Alox15 gene (7), suggesting that lipoxygenase activity is dispensable when cell death is initiated by direct inhibition of GPX4.
Reduced GSH was shown to inhibit lipoxygenases directly from 1 to 10 mM, which corresponds to the intracellular concentration of GSH under normal conditions (58, 59). System xc− inhibition and subsequent depletion of GSH is likely to exacerbate uncontrolled lipoxygenase activation compared with direct GPX4 inhibition without GSH depletion. Elaboration on the role of lipoxygenase within ferroptosis should provide a better understanding about the mechanism of lipoxygenase regulation by GSH and reveal the role of ferroptosis components in lipid signaling.
Methods
Affinity Purification of Cellular GFP-GPX4 Proteins.
Generation of stable cell lines.
Human GPX4 cDNA with a 3′ UTR region containing selenocysteine insertion sequence (GenBank accession no. NM_002085.3) was cloned into pBabe-puro retroviral expression plasmid as a GFP fusion form. Retroviruses were generated using PLAT-GP packaging cell line (Cell Biolabs, Inc.) and pCMV-VSV-G helper plasmid (60). G-401, a human renal carcinoma cell line, was cultured in six-well plates and infected with the retroviruses harboring expression cassette of indicated cDNAs. Two days later, cells were transferred to a fresh culture containing 1.5 µg/mL puromycin in the growth media to select for stably expressing clones.
Pull-down experiment.
An affinity purification experiment was performed as described previously (2). Briefly, G-401 cells stably expressing GFP-GPX4 protein were treated with 400 nM (1S, 3R)-RSL3-Fcn probe for 2 h followed by cell harvest and lysis. Cell lysates were precleared with Sepharose beads for 1 h and then incubated with antifluorescein antibody-conjugated Sepharose beads overnight at 4 °C. Unbound proteins were removed by spinning down the Sepharose beads, and the beads were washed three times with a washing buffer. SDS/PAGE gel sample buffer was added to packed beads and the mixture was heated at 95 °C for 5 min to release bound proteins. The supernatant after the heating was loaded on SDS/PAGE gel. The proteins were resolved on the gel and were subjected to Western blot analysis with anti-GFP antibody (sc-9996; Santa Cruz).
Analysis of Covalent Interaction Between RSL3 and GPX4.
Immunoprecipitation.
HT-1080 cells stably expressing GFP-GPX4 protein (2) were incubated with 0.5 µM (1S, 3R)-RSL3-fluorescein probe for 2 h. Cell lysates were prepared and incubated with either protein G beads or anti-GPX4 antibody protein-G beads at 4 °C overnight. The beads were washed twice with affinity purification buffer (50 mM Hepes, pH 7.4, 100 mM NaCl, and 1% Triton X-100) and twice with wash buffer (50 mM Hepes, pH 7.4, 500 mM NaCl, and 1% Triton X-100). The immunoprecipitated proteins were eluted off the beads by adding equal volume of 2× SDS sample buffer to the packed beads and boiling the mixture at 90 °C for 5 min. The eluted proteins were resolved by SDS/PAGE and processed for Western blotting with either anti-GFP antibody or antifluorescein antibody (200-002-037; Jackson ImmunoResearch Laboratories).
In vitro binding assay.
Bacterial expression vector pOE30-c-GPX4U46C was a generous gift from Hartmut Kuhn, University Medicine Berlin–Charité, Berlin. The 6xHis-GPX4U46C protein was expressed in E. coli and purified according to a published protocol (20). Twofold dilutions of (1S, 3R)-RSL3-fluorescein probe (highest concentration 800 ng) were incubated with 1.6 µg of 6xHis-GPX4U46C protein in each microtube for 30 min at room temperature. SDS sample buffer (2×) was added to the protein/probe complex and then the whole mixture was boiled at 90 °C for 5 min to disrupt any noncovalent interaction. The resulting samples were resolved on SDS/PAGE followed by Western blotting with either antifluorescein antibody or anti-GPX4 antibody (ab41787; Abcam).
Sample Preparation for Lipidomics Analysis.
Two million HT-1080 cells were seeded in 10-cm culture dishes. The next day, cells were treated with 5 mg/mL PE and incubated for 5 h before lipid extraction. A total of 3 mL of cold 100% (vol/vol) isopropanol was added to the cell monolayer to scrape cells. The resulting cell lysate/isopropanol mixture was transferred to a new 15-mL tube and centrifuged at 2,000 × g at 4 °C for 10 min. The cleared supernatant was transferred to a new tube and stored at – 20 °C for LC-MS/MS analysis.
Lipidomics Analysis.
We injected our lipid extract to SYNAPT G2-S (Waters), a qTOF mass spectrometer, and acquired MS1 and MS2 mass spectra in an unbiased and parallel manner (named as MSE technology). The raw mass spectra data were processed using TransOmics software (Waters) to detect and deconvolute meaningful mass peaks. For example, inconsistent and irregular mass peaks were excluded from further analysis. Statistical analysis was carried out under the same TransOmics software to quantify and determine differentially represented mass peaks between PE- and DMSO-treated samples (n = 12 for each condition). We requested CV < 30, fold-change > 2, Anova P value < 0.05, and elution time greater than 1 min in selecting the top 500 mass peaks in Dataset S1. For the top 14 differential mass peaks, we uploaded our qTOF mass data to METLIN (Scripps Center for Metabolomics) to identify metabolites with a mass tolerance value of 8 ppm.
Modulatory Profiling of Lipids.
The modulatory profiling of lipids was performed as described previously (4). The concentration of each lipid is listed here: OA, 80 µM; cardiolipin, 16.6 µM; cholesterol, 40 µM; ceramide, 37.2 µM; Lyso-PC, 80 µM; LA, 80 µM; and AA, 80 µM.
Inhibition of Lipoxygenase.
RNAi-mediated inhibition.
siRNA pools targeting individual ALOX isoforms were obtained from Dharmacon Technologies. On the day of reverse transfection, a mixture of 1 mL of Opti-MEM (Invitrogen), 6 μL of Lipofectamine RNAiMAX (Invitrogen), and 5 μL of 10 μM siRNA were prepared and transferred to each well of a six-well plate. The six-well plate was put in the tissue culture incubator for 20 min to allow the formation of transfection mixture. While the complex was forming, HT-1080 cells were trypsinized and the cell number was determined using ViCell (Trypan blue assay). Cells (n = 200,000) were prepared in 1 mL of growth media with 2× serum and then the cell suspension was transferred to each well containing 1 mL of the transfection mix. The six-well plate was returned to the incubator and the culture was grown for 2 d. Then, cells were trypsinized and reverse-transfected again for two additional days to ensure knockdown. After a second round of reverse transfection, cells were trypsinized and treated with lethal compounds to examine the effect of the knockdown on drug sensitivity. RNA was harvested from a population of cells for RT-qPCR analysis.
Small-molecule–based inhibition.
Lethal compounds were added to HT-1080 cells in the presence or absence of ALOX or COX inhibitors in a twofold dilution series. The concentrations of the inhibitors are listed here: cinnamyl-3,4-dihydroxya-cyanocinnamate (CDC), 20 μM; baicalein (BAI), 10 μM; PD-146176, 5 μM; AA-861, 2 μM; zileuton, 50 μM; and indomethacin (Indo), 200 μM. Cell viability was determined using Alamar Blue and percent growth inhibition was calculated as described previously (6).
GFP-GPX4 Translocation Assay.
A cDNA of ALOX5 (GenBank accession no. BC130332.1) was cloned into a pBabe-puro vector to express GFP fused ALOX5 (N-terminal GFP fusion) in cells. The plasmid was transfected into PLAT-GP cells (RV-103; Cell Biolabs) along with pVSV-G helper plasmid to produce retrovirus harboring the expression plasmid. Target cells (n = 0.2 million; BJeH, BJeHLT, BJeLR, or HT-1080 cells) were seeded in 10-cm tissue culture dishes and incubated in the tissue culture incubator for 1 d. Frozen stock of retrovirus solution was thawed at 37 °C for 2 min and polybrene (H9268; Sigma) was added at a final concentration of 8 μg/mL. Culture medium was replaced with virus/polybrene mix and the culture dish was incubated for 2 h with rocking every 30 min. After 2 h, 10 mL of growth media was added to culture dish and the culture was incubated further for 2 d. Cell lines stably expressing GFP-ALOX5 protein were selected using 1.5 μg/mL puromycin and used for the GFP-ALOX5 translocation assay.
Functional RNAi Screening.
Lentiviral shRNA library.
Our shRNA library was created by RNAi Consortium11 and consists of lentivirus solutions in 384-deepwell polypropylene plates (781270; Greiner). The library targets 1,006 human genes, including kinases, those similar to kinases, and some ancillary proteins. More information about the library is described in our previous publication (39).
Primary screening.
Assay plates were prepared by seeding 400 U-2-OS cells per well in 40 μL of growth media in black, clear-bottom, 384-well plates (3712; Corning). The next day, 40 μL of virus daughter plates were prepared by transferring 2 μL of virus stock solution from shRNA library plates and 4 μL of 10× polybrene solution to 34 μL of cell growth media in 384-well polypropylene plates (781280; Greiner). Whole growth media in the assay plates were replaced with 40 μL of virus/polybrene/media mixture from the virus daughter plates. Then, virus infection was carried out by centrifuging the assay plates for 1.5 h at 1,092 × g, 37 °C, and the assay plates were returned to a tissue culture incubator. Three days later, the culture media was replaced with fresh media containing DMSO or 10 μg/mL erastin. Erastin induces 100% cell killing at this concentration. The next day, Alamar Blue (DAL1100; Invitrogen) was added to the assay plates to determine cell viability. All liquid handling was carried out using a Biomek FX AP384 module (Beckman Coulter). Percent growth inhibition (%GI) was calculated from the following formula using the Alamar Blue readout:
where X is values from cells with shRNAs and erastin; N is the values from media, shRNAs, and erastin; and P is the values from cells, shRNAs, and DMSO. All experiments were performed in triplicate and the median percent growth inhibition value was taken for selecting primary hits to be analyzed.
Cellular Iron Staining.
HT1080 cells were seeded in six-well plates and infected with nontargeting shRNA or two shRNAs targeting PHKG2. Three days later, the cell monolayer was washed with PBS twice and stained with 10 nM of Calcein-AM (C3099; Invitrogen) in PBS by incubating the plate for 15 min in culture incubator. Cells were released with trypsin/EDTA, harvested in 2 mL PBS, and centrifuged at 216 × g for 5 min. The cell pellet was resuspended in 1 mL of PBS, the cell suspension was transferred to disposable FACS tubes, and the fluorescence profile of the sample was monitored using a FACSCalibur system (BD Biosciences).
Supplementary Material
Acknowledgments
We thank Matt Franklin (New York Structural Biology Center) for expression and purification of 6xHis-GPX4U46C protein and Andrew Baker (Waters Corp.) for analyzing lipidomics samples by LC-MS/MS. This work was supported by National Institute of Health Grants 5R01CA097061, 5R01GM085081, and R01CA161061 and New York Stem Cell Science Grant C026715 (to B.R.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603244113/-/DCSupplemental.
References
- 1.Dixon SJ, et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang WS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1-2):317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3(3):285–296. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 4.Wolpaw AJ, et al. Modulatory profiling identifies mechanisms of small molecule-induced cell death. Proc Natl Acad Sci USA. 2011;108(39):E771–E780. doi: 10.1073/pnas.1106149108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yagoda N, et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447(7146):864–868. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang WS, Stockwell BR. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem Biol. 2008;15(3):234–245. doi: 10.1016/j.chembiol.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedmann Angeli JP, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol. 2014;16(12):1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skouta R, et al. Ferrostatins inhibit oxidative lipid damage and cell death in diverse disease models. J Am Chem Soc. 2014;136(12):4551–4556. doi: 10.1021/ja411006a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Back SA, Gan X, Li Y, Rosenberg PA, Volpe JJ. Maturation-dependent vulnerability of oligodendrocytes to oxidative stress-induced death caused by glutathione depletion. J Neurosci. 1998;18(16):6241–6253. doi: 10.1523/JNEUROSCI.18-16-06241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volpe JJ. Neurobiology of periventricular leukomalacia in the premature infant. Pediatr Res. 2001;50(5):553–562. doi: 10.1203/00006450-200111000-00003. [DOI] [PubMed] [Google Scholar]
- 11.Boutaud O, Roberts LJ., 2nd Mechanism-based therapeutic approaches to rhabdomyolysis-induced renal failure. Free Radic Biol Med. 2011;51(5):1062–1067. doi: 10.1016/j.freeradbiomed.2010.10.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linkermann A, et al. Synchronized renal tubular cell death involves ferroptosis. Proc Natl Acad Sci USA. 2014;111(47):16836–16841. doi: 10.1073/pnas.1415518111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imai H, et al. Early embryonic lethality caused by targeted disruption of the mouse PHGPx gene. Biochem Biophys Res Commun. 2003;305(2):278–286. doi: 10.1016/s0006-291x(03)00734-4. [DOI] [PubMed] [Google Scholar]
- 14.Yant LJ, et al. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic Biol Med. 2003;34(4):496–502. doi: 10.1016/s0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
- 15.Dixon SJ, et al. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. eLife. 2014;3:e02523. doi: 10.7554/eLife.02523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Louandre C, et al. Iron-dependent cell death of hepatocellular carcinoma cells exposed to sorafenib. Int J Cancer. 2013;133(7):1732–1742. doi: 10.1002/ijc.28159. [DOI] [PubMed] [Google Scholar]
- 17.Mannes AM, Seiler A, Bosello V, Maiorino M, Conrad M. Cysteine mutant of mammalian GPx4 rescues cell death induced by disruption of the wild-type selenoenzyme. FASEB J. 2011;25(7):2135–2144. doi: 10.1096/fj.10-177147. [DOI] [PubMed] [Google Scholar]
- 18.Schnurr K, Borchert A, Gerth C, Anton M, Kuhn H. Bacterial and nonbacterial expression of wild-type and mutant human phospholipid hydroperoxide glutathione peroxidase and purification of the mutant enzyme in the milligram scale. Protein Expr Purif. 2000;19(3):403–410. doi: 10.1006/prep.2000.1262. [DOI] [PubMed] [Google Scholar]
- 19.Weïwer M, et al. Development of small-molecule probes that selectively kill cells induced to express mutant RAS. Bioorg Med Chem Lett. 2012;22(4):1822–1826. doi: 10.1016/j.bmcl.2011.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheerer P, et al. Structural basis for catalytic activity and enzyme polymerization of phospholipid hydroperoxide glutathione peroxidase-4 (GPx4) Biochemistry. 2007;46(31):9041–9049. doi: 10.1021/bi700840d. [DOI] [PubMed] [Google Scholar]
- 21.Krippendorff BF, Neuhaus R, Lienau P, Reichel A, Huisinga W. Mechanism-based inhibition: Deriving K(I) and k(inact) directly from time-dependent IC(50) values. J Biomol Screen. 2009;14(8):913–923. doi: 10.1177/1087057109336751. [DOI] [PubMed] [Google Scholar]
- 22.Strassburg K, et al. Quantitative profiling of oxylipins through comprehensive LC-MS/MS analysis: application in cardiac surgery. Anal Bioanal Chem. 2012;404(5):1413–1426. doi: 10.1007/s00216-012-6226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isaac G, McDonald S, Astarita G. 2011. Lipid separation using UPLC with charged surface hybrid technology (Waters Corp., Milford, MA)
- 24.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9(2):139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 25.Mathias S, Peña LA, Kolesnick RN. Signal transduction of stress via ceramide. Biochem J. 1998;335(Pt 3):465–480. doi: 10.1042/bj3350465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colles SM, Chisolm GM. Lysophosphatidylcholine-induced cellular injury in cultured fibroblasts involves oxidative events. J Lipid Res. 2000;41(8):1188–1198. [PubMed] [Google Scholar]
- 27.Liu J, et al. Cholesterol oxidase from Bordetella species promotes irreversible cell apoptosis in lung adenocarcinoma by cholesterol oxidation. Cell Death Dis. 2014;5:e1372. doi: 10.1038/cddis.2014.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orrenius S, Zhivotovsky B. Cardiolipin oxidation sets cytochrome c free. Nat Chem Biol. 2005;1(4):188–189. doi: 10.1038/nchembio0905-188. [DOI] [PubMed] [Google Scholar]
- 29.Shchepinov MS, et al. Isotopic reinforcement of essential polyunsaturated fatty acids diminishes nigrostriatal degeneration in a mouse model of Parkinson’s disease. Toxicol Lett. 2011;207(2):97–103. doi: 10.1016/j.toxlet.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 30.Hill S, et al. Small amounts of isotope-reinforced polyunsaturated fatty acids suppress lipid autoxidation. Free Radic Biol Med. 2012;53(4):893–906. doi: 10.1016/j.freeradbiomed.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haeggström JZ, Funk CD. Lipoxygenase and leukotriene pathways: biochemistry, biology, and roles in disease. Chem Rev. 2011;111(10):5866–5898. doi: 10.1021/cr200246d. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Wang H, Rosenberg PA. Vitamin K prevents oxidative cell death by inhibiting activation of 12-lipoxygenase in developing oligodendrocytes. J Neurosci Res. 2009;87(9):1997–2005. doi: 10.1002/jnr.22029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Maher P, Schubert D. A role for 12-lipoxygenase in nerve cell death caused by glutathione depletion. Neuron. 1997;19(2):453–463. doi: 10.1016/s0896-6273(00)80953-8. [DOI] [PubMed] [Google Scholar]
- 34.Maccarrone M, Melino G, Finazzi-Agrò A. Lipoxygenases and their involvement in programmed cell death. Cell Death Differ. 2001;8(8):776–784. doi: 10.1038/sj.cdd.4400908. [DOI] [PubMed] [Google Scholar]
- 35.Park HA, et al. Glutathione disulfide induces neural cell death via a 12-lipoxygenase pathway. Cell Death Differ. 2009;16(8):1167–1179. doi: 10.1038/cdd.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen XS, Funk CD. The N-terminal “beta-barrel” domain of 5-lipoxygenase is essential for nuclear membrane translocation. J Biol Chem. 2001;276(1):811–818. doi: 10.1074/jbc.M008203200. [DOI] [PubMed] [Google Scholar]
- 37.Kulkarni S, Das S, Funk CD, Murray D, Cho W. Molecular basis of the specific subcellular localization of the C2-like domain of 5-lipoxygenase. J Biol Chem. 2002;277(15):13167–13174. doi: 10.1074/jbc.M112393200. [DOI] [PubMed] [Google Scholar]
- 38.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124(6):1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 39.Yang WS, Stockwell BR. Inhibition of casein kinase 1-epsilon induces cancer-cell-selective, PERIOD2-dependent growth arrest. Genome Biol. 2008;9(6):R92. doi: 10.1186/gb-2008-9-6-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brushia RJ, Walsh DA. Phosphorylase kinase: the complexity of its regulation is reflected in the complexity of its structure. Front Biosci. 1999;4:D618–D641. doi: 10.2741/brushia. [DOI] [PubMed] [Google Scholar]
- 41.Elliott LH, et al. K252a is a potent and selective inhibitor of phosphorylase kinase. Biochem Biophys Res Commun. 1990;171(1):148–154. doi: 10.1016/0006-291x(90)91369-4. [DOI] [PubMed] [Google Scholar]
- 42.Rousset M, Chevalier G, Rousset JP, Dussaulx E, Zweibaum A. Presence and cell growth-related variations of glycogen in human colorectal adenocarcinoma cell lines in culture. Cancer Res. 1979;39(2 Pt 1):531–534. [PubMed] [Google Scholar]
- 43.Takahashi S, et al. Estimation of glycogen levels in human colorectal cancer tissue: Relationship with cell cycle and tumor outgrowth. J Gastroenterol. 1999;34(4):474–480. doi: 10.1007/s005350050299. [DOI] [PubMed] [Google Scholar]
- 44.Rousset M, Zweibaum A, Fogh J. Presence of glycogen and growth-related variations in 58 cultured human tumor cell lines of various tissue origins. Cancer Res. 1981;41(3):1165–1170. [PubMed] [Google Scholar]
- 45.Schnier JB, Nishi K, Monks A, Gorin FA, Bradbury EM. Inhibition of glycogen phosphorylase (GP) by CP-91,149 induces growth inhibition correlating with brain GP expression. Biochem Biophys Res Commun. 2003;309(1):126–134. doi: 10.1016/s0006-291x(03)01542-0. [DOI] [PubMed] [Google Scholar]
- 46.Klabunde T, et al. Acyl ureas as human liver glycogen phosphorylase inhibitors for the treatment of type 2 diabetes. J Med Chem. 2005;48(20):6178–6193. doi: 10.1021/jm049034y. [DOI] [PubMed] [Google Scholar]
- 47.Thomas F, et al. Calcein as a fluorescent probe for ferric iron. Application to iron nutrition in plant cells. J Biol Chem. 1999;274(19):13375–13383. doi: 10.1074/jbc.274.19.13375. [DOI] [PubMed] [Google Scholar]
- 48.Jiang L, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520(7545):57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jennis M, et al. An African-specific polymorphism in the TP53 gene impairs p53 tumor suppressor function in a mouse model. Genes Dev. 2016;30(8):918–930. doi: 10.1101/gad.275891.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu XM, et al. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007;5(1):e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zinoni F, Birkmann A, Stadtman TC, Böck A. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc Natl Acad Sci USA. 1986;83(13):4650–4654. doi: 10.1073/pnas.83.13.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arnér ES. Recombinant expression of mammalian selenocysteine-containing thioredoxin reductase and other selenoproteins in Escherichia coli. Methods Enzymol. 2002;347:226–235. doi: 10.1016/s0076-6879(02)47022-x. [DOI] [PubMed] [Google Scholar]
- 53.Arnér ES, Sarioglu H, Lottspeich F, Holmgren A, Böck A. High-level expression in Escherichia coli of selenocysteine-containing rat thioredoxin reductase utilizing gene fusions with engineered bacterial-type SECIS elements and co-expression with the selA, selB and selC genes. J Mol Biol. 1999;292(5):1003–1016. doi: 10.1006/jmbi.1999.3085. [DOI] [PubMed] [Google Scholar]
- 54.Cotticelli MG, Crabbe AM, Wilson RB, Shchepinov MS. Insights into the role of oxidative stress in the pathology of Friedreich ataxia using peroxidation resistant polyunsaturated fatty acids. Redox Biol. 2013;1:398–404. doi: 10.1016/j.redox.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Casañas-Sánchez V, et al. Addition of docosahexaenoic acid, but not arachidonic acid, activates glutathione and thioredoxin antioxidant systems in murine hippocampal HT22 cells: Potential implications in neuroprotection. J Neurochem. 2014;131(4):470–483. doi: 10.1111/jnc.12833. [DOI] [PubMed] [Google Scholar]
- 56.Wang H, et al. 12-Lipoxygenase plays a key role in cell death caused by glutathione depletion and arachidonic acid in rat oligodendrocytes. Eur J Neurosci. 2004;20(8):2049–2058. doi: 10.1111/j.1460-9568.2004.03650.x. [DOI] [PubMed] [Google Scholar]
- 57.Seiler A, et al. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metab. 2008;8(3):237–248. doi: 10.1016/j.cmet.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 58.Hagmann W, Kagawa D, Renaud C, Honn KV. Activity and protein distribution of 12-lipoxygenase in HEL cells: Induction of membrane-association by phorbol ester TPA, modulation of activity by glutathione and 13-HPODE, and Ca(2+)-dependent translocation to membranes. Prostaglandins. 1993;46(6):471–477. doi: 10.1016/0090-6980(93)90066-g. [DOI] [PubMed] [Google Scholar]
- 59.Shornick LP, Holtzman MJ. A cryptic, microsomal-type arachidonate 12-lipoxygenase is tonically inactivated by oxidation-reduction conditions in cultured epithelial cells. J Biol Chem. 1993;268(1):371–376. [PubMed] [Google Scholar]
- 60.Stewart SA, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9(4):493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Larraufie MH, et al. Incorporation of metabolically stable ketones into a small molecule probe to increase potency and water solubility. Bioorg Med Chem Lett. 2015;25(21):4787–4792. doi: 10.1016/j.bmcl.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.