Significance
Whether acquired epigenetic changes can escape the genome-wide epigenetic erasure in the primordial germ cells, which are the embryonic precursors of all types of germline cells and gametes, resulting in transgenerational transfer has been under debate. We have shown that an in vitro cell culture model of mouse primordial germ cells effectively recapitulates the process of germline epigenetic erasure, including DNA demethylation at both physiologically methylated and abnormally hypermethylated imprinting control regions. We also have identified examples of genomic repetitive sequences characterized by significant resistance to the genome-wide DNA demethylation process in mouse primordial germ cells and their cell culture models. Our study paves the way for mechanistic studies of transgenerational epigenetic inheritance using a cell culture model.
Keywords: PGCLC, epigenetic reprogramming, genetic imprinting, epimutation
Abstract
The genome-wide depletion of 5-methylcytosines (5meCs) caused by passive dilution through DNA synthesis without daughter strand methylation and active enzymatic processes resulting in replacement of 5meCs with unmethylated cytosines is a hallmark of primordial germ cells (PGCs). Although recent studies have shown that in vitro differentiation of pluripotent stem cells (PSCs) to PGC-like cells (PGCLCs) mimics the in vivo differentiation of epiblast cells to PGCs, how DNA methylation status of PGCLCs resembles the dynamics of 5meC erasure in embryonic PGCs remains controversial. Here, by differential detection of genome-wide 5meC and 5-hydroxymethylcytosine (5hmeC) distributions by deep sequencing, we show that PGCLCs derived from mouse PSCs recapitulated the process of genome-wide DNA demethylation in embryonic PGCs, including significant demethylation of imprint control regions (ICRs) associated with increased mRNA expression of the corresponding imprinted genes. Although 5hmeCs were also significantly diminished in PGCLCs, they retained greater amounts of 5hmeCs than intragonadal PGCs. The genomes of both PGCLCs and PGCs selectively retained both 5meCs and 5hmeCs at a small number of repeat sequences such as GSAT_MM, of which the significant retention of bisulfite-resistant cytosines was corroborated by reanalysis of previously published whole-genome bisulfite sequencing data for intragonadal PGCs. PSCs harboring abnormal hypermethylation at ICRs of the Dlk1-Gtl2-Dio3 imprinting cluster diminished these 5meCs upon differentiation to PGCLCs, resulting in transcriptional reactivation of the Gtl2 gene. These observations support the usefulness of PGCLCs in studying the germline epigenetic erasure including imprinted genes, epimutations, and erasure-resistant loci, which may be involved in transgenerational epigenetic inheritance.
Evidence is accumulating that parental experiences such as pain, nutritional restrictions, or exposure to toxic chemicals can be transmitted to subsequent generations via epigenetic alterations without mutations in the genomic DNA (gDNA) (1–3). Multigenerational transmission of a nongenetic phenotype is considered transgenerational when it is persistent beyond the epigenetic reprogramming in primordial germ cells (PGCs) (1, 2), potentially conveying illness including metabolic diseases, malignancies, reproductive defects, or behavioral alterations (2, 4, 5). However, this is still a controversial subject due partly to the lack of direct experimental demonstration of transgenerational epigenetic alterations escaping the epigenetic erasure in mammalian PGCs (2, 6, 7).
In early stage mouse embryos, a small cluster of Prdm1-positive PGCs consisting of about 40 cells arise in epiblast at embryonic day 7.25 (E7.25), and PGCs migrate toward the genital ridges while they are rapidly proliferating. By E12.5, about 25,000 PGCs settle in the genital ridges and cease cell division (8). Genome-wide gDNA demethylation is initiated in the migrating PGCs and completed in the intragonadal PGCs, decreasing the global CpG methylation level from 70% in E6.5 epiblast to about 10% in E13.5 PGCs (9). This massive genome-wide gDNA demethylation is critical for “resetting” the sex-specific epigenetic status of imprinted genes, which is important for normal development of fetuses in the subsequent generation, and it is achieved through passive dilution of 5-methylcytosines (5meCs) in the absence of the Dnmt1/Np95-dependent maintenance methylation of the daughter strands during DNA replication as well as multistep enzymatic processes resulting in replacement of 5meCs with unmethylated cytosines, which may involve 5-hydroxymethylcytosines (5hmeCs) as intermediates (9–14). A small fraction of genomic elements such as mouse intracisternal A particles (IAP) was reported to escape this global gDNA demethylation, and their possible roles in the transgenerational epigenetic inheritance have been proposed (2, 9, 15). On the other hand, a recent study detected aberrant 5meC distributions in the spermatogonial gDNA of mice prenatally exposed to endocrine disruptors, but these epimutations were not persistent in the subsequent generation beyond the germline epigenetic reprogramming (6). The fate of epimutations introduced in the reprogramming-resistant genomic elements still remains to be documented.
Recently, it has been shown that pluripotent stem cells (PSCs) such as embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs) can be differentiated into PGC-like cells (PGCLCs) in vitro (16). For example, Hayashi et al. produced PGCLCs from mouse PSCs via the generation of epiblast-like cells (EpiLCs) as intermediates (17, 18). To examine advantages and limitations of mouse PGCLCs as a cell culture model for studies on transgenerational epigenomics, we performed microarray-based transcriptomal profiling and deep-sequencing analyses of genomic 5meC and 5hmeC distributions in PGCLCs and compared these genomic characteristics with those of E12.5 mouse intragonadal PGCs. We show genome-wide dynamics of 5meC and 5hmeC erasure during PSC differentiation to PGCLCs via EpiLCs, demonstrating precise recapitulation of the DNA methylome, including previously known and unknown gDNA elements resistant to the global erasure of 5meCs and 5hmeCs. We also demonstrate that transcription-suppressing abnormal hypermethylation at the imprinting control region (ICR) of the Dlk1-Gtl2-Dio3 imprinting cluster in iPSCs was erased upon differentiation to PGCLCs to regain mRNA expression. These observations support the use of mouse PGCLCs for mechanistic studies of germline epigenetic reprogramming and transgenerational epigenetic inheritance as a valid model of embryonic PGCs.
Results
The SSEA1+/Integrin β3+/c-Kit+ Triple-Positive Mouse PGCLCs Resemble Early Stage PGCs in Marker mRNA Expression.
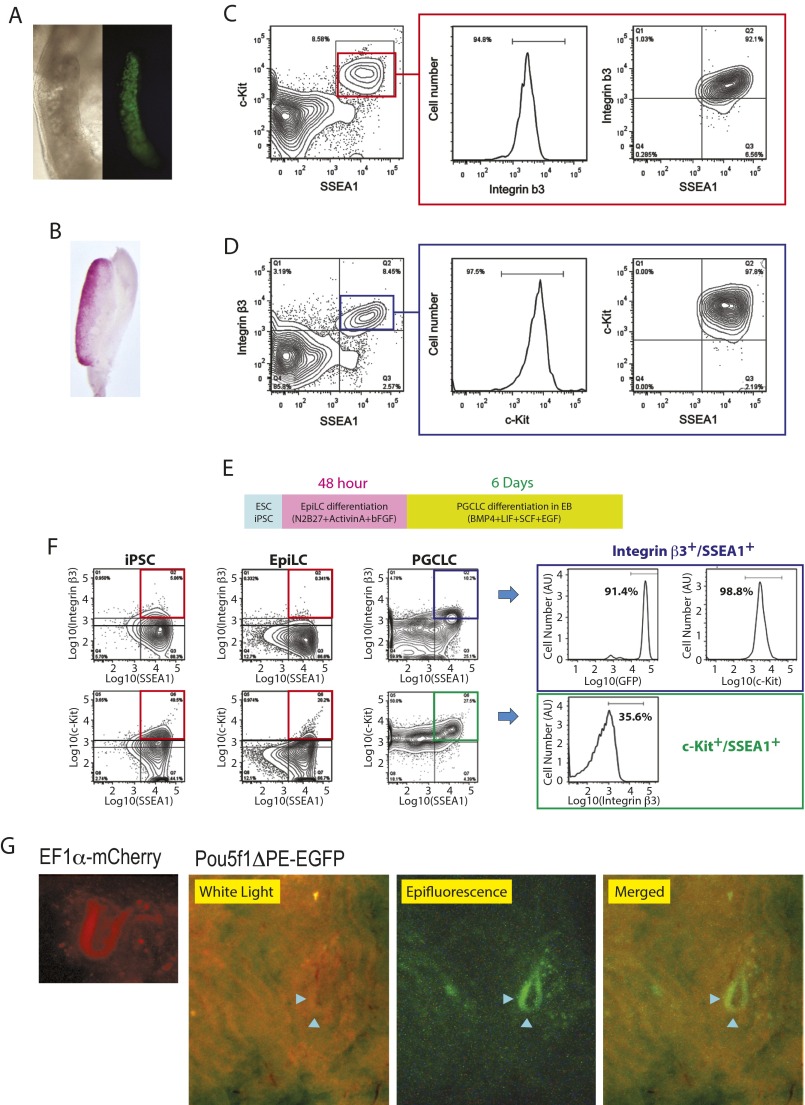
Mouse E12.5 intragonadal PGCs characterized by germline-specific transcriptional activation driven by the Pou5f1 distal enhancer/promoter (Fig. S1A) (19) and alkaline phosphatase activity (Fig. S1B) were examined for their surface-marker protein expression by FACS, which revealed their SSEA1+/Integrin β3+/c-Kit+ triple-positive status (Fig. S1 C and D). Following the protocol described by Hayashi et al. (17), we produced mouse EpiLCs and the day-6 PGCLCs from PSCs (Fig. S1E). More than 98% of PGCLCs enriched by FACS as SSEA1+/Integrin β3+ double-positive cells also strongly expressed c-Kit (Fig. S1F, Top row) whereas only 36% of SSEA1+/c-Kit+ double-positive cells were Integrin β3+-positive (Fig. S1F, Bottom row). In the present study, the SSEA1+/Integrin β3+ double-positive day-6 PGCLCs, which were almost triple-positive including c-Kit, were subjected to further analyses. When transplanted into mouse seminiferous tubules, PGCLCs visualized by EGFP expressed by the Pou5f1 distal enhancer/promoter [which is active in PGCLCs/PGCs (19) and spermatogonial stem cells (20)] or mCherry expressed by the human EF1 α promoter (also active in mouse germline cells) colonized in the lumen of the tubules (Fig. S1G), agreeing with the original report by Hayashi et al. about the capacity of PGCLCs to develop spermatogenic colonies as transplants in the tubules (17).
Fig. S1.
Characterization of PGCs and PGCLCs. (A–D) PGC isolation from E12.5 fetal male mouse gonads. (A) Microscopic images of PGCs expressing EGFP driven by the Pou5f1 promoter/distal enhancer (Left, phase contrast; Right, fluorescence). (B) Alkaline phosphatase staining of intragonadal PGCs. (C and D) FACS profiling of PGCs for cell-surface expression of SSEA1, Integrin β3, and c-Kit. (C) Expression of Integrin β3 in SSEA1+/c-Kit+ PGCs. (D) Expression of c-Kit in SSEA1+/Integrin β3+ PGCs. (E–G) PGCLC production from mouse PSCs. (E) Overall protocol. PSCs were first differentiated to EpiLCs as adherent cell culture for 48 h and then to PGCLCs in EBs for 6 d. (F) FACS profiling of mouse iPSCs, EpiLCs, and PGCLCs for expression of SSEA1, Integrin β3, and c-Kit. iPSCs express EGFP driven by the germline-active Pou5f1 distal enhancer/promoter. Boxes on the Right show expression of EGFP and c-Kit in Integrin β3+/SSEA1+ PGCLCs (Top) and Integrin β3 expression in c-Kit+/SSEA1+ PGCLCs (Bottom). (G) Colonization of PGCLCs in mouse testes. Mouse PGCLCs were labeled with germline-active human EF1α promoter (mCherry) or Pou5f1 distal enhancer/promoter (Pou5f1ΔPE-EGFP). The white light image shows seminiferous tubules, and the epifluorescence image shows an EGFP-positive segment of the tubules (pointed by arrowheads).
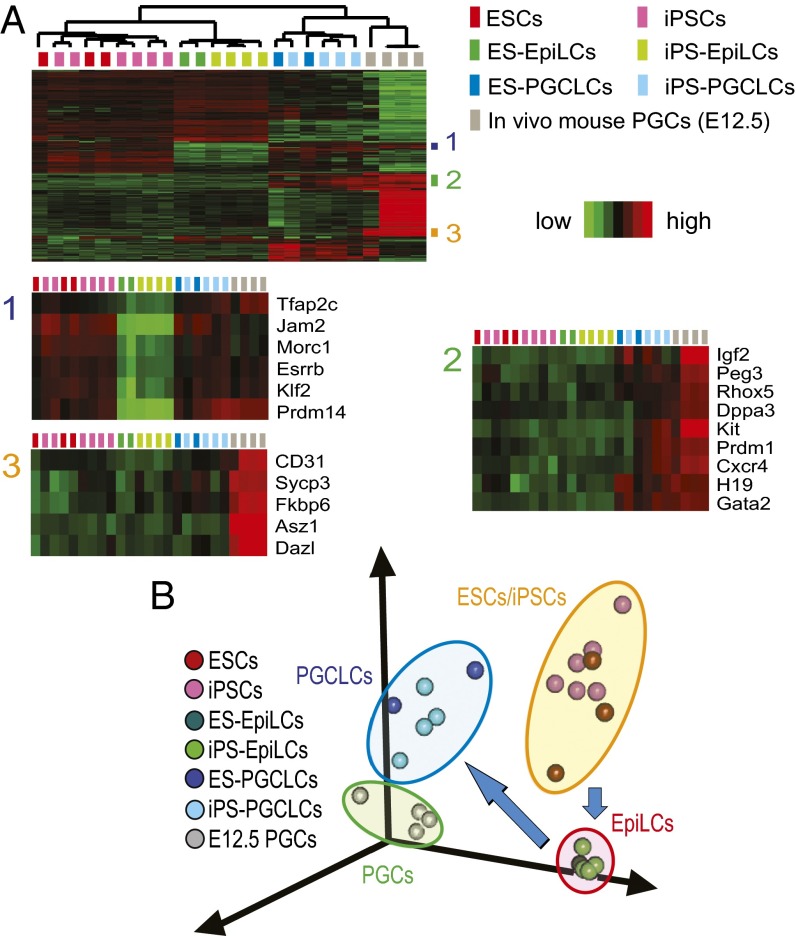
Unsupervised hierarchical clustering (Fig. 1A) and principal component analysis (PCA) (Fig. 1B) clearly separated transcriptomes along cell types—namely, PSCs, EpiLCs, PGCLCs, and intragonadal PGCs. Transcriptomes of PGCLCs were not separated along the types of PSCs from which they were derived (i.e., ESCs or iPSCs). The transcriptomes among the individual PSC clones showed significant heterogeneity and became remarkably homogeneous upon differentiation to EpiLCs, but diversified again among PGCLCs (Fig. 1B), suggesting that differentiation to EpiLC was a nearly deterministic process whereas commitment to PGCLC seemed stochastic. Among the genes induced upon EpiLC differentiation to PGCLCs, those belonging to clusters 1 and 2 in Fig. 1A were enriched with early markers of PGCs. Cluster 2 was also enriched with imprinted genes. Cluster 3 genes were more strongly expressed in intragonadal PGCs than in PGCLCs and enriched with markers of late-stage PGCs.
Fig. 1.
Transcriptomes of mouse PSCs, EpiLCs, PGCLCs, and in vivo PGCs. (A) Hierarchical clustering heatmap of differentially expressed genes. EpiLCs and PGCLCs are indicated with their precursor PSCs (e.g., ES-EpiLCs are EpiLCs derived from ESCs). The three gene clusters indicated in the Top heatmap are enlarged in the Bottom heatmaps. (B) PCA of transcriptomal changes during differentiation of PSCs to PGCLCs via EpiLCs.
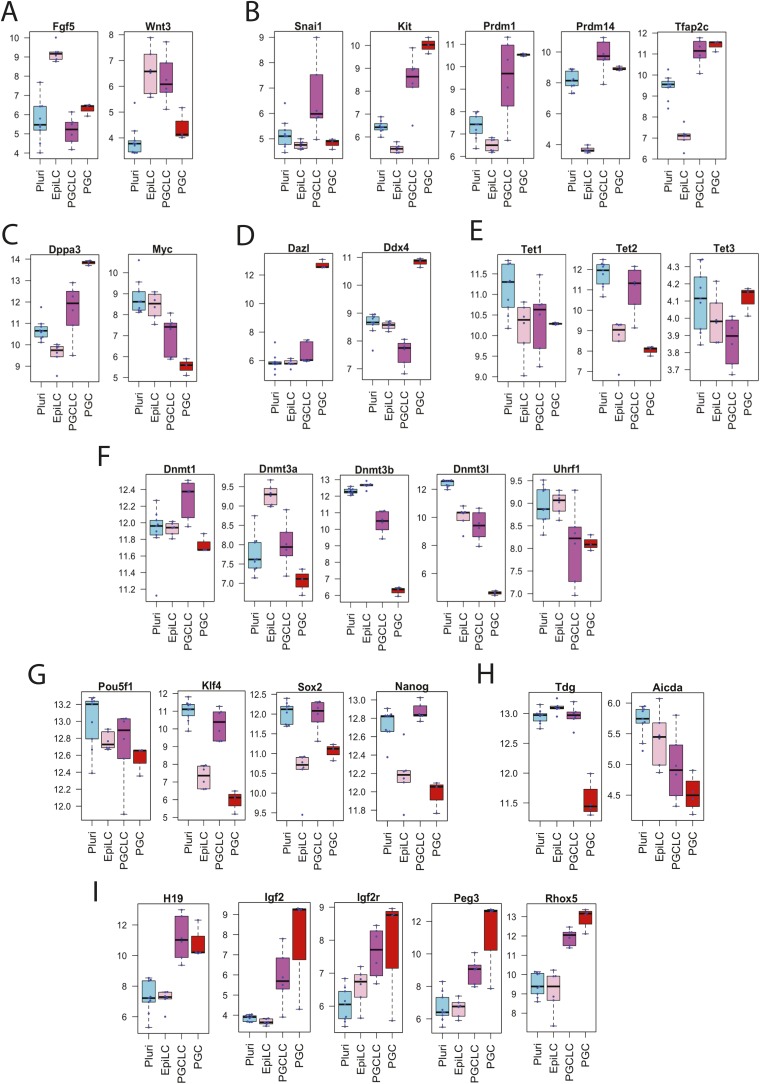
Expression of Fgf5 [an early stage EpiLC maker (17)] was strong in EpiLCs but reduced in PGCLCs whereas expression of Wnt3 [a late-stage EpiLC maker (17)] was maintained in both EpiLCs and PGCLCs (Fig. S2A). PGCLCs strongly expressed mRNA markers of committed and/or migrating PGCs (e.g., Prdm1, Prdm14, c-Kit, and Tfap2c, Fig. S2B). Induction of Dppa3 and suppression of c-Myc, which were reported to occur in PGCLCs after expression of the migrating PGC markers (17), were observed in our PGCLCs (Fig. S2C) whereas intragonadal PGC markers (Dazl or Ddx4, Fig. S2D) were not induced. Agreeing with a previous report that Snail1 was transiently expressed during EpiLC differentiation to PGCLCs but later suppressed when intragonadal PGC markers were induced (17), our PGCLCs expressed Snail1 but intragonadal PGCs did not (Fig. S2B). Our PGCLCs expressed all of the three Tet enzymes (Fig. S2E). Compared with EpiLCs, expression of the Dnmt3a and Dnmt3b de novo DNA methyltransferases as well as the Uhrf1/Np95 cofactor of Dnmt1 was reduced in PGCLCs whereas expression of the Dnmt1 maintenance DNA methyltransferase was maintained (Fig. S2F), agreeing with a previous study (17). Expression of the pluripotency genes Pou5f1, Klf4, Sox2, and Nanog (Fig. S2G) as well as Tdg and Aicda encoding thymine-DNA glycosylase and activation-induced cytidine deaminase, respectively, was stronger in PGCLCs than in intragonadal PGCs (Fig. S2H). Quality control analysis of microarray signal intensities confirmed the absence of significant batch effects that could have affected the above observations (Fig. S3A). Taken together, our transcriptomal profiling suggests that the differentiation status of our PGCLCs was comparable to the PGCLCs described by Hayashi et al. (17), presumably close to the migrating E8.5–E9.5 PGCs.
Fig. S2.
mRNA expression of marker genes in mouse PSCs, EpiLCs, PGCLCs, and E12.5 in vivo PGCs. Box plots represent median, first and third quartile, and 95% confidence intervals of mRNA expression (normalized intensity values) determined using Affymetrix microarray (n > 3). (A) Epiblast markers. (B–D) PGC markers for (B) early, (C) mid, and (D) late stages. (E) Tet enzymes. (F) DNA methyltransferases and the Uhrf1 cofactor of DNMT1. (G) Pluripotency genes. (H) Enzymes involved in active demethylation of DNA. (I) Imprinted genes.
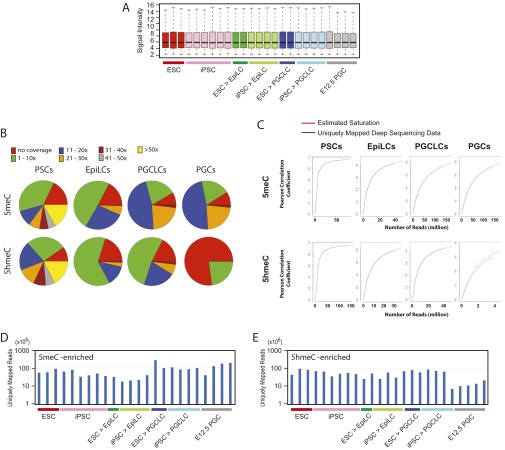
Fig. S3.
Microarray and deep-sequencing quality control data. (A) RMA-normalized signal intensities of Affymetrix microarray data shown in Fig. 1. Box plots are shown with the same color codes as in Fig. 1. The evenly distributed signal intensities across samples do not show signs of batch effects. The degrees of transcriptomal heterogeneity shown in the PCA analysis (Fig. 1B) are not directly explained by different intensities of normalized microarray data. (B) Coverage of CpG sites in the mouse mm9 reference genome sequences by deep-sequencing data for DNA methylation and hydroxymethylation (the seqCoverage function of the MEDIPS Bioconductor package). (C) Saturation analysis of deep-sequencing data (the saturation function of MEDIPS). Sufficient CpG coverage and saturation by deep-sequencing reads ensures appropriate interrogation of DNA methylation and hydroxymethylation. Note that strong CpG coverage and saturation do not immediately result in detection of DNA methylation and hydroxymethylation, which are dependent on formation of peaks over the background. The low CpG coverage or saturation of deep-sequencing reads for DNA hydroxymethylation in PGCs was due to the strong depletion of 5hmerCs in this type of cells. (D and E) Numbers of uniquely mapped deep-sequencing reads obtained for different types of cells for (D) DNA methylation and (E) DNA hydroxymethylation.
Erasure of 5meCs and 5hmeCs in PGCLCs.
To examine the epigenetic status of PGCLCs, we determined distributions of 5meCs and 5hmeCs in the genomes of mouse iPSCs, EpiLCs, PGCLCs, and E12.5 intragonadal PGCs by deep sequencing of gDNA fragments enriched for 5meCs using biotin-conjugated methylcytosine-binding protein 2 [MBD-sEq. (21)], which has no significant affinity to 5hmeCs (22), and gDNA fragments enriched for 5hmeCs by chemical labeling with no reactivity to 5meCs (23). Thus, in contrast to the bisulfite sequencing that cannot distinguish 5meCs and 5hmeCs (24), our approach permitted differential detection of gDNA fragments enriched with these two types of cytosine modifications. Deep-sequencing quality control assessments confirmed sufficient CpG site coverage and saturation in our analyses (Figs. S3 B–E and S4).
Fig. S4.
Distributions of aligned deep-sequencing quality scores along the read positions for (A) DNA methylation and (B) DNA hydroxymethylation (outputs of the FastQC quality control tool). The dark green areas were automatically truncated by the analytical software.
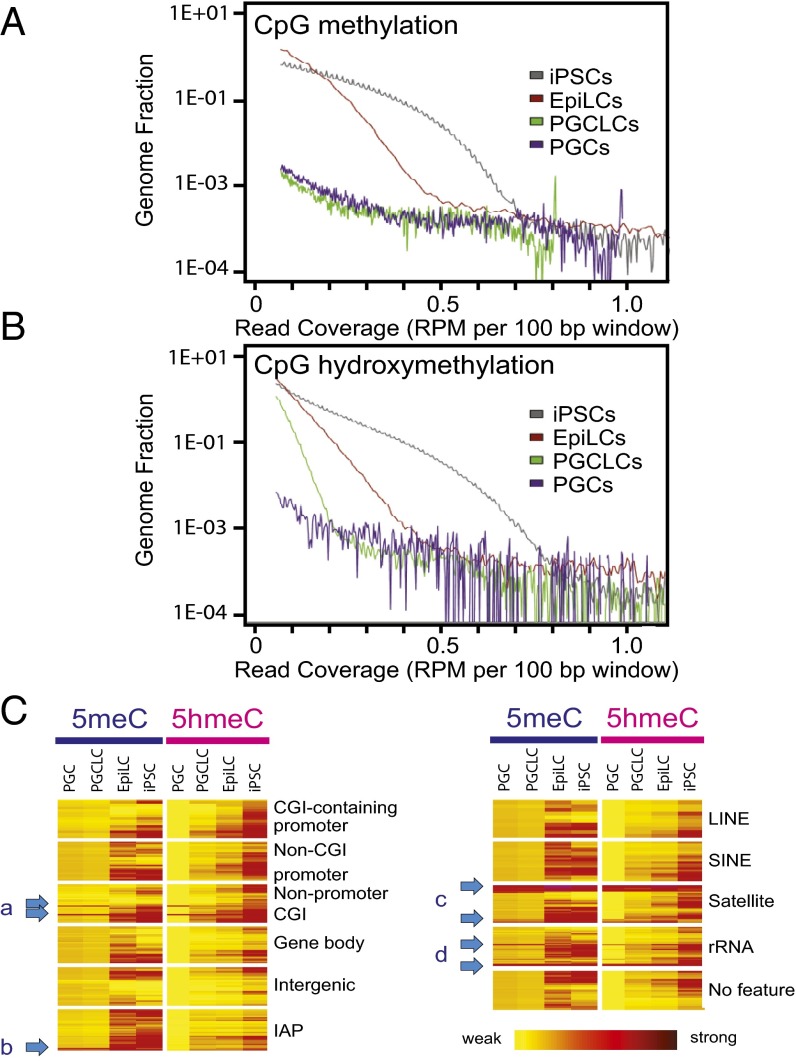
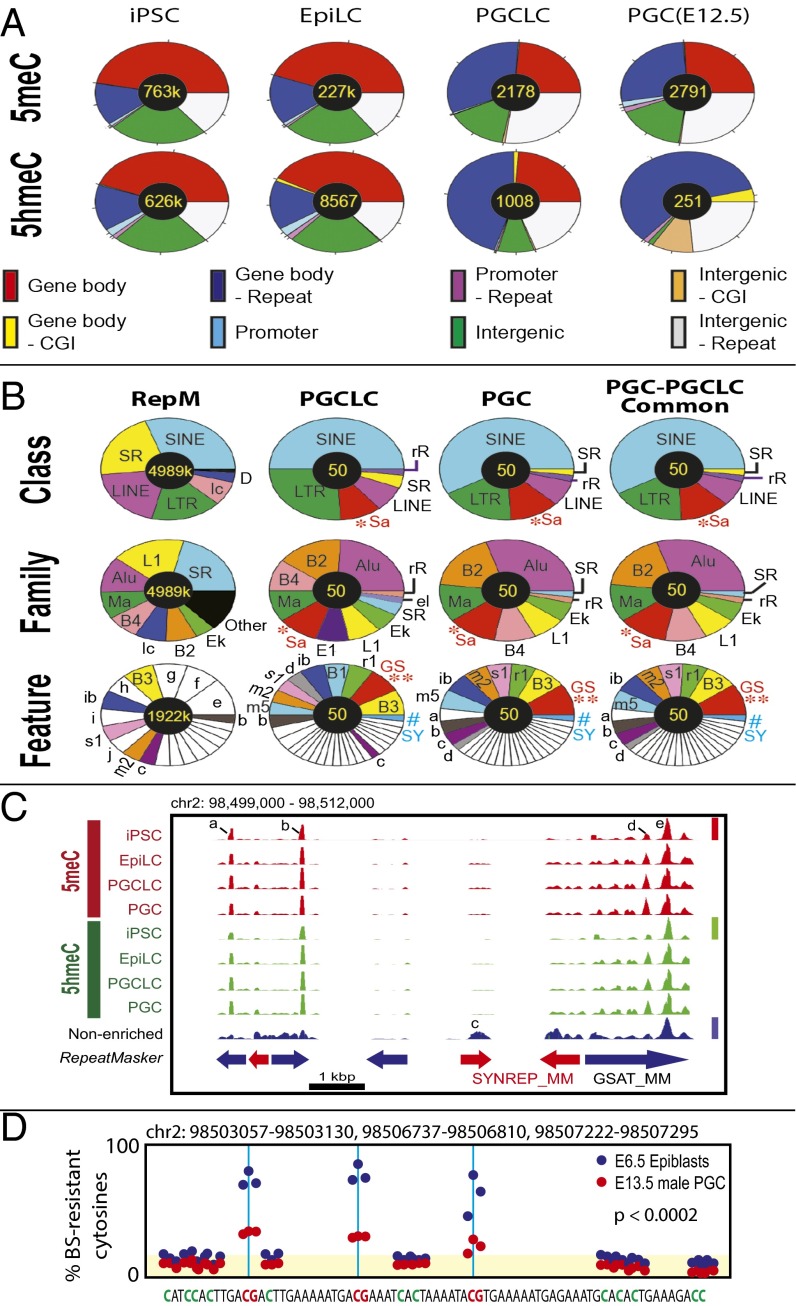
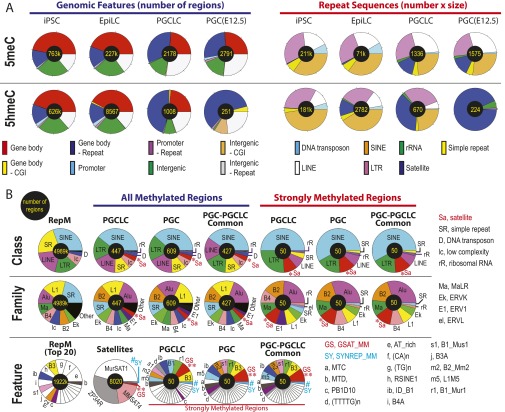
Distribution plot analyses revealed significant reduction in both 5meCs and 5hmeCs during differentiation of iPSCs to EpiLCs. EpiLC differentiation to PGCLCs further reduced 5meCs to a level that appeared comparable to E12.5 PGCs with the sensitivity of our 5meC detection method (Fig. 2A) whereas PGCLCs retained weak but significant amounts of 5hmeC-enriched gDNA segments compared with PGCs (Fig. 2B). Heatmaps of 5meC and 5hmeC distributions across the functional gDNA features revealed that a small fraction of gDNA elements at the nonpromoter CpG islands (Fig. 2 C, a), IAPs (Fig. 2 C, b), satellite repeats (Fig. 2 C, c), and rRNA genes (Fig. 2 C, d) concomitantly retained these epigenetic marks in both PGCLCs and PGCs. The contents of 5meCs detected by MBD-seq were indistinguishable between PGCLCs and PGCs across the gDNA features, whereas the contents of 5hmeCs were more significantly diminished in PGCs compared with PGCLCs (Fig. 2C). Detailed classification of 5meC-enriched gDNA fragments across genomic features revealed their strong enrichment in gene bodies and intergenic regions outside repetitive sequences in iPSCs and EpiLCs, whereas enrichment of these features was remarkably diminished in PGCLCs and PGCs (Fig. 3A and Fig. S5A). The 5meC enrichment profiles of PGCLCs and PGCs show significant similarities in both relative distributions across genomic features and total numbers of the 5meC-enriched regions (2,178 in PGCLCs vs. 2,791 in PGCs), and 91% of the 5meC-enriched regions detected in PGCLCs were also found in PGCs (Fig. S6). The 5hmeC enrichment profiles of iPSCs and EpiLCs were similar to 5meCs except that only 251 5hmeC-enriched regions (assigned mostly to repetitive elements) were found in PGCs (Fig. 3A and Fig. S5A). The majority of the repeat-containing, 5meC-enriched gDNA regions in PGCLCs and PGCs were found within the interspersed repeat classes such as SINEs, LINEs (short- and long-interspersed nuclear elements), or LTRs (which include the IAPs), approximately reflecting the genome-wide RepeatMasker registration profile of the mouse NCBI37/mm9 reference genome sequence (Fig. 3B and Fig. S5B). Interestingly, the satellite repeats (shown as *Sa) were overrepresented in all 5meC-enriched regions, and their proportion was increased further in the 50 regions with the highest relative methylation scores. Among the satellite sequences, the closely related GSAT_MM (shown as **GS) and SYNREP_MM (#SY) repeats were overrepresented.
Fig. 2.
Global reduction in gDNA 5meCs and 5hmeCs during mouse iPSC differentiation to PGCLCs. (A and B) Density distributions of (A) 5meCs and (B) 5hmeCs. The x axes represent densities of 5meCs or 5hmeCs in 100-bp windows, and the y axes indicate genome-wide frequencies. (C) Heatmaps of 5meC and 5hmeC densities across genomic features. Arrows a-d point to elements retaining 5meCs and/or 5hmeCs in PGCLCs/PGCs.
Fig. 3.
Genomic feature distributions of 5meCs and 5hmeCs in the genomic DNA of mouse iPSCs, EpiLCs, PGCLCs, and in vivo PGCs. (A) 5meC and 5hmeC distributions across genomic features. (B) Distributions of 5meCs across repeat sequences. RepM, genome-wide RepeatMasker-registered elements. Small elements (<5%) are left blank in pie charts. *Sa, satellite repeats; **GS, GSAT_MM; #SY, SYNREP_MM. Other keys of pie charts are defined in Fig. S5. (C) An example of deep-sequencing tracks showing 5meC and 5hmeC peaks at GSAT_MM and SYNREP_MM satellite repeats. Height of peaks reflects relative strength of DNA methylation across the four 5meC tracks (linearly scaled 0–1 between the baseline and the maximal methylation, red bar), DNA hydroxymethylation (four 5hmeC tracks, green bar), or nonenriched genome resequencing (blue bar); note that scaled value 1 is not equal to 100% methylation. Peaks a, b, and d are “informative” based on their enrichment over the nonenriched mouse genome resequencing track or changes between different types of cells. Peaks c and e are present in the nonenriched track and so are uninformative. (D) Reanalysis of the whole-genome bisulfite sequencing data generated by Seisenberger et al. (9) for a 74-nt sequence repeated three times in the GSAT_MM regions shown in C. Blue and red dots show percentage of bisulfite-resistant cytosines in E6.5 epiblasts and E13.5 PGCs, respectively. Yellow shade indicates the background levels of bisulfite-resistant cytosines in CpA, CpT, and CpC dinucleotides. The P values represent statistical significance between the CpG-context bisulfite-resistant cytosines over the background (t test).
Fig. S5.
Distributions of 5meCs and 5hmeCs across genomic features in mouse PSCs, PGCLCs, and in vivo PGCs. Numbers of total features are shown at the center of each pie chart. (A) 5meC and 5hmeC distributions in all genomic features (Left; reflecting numbers of regions only) and in repeat sequences (Right; reflecting products of numbers and sizes of regions). (B) Detailed distributions of 5meC in repeat sequences based on numbers of regions. RepM, profiles of the entire RpeatMasker elements. RepeatMasker features are integrated into families, which are further integrated into classes. Some of the small RepeatMasker features (<5%) are shown as blanks without labels.
Fig. S6.
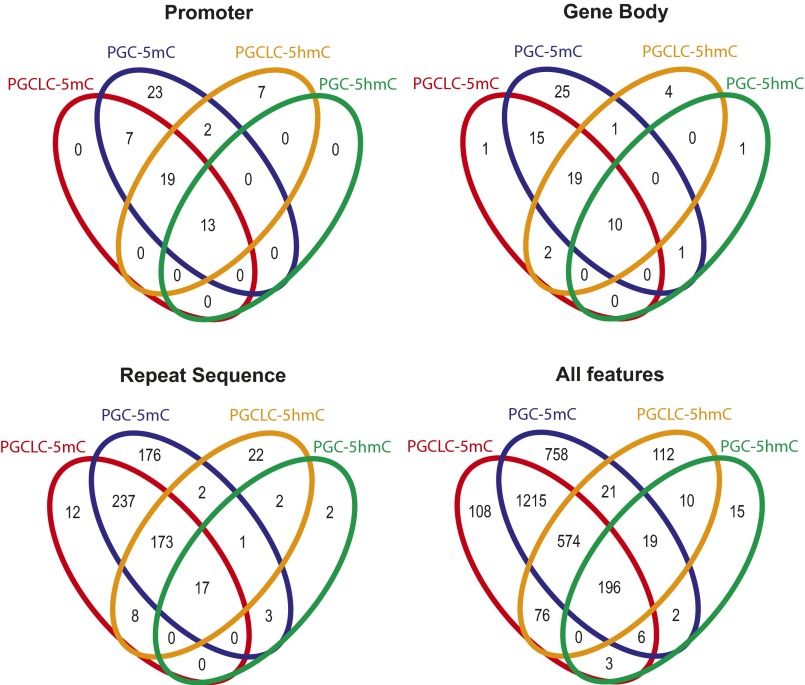
Venn diagrams representing overlapping locations of 5meCs and 5hmeCs between mouse PGCLCs and in vivo PGCs at distinct genomic features.
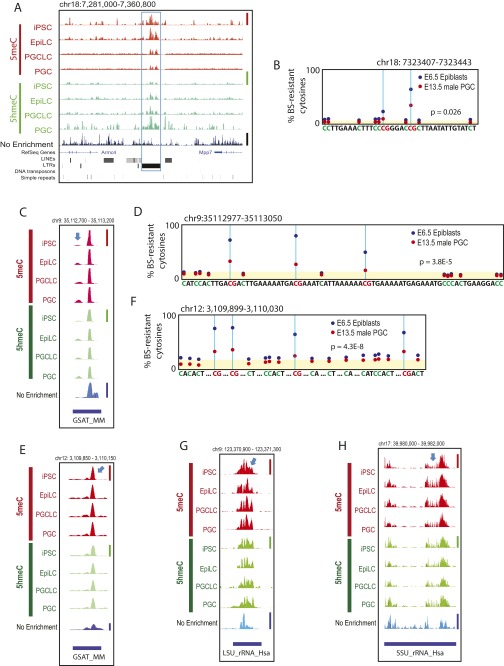
To obtain further evidence of 5meC retention at the repetitive elements, we performed visual inspections of deep-sequencing data generated in our present study, as well as the whole-genome bisulfite sequencing (WGBS) data of mouse E6.5 epiblasts and E13.5 male PGCs published by Seisenberger et al. (9). Fig. S7A shows an example of deep-sequencing tracks demonstrating significant retention of both 5meCs and 5hmeCs at a region containing IAPs in PGCLCs and PGCs. Fig. S7B shows the WGBS data corresponding to a part of the IAP-related 5meC/5hmeC-enriched region indicated in Fig. S7A, demonstrating significant retention of bisulfite-resistant cytosines (i.e., the sum of 5meCs and 5hmeCs) at two CpG sites in the gDNA of E13.5 male PGCs. Fig. 3 C and D shows similar analyses for a region rich in GSAT_MM and SYNREP_MM repeats. Although some 5meC/5hmeC peaks in the deep-sequencing tracks were not informative, as they were also evident in the nonenriched mouse genome resequencing track (peak e), several informative peaks (a, b, d) supported the presence of 5meC- and 5hmeC-enriched gDNA regions within GSAT_MM repeats (Fig. 3C). Inspection of the WGBS data for GSAT_MM repeats in the corresponding region identified three instances of an identical 74-nt sequence containing three CpG sites with significant retention of bisulfite-resistant cytosines in the gDNA of E13.5 male PGCs (Fig. 3D). On the other hand, the apparent lack of 5meC/5hmeC peaks at SYNREP_MM in Fig. 3C (peak c on the nonenriched track) left the 5meC/5hmeC retention in this element unconfirmed, possibly due to technical issues stemming from its up to 75% nucleotide base identity to GSAT_MM. Two additional examples of GSAT_MM retention of 5meC/5hmeC peaks and bisulfite-resistant cytosines are shown in Fig. S7 C–F. Agreeing with the retention of 5meCs and 5hmeCs at the ribosomal RNA gene shown in Fig. 2C (arrow d), visual inspection of deep-sequencing tracks at regions containing LSU_rRNA_Hsa and SSU_rRNA_Hsa ribosomal RNA genes revealed the presence of informative peaks (Fig. S7 G and H) although insufficient bisulfite conversion of the WGBS data for these regions precluded nucleotide base-resolution analysis. Interestingly, we observed a strong tendency for 5hmeC peaks to be closely associated with 5meC peaks (Fig. 3C and Fig. S7 A, C, E, G, and H) although the enrichment-based deep-sequencing approach did not provide relative amounts of 5hmeCs to 5meCs.
Fig. S7.
Examples of 5meC and 5hmeC retention in PGCLCs and PGCs. A, C, E, G, and H show deep-sequencing tracks at (A) an IAP, (C and E) GSAT_MM, (G) LSU_rRNA_Hsa, and (H) SSU_rRNA_Hsa. Informative peaks are indicated by blue arrows. Height of peaks reflects relative strength of DNA methylation across the four 5meC tracks (linearly scaled 0–1 between the baseline and the maximal methylation in the displayed area indicated by a red vertical bar at the right), DNA hydroxymethylation (four 5hmeC tracks, green vertical bar), or nonenriched genome resequencing coverage (nonenriched track, blue vertical bar); note that scaled value 1 is not equal to 100% methylation. (B, D, and F) Reanalysis of the whole-genome bisulfite sequencing data generated by Seisenberger et al. (9) for E6.5 mouse epiblasts and E13.5 male PGCs corresponding to sequencres shown in A, C, and E, respectively. Yellow shading indicates the background levels of bisulfite-resistant cytosines in CpA, CpT, and CpC dinucleotides. The P values represent statistical significance between the CpG-context bisulfite-resistant cytosines over the background (t test).
DNA Demethylation at the ICRs in PGCLCs.
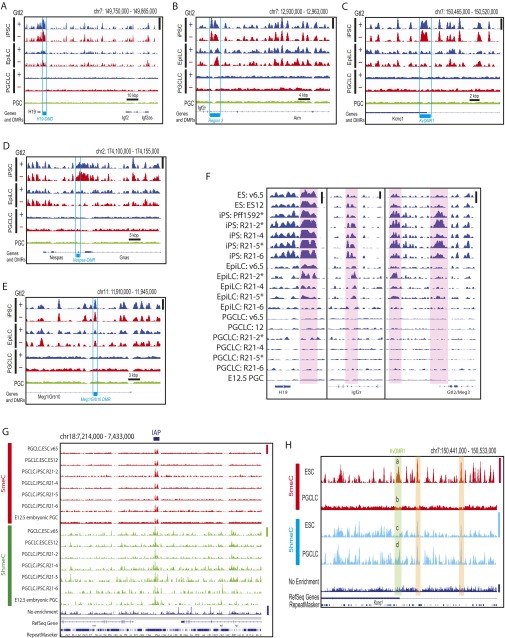
Demethylation of the ICRs is a hallmark of intragonadal PGCs (9, 12). Hayashi et al. reported highly limited ICR demethylation in their PGCLCs, the epigenetic status of which was hence presumed by the authors to be similar to E8.5–E9.5 migrating PGCs before initiation of the imprinting erasure (17, 18). In contrast, Zhou et al. recently reported more advanced ICR demethylation in PGCLCs, placing their epigenetic status close to E12.5 intragonadal PGCs (25). For all of the six ICRs examined, our deep-sequencing analysis showed progressive loss of 5meCs upon iPSC differentiation to EpiLCs and then to PGCLCs (Fig. 4A and Fig. S8 A–E). Expression of the mRNA transcripts for the corresponding imprinted genes increased in PGCLCs compared with PSCs or EpiLCs but still more weakly than in E12.5 intragonadal PGCs, suggesting that the epigenetic status of PGCLCs produced in our present study may be between E9.5 and E12.5 PGCs (Fig. S2I). Significant and progressive ICR demethylation was observed in all individual PSC-EpiLC-PGCLC differentiation experiments with no apparent differences among the PSC precursor clones (Fig. S8F). On the other hand, at the location of an IAP shown in Fig. S7A, 5meCs and 5hmeCs were retained in the genomes of all types of PGCLCs as well as E12.5 embryonic PGCs (Fig. S8G). Note that no 5meC or 5hmeC peak was detected in the genomes of PGCLCs or PGCs around the ICRs shown in Fig. S8 A–F due to the absence of IAP, GSAT_MM, LSU_rRNA_Hsa, or SSU_rRNA_Hsa repeat sequences. Interestingly, the ICR demethylation observed upon differentiation of PSCs to PGCLCs was often accompanied by increased DNA hydroxymethylation at the same region, whereas DNA hydroxymethylation outside the ICRs was typically diminished or unchanged upon PSC differentiation to PGCLC (Fig. S8H).
Fig. 4.
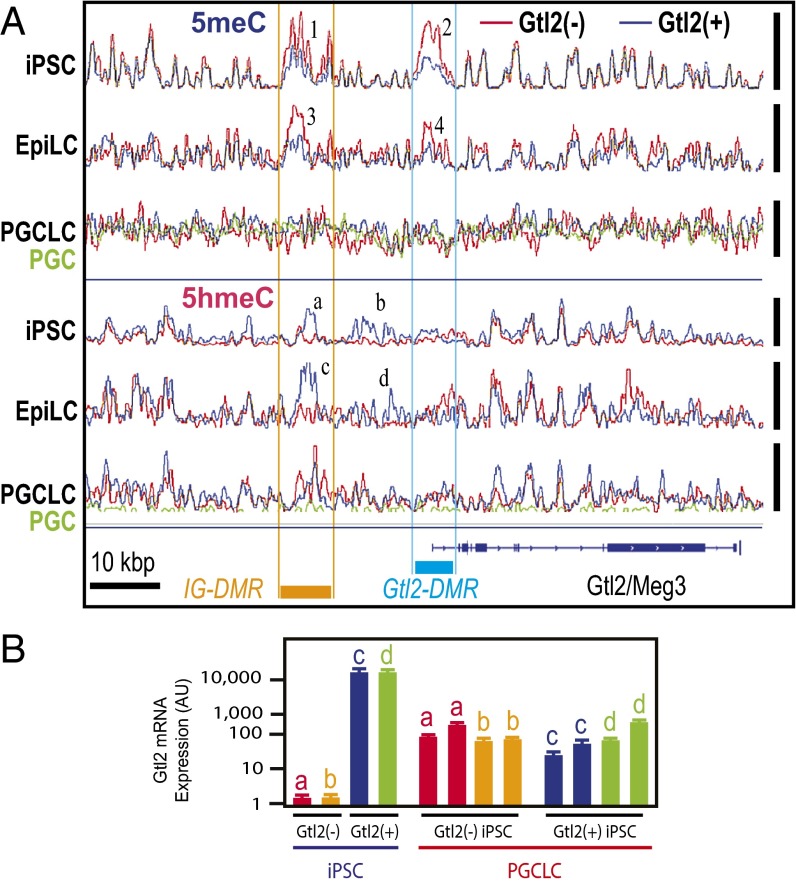
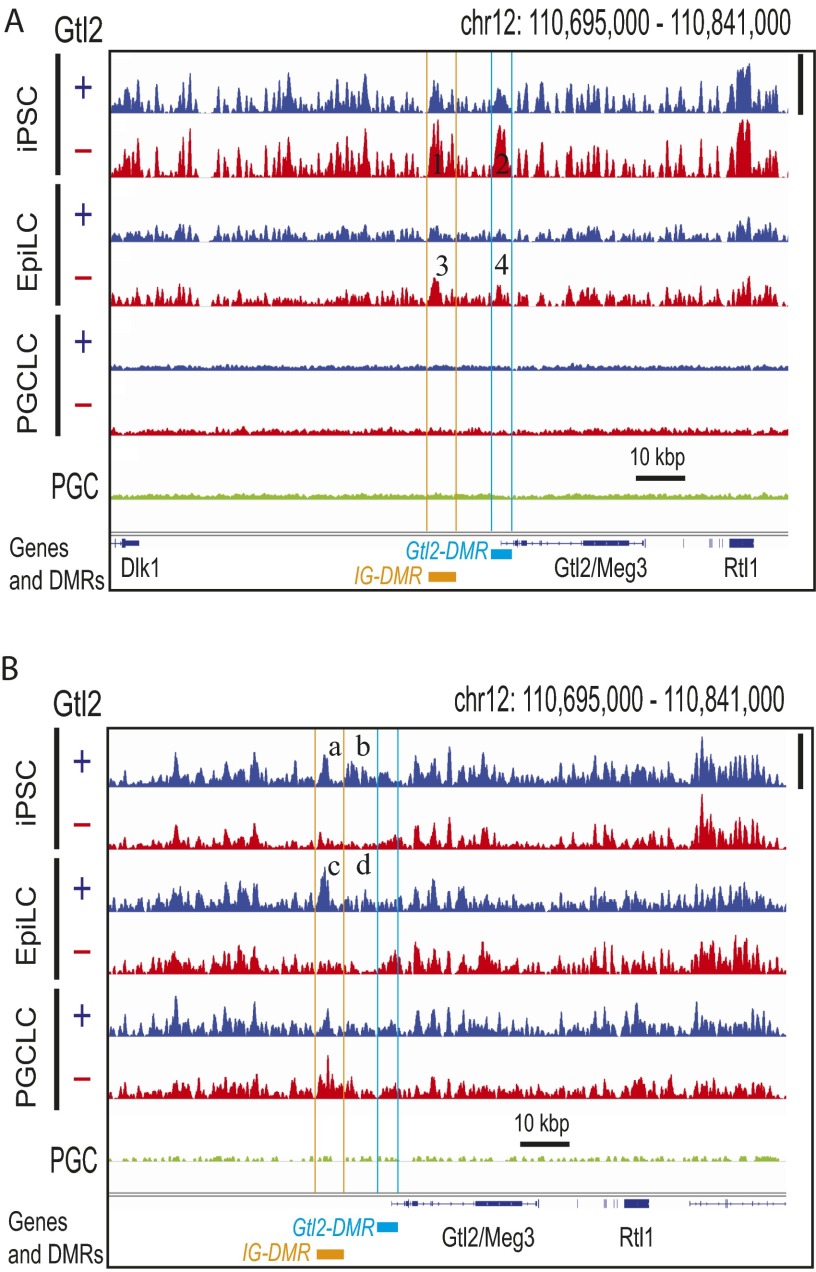
Erasure of DNA hypermethylation at the IG-DMR and Gtl2-DMR of Gtl2(−) iPSCs during differentiation to PGCLCs. (A) Superimposed deep-sequencing tracks of 5meCs (Top three tracks) and 5hmeCs (Bottom three tracks). Blue, red, and green traces represent Gtl2(+), Gtl2(−), and in vivo PGC, respectively, and all traces in each track are adjusted in a track-specific linear scale between the minimal and maximal methylation or hydroxymethylation in the displayed area shown with vertical bars at the right. The same data are displayed with fixed scales across tracks in Fig. S9. Orange and cyan bars indicate locations of IG-DMR and Gtl2-DMR, respectively. Numbers 1–4 show differential methylation between Gtl2(+) and Gtl2(−) iPSCs and EpiLCs at the DMRs. (a–d) Differential hydroxymethylation. (B) Expression of Gtl2 mRNA in independent clones of mouse Gtl2(−) iPSCs (a and b), Gtl2(+) iPSCs (c and d), and PGCLCs produced from them. Bars indicate qPCR data for Gtl2 mRNA expression normalized with Gapdh mRNA expression (n = 3, mean ± SEM).
Fig. S8.
Demethylation at the ICRs in PGCLCs. Deep-sequencing tracks of 5meCs at the ICRs in the genomes of Gtl2(+) and Gtl2(−) mouse iPS cells and their EpiLC and PGCLC derivatives are shown. (A) Igf2-H19, (B) Igf2r, (C) Kcnq1, (D) Gnas, and (E) Meg1/Grb10. (F) 5meC deep-sequencing tracks of the individual cell clones at the H19, Igf2r, and Gtl2/Meg3 ICRs (shaded in red). Asterisks indicate the Gtl2(×) iPSC clones, which show abnormal hypermethylation of the Gtl2/Meg3 ICRs, and their derivatives. Height of peaks in A–F reflects relative strength of DNA methylation across all of the tracks in each panel (or each subpanel of F), linearly scaled between the baseline and the maximal methylation in the displayed area shown with vertical bars at right. (G) Retention of DNA methylation and hydroxymethylation at an IAP location in PGCLCs and PGCs. Height of peaks reflects relative strength of DNA methylation across the seven 5meC tracks (linearly scaled between the baseline and the maximal methylation in the displayed area indicated with red vertical bar at right), seven DNA hydroxymethylation (5hmeC tracks, green vertical bar), or nonenriched genome resequencing coverage (Nonenriched track, blue vertical bar). IAP location is shown by horizontal bar at the top. (H) DNA methylation and hydroxymethylation profiles of the v6.5 mouse ESCs and their PGCLC derivatives at the ICR of the Kcnq1 imprinting cluster. Height of peaks reflects relative strength of DNA methylation across the two 5meC tracks (linearly scaled between the baseline and the maximal methylation in the displayed area indicated with red vertical bar at right), two 5hmeC tracks (cyan bar), or nonenriched genome resequencing track (blue bar). The ICR (KvDMR1) is shown with green shading, in which strong DNA methylation in the ESCs (a) was lost in PGCLCs (b) whereas DNA hydroxymethylation was enhanced upon ESC differentiation to PGCLC (compare peaks c and d). Two regions shaded with orange show loss of DNA methylation upon ESC differentiation to PGCLC without significant changes in DNA hydroxymethylation.
Erasure of Region-Specific Epimutations During iPSC Differentiation to PGCLCs.
We previously showed that generation of iPSCs by somatic cell reprogramming in the absence of sufficient vitamin C caused silencing of the Dlk1-Gtl2-Dio3 imprinting cluster, resulting in diminished pluripotency (26, 27). This silencing was associated with aberrant DNA hypermethylation of maternal IG-DMR (differentially methylated region) and Gtl2-DMR (26, 27). Taking advantage of this epimutation that is experimentally inducible in iPSCs, we examined whether aberrant, region-specific hypermethylation can be erased during iPSC differentiation to PGCLC. Reproducing our previously published bisulfite-pyrosequencing analysis (26), MBD-seq detected aberrant DNA hypermethylation at the IG-DMR and the Gtl2-DMR in mouse iPSCs (Fig. 4A and Fig. S9A). The accuracy of our 5meC profiling is supported by the nearly identical MBD-seq tracks of normal [Gtl2(+)] and silenced [Gtl2(−)] iPSCs except for the IG- and Gtl2-DMRs. Whereas these aberrant 5meC peaks were still observed in EpiLCs, they were not detected in PGCLCs. Concomitantly, Gtl2 mRNA expression, which was suppressed in Gtl2(−) iPSCs, was restored in PGCLCs to a level comparable to PGCLCs derived from Gtl2(+) iPSCs (Fig. 4B). Interestingly, the IG-DMR and the region between the IG- and the Gtl2-DMRs of Gtl2(−) iPSCs showed aberrant reduction in 5hmeC peaks (Fig. 4A and Fig. S9B), which were erased during iPSC differentiation to PGCLC. Thus, the aberrant DNA hypermethylation at the ICRs of the Dlk1-Gtl2-Dio3 imprinting cluster in iPSCs was erased upon differentiation to PGCLCs.
Fig. S9.
Erasure of DNA hypermethylation at the IG-DMR and Gtl2-DMR of Gtl2(−) iPSCs during differentiation to PGCLCs. Deep-sequencing tracks of 5meCs (A) and 5hmeCs (B) in the genomes of Gtl2(+) and Gtl2(−) mouse iPS cells and their EpiLC and PGCLC derivatives are shown. Height of peaks reflects relative strength of DNA methylation (A) or hydroxymethylation (B) across all seven tracks in each panel, linearly scaled between the baseline and the maximal methylation in the displayed area shown with vertical bars at right. Orange and cyan lines indicate locations of IG-DMR and Gtl2-DMR, respectively. Numbers 1–4 show differential methylation between Gtl2(+) and Gtl2(−) iPSCs and EpiLCs at the DMRs. (a–d) Differential hydroxymethylation. Note that regions b and d locate between the two DMRs.
Discussion
Transcriptomal and Epigenomic Characteristics of Mouse PGCLCs.
Following the protocol described by Hayashi et al. (17) with slight modifications, we generated SSEA1+/Integrin β3+/c-Kit+ triple-positive PGCLCs from mouse PSCs (Fig. S1). Transcriptomal profiling (Fig. 1 and Fig. S2) placed our PGCLCs isolated from 6-d culture embryoid bodies (EBs) in a status similar to the PGCLCs that Hayashi et al. obtained from EBs earlier than the 6-d culture but later than the 2-d culture (17). In a recent study, Zhou et al. generated mouse PGCLCs from 6-d culture EBs using a similar protocol (25) and observed a marker gene expression profile similar to the 6-d EB PGCLCs of Hayashi et al. (17). On the other hand, whereas Hayashi et al. observed only limited DNA demethylation at ICRs of the Igf2r, Snrpn, H19, and Kcnq1 imprinting clusters and so placed their PGCLS at a stage corresponding to E8.5–E9.5 migrating PGCs in mouse embryos [when the ICR demethylation in PGCs is not yet significant (9, 17, 18)], Zhou et al. reported more advanced ICR demethylation at the Snrpn and H19 imprinting clusters, placing their PGCLCs at a stage similar to E12.5 intragonadal mouse embryonic PGCs (25). In our present study, PGCLCs showed significant demethylation at all six ICRs examined (Dlk1-Meg3/Gtl2-Dio3, H19, Igf2r, Kcnq1, Nespas-Gnas, Meg1/Grb10) (Fig. 4 and Fig. S8) as well as global loss of 5meCs (Figs. 2 A and C and 3A). The progressive increase in mRNA expression of imprinted genes during PSC differentiation to PGCLC via EpiLC (Fig. S2I) may reflect release from monoallelic suppression by DNA methylation. The restoration of Gtl2 mRNA expression in PGCLCs derived from Gtl2(−) iPSCs from silencing due to the aberrant hypermethylation of the ICR of the Dlk1-Gtl2-Dio3 imprinting cluster [Fig. 4 and Fig. S9 (26)] further supports ICR demethylation in our PGCLCs. Although our results suggest the usefulness of mouse PGCLCs for mechanistic studies of the germline DNA demethylation including ICRs, it remains to be determined whether this in vitro model accurately represents a particular physiological status of embryonic PGCs. To achieve this goal, future studies should consider sensitivity, quantitativity, and specificity of the analytical methods. For example, PCR-based bisulfite sequencing may be insufficient for quantitative evaluation of ICR methylation (17, 18, 25). Specificity of bisulfite conversion (9, 17, 18, 25) is incomplete because it does not distinguish 5meCs from 5hmeCs. Whereas MBD-seq distinguishes 5meCs from 5hmeCs, in our present study this method did not robustly detect low levels of DNA methylation at the IG-DMR, Gtl2-DMR, or the Rtl1 in E12.5 PGCs, which was detected by Singh et al. using the Methylated CpG Island Recovery Assay (MIRA) and a custom-design microarray that targeted imprinted genes and IAP flanking regions (28). In contrast to MBD-seq using the 5meC-binding domain of human MBD2 for enrichment, MIRA uses heterodimers of MBD2b and MBD3L1, which has a significantly stronger affinity to 5meCs than MBD2 (29). Thus, the absence of 5meC in our study should be interpreted that DNA methylation was diminished to a level below the detection limit rather than complete depletion of 5meCs. Although the 5meC profiles of PGCLCs observed in the present study were indistinguishable from the profile of E12.5 embryonic PGCs, it remains to be determined whether weak DNA methylation in PGCLCs could be similar to earlier stage of PGCs.
Erasure of DNA Methylation in PGCLCs and PGCs.
The DNA methylomes of PSCs, EpiLCs, PGCLCs, and E12.5 PGCs using MBD-seq (Fig. 2) largely agreed with the gDNA demethylation dynamics in mouse embryonic germline cells determined by Seisenberger et al. using WGBS (9), reproducing significant retention of 5meCs at IAPs or nonpromoter CpG islands (CGIs) in PGCLCs and PGCs (Fig. 2A and Fig. S7A). MBD-seq also detected germline retention of 5meCs at repeat sequences GSAT_MM, LSU_rRNA_Hsa, and SSU_rRNA_Hsa (Fig. 3 A–C and Figs. S5 and S7 C, E, G, and H). Reanalysis of the WGBS data of Seisenberger et al. validated germline retention of 5meCs at GSAT_MMs (Fig. 3D and Fig. S7 D and F) as well as IAPs (Fig. S7B) although 5meC retention at other repeat elements was not validated due to insufficient bisulfite conversion of the WGBS data.
The importance of 5hmeCs in the active DNA demethylation and imprinting erasure in germline cells has been well recognized (12–14, 30, 31). In our present study, the abundant 5hmeCs in mouse iPSCs were dramatically lost during differentiation to PGCLCs via EpiLCs (Figs. 2 B and C and 3A). The 5meC content in PGCLCs and E12.5 intragonadal PGCs detected with the sensitivity of MBD-seq was largely comparable. However, PGCLCs retained about a four times greater number but relatively weak 5hmeC-enriched gDNA segments compared to PGCs (Figs. 2 and 3A and Fig. S5). Interestingly, 5meC-enriched gDNA fragments detected in the genomes of PGCs and PGCLSs were often coenriched with 5hmeCs (Fig. 3C and Fig. S7 A, C, E, G, and H). Genomic DNA regions strongly enriched with 5meCs in PSCs were typically enriched with 5hmeCs as well, and these 5hmeCs were often retained after differentiation to PGCLCs even when 5meCs were erased (Fig. S8H, orange shading). However, ICR of the Kncq1 imprinting cluster (KvDMR1) was strongly methylated in ESCs without coenrichment of 5hmeCs (Fig. S8H, a and c) whereas its 5hmeC content was augmented in PGCLCs and 5meCs were lost (Fig. S8H, b and d). In contrast, in iPSCs and EpiLCs, the normal ICRs of the Dlk1-Gtl2-Dio3 imprinting cluster (IG-DMR and Gtl2-DMR) were significantly enriched with 5hmeCs whereas aberrantly hypermethylated ICRs were deficient in 5hmeCs (Fig. 4A and Fig. S9). Taken together, these observations suggest that gDNA regions coenriched with 5meCs and 5hmeCs may be prone to demethylation, including 5meC-retaining regions in PGCLCs/PGCs.
Germline Epigenetic Erasure as a Barrier to Nongenetic Transgenerational Inheritance.
It has been proposed that a small fraction of genomic elements that escape the epigenetic erasure (such as IAPs or nonpromoter CGIs) may serve as vehicles of the transgenerational epigenetic inheritance (2, 9). However, a systematic examination recently reported by Iqbal et al. showed that transcriptional and DNA methylome aberrations introduced in spermatogonia of fetuses by in utero exposure to endocrine-disrupting chemicals were not persistent beyond the germline epigenetic erasure in a statistically significant manner even when the analysis was extended to IAPs (6). This negative but insightful observation may suggest the ability of PGCs to effectively repair epimutations or perhaps reflect technical challenges of identifying transgenerational epimutations that might occur stochastically within repetitive sequences. Taking advantage of the experimentally reproducible DNA hypermethylation at the otherwise demethylated maternal IG-DMR and Gtl2-DMR of the Dlk1-Gtl2-Dio3 imprinting cluster in mouse iPSCs (26, 27), our present study directly demonstrates significant reduction in this abnormal hypermethylation during iPSC differentiation to PGCLC (Fig. 4A and Fig. S9), which resulted in functional restoration of the Gtl2 imprinted mRNA expression (Fig. 4B). The ability of the PGCLC cell culture model to erase experimentally introduced epimutations will provide unique future opportunities to examine erasure, and possible retention, of various types of epimutations at specific gDNA locations during germline differentiation. It remains to be determined whether this PGCLC model can also be used to examine erasure of epimutations introduced outside ICRs and/or within repetitive elements, and the resolution power of this approach should be improved at the nucleotide base level because experience-induced changes in gametic gDNA methylation were reported to be specific to CpG sites, thus critically affecting gDNA binding to transcription factors (32, 33). It is also an interesting question as to whether or not apparently physiological epigenetic changes resulting from specific and regulated mechanisms (vs. stochastic, nonphysiological epimutations) are erased in the PGCLCs. The development of epigenome editing methods to introduce specific epimutations at targeted loci in the genome of iPSCs should provide unique opportunities to systematically evaluate the capabilities of PGCLCs to erase various types and locations of epigenetic changes or epimutations.
In summary, our present study has shown that mouse PGCLCs effectively recapitulate the genome-wide DNA demethylation events occurring in the intragonadal PGCs, including demethylation of ICRs. Reproducing previously reported 5meC retention at IAPs and nonpromoter CGIs in PGCs, we have identified additional 5meC-retaining genomic elements, including the GSAT_MM repeats. Deep-sequencing techniques that distinguish 5meCs and 5hmeCs have revealed coretention and dynamics of these epigenetic marks at ICRs and 5meC-retaining elements during PSC differentiation to PGCLCs. Finally, taking advantage of a region-specific epimutation experimentally introduced in iPSCs, our study has provided direct evidence that aberrant DNA hypermethylation at an ICR was diminished during the germline epigenetic reprogramming, resulting in functional restoration of the epigenetically silenced gene expression. These observations support the usefulness of mouse PGCLCs as a valuable cell culture model of embryonic PGCs for mechanistic studies of germline epigenetic reprogramming.
Materials and Methods
Experimental methods are described in SI Materials and Methods. The animal experiment protocol for the above procedures was reviewed and approved by the Institutional Animal Care and Use Committee of the Massachusetts General Hospital. The animal experiment protocol for the PGCLC transplantation was reviewed and approved by the Institutional Animal Care and Use Committee of the McGill University. Affymetrix microarray and deep-sequencing data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus and Sequence Read Archive databases (accession nos. GSE80983 and GSE81175).
SI Materials and Methods
Cell Culture.
Transcriptomal and epigenomic characteristics as well as pluripotency of ESCs and iPSCs [Gtl2(+) and Gtl2(−)] were described in our previous study (26). These PSCs [v6.5, ES12, ES15 (ESCs); Pff1592*, Pff1593, R21-2*, R21-4, R21-5*, and R21-6 (iPSCs; asterisks indicate Gtl2-negative clones)] were maintained in ESC medium [DMEM supplemented with 15% EScult-grade FCS and 1,000 U/mL recombinant mouse leukemia inhibitory factor (Stemcell Technologies), 2 mM l-glutamine, 1× penicillin–streptomycin, and 1× nonessential amino acids (Gibco), 40 nM 2-mercaptoethanol, and 50 μg/mL l-ascorbic acid (Sigma)]. Except for the ESC clone v6.5, which was provided by N. Bardeesy, Massachusetts General Hospital Center for Cancer Research, Boston, all PSCs examined in the present study shared the same genetic background of the male reprogrammable mouse strain harboring an inducible OKSM expression cassette [Jackson Laboratory stock no. 011001, B6;129S4-Col1a1tm1(tetO-Pou5f1,-Klf4,-Sox2,-Myc)Hoch].
Differentiation of Mouse PSCs to EpiLCs and PGCLCs.
EpiLCs and PGCLCs were generated from PSCs following the protocol described by Hayashi et al. [Fig. S1A (17, 34)] with modifications. PSCs (v6.5, ES12, R21-2, R21-4, R21-5, and R21-6) were differentiated to EpiLCs on fibronectin-coated dishes in modified N2B27 medium containing 40% (vol/vol) DMEM/F12 medium, 40% (vol/vol) Neurobasal medium, 0.8% B27 supplement, and 1% KSR (Thermo Fisher); 20 ng/mL activin A, and 12 ng/mL FGF2 (R&D Systems); 1× insulin–transferrin–selenium mix, 1 mM l-glutamine, and 1× penicillin–streptomycin (Gibco); and 8 pg/mL progesterone, 6 μg/mL putrescine, 40 nM 2-mercaptoethanol, 50 μg/mL l-ascorbic acid, and 20 μg/mL BSA (Sigma). Medium was changed once after 24 h of incubation, and EpiLCs were harvested after 48 h of induction. EpiLC cultures (derived from PSC clones v6.5, ES12, R21-2, R21-4, R21-5, and R21-6) were dissociated to single cells by TrypLE Express, and 30 million cells were casted into the Aggrewell 400 microwell plates (Stemcell Technologies) in PGCLC induction medium [Glasgow’s MEM supplemented with 15% KSR, 500 ng/mL recombinant human BMP4, 100 ng/mL stem cell factor, 50 ng/mL epidermal growth factor (R&D Systems), 1,000 U/mL recombinant mouse leukemia inhibitory factor, 1× nonessential amino acids, 2 mM l-glutamine, 50 μg/mL l-ascorbic acid, 100 nM 2-mercaptoethanol, and 1× penicillin–streptomycin] to form EBs at the density of 3,000 cells per microwell and 9,600 microwells per plate. After EBs were maintained in the PGCLC medium under a floating condition for 6 d, PGCLCs were isolated from TrypLE Express-dissociated, cell strainer-filtered EB cells using the FACSAriaII cell sorter (BD Biosciences) as SSEA1+/Integrin β3+ or SSEA1+/c-Kit+, forward scatter-gated single cells (Fig. S1B). Dead cells were excluded by preincubation with 7-AAD (eBioscience) before cell sorting. All fluorescence dye-conjugated antibodies were purchased from BD Biosciences. The typical yield of live SSEA1+/Integrin β3+ PGCLCs after FACS was ∼100,000–150,000. Batches with lower PGCLC yields (<50,000) were excluded from the study.
After the publication of the original protocol (17), the supplier of the recombinant mouse BMP8b protein used in the original study announced that the reagent (R&D Systems, catalog #1073-BP) was not BMP8b but actually was BMP8a (34). Although a revised protocol published by the original authors now includes BMP8a instead of BMP8b in their PGCLC induction medium (34), the PGCLC medium used in our present study did not contain either BMP8a or BMP8b. Our preliminary study did not detect any effects of the BMP8a reagent in the yield or characteristics of PGCLCs either (data not shown).
Isolation of Mouse Embryonic PGCs.
Embryonic PGCs were isolated either from the transgenic mice expressing EGFP under the germline-specific Pou5f1 promoter/distal enhancer [Jackson Laboratory stock no. 004654, B6;CBA-Tg(Pou5f1-EGFP)2Mnn/J (19)] or C57BL/6NCrl mice (Charles River). After timed mating and a plug were confirmed (defined as E0.5), gonads were dissected from E12.5 embryos before any morphological signs of gonadal sex differentiation, digested with TrypLE Express, and filtered through a 40-μm cell strainers to obtain single-cell suspension. The transgenic mouse gonadal cells were then directly subjected to FACS enrichment of the EGFP+ PGCs. The nontransgenic cells were stained using anti-SSEA1, anti–c-Kit, and/or anti-Integrin β3 antibodies for FACS enrichment of SSEA1+/c-Kit+/Integrin β3+ PGCs.
The typical yield of live PGCs after FACS was ∼50,000 per batch, each of which consisted of 6–10 embryos. Batches with lower PGC yields (<30,000) were excluded from the study.
Transplantation of PGCLCs into Seminiferous Tubules of Neonatal Mice.
PGCLCs were generated from ESCs and iPSCs expressing EGFP driven by the Pou5f1 promoter/distal enhancer (19) or the mCherry red fluorescence protein driven by a truncated human EF1α promoter [introduced by the lentiviral vector pLVX- EF1α-mCherry-C1 (Clontech)]. Pou5f1 is expressed in mouse spermatogonial stem cells (20), and the truncated human EF1α promoter is selectively active in mouse germline cells (35). The fluorescence protein-labeled PGCLCs were transplanted into testes of 129/SvEv × C57BL/6 F1 hybrid male pups the endogenous germline cells of which were depleted with exposure to busulfan at 8 days post partum (dpp) (5.4 × 106 PGCLCs per testis) using the technique described in our previous study (36). The recipient animals were returned to their littermates after surgery, and PGCLC colonization was evaluated using fluorescence microscope examination of dissected seminiferous tubules 4–5 mo after injection.
Transcriptomal Analysis.
Total RNA and genomic DNA were simultaneously prepared from the same batch of cells using the QIAGEN AllPrep kits (miniscale kit for PSCs and EpiLCs, and microscale kit for FACS-enriched PGCLCs and PGCs). RNA concentration and integrity were determined using Nanodrop (Thermo Fisher) or Agilent Bioanalyzer or Tapestation. Low-quality RNA (RNA integrity number < 9.0) was eliminated from the study.
Expression of specific mRNA transcripts was determined by real-time qPCR using the SuperScript double-stranded cDNA synthesis kit and TaqMan assays with Universal Master Mix II containing Uracil-N glycosylase (Thermo Fisher). qPCR amplification and data collection were performed using ABI7500 qPCR equipment (Thermo Fisher) and normalized with Gapdh mRNA TaqMan assay. The TaqMan Assay targets and probe IDs were as follows: Pou5f1, Mm0053917_g1; Sox2, Mm03053810_s1; Nanog, Mm0219550_s1; Klf4, Mm00516104_m1; Myc, Mm00487804_m1; Prdm1, Mm00476128_m1; Prdm14, Mm01237814_m1; Wnt3, Mm00437336_m1; Dppa3, Mm01184198_g1; Dnmt1, Mm01151063_m1; Dnmt3a, Mm00432881_m1; Dnmt3b, Mm01240113_m1; Gadph, 99999915_g1. Quantitation of the mouse Gtl2 mRNA transcripts was described in our previous study (26).
Transcriptomal profiling and data analysis were performed as we previously described (26). Transcriptome data were obtained using Affymetrix GeneChip microarray mouse 430 2.0 chips. Hybridization probes were synthesized using NuGEN Applause 3′-Amp System and Encore Biotin Module. Probe hybridization, wash, scanning, and normalized signal intensity calculation with the robust multiarray average (RMA) normalization were performed at the Microarray Core of the Harvard Partners Center for Genetics and Genomics. The absence of batch effect was confirmed by box-plot analysis of the RMA-normalized signal intensities (Fig. S3A). Hierarchical clustering and visualization were performed using Cluster and Java Treeview. Principal component analysis calculation and visualization were performed using R/Bioconductor packages pcaMethods and rgl, respectively.
Deep-Sequencing Determination of 5meCs and 5hmeCs.
Genome-wide gDNA methylation and hydroxymethylation profiles of mouse iPSCs, EpiLCs, PGCLCs, and E12.5 embryonic PGCs were determined by deep sequencing of 5meC- or 5hmeC-enriched DNA fragments. Genomic DNA was sonicated to 200–300 bp fragments using a Covaris S2 sonicator in AFA microtubes (10–200 ng DNA in 80 μL 0.1× Tris/HCl-EDTA buffer) at 4 °C, and DNA size distribution was determined using Bioanalyzer or Tapestation. DNA fragments were subjected to enrichment for 5meCs and 5hmeCs using biotin-conjugated methyl-CpG–binding domain (MBD) of the recombinant human methyl-CpG–binding domain protein 2 (MBD2) [MBD-sEq. (21)] and selective labeling of 5hmeCs with a biotinylated tag by two-step chemical reactions involving β-glucosyltransferase (23), respectively. Reagents and protocols for enrichment of 5meC- and 5hmeC-rich gDNA fragments were provided in the MethylMiner kit (Thermo Fisher) or the Hydroxymethyl Collector kit (Active Motif), respectively. To avoid sensitivity bias of enrichment reactions resulting from different amounts of input DNA, an equal amount (10 ng) of fragmented DNA was put into each enrichment reaction for both 5meCs and 5hmeCs for all cell types, and eluted DNA from multiple (typically 8–24) 10-ng scale reactions was pooled for ethanol precipitation to yield at least 0.5 ng 5meC-enriched gDNA fragments. For both the 5meC and 5hmeC enrichment kits, the manufacturer-recommended minimum amount of input DNA was 5 ng. Ten-nanogram input DNA has been shown to have sufficient molecular complexity for genome-wide ChIP-seq profiling of the mouse genome for common histone modifications (37). Successful enrichment was monitored by simultaneously performing positive control reactions using kit-provided reagents. The 5meC-rich DNA fragments were eluted in 200 μL per reaction with 2 M NaCl and then desalted/concentrated to 20 μL by ethanol precipitation with Pellet Paint coprecipitant fluorescence dye polymer (Millipore). The 5hmeC-rich DNA fragments were eluted in 50 μL per reaction elution buffer provided in the kit and subjected to ethanol precipitation.
The 5meC- or 5hmeC-enriched gDNA fragments were subjected to deep-sequencing library construction using the SOLiD 5500 Fragment Library Core Kit and the SOLiD EZ Bead emulsion PCR system (Thermo Fisher). Deep sequencing was performed using the SOLiD 5500XL deep sequencer (50 nt + 50 nt, paired-end), and the XSQ-format raw data were converted to the csfasta/QV.qual format using the XSQ tool provided by Life Technologies. The csfasta/QV.qual sequences were aligned to the NCBI37/mm9 mouse genome reference sequence to obtain the bam format aligned read data using the NovoalignCS color-space aligner. Sequencing operations were repeated, and sufficient amounts of uniquely mapped reads were obtained for each of the 10 data types—namely, ESC-5meC (324 million), ESC-5hmeC (381 million), iPSC-5meC (151 million), iPSC-5hmeC (200.2 million), EpiLC-5meC (127 million), EpiLC-5hmeC (203 million), PGCLC-5meC (692 million), PGCLC-5hmeC (458 million), E12.5 embryonic PGC-5meC (491 million), and E12.5 embryonic PGC-5hmeC (68 million). Numbers of uniquely mapped reads of individual batches of cells are summarized in Fig. S3 D and E. Note that the relatively smaller number of reads of the embryonic PGC 5hmeC-rich fragments was due to an extremely diminished number of the 5hmeC peaks in the genome, which was confirmed by repeated experiments with biological replicates of PGC specimens obtained from both the Pou5f1-EGFP transgenic mice (19) or the nontransgenic C57BL/6NCrl mice. Analyses of each cell culture type involved multiple independent subtypes—namely, five ESC clones, two Gtl2(+) iPSC clones, two Gtl2(−) iPSC clones, one ESC-derived EpiLC study, two Gtl2(+) iPSC-derived EpiLC studies, two Gtl2(−) iPSC-derived EpiLC studies, two ESC-derived PGCLC studies, two Gtl2(+) iPSC-derived PGCLC studies, and two Gtl2(−) iPSC-derived PGCLC studies. Each study consisted of at least three independently repeated experiments.
The bam format read data were examined using the fastQC deep-sequencing reads quality control tool (Simon Andrews, Babraham Institute; www.bioinformatics.babraham.ac.uk/projects/fastqc/) to exclude poor-quality reads from the study (Fig. S4). Quality control-passed bam format data were analyzed for 5meC and 5hmeC distributions in the NCBI37/mm9 mouse genome reference using the MEDIPS R/Bioconductor package (38) with the edgeR package as its statistics backend for differential methylation analysis. Results of control analyses provided by the MEDIPS package—namely, CpG coverage and saturation analyses—are presented as Fig. S3 B and C, respectively. Because we were aware of the assumption of MEDIPS that the global DNA (hydroxyl)methylation intensities among each dataset should be comparable for accurate differential 5meC or 5hmC calls, analyses of data obtained from PGCLCs and PGCs were performed with special precautions because of their far smaller numbers of the 5meC or 5hmeC peaks in the whole genome compared with PSCs or EpiLCs. To this end, results of analyses using MEDIPS were verified using the MACS deep-sequencing peak detection tool, which was originally developed for analysis of ChIP-seq data and adopted for the ENCODE project of ChIP-seq Roadmap database construction and manual inspection of the visualized 5meC and 5hmeC peaks using the Integrated Genomics Viewer after conversion to normalized wig or bigwig data format. The read peaks shown in the figures are normalized tdf file images unless specified otherwise. Peak detection cutoff was set as 10 times of the mean of all of the 100-bp windows assigned to the whole-genome sequence. Association of the detected peaks to known genomic features or RepeatMasker classes/families/functions was performed using MEDIPS functions, the ChIPseeker R/Bioconductor package, and the Ruby scripts developed in our laboratory. The aligned sequence data (bam format) of 5meC- or 5hmeC-enriched segments in the genomes of each cell type (i.e., PSCs, EpiLCs, PGCLCs, and PGCs) were merged using samtools and subjected to differential (hydroxy)methylation analysis using the MEDIPS.meth function. An aligned, whole-genome resequencing data file (bam format) for the genome of the C57BL/6 mouse was provided to the MEDIPS.meth function as no-enrichment input DNA. The locations of DNA (hydroxyl)methylation peaks determined by MEDIPS were compared with the mouse mm9 RepeatMasker database-registered locations of repeat sequences using Ruby scripts.
Reanalysis of WGBS Data.
The fastq-format Illumina deep-sequencing raw data of mouse WGBS deposited by Seisenberger et al. (9) were downloaded from the European Nucleotide Archive [ERX167039 and ERX167050 for E6.5 epiblasts (total 350 million reads) and ERX167042 for E13.5 male PGCs (244 million reads)] and subjected to alignment using the Bismark bisulfite mapper v0.14.4 with Bowtie2 as the backend aligner. The resulting bam files were subjected directly to manual inspection using the Integrative Genomics Viewer for counting bisulfite-converted and -resistant cytosines. To detect bisulfite-resistant CpG cytosines in the areas of interest determined by the MBD-seq analysis, we set the following criteria:
-
1.
For simplicity, we examined only the Watson strand of the double-stranded genomic DNA.
-
2.
Read coverage at a region of interest (∼150-bp window) is sufficient (greater than 30× at each base).
-
3.
Non-CpG cytosines (i.e., CpA, CpC, and CpT) within a region of interest are efficiently converted to thymidines by bisulfite (>80%).
-
4.
Non-CpG cytosines in a region of interest show no differences in bisulfite conversion rate between the E6.5 and E13.5 WGBS data unless there is a reasonable indication that they are also methylated or hydroxymethylated.
-
5.
CpG cytosines show significantly greater resistance to bisulfite conversion compared with the surrounding non-CpG cytosines. We expect strong resistance in the E6.5 WGBS data, and the question is whether these CpG cytosines are also significantly resistant to bisulfite conversion.
Statistical significance of the bisulfite-resistant CpG cytosines over the background of the bisulfite-sensitive CpA, CpT, and CpC cytosines in the genome of E13.5 maple PGCs was examined using Student’s t test.
Acknowledgments
We thank Haley Ellis and Shiomi (Misa) Yawata for technical assistance. This work was supported by Canadian Institutes of Health Research Grant MOP-130467 (to M.N.) and National Institutes of Health Grants HD058013 (to K.H.) and ES023316 and ES024861 (to T.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Affymetrix microarray and deep-sequencing data have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (accession nos. GSE80983 and GSE81175) and Sequence Read Archive (SRA) database (accession no. SRP074457).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610259113/-/DCSupplemental.
References
- 1.Prokopuk L, Western PS, Stringer JM. Transgenerational epigenetic inheritance: Adaptation through the germline epigenome? Epigenomics. 2015;7(5):829–846. doi: 10.2217/epi.15.36. [DOI] [PubMed] [Google Scholar]
- 2.Szyf M. Nongenetic inheritance and transgenerational epigenetics. Trends Mol Med. 2015;21(2):134–144. doi: 10.1016/j.molmed.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Alvarado S, et al. An epigenetic hypothesis for the genomic memory of pain. Front Cell Neurosci. 2015;9:88. doi: 10.3389/fncel.2015.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nilsson EE, Skinner MK. Environmentally induced epigenetic transgenerational inheritance of reproductive disease. Biol Reprod. 2015;93(6):145. doi: 10.1095/biolreprod.115.134817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohacek J, Mansuy IM. Molecular insights into transgenerational non-genetic inheritance of acquired behaviours. Nat Rev Genet. 2015;16(11):641–652. doi: 10.1038/nrg3964. [DOI] [PubMed] [Google Scholar]
- 6.Iqbal K, et al. Deleterious effects of endocrine disruptors are corrected in the mammalian germline by epigenome reprogramming. Genome Biol. 2015;16(1):59. doi: 10.1186/s13059-015-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heard E, Martienssen RA. Transgenerational epigenetic inheritance: Myths and mechanisms. Cell. 2014;157(1):95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saitou M, Yamaji M. Primordial germ cells in mice. Cold Spring Harb Perspect Biol. 2012;4(11):a008375. doi: 10.1101/cshperspect.a008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seisenberger S, et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012;48(6):849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohno R, et al. A replication-dependent passive mechanism modulates DNA demethylation in mouse primordial germ cells. Development. 2013;140(14):2892–2903. doi: 10.1242/dev.093229. [DOI] [PubMed] [Google Scholar]
- 11.Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J. 2013;32(3):340–353. doi: 10.1038/emboj.2012.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hackett JA, Zylicz JJ, Surani MA. Parallel mechanisms of epigenetic reprogramming in the germline. Trends Genet. 2012;28(4):164–174. doi: 10.1016/j.tig.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Kawasaki Y, et al. Active DNA demethylation is required for complete imprint erasure in primordial germ cells. Sci Rep. 2014;4:3658. doi: 10.1038/srep03658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamaguchi S, Shen L, Liu Y, Sendler D, Zhang Y. Role of Tet1 in erasure of genomic imprinting. Nature. 2013;504(7480):460–464. doi: 10.1038/nature12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guibert S, Forné T, Weber M. Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res. 2012;22(4):633–641. doi: 10.1101/gr.130997.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge W, Chen C, De Felici M, Shen W. In vitro differentiation of germ cells from stem cells: A comparison between primordial germ cells and in vitro derived primordial germ cell-like cells. Cell Death Dis. 2015;6(10):e1906. doi: 10.1038/cddis.2015.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146(4):519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 18.Nakaki F, et al. Induction of mouse germ-cell fate by transcription factors in vitro. Nature. 2013;501(7466):222–226. doi: 10.1038/nature12417. [DOI] [PubMed] [Google Scholar]
- 19.Szabó PE, Hübner K, Schöler H, Mann JR. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech Dev. 2002;115(1-2):157–160. doi: 10.1016/s0925-4773(02)00087-4. [DOI] [PubMed] [Google Scholar]
- 20.Dann CT, et al. Spermatogonial stem cell self-renewal requires OCT4, a factor downregulated during retinoic acid-induced differentiation. Stem Cells. 2008;26(11):2928–2937. doi: 10.1634/stemcells.2008-0134. [DOI] [PubMed] [Google Scholar]
- 21.Harris RA, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28(10):1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151(7):1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song C-X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29(1):68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y, et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5(1):e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Q, et al. Complete meiosis from embryonic stem cell-derived germ cells in vitro. Cell Stem Cell. 2016;18(3):330–340. doi: 10.1016/j.stem.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 26.Stadtfeld M, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465(7295):175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stadtfeld M, et al. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat Genet. 2012;44(4):398–405. doi: 10.1038/ng.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh P, et al. De novo DNA methylation in the male germ line occurs by default but is excluded at sites of H3K4 methylation. Cell Reports. 2013;4(1):205–219. doi: 10.1016/j.celrep.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rauch T, Li H, Wu X, Pfeifer GP. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66(16):7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- 30.Hackett JA, et al. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339(6118):448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaguchi S, et al. Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res. 2013;23(3):329–339. doi: 10.1038/cr.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szyf M, Tang Y-Y, Hill KG, Musci R. The dynamic epigenome and its implications for behavioral interventions: A role for epigenetics to inform disorder prevention and health promotion. Transl Behav Med. 2016;6(1):55–62. doi: 10.1007/s13142-016-0387-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weaver ICG, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 34.Hayashi K, Saitou M. Generation of eggs from mouse embryonic stem cells and induced pluripotent stem cells. Nat Protoc. 2013;8(8):1513–1524. doi: 10.1038/nprot.2013.090. [DOI] [PubMed] [Google Scholar]
- 35.Furuchi T, Masuko K, Nishimune Y, Obinata M, Matsui Y. Inhibition of testicular germ cell apoptosis and differentiation in mice misexpressing Bcl-2 in spermatogonia. Development. 1996;122(6):1703–1709. doi: 10.1242/dev.122.6.1703. [DOI] [PubMed] [Google Scholar]
- 36.Zohni K, Zhang X, Tan SL, Chan P, Nagano MC. The efficiency of male fertility restoration is dependent on the recovery kinetics of spermatogonial stem cells after cytotoxic treatment with busulfan in mice. Hum Reprod. 2012;27(1):44–53. doi: 10.1093/humrep/der357. [DOI] [PubMed] [Google Scholar]
- 37.Brind’Amour J, et al. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat Commun. 2015;6:6033. doi: 10.1038/ncomms7033. [DOI] [PubMed] [Google Scholar]
- 38.Lienhard M, Grimm C, Morkel M, Herwig R, Chavez L. MEDIPS: Genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics. 2014;30(2):284–286. doi: 10.1093/bioinformatics/btt650. [DOI] [PMC free article] [PubMed] [Google Scholar]