Significance
Generation of mechanical forces by molecular motors is essential for development. Previously, we showed that the microtubule motor kinesin-1 generates forces by sliding microtubules against each other. Here, we show that microtubule sliding by kinesin-1 is important for normal oocyte cytoplasmic rotation, a process required for efficient localization of mRNAs and proteins during oogenesis. Using recently developed imaging technologies (Maple3 photoconversion and SunTag), we discover a previously uncharacterized population of extremely stable microtubules immobilized at the oocyte cortex and demonstrate that free microtubules move against cortically anchored microtubules, generating forces that contribute to cytoplasmic streaming. Because kinesin-1–based sliding is highly conserved from Drosophila to humans, we propose that microtubule sliding is also important for cellular force generation in higher organisms.
Keywords: kinesin-1, microtubules, cytoplasmic streaming, Drosophila, axis determination
Abstract
Cytoplasmic streaming in Drosophila oocytes is a microtubule-based bulk cytoplasmic movement. Streaming efficiently circulates and localizes mRNAs and proteins deposited by the nurse cells across the oocyte. This movement is driven by kinesin-1, a major microtubule motor. Recently, we have shown that kinesin-1 heavy chain (KHC) can transport one microtubule on another microtubule, thus driving microtubule–microtubule sliding in multiple cell types. To study the role of microtubule sliding in oocyte cytoplasmic streaming, we used a Khc mutant that is deficient in microtubule sliding but able to transport a majority of cargoes. We demonstrated that streaming is reduced by genomic replacement of wild-type Khc with this sliding-deficient mutant. Streaming can be fully rescued by wild-type KHC and partially rescued by a chimeric motor that cannot move organelles but is active in microtubule sliding. Consistent with these data, we identified two populations of microtubules in fast-streaming oocytes: a network of stable microtubules anchored to the actin cortex and free cytoplasmic microtubules that moved in the ooplasm. We further demonstrated that the reduced streaming in sliding-deficient oocytes resulted in posterior determination defects. Together, we propose that kinesin-1 slides free cytoplasmic microtubules against cortically immobilized microtubules, generating forces that contribute to cytoplasmic streaming and are essential for the refinement of posterior determinants.
Force generation is essential for many different cellular functions, such as vesicle transport, spindle formation, cell migration, and cell shape change. Cytoskeletal filaments and molecular motors are major players that generate the forces required for these activities. For instance, kinesin-1 and microtubules have been shown to provide the mechanical forces required in multiple cellular contexts, such as organelle transport (1–4), ooplasmic streaming (5–9), and process formation (10–15).
Ooplasmic streaming is a bulk cytoplasmic movement that efficiently mixes the cytoplasm released from nurse cells through ring canals with ooplasm in the oocyte during Drosophila oogenesis. This streaming circulates and distributes maternally loaded mRNAs and proteins in oocytes for proper axis determination. Slow nondirectional streaming starts at stages 8–9, whereas fast directional streaming is initiated at stage 10B (5). Ooplasmic streaming has been shown to be dependent on microtubules and requires the activity of kinesin-1. Depolymerization of microtubules by colchicine (16) or knockout of the kinesin-1 heavy chain (KHC) gene (7, 8) completely abolishes streaming in the oocyte. It has been proposed that streaming of ooplasm is driven by the directional transport of cargoes along microtubules by kinesin-1 (8). Surprisingly, kinesin-1 light chain (KLC) is dispensable for streaming (7, 9), even though KLC is used as the adapter for many of the cargoes transported by kinesin-1. Based on these results, it has been proposed that kinesin-1 transports vesicles to drive ooplasmic streaming by an unknown KLC-independent mechanism (7). In contrast to kinesin-1’s function in streaming, dynein is not required for fast streaming; instead, it inhibits the shift from slow streaming to fast streaming (8, 9).
This KHC-dependent and KLC/dynein-independent streaming mechanism highly resembles another important function of KHC, called microtubule–microtubule sliding, studied by our group (10–15). In this process, KHC binds one microtubule through its ATP-independent C-terminal–binding site while walking on another microtubule using its ATP-dependent motor domain, as reported by Winding et al. (17). Sequence analysis of the C-terminal microtubule-binding site of KHC revealed a high degree of conservation across species; it contains multiple basic residues that are evolutionally conserved from Drosophila to humans (Fig. 1A) and binds to the acidic E-hook of tubulin through an electrostatic interaction (18, 19). Furthermore, we have shown that KHC-driven microtubule sliding is independent of KLC and cannot be inhibited by dynein knockdown or dynein inhibition (10, 12, 14, 15).
Fig. 1.
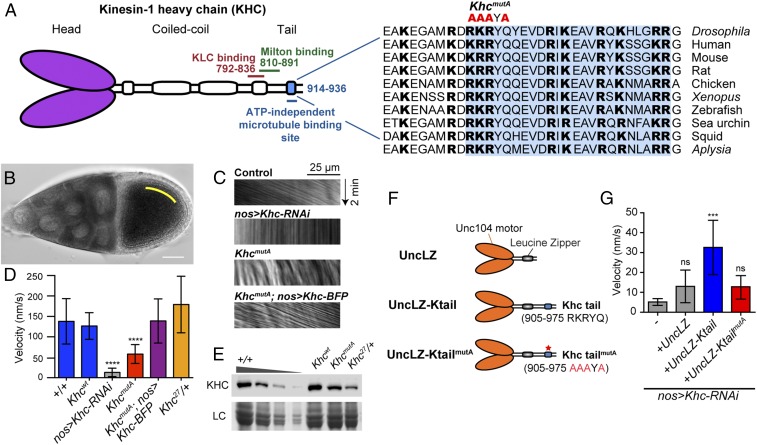
KHC C-terminal microtubule-binding site is important for ooplasmic streaming. (A) Schematic illustration of KHC and point mutations (R914A, K915A, R916A, and Q918A) made in the highly conserved C-terminal microtubule-binding site, which significantly decrease microtubule-binding affinity and microtubule sliding ability (17). The amino acid sequences of the C-terminal microtubule-binding site (shaded in blue) of KHC from multiple organisms are shown. The basic residues (highlighted in bold fonts) are highly conserved across species. (B) Example of streaming rate measurement in a control stage 10B oocyte (more details are provided in Materials and Methods). (C) Representative kymographs of vesicles in oocytes of different genotypes. Kymographs were generated along a line parallel to the flow of the streaming ooplasm (yellow line in B) from 2-min DIC movies (also Movies S1–S5). (D) Ooplasmic streaming rates in different genotypes. The rate of vesicle movement was quantified and used as a streaming readout. +/+: average = 138.0 nm⋅s−1, n = 20; Khcwt: average = 126.6 nm⋅s−1, n = 74, P = 0.4216 [Mann–Whitney test between +/+ and Khcwt, not significant (ns)]; nos>Khc-RNAi: average = 13.0 nm⋅s−1, n = 40, P < 0.0001 (Mann–Whitney test between +/+ and nos>Khc-RNAi, ****); KhcmutA: average = 58.0 nm⋅s−1, n = 69, P < 0.0001 (Mann–Whitney test between Khcwt and KhcmutA, ****); KhcmutA; nos>Khc-BFP: average = 138.9 nm⋅s−1, n = 31, P = 0.9181 (Mann–Whitney test between Khcwt and KhcmutA; nos>Khc-BFP, ns); Khc27/+, average = 179.3 nm⋅s−1, n = 25, P = 0.6209 (Mann–Whitney test between +/+ and Khc27/+, ns). The error bar represents the 95% confidence interval. (E) Western blot of KHC protein levels in ovary extracts from the genotypes assayed in C and D using a polyclonal antibody against the KHC motor domain (HD) (77). The +/+ ovary extract was titrated (100%, 50%, 25%, 10%) for comparison with Khcwt, KhcmutA, and Khc27/+ ovary extracts. Loading control (LC), Coomassie staining. Western blot signal intensities from multiple experiments were quantified using an ImageJ plug-in (Gel Analyzer) and normalized to the +/+ extract. This analysis showed that KhcmutA ovaries have 71.2 ± 1.4% KHC protein level as +/+ ovaries. Quantification of each sample (normalized to 1, average ± SD): 100% +/+, 1.00 ± 0.00; 50% +/+, 0.505 ± 0.094; 25% +/+, 0.152 ± 0.040; 10% +/+, 0.013 ± 0.004; Khcwt, 1.04 ± 0.02; KhcmutA, 0.712 ± 0.014; Khc27/+, 0.395 ± 0.107. (F) Schematic diagrams of transgenic constructs of chimeric motors used in G. (G) Streaming rates in KHC-depleted stage 10B oocytes with or without expression of chimeric Unc104 motor constructs (mean ± 95% confidence interval): nos>Khc-RNAi, 5.2 ± 1.7 nm⋅s−1, n = 36; nos>Khc-RNAi+UncLZ, 13.0 ± 8.2 nm⋅s−1, n = 41; nos>Khc-RNAi+UncLZ-Ktail, 32.6 ± 13.6 nm⋅s−1, n = 43; nos>Khc-RNAi+UncLZ-KtailmutA, 12.9 ± 5.9 nm⋅s−1, n = 46. Mann–Whitney tests were performed to assay the statistical significance between the following groups: nos>Khc-RNAi vs. nos>Khc-RNAi+UncLZ, P = 0.5572 (ns); nos>Khc-RNAi+UncLZ vs. nos>Khc-RNAi+UncLZ-Ktail, P = 0.0003 (***); nos>Khc-RNAi+UncLZ vs. nos>Khc-RNAi+UncLZ-KtailmutA, P = 0.5692 (ns) (also Movie S6).
Based on the fact that streaming and microtubule sliding both require KHC and are independent of KLC and dynein, we propose that KHC-dependent microtubule sliding plays an important role in ooplasmic streaming of Drosophila oocytes. In this study, we tested this hypothesis by using a recently characterized Khc mutant that inhibits microtubule sliding (17), and we demonstrated that microtubule sliding contributes to ooplasmic streaming. Furthermore, we ectopically induced microtubule sliding using a chimeric motor that slides microtubules but cannot transport organelles, and showed that the motor partially rescued ooplasmic streaming in KHC depleted oocytes. Consistent with these results, we observed two populations of microtubules, a network of extremely stable microtubules anchored to the cortex and cytoplasmic microtubules moving with the rest of the ooplasm. Furthermore, we showed that microtubule sliding is important to refine posterior determinants during body axis patterning. We propose that KHC slides free cytoplasmic microtubules against anchored cortical microtubules, providing an important contribution to ooplasmic streaming, and that this streaming ensures robust posterior determination.
Results
Microtubule Sliding by Kinesin-1 Contributes to Ooplasmic Streaming.
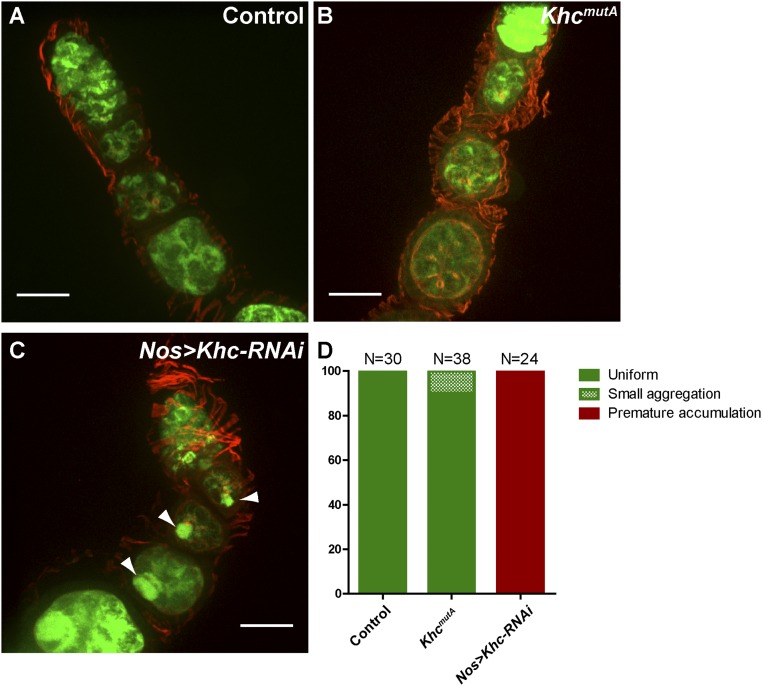
In an accompanying study by Winding et al. (17), we created a Khc endogenous knock-in mutant in which four conserved residues in the C-terminal microtubule-binding site were mutated to alanine (KhcmutA: R914A, K915A, R916A, and Q918A) (Fig. 1A). This mutant was based on a similar mutation of human kinesin-1 that greatly reduced the affinity of the tail to microtubules in vitro (18) (Fig. S1). We characterized the KhcmutA mutant and found that the ability of KHCmutA to drive microtubule sliding is significantly reduced (17). A majority of kinesin-1–driven cargo transport was not affected in both Drosophila S2 cells and primary cultured neurons expressing KHCmutA (17). A similar mutant in Caenorhabditis elegans kinesin-1, uncoordinated-116 (unc-116), also did not affect organelle transport (20). To test the ability of this mutant to transport cargoes in the germline, we assayed the distribution of mitochondria, well-known kinesin-1–dependent organelles (3), using a mitochondrial EGFP marker (21). We found that mitochondria were distributed correctly in stage 1–3 egg chambers of KhcmutA oocytes (Fig. S2). In contrast, Khc-RNAi oocytes displayed premature mitochondrial aggregation in the enlarged Balbiani body due to unbalanced bidirectional transport of mitochondria, consistent with previous reports using a Khc loss-of-function mutant (22). Together, these results indicate that KhcmutA is a hypomorphic allele for KHC-driven microtubule sliding that retains wild-type transport levels for many KHC-driven organelles. We reasoned that using this viable KhcmutA mutant could allow us to determine the role of KHC-driven microtubule sliding in ooplasmic streaming.
Fig. S1.
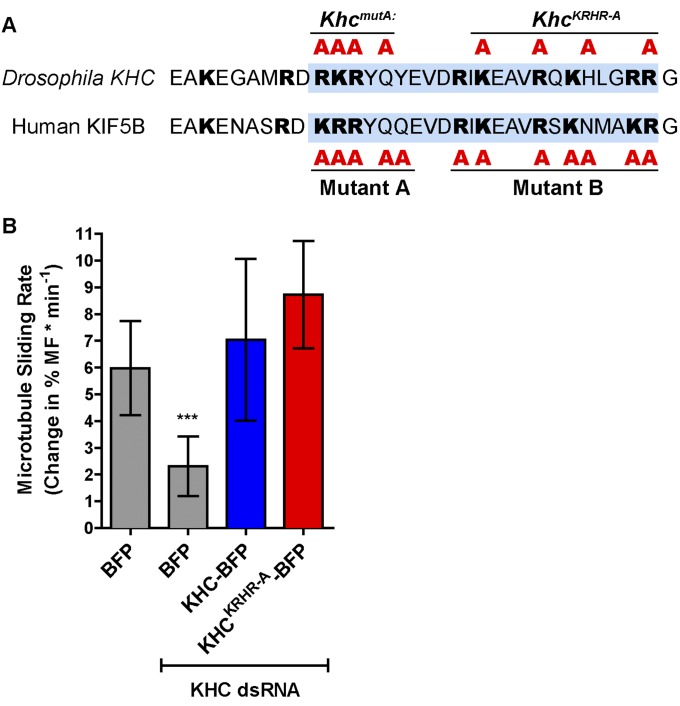
KhcKRHR-A mutations do not affect KHC-dependent microtubule sliding. (A) Schematic illustration of two types of tail mutations in the kinesin-1 C-terminal microtubule-binding site (blue shades) in Drosophila KHC and human KIF5B. Drosophila KHCmutA used in this study is similar to the human KIF5B tail mutant A (18), whereas a previously reported mutant KHCKRHR-A (53) is similar to the human KIF5B tail mutant B (18). Human KIF5B tail mutant B has been shown to have a much weaker effect on decreasing microtubule affinity than mutant A in vitro (mutant A, 20-fold decrease; mutant B, sixfold decrease) (18). (B) Microtubule sliding rates in Drosophila S2 cells of the following conditions (mean ± 95% confidence intervals): BFP, 5.98 ± 1.76% (n = 28); BFP+KHC 3′-UTR dsRNA, 2.31 ± 1.11% (n = 21); KHC-BFP+KHC 3′-UTR dsRNA, 7.04 ± 3.01% (n = 22); KHCKRHR-A-BFP+KHC 3′-UTR dsRNA, 8.72 ± 2.00% (n = 29). More details about the microtubule sliding assay and quantification are available in an accompanying study (17). Mann–Whitney tests were performed to assay the statistical significance between the following groups: BFP vs. BFP+KHC dsRNA, P = 0.0001 (***); BFP vs. KHC-BFP+KHC dsRNA, P = 0.8835 (ns); and KHC-BFP+KHC dsRNA vs. KHCKRHR-A-BFP+KHC dsRNA, P = 0.0665 (ns). ns, not significant.
Fig. S2.
KhcmutA ovaries display normal mitochondria localization in early stage egg chambers. (A) Mitochondria are localized uniformly in the cytoplasm of the germ cells of stage 1–3 egg chambers in control (nos>Mito-EGFP). (B) Mitochondrial localization is not affected in KhcmutA egg chambers (KhcmutA/KhcmutA; nos>Mito-EGFP). (C) Abnormal aggregation of mitochondria in oocytes (arrowheads) was seen in all Khc-RNAi ovaries (nos>Khc-RNAi+Mito-EGFP). Green, Mito-EGFP; red, rhodamine-conjugated phalloidin. Scale bars, 20 μm. (D) Summary of mitochondrial localization in genotypes shown in A–C. Control, n = 30; KhcmutA, n = 38; Nos>Khc-RNAi, n = 24. Tests (χ2) were performed (comparing uniform localization and aggregation, including small aggregation and premature accumulation): control vs. KhcmutA: P = 0.1243 (ns); control vs. nos>Khc-RNAi: P < 0.0001; KhcmutA vs. nos>Khc-RNAi: P < 0.0001.
To test whether kinesin-1–based microtubule sliding is important for streaming, we quantified ooplasmic streaming rates in different genotypes by measuring vesicle movement in differential interference contrast (DIC) time-lapse movies. Wild-type stage 10B oocytes (23, 24) displayed characteristic fast streaming at an average rate of ∼140 nm⋅s−1 (8) (Fig. 1 B–D and Movie S1). We knocked down KHC by RNAi in the germline under nanos-Gal4 (nos-Gal4) (25), which greatly reduced the KHC protein level (<14% of wild-type level) and led to a <1% embryo hatching rate. The lethality of embryos from nos>Khc-RNAi female flies was similar to the lethality of embryos produced by germline clones of the Khc protein-null allele, Khc27 (8, 26). The Khc-RNAi knockdown oocyte displayed very little ooplasmic streaming (Fig. 1 C and D and Movie S2), which is consistent with previous reports using Khc27 germline clones (7, 8). The ooplasm also displayed a stratified pattern, which is indicative of a failure to mix nurse cell cytoplasm and yolk-containing ooplasm (8) (Movie S2).
We then examined the streaming rates in Khc wild-type knock-in (Khcwt) and sliding-deficient KhcmutA knock-in ovaries. The streaming rates of Khcwt knock-in oocytes were similar to control oocytes (Fig. 1D), suggesting the knock-in strategy does not affect streaming. The streaming rates of KhcmutA knock-in oocytes were significantly decreased, with an average velocity at 46% of the Khcwt level and 42% of the control level (Fig. 1 C and D and Movie S3). This streaming defect observed in KhcmutA females can be fully rescued by expression of full-length wild-type KHC in the germline (KhcmutA; nos>Khc-BFP) (Fig. 1 C and D and Movie S4), resulting in an average velocity at 100% of the Khcwt level and 110% of the control level. These results indicate that the C-terminal microtubule-binding site of KHC is important for ooplasmic streaming and that the reduction of streaming is specifically caused by the Khc mutant and not by potential off-target effects from the knock-in procedure.
It is possible that the reduced ooplasmic streaming seen in KhcmutA oocytes is due to a decrease in KHC protein level caused by the mutations in the C terminus. Therefore, we measured the amount of KHC protein by Western blot using ovary extracts from wild-type, Khcwt, or KhcmutA flies. Although no KHC degradation products were observed, we found that KhcmutA knock-in ovaries had a slightly decreased KHC level (71% of the control level; Fig. 1E). To determine if this decrease contributes to the streaming defect observed in KhcmutA oocytes, we compared KHC levels and streaming rates in the oocytes of control (+/+) and Khc27 heterozygous (Khc27/+) flies. As expected, because Khc27 is a protein-null allele (26), Khc27/+ flies contain only 40% of the control KHC protein level by Western blot, which is less than the KHC level in KhcmutA flies (Fig. 1E). Nevertheless, the ooplasmic streaming rate of Khc27/+ oocytes is indistinguishable from the ooplasmic streaming rate of the control (Fig. 1 C and D and Movie S5). Therefore, we conclude that the decrease in KHC protein level is not the cause of the ooplasmic streaming defect in KhcmutA ovaries.
Because KhcmutA has a decreased ability to slide microtubules and displayed decreased cytoplasmic streaming, but is completely normal in its ability to move multiple cargoes along microtubules (17), we conclude that KHC-driven microtubule sliding is important for ooplasmic streaming.
Ectopic Microtubule Sliding Can Drive Ooplasmic Movement in KHC-Depleted Oocytes.
To further test whether microtubule sliding plays a role in ooplasmic streaming, we created a chimeric motor based on a construct described by Tomishige et al. (27). This construct encodes the motor domain of C. elegans kinesin-3 (Unc104) dimerized by a leucine zipper. We directly used this chimera, UncLZ, or fused it to the C-terminal tail of wild-type KHC (910–975 RKRYQ) or KHCmutA (910–975 AAAYA), creating UncLZ-Ktail and UncLZ-KtailmutA, respectively (Fig. 1F). These dimerized Unc104-LZ motors are similar to kinesin-1 in their velocity and run length (27, 28). UncLZ-Ktail can slide microtubules in S2 cells lacking endogenous kinesin-1, but cannot transport peroxisomes or mitochondria and lacks a majority of cargo adapter-binding domains (17). We expressed this chimeric motor in Khc-RNAi background and found that UncLZ-Ktail partially rescued streaming in Khc-RNAi oocytes (Movie S6), whereas neither UncLZ nor UncLZ-KtailmutA rescued streaming (Fig. 1G). We noticed that the rescue by the chimeric motor was incomplete; instead of organized global streaming, the rescue sometimes was local, was not as fast as wild type, and sometimes failed to rescue at all. This variability may be caused by impaired expression and/or localization of the chimeric motor under the nos-Gal4 driver, or because the stalk domain of the chimera is shorter than the stalk of KHC. Nevertheless, an ectopically expressed sliding-only motor partially rescues streaming, strongly indicating that microtubule sliding is an important driving force for ooplasmic streaming.
Overall, these experiments implicate microtubule sliding by kinesin-1 in ooplasmic streaming and demonstrate that neither organelle transport alone (KhcmutA) nor microtubule sliding alone (UncLZ-Ktail) can fully drive normal ooplasmic streaming. Based on these results, we conclude that both microtubule sliding and organelle transport by kinesin-1 are necessary to drive normal ooplasmic streaming.
A Previously Unidentified Population of Stable Cortically Anchored Microtubules in the Streaming Oocyte.
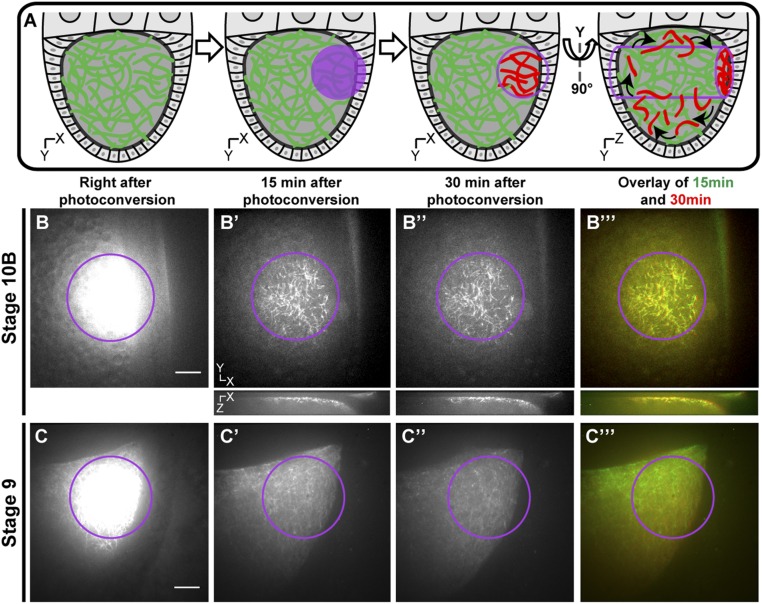
As we have previously shown in tissue culture cells and neurons, sliding occurs between two free microtubules; thus, microtubules move in both directions (10–15). However, oocyte streaming in stage 10B oocytes is usually unidirectional. One way that microtubule sliding can generate unidirectional streaming is to anchor one set of microtubules to immobile structures while allowing kinesin-1 to slide free cytoplasmic microtubules along the anchored ones. To search for a population of anchored microtubules, we used a photoconversion technique utilizing α-tubulin tagged with a photoconvertible tag, Maple3 (29, 30) (details are provided in Materials and Methods) under the nos driver. This technique allowed us to test for any potentially anchored microtubules in the photoconverted zone (the technique is illustrated in Fig. 2A). Immediately after photoconversion in an ∼30-μm-diameter circle, the intensity of the signal prevented us from identifying individual microtubules in the photoconversion area (Fig. 2B). After a few minutes, fast ooplasmic streaming removed unattached photoconverted microtubules as well as soluble tubulin subunits from the region, leaving behind a small population of microtubules that remained immotile in the photoconversion zone (Fig. 2B′). Remarkably, this population of microtubules did not move or depolymerize despite the vigorous streaming that occurred during the 30-min duration of the experiment (Fig. 2 B′–B′′′ and Movie S7). We examined the photoconverted signal in the x–z axis and found that this anchored microtubule network is adjacent to the oocyte cortex (Fig. 2 B′–B′′′, Bottom), suggesting that the stable microtubules are tightly attached to the cortex. Furthermore, in addition to their mechanical stability, we observed that these anchored microtubules do not exchange subunits with the cytoplasmic pool of (mostly nonphotoconverted) tubulin dimers, suggesting they are protected against depolymerization (compare Fig. 2B′ and 2B′′). Thus, we used photoconversion to reveal a population of superstable microtubules tightly anchored to the cortex of the streaming oocyte.
Fig. 2.
Visualization of stable cortically anchored microtubules in streaming oocytes by photoconversion. (A) Schematic cartoon illustrating the technique of microtubule photoconversion (photoconvertible Maple-αtub) to look for anchored microtubules in a streaming oocyte. (B–B′′′) Stage 10B oocyte expressing photoconvertible Maple-αtub (nos>tdMaple3-αtub84B) is photoconverted by 405 nm of light in the area indicated by the purple circle. Red channel images were taken at different time points after photoconversion. The x–y-axis images (Top) and x–z-axis images (Bottom) are shown. Note that a population of cortical microtubules can be observed 15 and 30 min after photoconversion. (B′′′) These microtubules do not change position, as demonstrated by the merged image (also Movie S7). (C–C′′′) Stage 9 oocyte expressing photoconvertible Maple-αtub (nos>tdMaple3-αtub84B) is photoconverted by 405 nm of light in the area indicated by the purple circle. Red channel images were taken at different time points after photoconversion. Note that the microtubule signal decreases over the 30-min time-lapse and that no cortical microtubules are observed (also Movie S8). Scale bars, 10 μm.
Fast streaming occurs in stage 10B to stage 12 oocytes, whereas earlier stage oocytes at stages 8–9 display much slower uncoordinated streaming (5, 8). We wondered if stable microtubules are recruited to the cortex right before the onset of fast streaming. To test this hypothesis, similar photoconversion experiments were performed in stage 8 to stage 9 oocytes. We found that photoconverted signal quickly moved outside the initial photoconversion zone and, importantly, no anchored microtubules could be detected (Fig. 2 C–C′′′ and Movie S8). This finding indicates that the stable cortical microtubule network forms later, consistent with previous reports describing high microtubule dynamics and active reorganization in the stage 9 egg chamber (31). This finding is also in agreement with the timing of fast ooplasmic streaming, suggesting that these stable microtubules might form to fulfill a functional role in streaming.
Sliding Between Cortically Anchored Microtubules and Free Cytoplasmic Microtubules Is Driven by Kinesin-1.
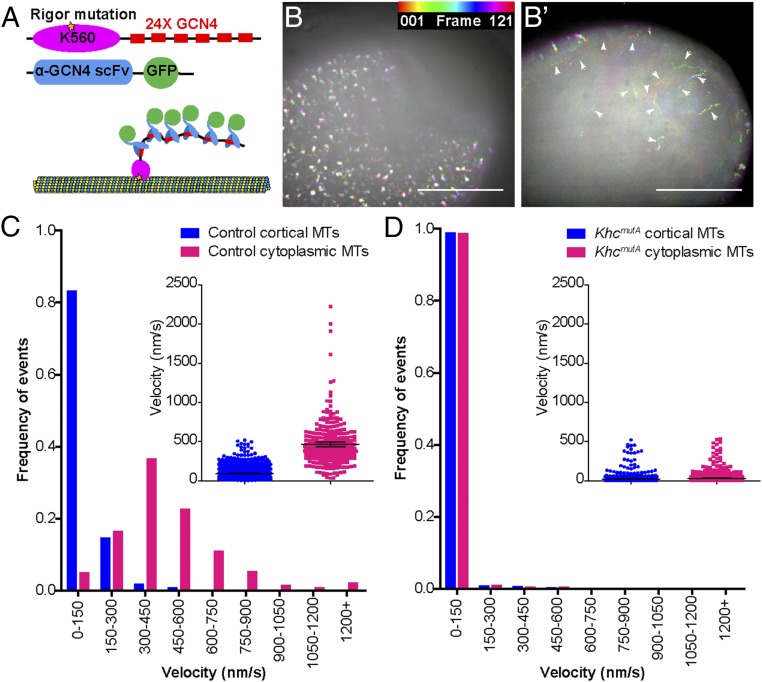
We next examined whether cytoplasmic microtubules can slide against these cortically anchored microtubules. To visualize microtubules with better resolution, we applied a newly developed microtubule probe, K560RigorE236A-SunTag (32) in oocytes. This probe contains the human kinesin-1 motor domain (residues 1–560) with a rigor mutation (K560RigorE236A) fused to 24 copies of a GCN4 peptide (Fig. 3A). The rigor mutation in the motor domain causes it to bind irreversibly to microtubules (33). When coexpressed with a GFP-tagged anti-GCN4 single-chain antibody, K560RigorE236A recruits up to 24 copies of GFP to a single position on a microtubule (the combination of these two constructs will be called SunTag-MT), thereby enabling long-term microtubule imaging in live oocytes (Fig. 3A). We generated transgenic flies expressing both SunTag-MT components under UASp control (details are provided in Materials and Methods). We found that oocytes expressing SunTag-MT under germline-specific nos-Gal4 displayed two populations of microtubules: (i) microtubules at the cortex, which display little movement (Fig. 3 B and C and Movie S9, Left), and (ii) cytoplasmic microtubules deeper in the oocyte, which move over long distances (Fig. 3 B′, white arrowheads, and C and Movie S9, Right). In contrast, both microtubule populations in KhcmutA oocytes display very little movement (Fig. 3D). Together, this SunTag-MT labeling allowed us to visualize two populations of microtubules: stable anchored microtubules that we first observed using photoconversion (Fig. 2) and free cytoplasmic microtubules that move with the ooplasm underneath the anchored microtubules. The free cytoplasmic microtubules moved at an average of ∼460 nm⋅s−1, which is consistent with kinesin-1 velocity in oocyte extracts (34–36). It should be noted that the velocities of cytoplasmic microtubules were greater than the streaming velocities observed in Fig. 1. However, streaming velocities are variable and depend on factors such as genetic background and distance from the cortex. GFP-excluding vesicles, observed in the background of SunTag-MT time-lapse movies, travel at approximately the same speed as the SunTag-MT punctae in the optical plane that was imaged. For this reason, we do not believe the difference in rates between Figs. 1 and 3 is biologically important, and it is likely attributable to different imaging conditions. The decrease in motility of cytoplasmic microtubules in KhcmutA oocytes demonstrated that the movement of free cytoplasmic microtubules is dependent on KHC’s ability to slide microtubules. Because microtubule sliding is severely disrupted in KhcmutA oocytes and some streaming still persists, we conclude that kinesin-1–based microtubule sliding contributes to oocyte streaming, but cannot account for the whole phenomenon. Ooplasmic streaming is likely driven by both kinesin-1–based organelle transport and microtubule sliding.
Fig. 3.
SunTag labeling of anchored and motile microtubules in the oocyte. (A) Illustration of SunTag-MT imaging technique. The human KHC motor domain with a rigor mutation (K560RigorE236A) is fused to 24 repeats of a GCN4 peptide. When coexpressed with a GFP-tagged anti-GCN4 single-chain antibody, K560RigorE236A binds a microtubule irreversibly and recruits many copies of GFP, creating a permanent fluorescent mark on the microtubule (32). (B and B′) Temporal hyperstacks of 10-min movies at two different focal planes of a fast-streaming oocyte expressing nos>SunTag-MT. Different frames are assigned different colors (color legend) and then projected into a single image, such that moving dots are depicted by a spectrum of color and immotile dots are white. (B) SunTag-MT dots at the cortex (cortical microtubules) displayed little movement. (B′) SunTag-MT dots underneath the cortex (cytoplasmic microtubules) displayed high motility (white arrowheads) (also Movie S9). Scale bars, 50 μm. The velocity of SunTag-MT dots on anchored cortical microtubules and free cytoplasmic microtubules in control (C) and KhcmutA (D) is shown. The average velocity (mean ± 95% confidence interval) of cortical dots and cytoplasmic dots in control oocytes was 91.67 ± 4.97 nm⋅s−1 (n = 958) and 460.6 ± 31.1 nm⋅s−1 (n = 294). The average velocity of cortical dots and cytoplasmic dots in KhcmutA oocytes was 22.95 ± 1.74 nm⋅s−1 (n = 1,834) and 31.46 ± 1.77 nm⋅s−1 (n = 1,951).
The Stable Cortically Anchored Microtubules Are Closely Associated with Cortical Actin.
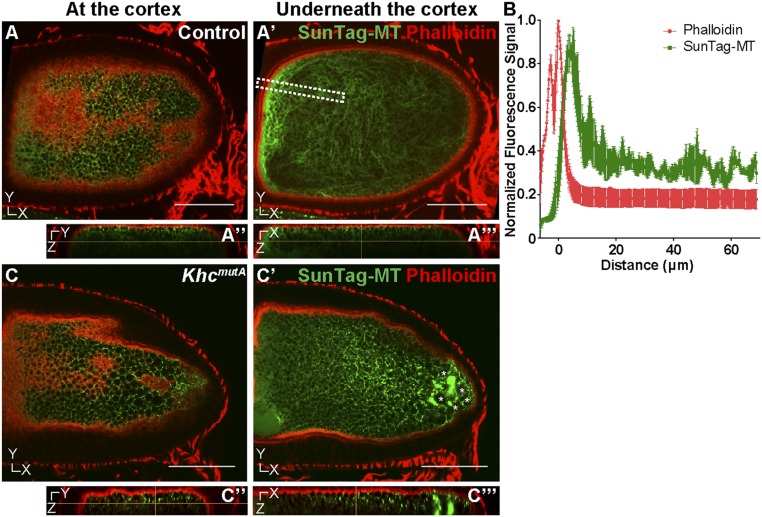
How are the stable microtubules anchored to the cortex? The oocyte cortex has a dense layer of F-actin that has been proposed to anchor mRNA and proteins (37). Therefore, we decided to examine the distribution of F-actin and microtubules in stage 10B oocytes. We performed dual labeling of microtubules by SunTag-MT and F-actin by phalloidin staining. When SunTag-MT is overexpressed, it decorates microtubules more heavily, and instead of individual SunTag-MT speckles, it labels microtubules along their length. We found that anchored microtubules are always located in close proximity to cortical F-actin (Fig. 4 A and B and Movie S10). Remarkably, cortical F-actin, like cortical microtubules, is extremely stable. We were unable to depolymerize it with very high concentrations of latrunculin B (up to 100 μM for 1 h); thus, cortical F-actin is capable of providing a platform for stable microtubule anchorage.
Fig. 4.
Stable immotile microtubules are closely associated with cortical actin. (A–A′′′) Dual labeling of microtubules by SunTag-MT and F-actin by phalloidin in a control oocyte. (A and A′) Microtubules were observed at the focal plane underneath the cortex. (A′′ and A′′′) The y–z-axis and x–z-axis views of microtubules and F-actin in the oocyte, respectively (also Movie S10). (B) Quantification of F-actin and SunTag-MT fluorescence signal within a specified area in multiple oocytes, shown as a white dashed box in A′. The position of the oocyte cortex was normalized as 0 μm on the x axis; thus, negative x values indicate the position outside the oocyte cortex, and positive x values indicate a position inside the oocyte. Error bar, SEM. (C–C′′′) Dual labeling of microtubules and F-actin in a KhcmutA oocyte. (C) Anchored microtubules at the F-actin–enriched cortex were not affected by the KhcmutA mutant. (C′) Suntag-MT aggregates (marked by asterisks) were seen underneath the actin cortex. (C′′ and C′′′) The y–z-axis and x–z-axis views of microtubules and F-actin in the oocyte, respectively (also Movie S11). Scale bars, 50 μm.
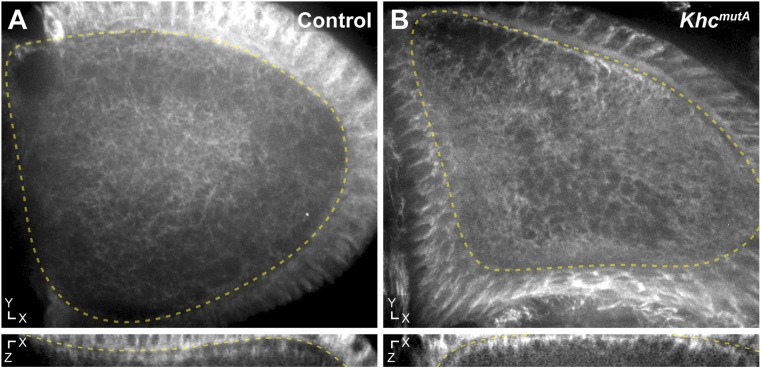
In KhcmutA oocytes, microtubules still remain closely associated with cortical F-actin (Fig. 4 C–C′′′ and Movie S11), suggesting that the C-terminal microtubule-binding site of KHC is not involved in cortical microtubule anchorage. It should be noted that some aggregation of the SunTag antibody was observed in the posterior of KhcmutA oocytes (asterisks in Fig. 4C′). To test whether this aggregation was an artifact of the labeling system, we stained oocytes using a silicon-rhodamine–conjugated microtubule probe (SiR-tubulin) (38). We found no aggregates in either control or KhcmutA oocytes using SiR-tubulin (Fig. S3), suggesting that these aggregates are artifacts of high SunTag-MT expression. Importantly, SiR-tubulin staining demonstrated that microtubules closely associate with the cortex in both control and KhcmutA oocytes (Fig. S3, Bottom). These two experiments demonstrate that mutation of the C-terminal microtubule-binding site of KHC does not affect cortical microtubule anchorage.
Fig. S3.
SiR-tubulin staining demonstrates that KhcmutA oocytes maintain cortical microtubule arrays. (A) Microtubule organization in a representative stage 10B control oocyte stained with SiR-tubulin. (B) Microtubule organization in a representative stage 10B KhcmutA oocyte. (A and B) The x–y axis (Top) and the x–z axis (Bottom) are depicted. Note that the KhcmutA oocyte maintains cortical microtubules, like the control oocyte. Dotted yellow lines indicate the oocyte cortex at the interface between the oocyte and follicle cells. Control, n = 9; KhcmutA, n = 6.
Together, we propose that the stable, cortically anchored microtubules are tethered to the actin-enriched cortex by microtubule–actin cross-linker(s), and the anchoring signal is activated immediately before the onset of fast streaming. KHC binds to cortically anchored microtubules and nonanchored cytoplasmic microtubules simultaneously using its motor domain and C-terminal tail, and slides nonanchored microtubules against anchored microtubules. This microtubule motility and kinesin-1–based organelle transport together generate the forces required to drive oocyte cytoplasmic streaming.
Microtubule Sliding-Driven Ooplasmic Streaming Is Important for the Refinement of Posterior Determinants.
Body axes of the Drosophila embryo are predetermined during mid-oogenesis. The anterior-posterior axis is defined by bicoid mRNA localized to the anterior side and oskar (osk) mRNA localized to the posterior pole of the oocyte, respectively (39–42). The posterior localization of osk mRNA is KHC-dependent, whereas the anterior bicoid mRNA is not (7, 26). During mid- to late oogenesis, ooplasmic streaming serves as an efficient procedure to mix and distribute the mRNAs and proteins from the nurse cell cytoplasm within the oocytes. Inhibition of streaming by Khc mutants results in stratified oocytes with distinct areas of transparent nurse cell cytoplasm and dark yolk-containing ooplasm (8) (Movie S2), which are always associated with defects in osk mRNA localization (7, 26). Two models have been proposed to explain the KHC-dependent osk mRNA localization: (i) osk mRNA is transported by kinesin-1 along microtubules to the posterior pole (7, 26) or (ii) osk mRNA is circulated by kinesin-1–driven ooplasmic streaming within the oocyte and is accumulated at the posterior pole by a localized anchor (16, 43).
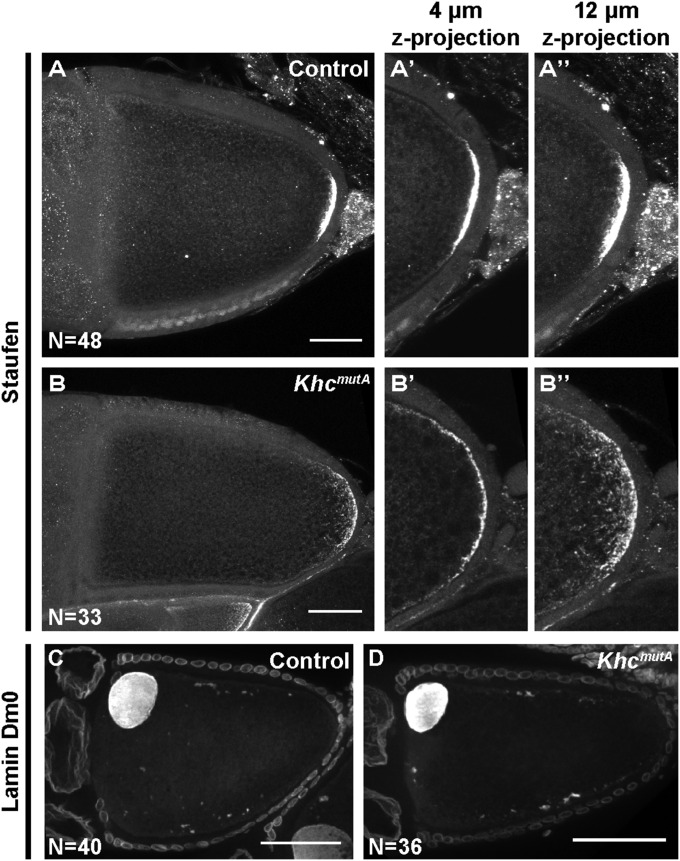
To distinguish between these two models, we decided to determine which function of KHC (organelle transport or microtubule sliding) is required for posterior determination by examining the localization of posterior determinants in KhcmutA oocytes. The protein Staufen colocalizes with osk mRNA throughout oogenesis and has served as a reliable marker of osk mRNA localization (7, 26). Therefore, we examined Staufen localization as a readout of posterior determination. In wild-type stage 10B oocytes, we observed a compact crescent of Staufen protein at the posterior pole (Fig. 5A), as previously reported (7, 26). In KhcmutA stage 10B oocytes, Staufen still localized to the posterior pole; however, the signal was more diffuse along the cortex compared with wild-type oocytes (Fig. 5B). Quantification showed that in KhcmutA oocytes, Staufen’s crescent at the cortex was significantly wider than control (mean ± SD: 37.5 ± 14.8 μm in control, 96.3 ± 30.0 μm in KhcmutA). This result suggests that both active transport along microtubules and ooplasmic streaming contribute to posterior determination. Because Staufen still localizes to the posterior pole in KhcmutA oocytes, direct transport by kinesin-1 is likely important for this initial localization. However, this experiment demonstrates that the contribution of microtubule sliding to ooplasmic streaming also serves as an important refining step in posterior determinant localization.
Fig. 5.
Kinesin-1–based microtubule sliding is important for refinement of posterior determination. Anti-Staufen staining in control (A) and KhcmutA (B) stage 10B oocytes. (A′–B′′) Zoomed-up posterior areas of (A and B): ∼4-μm z-projection (A′ and B′) and ∼12-μm z-projection (A′′ and B′′) to depict the x–y distribution (A′ and B′) as well as the z distribution (A′′ and B′′) of Staufen in control and KhcmutA stage 10B oocytes. Quantification of Staufen’s crescent length: 37.5 ± 14.8 μm in control (mean ± SD; n = 48); 96.3 ± 30.0 μm in KhcmutA (mean ± SD; n = 33); P < 0.0001, unpaired t test. The oocyte nucleus is labeled by anti-Lamin Dm0 staining in control (C) and KhcmutA (D) oocytes. No difference in nuclear positioning was observed between the two genotypes. Scale bars, 50 μm.
Besides anterior-posterior axis determination, dorsal-ventral patterning in Drosophila embryos is prespecified by the asymmetry of the oocyte (42). The oocyte nucleus becomes localized to the anterodorsal corner, and nucleus-associated gurken mRNA localization and translation define the dorsal-ventral axis by determining dorsal cell fates in lateral follicle cells (42, 44–48). KHC has been shown to be required for the proper nucleus position, and thus dorsal-ventral patterning (49), although the mechanism is not well understood. We used lamin labeling to examine oocyte nucleus position in control and KhcmutA oocytes, and found that in both samples, oocyte nuclei are tightly associated with the anterior cortex (Fig. 5 C and D; n = 40 and n = 36, respectively). We therefore conclude that kinesin-1–driven microtubule sliding is not involved in dorsal-ventral patterning.
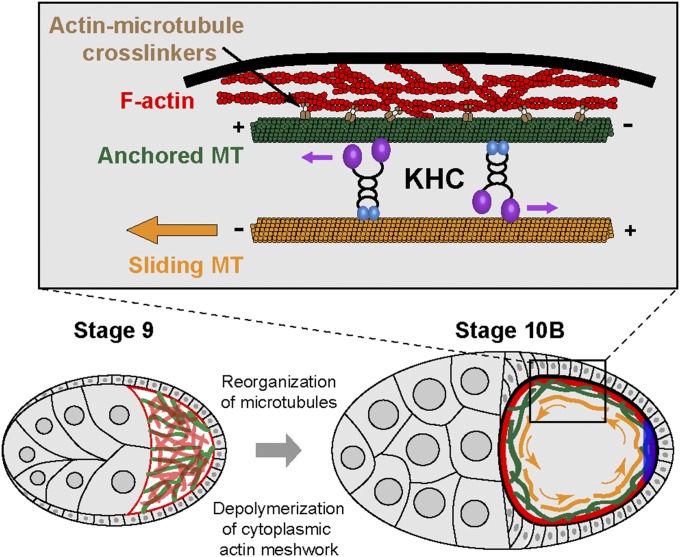
Overall, we propose that ooplasmic streaming is an essential step for refining the localization of posterior determinants. Streaming actively circulates any escaped posterior determinants along the oocyte cortex and allows them to accumulate at the posterior pole by some local anchorage (Fig. 6).
Fig. 6.
Model of kinesin-1–based microtubule sliding in ooplasmic streaming. In stage 9 oocytes, microtubules are oriented preferably with the plus-end toward the posterior and a dense cytoplasmic actin meshwork exists in the ooplasm. Between stage 9 and stage 10B, depolymerization of the cytoplasmic actin meshwork and active reorganization of microtubules occur. At stage 10B, a stable microtubule network is established at the actin cortex. We hypothesize that this stable network allows KHC to slide free cytoplasmic microtubules, which contributes to ooplasmic streaming. Simultaneously, subcortical microtubule arrays emerge (not depicted here to simplify the model), which support KHC-based organelle transport and generate ooplasmic flows. This combination of KHC-based organelle transport and microtubule sliding drives ooplasmic streaming to circulate axis determinants along the cortex, which facilitates the local anchorage for posterior determinants (blue crescent).
Discussion
Ooplasmic streaming is a vigorous cytoplasmic movement that is essential for efficient distribution of the ooplasm content and proper patterning of the oocyte. Because of its large size and complexity, understanding the mechanisms of basic developmental processes such as streaming is challenging in the oocyte. Previous studies have demonstrated that ooplasmic streaming depends on both microtubules and kinesin-1 (6–8). This streaming activity is preceded by a global redistribution of the microtubule network, from a strong concentration at the posterior pole during stage 7 to a gradient of microtubules starting at the anterior pole by stage 10A (50). At the onset of fast ooplasmic streaming in stage 10B, subcortical microtubule arrays form, which undulate with the cytoplasmic flows and eventually disappear as streaming ceases (50–52). Organelle transport by kinesin-1 along these subcortical microtubule arrays has been proposed to drive ooplasmic streaming, by exerting hydrodynamic forces on the surrounding cytoplasm (8, 52). However, surprisingly, the general kinesin-1 cargo adapter KLC and the microtubule minus-end motor dynein are dispensable for fast ooplasmic streaming (7–9). This independence of KLC and dynein is reminiscent of another function of kinesin-1, microtubule–microtubule sliding, which is also KLC- and dynein-independent (10).
Based on these observations, we wondered if microtubule sliding might play a role in ooplasmic streaming. Here, we reinvestigated the mechanism of ooplasmic streaming using recently developed molecular tools: a microtubule sliding-deficient Khc mutant (KhcmutA) and a microtubule sliding-competent chimeric motor that lacks major cargo transport domains (17). Our data show that sliding-deficient KhcmutA oocytes display a significant reduction in ooplasmic streaming and that expression of a sliding-competent chimeric motor in KHC-depleted oocytes results in a partial streaming rescue. These data demonstrate that the transport of microtubules themselves is important for the generation of cytoplasmic flows in the oocyte.
Microtubule sliding by kinesin-1 is bidirectional in both tissue culture cells and neurons (10, 12). This bidirectionality presents an apparent contradiction: How can bidirectional microtubule sliding contribute to the unidirectional flows observed in the oocyte? One possibility is that kinesin-1 slides free cytoplasmic microtubules along microtubules that are immobilized by an interaction with other cellular components. Before our work, stable immotile microtubules had not been reported in the oocyte, likely because these immotile microtubules are obscured by the much larger population of dynamic microtubules, and are therefore difficult to visualize. We used two recently developed imaging probes in our search for these immotile microtubules in the oocyte. Both techniques, photoconversion of Maple3-tagged tubulin (Fig. 2) and SunTag-MT labeling (Fig. 3), clearly demonstrated that the streaming oocyte contains a distinct population of cortical microtubules that do not move with the flowing ooplasm. These microtubules, unlike the bulk of cytoplasmic microtubules, do not lose subunits or move over the course of the experiment (over 30 min). This population of anchored stable microtubules makes them an excellent platform to slide free cytoplasmic microtubules, which can, in turn, help generate ooplasmic movement. Remarkably, this stable microtubule network appears at the actin-rich cortex in late stage 9 to stage 10A oocytes, immediately before fast streaming begins. Based on these observations, we propose that KHC-driven microtubule–microtubule sliding contributes to fast ooplasmic streaming and is required for proper posterior determination (Fig. 6). However, we found that KhcmutA oocytes display severely reduced microtubule sliding (Fig. 3) but only a partial reduction in streaming rates (Fig. 1). Additionally, microtubule sliding alone cannot fully rescue ooplasmic streaming (Fig. 1G). We conclude, based on these data, that both kinesin-1–based microtubule sliding and kinesin-1–based cargo transport drive normal ooplasmic streaming.
KHC in Ooplasmic Streaming.
Previous studies have established that KHC is required for ooplasmic streaming (7, 8). Follow-up papers addressed the role of KHC in streaming by replacing the wild-type protein with C-terminal truncations or point mutations (53, 54), and the authors concluded that the C-terminal tail of KHC is dispensable for fast streaming. Our work seems to be in apparent contradiction with their conclusions, so we reinvestigated their results. One study demonstrated that a KHC mutant with point mutations at K925A, R929A, H932A, and R936A in the C-terminal microtubule-binding site (KhcKRHR-A) is fully capable of driving ooplasmic streaming (53). However, direct biochemical analysis (18) suggests that a similar mutation in human kinesin-1 (mutant B; Fig. S1A) has a much weaker effect on microtubule affinity than the KhcmutA equivalent mutation (mutant A, Fig. S1A). Furthermore, we directly tested the ability of KHCKRHR-A to slide microtubules and found that it slides microtubules as actively as wild-type kinesin-1 in Drosophila S2 cells (Fig. S1B). Another study found that a truncated version of KHC, lacking the C-terminal tail, including the microtubule-binding site (KHC1–849), rescues fast streaming in Khc27 germline clones (54). Our analysis of this truncation in S2 cells agrees with that result, and shows local microtubule sliding in cells expressing the truncated protein. However, close examination of the truncated protein’s distribution shows that it forms aggregates in the cytoplasm and microtubule sliding is only rescued close to the aggregates. These aggregates presumably contain multiple motor domains of kinesin-1, and therefore can move microtubules in the vicinity of aggregates, accounting for localized rescue of streaming at stage 10B KHC1–849 oocytes (54).
Microtubule Organization in the Streaming Oocyte.
Microtubules are essential for ooplasmic streaming because their depolymerization completely abolishes streaming (6, 16, 51). Oocytes contain many different populations of microtubules that reorganize and change throughout different developmental stages. The microtubule network is initially polarized during early stages, with the majority of plus-ends at the posterior pole and minus-ends at the anterior pole (55, 56), but later becomes less organized, with minus-ends associating with the cortex (31, 57–61). Microtubules are also concentrated at the posterior pole during early stages, and these microtubules reorganize and relocate to the anterior pole during stages 8–10A, forming a gradient of microtubules from anterior to posterior. Next, a population of subcortical microtubules emerges in stage 10B oocytes (50, 51). These subcortical microtubules are highly dynamic and undulate and buckle with the flowing cytoplasm (8, 52). Ooplasmic streaming is thought to be driven by these subcortical microtubules, because they support long organelle transport events, which cause cytoplasmic movement, and because they appear precisely at the onset of fast streaming. Here, in addition to free cytoplasmic microtubules and the buckling subcortical microtubules, we observed a previously unidentified population of microtubules. By imaging advanced fluorescent probes live, we demonstrated that highly stable microtubules are immobilized and attached to the actin cortex. These microtubules must be different from the previously observed subcortical microtubules, because they (i) display no buckling or movement over time; (ii) do not exchange subunits with the soluble pool of tubulin, as has been demonstrated for the dynamic subcortical microtubules (52); and (iii) are physically closer to the cortex. However, like the subcortical microtubules, these newly identified stable microtubules are recruited immediately before fast streaming, suggesting a functional role in ooplasmic streaming. Furthermore, using the newly developed SunTag-MT probe (32), we were able to visualize both anchored cortical microtubules and freely moving cytoplasmic microtubules in live oocytes. The motility of these cytoplasmic microtubules was lost in microtubule sliding-deficient KhcmutA oocytes, likely resulting in the observed ooplasmic streaming defects.
Based on these data, we propose a two-step model for oocyte cytoplasmic streaming. First, the microtubule network must be reorganized in preparation for fast ooplasmic streaming. Stable, cortically anchored microtubules must form at the cortex, and subcortical microtubules involved in cargo transport must be released from the cortex (58, 59). Next, kinesin-1 slides free microtubules using the stable cortical microtubules as anchor points, whereas long kinesin-1–based cargo transport events occur along the dynamic subcortical microtubules. These combined kinesin-1 activities drive ooplasmic streaming. However, the complex relationship between free cytoplasmic microtubules, subcortical dynamic microtubules, and stable cortically anchored microtubules is unclear. We speculate that microtubule sliding by kinesin-1 may contribute to parallel alignment of subcortical microtubules, which is essential for efficient generation of cytoplasmic streams by moving organelles (52). However, microtubule sliding still no doubt contributes directly to ooplasmic streaming, because any moving cargoes including microtubules will exert hydrodynamic forces on the cytoplasm. Future studies must be conducted to more fully understand the role of microtubule sliding during oogenesis, as well as the relationship between the different populations of microtubules in the oocyte.
Dual Role of F-Actin in Streaming.
Besides microtubules, actin filaments play an important role in streaming. Two subsets of F-actin have been described in the oocyte cytoplasm during mid- to late oogenesis (stages 9–11): a transient cytoplasmic meshwork assembled at stage 9 and disassembled at stage 10B and a more stable layer of cortical actin filaments that persisted from stage 9 to stage 11 (62, 63). Actin has been thought to play an inhibitory role in ooplasmic streaming because depolymerization of F-actin by latrunculin A or cytochalasin D (64, 65) or mutations of the actin regulators chickadee, cappuccino, and spire (64–69) cause premature streaming. However, both of these drug treatments and actin regulator mutations abolish only the cytoplasmic actin meshwork in stage 9–10A oocytes, whereas the cortical actin filaments remain intact (62). We confirmed these data and found that even very high concentrations of latrunculin B (up to 100 μM) failed to decrease phalloidin staining at the cortex, suggesting that cortical actin filaments, just like immobilized cortical microtubules, are extremely stable during mid- to late oogenesis. Based on the close proximity between cortical actin and stable microtubules, and their relative distribution (Fig. 4), we propose that the actin cortex anchors the population of stable cortical microtubules, which are likely important for streaming. Therefore, actin filaments may have a dual role in streaming: The transient cytoplasmic actin meshwork physically inhibits premature streaming at early stages (stage 9 and before), whereas the stable actin cortex serves as a platform for anchored stable microtubules at later stages (stages 10B–11) (Fig. 6). We speculate that the actin meshwork at stage 9, in addition to blocking fast streaming, prevents early anchorage of cortical microtubules and formation of subcortical microtubules. Consistent with this idea, abnormal subcortical microtubule arrays were observed in stage 8–9 oocytes of cappuccino, chickadee, and spire mutants (64–66). Thus, in these stage 8–9 mutant oocytes, the absence of actin meshwork and the presence of both cortically anchored and subcortical microtubules, which support kinesin-1–based microtubule sliding and organelle transport, may induce premature streaming.
Streaming and Posterior Determinant Refinement.
In this study, we also examined the role of KHC-driven microtubule sliding in posterior determination and dorsal-ventral patterning in oocytes, both of which are dependent on kinesin-1 (26, 49). Using the sliding-deficient KhcmutA flies, we were able to determine that KHC-driven microtubule sliding is important for proper localization of posterior determinants but is dispensable for dorsal-ventral patterning. Considering the slow, uncoordinated streaming in stage 8–9 oocytes (5, 7, 8), we propose that posterior determinants are initially transported along the microtubule network and are enriched and anchored at the posterior side by a local anchor (61, 70, 71); in stage 10B–11 oocytes, fast streaming can recycle any escaped posterior determinants by moving them along the cortex and bringing them back to the posterior anchorage sites (Fig. 6). Consistent with this model, osk mRNA cortical localization is dependent on actin, but not on microtubules or KHC (58). Therefore, when ooplasmic streaming is decreased in KhcmutA flies, posterior determinants become more diffusely spread along the cortex (Fig. 5), suggesting that microtubule sliding by KHC is important for refinement of these posterior determinants.
In conclusion, we demonstrated that kinesin-1–based microtubule sliding contributes mechanical forces during oogenesis and, together with kinesin-1–based organelle transport, drives ooplasmic streaming. By using the newly characterized microtubule sliding-deficient KhcmutA and the ectopic microtubule sliding-driving chimeric motor, we demonstrated that both microtubule sliding and organelle transport by kinesin-1 contribute to ooplasmic streaming. We used advanced fluorescent probes to show that microtubule sliding occurs between immobilized cortical microtubules and motile cytoplasmic microtubules. Furthermore, we showed that streaming is important for proper localization of posterior determinants. Because the KHC motor domain and the C-terminal microtubule-binding site are both highly conserved from Drosophila to human (Fig. 1A), it would be interesting to investigate whether KHC-driven microtubule sliding helps provide mechanical force for cytoplasm movement in higher organisms.
Materials and Methods
Fly Stocks and Genetics.
Fly stocks and crosses were kept on standard cornmeal food supplemented with dry active yeast at room temperature (∼23 °C). The procedure of generating genomic knock-in of Khcwt and KhcmutA and transgenic lines of UASp-Khc-BFP, UASp-UncLZ, UASp-UncLZ-Ktail, and UASp-UncLZ-KtailmutA was described by Winding et al. (17).
To knock down KHC in the germline, a transgenic RNAi project (TRiP) line (Bloomington Stock Center no. 35409, encoding a short RNA hairpin against the Khc CDS 150–170 nt; third chromosome insertion at the attP2 site) was driven by nos-Gal4 [GAL4::VP16-nos.UTR, 3R 86B4 insertion (25)]. To rescue the KhcmutA phenotype, blue fluorescent protein (BFP)-tagged full-length KHC was driven by nos-Gal4 in KhcmutA background by crossing females of yw; KhcmutA/CyO; nos-Gal4 with males of yw; KhcmutA/CyO; UASp-Khc-BFP.
To examine mitochondria localization in KhcmutA background, females of yw; KhcmutA/CyO; nos-Gal4 were crossed with males of yw; KhcmutA/CyO; UASp-Mito-EGFP (III). To examine mitochondria localization in Khc-RNAi background, females of w; nos-Gal4 were crossed with males of w, UASp-Mito-EGFP (X); Khc-RNAi (III).
To examine the rescue of Unc104 chimeric motors, females of w; nos-Gal4 were crossed with the following males: (i) w; Khc-RNAi (III); (ii) w; UASp-UncLZ (II); Khc-RNAi (III); (iii) w; UASp-UncLZ-Ktail (II); Khc-RNAi (III); and (iv) w; UASp-UncLZ-KtailmutA (II); Khc-RNAi (III).
The photoconvertible Maple3 (29) was introduced into the pUASp vector as a tandem dimer by SpeI/EcoRI and EcoRI/NotI, and Drosophila α-tubulin 84B was inserted between NotI and XbaI. The pUASp-tdMaple3-αtub construct was then sent to BestGene for standard P-element–mediated germline transformation to generate transgenic flies. A third chromosome insertion was used in this study, driven by germline-specific nos-Gal4.
Anti-GCN4 single-chain Fv antibody fragments (scFv) tagged with superfolder GFP (sfGFP) and GB1 (32) were introduced into pUASp vector by KpnI/NotI (for short, UASp-scFv). Human kinesin-1 truncation (residues 1–560) carrying a rigor mutation (E236A) tagged with 24 repeats of the GCN4 peptide (32) was introduced into pUASp vector by SpeI/XbaI (for short, UASp-K560RigorE236A-SunTag). The constructs of UASp-scFv and UASp-K560RigorE236A-SunTag were then sent to BestGene for standard P-element–mediated germline transformation to generate transgenic flies. A second chromosome insertion of UASp-scFv and a third chromosome insertion of UASp-K560RigorE236A-SunTag were combined using standard double-balancing techniques and then driven by germline-specific nos-Gal4. To examine the SunTag-MT in KhcmutA background, females of yw; KhcmutA/CyO; nos-Gal4 were crossed with males of yw; KhcmutA/CyO; UASp-scFv, UASp-K560RigorE236A-SunTag (selected from a standard chromosome recombination between a third chromosome insertion of UASp-scFv and a third chromosome insertion of UASp-K560RigorE236A-SunTag).
Imaging of Ooplasmic Streaming.
Young mated female adults were fattened on dry active yeast for ∼48–72 h and dissected in Halocarbon oil 700 (Sigma). Dissection and mounting were done as previously described (72): Dissected ovaries were teased apart using 27-gauge × ½-inch syringe needles (BD PrecisionGlide Needles) in a Halocarbon oil drop on a 22-mm2 coverslip and mounted onto a glass slide with two additional 22-mm2 coverslips serving as spacers attached by silicon grease. Samples were imaged within 30 min after dissection on a Nikon Eclipse U2000 inverted stand with a 20× 0.75-N.A. lens. DIC images were acquired using Coolsnap CCD (Roger Scientific) every 2 s for 2 min, controlled by Nikon Elements software.
Visualization of Microtubules in Live Oocytes.
Ovary dissection and mounting were performed as described above. Samples were imaged immediately after dissection on a Nikon Eclipse U2000 inverted stand with a Yokogawa CSU10 spinning disk confocal head using Evolve EMCCD (Photometrics), controlled by Nikon Elements software. Photoconversion of tdMaple3-tagged microtubules was performed using 405-nm illumination from a Heliophor 89 North light source and confined by an adjustable pinhole (projected as an ∼30-μm-diameter circle in the sample plane) for 15 s. Z-sections were taken using a 100× 1.45-N.A. lens at certain time points after photoconversion with 0.5 μm per step. Time-lapse images of SunTag-MT samples were acquired with a 40× 1.30-N.A. lens at two different focal planes that are 5–10 μm apart (at the cortex and underneath the cortex, respectively). Images were analyzed in Fiji and DiaTrack 3.04 Pro (73, 74), with a maximum particle jump distance of 3.75 μm/s. Velocities were obtained using Analysis > Distribution of Speed (Track-Based) in DiaTrack 3.04 Pro. We then plotted these values in GraphPad Prism 5 and assembled them in Illustrator CS4.
Immunostaining of Drosophila Oocytes.
For Staufen and Lamin Dm0 staining, a standard fixation and staining protocol was used (75): Samples were dissected in 1× PBS and fixed in 4% (vol/vol) formaldehyde (EM grade, methanol-free) diluted in PBT (1× PBS + 0.1% Triton X-100), washed with PBT five times for 10 min each time, and blocked in 5% (vol/vol) normal goat serum-containing PBT for 1 h. Samples were then incubated with primary antibody at 4 °C overnight, washed with PBT five times for 10 min each time, incubated with secondary antibody at 4 °C overnight, and washed with PBT five times for 10 min each time before mounting. The following primary and secondary antibodies were used in this study: mouse anti-Staufen (1:50; a kind gift from Chris Q. Doe, University of Oregon, Eugene, OR), mouse anti-Lamin Dm0 (1:5, ADL84.12; Developmental Studies Hybridoma Bank, University of Iowa) (75, 76), and goat anti-mouse TRITC and goat anti-mouse FITC (1:100, both from Jackson ImmunoResearch). For mitochondrial labeling, samples were dissected in 1× PBS and fixed in 4% (vol/vol) formaldehyde (EM grade, methanol-free) diluted in PBT, washed with PBT five times for 10 min each time, stained with TRITC-conjugated phalloidin (1:50,000; Life Technologies) for 5 min, and mounted for imaging. For dual labeling of microtubules by SunTag and F-actin by phalloidin staining, a slightly modified protocol was used (50): nos>SunTag-MT or KhcmutA; nos>SunTag-MT samples were dissected in 1× PBS, fixed with 8% (vol/vol) formaldehyde (EM grade, methanol-free) diluted in 100 mM sodium cacodylate (pH 7.2), 100 mM sucrose, 40 mM potassium acetate, 10 mM sodium acetate, 10 mM Na3EGTA, 10 mM MgCl2 for 20 min, extracted with 1% Triton X-100 in 1× PBS for 1–1.5 h, washed with PBT five time for 10 min each time, and stained with TRITC-conjugated phalloidin (1:500; Life Technologies) for 10 min before mounting for imaging.
Dual labeling of SunTag-MT and phalloidin and Staufen staining were acquired on a Nikon A1-plus microscope at Northwestern University’s Nikon Imaging Center, with an S Fluor 40× 1.30-N.A. oil lens using Galvano scanning and DU4 Detector DU4 GaAsP CH2/3, controlled by Nikon Elements software. Lamin Dm0 staining and Mito-EGFP samples were imaged on a Nikon Eclipse U2000 inverted stand with a Yokogawa CSU10 spinning disk confocal head and 40× 1.30-N.A. lens and a 100× 1.45-N.A. lens, respectively. Images were acquired using Evolve EMCCD (Photometrics), controlled by Nikon Elements software.
Protein Sample Preparation for Western Blot.
Young mated female adults were fattened for ∼48–72 h and dissected in 1× dissecting saline [9.9 mM Hepes (pH 7.5), 137 mM NaCl, 5.4 mM KCl, 0.17 mM NaH2PO4, 0.22 mM KH2PO4, 3.3 mM glucose, 43.8 mM sucrose]. Ovaries were placed in lysis buffer on ice [50 mM Tris⋅HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 2 mM MgCl2, 1 mM PMSF, 1× CLP (10 μg/mL chymostatin, leupeptin, pepstatin A), 0.5% Triton X-100] and mechanically homogenized using pellet pestles in matching microtubes (74). Ovary homogenates were spun for 10 min at >20,000 × g at 4 °C. The supernatant was isolated, and bicinchoninic acid (BCA) assays were performed to determine protein concentration. Laemmli sample buffer [5×: 5% (vol/vol) SDS, 0.1 M Tris⋅Cl (pH 6.8), 1% β-mercaptoethanol, 50% (vol/vol) glycerol, 0.25% bromophenol blue] was added to ovary protein extracts, and samples were boiled for 5 min. Standard Western blotting techniques were used to probe extracts for kinesin-1 using a polyclonal antibody against the motor domain (HD) (77). Western blots were quantified using the Gel Analyzer ImageJ plug-in.
Quantification of Ooplasmic Streaming Rate.
Analysis was performed blindly using a custom Fiji plug-in to replace the names of streaming time-lapses with random numbers. Kymographs were generated and analyzed using the Multi Kymograph plug-in for Fiji (imagej.net/Multi_Kymograph). Two kymographs were generated on either side of each oocyte, parallel to the flow of the streaming ooplasm. The velocities of eight particles were quantified per oocyte and averaged to determine the streaming rate, a strategy used in a previous study (8). If an oocyte contained local or inconsistent streaming, kymographs were still generated on either side of the oocyte along active flows. If one side of the oocyte did not contain a flow, a kymograph was generated along the length of the cortex (where flows are typically seen in fast-streaming oocytes). Therefore, an oocyte that contains a local flow of wild-type velocity and other regions with little streaming would result in an average streaming velocity lower than an oocyte with true wild-type flows.
Quantification of Staufen Crescent Length.
Analysis was performed blindly using a custom Fiji plug-in to replace the names of micrographs with random numbers. The length of the Staufen posterior crescent was quantified using the segmented line tool in Fiji/ImageJ.
Staining Oocytes with SiR-Tubulin.
Ovaries were incubated with 20 μM SiR-tubulin (Cytoskeleton) (38) and 100 μΜ verapamil for 60 min in 100 μL of Schneider’s medium. Ovaries were teased apart in medium and mounted onto a glass slide. Samples were imaged immediately after dissection on a Nikon Eclipse U2000 inverted stand with a Yokogawa CSU10 spinning disk confocal head using Evolve EMCCD (Photometrics) and a 640-nm laser, controlled by Nikon Elements software. Z-sections were taken using a 40× 1.30-N.A. lens with 0.5 μm per step.
Supplementary Material
Acknowledgments
We thank Dr. Edwin L. Ferguson (The University of Chicago), Dr. Allan C. Spradling (Howard Hughes Medical Institute, Carnegie Institution), Dr. William M. Saxton (University of California, Santa Cruz), and the Bloomington Stock Center (NIH Grant P40OD018537) for fly stocks; Dr. Ronald D. Vale (University of California, San Francisco), Dr. Marvin E. Tanenbaum (Hubrecht Institute, Utrecht), and Dr. Xiaowei Zhuang (Harvard University) for DNA plasmid constructs; and Dr. Chris Q. Doe (University of Oregon) for antibodies used in this study. The monoclonal anti-Lamin Dm0 antibody developed by Dr. Paul A. Fisher (SUNY at Stony Brook) was obtained from the Developmental Studies Hybridoma Bank, created by the National Institute of Child Health and Human Development of the NIH and maintained at The University of Iowa. Our special thanks go to Dr. Joshua Z. Rappoport of the Center for Advanced Microscopy/Nikon Imaging Center at Northwestern University for assistance with microscopy and to Dr. Anthony P. Mahowald (The University of Chicago) for critical reading of the manuscript. We also thank all of the V.I.G. laboratory members for support, discussion, and suggestions. Research reported here was supported by the National Institute of General Medical Science under Award R01GM052111.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522424113/-/DCSupplemental.
References
- 1.Ling SC, Fahrner PS, Greenough WT, Gelfand VI. Transport of Drosophila fragile X mental retardation protein-containing ribonucleoprotein granules by kinesin-1 and cytoplasmic dynein. Proc Natl Acad Sci USA. 2004;101(50):17428–17433. doi: 10.1073/pnas.0408114101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ally S, Larson AG, Barlan K, Rice SE, Gelfand VI. Opposite-polarity motors activate one another to trigger cargo transport in live cells. J Cell Biol. 2009;187(7):1071–1082. doi: 10.1083/jcb.200908075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pilling AD, Horiuchi D, Lively CM, Saxton WM. Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol Biol Cell. 2006;17(4):2057–2068. doi: 10.1091/mbc.E05-06-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006;173(4):545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gutzeit HO, Koppa R. Time-lapse film analysis of cytoplasmic streaming during late oogenesis of Drosophila. J Embryol Exp Morphol. 1982;67:101–111. [Google Scholar]
- 6.Gutzeit H. The role of microtubules in the differentiation of ovarian follicles during vitellogenesis in Drosophila. Rouxs Arch Dev Biol. 1986;195(3):173–181. doi: 10.1007/BF02439435. [DOI] [PubMed] [Google Scholar]
- 7.Palacios IM, St Johnston D. Kinesin light chain-independent function of the Kinesin heavy chain in cytoplasmic streaming and posterior localisation in the Drosophila oocyte. Development. 2002;129(23):5473–5485. doi: 10.1242/dev.00119. [DOI] [PubMed] [Google Scholar]
- 8.Serbus LR, Cha BJ, Theurkauf WE, Saxton WM. Dynein and the actin cytoskeleton control kinesin-driven cytoplasmic streaming in Drosophila oocytes. Development. 2005;132(16):3743–3752. doi: 10.1242/dev.01956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mische S, Li MG, Serr M, Hays TS. Direct observation of regulated ribonucleoprotein transport across the nurse cell/oocyte boundary. Mol Biol Cell. 2007;18(6):2254–2263. doi: 10.1091/mbc.E06-10-0959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jolly AL, et al. Kinesin-1 heavy chain mediates microtubule sliding to drive changes in cell shape. Proc Natl Acad Sci USA. 2010;107(27):12151–12156. doi: 10.1073/pnas.1004736107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barlan K, Lu W, Gelfand VI. The microtubule-binding protein ensconsin is an essential cofactor of kinesin-1. Curr Biol. 2013;23(4):317–322. doi: 10.1016/j.cub.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu W, Fox P, Lakonishok M, Davidson MW, Gelfand VI. Initial neurite outgrowth in Drosophila neurons is driven by kinesin-powered microtubule sliding. Curr Biol. 2013;23(11):1018–1023. doi: 10.1016/j.cub.2013.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.del Castillo U, Lu W, Winding M, Lakonishok M, Gelfand VI. Pavarotti/MKLP1 regulates microtubule sliding and neurite outgrowth in Drosophila neurons. Curr Biol. 2015;25(2):200–205. doi: 10.1016/j.cub.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu W, Lakonishok M, Gelfand VI. Kinesin-1-powered microtubule sliding initiates axonal regeneration in Drosophila cultured neurons. Mol Biol Cell. 2015;26(7):1296–1307. doi: 10.1091/mbc.E14-10-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.del Castillo U, Winding M, Lu W, Gelfand VI. Interplay between kinesin-1 and cortical dynein during axonal outgrowth and microtubule organization in Drosophila neurons. eLife. 2015;4:e10140. doi: 10.7554/eLife.10140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glotzer JB, Saffrich R, Glotzer M, Ephrussi A. Cytoplasmic flows localize injected oskar RNA in Drosophila oocytes. Curr Biol. 1997;7(5):326–337. doi: 10.1016/s0960-9822(06)00156-4. [DOI] [PubMed] [Google Scholar]
- 17.Winding M, Kelliher MT, Lu W, Wildonger J, Gelfand VI. Role of kinesin-1–based microtubule sliding in Drosophila nervous system development. Proc Natl Acad Sci USA. 2016;113:E4985–E4994. doi: 10.1073/pnas.1522416113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seeger MA, Rice SE. Microtubule-associated protein-like binding of the kinesin-1 tail to microtubules. J Biol Chem. 2010;285(11):8155–8162. doi: 10.1074/jbc.M109.068247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hackney DD, Stock MF. Kinesin’s IAK tail domain inhibits initial microtubule-stimulated ADP release. Nat Cell Biol. 2000;2(5):257–260. doi: 10.1038/35010525. [DOI] [PubMed] [Google Scholar]
- 20.Yan J, et al. Kinesin-1 regulates dendrite microtubule polarity in Caenorhabditis elegans. eLife. 2013;2:e00133. doi: 10.7554/eLife.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cox RT, Spradling AC. A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development. 2003;130(8):1579–1590. doi: 10.1242/dev.00365. [DOI] [PubMed] [Google Scholar]
- 22.Cox RT, Spradling AC. Milton controls the early acquisition of mitochondria by Drosophila oocytes. Development. 2006;133(17):3371–3377. doi: 10.1242/dev.02514. [DOI] [PubMed] [Google Scholar]
- 23.Horne-Badovinac S, Bilder D. Mass transit: Epithelial morphogenesis in the Drosophila egg chamber. Dev Dyn. 2005;232(3):559–574. doi: 10.1002/dvdy.20286. [DOI] [PubMed] [Google Scholar]
- 24.Robinson DN, Cooley L. Genetic analysis of the actin cytoskeleton in the Drosophila ovary. Annu Rev Cell Dev Biol. 1997;13:147–170. doi: 10.1146/annurev.cellbio.13.1.147. [DOI] [PubMed] [Google Scholar]
- 25.Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8(4):243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- 26.Brendza RP, Serbus LR, Duffy JB, Saxton WM. A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science. 2000;289(5487):2120–2122. doi: 10.1126/science.289.5487.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomishige M, Klopfenstein DR, Vale RD. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 2002;297(5590):2263–2267. doi: 10.1126/science.1073386. [DOI] [PubMed] [Google Scholar]
- 28.Schnitzer MJ, Block SM. Kinesin hydrolyses one ATP per 8-nm step. Nature. 1997;388(6640):386–390. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- 29.Wang S, Moffitt JR, Dempsey GT, Xie XS, Zhuang X. Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proc Natl Acad Sci USA. 2014;111(23):8452–8457. doi: 10.1073/pnas.1406593111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robert A, Rossow MJ, Hookway C, Adam SA, Gelfand VI. Vimentin filament precursors exchange subunits in an ATP-dependent manner. Proc Natl Acad Sci USA. 2015;112(27):E3505–E3514. doi: 10.1073/pnas.1505303112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parton RM, et al. A PAR-1-dependent orientation gradient of dynamic microtubules directs posterior cargo transport in the Drosophila oocyte. J Cell Biol. 2011;194(1):121–135. doi: 10.1083/jcb.201103160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159(3):635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice S, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402(6763):778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 34.Blasius TL, Cai D, Jih GT, Toret CP, Verhey KJ. Two binding partners cooperate to activate the molecular motor Kinesin-1. J Cell Biol. 2007;176(1):11–17. doi: 10.1083/jcb.200605099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sung HH, et al. Drosophila ensconsin promotes productive recruitment of Kinesin-1 to microtubules. Dev Cell. 2008;15(6):866–876. doi: 10.1016/j.devcel.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Loiseau P, Davies T, Williams LS, Mishima M, Palacios IM. Drosophila PAT1 is required for Kinesin-1 to transport cargo and to maximize its motility. Development. 2010;137(16):2763–2772. doi: 10.1242/dev.048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Babu K, Cai Y, Bahri S, Yang X, Chia W. Roles of Bifocal, Homer, and F-actin in anchoring Oskar to the posterior cortex of Drosophila oocytes. Genes Dev. 2004;18(2):138–143. doi: 10.1101/gad.282604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lukinavičius G, et al. Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat Methods. 2014;11(7):731–733. doi: 10.1038/nmeth.2972. [DOI] [PubMed] [Google Scholar]
- 39.Lehmann R, Nüsslein-Volhard C. Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in Drosophila. Cell. 1986;47(1):141–152. doi: 10.1016/0092-8674(86)90375-2. [DOI] [PubMed] [Google Scholar]
- 40.Ephrussi A, Dickinson LK, Lehmann R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66(1):37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- 41.Berleth T, et al. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 1988;7(6):1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Eeden F, St Johnston D. The polarisation of the anterior-posterior and dorsal-ventral axes during Drosophila oogenesis. Curr Opin Genet Dev. 1999;9(4):396–404. doi: 10.1016/S0959-437X(99)80060-4. [DOI] [PubMed] [Google Scholar]
- 43.Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol. 2003;13(14):1159–1168. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 44.Neuman-Silberberg FS, Schüpbach T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell. 1993;75(1):165–174. [PubMed] [Google Scholar]
- 45.Neuman-Silberberg FS, Schupbach T. Dorsoventral axis formation in Drosophila depends on the correct dosage of the gene gurken. Development. 1994;120(9):2457–2463. doi: 10.1242/dev.120.9.2457. [DOI] [PubMed] [Google Scholar]
- 46.Neuman-Silberberg FS, Schüpbach T. The Drosophila TGF-alpha-like protein Gurken: Expression and cellular localization during Drosophila oogenesis. Mech Dev. 1996;59(2):105–113. doi: 10.1016/0925-4773(96)00567-9. [DOI] [PubMed] [Google Scholar]
- 47.Van Buskirk C, Schüpbach T. Versatility in signalling: Multiple responses to EGF receptor activation during Drosophila oogenesis. Trends Cell Biol. 1999;9(1):1–4. doi: 10.1016/s0962-8924(98)01413-5. [DOI] [PubMed] [Google Scholar]
- 48.Jaramillo AM, Weil TT, Goodhouse J, Gavis ER, Schupbach T. The dynamics of fluorescently labeled endogenous gurken mRNA in Drosophila. J Cell Sci. 2008;121(Pt 6):887–894. doi: 10.1242/jcs.019091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brendza RP, Serbus LR, Saxton WM, Duffy JB. Posterior localization of dynein and dorsal-ventral axis formation depend on kinesin in Drosophila oocytes. Curr Biol. 2002;12(17):1541–1545. doi: 10.1016/s0960-9822(02)01108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Theurkauf WE, Smiley S, Wong ML, Alberts BM. Reorganization of the cytoskeleton during Drosophila oogenesis: Implications for axis specification and intercellular transport. Development. 1992;115(4):923–936. doi: 10.1242/dev.115.4.923. [DOI] [PubMed] [Google Scholar]
- 51.Theurkauf WE. Microtubules and cytoplasm organization during Drosophila oogenesis. Dev Biol. 1994;165(2):352–360. doi: 10.1006/dbio.1994.1258. [DOI] [PubMed] [Google Scholar]
- 52.Monteith CE, et al. A mechanism for cytoplasmic streaming: Kinesin-driven alignment of microtubules and fast fluid flows. Biophys J. 2016;110(9):2053–2065. doi: 10.1016/j.bpj.2016.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moua P, Fullerton D, Serbus LR, Warrior R, Saxton WM. Kinesin-1 tail autoregulation and microtubule-binding regions function in saltatory transport but not ooplasmic streaming. Development. 2011;138(6):1087–1092. doi: 10.1242/dev.048645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams LS, Ganguly S, Loiseau P, Ng BF, Palacios IM. The auto-inhibitory domain and ATP-independent microtubule-binding region of Kinesin heavy chain are major functional domains for transport in the Drosophila germline. Development. 2014;141(1):176–186. doi: 10.1242/dev.097592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clark I, Giniger E, Ruohola-Baker H, Jan LY, Jan YN. Transient posterior localization of a kinesin fusion protein reflects anteroposterior polarity of the Drosophila oocyte. Curr Biol. 1994;4(4):289–300. doi: 10.1016/s0960-9822(00)00068-3. [DOI] [PubMed] [Google Scholar]
- 56.Clark IE, Jan LY, Jan YN. Reciprocal localization of Nod and kinesin fusion proteins indicates microtubule polarity in the Drosophila oocyte, epithelium, neuron and muscle. Development. 1997;124(2):461–470. doi: 10.1242/dev.124.2.461. [DOI] [PubMed] [Google Scholar]
- 57.Cha BJ, Koppetsch BS, Theurkauf WE. In vivo analysis of Drosophila bicoid mRNA localization reveals a novel microtubule-dependent axis specification pathway. Cell. 2001;106(1):35–46. doi: 10.1016/s0092-8674(01)00419-6. [DOI] [PubMed] [Google Scholar]
- 58.Cha BJ, Serbus LR, Koppetsch BS, Theurkauf WE. Kinesin I-dependent cortical exclusion restricts pole plasm to the oocyte posterior. Nat Cell Biol. 2002;4(8):592–598. doi: 10.1038/ncb832. [DOI] [PubMed] [Google Scholar]
- 59.Wang Y, Riechmann V. Microtubule anchoring by cortical actin bundles prevents streaming of the oocyte cytoplasm. Mech Dev. 2008;125(1-2):142–152. doi: 10.1016/j.mod.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 60.Sanghavi P, Lu S, Gonsalvez GB. A functional link between localized Oskar, dynamic microtubules, and endocytosis. Dev Biol. 2012;367(1):66–77. doi: 10.1016/j.ydbio.2012.04.024. [DOI] [PubMed] [Google Scholar]
- 61.Zimyanin VL, et al. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 2008;134(5):843–853. doi: 10.1016/j.cell.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dahlgaard K, Raposo AA, Niccoli T, St Johnston D. Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev Cell. 2007;13(4):539–553. doi: 10.1016/j.devcel.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quinlan ME. Direct interaction between two actin nucleators is required in Drosophila oogenesis. Development. 2013;140(21):4417–4425. doi: 10.1242/dev.097337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emmons S, et al. Cappuccino, a Drosophila maternal effect gene required for polarity of the egg and embryo, is related to the vertebrate limb deformity locus. Genes Dev. 1995;9(20):2482–2494. doi: 10.1101/gad.9.20.2482. [DOI] [PubMed] [Google Scholar]
- 65.Manseau L, Calley J, Phan H. Profilin is required for posterior patterning of the Drosophila oocyte. Development. 1996;122(7):2109–2116. doi: 10.1242/dev.122.7.2109. [DOI] [PubMed] [Google Scholar]
- 66.Theurkauf WE. Premature microtubule-dependent cytoplasmic streaming in cappuccino and spire mutant oocytes. Science. 1994;265(5181):2093–2096. doi: 10.1126/science.8091233. [DOI] [PubMed] [Google Scholar]
- 67.Verheyen EM, Cooley L. Profilin mutations disrupt multiple actin-dependent processes during Drosophila development. Development. 1994;120(4):717–728. doi: 10.1242/dev.120.4.717. [DOI] [PubMed] [Google Scholar]
- 68.Cooley L, Verheyen E, Ayers K. Chickadee encodes a profilin required for intercellular cytoplasm transport during Drosophila oogenesis. Cell. 1992;69(1):173–184. doi: 10.1016/0092-8674(92)90128-y. [DOI] [PubMed] [Google Scholar]
- 69.Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433(7024):382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- 70.Khuc Trong P, Doerflinger H, Dunkel J, St Johnston D, Goldstein RE. Cortical microtubule nucleation can organise the cytoskeleton of Drosophila oocytes to define the anteroposterior axis. eLife. 2015;4:e06088. doi: 10.7554/eLife.06088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nashchekin D, Fernandes AR, St. Johnston D. Patronin/Shot cortical foci assemble the noncentrosomal microtubule array that specifies the Drosophila anterior-posterior axis. Dev Cell. 2016;38(1):61–72. doi: 10.1016/j.devcel.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pokrywka NJ. Live imaging of GFP-labeled proteins in Drosophila oocytes. J Vis Exp. 2013;March 2013(73):e50044. doi: 10.3791/50044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vallotton P, Olivier S. Tri-track: Free software for large-scale particle tracking. Microsc Microanal. 2013;19(2):451–460. doi: 10.1017/S1431927612014328. [DOI] [PubMed] [Google Scholar]
- 74.Lu W, Del Castillo U, Gelfand VI. Organelle transport in cultured Drosophila cells: S2 cell line and primary neurons. J Vis Exp. 2013;(81):e50838. doi: 10.3791/50838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu W, et al. Niche-associated activation of rac promotes the asymmetric division of Drosophila female germline stem cells. PLoS Biol. 2012;10(7):e1001357. doi: 10.1371/journal.pbio.1001357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stuurman N, Maus N, Fisher PA. Interphase phosphorylation of the Drosophila nuclear lamin: Site-mapping using a monoclonal antibody. J Cell Sci. 1995;108(Pt 9):3137–3144. doi: 10.1242/jcs.108.9.3137. [DOI] [PubMed] [Google Scholar]
- 77.Rodionov VI, et al. Microtubule-dependent control of cell shape and pseudopodial activity is inhibited by the antibody to kinesin motor domain. J Cell Biol. 1993;123(6 Pt 2):1811–1820. doi: 10.1083/jcb.123.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.