Significance
This work provides direct evidence for the formation of an M-cluster on the assembly scaffold NifEN, establishing NifEN as the second known protein that houses a nitrogenase cofactor. A “half-on, half-off” scheme of cofactor biosynthesis can be proposed based on the outcome of this study, which suggests an asymmetric nature of the assembly sites in the seemingly equivalent αβ-halves of NifEN and a coordination of various biosynthetic events via a unique conformational switch on/off mechanism. The comparable substrate-reducing capabilities of NifEN and NifDK establish the former as a structural and functional homolog of the latter, providing a proof-of-concept for the feasibility of probing key catalytic features of NifDK via reconstruction of a NifDK equivalent on the basis of a “simplified” template, NifEN.
Keywords: nitrogenase, catalysis, assembly, functional homolog, NifEN
Abstract
NifEN is a biosynthetic scaffold for the cofactor of Mo-nitrogenase (designated the M-cluster). Previous studies have revealed the sequence and structural homology between NifEN and NifDK, the catalytic component of nitrogenase. However, direct proof for the functional homology between the two proteins has remained elusive. Here we show that, upon maturation of a cofactor precursor (designated the L-cluster) on NifEN, the cluster species extracted from NifEN is spectroscopically equivalent and functionally interchangeable with the native M-cluster extracted from NifDK. Both extracted clusters display nearly indistinguishable EPR features, X-ray absorption spectroscopy/extended X-ray absorption fine structure (XAS/EXAFS) spectra and reconstitution activities, firmly establishing the M-cluster–bound NifEN (designated NifENM) as the only protein other than NifDK to house the unique nitrogenase cofactor. Iron chelation experiments demonstrate a relocation of the cluster from the surface to its binding site within NifENM upon maturation, which parallels the insertion of M-cluster into an analogous binding site in NifDK, whereas metal analyses suggest an asymmetric conformation of NifENM with an M-cluster in one αβ-half and an empty cluster-binding site in the other αβ-half, which led to the proposal of a stepwise assembly mechanism of the M-cluster in the two αβ-dimers of NifEN. Perhaps most importantly, NifENM displays comparable ATP-independent substrate-reducing profiles to those of NifDK, which establishes the M-cluster–bound αβ-dimer of NifENM as a structural and functional mimic of one catalytic αβ-half of NifDK while suggesting the potential of this protein as a useful tool for further investigations of the mechanistic details of nitrogenase.
Nitrogenase plays a key role in global nitrogen cycle through its unique ability to reduce nitrogen to ammonia under ambient conditions. The best characterized, Mo-dependent nitrogenase uses a two-component system for substrate reduction, in which a specific reductase (designated the Fe protein) serves as the obligate electron donor for a catalytic component (designated the MoFe protein) during catalysis (1). The Fe protein (or NifH) is a γ2-dimer, which contains a [Fe4S4] cluster between the two subunits and an ATP-binding site within each subunit, whereas the MoFe protein (or NifDK) is an α2β2-tetramer, which contains a P-cluster ([Fe8S7]) between each α/β-subunit interface and an M-cluster (also termed the cofactor; [MoFe7S9C-homocitrate]) within each α-subunit (2–4). Catalysis of nitrogenase involves formation of a functional complex between the two component proteins of the Mo-nitrogenase (5), which permits ATP-dependent transfer of electrons from the [Fe4S4] cluster of NifH, via the P-cluster, to the M-cluster of NifDK and the subsequent reduction of substrate at the M-cluster site upon accumulation of a sufficient amount of electrons.
Arguably the most complex metallocluster identified to date, the M-cluster is assembled through the actions of a number of nif (nitrogen fixation) gene-encoded proteins. The final step of this process involves the maturation of a precursor of M-cluster on a scaffold protein, NifEN, followed by transfer of a fully assembled M-cluster to its final binding site in NifDK (6). Earlier genetic analyses revealed a significant degree of homology between the primary sequences of NifEN and NifDK, leading to the hypothesis that NifEN contains cluster-binding sites analogous to the P- and M-cluster sites in NifDK. Biochemical, EPR, XAS/EXAFS, X-ray emission spectroscopy (XES), and crystallographic studies provided strong support for this hypothesis, showing the presence of (i) a [Fe4S4] cluster at the α/β-subunit interface of NifEN, which can be regarded as an analog to P-cluster; and (ii) a [Fe8S9C] cluster on the α-subunit of NifEN, which represents an all-Fe precursor to M-cluster (7–10). This [Fe8S9C] precursor closely resembles the metal-sulfur core of M-cluster and, following the ATP-dependent, NifH-mediated insertion of Mo and homocitrate, it can be matured on NifEN and subsequently transferred to apo-NifDK, resulting in the formation of holo-NifDK (11–13).
The fact that the cluster matured on NifEN can be directly used for the reconstitution and activation of apo-NifDK suggests that it is a fully assembled form of M-cluster. Indeed, Mo K-edge XAS/EXAFS analyses of NifEN- and NifDK-bound M-cluster species demonstrated a considerable degree of homology between the bond distances (e.g., Mo-Fe and Mo-S) and the oxidation states (i.e., Mo) of the two clusters; however, they also revealed an asymmetric ligation of Mo and a loose Mo-O binding pattern in the NifEN-bound M-cluster compared with those in the NifDK-bound M-cluster, which likely originate from a difference in the protein environments of the M-clusters in NifEN and NifDK (13). A direct comparison between the two M-cluster species can only be achieved upon removal of the interfering protein environment, which will provide the definitive proof for the presence of a native cofactor on NifEN while suggesting this protein as a potential functional homolog of nitrogenase.
Here we show that the M-cluster extracted from NifEN is spectroscopically equivalent and functionally interchangeable with the native M-cluster extracted from NifDK. Our data indicate a stepwise assembly mechanism of the M-clusters in the two αβ-dimers of NifEN and demonstrate the capacity of NifEN as a structural and functional mimic of NifDK, suggesting the potential of this protein as a useful tool for comparative investigations of nitrognease mechanism.
Results and Discussion
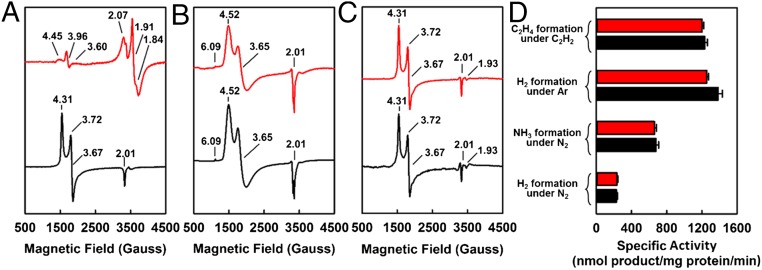
Using the same procedure as that for the extraction of the NifDK-bound M-cluster (14), the M-cluster was successfully extracted from NifEN into N-methylformamide (NMF), an organic solvent. Contrary to their respective protein bound counterparts (Fig. 1A, red and black), in the presence of 1,4-benzenedithiol, the M-clusters extracted from NifEN (designated M-clusterNifEN) and NifDK (designated M-clusterNifDK) display EPR features that are identical to each other, both showing a characteristic S = 3/2 signal at g = 4.52, 3.65, and 2.01, as well as an additional feature at g = 6.09 (Fig. 1B, red and black). The S = 3/2 signals of both solvent-extracted M-cluster species closely resemble a broadened S = 3/2 signal of the NifDK-bound M-cluster (Fig. 1A, black), whereas the g = 6.09 feature is believed to originate from the interactions between the M-cluster and the thiol groups in the solution (14). Incorporation of solvent-extracted M-clusterNifEN and M-clusterNifDK, respectively, into the apo-NifDK protein results in the formation of reconstituted NifDK proteins that display EPR features (Fig. 1C, red and black) and activities (Fig. 1D, red and black) that are nearly indistinguishable from each other. Together, these observations demonstrate that the M-cluster species extracted from NifEN and NifDK are spectroscopically equivalent and functionally interchangeable with each other.
Fig. 1.
EPR properties and reconstitution activities of M-clusters extracted from NifEN and NifDK. Shown are the EPR spectra of the M-clusterNifEN (red) and M-clusterNifDK (black) in the native protein environments (A), upon extraction into NMF and addition of 10 mM 1,4-benzenedithiol (B), or after incorporation into apo-NifDK (C), and the activities of NifDK upon reconstitution with the extracted M-clusterNifEN (red) and M-clusterNifDK (black) (D). All cluster and protein samples contained equimolar Mo, and all spectra were measured at 6 K. The g values are indicated. The S = 1/2 signal of NifENM (A, red) originates from the permanent [Fe4S4] clusters at the αβ-subunit interface of this protein (8).
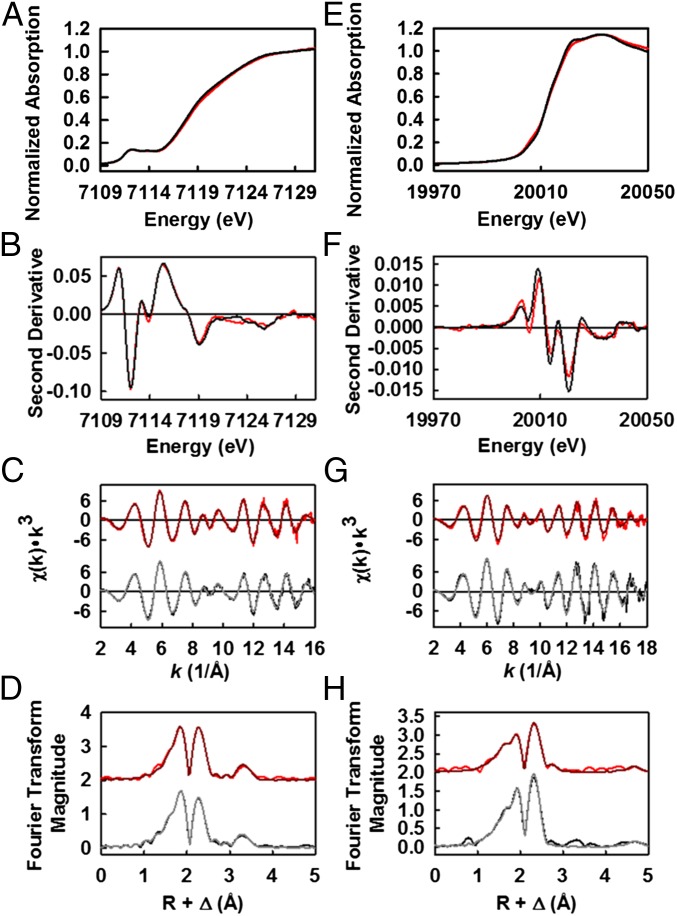
The significant degree of structural homology between the metal-sulfur cores of M-clusterNifEN and M-clusterNifDK is further illustrated by data derived from the Fe K-edge XAS/EXAFS analysis (Fig. 2 A–D). The preedge feature (15, 16) of Fe K-edge is highly conserved between M-clusterNifEN and M-clusterNifDK in terms of total preedge intensities, locations of the component peaks relative to incident photon energy, and ratios between the intensities of the two component peaks (Fig. 2 A and B). The rising edges (17) of the two spectra are also remarkably similar to each other (Fig. 2A), and the second derivative further reveals that the two spectra share a common inflection point at 7,118 eV (Fig. 2B). The close resemblance between the Fe K-edge spectra of M-clusterNifEN and M-clusterNifDK extends to the EXAFS regions (Fig. 2C). The first segment of the EXAFS data (k = 2–8 Å−1), dominated largely by contributions from S backscatterers, are nearly identical in the two spectra (11). Likewise, both spectra display the same phase and frequency in the key beat regions (k = 10) and overlap well between the high k components, where intensities from Fe backscatterers prevail (11). Given the high degree of similarity between the two EXAFS spectra, it is not surprising that their corresponding Fourier transforms have approximately the same maximum magnitude (Fig. 2D). Moreover, the location and integrated peak intensity of both the first and second peaks are highly conserved in the Fourier transforms of the two spectra, reaffirming the presence of nearly indistinguishable S and Fe backscattering shells in M-clusterNifEN and M-clusterNifDK (Fig. 2D).
Fig. 2.
XAS/EXAFS analyses of solvent-extracted M-clusters from NifEN and NifDK. Fe K-edge XAS spectra (A) and smoothed second derivatives (B) of NMF-extracted M-clusterNifEN (red) and M-clusterNifDK (black). Fe K-edge EXAFS (C) and Fourier transforms (D) of data (pink) and fits (red) of M-clusterNifEN, and those of data (gray) and fits (black) of M-clusterNifDK. Data have been normalized to one Fe absorber. Mo K-edge XAS spectra (E) and smoothed second derivatives (F) of NMF-extracted M-clusterNifEN (red) and M-clusterNifDK (black). Mo K-edge EXAFS (G) and Fourier transforms (H) of data (pink) and fits (red) of M-clusterNifEN, and those of data (gray) and fits (black) of M-clusterNifDK. Data have been normalized to one Mo absorber.
Further examination of the Mo K-edge spectra of M-clusterNifEN and M-clusterNifDK reveals the presence of highly similar coordination environments of Mo in both clusters. The two spectra closely resemble each other in the overall shapes of their rising edges (Fig. 2E), which is further illustrated by a nearly identical inflection point at ∼20,012 eV in the second derivatives of both spectra (Fig. 2F). The EXAFS regions of the two spectra are remarkably similar in phase and frequency and, most importantly, they are well-aligned at approximately k = 10, where S and Fe backscattering waves overlap (Fig. 2G). The first and second peaks of the Fourier transforms of the two EXAFS spectra, which are generated by the backscattering of S/light atoms and Fe atoms, respectively, are highly similar in terms of their line shapes and respective ratios (Fig. 2H) (18, 19). Notably, there is a conserved feature near 5.0 Å in the Fourier transforms of both spectra, which is attributed to a distant Fe atom in the opposite cubane and, therefore, is the hallmark of a well-ordered M-cluster (20). Together with the Fe K-edge data, these Mo K-edge data provide conclusive evidence that the M-cluster matured on NifEN is nearly indistinguishable in structure from the native M-cluster on NifDK, firmly establishing NifEN as the only protein scaffold other than NifDK that is capable of housing the unique nitrogenase cofactor.
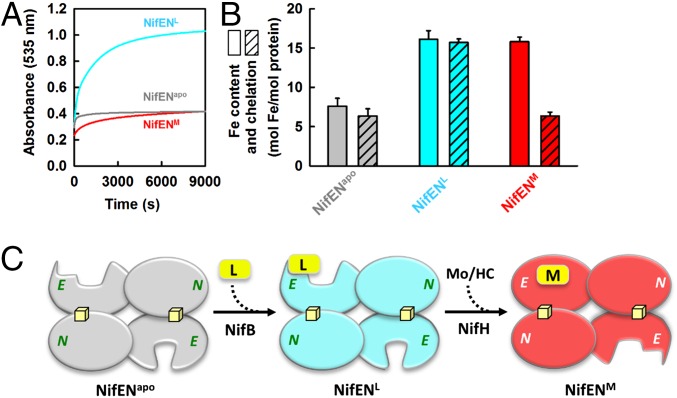
Compared with the L-cluster–bound NifEN (designated NifENL), the M-cluster–bound NifEN (designated NifENM) is much less susceptible to chelation by bathophenanthroline disulfonate (an iron chelator), indicating a conformational change upon conversion of L- to M-cluster on NifEN (Fig. 3A). The amount of Fe atoms chelated from NifENM (6.4 ± 0.9 mol of Fe per mol of protein) is comparable to that chelated from apo-NifEN (designated NifENapo; 6.4 ± 0.5 mol of Fe per mol of protein), whereas the amount of Fe atoms chelated from NifENL (15.7 ± 0.5 mol of Fe per mol of protein) is roughly twice of that chelated from NifENapo or NifENM (Fig. 3B). Given the presence of two solvent-accessible [Fe4S4] clusters—one at each αβ-subunit interface—in all three forms of NifEN, the Fe atoms chelated from NifENM and NifENapo could be assigned to the two subunit-bridging [Fe4S4] clusters (i.e., a total of eight chelation-accessible Fe atoms), whereas the Fe atoms chelated from NifENL could be assigned to the two subunit-bridging [Fe4S4] clusters and one [Fe8S9C] l-cluster (i.e., a total of 16 chelation-accessible Fe atoms). Although the solvent accessibility of the L-cluster is consistent with the near-surface–exposed location of this cluster in the crystal structure of NifENL (8), the apparent protection of the M-cluster from chelation suggests that the cluster is transferred from the surface of NifENM to its binding site within the protein upon maturation, a process that parallels the insertion of the M-cluster into an analogous binding site in apo-NifDK (21).
Fig. 3.
Conformational changes of NifEN upon cluster maturation. Rates of Fe chelation (A) and comparisons of total Fe content and amount of Fe chelation (B) from NifENL, NifENM, and NifENapo. The formation of bathophenanthroline disulfonate–Fe complex was monitored at 535 nm, and the amount of chelated Fe was calculated based on a molar extinction coefficient of 22,140 M−1⋅cm−1. (C) Proposed half-on/half-off model of cluster maturation on NifEN, which involves the maturation of one L-cluster into an M-cluster in one αβ-dimer (upper) that induces the necessary conformational changes of the other αβ-dimer (lower) for the attachment and maturation of a second L-cluster.
Interestingly, the Fe content of NifENL (16.1 ± 1.1 mol of Fe per mol of protein) or NifENM (15.8 ± 0.6 mol of Fe per mol of protein) would account for the presence of only one L- or M-cluster in one NifEN tetramer (Table 1 and Fig. 3B). Such a Fe composition has been consistently observed in different preparations of NifEN either before (i.e., NifENL) or after (i.e., NifENM) the cluster maturation process, suggesting the possibility of association of one L- or M-cluster to one αβ-dimer of NifEN and absence of either cluster species from the other αβ-dimer of this protein. A stepwise assembly scheme can be proposed based on this observation, which involves deposit of one L-cluster (by NifB) at the entrance of an “open” insertion path in the α-subunit (i.e., the NifE subunit) of one αβ-dimer, conversion of this L-cluster to an M-cluster upon insertion of Mo and homocitrate (by NifH), and insertion of the matured M-cluster into its binding site along the open insertion path in this α-subunit, which triggers a conformational change that opens up the “closed” insertion path in the α-subunit of the other αβ-dimer for the attachment and maturation of a second L-cluster (Fig. 3C). Interestingly, the apparent deviation of this half-on, half-off biosynthetic scheme of the cofactor from the crystallographic assignment of L-clusters in both αβ-dimers of NifEN (8) is mirrored by the deviation of a stepwise pattern of P-cluster assembly (22) from the crystallographic assignment of P-clusters in both αβ-dimers of NifDK (21), which reveals an asymmetric nature of the assembly sites in the seemingly equivalent αβ-halves of these homologous proteins while highlighting a significant degree of coordination of various biosynthetic events via a conformational switch on/off mechanism.
Table 1.
Metal contents of purified proteins and metal ratios of isolated clusters
| Sample | Metal content | Metal ratio, Fe/Mo | |
| Fe, mol metal/mol protein | Mo, mol metal/mol protein | ||
| Protein | |||
| NifENapo | 7.6 ± 1.1 | 0.03 ± 0.01 | — |
| NifENL | 16.1 ± 1.1 | 0.03 ± 0.01 | — |
| NifENM | 15.8 ± 0.6 | 1.12 ± 0.04 | — |
| NifDK | 32.3 ± 1.2 | 1.82 ± 0.10 | — |
| Cluster | |||
| M-clusterNifEN | — | — | 6.9 ± 0.4 |
| M-clusterNifDK | — | — | 7.3 ± 0.3 |
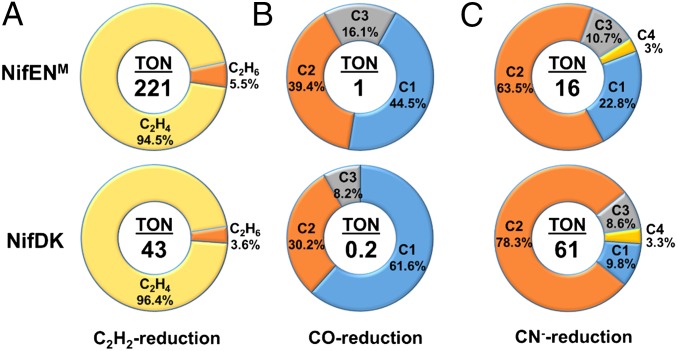
Consistent with its specialized function in cofactor assembly, despite the presence of a fully assembled M-cluster in one αβ-dimer of NifENM, this protein is unable to catalyze ATP-dependent reduction of substrates when combined with NifH, the obligate electron donor for NifDK during catalysis. In the presence of a strong reductant, europium (II) diethylenetriaminepentaacetic acid (EuII-DTPA; E0′ = ‒1.14 at pH 8), however, NifENM is capable of ATP-independent reduction of acetylene (C2H2), carbon monoxide (CO), and cyanide (CN−), showing carbon-based turnover numbers (TONs) of 221, 1, and 16, respectively, in these reactions (Fig. 4). Compared with NifDK, NifENM generates the same products in ATP-independent reduction of C2H2 [i.e., ethane (C2H4) and ethane (C2H6)], CO (i.e., C1–C3 alkenes and alkanes) and CN− (i.e., C1–C4 alkenes and alkanes), and it displays similar product distributions to NifDK in these EuII-DTPA–driven reactions (Fig. 4). However, NifENM differs somewhat from NifDK on the tendency of forming certain hydrocarbon products; moreover, NifENM is fivefold more active than NifDK in C2H2 and CO reduction, but fourfold less active than NifDK in CN− reduction (Fig. 4), which underlines the homology and distinction between the subunit and cluster compositions of these two proteins.
Fig. 4.
ATP-independent substrate-reducing activities of NifENM and NifDK. Shown are TONs and product distributions of C2H2 (A), CO (B), and CN− (C) reduction by NifENM and NifDK. TONs were calculated based on the total numbers of reduced carbon atoms that appeared in the products of C2H2, CO, and CN− reduction after 60 min, 6 h, and 60 min, respectively, per M-cluster on NifENM (one M-cluster per tetramer) or NifDK (two M-clusters per tetramer). The C1–C4 hydrocarbons generated in the reactions of CO and CN− reduction are as follows: C1, CH4; C2, C2H4, C2H6; C3, C3H6, C3H8; C4, C4H8, C4H10.
The observation of comparable yet distinct substrate-reducing profiles of NifENM and NifDK is exciting, because it establishes the αβ-dimer of NifENM as a structural and functional mimic of one of the two catalytic αβ-halves of NifDK and, thereby, provides a proof-of-concept for the feasibility of reconstructing a functional equivalent of NifDK on the basis of a simplified template, NifEN. Given the presence of a catalytically less competent [Fe4S4] cluster at the αβ-interface of NifENM, restoration of a P-cluster at this location of the protein may render it capable of ATP-dependent substrate reduction in one αβ-half and cofactor maturation in the other αβ-half. Additionally, the key residues surrounding the M-cluster site of NifENM, such as those involved in proton gating, could be altered for improved substrate-reducing activity of this protein. Together, these efforts could lead to the identification of key features for the catalytic activity of nitrogenase, which is crucial for elucidating the mechanistic details of this enzyme. Moreover, nitrogenase variants with unique catalytic properties could be identified along this line of work, which is important for exploring the biotechnological applicability of this important metalloenzyme.
Materials and Methods
M-Cluster Maturation/Extraction and Metal Analysis.
The L-cluster–bound conformation of NifEN (NifENL) was converted to an M-cluster–bound conformation of NifEN (NifENM) upon incubation of NifENL with NifH, ATP, MoO42−, and homocitrate (13). Subsequently, the M-cluster was extracted from 1.5 g of NifENM and 1.5 g of NifDK, respectively, into NMF by a previously described acid-treatment method (23). Iron and molybdenum were determined as published (24, 25). The Fe/Mo ratios of M-clusterNifEN and M-clusterNifDK are 6.9 and 7.3, respectively (Table 1).
Reconstitution Assays of Apo-NifDK.
Reconstitution of apo-NifDK (NifDKapo) by M-clusterNifEN or M-clusterNifDK was performed as described (14). The amount of M-cluster used to reconstitute NifDKapo was determined by titrating increasing amounts of isolated M-cluster for the maximum substrate-reducing activities. The products hydrogen (H2), ethene (C2H4), and ethane (C2H6) were analyzed as published (26), and ammonium ion (NH4+) was determined by a HPLCy fluorescence method (27).
EPR Spectroscopy.
All EPR samples were prepared in a Vacuum Atmospheres dry box at an oxygen level of less than 4 ppm. All NMF-extracted cofactor samples contained 2 mM dithionite (Na2S2O4) and 10 mM 1,4-benzenedithiol. The reconstituted NifDK samples were prepared by incubating 12.5 mg of NifDKapo with 60 nmol NMF-extracted M-cluster for 5 min at 30 °C in 2.5 mL of buffer containing 25 mM Tris⋅HCl (pH 8.0), 10% (vol/vol) glycerol, and 2 mM Na2S2O4. Subsequently, the reconstituted protein samples were concentrated and the excess cofactor was removed by gel filtration chromatography (Sephadex G-25; GE Healthcare). All protein samples were adjusted to a final concentration of 30 mg/mL and contained 10% (vol/vol) glycerol, 2 mM Na2S2O4, and 25 mM Tris⋅HCl (pH 8.0). Volume-calibrated, clear fused quartz EPR tubes (Wilmad-LabGlass) were used for EPR experiments. Spectra were collected in perpendicular mode by using a Bruker ESP 300 Ez spectrometer (Bruker) interfaced with an Oxford Instruments ESR-9002 liquid helium continuous-flow cryostat (Oxford Instruments). All spectra were recorded by using a gain of 5 × 104, a modulation frequency of 100 kHz, a modulation amplitude of 5 G, a microwave frequency of 9.62 GHz, and a power of 50 mW.
XAS Data Collection.
Collection of both the Fe and Mo K-edge XAS data of the M-clusterNifEN and M-clusterNifDK samples was undertaken at Beam Line 7-3 at Stanford Synchrotron Radiation Lightsource (SSRL), a 2.0 T 20-pole wiggler side station dedicated to structural molecular biology and especially suited to dilute protein XAS experiments with an ideal energy range of 5–25 keV. Beam line optics consist of a premonochromator flat bent Rh-coated mirror for collimation and higher harmonic rejection, followed by a Si(220) double-crystal monochromator. The SSRL synchrotron storage ring SPEAR3 was maintained at an energy level of 3 GeV and an operating current of 300 mA.
Protein samples were injected into Delrin cells (thickness, 2 mm; volume, 260 µL) with Kapton tape windows and flash frozen in liquid N2/pentane slush. Sample temperature during data collection was preserved at 10 K by using an Oxford Instruments CF1208 continuous flow liquid-He cryostat. The EXAFS spectra were measured via detection of Fe and Mo Kα fluorescence photons by an electronically windowed Canberra 30-element solid-state Ge detector with installed Soller slits. A 3-µm filter of either manganese (Fe K-edge) or zirconium (Mo K-edge) was placed between the sample and the fluorescence detector to attenuate the elastic/inelastic scattering and the Fe Kβ/Mo Kβ fluorescence. A metal foil standard, measured in tandem with the protein sample scans, was used to calibrate the Fe K-edge X-ray energy to a first inflection point of 7,111.2 eV and the Mo K-edge X-ray energy to a first inflection point of 20,003.9 eV. A total of 21 scans were taken at the Fe K-edge of both M-clusterNifEN and M-clusterNifDK, whereas a total of 20 and 19 scans were taken at the Mo K-edge of M-clusterNifEN and M-clusterNifDK, respectively.
XAS Data Analysis.
EXAFSPAK was used to generate an average file of the Fe and Mo K-edge spectra of each sample after eliminating abnormal channels and aberrant scans from the data (28); in particular, a three-point smoothing of the metal foil calibration spectrum was required to average the Mo K-edge data. Subsequently, PYSPLINE was used to subtract the first-order background from the data over the entire eV range, then to generate a spline function to model background absorption and normalize the EXAFS region. The data were normalized to have an edge jump of 1.0 at 7,130 eV for the Fe K-edge and 20,025 eV for the Mo K-edge (29). Several distinct spline functions were generated to model the background absorption and normalize the data. These functions were refined and compared before a unique four-region spline was chosen with 2, 3, 3 order polynomials over the postedge region for each data set (30). The Mo K-edge spectrum of M-clusterNifEN was deglitched at several points, with no effect observed in the corresponding Fourier transform.
Iron Chelation Assays.
Fe chelation assays were carried out to determine the cluster accessibility of NifENL, NifENM, and NifENapo. A solution containing 25 mM Tris⋅HCl (pH 8), 2 mM Na2S2O4, and a final concentration of 1 mg/mL of NifENL, NifENM, or NifENapo was prepared anaerobically and used to blank the UV/Vis spectrophotometer at 535 nm. Data collection was initiated immediately upon addition of 5 mM bathophenanthroline disulfonate. The amount of chelated Fe was calculated based on a molar attenuation coefficient (ε) of 22,140 cm−1⋅M−1 at 535 nm (31).
Acetylene Reduction Assays.
C2H2 reduction assays were conducted under 30% (vol/vol) C2H2 atmosphere in 10-mL vials. Each reaction mixture contained, in a total volume of 0.5 mL, 25 mM Tris⋅HCl (pH 8.0) and 1.0 mg of NifENM or NifDK. The reaction was initiated by addition of an aqueous solution of EuII-DTPA at a final concentration of 20 mM, followed by incubation at 30 °C in an isothermal waterbath shaker. Samples were taken at 60 min after the initiation of the reactions. The products C2H4 and C2H6 were determined by gas chromatography-flame ionization detection (GC-FID) analysis, in which 250 μL of the headspace of each sample was injected onto a Grace 5664PC column (3.2 mm diameter, 1.5 m length; Grace) that was held at 120 °C during the time of measurement.
Cyanide Reduction Assays.
CN− reduction assays were conducted under Ar atmosphere in 10-mL vials. Each reaction mixture contained, in a total volume of 1 mL, 25 mM Tris⋅HCl (pH 8.0), 100 mM NaCN, and 10 mg of NifENM or NifDK. The reactions were initiated by the addition of an aqueous solution of EuII-DTPA at a final concentration of 20 mM, followed by incubation at 30 °C in an isothermal waterbath shaker. Samples were taken at 60 min after the initiation of the reactions. The products methane (CH4), C2H4, C2H6, propene (C3H6), propane (C3H8), butene (C4H8), and butane (C4H10) were quantified by GC-FID analysis, in which 250 μL of the headspace of each sample was injected onto a Grace 5664PC column (3.2 mm diameter, 1.5 m length; Grace), which was held at 55 °C for 1 min, heated to 180 °C at 12.5 °C/min, and held at 180 °C for 2.6 min.
Carbon Monoxide Reduction Assays.
CO reduction assays were conducted under 100% (vol/vol) CO atmosphere in 10-mL vials. Each reaction mixture contained, in a total volume of 1 mL, 25 mM Tris⋅HCl (pH 8.0) and 10 mg of NifENM or NifDK. The reactions were initiated by addition of an aqueous solution of EuII-DTPA at a final concentration of 20 mM, followed by incubation at 30 °C in an isothermal waterbath shaker. Samples were taken at 6 h after the initiation of the reactions. Products CH4, C2H4, C2H6, C3H6, and C3H8 were quantified by GC-FID analysis, in which 250 μL of the headspace of each sample was injected onto a Grace 5664PC column (3.2 mm diameter, 1.5 m length; Grace), which was held at 55 °C for 1 min, heated to 180 °C at 12.5 °C/min, and held at 180 °C for 2.6 min.
Acknowledgments
This work was supported by NIH Grants P41GM103393 (to K.O.H.) and GM 67626 (to M.W.R.) and a Hellman Fellowship (to Y.H.). Use of the Stanford Synchrotron Radiation Lightsource (SSRL), Stanford Linear Accelerator Center National Accelerator Laboratory, is supported by the US Department of Energy (DOE), Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences, including Grant P41GM103393.
Footnotes
The authors declare no conflict of interest.
References
- 1.Burgess BK, Lowe DJ. Mechanism of molybdenum nitrogenase. Chem Rev. 1996;96(7):2983–3012. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 2.Einsle O, et al. Nitrogenase MoFe-protein at 1.16 A resolution: A central ligand in the FeMo-cofactor. Science. 2002;297(5587):1696–1700. doi: 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
- 3.Lancaster KM, et al. X-ray emission spectroscopy evidences a central carbon in the nitrogenase iron-molybdenum cofactor. Science. 2011;334(6058):974–977. doi: 10.1126/science.1206445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spatzal T, et al. Evidence for interstitial carbon in nitrogenase FeMo cofactor. Science. 2011;334(6058):940. doi: 10.1126/science.1214025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schindelin H, Kisker C, Schlessman JL, Howard JB, Rees DC. Structure of ADP x AIF4(-)-stabilized nitrogenase complex and its implications for signal transduction. Nature. 1997;387(6631):370–376. doi: 10.1038/387370a0. [DOI] [PubMed] [Google Scholar]
- 6.Hu Y, Ribbe MW. Biosynthesis of the metalloclusters of molybdenum nitrogenase. Microbiol Mol Biol Rev. 2011;75(4):664–677. doi: 10.1128/MMBR.05008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbett MC, et al. Structural insights into a protein-bound iron-molybdenum cofactor precursor. Proc Natl Acad Sci USA. 2006;103(5):1238–1243. doi: 10.1073/pnas.0507853103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaiser JT, Hu Y, Wiig JA, Rees DC, Ribbe MW. Structure of precursor-bound NifEN: A nitrogenase FeMo cofactor maturase/insertase. Science. 2011;331(6013):91–94. doi: 10.1126/science.1196954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fay AW, et al. Spectroscopic characterization of the isolated iron-molybdenum cofactor (FeMoco) precursor from the protein NifEN. Angew Chem Int Ed Engl. 2011;50(34):7787–7790. doi: 10.1002/anie.201102724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lancaster KM, Hu Y, Bergmann U, Ribbe MW, DeBeer S. X-ray spectroscopic observation of an interstitial carbide in NifEN-bound FeMoco precursor. J Am Chem Soc. 2013;135(2):610–612. doi: 10.1021/ja309254g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Y, et al. FeMo cofactor maturation on NifEN. Proc Natl Acad Sci USA. 2006;103(46):17119–17124. doi: 10.1073/pnas.0602647103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Y, et al. Nitrogenase Fe protein: A molybdate/homocitrate insertase. Proc Natl Acad Sci USA. 2006;103(46):17125–17130. doi: 10.1073/pnas.0602651103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshizawa JM, et al. Optimization of FeMoco maturation on NifEN. J Am Chem Soc. 2009;131(26):9321–9325. doi: 10.1021/ja9035225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fay AW, et al. Characterization of isolated nitrogenase FeVco. J Am Chem Soc. 2010;132(36):12612–12618. doi: 10.1021/ja1019657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solomon EI, Hanson MA. 1999. Bioinorganic spectroscopy. Inorganic Electronic Structure and Spectroscopy Volume II: Applications and Case Studies, eds Solomon EI, Lever ABP (John Wiley & Sons, New Jersey)
- 16.Westre TE, et al. A multiplet analysis of Fe K-edge 1s->3d pre-edge features of iron complexes. J Am Chem Soc. 1997;119:6297–6314. [Google Scholar]
- 17.Hasnain SS, Hodgson KO. Structure of metal centres in proteins at subatomic resolution. J Synchrotron Radiat. 1999;6:852–864. [Google Scholar]
- 18.Conradson SD, Burgess BK, Newton WE, Mortenson LE, Hodgson KO. Structural studies of the molybdenum site in the MoFe protein and its FeMo cofactor by EXAFS. J Am Chem Soc. 1987;109:7507–7515. [Google Scholar]
- 19.Conradson SD, et al. Structural insights from the Mo K-edge X-ray absorption near edge structure of the iron molybdenum protein of nitrogenase and its iron molybdenum cofactor by comparison with synthetic Fe-Mo-S clusters. J Am Chem Soc. 1985;107:7935–7940. [Google Scholar]
- 20.Liu HBI, et al. EXAFS studies of FeMo-cofactor and MoFe protein - Direct evidence for the long-range Mo-Fe-Fe interaction and cyanide binding to the Mo in FeMo-cofactor. J Am Chem Soc. 1994;116:2418–2423. [Google Scholar]
- 21.Schmid B, et al. Structure of a cofactor-deficient nitrogenase MoFe protein. Science. 2002;296(5566):352–356. doi: 10.1126/science.1070010. [DOI] [PubMed] [Google Scholar]
- 22.Lee CC, et al. Stepwise formation of P-cluster in nitrogenase MoFe protein. Proc Natl Acad Sci USA. 2009;106(44):18474–18478. doi: 10.1073/pnas.0909149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgess BK. The iron molybdenum cofactor of nitrogenase. Chem Rev. 1990;90:1377–1406. [Google Scholar]
- 24.Van de Bogart M, Beinert H. Micro methods for the quantitative determination of iron and copper in biological material. Anal Biochem. 1967;20(2):325–334. doi: 10.1016/0003-2697(67)90038-3. [DOI] [PubMed] [Google Scholar]
- 25.Clark LJ, Axley JH. Molybdenum determination in soils and rocks with dithiol. Anal Chem. 1955;27:2000–2003. [Google Scholar]
- 26.Gavini N, Burgess BK. FeMo cofactor synthesis by a nifH mutant with altered MgATP reactivity. J Biol Chem. 1992;267(29):21179–21186. [PubMed] [Google Scholar]
- 27.Corbin JL. Liquid chromatographic-fluorescence determination of ammonia from nitrogenase reactions: A 2-min assay. Appl Environ Microbiol. 1984;47(5):1027–1030. doi: 10.1128/aem.47.5.1027-1030.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.George GN. 1990. EXAFSPAK (Stanford Synchrotron Radiation Lightsource, Stanford, CA)
- 29.Tenderholt A. 2006. PYSPLINE (Stanford Synchrotron Radiation Lightsource, Stanford, CA)
- 30.Scott RA. Measurement of metal-ligand distances by EXAFS. Methods Enzymol. 1985;117:414–459. [Google Scholar]
- 31.Ribbe MW, Bursey EH, Burgess BK. Identification of an Fe protein residue (Glu146) of Azotobacter vinelandii nitrogenase that is specifically involved in FeMo cofactor insertion. J Biol Chem. 2000;275(23):17631–17638. doi: 10.1074/jbc.275.23.17631. [DOI] [PubMed] [Google Scholar]