Significance
M proteins are major virulence factors of the widespread bacterial pathogen group A Streptococcus. The sequences of M proteins are antigenically variable, but have in common the tendency to form α-helical coiled coil structures. Paradoxically, these same sequences also contain a substantial number of amino acids that destabilize coiled coils. We present an explanation for this paradox in finding that destabilizing residues are essential for conformational dynamics in the M protein, and that these dynamics are required for interaction with human fibrinogen. Our work leads to the general conclusion that destabilizing residues are conserved through evolution in M proteins to enable functional interactions that are necessary for pathogenesis.
Keywords: coiled coil, group A Streptococcus, M protein, fibrinogen, dynamics
Abstract
The sequences of M proteins, the major surface-associated virulence factors of the widespread bacterial pathogen group A Streptococcus, are antigenically variable but have in common a strong propensity to form coiled coils. Paradoxically, these sequences are also replete with coiled-coil destabilizing residues. These features are evident in the irregular coiled-coil structure and thermal instability of M proteins. We present an explanation for this paradox through studies of the B repeats of the medically important M1 protein. The B repeats are required for interaction of M1 with fibrinogen (Fg) and consequent proinflammatory activation. The B repeats sample multiple conformations, including intrinsically disordered, dissociated, as well as two alternate coiled-coil conformations: a Fg-nonbinding register 1 and a Fg-binding register 2. Stabilization of M1 in the Fg-nonbinding register 1 resulted in attenuation of Fg binding as expected, but counterintuitively, so did stabilization in the Fg-binding register 2. Strikingly, these register-stabilized M1 proteins gained the ability to bind Fg when they were destabilized by a chaotrope. These results indicate that M1 stability is antithetical to Fg interaction and that M1 conformational dynamics, as specified by destabilizing residues, are essential for interaction. A “capture-and-collapse” model of association accounts for these observations, in which M1 captures Fg through a dynamic conformation and then collapses into a register 2-coiled coil as a result of stabilization provided by binding energy. Our results support the general conclusion that destabilizing residues are evolutionarily conserved in M proteins to enable functional interactions necessary for pathogenesis.
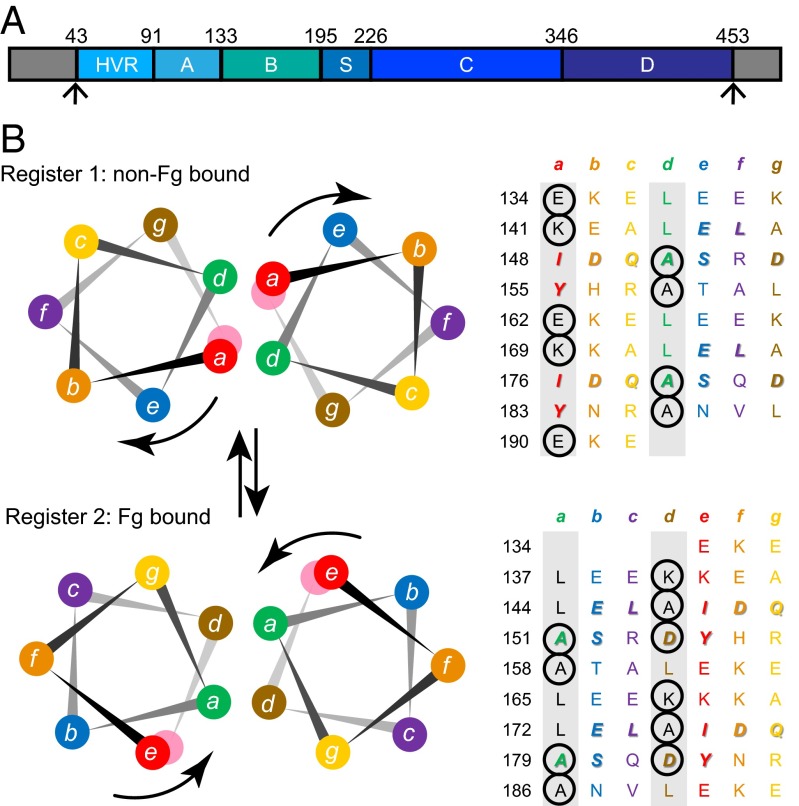
M proteins are the major surface-associated virulence factors of group A Streptococcus (GAS; Streptococcus pyogenes) and play a significant role in the diverse diseases caused by this widespread bacterial pathogen (1–3). M protein sequences are antigenically variable but have in common a strong propensity to form α-helical coiled coils (4). Paradoxically, these sequences are also replete with residues that destabilize coiled coils (5, 6). These destabilizing residues occur at the central a and d positions of the coiled-coil [abcdefg]n heptad repeat (Fig. 1). For dimeric α-helical coiled coils, as is the case for M proteins (7–9), the a and d positions are preferentially occupied by Val and Leu, respectively (10, 11). These small, hydrophobic residues typically engage in “knobs-into-hole” packing and form the hydrophobic core of the coiled-coil dimer. In contrast, the a and d positions of M proteins are often occupied by coiled-coil destabilizing residues (e.g., charged residues or alanines), and the heptad pattern sometimes contains short insertions or deletions (Fig. 1B) (5, 9). These sorts of destabilizing residues and heptad disruptions result in a highly irregular coiled-coil structure in M1 protein (9). The high proportion of coiled-coil destabilizing residues is also consistent with M proteins becoming less structured at physiological temperature (i.e., thermal instability) (5, 9, 12).
Fig. 1.
M1 is a nonideal coiled coil. (A) Schematic of the M1 protein, whose mature form consists of the A region, B repeats, S region, C repeats, and D region. The N-terminal gray region represents the cleaved signal sequence, and the C-terminal gray region the site cleaved by sortase A and attached to the peptidoglycan. (B) Helical wheel and heptad repeat schematic of the two registers of the B repeats of M1 protein. (Upper) register 1 and (Lower) register 2. Coiled-coil destabilizing residues at the a and d positions are circled. The color coding of residues is preserved in the two registers, and the arrows indicate the rotation of the helical face required to transition from one register to the other. The heptad a and d positions are shaded in gray. Residues observed to bind Fg in register 2 are italicized and bolded; the same residues are also italicized and bolded in register 1.
To address why M proteins have a high proportion of coiled-coil destabilizing residues, we investigated the M1 protein, which is the most structurally characterized M protein (8, 9). M1 serotype strains are globally prevalent and are the leading single cause of severe invasive GAS disease (13). In particular, we focused on the B repeats of M1 (Fig. 1), which confer binding to human fibrinogen (Fg) in an interaction that is key to phagocytic resistance and proinflammatory activation seen during septic shock (14, 15). The B repeats are composed of two 28-residue regions that differ slightly in sequence but are absolutely conserved in Fg-interacting residues. Remarkably, crystal structures have suggested that the B repeats exist in two different heptad registers, depending on whether they are free (register 1) or Fg-bound (register 2). In the crystal structure of free M1, the B repeats are splayed apart and engage in an antiparallel dimeric coiled coil with the B repeats of an adjacent M1 fragment, both occupying register 1 (9). Register 1 could also occur in the native parallel orientation of the M1 protein, in which case an irregular conformation would be expected as a result of charge–charge clashes and alanines in the a and d positions, as seen in the A region (9). The splaying of the B repeats observed in the crystal structure is likely en route to complete dissociation of M1 coiled-coil dimers into single chains, which has been observed at 37 °C (9). The B repeats sample at least one additional conformation. This is register 2, which is related to register 1 by a rotation of one helical face (Fig. 1). The two registers present different sets of exposed residues, and thus different possible interaction sites (8). Register 2 was observed in the crystal structure of Fg-bound M1, in which the B repeats form a structurally canonical dimeric parallel coiled coil (8). Coiled-coil destabilizing residues occur with near equal frequency in registers 1 and 2, suggesting that the unbound form of register 2 is also likely to have an irregular conformation. Further evidence for the existence of multiple registers comes from coiled-coil propensity analysis, which indicates that the B repeats are composed of short segments of register 1-preferring residues interspersed with short segments of register 2-preferring residues (Fig. S1).
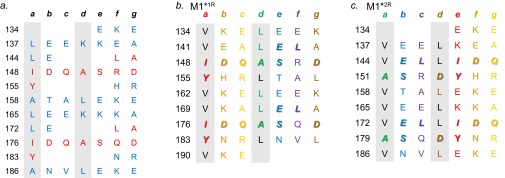
Fig. S1.
Idealization of the B repeats. (A) Heptad positions of residues in the B repeats as predicted by Coils (12). Residues that correspond to register 1 are in red, and those that correspond to register 2 are in blue. (B) Sequence of M1*1R, with idealizing mutations in black and depicted in register 1. Residues that contact Fg in register 2 are bolded and italicized. (C) Sequence of M1*2R, with idealizing mutations in black and depicted in register 2. Residues that contact Fg in register 2 are bolded and italicized.
To study the role of each register separately, we “idealized” the sequence of the B repeats, with the intention of stabilizing these repeats in either register 1 or 2. Idealization was achieved by substituting residues in the a position with Val and in the d position with Leu. These substitutions were made in register 1 (M1*1R), and separately in register 2 (M1*2R), while maintaining the Fg-contacting residues (Fig. S1). The resulting M1 mutant proteins maintain the residues that contact Fg (i.e., when M1 protein is in register 2). Prior work had shown that M1*1R was attenuated in Fg binding (9, 14), as would be expected for stabilization in the Fg-nonbinding register. We now report the surprising result that stabilization in the Fg-binding register 2 leads to an equal deficiency in Fg binding. Notably, both register 1- and register 2-stabilized M1 proteins became capable of binding Fg when destabilized by the chaotrope urea. These results indicate that structural stability in M1 protein is counter to interaction with Fg and suggest that dynamics in the coiled coil, as brought about by coiled-coil destabilizing residues, are necessary for this interaction. Furthermore, these studies support the general conclusion that destabilizing residues are evolutionarily conserved in M proteins to permit functional interactions that are necessary for pathogenesis.
Results
Idealizing the Coiled Coil Restricts the B Repeats to the Intended Conformation.
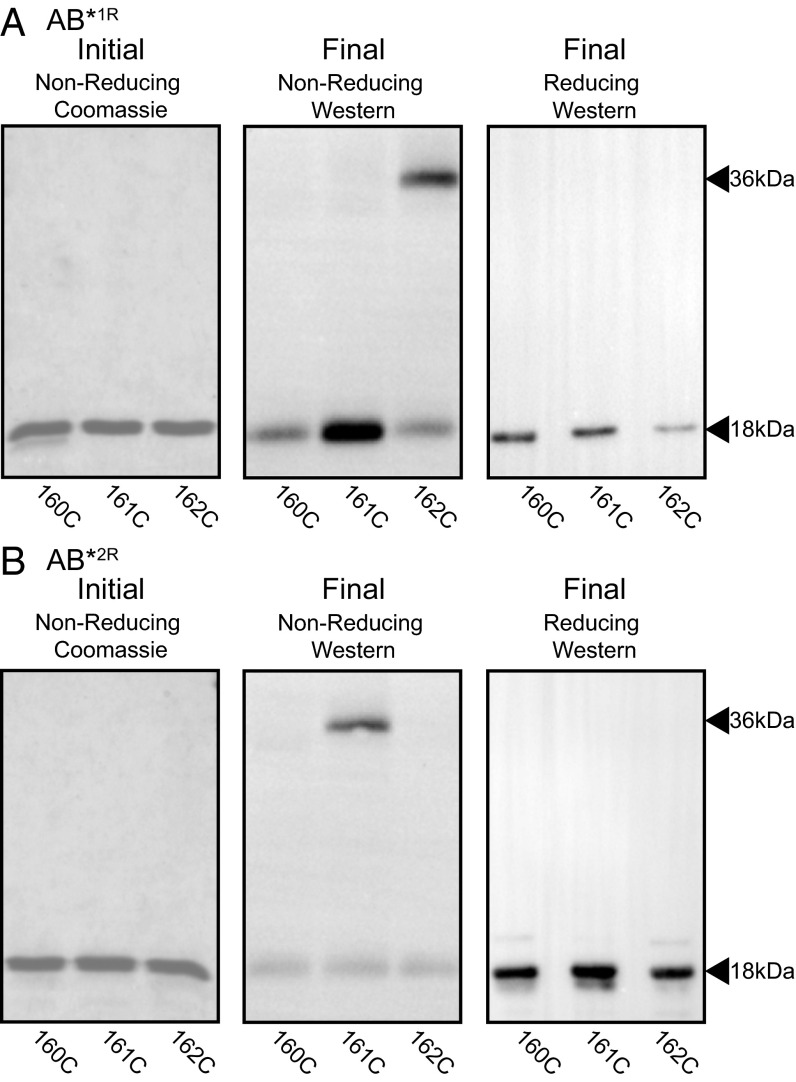
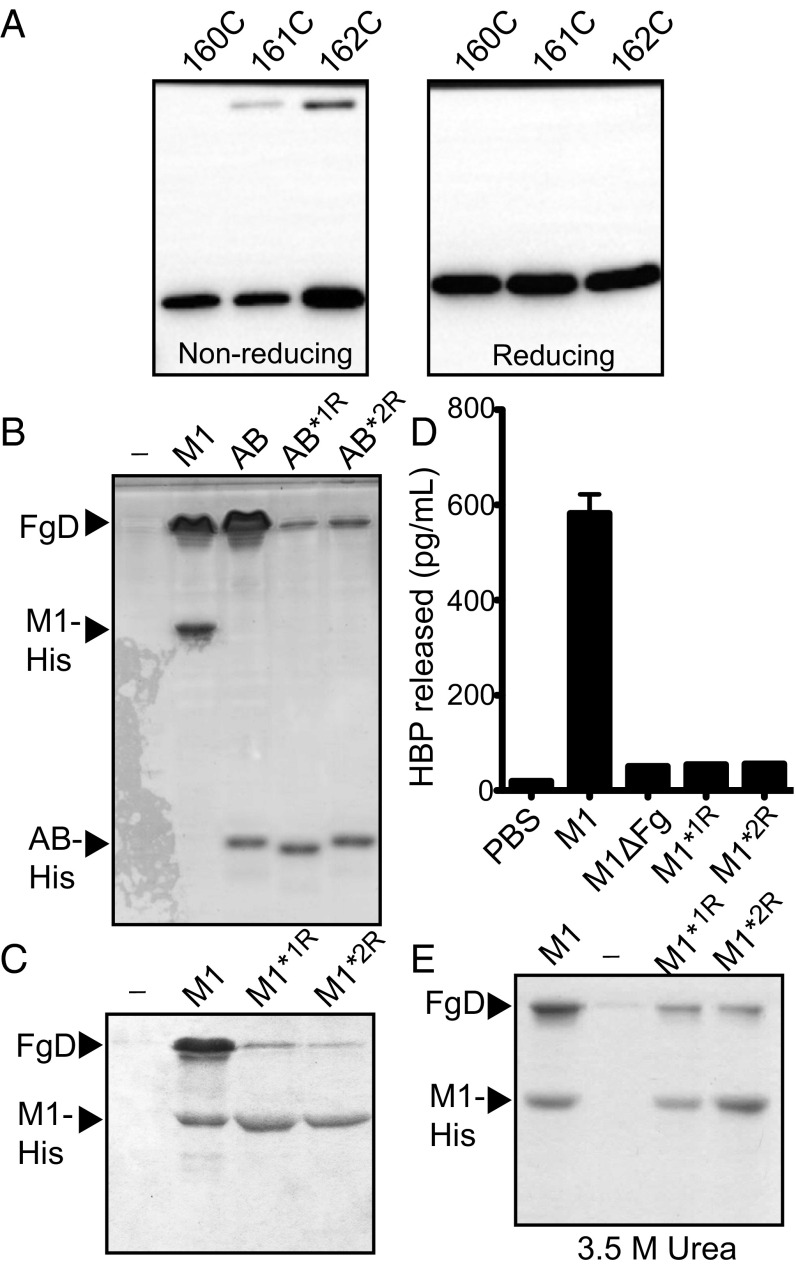
We first tested whether the register 1- and register 2-idealizing substitutions had resulted in molecules that adopted those particular registers. We used the AB fragment of the M1 protein (containing only the A region and B repeats) for this study (Fig. 1), as this fragment is more unstable than intact M1 protein, and thus provides a more stringent test of the mutations (9). An M1 protein fragment corresponding to the AB fragment is generated during conditions mimicking infection (15, 16). In addition, the AB fragment is sufficient to bind Fg (9, 17). To assess which conformation the AB fragment idealized in register 1 (AB*1R) or register 2 (AB*2R) assumed, cysteine substitutions were constructed as conformational probes (M1 protein contains no native cysteines). We inferred that the substituted cysteines would form intradimer disulfide bonds if they occupied the core a or d position, but not if they occupied exterior heptad positions. The position 162 substitution was created as a probe for register 1, as this residue occupies the core a heptad position in register 1, but the exterior e heptad position in register 2 (Fig. 1). The position 161 substitution was created as a probe for register 2, as this residue occupies the core d position in register 2, but the exterior g heptad position in register 1. Last, the position 160 substitution mutant was created as a negative control, as it occupies exterior heptad positions in both registers (f in register 1, c in register 2), and thus should not be capable of forming intradimer disulfide bonds.
The initial step in the experiment entailed the complete reduction of the versions of AB*1R and AB*2R each containing one of the three Cys substitutions (Fig. 2 A and B, Left). This was done to avoid confounding interdimer disulfide bonds that may form during expression or purification, and was achieved by heating the proteins to cause dissociation while incubating them in 100 mM DTT. The samples were then cooled and diluted in the absence of DTT to enable reassociation into dimers and the formation of intradimer disulfide bonds. Before SDS/PAGE, the samples were treated with iodoacetamide to block any remaining free cysteines that may yield artifactual disulfide bonds on denaturation. This set of experiments showed that disulfide-linked dimers formed for 162C, the probe for register 1, only in the case of AB*1R (Fig. 2A). Likewise, disulfide-linked dimers formed for 161C, the probe for register 2, only in the case of AB*2R (Fig. 2B). No disulfide-linked dimers were observed for 160C in either AB*1R or AB*2R, confirming that the disulfide bonds observed for 162C and 161C were indeed intradimer. These results indicate that the idealizing mutations restrained the B repeats to the intended registers and validated the use of cysteines as conformational probes.
Fig. 2.
Idealized mutants are restricted to the intended register. Formation of intradimer disulfide bonds by (A) AB*1R and (B) AB*2R containing 160C (control), 161C (register 2 probe), or 162C (register 1 probe) mutations. (Left, Initial) Proteins were initially heated and reduced and were visualized by Coomassie-stained, nonreducing SDS/PAGE. (Middle, Final) Proteins were then cooled and diluted without reducing agent. The formation of disulfide bonds was assessed by nonreducing SDS/PAGE, which was visualized by Western blot using anti-M1 polyclonal antibodies. (Right, Final) Same as the middle panel, except under reducing conditions.
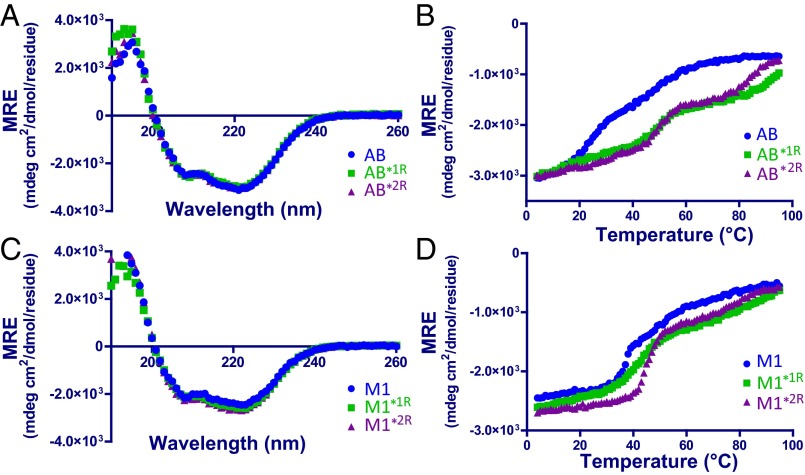
Idealization of AB in register 1 was previously shown to result in stabilization (9). However, it should be noted that in this prior case, the AB fragment used was also substituted with valines and leucines in Fg-contacting residues, and thus differed in detail from AB*1R. To assess whether idealization in register 1 for AB*1R and in register 2 for AB*2R resulted in stabilization, the α-helical content of these proteins, as determined by circular dichroism (CD), was tracked with increasing temperature. At 4 °C, the CD spectra of AB*1R and AB*2R were indistinguishable from that of wild-type AB (Fig. 3A), indicating that the stabilizing mutations had not changed secondary structure. Of particular note, both AB*1R and AB*2R maintained a much greater fraction of their α-helical content compared with wild-type AB at temperatures greater than 20 °C (Fig. 3B). Equally important was the fact that the melting profiles of AB*1R and AB*2R displayed cooperative transitions, whereas such a transition was not as evident for wild-type AB, which displayed a nearly monotonic loss of structure. Thus, idealization of the AB fragment in registers 1 and 2, while maintaining the Fg-contacting residues, yielded molecules that were more stable than wild-type.
Fig. 3.
Idealized mutants are more stable than wild-type. CD signal (mean residue ellipticity, MRE) at 4 °C as a function of wavelength (A and C) and at 222 nm as a function of temperature (B and D) of (Upper) AB and (Lower) intact M1 proteins. Wild-type is in blue, register 1-idealized in green, and register 2-idealized in purple.
B Repeats of M1 Protein Sample Multiple Conformations.
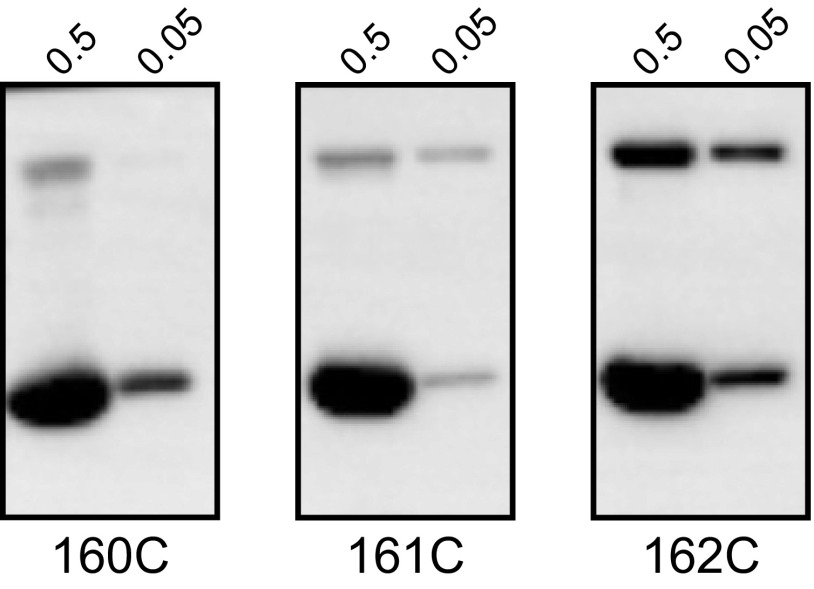
With the cysteine substitutions having been validated as conformational probes, a similar approach was applied to address the register of the B repeats in intact, wild-type M1 protein. The same cysteine substitutions were introduced into wild-type M1, and the procedure described earlier for the AB fragment was followed. Disulfide-linked dimers were evident for both the register 1 probe, 162C, and the register 2 probe, 161C, but not for the negative control, 160C (Fig. 4A). To further demonstrate that the disulfide bonds observed in M1 161C and 162C were intradimer and not interdimer, we carried out the same experiment with the M1 proteins at 10-fold higher concentration during the reassociation step (Fig. S2). Disulfide bonds were now evident for the negative control, 160C, indicating that interdimer disulfide bonds had formed at this 10-fold higher reassociation concentration. Accordingly, this result demonstrated that the disulfide bonds observed for the register 1 and 2 probes at the 10-fold lower reassociation concentration (as in Fig. 4A) were indeed intradimer, rather than interdimer, disulfide bonds. Overall, this set of experiments provided evidence that the B repeats in the context of intact, wild-type M1 protein are flexible and dynamic, sampling both registers 1 and 2.
Fig. 4.
M1 protein samples both registers, but when stabilized in either register, is attenuated in Fg interaction. (A) Disulfide bond formation in wild-type, His-tagged M1 protein containing A160C, L161C (register 2 probe), or E162C (register 1 probe) substitution mutations, as assessed by (Left) nonreducing and (Right) reducing SDS/PAGE and visualized by Western blot using an anti-His antibody. (B) Ni2+-NTA agarose coprecipitation assay for interaction of His-tagged AB proteins with FgD at 37 °C. Bound FgD was assessed through Coomassie-stained SDS/PAGE. His-tagged M1 and its bound FgD is included as a reference. (C) Ni2+-NTA agarose coprecipitation assay for interaction of His-tagged M1 proteins with FgD at 37 °C. Bound FgD was assessed through Coomassie-stained SDS/PAGE. (D) HBP released in whole blood that was incubated with M1 constructs, as assayed by ELISA. PBS with no bacteria was used as a negative control. Values represent the mean from two donors, with samples from each donor being measured in duplicate; the SD is shown. (E) Ni2+-NTA agarose coprecipitation assay for interaction of His-tagged M1 proteins with FgD at 37 °C carried out in the presence of 3.5 M urea. Bound FgD was assessed through Coomassie-stained SDS/PAGE.
Fig. S2.
Intradimer versus interdimer disulfide bond formation. Disulfide bond formation at 10-fold higher (0.5 mg/mL) or the same concentration (0.05 mg/mL) as in Fig. 4A, as assessed by nonreducing SDS/PAGE and visualized by Western blot using an anti-His antibody.
Stable Coiled Coils Are Not Competent to Bind Fg.
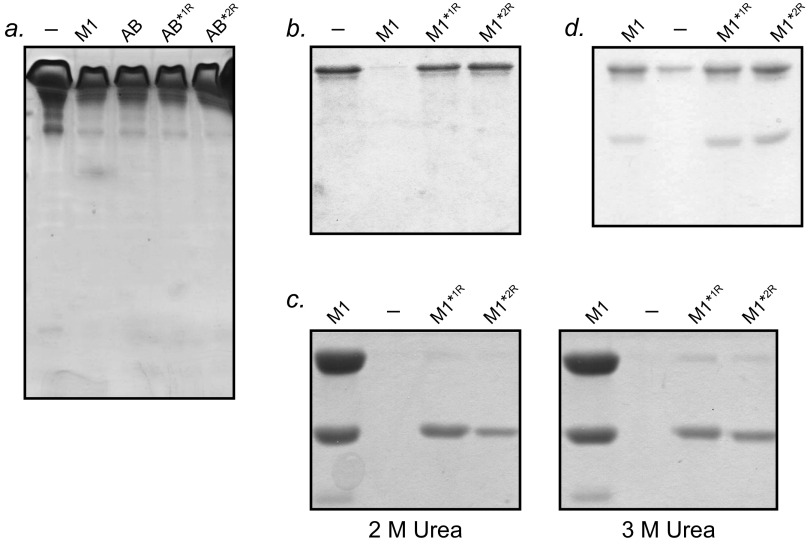
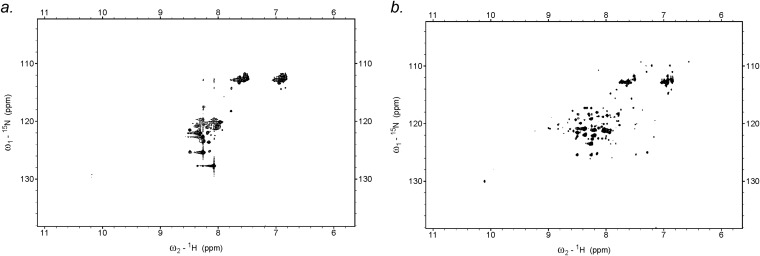
We next sought to verify that AB*2R, which is stabilized in the Fg-binding conformation, indeed binds Fg. A Ni2+-NTA agarose coprecipitation assay was carried out using Fg fragment D (FgD) and His-tagged AB proteins (Fig. 4B and Fig. S3A). AB*1R was greatly attenuated in Fg binding, as expected because of its stabilization in the Fg-nonbinding register, but counterintuitively, so too was AB*2R. Although the CD results indicated that AB*2R was a stable molecule, its unexpected deficiency in Fg binding compelled us to examine its structure further. We collected HSQC NMR spectra of AB*2R and AB (Fig. S4). The spectrum of AB showed broad peaks with very little dispersion (resonances confined to random coil region of ∼7–9 ppm), as is characteristic of intrinsically disordered regions (18, 19). In contrast, the spectrum of AB*2R showed clear dispersion and distinct peaks, indicative of a well-folded protein. This NMR result provides further evidence that AB is structurally unstable, whereas AB*2R is structurally stable, and suggests that the deficiency in Fg binding in AB*2R is a result of its increased stability and concomitant loss of dynamics.
Fig. S3.
Interaction with Fg. (A) Unbound proteins from Ni2+-NTA coprecipitation assay for interaction of His-tagged AB proteins with FgD, as shown in Fig. 4B. (B) Unbound proteins from Ni2+-NTA coprecipitation assay for interaction of His-tagged M1 proteins with FgD, as shown in Fig. 4C. (C) Ni2+-NTA agarose coprecipitation assay for interaction of His-tagged M1 proteins with FgD at 37 °C carried out in the presence of 2 M (Left) or 3 M (Right) urea. Bound FgD was assessed through Coomassie-stained SDS/PAGE. (D) Unbound proteins from Ni2+-NTA coprecipitation assay for interaction of His-tagged M1 proteins with FgD, as shown in Fig. 4E.
Fig. S4.
AB*2R is a structured protein. 1H-15N HSQC spectra of (A) AB (B) and AB*2R collected at 26 °C.
We next asked what effects idealization of register 2 has on intact M1 by introducing the register 2-stabilizing substitutions into the intact protein. M1, M1*1R, and M1*2R had identical secondary structures at 4 °C, and thermal melts of these proteins revealed a pattern similar to that observed for the AB fragment, with the idealized mutants maintaining greater α-helical content at higher temperatures (Fig. 3 C and D). M1*2R appeared to be the most stable of the three. Most pertinently, coprecipitation experiments with His-tagged versions of the M1 constructs also revealed significantly diminished binding to FgD by M1*2R, similar to the loss observed for M1*1R, compared with wild-type M1 (Fig. 4C and Fig. S3B).
To further verify loss of Fg binding, we examined whether the M1*1R or M1*2R register-stabilized proteins could form a proinflammatory, supramolecular network in conjunction with Fg. This network leads to the activation of neutrophils and consequent release of heparin binding protein (HBP), a vasodilator and clinical marker for sepsis (8, 15). When wild-type M1 protein was incubated with whole human blood, HBP was released, which is consistent with previous reports (15) (Fig. 4D). However, when M1*1R or M1*2R proteins, or an M1 protein with its Fg-binding sites deleted (labeled M1ΔFg, and equivalent to M1Δ98B1B2) (8), was incubated with whole human blood, no release of HBP was detected, consistent with the substantial decrease in Fg binding by the register-idealized mutants.
Fg Binding Promoted by Destabilization.
Because Fg binding was compromised by stabilization through idealization, we surmised that destabilization of the idealized M1 proteins should promote Fg binding. To test this, we explored the effect of the chaotrope urea on the binding of wild-type and register-stabilized M1 proteins to FgD. Coprecipitation assays were carried out as described earlier, except that urea was included. Two and 3 M urea had no effect on interaction: wild-type M1 protein bound FgD, but neither M1*1R nor M1*2R did (Fig. S3C). However, at 3.5 M urea, a significant effect was observed (Fig. 4E and Fig. S3D). At this chaotrope concentration, both M1*1R and M1*2R proteins bound FgD at levels approaching that of wild-type M1 protein. These results indicate that M1*1R and M1*2R have the capability to bind Fg, but only do so when they are destabilized. These results further indicate that the only deficiency in the register-stabilized M1 proteins with respect to Fg binding is increased stability.
Surface-Expressed Constructs Remain Incompetent to Bind Fg.
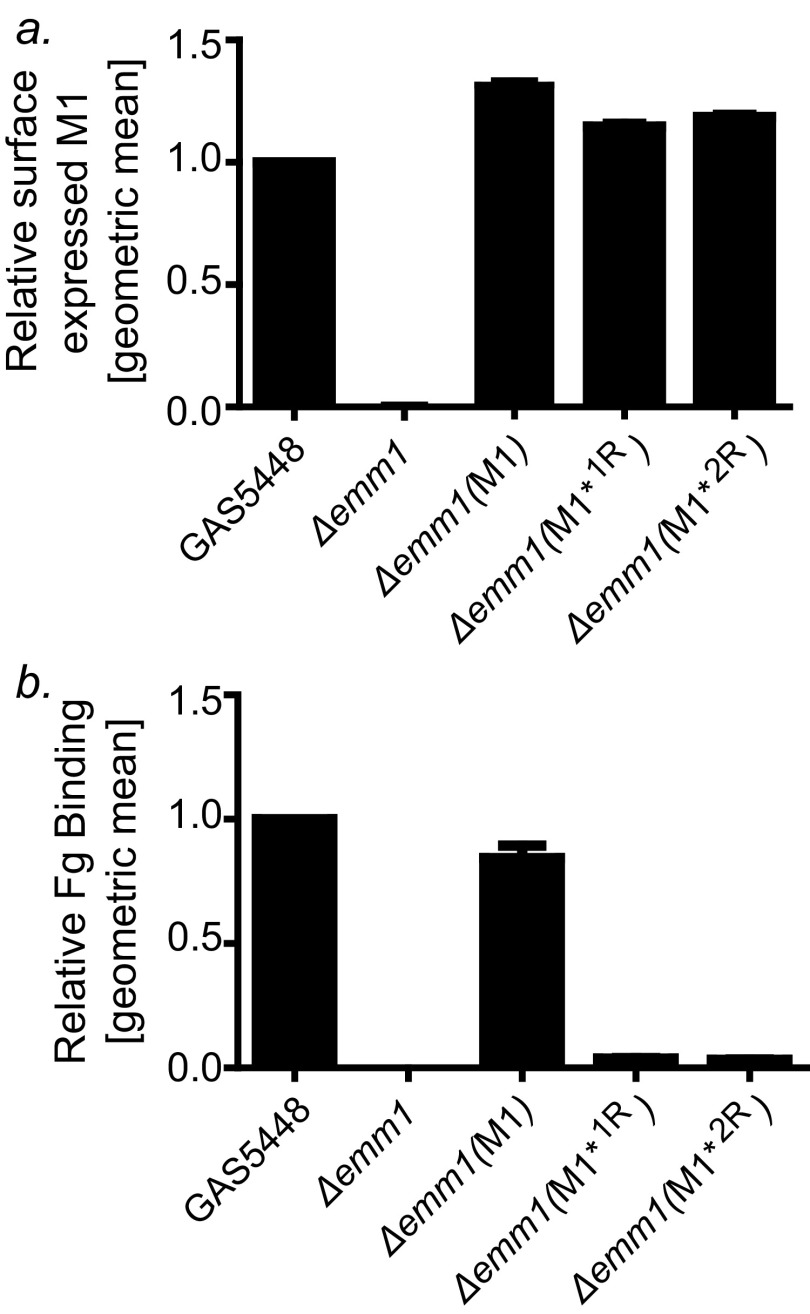
Last, we asked whether conformational dynamics were required for processes related to GAS cell surface-localization and cell surface-display of M1 protein. Genes encoding M1*1R and M1*2R were expressed in a GAS (Δemm1) mutant strain (20). The strains encoding these register-idealized variants expressed equivalent levels of surface-exposed M1 protein as the wild-type GAS 5448 parental strain, as determined by FACS analysis (Fig. S5A). Whereas the strain expressing wild-type M1 natively from the chromosome or from a complementing plasmid bound intact Fg, the strains expressing M1*1R or M1*2R did not, despite the M1–Fg interaction being potentially enhanced by avidity (Fig. S5B). These results show that there is no inherent barrier presented by coiled-coil stability for targeting to and display of M1 protein on the GAS cell surface. Furthermore, loss in dynamics of GAS surface-associated M1 protein was shown to block Fg binding in the context of the living bacterium.
Fig. S5.
Idealized M1 proteins on the GAS surface. (A) Surface expression of M1 protein by wild-type GAS 5448, GAS 5448 (Δemm1) carrying an empty plasmid or plasmids encoding wild-type M1, register 1-stabilized M1, or register 2-stabilized M1, as assayed by FACS, using a polyclonal anti-M1 protein antibody. The values are the means of three triplicates normalized to GAS 5448, with the SD shown. (B) Binding of FITC-labeled Fg to the same strains as in A as assayed by FACS. The values are normalized to the value for GAS 5448, with the SD shown.
Discussion
A paradoxical feature of M proteins is that they have sequences with strong propensities to form coiled coils, and yet these sequences are peppered with coiled-coil destabilizing residues (5). This unusual pattern is manifested in the instability of M protein coiled coils, with M proteins becoming less structured at physiological temperature (5, 9, 12). Here we present an explanation for this puzzling pattern in M protein sequences.
We showed that coiled-coil destabilizing residues in the B repeats of the M1 protein are required for interaction with Fg. This was accomplished by idealizing registers 1 and 2 such that Vals and Leus occupied the core heptad a and d positions in each register, while leaving untouched the two to three residues that contact FgD. These substitutions were verified by means of disulfide probes to restrain the unstable AB fragment of M1, as well as the more stable intact M1 protein to either register 1 or 2. The disulfide probes also furnished evidence that wild-type M1 protein sampled both registers 1 and 2, and prior experiments indicated that M1 protein also sampled a dissociated state (9). The current work extends these results to show through NMR that the AB fragment has characteristics of intrinsically disordered proteins (18, 19). Idealization of the B repeats yielded AB and M1 proteins that were more stable than their wild-type counterparts, as demonstrated through biophysical experiments. As expected, the AB and M1 proteins stabilized in the Fg-nonbinding register 1 were substantially reduced in Fg binding. The surprising result was that the AB and M1 proteins stabilized in the Fg-binding register 2 were likewise substantially attenuated in Fg binding. The B repeats in the crystal structure of the Fg-bound state of M1 have a canonical coiled-coil structure (8), and the idealizing mutations would tend to reinforce this canonical structure. The loss of binding in the register 2-idealized M1 protein suggested that increased stability was counter to Fg interaction. This hypothesis was definitively substantiated by the demonstration that both register 1- and register 2-stabilized M1 proteins became competent to bind Fg, once they were destabilized by the chaotrope urea. This result indicated that the only deficiency in the register-stabilized M1 proteins for Fg binding was their increased stability. Overall, these results support the conclusion that the coiled-coil destabilizing residues of M1 protein are required for promoting conformational dynamics, which are essential to interaction of M1 protein with Fg.
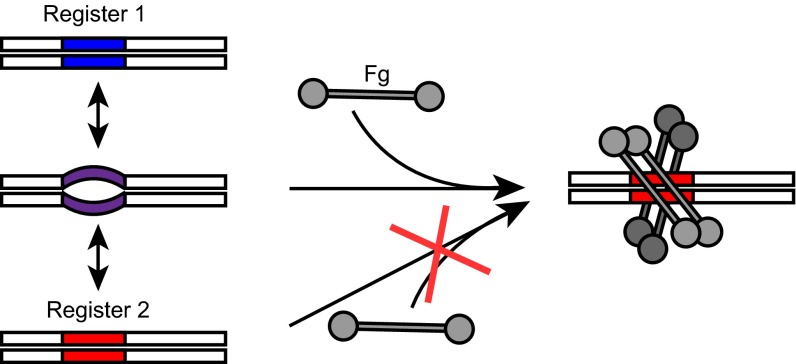
Why would dynamics be necessary for interaction? It seems evident that a perfect coiled coil would be limited in its conformational space, and therefore also in its ability to create specific binding interfaces. In contrast, a dynamic coiled coil would occupy greater conformational space, and therefore would be more likely to form specific binding sites. The observation that stabilization attenuates Fg binding is consistent with a “capture-and-collapse” model of binding (Fig. 5), as suggested for split inteins (21). The evidence indicates that the B repeats have multiple states, including intrinsically disordered, dissociated, as well as folded ones (i.e., registers 1 and 2). We surmise that the intrinsically disordered state initiates association with Fg (“capture”), and as a corollary that the B repeats in registers 1 or 2 are incompetent to do so. After this initial capture, the B repeats then “collapse” into register 2 as a result of stabilization of the coiled coil from energy provided by the Fg-binding event. This collapse is consistent with the observation that interaction of M proteins with their ligands results in stabilization of M proteins (5, 22).
Fig. 5.
Capture-and-collapse model of M1–Fg interaction. The B repeats of M1 protein are flexible and sample register 1 (blue), register 2 (red), dissociated, and intrinsically disordered (purple) states. The intrinsically disordered state captures Fg, after which the B repeats collapse into a register 2 coiled coil.
A similar example of competing coiled-coil registers has been documented in the cell cycle-regulated kinase Nek2 (23). The two registers in Nek2 were detected simultaneously in NMR spectra. However, unlike the case for M1 protein, the functional significance of competing dual registers in Nek2 is unknown. The authors speculated that the competing Nek2 registers may act as a “toggle” in signal transmission, analogous to the coiled coil in the HAMP domain of Af1503 and the microtubule-binding domain of dynein. In the former, a conformational shift in registers of a tetrameric coiled coil is necessary to transmit signals across a membrane (24, 25), and in the latter, helix sliding in the dynein coiled coil is required for communication between the ATPase and microtubule binding domains of dynein (26, 27). These toggle signaling mechanisms differ from the capture-and-collapse mechanism we propose for M1 protein. For the toggle mechanism, the two separate and competing registers are significant in and of themselves, each having a separate functional role. In contrast, for the capture-and-collapse mechanism of M1 protein, the two separate and competing registers are not significant in and of themselves, but instead the conformational dynamics that arise from the competing conformations are the significant aspect. In this regard, it is possible that Nek2 also belongs to the capture-and-collapse mechanism for a target that has not yet been identified.
In summary, the results presented here provide a general explanation for the puzzling prevalence of coiled-coil destabilizing residues in M proteins. These results support a model in which specific recognition by coiled coils relies on conformational dynamics brought about by destabilizing residues. We suggest that regions of the M protein that have ideal coiled coil sequences have structural purposes, and those that have nonideal coiled coil sequences have functional ones, especially in creating specific binding sites for host ligands. Although coiled coil structure is easily identified in primary sequence through heptad patterns, specific variations in that pattern can result in complex dynamics, thereby adding an additional level of intricacy and functionality to the coiled coil.
Materials and Methods
Cloning and DNA Manipulation.
Coding sequences for mature M1 protein (residues 42–453) and its AB fragment (residues 42–194) were cloned from GAS strain 5448 into pET28b. The constructs included an N-terminal His6-tag followed by a PreScission protease cleavage site or a C-terminal His6-tag. Single-site substitutions were introduced by QuikChange (Agilent Technologies). Idealization of the B repeats in register 2 was achieved by synthesizing a DNA fragment in which L137, L144, A158, L165, L172, and A186 were substituted with valine and K140, A147, K168, and A175 with leucine. This DNA fragment was incorporated into the coding sequence for mature M1 protein, as described previously (9).
Protein Expression and Purification.
Proteins were expressed in Escherichia coli BL21 (DE3) Gold (Agilent). Bacteria were grown with shaking in LB media at 37 °C to midlog phase and induced with 1 mM isopropyl-β-d-thiogalactopyranoside. Bacteria were grown further at 22 °C for 18 h with shaking and then centrifuged (2,000 × g, 30 min, 4 °C). Bacterial pellets were resuspended in lysis buffer (300 mM NaCl, 100 mM NaPi at pH 7.5, 0.5 mM phenylmethanesulfonyl fluoride; 20 mL per liter initial bacterial culture) containing 200 µg/mL lysozyme and 100 µg/mL DNase, and lysed using an EmulsiFlex-C5 (Avestin; 20,000 psi with three passes). Lysates were clarified by centrifugation (35,000 × g, 45 min, 4 °C), filtered through a 0.22 μm membrane (Millipore), and added to a Ni2+-NTA agarose column [room temperature (RT), 5 mL resin per liter of initial bacterial culture; Sigma] that had been pre-equilibrated with lysis buffer. The column was washed with 50 column volumes of wash buffer (500 mM NaCl, 50 mM NaPi at pH 7.5, 20 mM imidazole) and eluted with three column volumes of elution buffer (300 mM NaCl, 100 mM NaPi at pH 7.5, 500 mM imidazole). Purified proteins were dialyzed against 2 L buffer A (75 mM NaCl, 20 mM NaPi at pH 7.5) at 4 °C overnight (3500 MWCO tubing; Spectrum Laboratories). In cases in which the His6-tag was removed, eluates were incubated with PreScission Protease (0.5 mg/2 L initial bacterial culture) and 5 mM DTT, and proteins were further purified by Ni2+-NTA chromatography as described earlier, except that the flow-through was collected. Proteins were concentrated to 2–17 mg/mL by ultrafiltration (3500 MWCO membrane; Amicon), aliquoted, and flash frozen in liquid N2 for storage at −80 °C.
Cysteine Probes.
Cys substitution mutants of M1, AB*1R, and AB*2R were prepared by dilution to 0.5 mg/mL in buffer A containing 100 mM DTT and heating to 60 °C for 3 h. Samples were next diluted 1:10–1:100 in buffer A and incubated at RT for 3 h. Before analysis, samples were incubated with 50 mM iodoacetamide for 15 min at RT, resolved by SDS/PAGE, and visualized by Coomassie staining or Western blot. For Western blot analysis, samples were transferred from the polyacrylamide gel to a polyvinylidene fluoride membrane (Millipore) at 100 V for 40 min. Membranes were incubated in TBS (150 mM NaCl, 50 mM Tris at pH 8.0) for 10 min and then blocked with 5% (wt/vol) nonfat milk in TBS for 30 min. Membranes were incubated in TBS containing 0.05% Tween-20 (TBST) and 5% (wt/vol) nonfat milk, and mouse horseradish peroxidase-conjugated anti-His tag monoclonal antibody (1:2,000; Santa Cruz Biotechnologies 8036) or rabbit anti-M1 protein polyclonal antibody (1:500) at 4 °C overnight. Membranes were washed three times in TBST for 15 min each. Membranes incubated with anti-M1 protein antibodies were incubated in TBST containing 5% (wt/vol) nonfat milk, and horseradish peroxidase-conjugated goat anti-rabbit IgG (1:5,000; Santa Cruz Biotechnologies 2004) for 30 min at RT and washed as described earlier. SuperSignal West chemiluminescent substrate (Thermo Fisher Scientific) was used for detection, according to the manufacturer’s instructions.
CD Spectroscopy.
CD spectra were collected between 195 and 260 nm at 4 °C with 1-nm intervals, and between 4 °C and 95 °C at 222 nm with 1° intervals, using a quartz cell with a 1-cm pathlength on a model 202 spectrometer (Aviv Instruments) equipped with thermoelectric temperature control. For AB fragments, measurements were collected on samples at 9.8–12.2 µM, as determined by absorption, using a calculated absorption coefficient, in 40 mM NaF, 10 mM NaPi at pH 7.5. For intact M1, measurements were collected on samples at 3.6–5.7 µM in 40 mM NaF, 20 mM NaPi at pH 7.5. The CD signal from buffer alone was subtracted from the data before conversion to mean residue ellipticity.
NMR Spectroscopy.
AB and AB*2R were grown and expressed in minimal media supplemented with 15NH4Cl (Cambridge Isotopes) and purified as described earlier. Purified protein at 3 mg/mL was dialyzed into 75 mM NaCl, 20 mM Tris at pH 7.5, as described earlier, and supplemented with 10% (vol/vol) D2O. NMR experiments were recorded at 26 °C on a Varian VNMRS 800MHz spectrometer, equipped with a 5-mm triple-resonance coldprobe. 2D 1H-15N HSQC were recorded using standard pulse sequence included in the Varian BioPack. Data were processed using NMRPipe software.
Fg Binding Assay.
Fifty microliters of His6-tagged M1 or AB constructs at 20 µM were incubated for 1 h at 37 °C under rotation with 50 µL Ni2+-NTA agarose beads that had been pre-equilibrated with buffer A. Fifty microliters of 20 µM Fg fragment D (Calbiochem 341578) in buffer A, prepared as described previously (28), was added to the resin and incubated for 1 h at 37 °C with rotation. When urea was included, Ni2+-NTA agarose beads were added after prior incubation of M1 and FgD for 1 h at 37 °C, and samples were then further incubated at 4 °C with agitation for 1 h. Samples were centrifuged (500 × g, RT, 1 min) and the supernatant removed. The resin was washed with 1 mL buffer A containing 20 mM imidazole and centrifuged (500 × g, RT, 1 min), after which the supernatant was again removed. This washing step was repeated three times, and proteins bound to the resin were eluted with 30 µL buffer A containing 500 mM imidazole. The unbound fraction and elution samples were resolved and visualized by Coomassie-stained SDS/PAGE.
Heparin Binding Protein Release.
Human blood (100 μL) collected from healthy donors was diluted in PBS to 1.0 mL and incubated with 2 µM M proteins for 45 min. Samples were centrifuged (300 × g, RT, 15 min), and the supernatant was analyzed for HBP by sandwich ELISA, following the manufacturer’s instructions (MyBiosource.com Human Heparin Binding Protein ELISA kit).
Fg Binding by Complemented Strains.
Genes encoding M1*1R and M1*2R were cloned into pDCErm and transformed into electrocompetent GAS 5448 (Δemm1) bacteria, as previously described (20). Bacterial surface expression of M protein and surface binding of Fg was assayed through flow cytometry. For this, overnight growths of GAS were diluted into 10 mL THB and grown to midlog phase. Cultures were pelleted by centrifugation (3,000 × g, 10 min, 20 °C) and washed with 5 mL PBS. Bacteria were resuspended in PBS and incubated with either anti-M1 antibody or naive serum (1:1,000; 60 min, 4 °C), or 10 µg/mL FITC-labeled Fg generated with the Fluoreporter kit (Life Technologies). Bacteria were pelleted (3,000 × g, 10 min, 20 °C) and washed twice with PBS containing 0.1% BSA. Samples probed for M1 were incubated with the secondary antibody Alexa-488 anti-mouse IgG (1:500, 60 min, 4 °C; Life Technologies #A11001). Samples were pelleted by centrifugation (3,000 × g, 10 min, 20 °C) and washed as earlier. After the last wash, samples were resuspended in PBS and analyzed by flow cytometry, using a FACS Canto.
Acknowledgments
This work was supported by NIH Grant T32 GM008326 (to C.M.S.), NIH Grant T32 GM007240 (to C.Z.B.), American Heart Association Predoctoral Fellowship 14PRE18320032 (to C.Z.B.), NIH Grant R01 AI096837 (to P.G. and V.N.), and NIH Grant R01 AI077780 (to V.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606160113/-/DCSupplemental.
References
- 1.Walker MJ, et al. Disease manifestations and pathogenic mechanisms of Group A Streptococcus. Clin Microbiol Rev. 2014;27(2):264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghosh P. The nonideal coiled coil of M protein and its multifarious functions in pathogenesis. Adv Exp Med Biol. 2011;715:197–211. doi: 10.1007/978-94-007-0940-9_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole JN, Barnett TC, Nizet V, Walker MJ. Molecular insight into invasive group A streptococcal disease. Nat Rev Microbiol. 2011;9(10):724–736. doi: 10.1038/nrmicro2648. [DOI] [PubMed] [Google Scholar]
- 4.Hosein B, McCarty M, Fischetti VA. Amino acid sequence and physicochemical similarities between streptococcal M protein and mammalian tropomyosin. Proc Natl Acad Sci USA. 1979;76(8):3765–3768. doi: 10.1073/pnas.76.8.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nilson BH, et al. Structure and stability of protein H and the M1 protein from Streptococcus pyogenes. Implications for other surface proteins of gram-positive bacteria. Biochemistry. 1995;34(41):13688–13698. doi: 10.1021/bi00041a051. [DOI] [PubMed] [Google Scholar]
- 6.Cedervall T, Akesson P, Stenberg L, Herrmann A, Akerström B. Allosteric and temperature effects on the plasma protein binding by streptococcal M protein family members. Scand J Immunol. 1995;42(4):433–441. doi: 10.1111/j.1365-3083.1995.tb03677.x. [DOI] [PubMed] [Google Scholar]
- 7.André I, et al. Streptococcal M protein: Structural studies of the hypervariable region, free and bound to human C4BP. Biochemistry. 2006;45(14):4559–4568. doi: 10.1021/bi052455c. [DOI] [PubMed] [Google Scholar]
- 8.Macheboeuf P, et al. Streptococcal M1 protein constructs a pathological host fibrinogen network. Nature. 2011;472(7341):64–68. doi: 10.1038/nature09967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNamara C, et al. Coiled-coil irregularities and instabilities in group A Streptococcus M1 are required for virulence. Science. 2008;319(5868):1405–1408. doi: 10.1126/science.1154470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252(5009):1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 11.Woolfson DN, Alber T. Predicting oligomerization states of coiled coils. Protein Sci. 1995;4(8):1596–1607. doi: 10.1002/pro.5560040818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cedervall T, Johansson MU, Akerström B. Coiled-coil structure of group A streptococcal M proteins. Different temperature stability of class A and C proteins by hydrophobic-nonhydrophobic amino acid substitutions at heptad positions a and d. Biochemistry. 1997;36(16):4987–4994. doi: 10.1021/bi962971q. [DOI] [PubMed] [Google Scholar]
- 13.Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. Global emm type distribution of group A streptococci: Systematic review and implications for vaccine development. Lancet Infect Dis. 2009;9(10):611–616. doi: 10.1016/S1473-3099(09)70178-1. [DOI] [PubMed] [Google Scholar]
- 14.Uchiyama S, et al. Coiled-coil irregularities of the M1 protein structure promote M1-fibrinogen interaction and influence group A Streptococcus host cell interactions and virulence. J Mol Med (Berl) 2013;91(7):861–869. doi: 10.1007/s00109-013-1012-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herwald H, et al. M protein, a classical bacterial virulence determinant, forms complexes with fibrinogen that induce vascular leakage. Cell. 2004;116(3):367–379. doi: 10.1016/s0092-8674(04)00057-1. [DOI] [PubMed] [Google Scholar]
- 16.Berge A, Björck L. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J Biol Chem. 1995;270(17):9862–9867. doi: 10.1074/jbc.270.17.9862. [DOI] [PubMed] [Google Scholar]
- 17.Ringdahl U, et al. A role for the fibrinogen-binding regions of streptococcal M proteins in phagocytosis resistance. Mol Microbiol. 2000;37(6):1318–1326. doi: 10.1046/j.1365-2958.2000.02062.x. [DOI] [PubMed] [Google Scholar]
- 18.Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 19.Dyson HJ. Making sense of intrinsically disordered proteins. Biophys J. 2016;110(5):1013–1016. doi: 10.1016/j.bpj.2016.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauth X, et al. M1 protein allows Group A streptococcal survival in phagocyte extracellular traps through cathelicidin inhibition. J Innate Immun. 2009;1(3):202–214. doi: 10.1159/000203645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shah NH, Eryilmaz E, Cowburn D, Muir TW. Naturally split inteins assemble through a “capture and collapse” mechanism. J Am Chem Soc. 2013;135(49):18673–18681. doi: 10.1021/ja4104364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gubbe K, et al. C repeats of the streptococcal M1 protein achieve the human serum albumin binding ability by flanking regions which stabilize the coiled-coil conformation. Biochemistry. 1997;36(26):8107–8113. doi: 10.1021/bi962991s. [DOI] [PubMed] [Google Scholar]
- 23.Croasdale R, et al. An undecided coiled coil: The leucine zipper of Nek2 kinase exhibits atypical conformational exchange dynamics. J Biol Chem. 2011;286(31):27537–27547. doi: 10.1074/jbc.M110.196972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hulko M, et al. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126(5):929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 25.Ferris HU, et al. The mechanisms of HAMP-mediated signaling in transmembrane receptors. Structure. 2011;19(3):378–385. doi: 10.1016/j.str.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Kon T, et al. Helix sliding in the stalk coiled coil of dynein couples ATPase and microtubule binding. Nat Struct Mol Biol. 2009;16(3):325–333. doi: 10.1038/nsmb.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishikawa Y, et al. Structure of the entire stalk region of the Dynein motor domain. J Mol Biol. 2014;426(19):3232–3245. doi: 10.1016/j.jmb.2014.06.023. [DOI] [PubMed] [Google Scholar]
- 28.Everse SJ, Pelletier H, Doolittle RF. Crystallization of fragment D from human fibrinogen. Protein Sci. 1995;4(5):1013–1016. doi: 10.1002/pro.5560040523. [DOI] [PMC free article] [PubMed] [Google Scholar]