Abstract
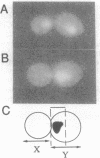
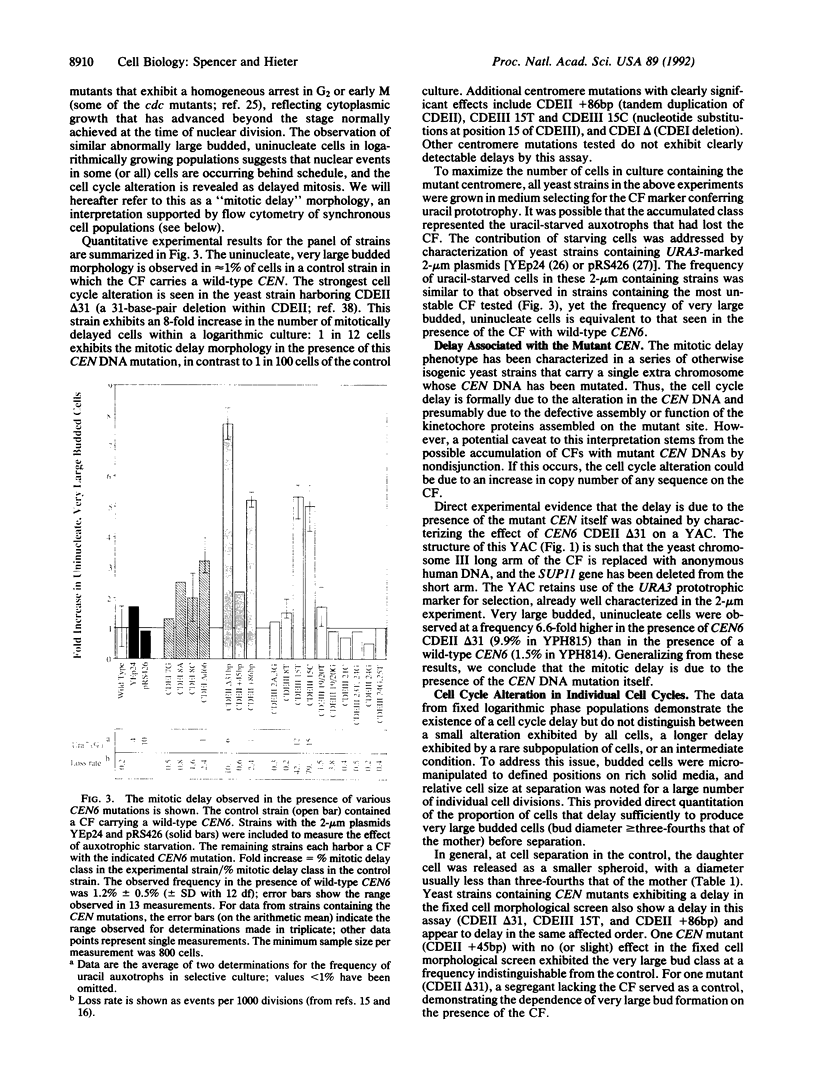
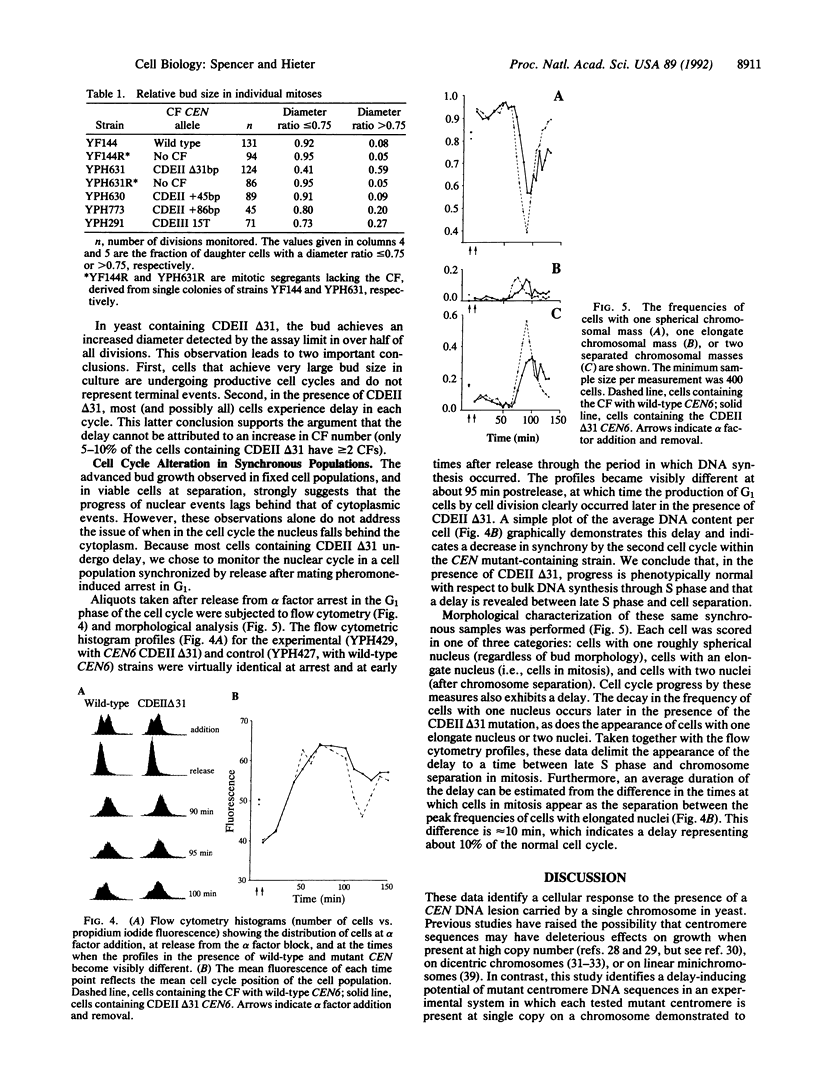
Cytological observations of animal cell mitoses have shown that the onset of anaphase is delayed when chromosome attachment to the spindle is spontaneously retarded or experimentally interrupted. This report demonstrates that a centromere DNA (CEN) mutation carried on a single chromosome can induce a cell cycle delay observed as retarded mitosis in the yeast Saccharomyces cerevisiae. A 31-base-pair deletion within centromere DNA element II (CDEII delta 31) that causes chromosome missegregation in only 1% of cell division elicited a dramatic mitotic delay phenotype. Other CEN DNA mutations, including mutations in centromere DNA elements I and III, similarly delayed mitosis. Single division pedigree analysis of strains containing the CDEII delta 31 CEN mutation indicated that most (and possibly all) cells experienced delay in each cell cycle and that the delay was not due to increased chromosome copy number. Furthermore, a synchronous population of cells containing the CDEII delta 31 mutation underwent DNA synthesis on schedule with wild-type kinetics, but subsequently exhibited late chromosomal separation and concomitant late cell separation. We speculate that this delay in cell cycle progression before the onset of anaphase provides a mechanism for the stabilization of chromosomes with defective kinetochore structure. Further, we suggest that the delay may be mediated by surveillance at a cell cycle checkpoint that monitors the completion of chromosomal attachment to the spindle.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker R. E., Masison D. C. Isolation of the gene encoding the Saccharomyces cerevisiae centromere-binding protein CP1. Mol Cell Biol. 1990 Jun;10(6):2458–2467. doi: 10.1128/mcb.10.6.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Cai M., Davis R. W. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990 May 4;61(3):437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- Chang F., Herskowitz I. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell. 1990 Nov 30;63(5):999–1011. doi: 10.1016/0092-8674(90)90503-7. [DOI] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992 Jan 2;110(1):119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Elion E. A., Grisafi P. L., Fink G. R. FUS3 encodes a cdc2+/CDC28-related kinase required for the transition from mitosis into conjugation. Cell. 1990 Feb 23;60(4):649–664. doi: 10.1016/0092-8674(90)90668-5. [DOI] [PubMed] [Google Scholar]
- Futcher B., Carbon J. Toxic effects of excess cloned centromeres. Mol Cell Biol. 1986 Jun;6(6):2213–2222. doi: 10.1128/mcb.6.6.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985 Jul;110(3):381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Weinert T. A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989 Nov 3;246(4930):629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hegemann J. H., Shero J. H., Cottarel G., Philippsen P., Hieter P. Mutational analysis of centromere DNA from chromosome VI of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jun;8(6):2523–2535. doi: 10.1128/mcb.8.6.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A., Bloom K. Acquisition and processing of a conditional dicentric chromosome in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Mar;9(3):1368–1370. doi: 10.1128/mcb.9.3.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M. A., Totis L., Roberts B. T. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991 Aug 9;66(3):507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Hutter K. J., Eipel H. E. Flow cytometric determinations of cellular substances in algae, bacteria, moulds and yeasts. Antonie Van Leeuwenhoek. 1978;44(3-4):269–282. doi: 10.1007/BF00394305. [DOI] [PubMed] [Google Scholar]
- Jehn B., Niedenthal R., Hegemann J. H. In vivo analysis of the Saccharomyces cerevisiae centromere CDEIII sequence: requirements for mitotic chromosome segregation. Mol Cell Biol. 1991 Oct;11(10):5212–5221. doi: 10.1128/mcb.11.10.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D., Rutledge L., Fitzgerald-Hayes M., Hartwell L. H. A genetic analysis of dicentric minichromosomes in Saccharomyces cerevisiae. Cell. 1987 Mar 13;48(5):801–812. doi: 10.1016/0092-8674(87)90077-8. [DOI] [PubMed] [Google Scholar]
- Lechner J., Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991 Feb 22;64(4):717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- Li R., Murray A. W. Feedback control of mitosis in budding yeast. Cell. 1991 Aug 9;66(3):519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Mellor J., Jiang W., Funk M., Rathjen J., Barnes C. A., Hinz T., Hegemann J. H., Philippsen P. CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 1990 Dec;9(12):4017–4026. doi: 10.1002/j.1460-2075.1990.tb07623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W. Coordinating cell cycle events. Cold Spring Harb Symp Quant Biol. 1991;56:399–408. doi: 10.1101/sqb.1991.056.01.047. [DOI] [PubMed] [Google Scholar]
- Murray A. W., Szostak J. W. Pedigree analysis of plasmid segregation in yeast. Cell. 1983 Oct;34(3):961–970. doi: 10.1016/0092-8674(83)90553-6. [DOI] [PubMed] [Google Scholar]
- Newlon C. S. Yeast chromosome replication and segregation. Microbiol Rev. 1988 Dec;52(4):568–601. doi: 10.1128/mr.52.4.568-601.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R. B. Chromosome micromanipulation. II. Induced reorientation and the experimental control of segregation in meiosis. Chromosoma. 1967;21(1):17–50. doi: 10.1007/BF00330545. [DOI] [PubMed] [Google Scholar]
- Niedenthal R., Stoll R., Hegemann J. H. In vivo characterization of the Saccharomyces cerevisiae centromere DNA element I, a binding site for the helix-loop-helix protein CPF1. Mol Cell Biol. 1991 Jul;11(7):3545–3553. doi: 10.1128/mcb.11.7.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. Universal control mechanism regulating onset of M-phase. Nature. 1990 Apr 5;344(6266):503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- Panzeri L., Landonio L., Stotz A., Philippsen P. Role of conserved sequence elements in yeast centromere DNA. EMBO J. 1985 Jul;4(7):1867–1874. doi: 10.1002/j.1460-2075.1985.tb03862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C. L. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int Rev Cytol. 1982;79:1–58. doi: 10.1016/s0074-7696(08)61672-1. [DOI] [PubMed] [Google Scholar]
- Runge K. W., Wellinger R. J., Zakian V. A. Effects of excess centromeres and excess telomeres on chromosome loss rates. Mol Cell Biol. 1991 Jun;11(6):2919–2928. doi: 10.1128/mcb.11.6.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears D. D., Hegemann J. H., Hieter P. Meiotic recombination and segregation of human-derived artificial chromosomes in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5296–5300. doi: 10.1073/pnas.89.12.5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shero J. H., Hieter P. A suppressor of a centromere DNA mutation encodes a putative protein kinase (MCK1). Genes Dev. 1991 Apr;5(4):549–560. doi: 10.1101/gad.5.4.549. [DOI] [PubMed] [Google Scholar]
- Shero J. H., Koval M., Spencer F., Palmer R. E., Hieter P., Koshland D. Analysis of chromosome segregation in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:749–773. doi: 10.1016/0076-6879(91)94057-j. [DOI] [PubMed] [Google Scholar]
- Smith D. R., Smyth A. P., Moir D. T. Amplification of large artificial chromosomes. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8242–8246. doi: 10.1073/pnas.87.21.8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H. Characterization of RAD9 of Saccharomyces cerevisiae and evidence that its function acts posttranslationally in cell cycle arrest after DNA damage. Mol Cell Biol. 1990 Dec;10(12):6554–6564. doi: 10.1128/mcb.10.12.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988 Jul 15;241(4863):317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- Zirkle R. E. Ultraviolet-microbeam irradiation of newt-cell cytoplasm: spindle destruction, false anaphase, and delay of true anaphase. Radiat Res. 1970 Mar;41(3):516–537. [PubMed] [Google Scholar]