Abstract
The Ebola virus (EBOV) GP gene encodes two glycoproteins. The major product is a soluble, dimeric glycoprotein termed sGP that is secreted abundantly. Despite the abundance of sGP during infection, little is known regarding its structure or functional role. A minor product, resulting from transcriptional editing, is the transmembrane-anchored, trimeric viral surface glycoprotein termed GP. GP mediates attachment to and entry into host cells, and is the intended target of antibody therapeutics. Because large portions of sequence are shared between GP and sGP, it has been hypothesized that sGP may potentially subvert the immune response or may contribute to pathogenicity. In this study, we present cryo-EM structures of GP and sGP in complex with GP-specific and GP/sGP cross-reactive antibodies undergoing human clinical trials. The structure of the sGP dimer presented here, in complex with both an sGP-specific antibody and a GP/sGP cross-reactive antibody, permits us to unambiguously assign the oligomeric arrangement of sGP and compare its structure and epitope presentation to those of GP. Further, we provide biophysical evaluation of naturally occurring GP/sGP mutations that fall within the footprints identified by our high-resolution structures. Taken together, our data provide a detailed and more complete picture of the accessible Ebolavirus glycoprotein landscape and a structural basis to evaluate patient and vaccine antibody responses toward differently structured products of the GP gene.
Summary
Zaire Ebola virus (EBOV) and related viruses of the family Filoviridae are highly lethal and have caused numerous outbreaks since emerging in 1967, including a sustained epidemic in West Africa from 2013–2015 (www.cdc.gov). No vaccines or therapeutics against EBOV are yet approved, although several have shown promise in animal models and have moved forward to human clinical trials1–3. The main target of these candidate vaccines and treatments is the viral-surface trimeric GP, which includes GP1 (receptor binding) and GP2 (viral fusion) subunits4,5. However, the major product of the GP gene is not viral-surface GP, but instead a secreted, dimeric glycoprotein termed sGP6. GP and sGP share 295 N-terminal amino acids, but have distinct C-terminal regions as a result of transcriptional editing6. The unique C terminus of sGP contains 65 amino acids and includes a Cys at position 306 that is critical for sGP dimerization7. In contrast, the unique C terminus of viral-surface GP contains 381 amino acids that assemble a large, heavily glycosylated mucin-like domain (MLD), the viral fusion machinery and a transmembrane domain8. Antibodies elicited during infection that cross-react to sGP and GP9 can potentially be absorbed by sGP, although the consequences of such are unclear10.
A candidate immunotherapeutic in clinical trials, ZMapp™, contains two GP-specific antibodies11,12 (c2G4 and c4G7) and one GP/sGP-cross reactive antibody13,14 (c13C6)15. The binding sites of each ZMapp™ component on EBOV GP have been generally described at low resolution (~20–25 Å)16,17, as well as by alanine scanning18, and the two GP-specific antibodies were found to compete. Detailed description and interpretation of differences between these epitopes, the structure of sGP, and reasons why c13C6 is effective have remained elusive.
Results and Discussion
CryoEM structures of EBOV GP in complex with ZMapp™ antibodies
In the work reported here, we used cryo-EM to study the fragments antigen binding (Fabs) of c2G4, c4G7 and c13C6 in complex with soluble, MLD-containing GP (GPΔTM) trimer and sGP dimer. The cryo-EM reconstructions of GP complexes (c13C6-c2G4-GP and c13C6-c4G7-GP) were resolved to ~4.3Å and ~4.9Å resolution, respectively (Fig. 1a,b, Supplementary Fig. 1–3, Supplementary Table 1). Both structures were solved using the fully glycosylated, MLD-containing GP (Fig. 1c, Supplementary Fig. 4)5,19 and the core of GPΔTM is similar to the MLD-deleted crystal structure (GPΔMuc) (Fig. 1d, Supplementary Table 7)4. We also resolved residues corresponding to the HR1-HR2 linker in GP2, which contains the GP1-GP2 disulfide linkages (Fig. 1d, Supplementary Fig. 5), a structural motif also observed in structures of Sudan virus (SUDV) GP20 and endosomally cleaved EBOV GP21. Further, at lower contour, density corresponding to a loop (residues 197–208) containing the cathepsin-cleavage site5,22, as well as additional portions of the glycan cap (278–302) were resolved (Supplementary Fig. 6). Our data suggest that the cathepsin-cleavage loop is well exposed and bridges over the internal fusion loop (IFL), similar to what has been observed in previous structures23. Finally, we observed density corresponding to five N-linked glycans in the core GP, linked to residues N228, N238, N257 and N268 in GP1 and N563 in GP2 (Fig. 1d, Supplementary Fig. 7).
Figure 1. Structural analyses of ZMapp™ - EBOV GP complexes.
a, Single particle cryo-EM reconstruction of antibodies c2G4 and c13C6 in complex with GPΔTM. There are three copies of each Fab per trimer. A single protomer from the map was segmented into c2G4 Fab (red), c13C6 Fab (blue), GP1 (cyan), and GP2 (yellow). b, Single particle cryo-EM reconstruction of antibodies c4G7 and c13C6 in complex with GPΔTM. There are two copies of each c13C6 Fab and three copies of each c4G7 Fab per trimer. A single protomer from the map was segmented into c4G7 Fab (orange), c13C6 Fab (blue), GP1 (cyan), and GP2 (yellow). c, Construct designs used for cryo-EM versus x-ray crystallography. Key differences are inclusion of the MLDs and all glycans in the proteins expressed for single particle cryo-EM. Predicted N-linked glycans are represented by magenta “Y” symbols. d, Overall structure of EBOV GP showing a side (above) and top (below) view relative to the viral membrane. Additional features not seen in the previous MLD-deleted structure include three N-linked glycans modeled at N238, N228 and N268, as well as the linker between HR1 and HR2. The putative location of the mucin-like domains, which were a part of the construct used for cryo-EM but unresolved in the maps, is indicated. SP = signal peptide (removed during processing) MPER = membrane proximal external region, TM = transmembrane region, CT = cytoplasmic tail, MLD = mucin-like domain. The TM and CT regions were not included in our constructs. Asterisks indicate glycans that were resolved in the crystal structure of EBOV GPΔMuc (PDB 3CSY).
c2G4 and c4G7 have overlapping footprints near the GP1/GP2 interface (Fig. 2a,b), similar to that of the human survivor mAb KZ524. KZ52 alone did not protect in non-human primate efficacy models24, yet c2G4 and c4G7, in combination with c13C6 did protect15. We delineated potential contacts between the Fab complementarity determining regions (CDRs) and GP by identifying amino acids of c2G4 and c4G7 residing in close proximity to complementary GP surfaces and compared them to those of KZ52 (Fig. 2d, Supplementary Fig. 8, Supplementary Table 2–3). Our results correspond well with a previous study that identified critical residues involved in the binding of ZMapp™ antibodies to GP (Supplementary Table 2)17. The common site of vulnerability for all three neutralizing antibodies (c2G4, c4G7 and KZ52) is the region surrounding the base of the IFL and the N terminus of GP2 and includes the residues V505, A507, P513, N550, Q551, D552, G553 and C556 (Fig. 2d,e). This region varies among the species of Ebolaviruses, possibly explaining the species-specificity of mAbs that bind to this region.
Figure 2. Details of the ZMapp™ antibody epitopes.
a, Epitope of antibody c2G4 bound to EBOV GPΔTM. The variable light chain is shown in dark red and the variable heavy chain in red-orange. GP2 is shown in yellow and GP1 in dark cyan. The major contacts are located at the base of the IFL and in HR1, which include residue Q508 (shown in green) and a glycan at N563. Contact regions between GP and c2G4 are resolved to ~3.9 Å (Supplementary Fig. 2). b, Epitopes and CDRs of antibody c4G7 bound to EBOV GPΔTM, with the variable light chain in dark orange and the variable heavy chain in yellow-orange. GP1 and GP2 are shown as in a). c4G7 is positioned differently from c2G4, more toward the IFL and the cathepsin loop. c4G7 also contacts part of GP1, below HR1 at the bottom of the GP chalice, in a manner similar to KZ52. Contact regions between GP and c4G7 are resolved to ~4.4 Å (Supplementary Fig. 2). c, Epitopes and CDRs of antibody c13C6 bound to EBOV GPΔTM (c2G4-c13C6 model) with variable light chain in dark blue and variable heavy chain in light blue. The major contacts are within the glycan cap, but some backbone contacts are made with the GP head. There is additional contact with glycans at N238 and N268. Contact regions between GP and c13C6 are resolved to ~4.1 Å (Supplementary Fig. 2). d, Epitopes of all three components of ZMapp™, including a comparison with antibody KZ52, which binds at the base of GP and overlaps with c2G4 and c4G7. For c13C6, contact residues are highlighted in shades of blue. For c2G4, c4G7 and KZ52, contact residues are highlighted in yellow (unique contact), orange (shared with one other base-binding mAb) or red (shared between all three base-binding mAbs). The asterisk indicates a known escape mutant for c2G4 and c4G7 at Q508. e, The GP1-GP2 interface epitope. A single protomer of the GP crystal structure PDB 3CSY is shown4. The boxed area is zoomed in on the right, with mAb contact residues colored as in panel d.
c2G4 binds primarily to GP2 in the core, including possible contact with a glycan attached to N563, while c4G7 has additional contacts in GP1 at the base of the trimer, similar to KZ52, consistent with previous Western Blot analysis (Fig. 2a,b,d)11. Notably, c4G7 makes potential contacts with the cathepsin loop via its heavy chain variable region (Supplementary Fig. 6). The heavy chain of c4G7 differs from the original mouse IgG2a that was used to generate c4G711. Chimerization of 4G7 was conducted without consideration of how the antibody framework might interact with GP. It is possible that the mouse IgG2a version of 4G7 had stronger interactions with GP mediated by the cathepsin loop, which were lost upon chimerization. Thus, structural evaluation prior to antibody engineering may mitigate loss of potentially favorable interactions.
We also determined CDR-GP contacts for c13C6 as outlined above (Fig. 2c,d, Supplementary Table 2)16. Overall, c13C6 binds to the top of the glycan cap (residues G264 to W275), which includes residues T270 and K272 previously identified by alanine scanning16. c13C6 makes close contact with glycans at N268 and N238 (Fig. 2c,d), both visible in the EM map (Fig. 1d, Supplementary Fig. 7). The c13C6 epitope also includes a portion of the GP1 head, which is contacted by the heavy chain framework (Fig. 2c,d). The loss of the majority of the c13C6 epitope upon cleavage in the endosome may explain why c13C6 exhibits less potent neutralization than other antibodies14.
CryoEM structure of EBOV sGP in complex with protective antibodies
Notably, c13C6 binds both GP and sGP. In order to study the relatively small sGP dimer (~70 kDa without glycans) by cryo-EM, we added Fabs to increase particle size and improve particle alignment25. Two Fabs were bound to sGP: c13C6 and BDBV91, which was isolated from a human survivor of Bundibugyo virus (BDBV). BDBV91 is non-neutralizing, binds sGP in a region distinct from c13C6, and cross-reacts among BDBV, EBOV and SUDV9.
The EBOV sGP-c13C6-BDBV91 ternary complex was resolved to ~5.5 Å globally averaged resolution. Within this complex however, sGP and the CDR-containing variable domains of the Fab (Fabv) are resolved better than 5 Å (Supplementary Fig. 1–2, 9, Supplementary Table 1). The apparent discrepancy arises because the constant Fab domains are inherently noisy in our cryo-EM reconstructions. In our complex, the constant domains constitute a third of the total mass. Our structure enables us to define the oligomeric arrangement of sGP and large portions shared with GPΔTM (Fig. 3a). The structure illustrates that the sGP homodimer has parallel topology, with the head and glycan cap regions that constitute the c13C6 epitopes arranged on opposite sides, so that the two copies of bound c13C6 Fabs face toward each other (Fig. 3a,b). In GP, the bound c13C6 Fabs instead face in a parallel direction (Fig. 1a,b). Dimerization of sGP is likely stabilized by a hydrophobic patch (Supplementary Fig. 10) as well as by C53-C53 (within sGP loop 1) and C306-C306 (within sGP loop 3) disulfide bonds on each side of the core, supporting previous biochemical evidence of a parallel intermolecular disulfide bond organization (Fig. 3b)26.
Figure 3. Cryo-EM structure of EBOV sGP in complex with c13C6 from ZMapp™ and BDBV human survivor mAb BDBV91.
a, Cryo-EM density map of EBOV sGP in complex with c13C6 and BDBV91 Fabs. sGP is in sea green (sGPA) and lime green (sGPB) with c13C6 Fabs in dark blue and BDBV91 Fabs in purple. There are two copies of each Fab bound per sGP dimer. Side (left) and top (right) views are shown. b, Subdomain assignments of sGP based on our model determined from the ~5.5 Å cryo-EM density map. A cartoon schematic of the sGP dimer is shown and corresponding domain positions are colored according to the figure below. Regions in white or black cross-hatch did not have any corresponding density, but were contained in our construct. The signal peptide (SP) in grey is excised from the construct during processing. Predicted disulfide bonds assignments are indicated between the two protomers and corresponding residues are labeled in red. Although we modeled an alanine residue for C53, we did not resolve the C53-C53 disulfide bond. c, Epitope details and CDR assignments of antibody BDBV91 bound to EBOV sGP, with the variable light chain in violet and the variable heavy chain in magenta. The sGP interface is colored as in panel b. The major contacts are formed between a single sGP protomer and CDRH3 and CDRL3. In the density map, contact regions between GP and BDBV91 (and between GP and c13C6) are resolved to ~4.4 Å (Supplementary Fig. 2). d, At hydrophobic pocket is formed at the dimer interface.and the long BDBV91 CDRH3 reaches into this pocket and contacts several residues in this region. Some side chain density between CDRH3 and sGP could be observed, including V92, K95, and F159.
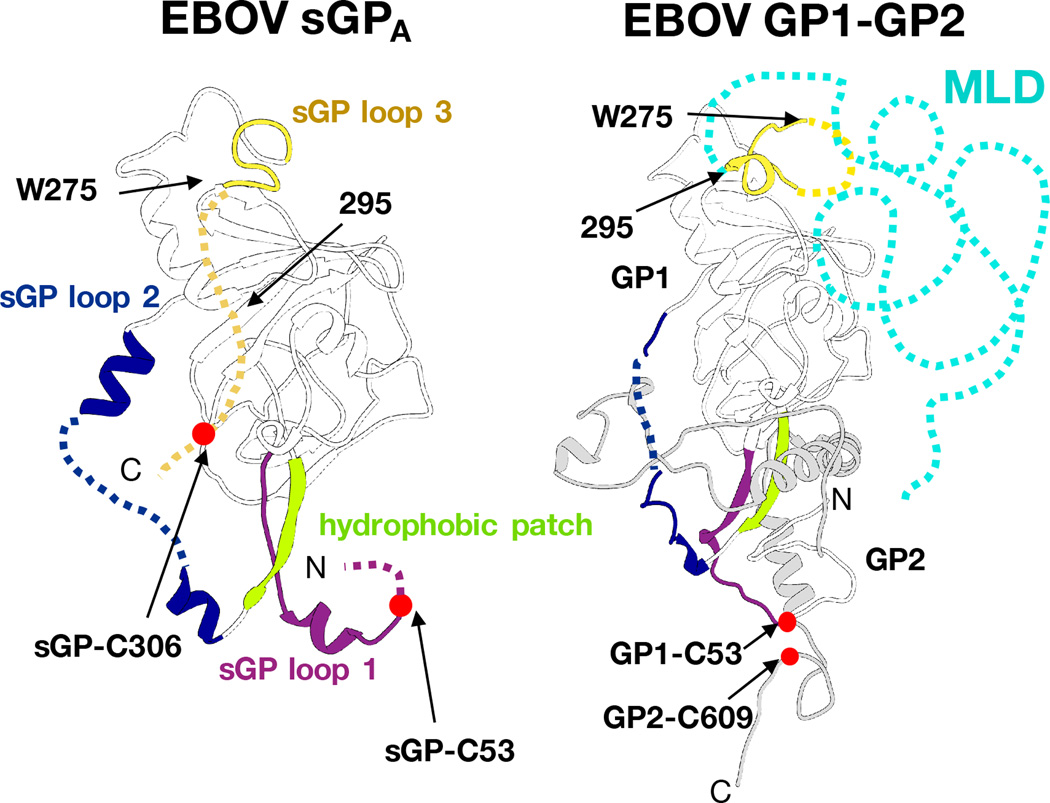
The protomers of GP and sGP are similar in tertiary structure, but differ in the sGP loop containing C53 (loop 1), the region containing the cathepsin cleavage site in GP (within sGP loop 2), and in the position and function of a hydrophobic patch, in which HR1 in GP forms a portion of the dimer interface in sGP (Fig. 4). Comparing GP1 and sGP side-by-side allows visualization of how protein products from identical genes, although different open reading frames, oligomerize to form distinct protein products and arrangement of shared epitopes (Supplementary Fig. 11).
Figure 4. Comparison of GP and sGP structures.
A single protomer from sGP (left) and GP (right) is shown in the same orientation. Domains with large structural divergence are colored the same in each corresponding structure, according to sub-domain assignments in Fig. 3b. In GP, the sGP loop1 (residues 53–70, magenta) manifests as the base of the GP1 core and the C53-C609 disulfide anchoring GP1 to GP2. The hydrophobic patch in sGP (residues 179–188, lime green) forms a hydrophobic clamp on HR1 in prefusion GP, while it comprises the dimer interface in sGP. sGP loop 2 (residues 189–214, dark blue) is the cathepsin cleavage loop in GP, which reaches across the outside of GP2 on the trimer, and is processed upon entry into the endosome. The different oligomeric contexts of sGP loop 2 and the GP cathepsin cleavage loop cause these regions to adopt distinct conformations. C-terminal of W275, the structures of GP and sGP diverge. Here, sGP terminates in sGP loop 3 (yellow), which contains the C306-C306 disulfide. However, in GP, these residues form the rest of the glycan cap and the beginning of the MLD. Beyond residue 295, sGP and GP also differ in their sequences; in GP, this region becomes the bulk of the MLD (in cyan).
The epitope of c13C6 is presented similarly on both sGP and GP (Fig. 2c). The epitope of BDBV91 spans across the sGP dimer interface (Fig. 3c, Supplementary Table 2), which likely explains its preferred binding of sGP over GP (Supplementary Fig. 12). This is in contrast to previous ELISA data indicating that BDBV91 could also bind the trimeric GP. Biolayer interferometry (BLI) enables a more precise measurement of binding than ELISA, which can suffer from artifacts such as the GP falling apart. Based on our structure BDBV91 is incapable of binding trimeric GP due to extensive clashing with oligomeric contacts in the trimer. CDRH3 of BDBV91 is buried in a hydrophobic pocket formed at the dimer interface, although it is biased toward one protomer (Fig. 3d). CDRL3 also makes interactions with the sGP hydrophobic patch on a single protomer (Fig. 3a, Supplementary Table 2). Interaction with the sGP dimer interface may impart the cross-reactivity of BDBV91 as these residues are conserved. Positive selection pressure to control EBOV sGP/GP expression, and the presence of sGP itself, have been observed during viral propagation in vitro and during an outbreak27,28, although, a definitive role for sGP has not yet been identified. It is intriguing, however, to speculate that the inclusion of an sGP-specific (and cross-reactive) antibody such as BDBV91 may be advantageous for the success of future pan-filoviral antibody therapeutics by lowering sGP concentrations, precluding viruses from any advantage that sGP may offer.
The different relative positions of the c13C6 Fabs when in complex with GP vs. sGP suggest they could play different roles in immune complex (IC) formation in viral infection. Analysis by composition gradient multi-angle light scattering (CG-MALS) (Supplementary Fig. 13, Supplementary Table 4) and negative stain EM (Supplementary Fig. 14a,b,c) reveals that one c13C6 IgG consistently binds to one GP trimer. In contrast, EM analysis of c13C6 IgG–sGP complexes reveals a variety of regular, multimeric, geometric shapes (Supplementary Fig. 14d,e). Formation of these ICs may be important for effectively activating Fc-receptor mediated immune responses29 and could be a key reason why c13C6 offers increased efficacy to ZMapp™12,13,15.
Evaluation of naturally occurring mutations on antibody binding
The consensus sequence for the 2014 EBOV Makona strain is 97% identical to the 1995 Kikwit strain at the amino acid level within the GP gene; however, 99 sequences from 78 individuals sampled during the 2014 outbreak revealed 35 non-synonymous substitutions in individuals27, nearly double the number of non-synonymous mutations compared to all other known outbreaks combined27. In regard to antibody therapeutics, even small changes in amino acid sequence can greatly affect binding kinetics, as antibodies are highly specific for particular sequences or structural motifs30. Therefore, we compiled 817 GP sequences from the 2014 EBOV Makona outbreak and identified mutations that occurred within the footprints identified here via cryoEM (Supplementary Fig. 15, Supplementary Table 5). Next, we performed site-directed mutagenesis on this residue subset followed by BLI to determine the effects of these changes on binding kinetics of the mAbs to EBOV GP (Supplementary Fig. 16, Supplementary Table 6). Additionally, we included known escape mutations as controls as well as knockout mutations previously identified by alanine scanning18,31,32.
There are far fewer naturally occurring mutations in the c2G4/c4G7 footprint than in the c13C6 footprint (one mutation and five mutations, respectively) (Supplementary Table 5). It may be that the base region in EBOV cannot tolerate major mutations due to its importance in viral fusion. Conversely, a large majority of mutations occur in GP1 within the glycan cap and mucin-like domains (Supplementary Table 5). We identified one mutation, E545D observed in one individual, which falls within the c4G7 footprint (Supplementary Fig. 15, Supplementary Table 5). We analyzed this E545D mutation, N550A, a known alanine scanning mutant that decreases c2G4 binding, D552A, a known alanine scanning mutant that decreases c4G7 binding, and Q508R, a known escape mutant for c2G4 and c4G712,18. As expected, N550A completed knocked out c2G4 binding (no binding, N.B.) and also lowered c4G7 binding to <20% of WT activity (Fig. 5a,b, Supplementary Table 6). D552A completely knocked out c4G7 binding and lowered c2G4 binding to ~30% of WT activity (Fig. 5a,b, Supplementary Table 6). The Q508R mutation knocked out binding of both c2G4 and c4G7 (Fig. 5a,b, Supplementary Table 6). In contrast, the naturally occurring mutation, E545D, did not significantly affect c2G4 binding. Interestingly though, E545D enhanced c4G7 binding (~120% WT activity) (Fig. 5a,b, Supplementary Table 6), and decreased the off-rate.
Figure 5. Effects of naturally occurring mutations that appeared in the 2014 EBOV outbreak on antibody binding within identified GP and sGP footprints.
a–c, We identified several mutations in GP sequences of viruses sequenced during the 2014 EBOV outbreak that fell within the ZMapp™ footprints identified in our cryoEM reconstructions. To evaluate their effects on antibody binding, we performed site-directed mutagenesis followed by biolayer interferometry (BLI). Our BLI results were normalized to WT binding Kd values and graphed. We also included negative controls identified by alanine scanning or known escape mutants, in order to validate previous work. d–e, For sGP, we identified a single naturally occurring mutation that fell within the BDBV91 footprint (D150A), and we also identified key residues from our model to manipulate BDBV91 binding. Experiments were performed in triplicate (technical replicates, using a single preparation of GP). Error bars represent standard deviation from the mean.
Five mutations accumulated in the c13C6 footprint, from 43 instances, including E229K (1 individual), T230A (37 individuals), L239S (2 individuals) and T240N (1 individual) (Supplementary Fig. 15, Supplementary Table 5). The glycan cap of GP is not highly conserved among filoviruses, and therefore may be capable of tolerating a higher frequency of mutation. Nevertheless, it is a key site of vulnerability for both neutralizing and protective antibodies9. Notably, two of these mutations fell within predicted N-linked glycan sequons, at N228 and N238 (Supplementary Table 5–6). While the construct used for cryoEM contained a T230V mutation, which was previously introduced to improve X-ray diffraction, the WT construct used here for binding studies retained the glycan at N22819.
We analyzed these GP mutants, as well as T270A, a known alanine-scanning mutant for c13C618, and W275L, a known escape mutant for the overlapping mAb c1H312,16. As expected, T270A completely knocked out c13C6 binding (<1% WT activity), while W275L significantly lowered but did not completely knock out c13C6 binding (~55% WT activity) (Fig. 5c, Supplementary Table 6). The mutations T230A (found in 37 individuals) and E229K had no substantial effect on binding, but L239S lowered binding to ~66% of WT activity (Fig. 5c, Supplementary Table S6). T240N knocked out c13C6 binding completely (<2% WT activity) (Fig. 5c, Supplementary Table 6). As noted above, this mutation falls within the coding region for a glycan at N238, which is near the binding site for c13C6 (Fig. 2c). It is unclear, however, if direct contact of c13C6 to this glycan is essential or if the glycan is necessary for proper folding of the glycan cap. Nevertheless, at least one known individual was infected with a virus that incorporated this mutation, which would have made c13C6 unable to bind to this viral GP. Further, two individuals were infected with viruses containing L239S mutations, which we show significantly decreased c13C6 binding, and therefore may also diminish ZMapp™ efficacy.
To further validate our conclusions regarding the newly identified sGP epitope of the sGP-specific mAb BDBV91, we performed site-directed mutagenesis on sGP. We chose three mutations: V92L, which makes three CDR contacts; F159S, which falls within the center of the hydrophobic patch near CDRH3 (Fig. 3d, Supplementary Table 2); and Q188R, which also makes three CDR contacts (Fig. 3d, Supplementary Table 2). Additionally, we chose D150A, which is a naturally occurring mutation (4 individuals) near the BDBV91 binding site (Supplementary Table 5). As a control, we checked c13C6 binding against all mutations, which we hypothesized would not change significantly as the c13C6 epitope on sGP is far removed from the BDBV91 epitope.
Although none of our mutations completely knocked out binding, we did observe significant changes in binding compared to WT sGP with several of the mutations we created (Fig. 5d,e, Supplementary Table 6). While V92L is a somewhat conservative mutation, binding is still lowered to <50% of WT, although c13C6 binding is lowered as well (~70% of WT) (Fig. 5d,e, Supplementary Table 6). V92 is part of the aforementioned pocket that accommodates BDBV91 CDRH3 interaction (Fig. 4d, Supplementary Fig. 15). The Q188R mutation also substantially lowered BDBV91 binding (~50% WT binding), while not affecting c13C6 binding. Q188 resides in the part of the BDBV91 binding site contributed by the opposite sGP monomer and which interacts with CDRL2 of BDBV91 (Fig. 4c, Supplementary Fig. 15). Interestingly, F159S enhanced BDBV91 binding (~150% WT), while lowering c13C6 binding (~60% WT) (Fig. 5d,e, Supplementary Table 6). The change in hydrophobicity associated with the F159S mutation may open up the sGP dimer pocket, slightly disrupting c13C6 binding, but enabling increased contacts within the BDBV91 epitope. Conversely, the naturally occurring mutation D150A lowers BDBV91 binding (~50% WT), perhaps by slightly increasing hydrophobicity and occluding the dimer interface (Fig. 5d,e, Supplementary Table 6).
Together, our structures identify key contacts made by GP-specific (c2G4 and c4G7) and sGP/GP-cross-reactive (c13C6) therapeutic antibodies being evaluated in ongoing clinical trials, as well as a novel sGP-specific and cross-reactive antibody (BDBV91). Further, these structures provide a basis to analyze naturally occurring mutations during the 2014 EBOV outbreak, and aid future therapeutic development. Surveillance of future outbreaks can also be mapped onto our structures to predict the efficacy of ZMapp™. We also present the first structure of sGP, an antigen that predominates in natural infection. Taken together, our structures offer a detailed and more complete picture of the accessible protein landscape of Ebolaviruses and reveal how two products of the same gene diverge in their quaternary assembly, thus giving rise to distinct functions and epitope presentation (Fig. 6). While the effects of copious sGP production on pathogenesis and immune response are not fully understood, certainly, the effect of sGP on antibody elicitation-and-absorption must be considered in future immunotherapeutic and vaccine design and testing.
Figure 6. Differential presentation of GP/sGP epitopes during ebolavirus infection.
During infection, both sGP and GP are produced and secreted from the Golgi apparatus. While sGP is secreted in large abundance into the serum, GP is anchored to the cell membrane that forms the enveloped surface of budding virions. Antibodies detailed in this study demonstrate a portion of the antigenic landscape available during an active infection, including a GP-specific epitope at the base of GP (c2G4 and c4G7), sGP/GP cross reactive epitopes at the top of GP and sides of sGP in the glycan cap (c13C6), and an sGP-specific epitope at the sGP dimer interface (BDBV91).
Materials and Methods
Expression and purification of EBOV GP and sGP
Recombinant EBOV GP ectodomains containing the mucin-like domains and lacking the transmembrane region (EBOV GPΔTM) were expressed and purified as described previously16. EBOV sGP was expressed and purified similarly to EBOV GP. The construct for sGP was truncated at residue P314 and cloned into the vector pMT-puro with a thrombin cut site and double strep tag at the C terminus. The truncation was originally made to optimize expression and to facilitate crystallization by removing the furin cleavage site. Stable cell lines were then created by transfection of Drosophila Schneider 2 cells (Invitrogen) and selection with 6 µg/mL puromycin. Secreted proteins were then purified the same as GPΔTM.
Antibody purification and Fab production
Plant-derived mAbs were generously provided by Mapp Biopharmaceuticals and Kentucky Bioprocessing, and engineered, expressed and purified as previously described33. BDBV91 was produced by a previously described human hybridoma cell line, derived from B cells from a Bundibugyo virus survivor, as previously described34. Briefly, hybridoma cells were cultured in serum-free medium (Gibco #12045-076). After 21 days, supernatants were clarified by centrifugation and sterile filtered using 0.2-µm pore size filter devices; IgG was purified by Protein G chromatography from filtered supernatants. Fabs were generated by optimized papain digestion followed by Protein A and size exclusion chromatography (SEC) purification, as previously described16. Fabs were added in 10-Molar excess and allowed to bind overnight at 4°C before complexes were purified by SEC.
Site-directed mutagenesis
To generate desired point mutations, site-directed mutagenesis was performed using the Q5 site-directed mutagenesis kit from New England BioLabs. Primers were designed with desired mutations using NEBaseChanger™. A construct of GPΔMuc in pDisplay (Thermo Fisher) with a C-terminal double-strep tag was utilized as a template. GPΔMuc was utilized for binding studies because of higher protein expression. After confirmation of integrated mutations by sequencing (Eton Biosciences), we transiently transfect 500 mL of 70% confluent HEK 293T (low passage, from Invitrogen) cells grown in DMEM (Invitrogen) supplemented with 5% (v/v) fetal bovine serum (FBS). No further authentication of the cell lines was carried out and they were not tested for mycoplasma contamination. Cells were prepared in HYPERFlasks (Corning), which have a growth area of 1,720 cm2. Transfection reagent was prepared by adding 227 µg DNA and 681 µg polyethylenimine (PEI) to 25 mL of Dulbecco’s phosphate-buffered saline (DPBS) and vigorously shaking. The solution was allowed to incubate at RT for 10–15 min. Next, 25 mL of media was gently removed from the top of HYPERFlasks, replaced with 25 mL of transfection reagent and gently mixed by rotating the flasks. Cells were allowed to express for 72h at 37°C in 8% CO2. The supernatant was then filtered using 0.2 µm filters and 300 µL of BioLock (iba LifeSciences) was added to bind any biotin in the media, per manufacturer instructions. Strep-tagged proteins were then purified as described above.
Biolayer interferometry
GP/sGP binding studies were performed using the Octet RED96 system (ForteBio) at 25°C. SA biosensors were first equilibrated in 10X Kinetics Buffer (KB) containing 1X phosphate buffered saline (PBS, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2HPO4, pH 7.4), 0.1% w/v bovine serum albumin (BSA) and 0.02% (v/v) Tween-20. BDBV and EBOV GPΔTM or sGP with a double strep-tag diluted to a concentration of 100 mg/mL in 10X KB were then loaded onto SA biosensors to ~0.8 nm absorbance. A short wash step in 10X KB followed by a baseline step were then performed. GP-bound biosensors were then dipped into solutions of antibodies at 1 µM concentration for 60 s and then into 10X KB alone for 60 s. No antibody controls were also performed and subtracted from the data. There was no detectable antibody binding to the sensors themselves.
Mutagenesis binding were also performed using BLI at 25°C. SA biosensors were first equilibrated in 10X KB containing 1X DPBS, 0.1% w/v BSA and 0.02% (v/v) Tween-20. EBOV GPΔMuc with a double strep-tag diluted to a concentration of 25 µg/mL in 10X KB were then loaded onto SA biosensors to ~1–2 nm absorbance. A short wash step in 10X KB followed by a baseline step was then performed. GP-bound biosensors were then dipped into 2X serially diluted solutions of Fab from 1 µM to ~30 nM concentrations for 60 s and then into 10X KB alone for 60 s. No antibody controls were also performed and subtracted from the data. There was no detectable antibody binding to the SA biosensors themselves. Binding experiments were performed in triplicate. Binding curves were fitted globally using a 1:1 binding equation with the OctetRED software. All Kd values were normalized to respective WT samples. For samples with very low Kd values, experiments were repeated with Fabs 2X serially diluted from 10 µM to 1 µM to achieve a more accurate binding constant.
Size exclusion chromatography-multi angle light scattering and protein conjugate analysis
Size exclusion chromatography-multi angle light scattering (SEC-MALS) experiments were performed using an S200 10/30 column (GE Healthcare), and an AKTA purifier FPLC in-line with the miniDAWN TREOS and Optilab T-rEX system. Prior to CG-MALS analysis, protein solutions were filtered to 0.1 µM using Anotop (Whatman) syringe filters. All experiments were performed in 20 mM Tris, 150 mM NaCl, pH 7.4 (1X TBS). Data collection and analysis were performed with ASTRA software. Protein conjugate analysis in ASTRA was used to determine the protein fraction for GPdTM and a corrected extinction coefficient and dn/dc to account for the glycan content. An extinction coefficient of 1.5 and dn/dc of 0.185 was used to determine the concentration of 13C6.
Composition gradient-multi angle light scattering
Composition gradient-multi angle light scattering (CG-MALS) experiments were completed with a Calypso II (Wyatt) composition gradient system to prepare different compositions of buffer, glycoprotein and antibody and deliver to the miniDAWN TREOS and an online UV detector. The extinction coefficient obtained from the SEC-MALS experiment was used to measure the concentration of the glycoprotein during CG-MALS experiments. Polycarbonate (Millipore) filter membranes with 0.1 µM pore size were installed in the Calypso for sample and buffer filtration. GPdTM was diluted to a stock concentration of 15–25 µg/mL in TBS and was filtered to 0.1 µM. The IgG was diluted to a stock concentration of 35–50 µg/mL in TBS and was filtered with a 0.02 µM filter. The automated Calypso method consisted of a dual-component “crossover” gradient to assess hetero-association between the GPdTM and IgG. For each composition, 0.7 mL of protein solution was injected into the UV and MALS detectors and allowed to come to equilibrium within the MALS flow cell and the flow stopped for 900 s. Data collection and analysis were performed with the CALYPSO software. GPdTM-13C6 association was measured in triplicate with three different preparations of GPdTM.
Negative stain electron microscopy
To examine the formation of immune complexes with the intact c13C6 IgG, we utilized negative stain EM. c13C6 IgG was added in excess of GP or sGP and allowed to incubate overnight at 4° C. Complexes were then visualized as previously described16.
Cryo sample preparation for Electron Microscopy
5 µL of the purified complex was incubated with 1 µL of DDM solution at 1.8 mM. At 4°C, 3 µL of this solution was applied on the carbon face a CF-2/2–4C C-Flat grid (Electron Microscopy Sciences, Protochips, Inc.) which had been plasma cleaned for 5 seconds using a mixture of Ar/O2 (Gatan Solarus 950 Plasma system). The grid was then manually blotted and immediately plunged into liquid ethane using a manual freeze plunger.
Cryo-electron microscopy data collection
Micrographs were collected via the Leginon interface on a FEI Titan Krios operating at 300 KeV mounted with a Gatan K2 direct electron detector35. Each micrograph was collected in the counting mode at 22,500× nominal magnification resulting in a calibrated pixel size of 1.31 Å/pixel at the object level. A dose rate of ~10 e-/pix/sec was used; exposure time was 200 ms per frame. The data collection for c2G4/c13C6/GPdTM complex resulted in a total of 2526 micrographs. Total dose for these micrographs was 57 e-/Å2. The nominal defocus range used was −1.0 to −3.0µm. The data for the c4G7/c13C6/GPdTM complex was collected over two sessions. Total dose for 1101 micrographs was 57 e-/Å2 and for 1454 micrographs 45 e-/Å2. The nominal defocus range used was −1.0 to −4.0µm. The data collection for BDBV91/c13C6/sGP complex resulted in a total of 2048 micrographs. The total dose for these micrographs was 57 e-/Å2. The nominal defocus range used was −1.0 to −2.5µm.
Electron Microscopy Data processing
All of the collected frames were motion-corrected36. CTF estimation was carried out using CTFFind335, and particles were picked employing a difference-of-Gaussians approach37. Reference-free classification was performed in RELION version 1.4b1. After 2D classification, selected particles were refined against a density map at 60A resolution simulated from the GP1-GP2 parts of PDB ID 3CSY. 3D classification was then performed in RELION version 1.4b1 resulting in four-six classes within each data set38,39. Stable classes were selected for further processing followed by various steps of additional 3D classification. Stoichiometric classes were selected for further refinement followed by particle polishing and refinement of the resulting realigned, B-factor-weighed and signal-integrated particles were refined in their respective classes. The final resolutions were 4.3Å (c2G4/c13C6/GPdTM), 4.9Å (c4G7/c13C6/GPdTM) and 5.5Å (BDBV91/c13C6/sGP) at an FSC cutoff of 0.143. FSC was calculated using a soft-edged mask with a Gaussian falloff, encompassing the entire structure, including the Fab constant regions.
Model Building and Refinement
Initial models of the Fv region of the c13C6, c2G4, c4G7, and BDBV91 Fabs were generated using the ROSIE Rosetta antibody protocol server40,41. An initial model of EBOV GPΔTM was generated using the Modeller homology modeling tool in UCSF Chimera and the GP portion of 3CSY as a template4,42,43. An initial model of c2G4/c13C6/GPΔTM was generated by docking the initial GPΔTM model and the initial Fv models of c13C6 and c2G4 into the c2G4/c13C6/GPΔTM Class 6 EM map. The resulting c2G4/c13C6/GPΔTM model was iteratively refined and manually rebuilt with C3 symmetry imposed using Rosetta density-guided iterative local refinement and Coot44,45. The final refinement was conducted with RosettaRelax using the c2G4/c13C6/GPΔTM All Classes EM map as a spatial constraint while imposing C3 symmetry46.
An initial model of the c4G7/GPΔTM structure was generated by docking the GPΔTM from the final c2G4/c13C6/GPΔTM refinement into the C3 symmetric c4G7/c13C6/GPΔTM All Classes map along with the c4G7 Fv initial model. The resulting model was refined using RosettaRelax with C3 symmetry imposed. The best output model was docked into the asymmetric c4G7/c13C6/GPΔTM All Classes map along with two copies of the c13C6 Fv regions from the final c2G4/c13C6/GPΔTM structure. The resulting model was refined using RosettaRelax without imposing symmetry.
An initial model of BDBV91/c13C6/sGP was generated by docking the c13C6 Fv and GP1 regions from the final c2G4/c13C6/GPΔTM structure and the initial BDBV91 Fv model into the C2 symmetric BDBV91/c13C6/sGP Class 2 map and refined using RosettaRelax with C2 symmetry imposed.
In all cases, the “best” output model from Rosetta refinements was selected based optimizing the Rosetta non-density weighted energy score, the MolProbity score, and the agreement of the model to the EM density as measured by the EMRinger score47,48.
Energy minimized GlcNAc2Man3 glycans were added to the final c2G4/c13C6/GPΔTM protein structure using GlyProt49. The glycan torsion angles were manually adjusted to fit into density and residues without supporting density were removed. The glycans were further refined into the density using Coot with torsional constraints enabled. The final glycan structures and glycan torsion angles for the c2G4/c13C6/GPΔTM structure were validated with Privateer and CARP respectively50,51. Glycans from the c2G4/c13C6/GPΔTM structure were used as starting models for the c4G7/c13C6/GPΔTM structures. Glycans at sites that did not have supporting density were removed. The BDBV91/c13C6/sGP density did not support the modeling of glycans.
Supplementary Material
Acknowledgments
We thank Kelsi Swope and Josh Morton of Kentucky Bioprocessing for preparing and sending all ZMapp™ IgGs. We thank Travis Nieusma and Bill Anderson of TSRI for assistance with loading cryo samples, data collection, and running the microscopy facility. We thank Benjamin Barad for providing the scripts for EMRinger. This work was supported by R01 AI067927 “Ebola Viral Glycoproteins: Structural Analysis”, the NIH/National Institute of Allergy and Infectious Diseases Center for Excellence in Translational Research Grant U19AI109762 “Consortium for Immunotherapeutics Against Viral Hemorrhagic Fevers” and U19AI109711 “Advancement of Treatments for Ebola and Marburg Virus Infections”. C.D.M was supported by a predoctoral fellowship from the National Science Foundation. This is manuscript number 29260 from The Scripps Research Institute.
Footnotes
Author contributions. J.P., C.D.M., E.O.S. and A.B.W. designed the research plan; M.L.F. designed the protein expression constructs for GP and sGP. A.F. and J.E.C. isolated and purified BDBV91. C.D.M. and H.L.T. expressed and purified proteins. K.M.H and C.D.M. performed biophysical experiments. J.P., C.D.M. and H.L.T. prepared protein complexes. J.P., C.D.M and N.dV. froze grids. J.P. and N.dV. collected cryo data. J.P. processed the cryo data. C.D.M. collected and processed negative stain data. C.C. performed structure modeling and refinement. J.P., C.D.M., C.C., K.M.H, M.L.F, E.O.S. and A.B.W analyzed data. J.P., C.D.M., C.C., E.O.S. and A.B.W. wrote the manuscript. K.G.A. provided Makona outbreak sequence information.
Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-8240, EMD-8241 and EMD-8242. Models built from the EM data have been deposited in the Protein Data Bank under accession numbers 5KEL, 5KEM and 5KEN. All figures were prepared using UCSF Chimera. Larry Zeitlin is a co-owner of Mapp Biopharmaceutical, Inc.
References
- 1.Henao-Restrepo AM, et al. Efficacy and effectiveness of an rVSV-vectored vaccine expressing Ebola surface glycoprotein: interim results from the Guinea ring vaccination cluster-randomised trial. Lancet. 2015;386:857–866. doi: 10.1016/S0140-6736(15)61117-5. [DOI] [PubMed] [Google Scholar]
- 2.Stanley DA, et al. Chimpanzee adenovirus vaccine generates acute and durable protective immunity against ebolavirus challenge. Nature medicine. 2014;20:1126–1129. doi: 10.1038/nm.3702. [DOI] [PubMed] [Google Scholar]
- 3.Tapia MD, et al. Use of ChAd3-EBO-Z Ebola virus vaccine in Malian and US adults, and boosting of Malian adults with MVA-BN-Filo: a phase 1, single-blind, randomised trial, a phase 1b, open-label and double-blind, dose-escalation trial, and a nested, randomised, double-blind, placebo-controlled trial. Lancet Infect Dis. 2016;16:31–42. doi: 10.1016/S1473-3099(15)00362-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JE, et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JE, Saphire EO. Ebolavirus glycoprotein structure and mechanism of entry. Future virology. 2009;4:621–635. doi: 10.2217/fvl.09.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchez A, Trappier SG, Mahy BW, Peters CJ, Nichol ST. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:3602–3607. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volchkova VA, Feldmann H, Klenk HD, Volchkov VE. The nonstructural small glycoprotein sGP of Ebola virus is secreted as an antiparallel-orientated homodimer. Virology. 1998;250:408–414. doi: 10.1006/viro.1998.9389. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez A, et al. Biochemical analysis of the secreted and virion glycoproteins of Ebola virus. Journal of virology. 1998;72:6442–6447. doi: 10.1128/jvi.72.8.6442-6447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flyak AI, et al. Cross-reactive and potent neutralizing antibody responses in human survivors of natural ebolavirus infection. Cell. 2016;164:392–405. doi: 10.1016/j.cell.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de La Vega MA, Wong G, Kobinger GP, Qiu X. The multiple roles of sGP in Ebola pathogenesis. Viral immunology. 2015;28:3–9. doi: 10.1089/vim.2014.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu X, et al. Characterization of Zaire ebolavirus glycoprotein-specific monoclonal antibodies. Clinical immunology. 2011;141:218–227. doi: 10.1016/j.clim.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Qiu X, et al. Successful treatment of ebola virus-infected cynomolgus macaques with monoclonal antibodies. Science translational medicine. 2012;4:138ra181. doi: 10.1126/scitranslmed.3003876. [DOI] [PubMed] [Google Scholar]
- 13.Olinger GG, Jr, et al. Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18030–18035. doi: 10.1073/pnas.1213709109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson JA, et al. Epitopes involved in antibody-mediated protection from Ebola virus. Science. 2000;287:1664–1666. doi: 10.1126/science.287.5458.1664. [DOI] [PubMed] [Google Scholar]
- 15.Qiu X, et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murin CD, et al. Structures of protective antibodies reveal sites of vulnerability on Ebola virus. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:17182–17187. doi: 10.1073/pnas.1414164111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran EE, et al. Mapping of Ebolavirus Neutralization by Monoclonal Antibodies in the ZMapp Cocktail Using Cryo-Electron Tomography and Studies of Cellular Entry. Journal of virology. 2016 doi: 10.1128/JVI.00406-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davidson E, et al. Mechanism of Binding to Ebola Virus Glycoprotein by the ZMapp, ZMAb, and MB-003 Cocktail Antibodies. Journal of virology. 2015;89:10982–10992. doi: 10.1128/JVI.01490-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JE, et al. Techniques and tactics used in determining the structure of the trimeric ebolavirus glycoprotein. Acta Crystallogr D. 2009;65:1162–1180. doi: 10.1107/S0907444909032314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dias JM, et al. A shared structural solution for neutralizing ebolaviruses. Nature structural & molecular biology. 2011;18:1424–1427. doi: 10.1038/nsmb.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bornholdt ZA, et al. Host-Primed Ebola Virus GP Exposes a Hydrophobic NPC1 Receptor-Binding Pocket, Revealing a Target for Broadly Neutralizing Antibodies. MBio. 2015;7 doi: 10.1128/mBio.02154-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brecher M, et al. Cathepsin cleavage potentiates the Ebola virus glycoprotein to undergo a subsequent fusion-relevant conformational change. Journal of virology. 2012;86:364–372. doi: 10.1128/JVI.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Misasi J, et al. Structural and molecular basis for Ebola virus neutralization by protective human antibodies. Science. 2016;351:1343–6. doi: 10.1126/science.aad6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oswald WB, et al. Neutralizing antibody fails to impact the course of Ebola virus infection in monkeys. PLoS pathogens. 2007;3:e9. doi: 10.1371/journal.ppat.0030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu S, et al. Fabs enable single particle cryoEM studies of small proteins. Structure. 2012;20:582–592. doi: 10.1016/j.str.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrientos LG, Martin AM, Rollin PE, Sanchez A. Disulfide bond assignment of the Ebola virus secreted glycoprotein SGP. Biochemical and biophysical research communications. 2004;323:696–702. doi: 10.1016/j.bbrc.2004.08.148. [DOI] [PubMed] [Google Scholar]
- 27.Gire SK, et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345:1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volchkova VA, Dolnik O, Martinez MJ, Reynard O, Volchkov VE. Genomic RNA editing and its impact on Ebola virus adaptation during serial passages in cell culture and infection of guinea pigs. The Journal of infectious diseases. 2011;204(Suppl 3):S941–S946. doi: 10.1093/infdis/jir321. [DOI] [PubMed] [Google Scholar]
- 29.Pincetic A, et al. Type I and type II Fc receptors regulate innate and adaptive immunity. Nature immunology. 2014;15:707–716. doi: 10.1038/ni.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kugelman JR, et al. Monitoring of Ebola Virus Makona Evolution through Establishment of Advanced Genomic Capability in Liberia. Emerg Infect Dis. 2015;21:1135–1143. doi: 10.3201/eid2107.150522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Audet J, et al. Molecular characterization of the monoclonal antibodies composing ZMAb: a protective cocktail against Ebola virus. Scientific reports. 2014;4:6881. doi: 10.1038/srep06881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu X, et al. Sustained protection against Ebola virus infection following treatment of infected nonhuman primates with ZMAb. Scientific reports. 2013;3:3365. doi: 10.1038/srep03365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeitlin L, et al. Enhanced potency of a fucose-free monoclonal antibody being developed as an Ebola virus immunoprotectant. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20690–20694. doi: 10.1073/pnas.1108360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flyak AI, et al. Mechanism of human antibody-mediated neutralization of Marburg virus. Cell. 2015;160:893–903. doi: 10.1016/j.cell.2015.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of structural biology. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 37.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. Journal of structural biology. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyskov S, et al. Serverification of molecular modeling applications: the Rosetta Online Server that Includes Everyone (ROSIE) PloS one. 2013;8:e63906. doi: 10.1371/journal.pone.0063906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sivasubramanian A, Sircar A, Chaudhury S, Gray JJ. Toward high-resolution homology modeling of antibody Fv regions and application to antibody-antigen docking. Proteins. 2009;74:497–514. doi: 10.1002/prot.22309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Webb B, Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Bioinformatics. 2014;47:561–532. doi: 10.1002/0471250953.bi0506s47. [DOI] [PubMed] [Google Scholar]
- 44.DiMaio F, et al. Atomic-accuracy models from 4.5-A cryo-electron microscopy data with density-guided iterative local refinement. Nat Methods. 2015;12:361–365. doi: 10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica. Section D, Biological crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D. Refinement of protein structures into low-resolution density maps using rosetta. Journal of molecular biology. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barad BA, et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bohne-Lang A, von der Lieth CW. GlyProt: in silico glycosylation of proteins. Nucleic Acids Res. 2005;33:W214–W219. doi: 10.1093/nar/gki385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Agirre J, et al. Privateer: software for the conformational validation of carbohydrate structures. Nature structural & molecular biology. 2015;22:833–834. doi: 10.1038/nsmb.3115. [DOI] [PubMed] [Google Scholar]
- 51.Lutteke T, Frank M, von der Lieth CW. Carbohydrate Structure Suite (CSS): analysis of carbohydrate 3D structures derived from the PDB. Nucleic Acids Res. 2005;33:D242–D246. doi: 10.1093/nar/gki013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.