Abstract
Objective
To evaluate factors associated with HIV tropism among Black men who have sex with men (MSM) in the United States enrolled in a clinical study (HIV Prevention Trials Network 061).
Methods
HIV tropism was analyzed using a phenotypic assay (Trofile assay, Monogram Biosciences). Samples were analyzed from 43 men who were HIV infected at enrollment and reported either exclusive insertive intercourse or exclusive receptive intercourse; samples were also analyzed from 20 men who were HIV uninfected at enrollment and seroconverted during the study. Clonal analysis of individual viral variants was performed for seroconverters who had dual/mixed viruses.
Results
Dual/mixed viruses were detected in samples from 11 (26%) of the 43 HIV-infected men analyzed at the enrollment visit; HIV tropism did not differ between those reporting exclusive insertive vs. receptive intercourse. Dual/mixed viruses were also detected in five (25%) of the 20 seroconverters. Dual/mixed viruses were associated with lower CD4 cell counts. Seroconverters with dual/mixed viruses had dual-tropic viruses only or mixed populations of CCR5− and dual-tropic viruses.
Conclusions
Dual/mixed viruses were frequently detected among Black MSM in this study, including seroconverters. Further studies are needed to understand factors driving transmission and selection of CXCR4− and dual-tropic viruses among Black MSM.
Keywords: HIV, tropism, Black, men who have sex with men, dual/mixed, CXCR4, dual-tropic
INTRODUCTION
HIV entry into cells is mediated by interactions between the viral envelope (Env), CD4 receptor, and CCR5 or CXCR4 coreceptors. HIV tropism is determined by coreceptor usage; viruses can use CCR5 exclusively (R5), CXCR4 exclusively (X4), or both coreceptors (dual-tropic). Viral populations can be comprised of solely R5 or X4 viruses or include dual/mixed (DM) viruses. Individuals with X4/DM viruses often exhibit accelerated disease progression.1,2 While R5 viruses usually predominate early in infection, X4/DM viruses have been documented in up to 20% of recently-infected individuals.1,3–5 The predominance of R5 viruses during primary HIV infection is thought to result from selective advantages that favor transmission and/or replication of R5 strains.6 The prevalence of X4/DM viruses in recent infection is higher among people who inject drugs than among those with sexually-acquired HIV infection.2,3,7 This suggests that the absence of a mucosal barrier may facilitate transmission of X4/DM strains. Sexual practices (e.g., insertive vs. receptive intercourse among men who have sex with men [MSM]) may also differentially impact transmission of X4/DM viruses. These issues are relevant to HIV prevention efforts, since the CCR5 coreceptor antagonist, maraviroc, is being evaluated for use in pre-exposure prophylaxis (PrEP).8,9
In this report, we explored whether sexual practices were associated with coreceptor tropism among HIV-infected MSM in the United States (US) who were enrolled in a clinical study (HIV Prevention Trials Network [HPTN] 061; NCT 00951249). This study assessed the feasibility and acceptability of a multi-component intervention to reduce HIV incidence among Black MSM who were at a high risk of HIV acquisition or transmission.10,11 Annual HIV incidence in the cohort was high (3.0% overall, 95% confidence interval [CI]: 2.0–4.4%; 5.9% among men ≤30 years old, 95% CI: 3.6–9.1%).10 We analyzed HIV tropism for the subset of study participants in HPTN 061 who were HIV infected at enrollment and reported either exclusive insertive intercourse or exclusive receptive intercourse, and among men who seroconverted during the study.
METHODS
Study cohort
HPTN 061 enrolled 1,553 Black MSM in six US cities (Atlanta, Boston, Los Angeles, New York City, San Francisco, and Washington, DC; enrollment period: 2009–2010).10,11 Briefly, men were recruited from the community or were referred as sexual network partners. Participants were eligible for the study if they reported at least one instance of unprotected anal intercourse with a man in the prior six months. Both HIV-uninfected and HIV-infected men were enrolled; the number of HIV-infected men in HIV care was capped at 10 men per study site. HIV testing was performed at enrollment for all participants and at study visits 6 and 12 months after enrollment for men who were uninfected at enrollment. CD4 cell count and HIV viral load were measured for men with HIV infection. All participants were tested for sexually transmitted infections. Additional testing was performed retrospectively by the HPTN Laboratory Center (Johns Hopkins University, Baltimore, MD). Participants also completed behavioral assessments via audio computer-assisted self-interview (ACASI) and answered social and sexual network questionnaires with an interviewer at each visit. The ACASI asked participants to provide detailed information on sexual practices. Insertive intercourse was defined as insertive vaginal or anal intercourse with female, male, or transgender female/male partners; receptive intercourse was defined as receptive anal intercourse with male or transgender male partners.
Analysis of HIV tropism
HIV tropism testing was performed retrospectively using samples from two groups of men: (1) men who were HIV infected at enrollment and reported either exclusive insertive intercourse or exclusive receptive intercourse in the previous six months, and (2) men who seroconverted during the study. All samples tested had a viral load ≥1,000 copies/mL. Tropism testing was performed using the Trofile assay (Monogram Biosciences, San Francisco, CA). Trofile is a recombinant phenotypic assay for determining HIV coreceptor usage.12,13 Briefly, full-length env sequences from each sample are amplified and cloned into expression vectors. Pseudoviruses are prepared by cotransfecting human embryonic kidney cell cultures with the env expression vectors and a replication-defective HIV genomic vector containing a luciferase reporter gene. HIV coreceptor tropism is determined by infecting CCR5+ and CXCR4+ cells in the presence and absence of CCR5 and CXCR4 inhibitors. Viral replication is quantified by measuring luciferase activity (reported as relative light units, which reflect the level of luciferase activity).13
Analysis of individual viral variants (clonal analysis) was performed using samples from seroconverters who had DM viruses. HIV tropism was determined for 16–20 env clones from the viral populations of each individual using Trofile, and the V3 loop of each clone was sequenced using standard dideoxy-chain termination.14 In addition to phenotypic tropism testing, HIV tropism was predicted using an algorithm based on V3 region sequences (the geno2pheno[coreceptor] algorithm, version 2.5).15 The European guidelines for HIV-1 tropism testing recommend using a false positive rate (FPR) of ≤10% to predict X4 tropism.16
Statistical methods
Correlates of HIV tropism were analyzed using Fisher’s exact and chi-square tests for univariate analyses. SAS, version 9.2 (SAS Institute, Cary, NC) was used for these analyses. P values <0.05 were considered to be statistically significant.
Ethical considerations
Written informed consent was obtained from all participants in the HPTN 061 study. The institutional review boards at each participating institution approved the study.
RESULTS
In HPTN 061, 147 (42%) of the 348 men who were HIV-infected at enrollment had a viral load ≥1,000 copies/mL. HIV tropism was analyzed in samples from 51 of those men: 32 (63%) reported exclusive insertive intercourse and 19 (37%) reported exclusive receptive anal intercourse (see Methods). The remaining 96 men either did not disclose their sexual practices or reported both insertive and receptive intercourse; these men were not included in the analysis. Samples from the first HIV-positive visit were also analyzed for 21 (75%) of the 28 men who seroconverted during HPTN 061; the remaining seven men included six who had viral loads <1,000 copies/mL at the first HIV-positive visit17 and one who had no sample available for testing. HIV tropism was determined for 63 of the 72 men (Table 1): 43 who were HIV infected at enrollment and 20 seroconverters. The 43 men who were HIV infected at enrollment had a median age of 41 years (range: 18–61), a median CD4 cell count of 354 cells/mm3 (range: 20–1,849), and a median viral load of 22,551 copies/mL (range: 1,716–625,171). The 20 seroconverters had a median age of 22 years (range: 18–48), a median CD4 cell count of 563 cells/mm3 (range: 151–771), and a median viral load of 85,185 copies/mL (range: 5,933–1,732,182). All 63 men included in this study were infected with HIV-1 subtype B.18 HIV tropism was not determined for nine samples due to assay failure, including samples from five men who reported exclusive insertive intercourse, three who reported exclusive receptive intercourse, and one seroconverter.
Table 1.
Association of HIV tropism with demographic, clinical, and behavioral factors.
| Characteristic | Total 63 |
DM 16 |
R5 47 |
P value | |
|---|---|---|---|---|---|
| City | Atlanta | 12 (19%) | 3 (19%) | 9 (19%) | 0.26 |
| Boston | 5 (8%) | 1 (6%) | 4 (9%) | ||
| Los Angeles | 24 (38%) | 7 (44%) | 17 (36%) | ||
| New York City | 9 (14%) | 1 (6%) | 8 (17%) | ||
| San Francisco | 7 (11%) | 4 (25%) | 3 (6%) | ||
| Washington, DC | 6 (10%) | 0 (0%) | 6 (13%) | ||
| Age | ≤30 years | 27 (43%) | 7 (44%) | 20 (43%) | 0.93 |
| >30 years | 36 (57%) | 9 (56%) | 27 (57%) | ||
| Sexual identity | Homosexual/gay | 30 (48%) | 9 (56%) | 21 (45%) | 0.76 |
| Bisexual | 16 (25%) | 3 (19%) | 13 (28%) | ||
| Other | 17 (27%) | 4 (25%) | 13 (28%) | ||
| Education | High school or less | 34 (54%) | 9 (56%) | 25 (53%) | 0.83 |
| At least some college | 29 (46%) | 7 (44%) | 22 (47%) | ||
| Household income | ≤$30,000/year | 51 (81%) | 12 (75%) | 39 (83%) | 0.48 |
| >$30,000/year | 12 (19%) | 4 (25%) | 8 (17%) | ||
| Employment status | Employed | 15 (24%) | 5 (31%) | 10 (21%) | 0.50 |
| Unemployed | 48 (76%) | 11 (69%) | 37 (79%) | ||
| Student status | Student | 18 (29%) | 5 (31%) | 13 (28%) | 0.76 |
| Non-student | 45 (71%) | 11 (69%) | 34 (72%) | ||
| Circumcision status | Circumcised | 51 (81%) | 13 (81%) | 38 (81%) | 1.00 |
| Uncircumcised | 12 (19%) | 3 (19%) | 9 (19%) | ||
| STI at enrollment | Yes | 13 (21%) | 2 (13%) | 11 (23%) | 0.49 |
| No | 50 (79%) | 14 (88%) | 36 (77%) | ||
| Median viral load (copies/mL) | 22,551 | 56,152 | 20,236 | 0.28 | |
| Median CD4 count (cells/mm3) | 357 | 184 | 386 | 0.019 | |
| In prior six months: | |||||
| Number of male partners | 0–1 | 13 (21%) | 5 (31%) | 8 (17%) | 0.29 |
| >1 | 50 (79%) | 11 (69%) | 39 (83%) | ||
| Gender of partners | Male only | 42 (67%) | 9 (56%) | 33 (70%) | 0.31 |
| Male and female | 21 (33%) | 7 (44%) | 14 (30%) | ||
| Had a new male partner | Yes | 51 (81%) | 11 (69%) | 40 (85%) | 0.16 |
| No | 12 (19%) | 5 (31%) | 7 (15%) | ||
| Race/ethnicity of partners | All Black | 41 (65%) | 10 (63%) | 31 (66%) | 0.89 |
| Some Black | 18 (29%) | 5 (31%) | 13 (28%) | ||
| None Black | 4 (6%) | 1 (6%) | 3 (6%) | ||
| Unprotected receptive anal intercourse | Yes | 31 (49%) | 6 (38%) | 25 (53%) | 0.28 |
| No | 32 (51%) | 10 (63%) | 22 (47%) | ||
| Unprotected insertive anal intercourse | Yes | 30 (48%) | 8 (50%) | 22 (47%) | 0.83 |
| No | 33 (52%) | 8 (50%) | 25 (53%) | ||
| Received money/goods for sex | Yes | 13 (21%) | 3 (19%) | 10 (21%) | 1.00 |
| No | 50 (79%) | 13 (81%) | 37 (79%) | ||
| Provided money/goods for sex | Yes | 7 (11%) | 1 (6%) | 6 (13%) | 0.67 |
| No | 56 (89%) | 15 (94%) | 41 (87%) | ||
| Alcohol problema | Yes | 14 (22%) | 4 (25%) | 10 (21%) | 0.74 |
| No | 49 (78%) | 12 (75%) | 37 (79%) | ||
| Substance useb | Yes | 41 (65%) | 11 (69%) | 30 (64%) | 0.72 |
| No | 22 (35%) | 5 (31%) | 17 (36%) | ||
The table shows associations between HIV tropism and demographic, clinical, and behavioral factors for all 63 men with HIV tropism data, including 43 men who were HIV-infected at enrollment and 20 HIV seroconverters. P values <0.05 are bolded. Collection of demographic and behavioral data has been previously described in detail.10,11 Abbreviations: DM: dual/mixed viruses; R5: CCR5-using viruses; STI, sexually transmitted infection.
Alcohol use was defined as having a score ≥8 using the Alcohol Use Disorders Identification Test (AUDIT).
Substance use included inhaled nitrates, smoked and powder cocaine, methamphetamine, heroin, nonprescription drug use (Oxycontin, Vicodin, or Xanax), or any other hallucinogens. Injection drug use was reported in only four (6%) of 63 men included in this analysis. This included three men who were HIV infected at enrollment (two with R5 viruses and one with DM viruses) and one seroconverter (with R5 viruses).
DM viruses were detected in 11 (26%) of 43 men who were HIV infected at enrollment, including eight (30%) of 27 men who reported exclusive insertive intercourse and three (19%) of 16 men who reported exclusive receptive intercourse (P=0.49; data not shown). DM viruses were also detected in 5 (25%) of 20 seroconverters. One of the 20 seroconverters reported exclusive insertive intercourse and four reported exclusive receptive intercourse; the remaining 15 seroconverters reported both insertive and receptive intercourse, or did not report their sexual practices. These data were too limited to assess associations between HIV tropism and sexual practices among the seroconverters.
Since the proportion of men with DM viruses was similar among the HIV-infected men at enrollment and the seroconverters, these groups were combined to analyze factors associated with detection of DM viruses. Overall, men who had DM viruses had lower median CD4 cell counts compared to men who had R5 viruses (184 cells/mm3 vs. 386 cells/mm3; P=0.019; Table 1). All five seroconverters who had DM viruses had CD4 cell counts <350 cells/mm3 (median: 254 cells/mm3, range: 151–335, Table 2); in contrast, the median CD4 cell count for the seroconverters with R5 viruses was 604 cells/mm3 (range: 281–771). None of the other factors evaluated was associated with detection of DM viruses in this study.
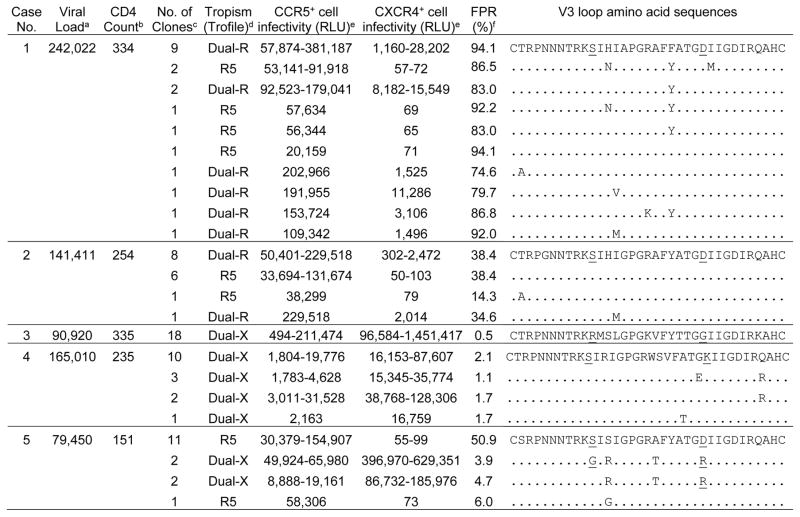
Table 2.
Coreceptor usage and V3 loop sequences of env clones from HIV seroconverters with dual/mixed viruses.
The table shows results of env clonal analyses for five HIV seroconverters with DM viruses. Amino acid positions 11 and 25 in the env V3 loop are underlined. Abbreviations: no.: number; RLU: relative light units of luciferase output from the Trofile assay; FPR: false-positive rate for geno2pheno[coreceptor].
Viral load was measured as copies/mL.
CD4 cell count was measured as cells/mm3.
The total number of clones in each individual with the indicated V3 loop sequence.
HIV tropism for each env clone was determined using the Trofile Assay. Clones were identified as CCR5-using (R5), CXCR4-using (X4), or dual-tropic virus. Dual-tropic viruses were further classified as dual-R or dual-X according to their CXCR4+ cell infectivity and V3 loop sequence.
The minimum to maximum RLU is shown in cases with multiple distinct clones.
A false positive rate (FPR) of 10% was used to predict X4 tropism using an algorithm based on V3 region sequences (geno2pheno[coreceptor]). A FPR ≤10% predicts X4 tropism; a FPR >10% predicts R5 tropism.
We analyzed env clones from the five seroconverters who had DM viruses to determine whether their viral populations were comprised solely of dual-tropic viruses or mixtures of R5, X4, and dual-tropic viruses (Table 2). Dual-tropic viruses were further classified as either dual-R or dual-X in these analyses.14,19 Dual-R viruses infect CXCR4+ cells poorly and have env V3 loop sequences similar or identical to R5 viruses from the same individual, while dual-X viruses infect CXCR4+ cells efficiently and have env V3 loop sequences distinct from R5 viruses from the same individual. None of the five men had X4 viruses detected. Mixed populations of R5 and dual-R viruses were detected in two of the five men (Cases 1 and 2, Table 2); all of these viruses were predicted to be R5 viruses by the geno2pheno algorithm. There was a major subpopulation of dual-R viruses in Case 1, which had V3 sequences identical to those in one R5 clone. In Case 2, there were two proportionate subpopulations of R5 and dual-R viruses, which had identical V3 sequences. Dual-X viruses were detected in the remaining three men (Cases 3–5); all of the dual-X viruses were predicted to be X4 viruses by the geno2pheno algorithm. All clones from Cases 3 and 4 were dual-X and had identical (Case 3) or nearly identical (Case 4) V3 sequences. The viral population in Case 5 included a major subpopulation of R5 and minor subpopulations of dual-X viruses; one R5 clone was predicted to be a X4 virus by the geno2pheno algorithm.
DISCUSSION
We analyzed HIV-infected men enrolled in HPTN 061 to explore whether sexual practices were associated with HIV tropism. We did not observe a significant difference in HIV tropism among men who reported exclusive insertive vs. receptive intercourse at study enrollment. We note that the sexual practices of the men included in this analysis may have been different at the time of HIV infection than those reported for the six months preceding enrollment; self-report of sexual practices may also be unreliable in some cases. In addition, HIV evolution and superinfection could also impact the tropism of the viral population over time. Overall, DM viruses were detected among 26% of the HIV-infected men at enrollment, which is consistent with other studies that found X4/DM viruses in 20% of individuals with chronic HIV infection.1
Twenty-five percent of the seroconverters in HPTN 061 also had DM viruses. In other studies, X4/DM viruses have been observed in 2–20% of individuals with recent HIV infection.1,3–5 It is difficult to compare the proportion of individuals with X4/DM viruses in different studies, since the sensitivity for detecting X4/DM viruses depends on the method used for tropism testing. In this study, we used a clinically-validated, phenotypic tropism assay with enhanced sensitivity (Trofile), which can detect X4/DM variants present in 0.3% of the viral population.13 Most genotypic tropism assays use bulk (population) Sanger sequencing; these methods typically detect minority variants present at 15–20%.20 The sensitivity and specificity of genotypic tropism assays varies depending on the algorithms and cut-offs used to analyze V3 loop sequences.20 Determinants outside the V3 loop may also impact HIV tropism.14,21 Those regions are included in the recombinant viruses analyzed in the Trofile assay but not in genotypic tropism assays. In two of the five seroconverters in this study, dual-tropic env clones were identified that had V3 loop sequences identical to those in R5-tropic clones from the same samples. Notably, HIV tropism was misclassified in these cases using a genotypic assay based on env V3 sequencing (the geno2pheno algorithm).
In HPTN 061, men with DM viruses had significantly lower CD4 cell counts, consistent with findings from other studies.1 The five seroconverters with DM viruses also had low CD4 cell counts (median: 254 cells/mm3, range: 151–335). Other studies did not find a significant difference in CD4 cell count between individuals with X4/DM vs. R5 viruses early in infection,2,7,22 although those with X4/DM viruses had more pronounced CD4 cell count decline over time in two of those studies.2,7 In another study, a lower mean CD4 cell count was observed among seroconverters with DM vs. R5 viruses (450 vs. 629 cells/mm3).5 Low CD4 cell counts after seroconversion have been associated with faster HIV disease progression, independent of HIV tropism.23,24 In addition, individuals starting antiretroviral treatment (ART) with CD4 cell counts <500 cells/mm3 may not achieve the same degree of CD4 cell count recovery as those starting ART at higher CD4+ cell counts.25
The CCR5 coreceptor antagonist, maraviroc, is currently approved for ART in the US,26 and clinical trials are exploring its use for PrEP.8,9 Coreceptor tropism testing is recommended before using maraviroc for ART, since maraviroc may select for X4 and dual-tropic viruses.27 In HPTN 061, the seroconverters with DM viruses had viral populations that included either dual-tropic viruses or mixtures of R5 and dual-tropic viruses; the efficiency of CXCR4 use varied among the dual-tropic clones from each individual. Several studies suggest that maraviroc can inhibit dual-R viruses in vitro.28–30 Two of the five seroconverters who had DM viruses in our study had mixtures of R5 and dual-R viruses, which may have been susceptible to maraviroc. However, the remaining three seroconverters had dual-X viruses, which may not be inhibited by maraviroc. Further studies are needed to determine the susceptibility of dual-tropic viruses to maraviroc. In addition, maraviroc has been shown to rapidly select for minority dual-X populations.28 HIV drug resistance remains a concern for individuals who become HIV infected while taking antiretroviral drugs for PrEP. In this study, both R5 and dual-X viruses were detected in one man. Exposure to maraviroc in individuals with this tropism pattern could lead to selection of dual-X viruses, which could impact disease progression.
Black MSM in the US are disproportionately affected by HIV. Infrequent HIV testing prior to enrollment and late HIV diagnosis were common in the HPTN 061 cohort.31 X4/DM viruses have been associated with increases in viral load, more rapid CD4 cell decline, and faster disease progression.1,2 The high prevalence of X4/DM viruses among men in HPTN 061 highlights an urgent need to increase HIV testing frequency in this population. Further research is also needed to identify factors driving transmission and selection of X4/DM viruses and to evaluate potential associations of HIV tropism with disease progression and treatment outcomes among Black MSM.
Acknowledgments
The authors thank the staff at the HPTN study sites for their assistance and the HPTN 061 study participants for their contributions.
Footnotes
Ethical considerations:
Written informed consent was obtained from all participants in the HPTN 061 study. The institutional review boards at each participating institution approved the study.
Clinical Trials Registration Number: NCT 00951249
Author Contributions:
All authors contributed to manuscript preparation. Additional author roles are listed below.
| Iris Chen | Analyzed HIV tropism data; contributed to study design; drafted the manuscript |
| Wei Huang | Performed tropism testing and sequence analysis; reviewed test results |
| Matthew B. Connor | Data analyst; performed statistical analyses |
| Arne Frantzell | Performed tropism testing and sequence analysis; reviewed test results |
| Vanessa Cummings | HPTN Network Laboratory Quality Assurance/Quality Control Representative for HPTN 061; assisted with sample management |
| Geetha G. Beauchamp | Statistician; reviewed statistical analyses |
| Sam Griffith | Project coordinator for HPTN 061 |
| Sheldon D. Fields | Chair of the HPTN 061 Black Caucus |
| Hyman M. Scott | Co-investigator for the HPTN 061 site in San Francisco |
| Steven Shoptaw | Principal Investigator for the HPTN 061 site in Los Angeles |
| Carlos del Rio | Principal Investigator for the HPTN 061 site in Atlanta |
| Manya Magnus | Principal Investigator for the HPTN 061 site in Washington, DC |
| Sharon Mannheimer | Principal Investigator for one HPTN 061 site in New York City |
| Hong Van Tieu | Co-investigator for one HPTN 061 site in New York City |
| Darrell P. Wheeler | Protocol Co-chair for HPTN 061 |
| Kenneth H. Mayer | Protocol Co-chair for HPTN 061; Principal Investigator for the HPTN 061 site in Boston |
| Beryl A. Koblin | Protocol Chair for HPTN 061 |
| Susan H. Eshleman | Responsible for study design, analyzed data; drafted the manuscript |
Declarations of Interest and Financial Support:
The authors report no declarations of interest, with the following exceptions: Wei Huang and Arne Frantzell are employees and shareholders of Monogram Biosciences.
The HIV Prevention Trials Network (HPTN) is supported by the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Child Health and Human Development (NICH/HD), National Institute of Drug Abuse (NIDA) and the National Institute of Mental Health (NIMH), Office of AIDS Research, National Institutes of Health (NIH), Department of Health and Human Services under Grant UM1-AI068613.
Contributor Information
Iris Chen, Email: ichen@jhmi.edu, Dept. of Pathology, Johns Hopkins University School of Medicine, 720 Rutland Ave, Ross Building, Room 646, Baltimore, MD 21205, USA, (410) 614-6498.
Wei Huang, Email: huangw@labcorp.com, Monogram Biosciences, 345 Oyster Point Blvd, South San Francisco, CA 94080, USA, (650) 866-7429.
Matthew B. Connor, Email: matt.connor.mail@gmail.com, Vaccine and Infectious Disease Division, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, Seattle, WA 98109, USA, (210) 725-4161
Arne Frantzell, Email: frantza@labcorp.com, Monogram Biosciences, 345 Oyster Point Blvd, South San Francisco, CA 94080, USA, (615) 624-4284.
Vanessa Cummings, Email: vcummin1@jhmi.edu, Dept. of Pathology, Johns Hopkins University School of Medicine, 600 N. Wolfe St, Baltimore, MD 21205, USA, (410) 614-0479.
Geetha G. Beauchamp, Email: geetha@scharp.org, Vaccine and Infectious Disease Division, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, Seattle, WA 98109, USA, (206) 667-6167
Sam Griffith, Email: sgriffith@fhi360.org, Science Facilitation Department, FHI 360, 359 Blackwell St, Suite 200, Durham, NC 27703, USA, (919) 544-7040.
Sheldon D. Fields, Email: sdfields@fiu.edu, Mervyn M. Dymally School of Nursing, Charles R. Drew University of Medicine and Science, 1731 East 120th St, Los Angeles, CA 90059, USA, (323) 568-3304
Hyman M. Scott, Email: hyman.scott@sfdph.org, Bridge HIV, San Francisco Department of Public Health, 25 Van Ness Ave, San Francisco, CA 94102, USA, (415) 437-7483
Steven Shoptaw, Email: sshoptaw@mednet.ucla.edu, Dept. of Family Medicine, University of California Los Angeles, 10880 Wilshire Blvd, Suite 1800, Los Angeles, CA 90024 USA, (310) 740-7021.
Carlos del Rio, Email: cdelrio@emory.edu, Dept. of Global Health, Emory University Rollins School of Public Health, 1518 Clifton Rd NE, Atlanta, GA 30322, USA, (404) 727-7112.
Manya Magnus, Email: manyadm@gwu.edu, Dept. of Epidemiology and Biostatistics, The George Washington University, 950 New Hampshire Ave NW, Suite 507, Washington, DC 22305, USA, (202) 994-3024.
Sharon Mannheimer, Email: sbm20@columbia.edu, Dept. of Medicine, Harlem Hospital, Columbia University, Mailman School of Public Health, 506 Lenox Ave, Room 3101A, New York, NY 10037, USA, (212) 939-2940.
Hong-Van Tieu, Email: htieu@nybloodcenter.org, Laboratory of Infectious Disease Prevention, Lindsley F. Kimball Research Institute, New York Blood Center, 310 E 67th St, Suite 3-110, New York, NY 10065, USA, (212) 570-3081.
Darrell P. Wheeler, Email: dwheeler@luc.edu, School of Social Welfare, University at Albany, State University of New York, 13 Western Ave, Albany, NY 12222, USA, (518) 443-5324
Kenneth H. Mayer, Email: kmayer@fenwayhealth.org, The Fenway Institute, Fenway Health/Infectious Disease Division, Beth Israel Deaconess Medical Center/Department of Medicine, Harvard Medical School, 1340 Boylston St, 8th Floor, Boston, MA 02215, USA, (617) 927-6087
Beryl A. Koblin, Email: bkoblin@nybloodcenter.org, Laboratory of Infectious Disease Prevention, Lindsley F. Kimball Research Institute, New York Blood Center, 310 E 67th Street, New York, NY 10065, USA, (212) 570-3105
Susan H. Eshleman, Email: seshlem@jhmi.edu, Dept. of Pathology, Johns Hopkins University School of Medicine, 720 Rutland Ave, Ross Building, Room 646, Baltimore, MD, USA 21205, (410) 614-4734
References
- 1.Verhofstede C, Nijhuis M, Vandekerckhove L. Correlation of coreceptor usage and disease progression. Curr Opin HIV AIDS. 2012;7(5):432–439. doi: 10.1097/COH.0b013e328356f6f2. [DOI] [PubMed] [Google Scholar]
- 2.Raymond S, Delobel P, Mavigner M, Cazabat M, Encinas S, Souyris C, et al. CXCR4-using viruses in plasma and peripheral blood mononuclear cells during primary HIV-1 infection and impact on disease progression. AIDS. 2010;24(15):2305–2312. doi: 10.1097/QAD.0b013e32833e50bb. [DOI] [PubMed] [Google Scholar]
- 3.Chalmet K, Dauwe K, Foquet L, Baatz F, Seguin-Devaux C, Van Der Gucht B, et al. Presence of CXCR4-using HIV-1 in patients with recently diagnosed infection: correlates and evidence for transmission. J Infect Dis. 2012;205(2):174–184. doi: 10.1093/infdis/jir714. [DOI] [PubMed] [Google Scholar]
- 4.Sierra-Enguita R, Rodriguez C, Aguilera A, Gutierrez F, Eiros JM, Caballero E, et al. X4 tropic viruses are on the rise in recent HIV-1 seroconverters in Spain. Aids. 2014;28(11):1603–1609. doi: 10.1097/QAD.0000000000000269. [DOI] [PubMed] [Google Scholar]
- 5.de Mendoza C, Van Baelen K, Poveda E, Rondelez E, Zahonero N, Stuyver L, et al. Performance of a population-based HIV-1 tropism phenotypic assay and correlation with V3 genotypic prediction tools in recent HIV-1 seroconverters. J Acquir Immune Defic Syndr. 2008;48(3):241–244. doi: 10.1097/QAI.0b013e3181734f0e. [DOI] [PubMed] [Google Scholar]
- 6.Schuitemaker H, van’t Wout AB, Lusso P. Clinical significance of HIV-1 coreceptor usage. J Transl Med. 2011;9(Suppl 1):S5. doi: 10.1186/1479-5876-9-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Mendoza C, Rodriguez C, Garcia F, Eiros JM, Ruiz L, Caballero E, et al. Prevalence of X4 tropic viruses in patients recently infected with HIV-1 and lack of association with transmission of drug resistance. J Antimicrob Chemother. 2007;59(4):698–704. doi: 10.1093/jac/dkm012. [DOI] [PubMed] [Google Scholar]
- 8.Chen BA, Panther L, Marzinke MA, Hendrix CW, Hoesley CJ, van der Straten A, et al. Phase 1 safety, pharmacokinetics, and pharmacodynamics of dapivirine and maraviroc vaginal rings: a double-blind randomized trial. J Acquir Immune Defic Syndr. 2015;70(3):242–249. doi: 10.1097/QAI.0000000000000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gulick RM, Wilkin TJ, Chen YQ, Landovitz RJ, Amico KR, Young AM, et al. HPTN 069/ACTG 5305: phase II study of maraviroc-based regimens for HIV PrEP in MSM. Conf. on Retroviruses and Opportunistic Infections; Boston, MA. February 22–25, 2016. [Google Scholar]
- 10.Koblin BA, Mayer KH, Eshleman SH, Wang L, Mannheimer S, Del Rio C, et al. Correlates of HIV acquisition in a cohort of Black men who have sex with men in the United States: HIV Prevention Trials Network (HPTN) 061. PLoS One. 2013;8(7):e70413. doi: 10.1371/journal.pone.0070413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer KH, Wang L, Koblin B, Mannheimer S, Magnus M, del Rio C, et al. Concomitant socioeconomic, behavioral, and biological factors associated with the disproportionate HIV infection burden among Black men who have sex with men in 6 U.S. Cities. PLoS One. 2014;9(1):e87298. doi: 10.1371/journal.pone.0087298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitcomb JM, Huang W, Fransen S, Limoli K, Toma J, Wrin T, et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob Agents Chemother. 2007;51(2):566–575. doi: 10.1128/AAC.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reeves JD, Coakley E, Petropoulos CJ, Whitcomb JM. An enhanced sensitivity Trofile HIV coreceptor tropism assay for selecting patients for therapy with entry inhibitors targeting CCR5: a review of analytical and clinical studies. J Viral Entry. 2009;3(3):94–102. [Google Scholar]
- 14.Huang W, Eshleman SH, Toma J, Fransen S, Stawiski E, Paxinos EE, et al. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: high prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J Virol. 2007;81(15):7885–7893. doi: 10.1128/JVI.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lengauer T, Sander O, Sierra S, Thielen A, Kaiser R. Bioinformatics prediction of HIV coreceptor usage. Nat Biotechnol. 2007;25(12):1407–1410. doi: 10.1038/nbt1371. [DOI] [PubMed] [Google Scholar]
- 16.Vandekerckhove LP, Wensing AM, Kaiser R, Brun-Vezinet F, Clotet B, De Luca A, et al. European guidelines on the clinical management of HIV-1 tropism testing. Lancet Infect Dis. 2011;11(5):394–407. doi: 10.1016/S1473-3099(10)70319-4. [DOI] [PubMed] [Google Scholar]
- 17.Chen I, Cummings V, Fogel JM, Marzinke MA, Clarke W, Connor MB, et al. Low-level viremia early in HIV infection. J Acquir Immune Defic Syndr. 2014;67(4):405–408. doi: 10.1097/QAI.0000000000000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen I, Connor MB, Clarke W, Marzinke MA, Cummings V, Breaud A, et al. Antiretroviral drug use and HIV drug resistance among HIV-Infected Black men who have sex with men: HIV Prevention Trials Network 061. J Acquir Immune Defic Syndr. 2015;69(4):446–452. doi: 10.1097/QAI.0000000000000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang W, Toma J, Stawiski E, Fransen S, Wrin T, Parkin N, et al. Characterization of human immunodeficiency virus type 1 populations containing CXCR4-using variants from recently infected individuals. AIDS Res Hum Retroviruses. 2009;25(8):795–802. doi: 10.1089/aid.2008.0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obermeier M, Symons J, Wensing AM. HIV population genotypic tropism testing and its clinical significance. Curr Opin HIV AIDS. 2012;7(5):470–477. doi: 10.1097/COH.0b013e328356eaa7. [DOI] [PubMed] [Google Scholar]
- 21.Lin NH, Becerril C, Giguel F, Novitsky V, Moyo S, Makhema J, et al. Env sequence determinants in CXCR4-using human immunodeficiency virus type-1 subtype C. Virology. 2012;433(2):296–307. doi: 10.1016/j.virol.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frange P, Galimand J, Goujard C, Deveau C, Ghosn J, Rouzioux C, et al. High frequency of X4/DM-tropic viruses in PBMC samples from patients with primary HIV-1 subtype-B infection in 1996–2007: the French ANRS CO06 PRIMO Cohort Study. J Antimicrob Chemother. 2009;64(1):135–141. doi: 10.1093/jac/dkp151. [DOI] [PubMed] [Google Scholar]
- 23.Audigé A, Taffé P, Rickenbach M, Battegay M, Vernazza P, Nadal D, et al. Low postseroconversion CD4 count and rapid decrease of CD4 density identify HIV+ fast progressors. AIDS Res Hum Retroviruses. 2010;26(9):997–1005. doi: 10.1089/aid.2009.0263. [DOI] [PubMed] [Google Scholar]
- 24.Olson AD, Guiguet M, Zangerle R, Gill J, Perez-Hoyos S, Lodi S, et al. Evaluation of rapid progressors in HIV infection as an extreme phenotype. J Acquir Immune Defic Syndr. 2014;67(1):15–21. doi: 10.1097/QAI.0000000000000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le T, Wright EJ, Smith DM, He W, Catano G, Okulicz JF, et al. Enhanced CD4+ T-cell recovery with earlier HIV-1 antiretroviral therapy. N Engl J Med. 2013;368(3):218–230. doi: 10.1056/NEJMoa1110187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.U.S. Food and Drug Administration. [Accessed December 8, 2015];Antiretroviral drugs used in the treatment of HIV infection. 2015 http://www.fda.gov/ForPatients/Illness/HIVAIDS/Treatment/ucm118915.htm.
- 27.Cooper DA, Heera J, Goodrich J, Tawadrous M, Saag M, Dejesus E, et al. Maraviroc versus efavirenz, both in combination with zidovudine-lamivudine, for the treatment of antiretroviral-naive subjects with CCR5-tropic HIV-1 infection. J Infect Dis. 2010;201(6):803–813. doi: 10.1086/650697. [DOI] [PubMed] [Google Scholar]
- 28.Symons J, van Lelyveld SF, Hoepelman AI, van Ham PM, de Jong D, Wensing AM, et al. Maraviroc is able to inhibit dual-R5 viruses in a dual/mixed HIV-1-infected patient. J Antimicrob Chemother. 2011;66(4):890–895. doi: 10.1093/jac/dkq535. [DOI] [PubMed] [Google Scholar]
- 29.Svicher V, Balestra E, Cento V, Sarmati L, Dori L, Vandenbroucke I, et al. HIV-1 dual/mixed tropic isolates show different genetic and phenotypic characteristics and response to maraviroc in vitro. Antiviral Res. 2011;90(1):42–53. doi: 10.1016/j.antiviral.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Surdo M, Alteri C, Puertas MC, Saccomandi P, Parrotta L, Swenson L, et al. Effect of maraviroc on non-R5 tropic HIV-1: refined analysis of subjects from the phase IIb study A4001029. Clin Microbiol Infect. 2015;21(1):103e101–106. doi: 10.1016/j.cmi.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 31.Mannheimer SB, Wang L, Wilton L, Van Tieu H, Del Rio C, Buchbinder S, et al. Infrequent HIV testing and late HIV diagnosis are common among a cohort of black men who have sex with men in 6 US cities. J Acquir Immune Defic Syndr. 2014;67(4):438–445. doi: 10.1097/QAI.0000000000000334. [DOI] [PMC free article] [PubMed] [Google Scholar]