Abstract
Hypertension is one of the most common complex genetic disorders. We have described previously 38 single nucleotide polymorphisms (SNPs) with suggestive association with hypertension in Japanese individuals. In this study we extend our previous findings by analyzing a large sample of Japanese individuals (n=14 105) for the most associated SNPs. We also conducted replication analyses in Japanese of susceptibility loci for hypertension identified recently from genome-wide association studies of European ancestries. Association analysis revealed significant association of the ATP2B1 rs2070759 polymorphism with hypertension (P=5.3×10−5; allelic odds ratio: 1.17 [95% CI: 1.09 to 1.26]). Additional SNPs in ATP2B1 were subsequently genotyped, and the most significant association was with rs11105378 (odds ratio: 1.31 [95% CI: 1.21 to 1.42]; P=4.1×10−11). Association of rs11105378 with hypertension was cross-validated by replication analysis with the Global Blood Pressure Genetics consortium data set (odds ratio: 1.13 [95% CI: 1.05 to 1.21]; P=5.9×10−4). Mean adjusted systolic blood pressure was highly significantly associated with the same SNP in a meta-analysis with individuals of European descent (P=1.4×10−18). ATP2B1 mRNA expression levels in umbilical artery smooth muscle cells were found to be significantly different among rs11105378 genotypes. Seven SNPs discovered in published genome-wide association studies were also genotyped in the Japanese population. In the combined analysis with replicated 3 genes, FGF5 rs1458038, CYP17A1, rs1004467, and CSK rs1378942, odds ratio of the highest risk group was 2.27 (95% CI: 1.65 to 3.12; P=4.6×10−7) compared with the lower risk group. In summary, this study confirmed common genetic variation in ATP2B1, as well as FGF5, CYP17A1, and CSK, to be associated with blood pressure levels and risk of hypertension.
Keywords: hypertension, genetic variation, ATP2B1, Millennium Genome Project, Global BPgen
Because of its large impact on a number of cardiovascular diseases, hypertension is a major contributor to global health burden. Because hypertension is one of the most prevalent complex genetic disorders, with a heritability of ≤60% based on the estimation by 24-hour blood pressure (BP) readings,1 numerous studies, including recent genome-wide association studies (GWAS),2–6 have attempted to identify genetic variation associated with human BP levels. Except for rare mendelian forms of hypertension,7 the estimated effects of each genetic factor on BP levels have been found to be small in the general population (typically <1.0 mm Hg on systolic BP [SBP] and <0.5 mm Hg on diastolic BP [DBP] per risk allele). However, multiple risk alleles are known to have a cumulative impact on several complex traits, including BP and hypertension risk.3 In addition, it is anticipated that identification of novel susceptibility genes would lead to further understanding of disease pathogenesis.
As a part of a series of nationally based cooperative projects, the Millennium Genome Project (Millennium GPJ), we conducted multiple candidate gene analyses to identify susceptible genes and polymorphisms for hypertension. In a previously reported study,6 we focused on 307 genes, which were genes encoding components of signal transduction pathways potentially related to BP regulation, including receptors, soluble carrier proteins, binding proteins, channels, enzymes, and G proteins. That study identified 38 single nucleotide polymorphisms (SNPs) as suggestively associated with hypertension by analysis of 758 hypertensive patients and 726 normotensive controls.6 To extend our previous study, we have now genotyped all 38 of the SNPs in a replication panel composed of 1929 hypertensives and 1993 normotensives and have taken forward validated SNPs with further genotyping in a large Japanese genetic epidemiological cohort sample (n=14 105). An in silico validation analysis of our most promising loci was performed using the Global Blood Pressure Genetics (Global BPgen) consortium data set, a large-scale GWAS of samples of European descent.2 Furthermore, we also conducted a replication analysis of recent European GWAS-derived susceptible loci for hypertension from Global BPgen2 and CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) GWAS3 in a Japanese large-scale general population sample (Figure S1, available in the online Data Supplement at http://hyper.ahajournals.org).
Methods
Case and Control Subjects (Screening Panel)
Details of the screening panel subjects have been described previously.6 Briefly, hypertensive patients and normotensive controls were recruited in the Asahikawa, Tokyo, Osaka, and Hiroshima regions of Japan according to the following criteria. Hypertensive subjects (n=758) had a previous diagnosis of hypertension at between 30 and 59 years of age and were either being treated with antihypertensive medication or had a SBP >160 mm Hg and/or DBP >100 mm Hg. They had a family history of hypertension in their parents and/or siblings and were not obese (body mass index [BMI] <25 kg/m2). Normotensive controls (n=726) aged >45 years were recruited from the same regions. These individuals have never been treated with antihypertensive medications, and their SBP was <120 mmHg and DBP <80 mmHg. They had no family history of hypertension. All of the subjects were unrelated and were native Japanese.
Cohort-Based Population Samples
Seven independent study cohorts for cardiovascular diseases and related risk factors were combined to compose a large-scale Japanese genetic epidemiological population sample of 14 105. The Ohasama, Shigaraki, Takashima, Suita, and Nomura studies are general population-based genetic epidemiological studies. The study subjects were recruited via a medical checkup process for community residents. The 2 other cohorts, Yokohama and Matsuyama, are derived from employees of large manufacturing industries. The clinical parameters used in this study were obtained from personal health records during annual medical checkups. Further details of the study cohorts are described in the online Data Supplement.
Nested Case and Control Subjects Derived From the Cohort-Based Sample (Replication Panel)
Hypertensive cases and normotensive controls were chosen from the cohort-based population samples described above (n=11 569; the Suita study was excluded because of ethical issues). The selection criteria of the hypertensive and normotensive subjects were as follows: hypertensive subjects (n=1929) aged ≤64 years and either treatment with antihypertensive medication and/or SBP >160 mm Hg and/or DBP >90 mm Hg; normotensive subjects (n=1993) aged ≥40 years and having SBP <120 mm Hg and DBP <80 mm Hg; and no current use of antihypertensive medication and free from any history of cardiovascular disease.
Global BPgen (In Silico) Analyses
To investigate cross-validation of the most promising SNPs, we obtained results for 4 SNPs in the ATP2B1 gene from the Global BPgen consortium, a study that is composed of 17 GWAS studies with 34 433 individuals of European descent. A detailed description of the study design and phenotype measurement for all of the cohorts has been reported previously.2
Validation of Published BP Polymorphisms in the Japanese Millennium Cohort
Thirteen loci have been identified recently and robustly validated for association with BP and hypertension in recent large-scale GWAS of European samples, by the Global BPgen consortium2 and the CHARGE consortium.3 From the associated SNPs reported at these 13 loci, we selected SNPs expected to have minor allele frequencies in Japanese samples >0.10, based on the HapMap database (JPT only, Public Release No. 27)8: FGF5 rs1458038, CYP17A1 rs1004467, CSK rs1378942, PLCD3 rs12946454, PLEKHA7 rs381815, ULK4 rs9815354, and CSK-ULK3 rs6495122. These 7 SNPs were genotyped in the Japanese population-based cohort sample to test whether the same associations exist in samples of Japanese ancestry.
Genotyping
Genomic DNA was extracted from peripheral blood. All of the SNPs were analyzed by TaqMan probe assays (Applied Biosystems Co, Ltd) using commercially available primers and probes purchased from the Assay-on-Demand system. The fluorescence level of PCR products was measured using an ABI PRISM 7900HT sequence detector.
Ethical Considerations
All of the study procedures were approved by the ethics committee of each university or research institute. Written informed consent was obtained from all of the participating subjects.
Ex Vivo Expression Analysis of ATP2B1 mRNA
Umbilical artery smooth muscle cells were isolated from umbilical cords obtained at delivery (n=34). Expression levels of ATP2B1 mRNA were analyzed by RT-PCR using a relative quantification method. Further details of the ex vivo expression analysis are described in the online Data Supplement.
Statistical Analysis
At each SNP, frequency differences in each genotype among hypertensive and normotensive subjects were assessed using a χ2 test. Linkage disequilibrium (LD) coefficients were calculated using the Haploview software (Broad Institute).9 Adjusted odds ratios for hypertension, as well as coefficients and SEs for SBP and DBP, were calculated using logistic and linear multiple regression analysis, adjusting for sex, age, age2, BMI, and cohort variables, using additive (1 degree of freedom) and genotypic (2 degrees of freedom) genetic models. Adjustment for treatment with antihypertensive medication was achieved by adding fixed constants to measured values (+15 mm Hg for SBP and +10 mm Hg for DBP).10 The Global BPgen data and statistical methods have been described elsewhere.2 Meta-analysis was performed assuming fixed effects and using inverse variance weights. An unweighted genetic risk score based on 4 SNPs (ATP2B1 rs1105378, FGF5 rs1458038, CYP17A1 rs1004467, and CSK rs1378942) was calculated by adding the number of risk alleles showing higher BP values. Risk allele of each SNP was defined as follows: ATP2B1, C allele; FGF5, T allele; CYP17A1, A allele; and CSK, C allele. The CSK-ULK3 SNP rs6495122 showing positive association with BP trait and hypertension was not included in the calculation of genetic risk score, because the strong LD with the CSK SNP rs1378942 (D′=0.884; r2=0.731) is most parsimoniously explained by both SNPs tagging a single risk variant. Differences in mRNA expression levels among the ATP2B1 rs1105378 genotype were assessed by ANOVA. The statistical analyses were performed using a commercially available statistical software package (JMP version 8, SAS Institute).
Results
Replication Genotyping
The clinical characteristics of the replication panel chosen from the cohort-based population samples (Table S1, available in the online Data Supplement) are shown in Table S2. Stringent case and control definitions, corresponding with the extreme upper ≈17% and lower ≈17% of the general population, were used to maximize power for fixed genotyping costs.11 Thirty-six SNPs were successfully genotyped, and results for all of the SNPs are shown in Table S3. Significant association was observed for the ATP2B1 rs2070759 polymorphism located in intron 8 (P=4.4×10−4; allele odds ratio [OR]: 1.18 [95% CI: 1.07 to 1.29]). Several other SNPs also showed marginally significant association; however, the P values did not reach statistical significance after application of Bonferroni correction for multiple comparisons (threshold: 0.05/36=0.0014; Table S3; we note that no other SNPs are significant if the less conservative Benjamini-Hochberg procedure is used to control the false discovery rate at 0.05). Although, the replication results in the less-strict nested case-control sample chosen from the same population sample have been reported in our previous article,6 the association was recalculated to narrow down the SNPs to be applied to the following dense SNP analysis.
Dense SNP Analysis of the ATP2B1 Gene
To more precisely identify the SNP or SNPs increasing susceptibility for hypertension, we performed “de novo” genotyping of a dense SNP panel around marker rs2070759 in individuals from the original screening panel (Table S4).6 Forty-one tag SNPs located in a 167-kb region around rs2070759 were selected using the HapMap database (Table S5).8 Among the 27 SNPs polymorphic in our Japanese sample, the most significant association was observed with rs11105378; this yielded an allelic P value of 6.3×10−5 (OR: 1.37 [95% CI: 1.17 to 1.60]; Table 1 and Figure S2).
Table 1.
Association of ATP2B1 SNPs With Hypertension in the Screening and Replication Panels
| Screening Panel | Replication Panel | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Genotype | Genotype Frequency |
HWE | Call Rate |
Odds (P) | Genotype Frequency |
HWE | Call Rate |
Odds (P) | Overall Odds (P) | |||||
| rs1401982 | AA/AG/GG | HT | 318 | 328 | 92 | 0.603 | 96.3 | 1.28 (0.001) | 825 | 833 | 247 | 0.108 | 98.7 | 1.25 (3.0×10−6) | 1.26 (1.5×10−8) |
| NT | 249 | 324 | 118 | 0.474 | 699 | 961 | 305 | 0.397 | |||||||
| rs2681472 | AA/AG/GG | HT | 335 | 321 | 90 | 0.334 | 97.8 | 1.26 (0.003) | 846 | 832 | 242 | 0.095 | 99.5 | 1.26 (1.0×10−6) | 1.26 (8.7×10−9) |
| NT | 267 | 328 | 111 | 0.539 | 715 | 966 | 303 | 0.431 | |||||||
| rs2070759 | GG/GT/TT | HT | 216 | 379 | 151 | 0.515 | 97.6 | 1.16 (0.045) | 582 | 896 | 399 | 0.118 | 97.2 | 1.18 (4.4×10−4) | 1.17 (5.3×10−5) |
| NT | 186 | 341 | 175 | 0.454 | 507 | 956 | 474 | 0.579 | |||||||
| rs11105364 | TT/TG/GG | HT | 335 | 322 | 88 | 0.432 | 97.2 | 1.29 (0.001) | 846 | 834 | 236 | 0.171 | 99.3 | 1.25 (2.4×10−6) | 1.26 (4.1×10−9) |
| NT | 261 | 323 | 113 | 0.438 | 729 | 947 | 303 | 0.874 | |||||||
| rs11105378 | CC/CT/TT | HT | 359 | 301 | 76 | 0.276 | 97.3 | 1.37 (6.3×10−5) | 868 | 821 | 217 | 0.280 | 98.8 | 1.28 (1.4×10−7) | 1.31 (4.1 ×10−11) |
| NT | 280 | 320 | 108 | 0.295 | 746 | 922 | 300 | 0.586 | |||||||
The screening panel is composed of 758 middle age– onset severe hypertensive patients and 726 middle-aged to elderly evidently normotensive controls (Table S4). The replication panel consists of 1929 hypertensive cases, and 1993 normotensive controls selected from 11 569 cohort sample were enrolled (Table S2). ORs and P values for allelic model are shown.
The most associated SNP and the 4 others from the dense SNP analyses were subsequently genotyped in the replication panel. Significant association of rs11105378 was confirmed in the replication panel with an allelic P value of 1.4×10−7 (OR: 1.28 [95% CI: 1.17 to 1.41]; Table 1). Meta-analysis of both study panels indicated significant association (P=4.1×10−11; OR: 1.31 [95% CI: 1.21 to 1.42]) and confirmed that the strongest association is seen for rs11105378. The D′ and r2 measures of LD between rs2070759 and rs11105378 were 0.92 and 0.48, respectively. Other SNPs, rs1401982 (D′=0.99; r2=0.64), rs2681472 (D′=0.99; r2=0.61), rs11105364 (D′=0.97; r2=0.59), located within the same LD block, were also significantly associated with hypertension (Table 1). The strong LD between associated SNPs suggests a single true association signal in this region.
We examined for possible association of SNPs in the ATP2B4 gene, a well-investigated isoform of the ATP2B1 gene, with hypertension in the screening panel. We observed no significant correlation with the 17 SNPs analyzed, which were selected using the HapMap database (Table S6).
Population-Based Meta-Analyses of ATP2B1 SNPs
The complete Japanese population-based sample was subsequently genotyped for the 4 most significant SNPs in ATP2B1. To further validate and get more precise effect size estimates in Japanese, for this analysis, hypertensive cases were defined as individuals with treatment with antihypertensive medication, SBP ≥140 mm Hg, or DBP ≥90 mm Hg. The ORs for the 4 SNPs were all extremely similar (ranging from 1.19 to 1.21 under the additive model adjusted for age, age2, sex, BMI, and cohort variables; see Table S7). These associations were replicated in the Global BPgen subjects of European descent; the pooled analysis demonstrated increased significance (rs1105378: OR: 1.17 [95% CI: 1.11 to 1.23]; P=7.0×10−10), as expected for a larger total sample size (n=28 866; Table S7).
We next evaluated the effect of the most associated SNP, rs11105378, on BP levels in the Millennium GPJ cohort (Table 2). We adjusted for several covariates that are associated with BP phenotypes: age (r=0.362; P<0.001 for SBP), BMI (r=0.275; P<0.001), and sex (male: 131.7±18.2; female: 128.6±20.8 mm Hg; P<0.001). In multiple regression analysis for BP levels, including also cohort indicator variables as covariates, the results for a 2-degree-of-freedom test with the TT genotype as a reference identified both the TC genotype (coefficient=+1.66 mm Hg; P=2.2×10−4) and CC genotype (+2.47 mm Hg; P=4.9×10−8) as independent determinants for SBP after adjustment. The TC (+0.91 mm Hg; P=8.0×10−4) and CC genotypes (+1.32 mm Hg; P=1.8×10−6) were also independently associated with DBP levels. We depict the covariate adjusted mean BP levels by rs11105378 genotype in Figure S3. Results of each cohort separately are summarized in Table S8. We next performed a meta-analysis of data from the Millennium GPJ and 2 large epidemiological studies (Global BPgen and CHARGE; Table 2). Results show the per-allele differences in SBP and DBP to be ≈1.0 and 0.5 mm Hg, respectively.
Table 2.
Meta-Analysis of ATP2B1 SNPs With BP Traits
| Millennium GPJ | Global BPgen | CHARGE* | Pooled | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Coded Allele |
n (Frequency) |
Coefficient (SE), mm Hg |
P | n (Frequency) |
Coefficient (SE), mm Hg |
P | n (Frequency) |
Coefficient (SE), mm Hg |
P | Coefficient (95% CI), mm Hg |
P |
| SBP | ||||||||||||
| rs1401982 | G | 13 944 (0.376) |
−1.22 (0.23) |
1.8×10−7 | 33 885 (0.385) |
−0.30 (0.13) |
0.022 | −0.52 (−0.74 to −0.30) |
3.9×10−6 | |||
| rs2681472 | G | 14 032 (0.373) |
−1.33 (0.23) |
1.2×10−8 | 33 803 (0.158) |
−0.62 (0.18) |
5.2×10−4 | 0.17 | −1.29 (0.19) |
3.5×10−11 | −1.03 (−1.26 to −0.81) |
9.9×10−20 |
| rs11105364 | G | 14 013 (0.364) |
−1.34 (0.23) |
8.9×10−9 | 33 877 (0.179) |
−0.60 (0.18) |
7.4×10−4 | 0.17 | −1.30 (0.19) |
4.8×10−11 | −1.03 (−1.25 to −0.81) |
1.2×10−19 |
| rs11105378 | T | 13 948 (0.360) |
−1.33 (0.23) |
1.5×10−8 | 33 171 (0.158) |
−0.59 (0.18) |
0.001 | 0.16 | −1.31 (0.20) |
9.1×10−11 | −1.02 (−1.24 to −0.79) |
1.4×10−18 |
| DBP | ||||||||||||
| rs1401982 | G | 13 944 (0.376) |
−0.72 (0.14) |
2.0×10−7 | 33 898 (0.392) |
−0.18 (0.09) |
0.041 | −0.34 (−0.49 to −0.19) |
8.1×10−6 | |||
| rs2681472 | G | 14 032 (0.373) |
−0.65 (0.14) |
2.7×10−6 | 33 829 (0.157) |
−0.35 (0.12) |
0.003 | 0.17 | −0.64 (0.11) |
3.7×10−8 | −0.54 (−0.68 to −0.41) |
9.7×10−15 |
| rs11105364 | G | 14 013 (0.364) |
−0.70 (0.14) |
4.5×10−7 | 33 898 (0.158) |
−0.34 (0.12) |
0.004 | 0.17 | −0.63 (0.12) |
1.2×10−7 | −0.54 (−0.68 to −0.40) |
7.5×10−14 |
| rs11105378 | T | 13 948 (0.360) |
−0.70 (0.14) |
5.4×10−7 | 33 183 (0.158) |
−0.33 (0.12) |
0.005 | 0.16 | −0.62 (0.12) |
3.1×10−7 | −0.54 (−0.68 to −0.39) |
1.6×10−13 |
Coefficients and SE for SBP and DBP were calculated under the additive model using multiple regression analysis adjusted for age, age2, sex, and BMI. In both Millennium GPJ and Global BPgen, adjustment for treatment with antihypertensive medication was achieved by adding fixed constants to measured values (+15 mm Hg for SBP and +10 mm Hg for DBP).2 In the Japanese Millennium GPJ and also for some cohorts within Global BPgen, cohort variables were also adjusted to avoid residual population stratification.
Results of the CHARGE Study were obtained from the published article.3
Genotype-Specific Differences in Ex Vivo Expression of ATP2B1 mRNA
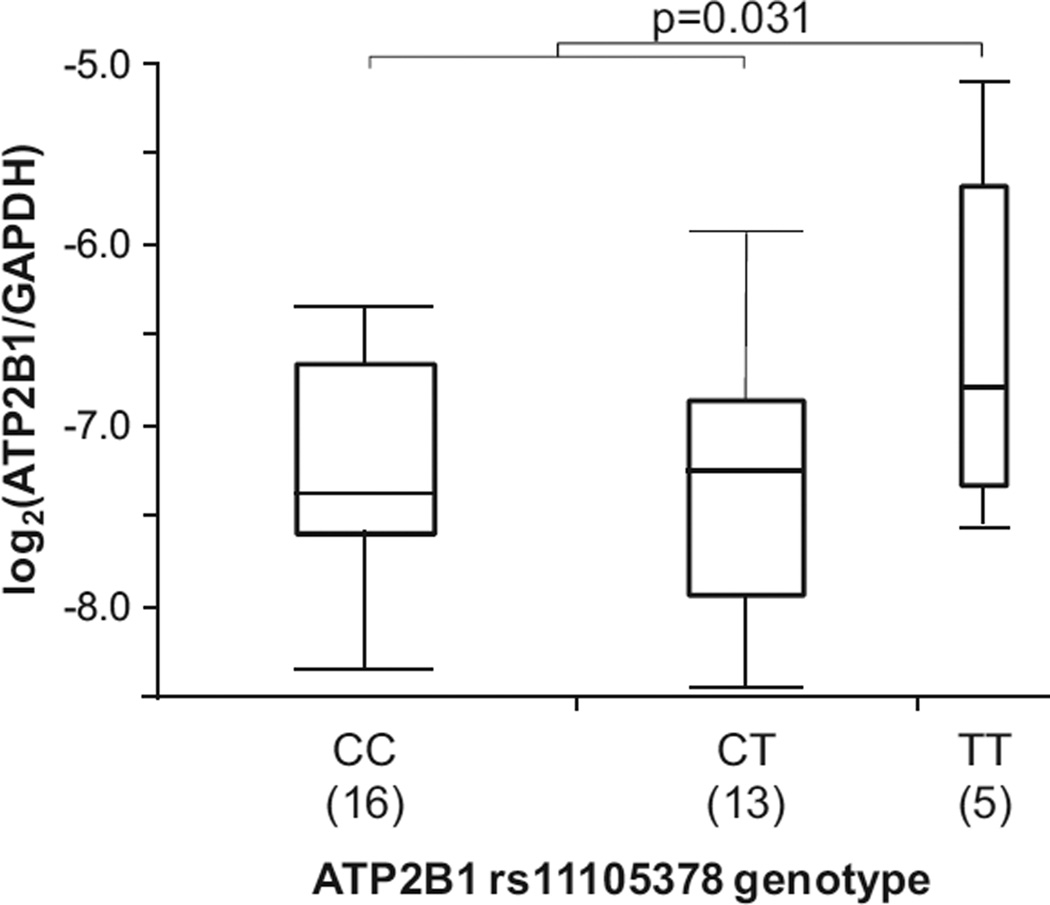
Differences in ATP2B1 mRNA expression in umbilical artery smooth muscle cells among rs11105738 genotype are shown in Figure 1. Assuming a recessive genetic model, cells homozygous for T allele showed significantly higher levels of ATP2B1 mRNA as compared with cells carrying 1 or 2 C alleles (P=0.031; see Figure 1). Under an additive genetic model, the overall P value was marginally significant (P=0.091).
Figure 1.
Ex vivo expression analysis of ATP2B1 mRNA. Graphs depict the log2 relative expression levels of the ATP2B1 mRNA in umbilical artery smooth muscle cells obtained by normalizing to GAPDH. Genotype of ATP2B1 rs11105378 of each sample was analyzed by direct sequencing using isolated genomic DNA from umbilical artery smooth muscle cells.
Replication Analysis of European GWAS-Derived Susceptible SNPs in Japanese
We next conducted a replication analysis in the Millennium GPJ, in which we tested associated SNPs identified in recent large-scale European GWAS by the Global BPgen2 and the CHARGE consortia.3 From the 7 most promising SNPs of which the minor allele frequency in Japanese was >0.10 based on the HapMap database, 4 SNPs, namely, FGF5 rs1458038, CYP17A1 rs1004467, CSK rs1378942, and CSK-ULK3 rs6495122, showed significant association in either binary trait analyses (Tables S9) or quantitative trait analysis (Table 3 and S10). The most significant association was observed with FGF5 rs1458038; this yielded a P value of 1.6×10−8 (+1.33 mm Hg) with SBP and 1.8×10−7 (+0.73 mm Hg) with DBP in the Millennium GPJ cohort, and the effect size was greater than that of Europeans (Table 3). Meta-analysis of both study panels with data from Global BPgen indicated further significant associations.
Table 3.
Meta-Analysis of SNPs With BP Traits
| Millennium GPJ | Global BPgen | Pooled | |||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | Coded Allele |
n (Frequency) |
Coefficient (SE), mm Hg |
P | n (Frequency) |
Coefficient (SE), mm Hg |
P | Coefficient (95% CI), mm Hg |
P |
| Systolic BP | |||||||||
| FGF5 | T | 13 826 | 1.33 | 1.6×10−8 | 30 850 | 0.62 | 1.6×10−6 | 0.81 | 1.1×10−11 |
| rs1458038 | (0.343) | (0.23) | (0.275) | (0.14) | (0.58 to 1.05) | ||||
| CYP17A1 | A | 14 007 | 0.89 | 2.3×10−4 | 33 735 | 0.94 | 1.0×10−5 | 0.92 | 6.2×10−9 |
| rs1004467 | (0.680) | (0.24) | (0.901) | (0.21) | (0.61 to 1.23) | ||||
| CSK | C | 13 920 | 0.77 | 0.007 | 34 126 | 0.62 | 2.4×10−6 | 0.65 | 4.2×10−8 |
| rs1378942 | (0.803) | (0.28) | (0.36) | (0.13) | (0.42 to 0.88) | ||||
| PLCD3 | T | 14 003 | 0.11 | 0.703 | 32 120 | 0.68 | 3.9×10−6 | 0.57 | 2.5×10−5 |
| rs12946454 | (0.831) | (0.30) | (0.28) | (0.15) | (0.30 to 0.83) | ||||
| PLEKHA7 | T | 14 030 | 0.11 | 0.687 | 33 706 | 0.52 | 2.6×10−4 | 0.44 | 4.7×10−4 |
| rs381815 | (0.199) | (0.28) | (0.26) | (0.14) | (0.19 to 0.68) | ||||
| CSK-ULK3 | A | 14 014 | 0.68 | 0.017 | 33 308 | 0.47 | 2.4×10−4 | 0.51 | 1.7×10−5 |
| rs6495122 | (0.812) | (0.28) | (0.45) | (0.13) | (0.28 to 0.74) | ||||
| ULK4 | A | 13 976 | −0.67 | 0.059 | 32 034 | 0.17 | 0.297 | 0.01 | 0.950 |
| rs9815354 | (0.116) | (0.35) | (0.18) | (0.17) | (−0.29 to 0.31) | ||||
| DBP | |||||||||
| FGF5 | T | 13 826 | 0.73 | 1.8×10−7 | 30 850 | 0.55 | 1.5×10−8 | 0.61 | 6.1×10−14 |
| rs1458038 | (0.343) | (0.14) | (0.275) | (0.10) | (0.45 to 0.77) | ||||
| CYP17A1 | A | 14 007 | 0.29 | 0.047 | 33 735 | 0.40 | 5.4×10−3 | 0.35 | 4.9×10−4 |
| rs1004467 | (0.680) | (0.14) | (0.901) | (0.14) | (0.15 to 0.54) | ||||
| CSK | C | 13 920 | 0.41 | 0.015 | 34 126 | 0.48 | 5.9×10−8 | 0.46 | 5.2×10−9 |
| rs1378942 | (0.803) | (0.17) | (0.36) | (0.09) | (0.31 to 0.62) | ||||
| PLCD3 | T | 14 003 | 0.14 | 0.426 | 32 120 | 0.34 | 5.7×10−4 | 0.30 | 1.9×10−4 |
| rs12946454 | (0.831) | (0.18) | (0.28) | (0.09) | (0.14 to 0.46) | ||||
| PLEKHA7 | T | 14 030 | 0.13 | 0.437 | 33 706 | 0.23 | 0.014 | 0.20 | 0.018 |
| rs381815 | (0.199) | (0.17) | (0.26) | (0.10) | (0.04 to 0.37) | ||||
| CSK-ULK3 | A | 14 014 | 0.38 | 0.027 | 33 308 | 0.35 | 4.2×10−5 | 0.36 | 7.4×10−6 |
| rs6495122 | (0.812) | (0.17) | (0.45) | (0.09) | (0.20 to 0.51) | ||||
| ULK4 | A | 13 976 | 0.21 | 0.325 | 32 034 | 0.40 | 2.9×10−4 | 0.36 | 2.3×10−4 |
| rs9815354 | (0.116) | (0.21) | (0.18) | (0.11) | (0.17 to 0.55) | ||||
Multiple Regression Analysis for BP Trait and Hypertension in Japanese
To clarify whether the 4 susceptibility SNPs (ATP2B1, FGF5, CYP17A1, and CSK) were independently associated with BP traits and hypertension, multiple regression analysis was performed with possible covariates (Table S11). After adjustment for age, age2, sex, BMI, and drinking habits, this analysis confirmed that all 4 of the SNPs were independent determinants for both BP traits and hypertension.
Combined Effect of Risk Genotypes on Hypertension
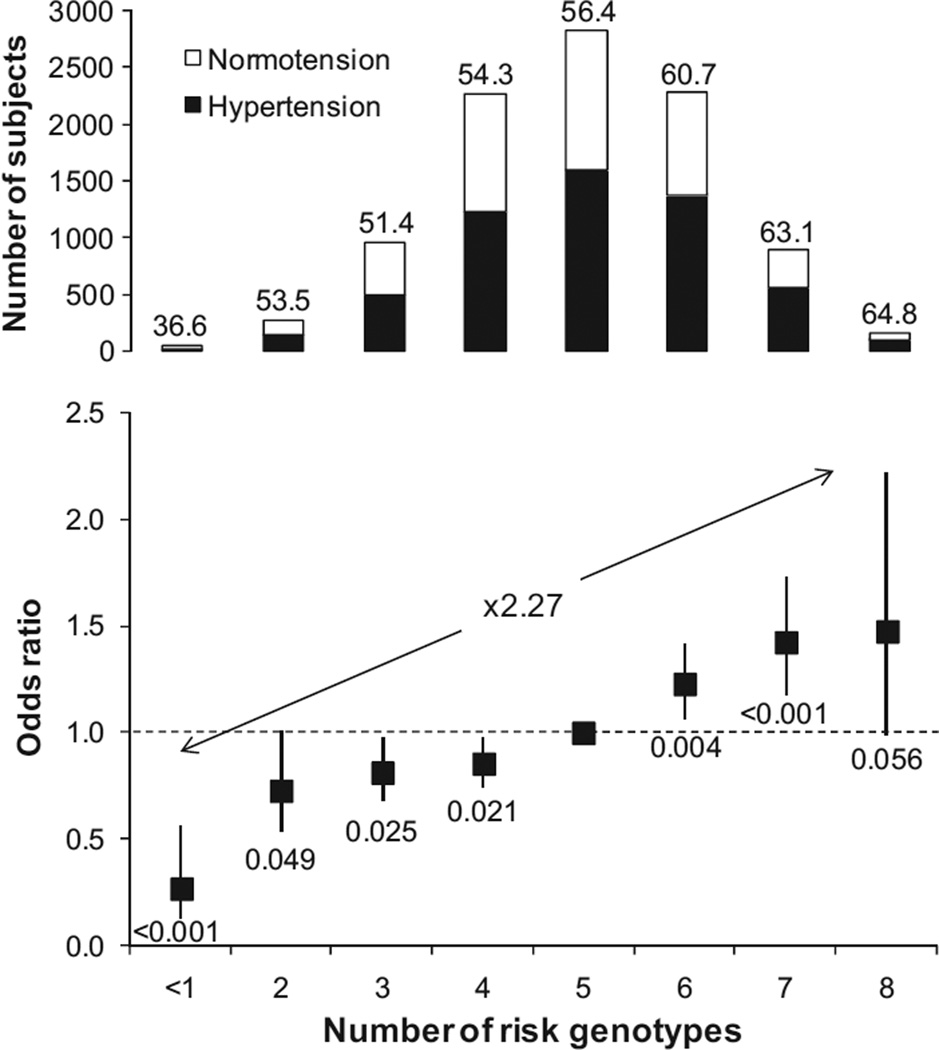
A risk score for 4 susceptible genotypes was calculated to evaluate their combined effects on hypertension. ORs associated with increasing number of risk genotypes in a covariates adjusted logistic regression model are depicted in Figure 2 (overall P value was 5.4×10−5). Compared with the reference group (5 risk genotypes), individuals carrying 7 or 8 risk genotypes had higher risk (OR: 1.43 [95% CI: 1.20 to 1.72]; P=1.0×10−4) in contrast to the lower OR of individuals with ≤2 risk genotypes (OR: 0.63 [95% CI: 0.47 to 0.85]; P=0.020). The OR of the high-risk group was raised to 2.27 (95% CI: 1.65 to 3.12; P=4.6×10−7) compared with the lowest risk group. Adjusted per-allele OR for hypertension was 1.17 (95% CI: 1.12 to 1.21; P=4.0×10−15). The distribution of the Japanese population sample among the number of risk genotypes is shown in Figure S4.
Figure 2.
ORs for hypertension according to the number of risk genotypes Number of risk genotype was calculated by the following 4 SNPs: ATP2B1 rs1105378, FGF5 rs1458038, CYP17A1, rs1004467, and CSK rs1378942. Hypertensive subjects were defined as being treated with antihypertensive medication, SBP ≥140 mm Hg, or DBP ≥90 mm Hg; normotensive subjects were defined as all not treated with antihypertensive medication, SBP ≤120 mm Hg, and DBP ≤85 mm Hg.2 Adjusted OR for hypertension and BP levels were calculated using logistic and linear multiple regression analysis, adjusting for sex, age, age2, BMI, and cohort variables. Frequency of hypertension and P values for the hypertension odds are shown in the top of column and the bottom of square, respectively.
Discussion
The present study has identified SNPs located upstream or within the ATP2B1 gene as strong susceptibility polymorphisms for hypertension in Japanese. These are findings that have also been reported recently in individuals of European descent3 and in Koreans.4 Although numerous studies have attempted to identify genetic markers for hypertension over the past 2 decades, there has been little cross-validation of loci in different ethnic groups so far except for mendelian forms of hypertension. The SNPs in ATP2B1 identified in this study showed significant association in large-scale studies in populations with different ancestries and using different discovery approaches, including GWAS in the CHARGE consortium and the Korean study and an independent candidate gene analysis in our present study. Similar findings in different ethnic groups with different methods further strengthen these findings and indicate the ATP2B1 gene region as a susceptibility locus of likely global significance for BP variation and development of hypertension. Two replication results very recently reported by another Japanese group12 and a Korean group13 also indicated the disease susceptibility of ATP2B1 SNPs located in the same LD block.
No biological data have been provided whether SNP rs1105378 or other SNPs in strong LD have any effect on the transcriptional activity or transcriptional regulation of the ATP2B1 gene. Furthermore, although alternative splicing has been found to generate several variants of ATP2B1 mRNA,14 the SNP associations that we have observed do not shed light on whether this is a potential mechanism for affecting BP. Our data first showed that the effect of SNPs on ATP2B1 gene expression levels is a potential mechanism by which disease-associated SNP alleles cause the phenotypic changes. Changes in the ATP2B1 gene product levels are involved in BP regulation. We found no microRNA harboring rs11105378 in the miRBase database.15
The ATP2B1 (so-called PMAC1) gene encodes the plasma membrane calcium ATPase isoform 1, which removes bivalent calcium ions from eukaryotic cells against very large concentration gradients and plays a critical role in intracellular calcium homeostasis. Although pathophysiological implications of ATP2B1 gene products on the development of hypertension are uncertain, it has been reported that inhibition of ATP2B1 by the selective inhibitor caloxin 2A1 showed endothelium-dependent relaxation of rat aorta by increasing cytosolic Ca2+ concentration and consequent activation of endothelial NO synthase.16 Other information on the role of ATP2B1 has been obtained from experiments using bladder smooth muscle cells: contractility measurements on these cells have documented the important role of ATP2B1 in the extrusion of Ca2+ after carbachol stimulation or depolarization with potassium chloride.17 These reports suggest altered vascular reactivity as a plausible explanation for disease susceptibility of ATP2B1 gene.
In mammals, calcium ATPase isoforms are encoded by ≥4 separate genes (ATP2B1 to ATP2B4).18 It has been reported that overexpression of the human ATP2B4 gene in arterial smooth muscle cells in mice increases vascular reactivity and BP partly because of negative regulation of neuronal NO synthase.19 We, therefore, examined the possible association of ATP2B4 gene polymorphisms with hypertension by using the screening panel. However, no significant correlation was observed in the 17 SNPs analyzed, which were selected by reference to the HapMap database. The pathophysiological association of plasma membrane Ca2+ pump with BP regulation may be isoform specific.
Numerous studies, including the recent GWAS,3–6 have attempted to identify genetic variations associated with human BP levels. At present, it is not clear to what extent findings from GWAS in one population can be extrapolated to other populations with different lifestyles and genetic background. However, the present study provides a cross-validation of 4 of 7 SNPs (most likely representing 3 of 6 independent signals) derived from European GWAS. Replication studies in other Japanese12 and Korean13 populations also reported the cross-validation of European GWAS-derived SNP. Conservation of susceptible loci for hypertension was independent of ethnic background. This finding suggests an existence of unidentified common etiology of essential hypertension in relation to the susceptible genes and their physiological pathways.
Although individual common genetic variants confer a modest risk of hypertension, their combination showed a large impact on hypertension. The genetic risk score was associated with ≤2.27-times greater odds for hypertension. Similar observations have been found in other common diseases and multifactorial phenotypes, including, for example, type 2 diabetes mellitus,20 serum lipid levels,21 and serum uric acid levels.22 We reported previously that the findings of the cross-sectional analysis revealed a similar association in the longitudinal analysis23; the fat mass and obesity-associated gene polymorphism was an independent risk factor for the future development of obesity after adjustment for possible confounding factors. The present cross-sectional study cannot address the question of whether the ATP2B1 polymorphism and other susceptible variants predict future development of hypertension. However, recent articles investigating a prognostic significance of susceptible variants for type 2 diabetes mellitus24 and cardiovascular disease25 showed poor predictive performance of common variants in spite of the high OR observed in subjects carrying multiple risk alleles. A small proportion of the genetically high-risk persons attributed to independent inheritance of risk alleles may make it difficult to discriminate intermediate-risk persons. Genetic information may be most useful to identify a high-risk individual’s need for early intervention.
Several definitions of hypertension were used in this study to explore susceptible SNPs with modest effects and to further validate the susceptibility. Since it was expected to be underpowered to detect the effects of common variants in a dichotomized analysis with slightly elevated BP, subjects with high normal BP were excluded from the 65 347 case-control analyses. All of the alleles associated with hypertension in a dichotomized analysis (Table S7) were also associated with BP levels (Table 2). Our methodology may, thus, be appropriate to identify susceptible variants for hypertension.
Perspectives
We have identified SNPs located in the ATP2B1 gene region as susceptibility loci for hypertension in Japanese using a multistage association study, an association that has now been confirmed across different ethnic groups. Differences in the ex vivo ATP2B1 mRNA expression levels further supported the disease susceptibility of SNP rs1110578. We also replicated the susceptibility of the European GWAS-derived SNPs in Japanese. Because hypertension is a trait that is preventable by dietary and exercise interventions, early detection of at-risk populations using genetic information may be useful in preventing future hypertension-related diseases.
Supplementary Material
Acknowledgments
We greatly appreciate the efforts of Drs Sumio Sugano and Shoji Tsuji in planning and organization of this study. We thank Drs Hirohito Metoki, Masahiro Kikuya, Takuo Hirose, Kei Asayama, Ken Sugimoto, Kei Kamide, Mitsuru Ohishi, Ryuichi Morishita, Hiromi Rakugi, Yasuyuki Nakamura, Shinji Tamaki, Kenji Matsui, Tanvir Chowdhury Turin, Nahid Rumana, Tadashi Shiwa, Momoko Ogawa, Keisuke Yatsu, Sanae Saka, Nobuko Miyazaki, and Iimori-Tachibana-Rieko for their continued support in this research.
Sources of Funding
This work was supported by Grants for Scientific Research (Priority Areas “Medical Genome Science [Millennium Genome Project]” and “Applied Genomics,” Leading Project for Personalized Medicine, and Scientific Research 20390185, 21390099, 19659163, 16790336, 12204008, 15790293, 16590433, 17790381, 17790381, 18390192, 18590265, 18590587, 18590811, 19590929, 19650188, 19790423, 17390186, 20390184, and 21390223) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan; a Grants-in-Aid (H15-longevity-005, H17-longevity-003, H16-kenko-001, H18-longevity (kokusai), H11-longevity-020, H17-kenkou-007, H17-pharmaco-common-003, H18-Junkankitou[Seishuu]-Ippan-012, and H20-Junkankitou[Seishuu]-Ippan-009, 013) from the Ministry of Health, Labor and Welfare, Health and Labor Sciences Research Grants, Japan; a Science and Technology Incubation Program in Advanced Regions, Japan Science and Technology Agency; the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation; a Grant-in-Aid from the Japan Society for the Promotion of Science fellows (16.54041, 18.54042, 19.7152, 20.7198, 20.7477, and 20.54043), Tokyo, Japan; Health Science Research Grants and Medical Technology Evaluation Research Grants from the Ministry of Health, Labor and Welfare, Japan; the Japan Atherosclerosis Prevention Fund; the Uehara Memorial Foundation; the Takeda Medical Research Foundation; National Cardiovascular Research grants; Biomedical Innovation grants; and the Japan Research Foundation for Clinical Pharmacology.
Footnotes
Full author list of the Global BPgen consortium is given in the online Data Supplement.
Disclosures
Several authors (Y.T., K.K., Y.Ki., N.H., J.N., S.U., H.U., and T.Mik.) have been named as inventors on a patent application by Ehime University, Shiga University of Medical Science, and Yokohama City University in work related to this study.
References
- 1.Kotchen TA, Kotchen JM, Grim CE, George V, Kaldunski ML, Cowley AW, Hamet P, Chelius TH. Genetic determinants of hypertension: identification of candidate phenotypes. Hypertension. 2000;36:7–13. doi: 10.1161/01.hyp.36.1.7. [DOI] [PubMed] [Google Scholar]
- 2.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, Papadakis K, Voight BF, Scott LJ, Zhang F, Farrall M, Tanaka T, Wallace C, Chambers JC, Khaw KT, Nilsson P, van der Harst P, Polidoro S, Grobbee DE, Onland-Moret NC, Bots ML, Wain LV, Elliott KS, Teumer A, Luan J, Lucas G, Kuusisto J, Burton PR, Hadley D, McArdle WL, for the Wellcome Trust Case Control Consortium. Brown M, Dominiczak A, Newhouse SJ, Samani NJ, Webster J, Zeggini E, Beckmann JS, Bergmann S, Lim N, Song K, Vollenweider P, Waeber G, Waterworth DM, Yuan X, Groop L, Orho-Melander M, Allione A, Di Gregorio A, Guarrera S, Panico S, Ricceri F, Romanazzi V, Sacerdote C, Vineis P, Barroso I, Sandhu MS, Luben RN, Crawford GJ, Jousilahti P, Perola M, Boehnke M, Bonnycastle LL, Collins FS, Jackson AU, Mohlke KL, Stringham HM, Valle TT, Willer CJ, Bergman RN, Morken MA, Döring A, Gieger C, Illig T, Meitinger T, Org E, Pfeufer A, Wichmann HE, Kathiresan S, Marrugat J, O’Donnell CJ, Schwartz SM, Siscovick DS, Subirana I, Freimer NB, Hartikainen AL, McCarthy MI, O’Reilly PF, Peltonen L, Pouta A, de Jong PE, Snieder H, van Gilst WH, Clarke R, Goel A, Hamsten A, Peden JF, Seedorf U, Syvänen AC, Tognoni G, Lakatta EG, Sanna S, Scheet P, Schlessinger D, Scuteri A, Dörr M, Ernst F, Felix SB, Homuth G, Lorbeer R, Reffelmann T, Rettig R, Völker U, Galan P, Gut IG, Hercberg S, Lathrop GM, Zelenika D, Deloukas P, Soranzo N, Williams FM, Zhai G, Salomaa V, Laakso M, Elosua R, Forouhi NG, Völzke H, Uiterwaal CS, van der Schouw YT, Numans ME, Matullo G, Navis G, Berglund G, Bingham SA, Kooner JS, Connell JM, Bandinelli S, Ferrucci L, Watkins H, Spector TD, Tuomilehto J, Altshuler D, Strachan DP, Laan M, Meneton P, Wareham NJ, Uda M, Jarvelin MR, Mooser V, Melander O, Loos RJ, Elliott P, Abecasis GR, Caulfield M, Munroe PB. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, Aulchenko Y, Lumley T, Köttgen A, Vasan RS, Rivadeneira F, Eiriksdottir G, Guo X, Arking DE, Mitchell GF, Mattace-Raso FU, Smith AV, Taylor K, Scharpf RB, Hwang SJ, Sijbrands EJ, Bis J, Harris TB, Ganesh SK, O’Donnell CJ, Hofman A, Rotter JI, Coresh J, Benjamin EJ, Uitterlinden AG, Heiss G, Fox CS, Witteman JC, Boerwinkle E, Wang TJ, Gudnason V, Larson MG, Chakravarti A, Psaty BM, van Duijn CM. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban HJ, Yoon D, Lee MH, Kim DJ, Park M, Cha SH, Kim JW, Han BG, Min H, Ahn Y, Park MS, Han HR, Jang HY, Cho EY, Lee JE, Cho NH, Shin C, Park T, Park JW, Lee JK, Cardon L, Clarke G, McCarthy MI, Lee JY, Lee JK, Oh B, Kim HL. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41:527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 5.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohara K, Tabara Y, Nakura J, Imai Y, Ohkubo T, Hata A, Soma M, Nakayama T, Umemura S, Hirawa N, Ueshima H, Kita Y, Ogihara T, Katsuya T, Takahashi N, Tokunaga K, Miki T. Identification of hypertension-susceptibility genes and pathways by a systemic multiple candidate gene approach: the millennium genome project for hypertension. Hypertens Res. 2008;31:203–212. doi: 10.1291/hypres.31.203. [DOI] [PubMed] [Google Scholar]
- 7.Lifton RP. Molecular genetics of human blood pressure variation. Science. 1996;272:676–680. doi: 10.1126/science.272.5262.676. [DOI] [PubMed] [Google Scholar]
- 8.International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 9.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 10.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med. 2005;24:2911–2935. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- 11.Xiong M, Fan R, Jin L. Linkage disequilibrium mapping of quantitative trait loci under truncation selection. Hum Hered. 2002;53:158–172. doi: 10.1159/000064978. [DOI] [PubMed] [Google Scholar]
- 12.Takeuchi F, Isono M, Katsuya T, Yamamoto K, Yokota M, Sugiyama T, Nabika T, Fujioka A, Ohnaka K, Asano H, Yamori Y, Yamaguchi S, Kobayashi S, Takayanagi R, Ogihara T, Kato N. Blood pressure and hypertension are associated with 7 loci in the Japanese population. Circulation. 2010;121:2302–2309. doi: 10.1161/CIRCULATIONAHA.109.904664. [DOI] [PubMed] [Google Scholar]
- 13.Hong KW, Jin HS, Lim JE, Kim S, Go MJ, Oh B. Recapitulation of two genomewide association studies on blood pressure and essential hypertension in the Korean population. J Hum Genet. 2010;55:336–341. doi: 10.1038/jhg.2010.31. [DOI] [PubMed] [Google Scholar]
- 14.Keeton TP, Burk SE, Shull GE. Alternative splicing of exons encoding the calmodulin-binding domains and C termini of plasma membrane Ca(2+)-ATPase isoforms 1, 2, 3, and 4. J Biol Chem. 1993;268:2740–2748. [PubMed] [Google Scholar]
- 15.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhary J, Walia M, Matharu J, Escher E, Grover AK. Caloxin: a novel plasma membrane Ca2+ pump inhibitor. Am J Physiol Cell Physiol. 2001;280:C1027–C1030. doi: 10.1152/ajpcell.2001.280.4.C1027. [DOI] [PubMed] [Google Scholar]
- 17.Liu L, Ishida Y, Okunade G, Shull GE, Paul RJ. Role of plasma membrane Ca2+-ATPase in contraction-relaxation processes of the bladder: evidence from PMCA gene-ablated mice. Am J Physiol Cell Physiol. 2006;290:C1239–C1247. doi: 10.1152/ajpcell.00440.2005. [DOI] [PubMed] [Google Scholar]
- 18.Carafoli E. The Ca2+ pump of the plasma membrane. J Biol Chem. 1992;267:2115–2118. [PubMed] [Google Scholar]
- 19.Gros R, Afroze T, You XM, Kabir G, Van Wert R, Kalair W, Hoque AE, Mungrue IN, Husain M. Plasma membrane calcium ATPase overexpression in arterial smooth muscle increases vasomotor responsiveness and blood pressure. Circ Res. 2003;93:614–621. doi: 10.1161/01.RES.0000092142.19896.D9. [DOI] [PubMed] [Google Scholar]
- 20.Lango H, UK Type 2 Diabetes Genetics Consortium. Palmer CN, Morris AD, Zeggini E, Hattersley AT, McCarthy MI, Frayling TM, Weedon MN. Assessing the combined impact of 18 common genetic variants of modest effect sizes on type 2 diabetes risk. Diabetes. 2008;57:3129–3135. doi: 10.2337/db08-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, Voight BF, Bonnycastle LL, Jackson AU, Crawford G, Surti A, Guiducci C, Burtt NP, Parish S, Clarke R, Zelenika D, Kubalanza KA, Morken MA, Scott LJ, Stringham HM, Galan P, Swift AJ, Kuusisto J, Bergman RN, Sundvall J, Laakso M, Ferrucci L, Scheet P, Sanna S, Uda M, Yang Q, Lunetta KL, Dupuis J, de Bakker PI, O’Donnell CJ, Chambers JC, Kooner JS, Hercberg S, Meneton P, Lakatta EG, Scuteri A, Schlessinger D, Tuomilehto J, Collins FS, Groop L, Altshuler D, Collins R, Lathrop GM, Melander O, Salomaa V, Peltonen L, Orho-Melander M, Ordovas JM, Boehnke M, Abecasis GR, Mohlke KL, Cupples LA. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dehghan A, Köttgen A, Yang Q, Hwang SJ, Kao WL, Rivadeneira F, Boerwinkle E, Levy D, Hofman A, Astor BC, Benjamin EJ, van Duijn CM, Witteman JC, Coresh J, Fox CS. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372:1953–1961. doi: 10.1016/S0140-6736(08)61343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabara Y, Osawa H, Guo H, Kawamoto R, Onuma H, Shimizu I, Takara Y, Nishida W, Yamamoto M, Makino H, Kohara K, Miki T. Prognostic significance of FTO genotype in the development of obesity in Japanese: the J-SHIPP study. Int J Obes (Lond) 2009;33:1243–1248. doi: 10.1038/ijo.2009.161. [DOI] [PubMed] [Google Scholar]
- 24.Talmud PJ, Hingorani AD, Cooper JA, Marmot MG, Brunner EJ, Kumari M, Kivimäki M, Humphries SE. Utility of genetic and non-genetic risk factors in prediction of type 2 diabetes: Whitehall II prospective cohort study. BMJ. 2010;340:b4838. doi: 10.1136/bmj.b4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paynter NP, Chasman DI, Buring JE, Shiffman D, Cook NR, Ridker PM. Cardiovascular disease risk prediction with and without knowledge of genetic variation at chromosome 9p21.3. Ann Intern Med. 2009;150:65–72. doi: 10.7326/0003-4819-150-2-200901200-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.