Abstract
Pathogens have evolved a myriad of ways to abrogate and manipulate the host response to infections. Of the various mechanisms involved, pathogen-encoded and sometimes host-encoded proteases are an important category of virulence factors that cause robust changes on the host response by targeting key proteins along signaling cascades. The nuclear factor kappaB (NF-κB) signaling pathway is a crucial regulatory mechanism for the cell, controlling the expression of survival, immune, and proliferation genes. Proteases from pathogens of almost all types have been demonstrated to target and cleave members of the NF-κB signaling pathway at nearly every level. This review provides discussion of proteases targeting the most abundant NF-κB subunit, p65, and the impact of protease-mediated p65 cleavage on the immune responses and survival of the infected host cell. After examining various examples of protease interference, it becomes evident that the cleavage fragments produced by pathogen-driven proteolytic processing should be further characterized to determine whether they have novel and unique functions within the cell. The selective targeting of p65 and its effect on gene transcription reveals unique mechanisms by which pathogens acutely alter their microenvironment and further research may open new opportunities for novel therapeutics to combat pathogens.
Epithelial cells and innate immune cells stand as the first line of defense against tissue damage and invading pathogens at mucosal surfaces. Encoding a variety of pattern recognition receptors (PRRs) that are capable of responding to an even greater variety of pathogen-associated molecular patterns (PAMPs), mucosal epithelial cells initiate immune responses. Stimulation of PRRs results in the activation of signaling cascades, through adaptor proteins, that transduce the signal to the transcription factor level. Most commonly, key transcription factors, including nuclear factor kappaB (NF-κB), activator protein 1 (AP-1), cAMP response element-binding protein (CREB), and interferon-regulatory factor (IRF), are the targets of these signaling pathways, resulting in the transactivation of an array of target genes. The activation of inflammatory and anti-apoptotic proteins ultimately alerts the immune system of invasion and induces the recruitment of leukocytes to the site of infection. Cell surface receptors, such as toll-like receptors (TLRs), stimulate the cellular signaling and activation of NF-κB when the corresponding ligand binds its receptor. The NF-κB signaling pathways are crucial for the host response, as they serve as a master regulator and orchestrator of both the innate and adaptive immune responses (Finlay & McFadden, 2006, Le Negrate, 2012), making it a key target for pathogen interference (Thaker, 2014, Vallabhapurapu & Karin, 2009). A great number of previous studies have elucidated the mechanism by which the NF-κB pathways are regulated in normal and disease conditions (Hayden & Ghosh, 2012, Lenardo & Baltimore, 1989). Typically, NF-κB activation is initiated by ligand binding to a membrane bound cytokine or PRR receptor whose signal is transduced by the adaptor protein myeloid differentiation primary response gene 88 (MyD88) and others until the IκB kinase (IKK) complex is activated. The IKK beta subunit (IKKβ) phosphorylates and marks IκB for proteasomal degradation, which in turn unmasks the nuclear localization signal (NLS) of NF-κB thus liberating the NF-κB complex for translocation into the nucleus. Once in the nucleus, the NF-κB complex binds to a variety of specific double-stranded DNA sequences, termed κB sites, in the genome and recruits the general transcriptional machinery to induce the transcription of target genes (Lenardo & Baltimore, 1989), including those controlling immune responses and survival of host cells.
Besides the best-characterized Rel family proteins, i.e. p65 (also named RelA), RelB, c-Rel, p50, and p52, other non-Rel protein subunits in NF-κB complexes play an important role in optimizing the binding of target DNA and transcription of target genes. Ribosomal protein S3 (RPS3) was identified as the first ‘specifier’ subunit of NF-κB that confers the promoter selectivity and transcriptional specificity of NF-κB. RPS3 occurs along with p65 and IκBα in a portion, but not all, of the inhibitory NF-κB complexes in the cytoplasm (Wan et al. unpublished results). Once activated, RPS3 translocates, independent of p65, into the nucleus where it increases the affinity of p65/p50 to particular κB sites (Wan et al., 2007, Wan et al., 2011). RPS3 functions as a ‘specifier’ of NF-κB gene transcription, shedding light on a major lingering question within the field regarding promoter selectivity and transcriptional specificity, especially considering how diverse stimuli generate specific responses amongst the large number of κB binding sites scattered throughout the genome. Moreover, this additional binding partner strengthens the association of the p65/p50 dimer at the particular κB sites of a growing list of genes that are now considered RPS3-dependent genes, allowing the activation of only a subset of the vast potential p65-dependent genes (Wan et al., 2007, Wan et al., 2011, Sen et al., 2012). Since then, other specifiers have been attributed to helping hone the activation of NF-κB-dependent genes. Src-associated substrate during mitosis of 68 kDa (Sam68) has been discovered as another non-Rel subunit of p65, functioning in the nucleus to mediate the binding of NF-κB to the CD25 promoter in activated T cells (Fu et al., 2013), and Kruppel-like factor 6 (KLF6) has been identified as another nuclear co-activator of NF-κB for a subset of genes (Zhang et al., 2014). The discovery of these specifier proteins not only serves to answer the question of promoter selectivity but also emphasize that while NF-κB dimer binding is required at the promoter, it is not sufficient to induce the transcription of particular genes. The specifier proteins function to promote the binding and enhance the efficiency of transcriptional activation of particular genes over others. The function of the currently known specifier proteins in particular inflammatory settings or specific tissue/cell types remains unknown. The existence of cell type specific responses to similar stimuli is well recognized and exemplified by the discussion of how gut epithelial cells tend to response to infection with a mainly anti-apoptotic profile (and some inflammatory responses), whereas the infiltrating immune cells build the majority of the inflammatory response (Spehlmann & Eckmann, 2009, Eckmann & Neish, 2011). Understanding the cell type specific responses to stimuli is particularly important when many pathogens have evolved preferences for infecting particular cell types which presumably aides their survival within the host.
As a result of the many host protective mechanisms, pathogens have co-evolved many strategies to overcome these host defense mechanisms. Encoding virulence factors, such as toxins, adhesins, secretion systems, decoy receptors, kinases, proteases, and compounds, that modulate host immune responses have resulted in the ability to greatly manipulate the host response (Casadevall & Pirofski, 2009). Pathogens express an array of virulence factors that change as the pathogen progresses through its life cycle/infection cycle. These factors target all aspects of the cellular immune responses to infection, however those that target proteins where intracellular signaling cascades converge are arguably the most challenging for host cells to overcome. This is likely because many surface receptors rely on a smaller pool of adaptor proteins, that if disrupted, prevent the relay of signals from the outside of the cell to the nucleus (Silke & Hartland, 2013), therefore the pathogen does not need to encode a virulence factor to target all cellular receptors, when it can target a common adaptor protein to turn off the cellular signaling cascades. Whether the pathogen is intracellular or extracellular and/or encodes a secretion system capable of translocating proteins into a host cell, it can gain access to the major signaling junctions employed by the mammalian host to orchestrate immune responses (Brodsky & Medzhitov, 2009). Due to the convergence of many PRR signaling cascades on proteins such as IKK and mitogen activated protein kinase (MAPK), it is more effective for pathogens to target the intracellular activator than attempt to block all the extracellular receptors (Silke & Hartland, 2013). With this strategy, pathogens can exact robust effects on cell signaling while encoding fewer effectors. Along with targeting a myriad of signaling pathways, pathogens also encode virulence factors with redundant functions in an effort to abrogate the host-signaling cascade. Many pathogens have acquired sophisticated mechanisms to directly interfere with the host NF-κB signaling pathways through regulating or mimicking host proteins to their own advantage, highlighting the importance of NF-κB signaling. Bacteria, viruses, and parasites encode virulence factors that operate at multiple levels of the sequential process of NF-κB signaling using redundancy to ensure blockade of the pathway, demonstrating how crucial inhibiting this particular pathway is for pathogen survival. Of note, deubiquitinating enzymes (Ndubaku & Tsui, 2015), kinases (Krachler et al., 2011), proteases, and other encoded proteins are key virulence factors employed by pathogens to modulate NF-κB signaling in host cells. Of these, proteases are of particular interest because they alter the inherent structure of host proteins, thus preventing their native functions, although the cleaved fragments may play additional roles within the cell. This review will focus on the targeting of the NF-κB signaling pathway by intracellular proteases encoded by various pathogens. The regulation of NF-κB and the activation of downstream gene targets remain of great interest in host-pathogen interactions, but understanding how pathogens are manipulating these interactions to abrogate intracellular NF-κB signaling and dampen immune responses is paramount.
Targeting the upstream signaling molecules in the NF-κB pathway
Upon receptor engagement, the activation of adaptor proteins occurs to start the NF-κB signal transduction cascade in the cytoplasm that leads to changes at the transcriptional level of specific NF-κB target genes in the nucleus. The NF-κB signaling cascade contains many intermediate molecules (as well as important post-translational modifications) that are subject to pathogen intervention, in particular pathogen-encoded protease virulence factors. The Toll–interleukin 1 receptor (TIR) domain-containing proteins TRIF and MyD88, both of which are important adaptor proteins residing just inside the cellular membrane, were reported to be targeted by the NS3/4A protease encoded by Hepatitis C virus (HCV) (Li et al., 2005a) and the 3C protease encoded by enterovirus 68 (Xiang et al., 2014), whereas infection by the bacterium Yersinia enterocolitica activates human Caspase 3 to cleave these host proteins (Novikova et al., 2014). Targeting TRIF prevents signal transduction from TLR3 and TLR4, whereas abrogating MyD88 blocks signal transduction from most surface stimulation, as it is employed by all TLRs (except TLR3) and some cytokine and chemokine receptors (Kawai & Akira, 2010, Piras & Selvarajoo, 2014). Thus the protease-mediated cleavage of TRIF and MyD88 has an extensive effect on downstream pathways, as evidenced by the lack of NF-κB activation during HCV (Li et al., 2005a), enterovirus 68 (Xiang et al., 2014), and Y. enterocolitica (Novikova et al., 2014) infections. Of note, besides the NF-κB signaling pathway, the NS3/4A and 3C proteases also block the activation of the interferon response by targeting TRIF (Kawai & Akira, 2010, Piras & Selvarajoo, 2014). As a result, these pathogens have robust effects on host defense with minimal energy input, by encoding a single protease that can target the nexus of multiple signaling pathways. Moreover, HCV is especially adept in this respect, as the same protease also targets the intracellular adaptor mitochondrial antiviral-signaling protein (MAVS) (Li et al., 2005b, Meylan et al., 2005) that is crucial for RIG-I helicase-initiated activation of NF-κB and interferon responses (Yoneyama & Fujita, 2007). Moreover, the porcine reproductive and respiratory syndrome virus (PRRSV) encoded protease NSP4 has been shown to cleave NEMO/IKKγ (Huang et al., 2014), a regulatory subunit of the IKK complex that serves as the master NF-κB regulator that is activated by the stimulation of various cell surface molecules. Furthermore, the 3C protease encoded by Coxsackievirus has been demonstrated to cleave IκBα (Saura et al., 2007, Zaragoza et al., 2006), which sequesters NF-κB in the cytoplasm. Interestingly, the cleavage of IκBα by Coxsackievirus 3C protease does not completely degrade IκBα, something that would lead to the full liberation and constitutive activation of NF-κB. Instead, the proteolytic fragment, remains bound to and translocates into the nucleus with NF-κB complexes, dominantly interfering with NF-κB DNA binding capability, thus dampening the NF-κB activation in host cells during Coxsackievirus infection (Saura et al., 2007, Zaragoza et al., 2006). These pathogen-encoded proteases and their proposed target proteins in host cells are summarized in Table 1. By targeting the upstream nodes of this signaling cascade, these proteases are capable of causing a global suppression of the NF-κB signaling pathway specifically, as well as other signaling cascades that crosstalk via some of these adaptor molecules.
Table 1.
Pathogen- and host-encoded proteases targeting NF-κB signaling molecules, excluding p65, during infections.
| Pathogen | Protease | Protease category | Host target | Cleavage site(s) | Cleavage verification | Reference |
|---|---|---|---|---|---|---|
| HCV | NS3/4A | Serine Protease | TRIF | C372/S373 | recombinant, in vitro | Li et al. 2005a |
| EV68 | 3C | Serine Protease | TRIF | Q312/S313, Q653/S654 |
in vitro | Xiang et al. 2014 |
| Y. enterocolitica | Caspase-3 (host) |
Cysteine Protease | MyD88 | D135/S136 | recombinant, in vitro | Novikova et al. 2014 |
| HCV | NS3/4A | Serine Protease | CARDIF/MAVS | C508/H509 | in vitro | Meylan et al. 2005; Li et al. 2005b |
| PRRSV | NSP4 | Serine Protease | NEMO | E349/S350 | in vitro | Huang et al. 2014 |
| CVB3 | 3Cpro | Serine Protease | IκBα | Q249/G250 | recombinant, in vitro | Zaragoza et al. 2006 |
| EPEC | NleC | Zinc Metalloprotease | p50 | Not characterized | in vitro |
Pearson et al. 2011; Muhlen et al. 2011 |
| EPEC | NleC | Zinc Metalloprotease | p50 | C61/E62 | recombinant | Turco & Sousa 2014 |
| EHEC | NleC | Zinc Metalloprotease | RelB | C144/E145 | recombinant | Turco & Sousa 2014 |
EPEC, Enteropathogenic E. coli; EHEC, Enterohemorrhagic E. coli; CVB3, Coxsackievirus B3; HCV, Hepatitis C virus; EV68, Enterovirus 68; PRRSV, Porcine reproductive and respiratory syndrome virus; Y. enterocolitica, Yersinia enterocolitica; recombinant, purified protein assays; in vitro, cell culture/transfection assays; in vivo, animal studies.
p65, a major target of pathogen-encoded proteases
In addition to targeting various key molecules upstream of the NF-κB signaling pathway, pathogens have acquired strategies to interfere with NF-κB, in particular the Rel subunits that are directly involved in binding to DNA promoters of various target genes. Although there is some cell type variability, the most abundant NF-κB species in a cell consists of p65, p50, and other proteins (Wan et al., 2007). The p65 protein also possesses a transactivation domain (TAD) that is crucial for recruiting the general transcriptional machinery for the transactivation of target genes (Wan & Lenardo, 2010). These features make p65 unique among Rel family proteins and most frequently modulated by many pathogens including bacteria, parasites, and viruses.
Bacteria encoded proteases
Pathogenic bacteria encoding a membrane-bound protein complex, termed the type III secretion system (T3SS), such as the attaching/effacing (A/E) pathogens including enteropathogenic Escherichia coli (EPEC), enterohemorrhagic E. coli (EHEC), and the mouse pathogen Citrobacter rodentium, are prime examples of pathogens that interact with host intracellular signaling cascades to alter their microenvironment (Hartland & Leong, 2013, Shames & Finlay, 2012, Silke & Hartland, 2013, Wong et al., 2011). EHEC, EPEC, and C. rodentium encode a long list of virulence proteins (effectors) that are delivered into host cells during infection via the T3SS. These effectors target multiple cellular pathways present within the cell and function to manipulate the host cellular signaling, often with multiple effectors targeting the same pathway (reviewed in (Wong et al., 2011)). As such, the non-LEE-encoded (Nle) effectors NleB, NleE, NleH1, and NleC all target the NF-κB signaling pathway, of which NleB blocks the signal transduction between the TNF receptor-associated factor 2 (TRAF2) and NF-κB upstream proteins by preventing the ubiquitination of TRAF2 (Pham et al., 2013); NleE modifies TGF-β Activated Kinase 1 Binding Protein 2/3 (TAB2/3) that transduces the activating signal from a cytokine receptor to TGF-β Activated Kinase 1 (TAK1) or IKK kinases (Yao et al., 2014); NleH1 inhibits the phosphorylation of RPS3 thereby preventing its nuclear translocation and ‘specifier’ activity (Gao et al., 2009). Using multiple effectors to block NF-κB signaling allows the pathogen to regulate its microenvironment and ensure particular host signal transduction pathways are blocked. Another effector targeting NF-κB is the metalloprotease NleC, which was shown to cleave mainly p65 (Baruch et al., 2011, Hodgson et al., 2015, Li et al., 2014, Muhlen et al., 2011, Pearson et al., 2011, Turco & Sousa, 2014, Yen et al., 2010), although p300 was also proposed (Shames et al., 2011), in host cells. As a canonical zinc metalloprotease with the conserved HEXXH motif, NleC is well conserved among EHEC, EPEC, and C. rodentium (Turco & Sousa, 2014). NleC was first described as an important effector for the infection-mediated suppression of interleukin-8 (IL-8) in host cells (Yen et al., 2010, Sham et al., 2011) and subsequently revealed by multiple groups to cleave p65 in order to suppress host inflammatory responses (Baruch et al., 2011, Hodgson et al., 2015, Li et al., 2014, Muhlen et al., 2011, Pearson et al., 2011, Turco & Sousa, 2014, Yen et al., 2010). Moreover, the molecular details about the interaction between p65 and NleC have also been illustrated by recent crystal structural analyses showing that the complimentary electrostatic interactions surrounding the catalytic zinc binding site of NleC and the DNA binding loop of p65 (Li et al., 2014) and that NleC structurally and chemically mimics DNA to exploit the DNA binding motifs of p65 (Turco & Sousa, 2014). Although different cell lines and pathogens were utilized, the cleavage sites of p65 by NleC from previous studies appear to occur between proline 10 and alanine 11 (P10/A11) (Yen et al., 2010), cysteine 38 and glutamic acid 39 (C38/E39) (Baruch et al., 2011, Li et al., 2014, Turco & Sousa, 2014), or both with C38/E39 being the primary site (Hodgson et al., 2015). More importantly, the C38/E39 cleavage site on p65 is of particular interest as C38 sulfahydration was recently proposed to be a crucial post-translational modification to stabilize the p65-RPS3 interaction, thus facilitating RPS3/p65-dependent NF-κB target gene transcription (Sen et al., 2012).
The gram-negative bacterium Photobacterium damselae piscicida (Phdp) that causes fish pasteurellosis, a disease characterized by septicemia and high mortality is a considerable threat for mariculture (Barnes et al., 2005, Romalde, 2002). Phdp encodes an A-B toxin termed AIP56, whose N-terminus is homologous to NleC, maintaining the conserved HEIVH zinc binding domain, whereas the C-terminus serves as an internalization mechanism for the secreted AIP56 to enter target cells (Silva et al., 2013), macrophages and neutrophils (do Vale et al., 2007, do Vale et al., 2003). During Phdp infection, macrophages and neutrophils, the primary cell type employed by the immune system to clear apoptotic cells before secondary necrosis, undergo AIP56-dependent apoptosis (do Vale et al., 2007, Silva et al., 2013), causing the release of their cytotoxic components thus resulting in an inflammatory response (do Vale et al., 2007, do Vale et al., 2003). Given the homology between NleC and AIP56, it is not surprising that AIP56 also targets the same Cys/Glu residues, i.e. C39/E40 in fish, within the Rel homology domain (RHD) of p65 for cleavage. The apoptotic effect executed by AIP56 is thought to result from the abrogated gene transcription of anti-apoptotic genes normally conferred by NF-κB/p65 (Silva et al., 2013). In this setting, blocking the NF-κB-mediated anti-apoptotic gene transcription induces an overwhelming immune response and severe immunopathology in fish hosts subsequent to Phdp infection.
The well known intracellular bacteria, Chlamydia trachomatis, is the primary cause of preventable blindness worldwide as well as the most commonly found sexually-transmitted disease in the United States (Brunham & Rey-Ladino, 2005), which makes it a pathogen of intense clinical interest. Although the murine infection model nicely recapitulates the acute genital tract infection observed in women, spontaneous clearance and life long immunity make the mouse disease differ from the human disease in which infections can last several months and re-infection is common (Morrison & Caldwell, 2002). As an intracellular pathogen, C. trachomatis employs several proteases at various life stages to regulate host-cellular signaling. CT441 is a tail-specific protease (Tsp) containing a PDZ (postsynaptic density protein [PSD95], Drosophila disc large tumor suppressor [Dlg1], and zonula occludens-1 protein [zo-1]) domain and conserved serine and lysine residues that serve as the catalytic unit. CT441 shares approximately 70% sequence identity around the catalytic unit with other PDZ domain-containing Tsp proteases (Lad et al., 2007b). Lad and colleagues reported that CT441 cleaves human p65, however the murine protein is resistant to cleavage, due to several differences in their amino acid sequences between residues 331–359 (Lad et al., 2007b). Moreover, swapping the residues 331–359 between human and murine p65 proteins renders the human p65 resistant to CT441 cleavage. Of note, selectively cleaving only the human protein versus the mouse protein could serve as a key-determining factor explaining the disease difference between the two species. It has been speculated that without cleaving p65, the mouse immune response may be induced early enough to prevent widespread dissemination of the Chlamydia bacteria, leading to an accelerated clearance. Lad and colleagues also demonstrated that expression of CT441 not only induced cleavage of human p65 but also prevented NF-κB activation, as illustrated by a lack of IκBα degradation and p65 nuclear translocation (Lad et al., 2007a). Cleavage of p65 could result in the lack of p65 nuclear translocation; moreover, the cleaved N-terminus of p65, defined as p40, was proposed to directly interact with IκBα, adding additional dominant negative regulation to NF-κB activation. The p40-IκBα interaction was hypothesized to protect IκBα from degradation, although the protective mechanism is unclear and there exists the possibility that another effector encoded by C. trachomatis has a role in protecting IκBα from degradation (Lad et al., 2007a). Indeed, targeting the same signaling pathway via multiple strategies is not uncommon for many pathogens, as such redundancy ensures full manipulation of the host microenvironment to the pathogens’ advantage. It remains to be determined whether the lack of IκBα degradation and subsequent NF-κB activation are also restricted to human infection, but not in the murine infection. Further inquiry would further clarify the cause of the IκBα protection being dependent on the presence of the p40 cleavage product, since only the human p65 can be cleaved to generate the p40 product.
Parasite encoded proteases
The Leishmania parasite is responsible for the infection of more than 2 million people around the world every year (de Vries et al., 2015). Spread by the Phlebotomus sandfly, this vector borne parasite has many life stages which vary in their expression and activation of various proteins, constantly providing a moving target for treatment and vaccine development (Jain & Jain, 2015). It was reported that the Leishmania metalloprotease gp63 cleaves p65 and produces a stable truncated form of p65, termed p35RelA, which executes a functional role within the cell (Gregory et al., 2008a). The p35 fragment retains the nuclear translocation ability of the full-length protein where it heterodimerizes with p50 and binds DNA. Surprisingly, despite both proteins lacking a TAD domain, the p35 truncation and p50 heterodimer are still capable of turning on the expression of a particular set of chemokines (Gregory et al., 2008a). This indicates that beyond NF-κB, additional transcriptional factor(s) that possess a TAD may be involved in the transcription of these chemokine genes, considering that it is known that NF-κB couples with other transcription factors such as AP-1 and STATs to form a higher-order enhanceosome for optimizing the gene expression in immune responses (Fan et al., 2010, Fujioka et al., 2004, Neff et al., 2001). Previous studies showed that infection with Leishmania parasites resulted in the failed activation of macrophages and proinflammatory signaling. Moreover, infected macrophages have also been revealed to be defective in reactive oxygen species (ROS) production and chemotaxis (Gregory et al., 2008b, Olivier et al., 2005). However, it remains unknown how the expression of certain chemokines, such as macrophage inflammatory protein 1α (MIP-1α), MIP-1β, and Regulated on Activation, Normal T Cell Expressed and Secreted (RANTES) are induced during Leishmania infection (Dasgupta et al., 2003, Olivier et al., 2005). In particular the transactivation of these chemokines is a survival strategy for the parasite, as the recruitment of more macrophages to the infection site allows the parasite to move into a new host cell and continue the infection cycle (Dasgupta et al., 2003). Gregory and colleagues demonstrated that the cleaved p35 fragment induced chemokine gene transcription, however, little more mechanistic detail beyond DNA binding was offered. The authors speculated that the remaining molecules of the p35 fragment present within the nucleus could be acting in a dominant negative capacity to block the transactivation of other NF-κB target genes, such as Nos2, encoding inducible nitric oxide synthase that would be detrimental to the parasite (Gregory et al., 2008a). In this way, the p35 fragment may be acting in a repressive manner, similar to how p50 homodimers are thought to act, as they also lack the TAD domain. Hence, gp63 induced cleavage of p65 provides the parasite with the ability to acutely modify its microenvironment by regulating NF-κB target gene transcription.
Virus-encoded proteases
Viruses also encode various virulence factors that allow them to manipulate the crucial signaling pathways in host cells, which is more critical for viruses than other pathogens as viruses solely rely on host cells for replication. For instance, all members of the picornavirus family encode a 3C cysteine protease that processes the viral RNA encoded polyprotein (reviewed in (Palmenberg, 1990)). Among them, it has been demonstrated that poliovirus, as well as a few others, targets NF-κB p65 in order to shut down aspects of the innate immune response in host cells (Neznanov et al., 2005). Poliovirus infects humans via the gastrointestinal tract and enters the bloodstream via the local lymphoid tissue. Most infections are asymptomatic or mild; however in a subset of individuals the virus eventually infects cells within the central nervous systems (CNS) where it causes the characteristic paralysis known as poliomyelitis. In vitro, poliovirus infection of host cells causes a short-lived activation of NF-κB, as demonstrated by nuclear translocation of p65/p50 and IκBα degradation peaking at about 4 hours post infection (Neznanov et al., 2005). Interestingly, around 3 hours post infection, the appearance of an N-terminal truncation of p65, termed p65ΔC, occurs simultaneously with the translation and detection of the poliovirus-encoded 3C protease (Neznanov et al., 2005). It was further demonstrated that the cleavage product is a result of the 3C protease and not the induction of apoptosis, which is also supported by the careful control over the timing and induction of apoptosis that poliovirus infection is able to exert on the cell (Agol et al., 2000, Levkau et al., 1999). Indeed, the control of apoptosis and the initial activation of the NF-κB signaling pathway, which causes the expression of some anti-apoptotic genes, allow the host cell to survive long enough for the virus to replicate its genome. While poliovirus is a well-studied example of the picornavirus family, Neznanov and colleagues also showed that other representatives of the family encode 3C, such as ECHO-1 and rhino-14, and can also cleave p65 (Neznanov et al., 2005). Elucidating the spatial-temporal regulation of these virulence factors is needed in future studies to allow for a deeper understanding of how these many strategies and regulatory pathways interact within host cells.
Host-encoded proteases benefiting pathogens
Besides interfering with host signaling pathways directly with pathogen-encoded proteases, pathogens take advantage of host protein functions (NF-κB being one of them) to serve their own needs. This is best exemplified by the human immunodeficiency virus (HIV). Indeed, the HIV genome encodes and requires NF-κB binding sites to boost the transcription of certain viral proteins (Alcami et al., 1995, Bachu et al., 2012, Coiras et al., 2008, Perkins et al., 1993, Zhang et al., 2012). Since HIV primarily infects and replicates in activated T cells, Coiras and colleagues characterized the effects of T cell activation on the NF-κB signaling pathway (Coiras et al., 2008). Interestingly, they uncovered that immediately after the phorbol myristate acetate (PMA) or phytohemagglutinin (HA) stimulation of T cells, an apoptosis-unrelated, low-level activation of Caspase-3 leads to proteolytic processing of p65. The group further characterized the fate of the cleaved C-terminal product of p65, containing amino acids 97–551 and termed ΔNH2p65. They demonstrated that ΔNH2p65 bound to IκBα and caused constitutive activation of p65 by protecting the full-length p65 from IκBα inhibition, ultimately leading to an increase in HIV viral replication (Coiras et al., 2008). While the HIV-encoded proteins themselves are not causing the cleavage of p65, the virus’s preference for a cell type that innately cleaves a low level of p65 upon activation, is indicative of the advantage HIV gains by replicating in such a host cell. The careful balance between activating Caspase-3 at low levels to cleave some of the p65 molecules within T cells and inducing extensive apoptosis should be highlighted here as too much Caspase-3 activity could certainly backfire on the virus. Anti-apoptotic genes would still be transactivated in such an infected cell, as most of the p65 molecules in the cell are left intact and constitutive NF-κB activation would induce their expression, which therefore results in the maintenance of an infected cell with enough host signaling left intact to produce more viral copies and propagate the infection.
NF-κB/p65 cleavage: global versus selective inhibition?
As supported by the previous cited studies, it is apparent that pathogen-encoded protease mediated cleavage of a variety of signaling molecules in host cells, especially the NF-κB signaling pathway (as summarized in Table 2), executes a dramatic and robust impact on the host immune responses. In most cases, it is proposed that the cleavage of a single key molecule in the NF-κB signaling pathway, along with other encoded virulence factors, is sufficient to block the signal transduction necessary to activate NF-κB target genes, especially proinflammatory cytokine/chemokine genes, thus modulating the global host immune responses to the pathogen’s advantage and allowing the persistence of the infection (Figure 1). This explanation, at first glance, appears to be a reasonable incorporation of observations made during pathogen infection; however, it falls short in clarifying the dramatic impact on host inflammatory cytokine/chemokine gene expression when p65 is cleaved during the bona fide pathogen infections. A great number of studies have established that NF-κB is acutely and robustly activated during pathogen infections, as illustrated by a rapid nuclear translocation of the majority of p65, as well as the other Rel proteins, in host cells (Hayden & Ghosh, 2012, Li & Verma, 2002, Pott & Hornef, 2012, Sun & Andersson, 2002, Thaker, 2014). This is further emphasized by the need to encode a series of virulence factors that target multiple levels of the same signaling pathway as has been demonstrated by the A/E pathogens (Wong et al., 2011). If each p65 molecule in the host cell makes an equal contribution to the transactivation of NF-κB-targeted genes, pathogen-encoded proteases would have to cleave the majority, or even the entirety of the p65 pool in the cell to reach a substantial suppression of cytokine/chemokine gene transcription. However, most of the reported protease-mediated cleavage events in infected host cells have been characterized qualitatively, focusing on the identification of target host protein(s), but lacking further definition quantitatively. Even under conditions such as ectopic expression in host cells and in vitro protease assays with recombinant proteins, only a percentage of the entire p65 molecule pool in host cells is cleaved by the pathogen-encoded protease in question, leaving a substantial amount of intact full-length p65 (Baruch et al., 2011, Coiras et al., 2008, Hodgson et al., 2015, Li et al., 2014, Muhlen et al., 2011, Silva et al., 2013, Turco & Sousa, 2014, Yen et al., 2010). Unsurprisingly, the amount of p65 cleavage is even lower under the pathophysiological conditions of pathogen infection in vivo where the expression level of the protease is likely lower (Deng et al., 2010). Hence, it remains puzzling how incomplete cleavage of p65 by many pathogen-encoded proteases has a disproportionally dramatic impact on NF-κB-mediated gene transcription and the global inflammatory response. Even in scenarios where pathogens encode multiple effectors to target one pathway, knockout of just the protease, as in the case of NleC, is capable of dramatically changing the NF-κB activation status. Therefore, redundancy in effectors is unable to fully explain the effects observed when incomplete cleavage occurs.
Table 2.
Pathogen- and host-encoded proteases targeting p65 during infections.
| Pathogen | Protease | Protease category | p65 cleavage site(s) |
Cleavage verification |
Fate and function of p65 fragment |
Proposed mechanism for NF-κB inhibition |
Reference |
|---|---|---|---|---|---|---|---|
| EPEC, EHEC | NleC | Zinc Metalloprotease | P10/A11 | recombinant in vitro |
Undetermined | Global | Yen et al. 2011 |
| EPEC | NleC | Zinc Metalloprotease | Not characterized | in vitro | Undetermined | Global | Muhlen et al. 2011 |
| EPEC | NleC | Zinc Metalloprotease | Not characterized | in vitro | Undetermined | Global | Pearson et al. 2011 |
| EPEC | NleC | Zinc Metalloprotease | C38/E39 | recombinant in vitro |
Undetermined | Global | Baruch et al. 2011 |
| EPEC, C. rodentium |
NleC | Zinc Metalloprotease | Not characterized |
in vitro, in vivo |
Undetermined | Global | Sham et al. 2011 |
| EHEC | NleC | Zinc Metalloprotease | C38/E39 | recombinant | Undetermined | Undetermined | Li et al. 2011; Turco & Sousa 2014 |
| EPEC, C. rodentium |
NleC | Zinc Metalloprotease | P10/A11, C38/E39 |
in vitro, in vivo |
N-terminal fragment binds RPS3 |
Amplification | Hodgson et al. 2015 |
| Phdp | AIP56 | Zinc Metalloprotease | C39/E40 (fish p65) | recombinant in vitro |
Undetermined | Global | Silva et al. 2015 |
| C. trachomatis | CT441 | TSP Protease | F351/T352 | in vitro | p40 product binds to IκBα |
Amplification | Lad et al. 2007a; Lad et al. 2007b |
| Leishmania | Gp63 | Metalloprotease | C-terminus | in vitro | p35 fragment binds DNA with p50 |
Amplification | Gregory et al. 2008a |
| Poliovirus | 3C | Cysteine Protease | Q480/G481 | in vitro | Undetermined | Global | Neznanov et al. 2005 |
| HIV-1 | Caspase-3 (host) |
Cysteine Protease | D97/G98 | recombinant in vitro |
binds p50/IκBα | Amplification | Coiras et al. 2008 |
EPEC, Enteropathogenic E. coli; EHEC, Enterohemorrhagic E. coli; Phdp, Photobacterium damselae piscicida; C. trachomatis, Chlamydia trachomatis; HIV, human immunodeficiency virus type 1; recombinant, purified protein assays; in vitro, cell culture/transfection assays; in vivo, animal studies. Global, general blockade of NF-κB signaling either by cleavage alone or in combination with other virulence factors; Amplification, cleaved fragment functions to further the effect of cleaving p65.
Figure 1.
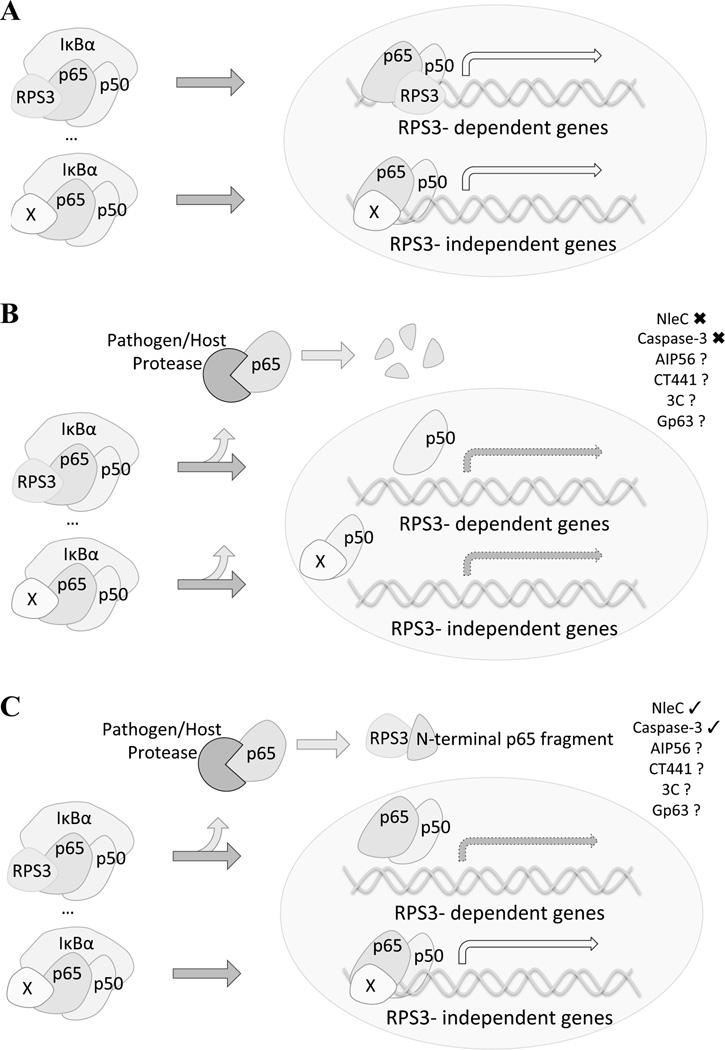
Comparison of global and selective inhibition interpretation models of pathogen-encoded protease mediated NF-κB interference. Under infection conditions, NF-κB activation results in p65 nuclear translocation and the transactivation of a wide array of target genes, a subset of which require the aid of known specifier proteins, such as RPS3 and Sam68. This leads to a coordinated cellular responses and clearance of infection (A). Pathogen-encoded proteases can have devastating, widespread effects (global, B) or they can inhibit specific aspects of a pathway. Complete cleavage of p65 interrupts the signal transduction cascade, preventing all downstream outcomes in a global fashion. However, partial proteolytic processing of p65 can result in selective inhibition (C), as there are still some full length and functioning p65 molecules within the cell. In the experimentally verified (checked) examples of NleC-mediated cleavage of p65, during A/E pathogen infections, an N-terminal fragment is produced that sequesters RPS3, preventing the activation of that arm of the NF-κB signaling pathway. This inhibition thus results in the amplification of the effect of cleaving only a percentage of the p65 cellular pool. It is likely, yet remains to be verified experimentally (question marked), that a similar mechanism is employed by other pathogens. EPEC/EHEC/C. rodentium- NleC; Phdp- AIP56; C. trachomatis- CT441; Poliovirus- 3C; Leishmania spp.- Gp63; Host- Caspase-3.
Of note, recent progress in elucidating the mechanisms that control the promoter selectivity and transcriptional specificity of NF-κB provides new insights into this apparent inconsistency between the protease-mediated p65 cleavage and the impact on NF-κB-conferred inflammatory gene expression and inflammatory response in host cells. This is best exemplified by our recent study on how the afore-mentioned protease effector NleC interferes with the NF-κB ‘specifier’ component RPS3 and the selective activation of RPS3-dependent, NF-κB target gene transcription during EPEC and C. rodentium infections by cleaving p65 (Hodgson et al., 2015). NleC was recently revealed as an important effector functioning to dampen the secretion of inflammatory cytokines in A/E pathogen-infected cells by cleaving p65 (Baruch et al., 2011, Li et al., 2014, Muhlen et al., 2011, Pearson et al., 2011, Shames & Finlay, 2011, Turco & Sousa, 2014, Yen et al., 2010). Injected via the T3SS into host cells at low abundance, relative to other effector molecules (Deng et al., 2010), NleC cleaves only a small percentage of the p65 molecules in infected host cells, even when ectopically expressed (Baruch et al., 2011, Muhlen et al., 2011, Yen et al., 2010). For many pathogen-encoded proteases it remains uncommon to further characterize the fate and possible function of the cleaved fragments, however, our group uncovered that the newly produced fragments had a further functional role within the cell. The N-terminal fragment of p65, termed as p651–38 (Hodgson et al., 2015), generated by NleC cleavage, executes a unique function by interfering with the cytoplasmic RPS3/p65 interaction and inhibiting the nuclear translocation of RPS3, but not the remaining uncleaved p65. It is noteworthy that RPS3 occurs along with p65 and IκBα in a portion, but not all, of inhibitory NF-κB complexes in the cytoplasm (unpublished observation) and that the nuclear translocation of RPS3 is crucial for the transactivation of the RPS3/p65-dependent NF-κB target genes (Gao & Hardwidge, 2011, Gao et al., 2009, Sen et al., 2012, Wan et al., 2007, Wan et al., 2011, Wier et al., 2012). By interfering with and preventing RPS3 from nuclear translocation with the cleaved p651–38 fragment, the NleC protease is not required to cleave all or even most of the p65 molecules present within the cell, since disrupting the limited NF-κB complexes containing both RPS3 and p65 is sufficient to alter the RPS3-dependent NF-κB gene transcription (Hodgson et al., 2015, Sen et al., 2012, Wan et al., 2007, Wier et al., 2012). It should be noted that aside from the NleC cleaved p651–38, other effectors introduced into the host cell during infections will limit the amount of active p65 and NleH will also specifically target p65/RPS3, creating redundancy. The RPS3-conferred NF-κB promoter selectivity provides a novel avenue for pathogen interference, effectively “amplifying” the effect of cleaving a small percentage of p65 in A/E pathogen-infected host cells in order to sequester RPS3 in the cytoplasm (Figure 1). In support of the interrupted RPS3/p65 model, our previous proof-of-concept study demonstrated that an ectopically expressed N-terminal truncation of p65 is capable of blocking the RPS3/p65 interaction and altering NF-κB activity (Wier et al., 2012). Moreover, this work demonstrated that a host Caspase-3 cleaved p651–97 fragment also functions to block the RPS3/p65 interaction. The power of amplifying the effects of NleC lies in the documented importance of the RPS3/p65-mediated NF-κB signaling for host defense against A/E pathogen infections (Gao et al., 2009, Hodgson et al., 2015, Holmes et al., 2012, Perkins, 2012, Pham et al., 2013, Pham et al., 2012, Wan et al., 2007, Wan et al., 2011). In comparison to the global model, this selective inhibition strategy proposes a more targeted strategy for a pathogen to exert a robust impact on host signaling pathways and inflammatory response with one virulence factor, and the NleC-mediated p65 cleavage serves as a biologically relevant example of this amplification mechanism, though it is likely not the only scenario where this amplification strategy occurs.
Although not extensively examined in the context of other protease-mediated p65 cleavage, the RPS3-dependent selective inhibition mechanism could also be implicated during other pathogenic infections, particularly when proteases create N-terminal fragments that maintain the p65/RPS3 binding interaction. The sulfahydration modification on the C38 of p65 mediates and stabilizes the interaction with RPS3 (Perkins, 2012, Sen et al., 2012); in this way, p65 fragments could maintain their affinity for RPS3 as long as that modification is preserved. The homology between AIP56 and NleC and the similar cleavage site at the N-terminus of p65 indicate that Phdp could employ a similar strategy to suppress host inflammatory response (Silva et al., 2013). Moreover, AIP56 also inhibited the gene expression of certain anti-apoptotic genes, which have been revealed to rely on RPS3/p65 for their transcriptional activation, such as Birc3 (encoding cellular inhibitor of apoptosis protein-2, cIAP2) and Bcl2l1 (encoding B-cell lymphoma-extra large, Bcl-XL) (Sen et al., 2012). Hence, it would be logical to hypothesize that the p65 cleavage product produced by AIP56 is also able to maintain its interaction with RPS3, sequestering RPS3 away from full-length p65 in immune cells, as demonstrated in C. rodentium-infected colon epithelial cells (Hodgson et al., 2015). As demonstrated in the in vitro experiments (Hodgson et al., 2015), the interaction between the AIP56-cleaved p65 fragment and RPS3 would prevent the nuclear accumulation of RPS3. Such an interaction in Phdp-infected neutrophils and macrophages would still need to be confirmed with further experiments, as there may be cell type specific effects and the importance of RPS3/p65 signaling has yet to be confirmed in these cells. Moreover, the p65ΔC fragment generated by 3C protease cleavage during poliovirus, ECHO-1, and rhino-14 infections (Neznanov et al., 2005) and the p65 N-terminal fragment generated by the activated host Caspase-3 during HIV infection (Coiras et al., 2008), the Chlamydia CT441 protease (Lad et al., 2007a), and the Leishmania GP63 protease (Dasgupta et al., 2003, Gregory et al., 2008a) could be capable of blocking the RPS3-p65 interaction, although the fate or function of these N-terminal fragments of p65 have not been well defined yet with regards to RPS3. In particular, we demonstrated previously that truncated p65 fragments as large as 21–186 (Wier et al., 2012) and 1–311 (Wan et al., 2007) are capable of maintaining their binding to RPS3. Though the most profound RPS3 interference was observed with the p6521–186 fragment, the ability of additional larger N-terminal p65 fragments to interfere with the RPS3/p65 interaction and function remains to be elucidated (Wier et al., 2012). Moreover, Johannes et al. speculated at the possibility of p65 cleavage occurring to block the anti-apoptotic response during picornavirus infection, noting that some NF-κB target genes, e.g. IL-6 and IL-8 (Johannes et al., 1999), appeared unaffected while others, including IκBα, were never reset to the steady state levels (Neznanov et al., 2005). Overall this suggests that p65 cleavage modulates the transcriptional specificity of NF-κB to some degree during these infections. While the entire repertoire of RPS3/p65 dependent genes in distinct cell type(s) has not yet been defined, exploiting systems that show divergent activation and expression of NF-κB target genes will further define this cohort of genes or identify other specifier proteins. The evidence that not only NleC but also other pathogen-encoded proteases generate N-terminal p65 fragments with potential interactions with RPS3 indicates that p65 fragment interference could be a widespread virulence strategy for pathogens in order to abrogate RPS3-dependent NF-κB activation in host cells. It is therefore worthy to further examine whether the cleaved p65 fragments manipulate RPS3, or other specifier proteins, for inhibition or activation of a subset of NF-κB target genes in future studies, in particular when the pathogen-encoded proteases are unable to cleave the majority of the cellular pool of p65. Moreover, when cleavage of a target protein is recognized within host cells, deeper consideration of how the pathogen amplifies its efforts may uncover previously underappreciated protein interactions and gene regulation strategies.
CONCLUSION
Emerging evidence highlights pathogen-encoded proteases as critical virulence factors that can have dramatic effects on the course of the infection and the ability of the host to detect and respond to infection. In this review, we have discussed how targeting the NF-κB signaling pathway, specifically protease-mediated cleavage of p65, has allowed various pathogens to achieve control over their microenvironment. Future studies to characterize proteases would benefit from overcoming some major obstacles, such as 1) confirming their interactions with target proteins in vivo as opposed to exclusively relying on ectopic or recombinant expression systems; 2) the development of a unified system to measure protease activity would allow for greater control, consistency, and comparison between different research groups; and 3) the development of more sensitive detection methods to study the spatial-temporal activity, localization, and induction of pathogen-encoded proteases (and other virulence factors) during infections. It is likely that recombinant and overexpression systems may lead to the over-identification of target substrates as they rely on purified proteins mixed at concentrations that may not be biologically relevant. Moreover, interactions detected in vitro may not be relevant in the context of in vivo infection as these proteins may not function in proximity of each other for the cleavage to be pathophysiologically relevant. The zinc metalloprotease NleC is an example of such a situation, where multiple groups, including our own, have reported differences in the target substrates. While most of the groups employed an NleC ectopic expression system (Baruch et al., 2011, Muhlen et al., 2011, Yen et al., 2010) or purified NleC recombinant proteins (Li et al., 2014, Turco & Sousa, 2014), our group was unable to detect cleavage of targets outside of p65 when examining endogenous protein levels (Hodgson et al., 2015). A similar discussion has occurred in reference to the Chlamydial proteases CT441 (Lad et al., 2007a, Lad et al., 2007b) and CPAF (Christian et al., 2010, Zhong, 2011), further demonstrating the need to resolve the difference in proposed target substrates. To further help clarify the results between research groups, a standard unit of protease activity would work in much the same way to unify the results from multiple studies. Variations in the methods and buffers employed for protein purification may account for the discrepancy in substrate specificity and cleavage efficiency reported by different groups. The field would arguably benefit the most by the improvement of the tools used to study the spatial-temporal regulation of virulence factors and their functions in vivo. Novel methods for more sensitive detection of low abundant proteins and their interacting partners within host cells would allow for a deeper understanding of how pathogens manipulate host cell signaling pathways.
Determining the fate of cleaved fragments generated by pathogen-encoded proteases or host proteases to elucidate any further activity would also significantly benefit the field, as exampled by the NleC and Chlamydia CT441 studies (Hodgson et al., 2015, Lad et al., 2007a, Lad et al., 2007b). The targeting and cleavage of host proteins is an advantage that pathogens have acquired, enhancing their odds of survival. We have highlighted events in which this advantage has resulted in the regulation of a cellular pathway in a very precise and particular manner, which would have gone unnoticed without the full characterization of the cleaved fragments role in the host cell. RPS3 selective targeting serves as a mechanism by which aspects of a signaling hub, where multiple pathways converge, are targeted without disrupting the entirety of hub activity (Brodsky & Medzhitov, 2009). By targeting the RPS3 branch, pathogens are specifically blocking inflammatory and anti-apoptotic signaling within host cells. Other NF-κB specifiers have already been identified, such as Sam68 (Fu et al., 2013) and KLF6 (Zhang et al., 2014), and their functions could also serve as targets for selective inhibition by pathogens in a cell type specific manner. Our model demonstrates that as the promoter selectivity and specificity of NF-κB regulation is further elucidated, there are opportunities to uncover further virulence strategies by pathogens that have been until now underappreciated. Advancements in the tools available and a deeper understanding of the cell types and tissues where specifier proteins are playing a role in regulating NF-κB target genes would also add significantly to the study of host-pathogen interactions. The development of conventional and/or conditional knockout mice lacking these specifier proteins would provide further insight as to the importance of infection site (target cell type), pathogen-mediated blockade of a signaling pathway, and the effect this has on the subsequent host responses to infections. As discussed briefly above, the hypothesis that anti-apoptotic responses dominate the tissue response, whereas survival and inflammatory responses dominate the infiltrating immune cell response can be teased apart through the creation and use of lineage specific knockout mice (i.e. conditional RPS3 ablation driven by the Villin-Cre transgene to knockout RPS3 in intestinal epithelial cells). Such innovations would clarify the crosstalk and redundancy among virulence factors, opening the field for the development of more targeted and specific therapeutics to combat pathogens.
Acknowledgments
We apologize to those who also made important contributions to the issues discussed and could not be cited due to space limitations. We thank the members of the Wan lab for critically reading the manuscript. A.H. is a Hopkins Sommer Scholar. The research in the authors’ laboratory is supported by Research Scholar Grant RSG-13-052-01-MPC from the American Cancer Society and R01GM111682 grant from the National Institutes of Health.
REFERENCES
- Agol VI, Belov GA, Bienz K, Egger D, Kolesnikova MS, Romanova LI, Sladkova LV, Tolskaya EA. Competing death programs in poliovirus-infected cells: commitment switch in the middle of the infectious cycle. Journal of virology. 2000;74:5534–5541. doi: 10.1128/jvi.74.12.5534-5541.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcami J, Lain de Lera T, Folgueira L, Pedraza MA, Jacque JM, Bachelerie F, Noriega AR, Hay RT, Harrich D, Gaynor RB, et al. Absolute dependence on kappa B responsive elements for initiation and Tat-mediated amplification of HIV transcription in blood CD4 T lymphocytes. The EMBO journal. 1995;14:1552–1560. doi: 10.1002/j.1460-2075.1995.tb07141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachu M, Yalla S, Asokan M, Verma A, Neogi U, Sharma S, Murali RV, Mukthey AB, Bhatt R, Chatterjee S, Rajan RE, Cheedarla N, Yadavalli VS, Mahadevan A, Shankar SK, Rajagopalan N, Shet A, Saravanan S, Balakrishnan P, Solomon S, Vajpayee M, Satish KS, Kundu TK, Jeang KT, Ranga U. Multiple NF-kappaB sites in HIV-1 subtype C long terminal repeat confer superior magnitude of transcription and thereby the enhanced viral predominance. The Journal of biological chemistry. 2012;287:44714–44735. doi: 10.1074/jbc.M112.397158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes AC, dos Santos NM, Ellis AE. Update on bacterial vaccines: Photobacterium damselae subsp. piscicida. Developments in biologicals. 2005;121:75–84. [PubMed] [Google Scholar]
- Baruch K, Gur-Arie L, Nadler C, Koby S, Yerushalmi G, Ben-Neriah Y, Yogev O, Shaulian E, Guttman C, Zarivach R, Rosenshine I. Metalloprotease type III effectors that specifically cleave JNK and NF-kappaB. The EMBO journal. 2011;30:221–231. doi: 10.1038/emboj.2010.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky IE, Medzhitov R. Targeting of immune signalling networks by bacterial pathogens. Nature cell biology. 2009;11:521–526. doi: 10.1038/ncb0509-521. [DOI] [PubMed] [Google Scholar]
- Brunham RC, Rey-Ladino J. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nature reviews. Immunology. 2005;5:149–161. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- Casadevall A, Pirofski LA. Virulence factors and their mechanisms of action: the view from a damage-response framework. Journal of water and health. 2009;7(Suppl 1):S2–S18. doi: 10.2166/wh.2009.036. [DOI] [PubMed] [Google Scholar]
- Christian J, Vier J, Paschen SA, Hacker G. Cleavage of the NF-kappaB family protein p65/RelA by the chlamydial protease-like activity factor (CPAF) impairs proinflammatory signaling in cells infected with Chlamydiae. The Journal of biological chemistry. 2010;285:41320–41327. doi: 10.1074/jbc.M110.152280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coiras M, Lopez-Huertas MR, Mateos E, Alcami J. Caspase-3-mediated cleavage of p65/RelA results in a carboxy-terminal fragment that inhibits IkappaBalpha and enhances HIV-1 replication in human T lymphocytes. Retrovirology. 2008;5:109. doi: 10.1186/1742-4690-5-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta B, Roychoudhury K, Ganguly S, Akbar MA, Das P, Roy S. Infection of human mononuclear phagocytes and macrophage-like THP1 cells with Leishmania donovani results in modulation of expression of a subset of chemokines and a chemokine receptor. Scandinavian journal of immunology. 2003;57:366–374. doi: 10.1046/j.1365-3083.2003.01227.x. [DOI] [PubMed] [Google Scholar]
- de Vries HJ, Reedijk SH, Schallig HD. Cutaneous leishmaniasis: recent developments in diagnosis and management. American journal of clinical dermatology. 2015;16:99–109. doi: 10.1007/s40257-015-0114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, de Hoog CL, Yu HB, Li Y, Croxen MA, Thomas NA, Puente JL, Foster LJ, Finlay BB. A comprehensive proteomic analysis of the type III secretome of Citrobacter rodentium. The Journal of biological chemistry. 2010;285:6790–6800. doi: 10.1074/jbc.M109.086603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Vale A, Costa-Ramos C, Silva A, Silva DS, Gartner F, dos Santos NM, Silva MT. Systemic macrophage and neutrophil destruction by secondary necrosis induced by a bacterial exotoxin in a Gram-negative septicaemia. Cellular microbiology. 2007;9:988–1003. doi: 10.1111/j.1462-5822.2006.00846.x. [DOI] [PubMed] [Google Scholar]
- do Vale A, Marques F, Silva MT. Apoptosis of sea bass (Dicentrarchus labrax L.) neutrophils and macrophages induced by experimental infection with Photobacterium damselae subsp. piscicida. Fish & shellfish immunology. 2003;15:129–144. doi: 10.1016/s1050-4648(02)00144-4. [DOI] [PubMed] [Google Scholar]
- Eckmann L, Neish AS. NF-kappaB and mucosal homeostasis. Curr Top Microbiol Immunol. 2011;349:145–158. doi: 10.1007/82_2010_103. [DOI] [PubMed] [Google Scholar]
- Fan J, Zeller K, Chen YC, Watkins T, Barnes KC, Becker KG, Dang CV, Cheadle C. Time-dependent c-Myc transactomes mapped by Array-based nuclear run-on reveal transcriptional modules in human B cells. PloS one. 2010;5:e9691. doi: 10.1371/journal.pone.0009691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BB, McFadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124:767–782. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Fu K, Sun X, Zheng W, Wier EM, Hodgson A, Tran DQ, Richard S, Wan F. Sam68 modulates the promoter specificity of NF-kappaB and mediates expression of CD25 in activated T cells. Nature communications. 2013;4:1909. doi: 10.1038/ncomms2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Niu J, Schmidt C, Sclabas GM, Peng B, Uwagawa T, Li Z, Evans DB, Abbruzzese JL, Chiao PJ. NF-kappaB and AP-1 connection: mechanism of NF-kappaB-dependent regulation of AP-1 activity. Mol Cell Biol. 2004;24:7806–7819. doi: 10.1128/MCB.24.17.7806-7819.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Hardwidge PR. Ribosomal protein s3: a multifunctional target of attaching/effacing bacterial pathogens. Frontiers in microbiology. 2011;2:137. doi: 10.3389/fmicb.2011.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wan F, Mateo K, Callegari E, Wang D, Deng W, Puente J, Li F, Chaussee MS, Finlay BB, Lenardo MJ, Hardwidge PR. Bacterial effector binding to ribosomal protein s3 subverts NF-kappaB function. PLoS pathogens. 2009;5:e1000708. doi: 10.1371/journal.ppat.1000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory DJ, Godbout M, Contreras I, Forget G, Olivier M. A novel form of NF-kappaB is induced by Leishmania infection: involvement in macrophage gene expression. European journal of immunology. 2008a;38:1071–1081. doi: 10.1002/eji.200737586. [DOI] [PubMed] [Google Scholar]
- Gregory DJ, Sladek R, Olivier M, Matlashewski G. Comparison of the effects of Leishmania major or Leishmania donovani infection on macrophage gene expression. Infection and immunity. 2008b;76:1186–1192. doi: 10.1128/IAI.01320-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartland EL, Leong JM. Enteropathogenic and enterohemorrhagic E. coli: ecology, pathogenesis, and evolution. Frontiers in cellular and infection microbiology. 2013;3:15. doi: 10.3389/fcimb.2013.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes & development. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson A, Wier EM, Fu K, Sun X, Yu H, Zheng W, Sham HP, Johnson K, Bailey S, Vallance BA, Wan F. Metalloprotease NleC suppresses host NF-kappaB/inflammatory responses by cleaving p65 and interfering with the p65/RPS3 interaction. PLoS pathogens. 2015;11:e1004705. doi: 10.1371/journal.ppat.1004705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A, Lindestam Arlehamn CS, Wang D, Mitchell TJ, Evans TJ, Roe AJ. Expression and regulation of the Escherichia coli O157:H7 effector proteins NleH1 and NleH2. PloS one. 2012;7:e33408. doi: 10.1371/journal.pone.0033408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Zhang Q, Guo XK, Yu ZB, Xu AT, Tang J, Feng WH. Porcine reproductive and respiratory syndrome virus nonstructural protein 4 antagonizes beta interferon expression by targeting the NF-kappaB essential modulator. Journal of virology. 2014;88:10934–10945. doi: 10.1128/JVI.01396-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain K, Jain NK. Vaccines for visceral leishmaniasis: A review. Journal of immunological methods. 2015;422:1–12. doi: 10.1016/j.jim.2015.03.017. [DOI] [PubMed] [Google Scholar]
- Johannes G, Carter MS, Eisen MB, Brown PO, Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature immunology. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Krachler AM, Woolery AR, Orth K. Manipulation of kinase signaling by bacterial pathogens. The Journal of cell biology. 2011;195:1083–1092. doi: 10.1083/jcb.201107132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad SP, Li J, da Silva Correia J, Pan Q, Gadwal S, Ulevitch RJ, Li E. Cleavage of p65/RelA of the NF-kappaB pathway by Chlamydia. Proceedings of the National Academy of Sciences of the United States of America. 2007a;104:2933–2938. doi: 10.1073/pnas.0608393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad SP, Yang G, Scott DA, Wang G, Nair P, Mathison J, Reddy VS, Li E. Chlamydial CT441 is a PDZ domain-containing tail-specific protease that interferes with the NF-kappaB pathway of immune response. Journal of bacteriology. 2007b;189:6619–6625. doi: 10.1128/JB.00429-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Negrate G. Subversion of innate immune responses by bacterial hindrance of NF-kappaB pathway. Cellular microbiology. 2012;14:155–167. doi: 10.1111/j.1462-5822.2011.01719.x. [DOI] [PubMed] [Google Scholar]
- Lenardo MJ, Baltimore D. NF-kappa B: a pleiotropic mediator of inducible and tissue-specific gene control. Cell. 1989;58:227–229. doi: 10.1016/0092-8674(89)90833-7. [DOI] [PubMed] [Google Scholar]
- Levkau B, Scatena M, Giachelli CM, Ross R, Raines EW. Apoptosis overrides survival signals through a caspase-mediated dominant-negative NF-kappa B loop. Nature cell biology. 1999;1:227–233. doi: 10.1038/12050. [DOI] [PubMed] [Google Scholar]
- Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M, Jr, Lemon SM. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proceedings of the National Academy of Sciences of the United States of America. 2005a;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Verma IM. NF-kappaB regulation in the immune system. Nature reviews. Immunology. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- Li W, Liu Y, Sheng X, Yin P, Hu F, Liu Y, Chen C, Li Q, Yan C, Wang J. Structure and mechanism of a type III secretion protease, NleC. Acta crystallographica. Section D, Biological crystallography. 2014;70:40–47. doi: 10.1107/S1399004713024619. [DOI] [PubMed] [Google Scholar]
- Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proceedings of the National Academy of Sciences of the United States of America. 2005b;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Morrison RP, Caldwell HD. Immunity to murine chlamydial genital infection. Infection and immunity. 2002;70:2741–2751. doi: 10.1128/IAI.70.6.2741-2751.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlen S, Ruchaud-Sparagano MH, Kenny B. Proteasome-independent degradation of canonical NFkappaB complex components by the NleC protein of pathogenic Escherichia coli. The Journal of biological chemistry. 2011;286:5100–5107. doi: 10.1074/jbc.M110.172254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndubaku C, Tsui V. Inhibiting the deubiquitinating enzymes (DUBs) Journal of medicinal chemistry. 2015;58:1581–1595. doi: 10.1021/jm501061a. [DOI] [PubMed] [Google Scholar]
- Neff L, Zeisel M, Sibilia J, Scholler-Guinard M, Klein JP, Wachsmann D. NF-kappaB and the MAP kinases/AP-1 pathways are both involved in interleukin-6 and interleukin-8 expression in fibroblast-like synoviocytes stimulated by protein I/II, a modulin from oral streptococci. Cellular microbiology. 2001;3:703–712. doi: 10.1046/j.1462-5822.2001.00148.x. [DOI] [PubMed] [Google Scholar]
- Neznanov N, Chumakov KM, Neznanova L, Almasan A, Banerjee AK, Gudkov AV. Proteolytic cleavage of the p65-RelA subunit of NF-kappaB during poliovirus infection. The Journal of biological chemistry. 2005;280:24153–24158. doi: 10.1074/jbc.M502303200. [DOI] [PubMed] [Google Scholar]
- Novikova L, Czymmeck N, Deuretzbacher A, Buck F, Richter K, Weber AN, Aepfelbacher M, Ruckdeschel K. Cell death triggered by Yersinia enterocolitica identifies processing of the proinflammatory signal adapter MyD88 as a general event in the execution of apoptosis. Journal of immunology. 2014;192:1209–1219. doi: 10.4049/jimmunol.1203464. [DOI] [PubMed] [Google Scholar]
- Olivier M, Gregory DJ, Forget G. Subversion mechanisms by which Leishmania parasites can escape the host immune response: a signaling point of view. Clinical microbiology reviews. 2005;18:293–305. doi: 10.1128/CMR.18.2.293-305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmenberg AC. Proteolytic processing of picornaviral polyprotein. Annual review of microbiology. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- Pearson JS, Riedmaier P, Marches O, Frankel G, Hartland EL. A type III effector protease NleC from enteropathogenic Escherichia coli targets NF-kappaB for degradation. Molecular microbiology. 2011;80:219–230. doi: 10.1111/j.1365-2958.2011.07568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins ND. Cysteine 38 holds the key to NF-kappaB activation. Molecular cell. 2012;45:1–3. doi: 10.1016/j.molcel.2011.12.023. [DOI] [PubMed] [Google Scholar]
- Perkins ND, Edwards NL, Duckett CS, Agranoff AB, Schmid RM, Nabel GJ. A cooperative interaction between NF-kappa B and Sp1 is required for HIV-1 enhancer activation. The EMBO journal. 1993;12:3551–3558. doi: 10.1002/j.1460-2075.1993.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham TH, Gao X, Singh G, Hardwidge PR. Escherichia coli virulence protein NleH1 interaction with the v-Crk sarcoma virus CT10 oncogene-like protein (CRKL) governs NleH1 inhibition of the ribosomal protein S3 (RPS3)/nuclear factor kappaB (NF-kappaB) pathway. The Journal of biological chemistry. 2013;288:34567–34574. doi: 10.1074/jbc.M113.512376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham TH, Gao X, Tsai K, Olsen R, Wan F, Hardwidge PR. Functional differences and interactions between the Escherichia coli type III secretion system effectors NleH1 and NleH2. Infection and immunity. 2012;80:2133–2140. doi: 10.1128/IAI.06358-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras V, Selvarajoo K. Beyond MyD88 and TRIF Pathways in Toll-Like Receptor Signaling. Frontiers in immunology. 2014;5:70. doi: 10.3389/fimmu.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott J, Hornef M. Innate immune signalling at the intestinal epithelium in homeostasis and disease. EMBO reports. 2012;13:684–698. doi: 10.1038/embor.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romalde LJ. Photobacterium damselae subsp. piscicida: an integrated view of a bacterial fish pathogen. International microbiology : the official journal of the Spanish Society for Microbiology. 2002;5:3–9. doi: 10.1007/s10123-002-0051-6. [DOI] [PubMed] [Google Scholar]
- Saura M, Lizarbe TR, Rama-Pacheco C, Lowenstein CJ, Zaragoza C. Inhibitor of NF kappa B alpha is a host sensor of coxsackievirus infection. Cell cycle. 2007;6:503–506. doi: 10.4161/cc.6.5.3918. [DOI] [PubMed] [Google Scholar]
- Sen N, Paul BD, Gadalla MM, Mustafa AK, Sen T, Xu R, Kim S, Snyder SH. Hydrogen sulfide-linked sulfhydration of NF-kappaB mediates its antiapoptotic actions. Molecular cell. 2012;45:13–24. doi: 10.1016/j.molcel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham HP, Shames SR, Croxen MA, Ma C, Chan JM, Khan MA, Wickham ME, Deng W, Finlay BB, Vallance BA. Attaching and effacing bacterial effector NleC suppresses epithelial inflammatory responses by inhibiting NF-kappaB and p38 mitogen-activated protein kinase activation. Infection and immunity. 2011;79:3552–3562. doi: 10.1128/IAI.05033-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shames SR, Bhavsar AP, Croxen MA, Law RJ, Mak SH, Deng W, Li Y, Bidshari R, de Hoog CL, Foster LJ, Finlay BB. The pathogenic Escherichia coli type III secreted protease NleC degrades the host acetyltransferase p300. Cellular microbiology. 2011;13:1542–1557. doi: 10.1111/j.1462-5822.2011.01640.x. [DOI] [PubMed] [Google Scholar]
- Shames SR, Finlay BB. Proteolytic Cleavage of NF-kappaB p65: A Novel Mechanism for Subversion of Innate Immune Signaling by Pathogenic E. Coli. Frontiers in microbiology. 2011;2:38. doi: 10.3389/fmicb.2011.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shames SR, Finlay BB. Bacterial effector interplay: a new way to view effector function. Trends in microbiology. 2012;20:214–219. doi: 10.1016/j.tim.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Silke J, Hartland EL. Masters, marionettes and modulators: intersection of pathogen virulence factors and mammalian death receptor signaling. Current opinion in immunology. 2013;25:436–440. doi: 10.1016/j.coi.2013.05.011. [DOI] [PubMed] [Google Scholar]
- Silva DS, Pereira LM, Moreira AR, Ferreira-da-Silva F, Brito RM, Faria TQ, Zornetta I, Montecucco C, Oliveira P, Azevedo JE, Pereira PJ, Macedo-Ribeiro S, do Vale A, dos Santos NM. The apoptogenic toxin AIP56 is a metalloprotease A-B toxin that cleaves NF-kappab P65. PLoS pathogens. 2013;9:e1003128. doi: 10.1371/journal.ppat.1003128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spehlmann ME, Eckmann L. Nuclear factor-kappa B in intestinal protection and destruction. Curr Opin Gastroenterol. 2009;25:92–99. doi: 10.1097/MOG.0b013e328324f857. [DOI] [PubMed] [Google Scholar]
- Sun Z, Andersson R. NF-kappaB activation and inhibition: a review. Shock. 2002;18:99–106. doi: 10.1097/00024382-200208000-00001. [DOI] [PubMed] [Google Scholar]
- Thaker YRR, Christopher E. Distinct NF-kB activation pathways engaged by T-cell receptor and co-receptor CD28 on T-cells. Inflammation and Cell Signaling. 2014:1. [Google Scholar]
- Turco MM, Sousa MC. The structure and specificity of the type III secretion system effector NleC suggest a DNA mimicry mechanism of substrate recognition. Biochemistry. 2014;53:5131–5139. doi: 10.1021/bi500593e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annual review of immunology. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- Wan F, Anderson DE, Barnitz RA, Snow A, Bidere N, Zheng L, Hegde V, Lam LT, Staudt LM, Levens D, Deutsch WA, Lenardo MJ. Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell. 2007;131:927–939. doi: 10.1016/j.cell.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Wan F, Lenardo MJ. The nuclear signaling of NF-kappaB: current knowledge, new insights, and future perspectives. Cell research. 2010;20:24–33. doi: 10.1038/cr.2009.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan F, Weaver A, Gao X, Bern M, Hardwidge PR, Lenardo MJ. IKKbeta phosphorylation regulates RPS3 nuclear translocation and NF-kappaB function during infection with Escherichia coli strain O157:H7. Nature immunology. 2011;12:335–343. doi: 10.1038/ni.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wier EM, Neighoff J, Sun X, Fu K, Wan F. Identification of an N-terminal truncation of the NF-kappaB p65 subunit that specifically modulates ribosomal protein S3-dependent NF-kappaB gene expression. The Journal of biological chemistry. 2012;287:43019–43029. doi: 10.1074/jbc.M112.388694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AR, Pearson JS, Bright MD, Munera D, Robinson KS, Lee SF, Frankel G, Hartland EL. Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Molecular microbiology. 2011;80:1420–1438. doi: 10.1111/j.1365-2958.2011.07661.x. [DOI] [PubMed] [Google Scholar]
- Xiang Z, Li L, Lei X, Zhou H, Zhou Z, He B, Wang J. Enterovirus 68 3C protease cleaves TRIF to attenuate antiviral responses mediated by Toll-like receptor 3. Journal of virology. 2014;88:6650–6659. doi: 10.1128/JVI.03138-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Q, Zhang L, Wan X, Chen J, Hu L, Ding X, Li L, Karar J, Peng H, Chen S, Huang N, Rauscher FJ, 3rd, Shao F. Structure and specificity of the bacterial cysteine methyltransferase effector NleE suggests a novel substrate in human DNA repair pathway. PLoS pathogens. 2014;10:e1004522. doi: 10.1371/journal.ppat.1004522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen H, Ooka T, Iguchi A, Hayashi T, Sugimoto N, Tobe T. NleC, a type III secretion protease, compromises NF-kappaB activation by targeting p65/RelA. PLoS pathogens. 2010;6:e1001231. doi: 10.1371/journal.ppat.1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T. Function of RIG-I-like receptors in antiviral innate immunity. The Journal of biological chemistry. 2007;282:15315–15318. doi: 10.1074/jbc.R700007200. [DOI] [PubMed] [Google Scholar]
- Zaragoza C, Saura M, Padalko EY, Lopez-Rivera E, Lizarbe TR, Lamas S, Lowenstein CJ. Viral protease cleavage of inhibitor of kappaBalpha triggers host cell apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19051–19056. doi: 10.1073/pnas.0606019103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Clausell A, Robinson T, Yin J, Chen E, Johnson L, Weiss G, Sabbaj S, Lowe RM, Wagner FH, Goepfert PA, Kutsch O, Cron RQ. Host factor transcriptional regulation contributes to preferential expression of HIV type 1 in IL-4-producing CD4 T cells. Journal of immunology. 2012;189:2746–2757. doi: 10.4049/jimmunol.1103129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lei CQ, Hu YH, Xia T, Li M, Zhong B, Shu HB. Kruppel-like factor 6 is a co-activator of NF-kappaB that mediates p65-dependent transcription of selected downstream genes. The Journal of biological chemistry. 2014;289:12876–12885. doi: 10.1074/jbc.M113.535831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G. Chlamydia trachomatis secretion of proteases for manipulating host signaling pathways. Frontiers in microbiology. 2011;2:14. doi: 10.3389/fmicb.2011.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]