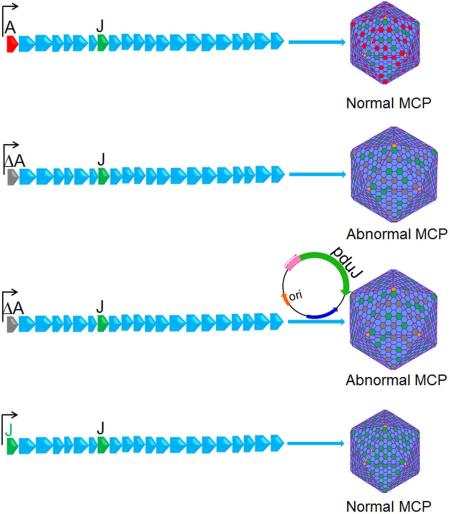
Summary
Bacterial microcompartments (MCPs) are complex organelles that consist of metabolic enzymes encapsulated within a protein shell. In this study, we investigate the function of the PduJ MCP shell protein. PduJ is 80% identical in amino acid sequence to PduA and both are major shell proteins of the 1,2-propanediol (1,2-PD) utilization (Pdu) MCP of Salmonella. Prior studies showed that PduA mediates the transport of 1,2-PD (the substrate) into the Pdu MCP. Surprisingly, however, results presented here establish that PduJ has no role 1,2-PD transport. The crystal structure revealed that PduJ was nearly identical to that of PduA and, hence, offered no explanation for their differential functions. Interestingly, however, when a pduJ gene was placed at the pduA chromosomal locus, the PduJ protein acquired a new function, the ability to mediate 1,2-PD transport into the Pdu MCP. To our knowledge, these are the first studies to show that that gene location can determine the function of a MCP shell protein. We propose that gene location dictates protein-protein interactions essential to the function of the MCP shell.
Keywords: Microcompartments; 1,2-Propanediol; Salmonella; B12; Carboxysome
Graphical abstract
Introduction
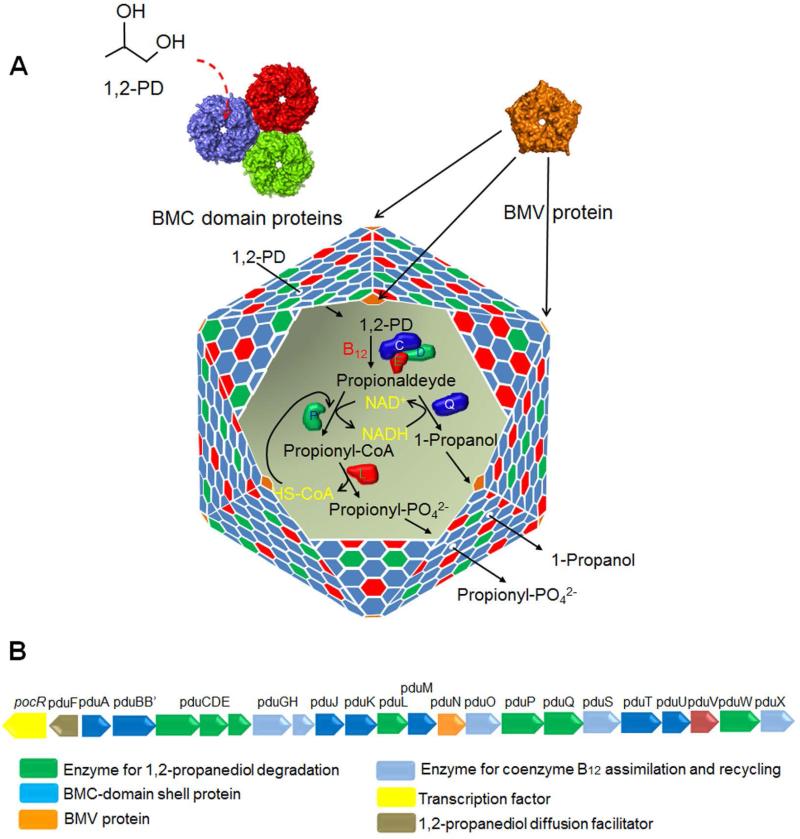
Many bacteria use proteinaceous organelles known as microcompartments (MCPs) to optimize metabolic processes (Bobik et al., 2015, Rae et al., 2013, Chowdhury et al., 2014, Abdul-Rahman et al., 2013). MCPs are polyhedral in shape, about the size of a large virus (100-200 nm in diameter), and consist of metabolic enzymes encapsulated within a selectively permeable protein shell. The functions of MCPs are to accelerate catalysis, block wasteful by-reactions, and sequester toxic or volatile metabolic intermediates in order to minimize cellular damage and/or loss of valuable carbon (Fig 1A) (Havemann et al., 2002, Sampson & Bobik, 2008, Penrod & Roth, 2006, Price & Badger, 1989). Bioinformatic analyses indicate that MCPs are produced by hundreds of different bacterial species ranging across 23 different phyla and that they are used to enhance seven or more different metabolic processes (Abdul-Rahman et al., 2013, Jorda et al., 2013). The three best-studied MCPs are carboxysomes, which are used for autotrophic carbon fixation (Rae et al., 2013) and the Pdu and Eut MCPs which are involved in the coenzyme B12-dependent catabolism of 1,2-propanediol (1,2-PD) and ethanolamine, respectively (Chowdhury et al., 2014). Other MCPs are proposed to be involved in ethanol degradation (Seedorf et al., 2008), choline catabolism (Craciun & Balskus, 2012, Kuehl et al., 2014), 1,2-PD degradation initiated by a glycyl radical enzyme (rather that a B12-dependent enzyme) (Petit et al., 2013, Scott et al., 2006) and in the degradation of algal polysaccharides (Erbilgin et al., 2014).
Figure 1. Model for the Pdu MCP of Salmonella.
(A) The mosaic structure of the Pdu MCP where different BMC-domain proteins tile together forming facets of the shell. A pentameric non-BMC domain protein (BMV) caps the vertices of the polyhedral structure. The central pore of PduA, a BMC domain protein plays a pivotal role in the selective transport of 1, 2-PD (the substrate) across the shell. A primary reason for the encapsulation of enzymes for 1,2-PD degradation is to sequester a toxic intermediate (propionaldehyde). (B) The organization of pdu operon which encodes the known genes of the Pdu MCP including seven BMC-domain shell proteins and 1,2-PD degradative enzymes. The details of the functions of these genes were reviewed recently (Bobik et al., 2015, Chowdhury et al., 2014)
Although MCPs are functionally distinct in terms of metabolism due to variation in their encapsulated enzymes, all have related protein shells (Yeates et al., 2013, Chowdhury et al., 2014). MCP shells are primarily composed of a family of proteins known as bacterial microcompartment (BMC) domain proteins (Kerfeld et al., 2005, Tanaka et al., 2008, Tanaka et al., 2009, Yeates et al., 2013). Most MCP shells are built from several thousand BMC-domain proteins of 5-10 different types (Chowdhury et al., 2014). A number of BMC-domain proteins pack tightly side-by-side into extended two-dimensional layers and different types of BMC-domain proteins have edges with complementary shapes. Hence, the protein shells of bacterial MCPs are thought to be composed primarily of mixed sheets of BMC-domain proteins (Fig. 1A) (Crowley et al., 2010, Heldt et al., 2009, Klein et al., 2009, Sagermann et al., 2009). Nearly all MCPs also include at least one bacterial microcompartment vertex (BMV) protein (Tanaka et al., 2008, Wheatley et al., 2013). BMV proteins are pentamers (unrelated to BMC-domain proteins) that cap the vertices of MCPs and help close the polyhedral structure (Fig. 1A) (Tanaka et al., 2008, Wheatley et al., 2013).
MCP function requires that the substrates and cofactors needed by the encapsulated enzymes cross the protein shell and enter the MCP interior (Cheng et al., 2008). However, the MCP must also restrict the egress of certain toxic/volatile intermediates (Havemann et al., 2002, Sampson & Bobik, 2008, Penrod & Roth, 2006, Price & Badger, 1989). Studies indicate that BMC-domain proteins provide the basis for a selectively permeable protein shell that fulfills these requirements (Crowley et al., 2010, Kerfeld et al., 2005). In the case of the Pdu MCP, crystallographic studies showed that the PduA shell protein tiles into tightly packed protein sheets whose most notable openings are central pores at the six-fold axis of the PduA hexamer (Crowley et al., 2010). These pores are lined with numerous hydrogen-bond donors and acceptors suggesting they might preferentially allow the movement of 1,2-PD (the substrate) compared to propionaldehyde (a toxic intermediate) which is less polar (Fig 1A) (Crowley et al., 2010, Crowley et al., 2008). Importantly, Chowdhury and co-workers recently demonstrated selective permeability; structure-guided mutagenesis showed that the PduA pore acts as a major route for entry 1,2-PD into the Pdu MCP while restricting the egress of a propionaldehyde (Chowdhury et al., 2015, Crowley et al., 2010). Interestingly, crystallographic studies have suggested that tandem-BMC domain proteins such as PduB might have an allosterically-gated pores that allow the entry of bulkier enzymatic cofactors (Klein et al., 2009, Sagermann et al., 2009, Tanaka et al., 2010, Cai et al., 2013, Thompson et al., 2015). Several tandem-BMC domain proteins have been crystallized in alternate conformations that have either a large central pore or one that is occluded suggestive of a gate (Klein et al., 2009, Sagermann et al., 2009, Tanaka et al., 2010, Cai et al., 2013). In addition, the EutL tandem-BMC-domain protein has been shown to bind ethanolamine (the substrate of the Eut MCP) in a manner that indicates allosteric regulation of its pore (Thompson et al., 2015). It is also notable that the PduT shell protein has a [4Fe-4S] cluster where the central pore would typically be found (Crowley et al., 2010, Pang et al., 2011). It has been suggested that PduT might act as an electron conduit or in the transport of Fe-S clusters between the cytoplasm of the cell and the MCP lumen (Crowley et al., 2010, Crowley et al., 2008).
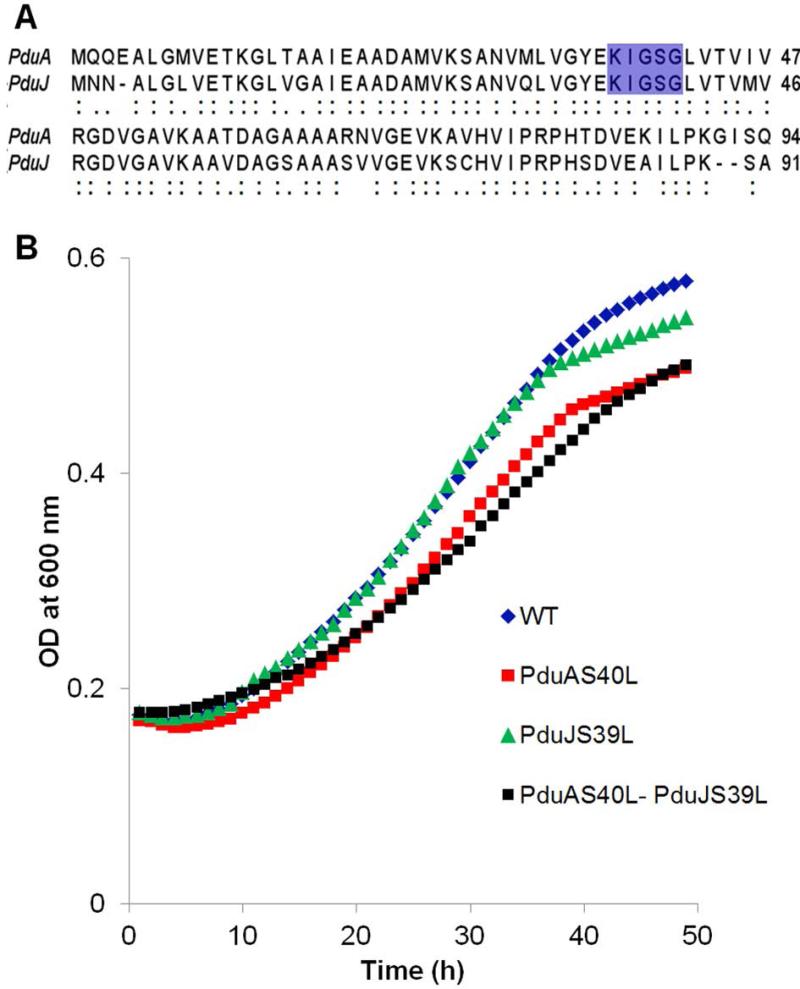
The PduJ and PduA proteins are both major components of the shell of the Pdu MCP which is used by Salmonella and many other species for the catabolism of 1,2-PD as a carbon and energy source (Havemann & Bobik, 2003). The function of the Pdu MCP is to sequester propionaldehyde to prevent toxicity during 1,2-PD metabolism (Fig. 1A) (Havemann et al., 2002, Sampson & Bobik, 2008). As mentioned above, PduA provides a major route for the selective entry of 1,2-PD into the Pdu MCP (Chowdhury et al., 2015). Sequence alignments show that PduJ and PduA are 80% identical in amino acid sequence and have identical pore-lining residues (Fig. 2A). Hence, it would seem reasonable the PduJ and PduA have similar functions. Surprisingly, however, results presented here show that unlike PduA, the PduJ protein has no role in 1,2-PD transport. We also present the crystal structure of PduJ and show that the pore structures of PduA and PduJ are highly conserved. Lastly, we show that the function of the PduJ protein depends on the chromosomal position of its encoding gene. If a pduJ gene is placed in the chromosomal position normally occupied by pduA, the PduJ protein can then mediate 1,2-PD transport into the Pdu MCP. Based on these findings, we propose a model in which the specific location of the PduJ protein within MCP shell determines whether it can function in 1,2-PD transport.
Figure 2. (A) Differential effects of analogous pore mutations in PduA and PduJ on growth of Salmonella on 1,2-PD at limiting B12.
(A) Pairwise sequence alignment showing high level of identity between PduA and PduJ. The conserved pore-lining residues are highlighted in blue. In PduA, S40 is a key pore-forming residue that plays a crucial role in 1, 2-PD transport. The analogous residue in PduJ is S39. (B) Growth of PduA S40L and PduJ S39L pore mutants with respect to wild-type (WT). Growth assays were repeated at least three times in triplicate or more and representative curves are shown.
Results
A PduJ pore mutant grows normally on 1,2-PD minimal medium
Recent studies showed that the pore structure of the PduA shell protein allows for the selective influx of substrate (1,2-PD) into the Pdu MCP (Chowdhury et al., 2015). In these studies, a PduA S40L pore mutant was shown to be impaired for 1,2-PD transport across the MCP shell (Chowdhury et al., 2015). Because the pore region of PduJ is identical in sequence to PduA (Fig. 2A), we tested whether the PduJ pore might also play a role in 1,2-PD uptake into the Pdu MCP. A chromosomal PduJ S39L mutant was constructed (S39 of PduJ corresponds to S40 of PduA). However, in contrast to a PduA S40L mutant which grows slowly in 1,2-PD minimal medium at limiting B12 due to impaired 1,2-PD transport (Chowdhury et al., 2015), a PduJ S39L mutant grew similarly to wild-type (Fig. 2B). The doubling times were as follows: wild-type (17.6 ±1.1 h); PduA S40L (22.8 ± 0.6 h); and PduJ S39L (16.4 ± 0.8 h). We also tested growth of a PduJ S39L PduA S40L double mutant, and found that it grew similarly to PduA S40L (Fig. 2B) (the doubling time of the PduJ S39L PduA S40L double mutant was 23.1 ± 0.6 h). The error shown for doubling times is one standard deviation from the mean and in all cases was determined from at least three biological replicates measured in triplicate. The similar growth pattern of PduJ S39L to wild-type tentatively suggests that PduJ does not mediate 1,2-PD transport despite the fact that the pore region of PduJ is identical in sequence to that of PduA. Thus, despite their high sequence identity, PduJ seemed to have a different function than PduA.
MCPs isolated from the PduJ S39L pore mutant exhibited diol dehydratase activity similar to wild-type
Diol dehydratase (DDH) is an MCP lumen enzyme that catalyzes the first step of 1,2-PD degradation (the conversion of 1,2-PD to propionaldehyde); hence, its activity depends on the diffusion of 1,2-PD across the MCP shell. Previous studies showed that MCPs purified from a PduA S40L mutant exhibited lower DDH activity compared to wild-type due to impaired diffusion of 1,2-PD across the MCP shell (Chowdhury et al., 2015). Therefore, as a second test to assess whether the PduJ pore has a role in 1,2-PD transport, we assayed DDH activity of MCPs purified from a PduJ S39L mutant. PduJ S39L MCPs were purified as described (Sinha et al., 2012) with a yield > 90% compared to wild-type. In addition, transmission electron microscopy showed no gross abnormalities in the size and shape of PduJ S39L MCPs (Fig. S1 and S2) and SDS-PAGE followed by densitometry showed no obvious changes in the major proteins bands compared to wild-type MCPs (Fig. S3). However, unlike PduA S40L MCPs which had substantially reduced DDH activity, the DDH activity of PduJ S39L MCPs was indistinguishable from wild-type (Table 1). In addition, MCPs purified from the PduA S40L PduJ S39L double mutant had DDH activity similar to the PduA S40L single mutant further indicating that the PduJ S39L mutant did not restrict 1,2-PD transport (Table 1). As a control, dialysis and sonication were used to disrupt the shells of each type of MCP to allow free diffusion of 1,2-PD to DDH (Chowdhury et al., 2015). After their shells were broken, wild-type, PduJ S39L, PduA S40L and PduA S40L PduJ S39L (double mutant) MCPs all had similar DDH activity (Table 1) showing that DDH was normally recruited and incorporated into the mutant MCPs. Thus, DDH assays supported the growth studies described above and provided further evidence that in contrast to PduA, PduJ has no role in 1,2-PD influx into the Pdu MCP.
Table 1.
Effect of pore mutations in PduA and PduJ on the DDH activities of purified Pdu MCPs.
| MCPs from Pdu Mutants | Specific Activity (μmol min−1mg−1) |
|---|---|
| Wild type (WT) | 28.2 ± 1.2 |
| PduJ S39L | 27.6 ± 0.8 |
| PduA S40L | 14.8 ± 1.5* |
| PduA S40L-PduJ S39L | 14.4 ± 1.1* |
| WT-Broken MCP | 34.8 ± 1.5* |
| PduJ S39L-Broken MCP | 34.2 ± 0.8* |
| PduA S40L-Broken MCP | 33.8 ± 0.3* |
| PduA S40L-PduJ S39L-Broken MCP | 34.2 ± 0.8* |
The DDH activities shown are based on at least three biological replicates measured in triplicate. The error estimate shown is ± one standard deviation.
p-value <0.0001 compared to wild-type.
The structure of PduJ is nearly identical to that of PduA
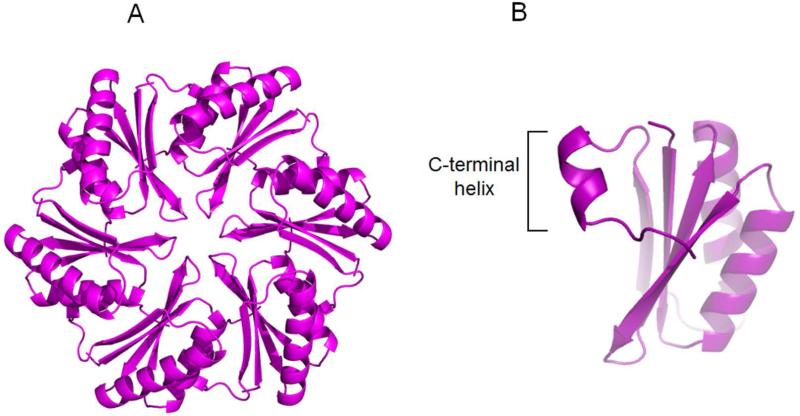
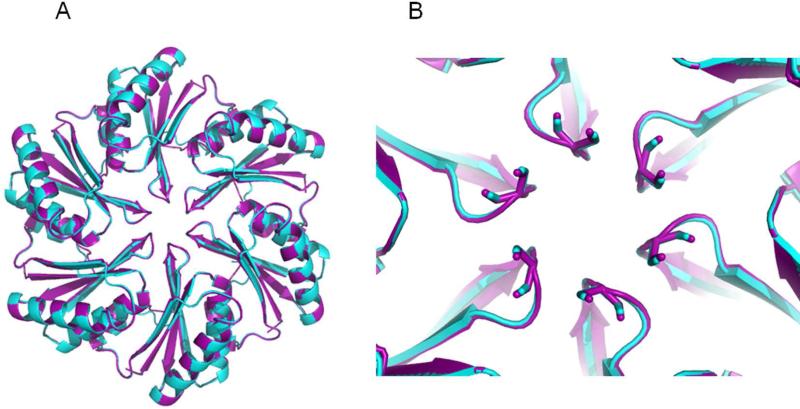
The dissimilar transport behavior of PduA and PduJ, despite identical amino acid sequences in their pore regions, was unexpected. One possibility was that PduJ and PduA have significant structural differences despite their high sequence identity (80%). To examine this, PduJ K25A was crystallized in space group P6 and its structure was solved at 1.5Å resolution. The K25A mutant was used to reduce crystallographic problems that result from edge-to-edge aggregation of BMC hexamers (Sinha et al., 2014). The structure was refined using PHENIX and manual manipulation using COOT, and BUSTER 2.10.0. Protein geometry analysis revealed no Ramachandran outliers, with 98.9% residues in favored regions. The Molprobity clash score after adding hydrogens was 0.78. Data collection statistics are shown in Table S1. In the crystal structure, PduJ K25A forms a hexamer, adopting the canonical BMC hexamer conformation (PDBID 5D6V, Fig. 3A). In the PduJ structure, the C-terminal region is well-ordered and forms a short alpha helix whereas this segment is disordered and not resolved in the PduA structure (Fig. 3B). Structural alignment of the PduJ hexamer with the PduA hexamer (PDBID 3NGK) gives a backbone r.m.s.d. deviation of 0.28Å (Fig. 4A). The pore region of PduJ appears identical to that of the PduA (Fig. 4B). Aligning all atoms of the pore regions (PduJ residues I37-L41 and PduA residues I38-L42) of the monomers gives an r.m.s.d of 0.185Å. Also, the PduJ pore has a similar pore diameter of 5.7Å to that of PduA, 5.6Å (Fig. 4B). Thus, there are no obvious structural differences between PduA and PduJ that could account for the dissimilar transport behavior.
Figure 3. Crystal structure of PduJ K25A (PDBID 5D6V).
(A) Cartoon representation from cytosolic face. (B) The C-terminal alpha helix of PduJ is shown which is well-ordered, unlike PduA.
Figure 4. Structural alignment of PduA and PduJ.
(A) PduJ hexamer (purple) and PduA hexamer (cyan). (B) Close up view of the aligned pore regions of PduA and PduJ.
pduA can complement the growth phenotype of a pduJ deletion mutant but not vice versa
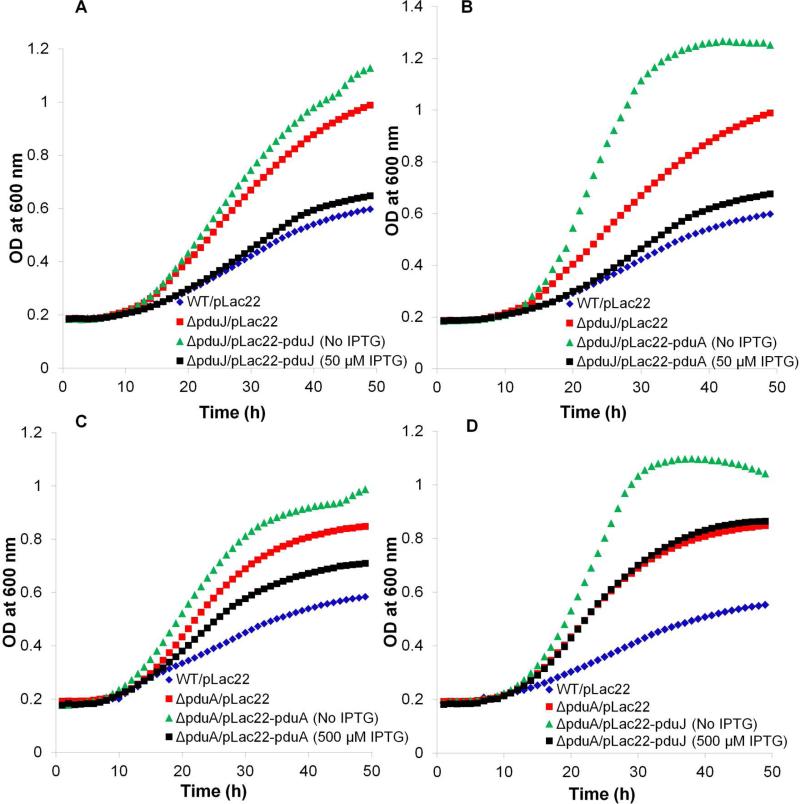
Prior studies showed that a pduA deletion mutant grows faster than wild-type at limiting levels of B12 (20 nM) (Cheng et al., 2011, Sinha et al., 2014). This phenotype results from a damaged MCP shell which allows increased access of the lumen enzymes to their substrates (Sinha et al., 2014). Hence, growth rate at limiting B12 provides a test for the structural integrity of the Pdu MCP (Cheng et al., 2011). Using this test, we examined whether PduJ and PduA are interchangeable for MCP assembly. Control experiments (complementation tests) showed that the fast-growth phenotype of ΔpduJ was restored to wild-type levels by expression of pduJ from plasmid pLac22 at ~50 μM IPTG (Fig. 5A, Table 2). We also found that production of PduA using 50 μM IPTG largely corrected the fast-growth phenotype of a pduJ deletion mutant (Fig. 5B, Table 2). In contrast, a pduA deletion was only partially complemented by expression of PduA at optimal IPTG levels (Fig. 5C, Table 2) as reported previously (Cheng et al., 2011), and was not complemented at all by production of PduJ from pLac22 at any IPTG concentration tested (Fig. 5D, Table 2). Thus, with respect to MCP function, PduA can replace PduJ but not vice versa.
Figure 5. Complementation and cross-complementation studies with ΔpduA and ΔpduJ mutants.
(A) Complementation of ΔpduJ with pduJ. (B) Complementation of ΔpduJ by pduA. (C) Partial complementation of ΔpduA by pduA. (D) Attempted complementation of ΔpduA by pduJ. Gene expression from pLac22 was induced by adding IPTG at 0, 10, 20, 30, 50, 100, 200, 500 μM or 1 mM). The results shown in the figure are for the IPTG level at which optimal complementation occurred. Other concentrations of IPTG gave resulted in similar or worse complementation. All growth assays were done at least three times and in triplicate and a representative outcome is shown.
Table 2.
Complementation and cross-complementation of pduA and pduJ mutants.
| Pdu mutants | Doubling time (h) |
|---|---|
| WT/pLac22 | 19.4 ± 0.8 |
| ΔpduA/pLac22 | 10.5 ± 0.8* |
| ΔpduA/pLac22-pduA (induced with 500 μM IPTG) | 14.2 ± 0.3* |
| ΔpduJ/pLac22 | 10.1 ± 0.7* |
| ΔpduJ/pLac22-pduJ (induced with 50 μM IPTG) | 18.1 ± 0.5 |
| ΔpduA/pLac22-pduJ (induced with 500 μM IPTG) | 9.5 ± 1.2* |
| ΔpduJ/pLac22-pduA (induced with 50 μM IPTG) | 17.1± 1.1 |
| ΔpduA/pLac22-pduA (uninduced) | 9.5 ± 0.7* |
| ΔpduJ/pLac22-pduJ (uninduced) | 8.8 ± 1.1* |
Doubling times were calculated from at least three biological replicates measured in triplicate. The error estimate shown is ± one standard deviation
p-value <0.001 compared to wild-type.
We note that the growth phenotypes certain strains (ΔpduA/pLac22-pduA and ΔpduJ/pLac22-pduJ) were more severe (faster growth) without added IPTG (Fig. 5A, 5C). We speculate that a very low level of expression from pLac22 plasmid might have an antagonistic effect on MCP assembly in these particular cases.
pduA can complement the DDH phenotype of a ΔpduJ mutant but not vice versa
MCPs purified from a pduJ deletion mutant show higher DDH activity than wild-type MCPs because the MCP shell was disrupted allowing DDH greater access to its substrate 1,2-PD (Table 3) (Sinha et al., 2014). Ectopic production of either PduJ or PduA reduced the DDH activity of purified ΔpduJ MCPs substantially closer to wild-type levels (Table 3) consistent with growth studies (Fig. 5). The ectopic production of PduA and PduJ from pLac22 was confirmed by SDS PAGE of purified MCPs (Fig. S3). In contrast, production of PduJ from pLac22 had no effect on the DDH activity of MCPs purified from a pduA deletion (Table 3). Thus, in vitro assays with purified MCPs also indicate that PduA can substantially substitute for PduJ, but not vice versa.
Table 3.
Effect of complementation on DDH activity
| MCPs from Pdu Mutants | Specific Activity (μmol min−1mg−1) |
|---|---|
| WT/pLac22 | 27.3 ± 0.8 |
| ΔpduJ/pLac22 | 33.7 ± 0.7 |
| ΔpduJ/pLac22-pduJ | 27.8 ± 0.7 |
| ΔpduA/pLac22-pduA | 29.5 ± 0.8* |
| ΔpduA/pLac22 | 34.8 ± 1.1 |
| ΔpduA/pLac22-pduJ | 34.5 ± 0.9* |
| ΔpduJ/pLac22-pduA | 28.7 ± 0.7 |
The DDH activities are based on at least three biological replicates measured in triplicate. The error estimate shown is ± one standard deviation.
p-value <0.001 compared to wild-type/pLac22.
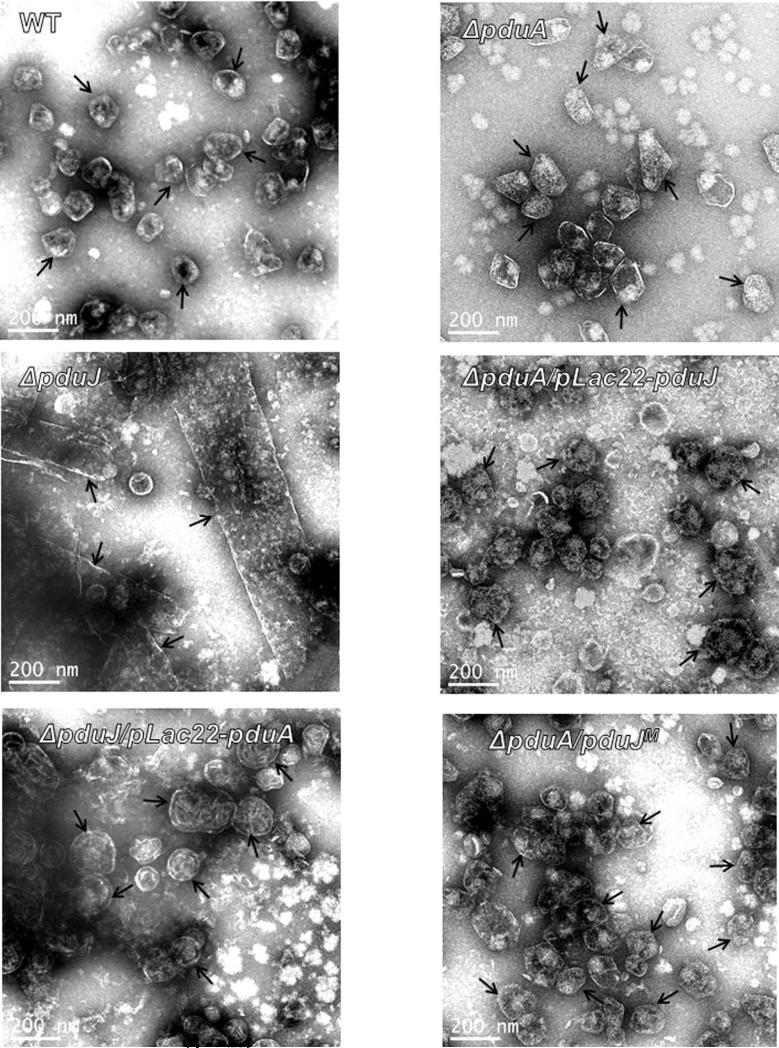
The aberrant morphology of ΔpduJ MCPs is partially corrected by expression of PduA from a plasmid
A pduJ deletion mutant produced elongated MCPs (Fig. 6) as was observed earlier (Cheng et al., 2011, Sinha et al., 2014). The structure of ΔpduJ MCPs was partly corrected by production of PduA from a plasmid (Fig. 6). Upon expression of PduA, the elongated MCPs produced by a ΔpduJ mutant assumed a rounded shape more similar to wild-type although they had a somewhat more wrinkled or folded appearance (Fig. 6). A ΔpduA mutant forms slightly enlarged MCPs (Fig. 6 and S2) as was previously shown (Cheng et al., 2011). PduJ expressed from a plasmid did not observably change the MCP morphology of ΔpduA mutant although any cross complementation would be difficult to discern since ΔpduA MCPs are only slightly larger than wild-type MCPs (Fig. 6 and S2). Nonetheless, overall, three lines of evidence (described above) indicate that PduA can substitute for PduJ to a large extent but not vice versa. This emphasizes the question, why can PduA functionally substitute for PduJ but not vice versa?
Figure 6. Transmission electron micrographs of purified MCPs from PduA and PduJ deletion mutants as compared to wild-type.
WT is wild-type Salmonella. A pduA deletion mutant formed somewhat enlarged MCPs (Sinha et al., 2014) while a ΔpduJ mutant formed elongated MCPs.
A pduJ gene can complement a pduA deletion if placed at the pduA chromosomal locus
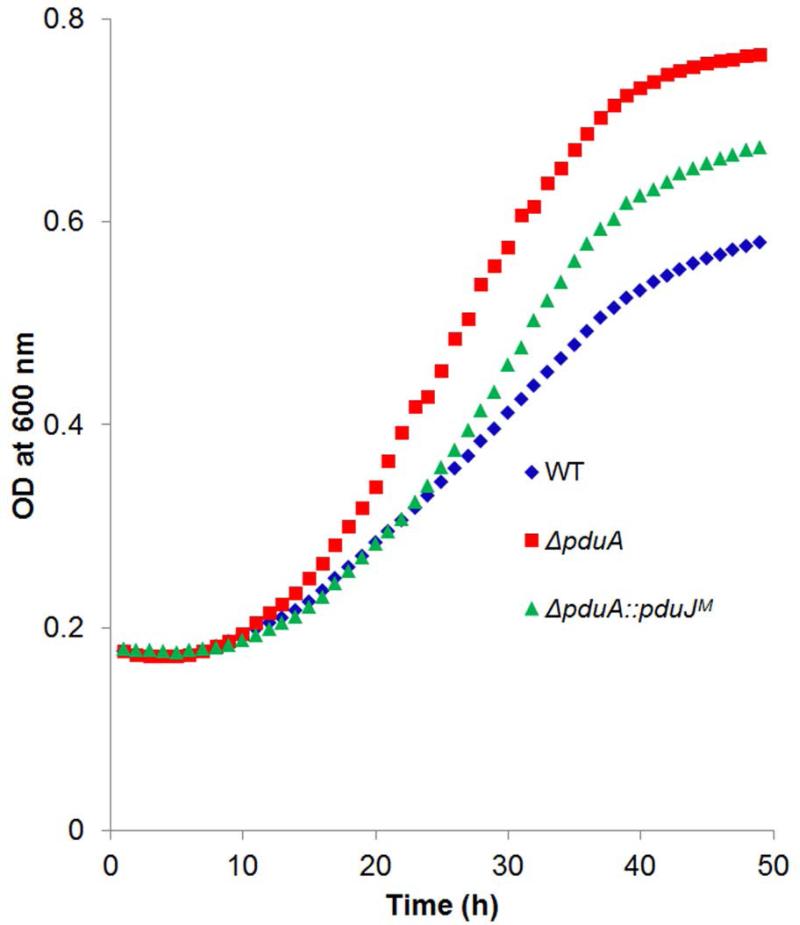
In order to examine the effects of genomic position, the chromosomal copy of the pduA gene was replaced with a pduJ gene that had altered codon usage (to block recombination with the native copy of pduJ) but produced a wild-type PduJ protein. We refer to the resulting mutant as ΔpduA::pduJM (M for modified). Studies showed that the growth rate of ΔpduA::pduJM mutant on 1,2-PD minimal medium with limiting B12 was substantially closer to wild-type compared to the pduA deletion mutant whose growth rate is markedly increased due to increased permeability of the MCP shell. The doubling times were as follows: wild-type, 17.6 ± 1.1 h; ΔpduA::pduJM, 14.5 ± 0.2 h; and ΔpduA was 9.1 ± 0.8 h. (Fig. 7). The error shown for doubling times is one standard deviation from the mean and in all cases was determined from at least three biological replicates measured in triplicate. Hence, these growth tests suggested that PduJ partially replaced PduA when produced from the pduA chromosomal position. This contrasted with tests in which PduJ was produced from a plasmid where no observable correction of the ΔpduA phenotype was observed at any IPTG level used (0, 10, 20, 30, 50, 100, 200, 500 μM or 1 mM) (Fig. 5D).
Figure 7. Growth phenotypes of wild-type, ΔpduA and ΔpduA::pduJM at limiting B12.
WT is wild-type Salmonella. ΔpduA::pduJM is a mutant were the chromosomal copy of pduA has been replaced by a recoded pduJ gene.
We also purified MCPs from a ΔpduA::pduJM mutant and found that they had close to wild-type levels of DDH activity (~29 μmol min−1mg−1) (Table 4). This is in contrast to MCPs purified from the ΔpduA mutant which had increased levels of DDH activity (~34 μmol min−1mg−1) (Table 4) due to a damaged shell. Furthermore, control experiments showed that when MCPs of ΔpduA::pduJM mutant were broken by dialysis followed by sonication, the DDH activity the mutant was similar to that of broken wild-type MCPs. This showed that the amount of DDH encapsulated in the PduJM MCPs were similar to the parent strain. We also note that the PduJM MCPs were purified with ~90% yield compared to wild-type MCPs. Transmission electron microscopy did not reveal any obvious morphological defect in the PduJM MCPs (Fig. S1 and S2) and SDS-PAGE followed by densitometry indicated that the major MCP proteins were present at near wild-type levels (minor proteins are not detected by SDS-PAGE) (Fig. S4). The near wild-type levels of DDH activity in purified ΔpduA::pduJM MCPs further supported the idea the pduJ can replace pduA when expressed from the chromosomal position of pduA.
Table 4.
DDH activity upon change in chromosomal position of PduJ
| MCPs from Pdu Mutants | Specific Activity (μmol min−1mg−1) |
|---|---|
| Wild type (WT) | 28.2 ± 1.2 |
| ΔpduA::pduJM | 29.3 ± 0.6 |
| ΔpduA | 34.3 ± 1.2 |
| WT-Broken MCP | 33.7 ± 0.8 |
| ΔpduA::pduJM-Broken MCP | 34.2 ± 1.2 |
| ΔpduA::pduJMS39L | 18.8 ± 1.5* |
| ΔpduA::pduJMS39L-Broken MCP | 33.4 ± 0.6 |
DDH activities are based on at least three biological replicates measured in triplicate. The error estimate shown is ± one standard deviation.
p-value <0.001 compared to the parent, ΔpduA::pduJM.
The pore of PduJM functions in 1,2-PD transport
Prior studies showed that the PduA shell protein functions in 1,2-PD transport and that a PduA S40L pore mutation impairs movement of 1,2-PD into the MCP lumen (Chowdhury et al., 2015). To determine if a change in chromosomal position allowed the PduJ pore to function in 1,2-PD transport, we mutated serine 39 to leucine in PduJM to produce ΔpduA::pduJM S39L. Changing the pore lining serine to leucine (S39L) in PduJM resulted in a slower growth rate on 1,2-PD which is indicative of impaired 1,2-PD transport into the Pdu MCP (Fig. S5) as previously described for a PduA S40L mutant (Chowdhury et al., 2015). This is in contrast to S39L mutation in pduJ located at its wild-type chromosomal position where no growth defect was observed (Fig. 2B).
As a second test of the pore function in PduJM, we purified PduJM S39L MCPs and measured their DDH activity. The PduJM S39L MCPs had lower DDH activity (18.8 ± 1.5 μmol min−1 mg−1) than purified wild-type or PduJM MCPs (28.2 ± 1.2 and 29.3 ± 0.6 μmol min−1 mg−1) (Table 4, line 6). Controls showed that when the PduJM S39L MCPs were broken, their DDH activity was similar to that of broken wild-type showing normal DDH targeting the mutant MCPs (Table 4, line 7). Thus, the reduced DDH activity of the PduJM S39L MCPs is indicative of impaired pore function as previously described for PduA S40L (Chowdhury et al., 2015). These results contrast markedly to those obtained for the PduJ S39L mutation (the pduJ gene encoding the S39L mutation is at its wild-type chromosomal location) which had no significant effect on DDH activity (Table 1). Together, these studies provide strong evidence that the function of PduJ changes depending on the location of its encoding gene.
As a further control, we used LC-MS/MS to verify incorporation of both PduJ and PduJM S39L into purified MCPs isolated from the pduA::pduJM S39L mutant. A number of peptides with m/z values in good agreement with predictions from the protein sequence confirmed the presence of both PduJ and PduJM S39L (Table S2) in purified MCPs.
Discussion
Bacterial MCPs are distinguished from other known organelles in having a protein shell rather than a lipid membrane as a permeability barrier. MCP shells are generally composed of 5-10 different types of BMC-domain proteins that are functionally diversified to allow the selective transport of metabolites and perhaps to satisfy different architectural requirements (Chowdhuryet al., 2014, Rae et al., 2013, Yeates et al., 2013). Regardless of the structural diversification, nearly all BMC domain proteins have a conserved flat hexagonal structure (Chowdhury et al., 2014, Rae et al., 2013, Yeates et al., 2013), and it is thought that shape complementarity along the edges allows different types of BMC-domain proteins to tessellate edge-to-edge to form the mixed protein sheets that comprise the facets of the shell (Fig. 1A) (Chowdhury et al., 2014, Raeet al., 2013, Yeates et al., 2013). This idea is supported by alanine scanning mutagenesis studies that showed a conserved N-R-K triad found along the edges of BMC-domain proteins is essential in sheet formation and MCP assembly (Sinha et al., 2014). The structural similarity and edge complementarity among varied BMC domain proteins raises the question of the rules that determine the incorporation of particular shell proteins into the MCP. Is the shell a random assembly of BMC-domain proteins, or do particular BMC proteins occupy specific positions within the polyhedral shell? Modeling suggested that mixed sheets of the PduA and PduB proteins would allow their edges to align more precisely resulting in greater stability (Pang et al., 2012). Thus, minor variations along the edges of BMC-domain proteins might guide assembly. In addition, prior studies showed that N-terminal extensions are necessary and sufficient for targeting enzymes to the MCP lumen and that these extensions interact with the termini of shell proteins (Fan et al., 2010, Fan et al., 2012). Hence, it was proposed that the cargo enzymes might play a role in organizing the shell. In the case of the carboxysome, a scaffold protein (CcmN) links the cargo proteins to the shell and might serve a role in shell assembly (Cameron et al., 2013, Cot et al., 2008, Kinney et al., 2012). On the other hand, studies have suggested that empty MCP shells can be produced by recombinant strains (although their organization is uncertain) (Parsons et al., 2010). In addition, prior studies have indicated that certain hexameric BMC domain shell proteins are interchangeable (Cai et al., 2015). Hexameric β-carboxysomal CcmK4 was shown to structurally complement a ΔccmK2 mutant of Synechococcus elongatus PCC 7942 (Cai et al., 2015). Moreover; a chimeric carboxysomal shell was also constructed by replacing β-carboxysomal shell protein CcmK2 with the α-carboxysomal shell protein CsoS1. In the chimeric carboxysomal MCP the α-carboxysomal BMC hexamer fit well into a β-carboxysome shell because of their structural similarity. In the present study, we found that PduJ was unable to compensate for the structural defects in a ΔpduA mutant, when expressed from a plasmid even though PduJ and PduA are very closely related in amino acid sequence (80% identical). Several explanations are possible for this finding: 1) plasmid expression might result in improper protein levels; 2) the difference in amino acid sequence between PduJ and PduA might dictate interactions with other MCP proteins or result in structural variation; or 3) interactions with proteins encoded by adjacent chromosomal genes might be necessary for proper PduA function. It seems unlikely to us that protein production levels are the cause of the observed behavior. The vector used in these studies (pLac22) has a low level of expression without IPTG (Warren et al., 2000), and in our prior studies, pLac22 was successfully used in complementation tests with a number of different Pdu proteins including major and minor shell proteins as well as proteins whose dosage is critical to MCP assembly (Cheng et al., 2011). We also found that PduA itself did not fully complement a ΔpduA mutant when produced from a plasmid (Fig. 5C) as was shown previously (Cheng et al., 2011). Furthermore, partial complementation (the best result obtained) occurred at an intermediate level of IPTG suggesting that higher or lower levels of PduA expression would not have resulted in improved complementation. Thus, these results suggested that the sequence variation between PduA and PduJ does not fully account for the inability of ectopically produced PduJ to correct a ΔpduA mutation. Moreover, we determined the crystal structure of PduJ and found it to be nearly identical to the structure of PduA (Crowley et al., 2010, Sinha et al., 2014). On the other hand, we found that defects in MCP function that result from a ΔpduA mutation were largely corrected by replacing pduA with pduJ chromosomally. The growth phenotype and the MCP morphology of a ΔpduA::pduJM mutant were similar to wild-type (Fig. 6, 7 and S2), and the DDH activity of MCPs purified from a ΔpduA::pduJM mutant was close to normal (Table 4). In addition, we showed that PduJ assumed a new function when expressed from the pduA locus. PduJ did not mediate 1,2-PD transport into MCPs when produced from its native chromosomal locus (Fig. 2B, Table 1), but it did so when produced from the pduA locus (Fig. S5). We propose that when PduJ is produced from the pduA locus, it interacts with new protein partners in a manner that changes its function. The pdu operon is transcribed as a polycistronic message (Fig. 1B) (Bobik et al., 1999). The juxtaposition of the pduA and pduB genes will result in co-localization of their protein products in time and space during and immediately following translation and this might promote protein-protein interactions needed for proper MCP shell assembly. Thus, high sequence similarity allows PduJ to replace PduA and assume its transport function when expressed from the pduA locus but not when expressed from its native chromosomal position or from a plasmid. When PduJ is expressed from its native chromosomal position, it is apparently incorporated into the MCPs in a fashion that does not allow it to mediate 1,2-PD transport. The fact that PduJ changes function depending on the chromosomal position of its encoding gene strongly supports this model. At the present time it is unclear whether the observed phenomena reflect context-dependent differences in local protein conformation (e.g. in the pore of PduJ) or more global differences in the way subunits are arranged in the fully-assembled MCP. Precedence for gene location influencing protein assembly was recently described for bacterial luciferase (LuxAB) (Shieh et al., 2015). In this case, LuxA and LuxB were shown to assemble close to the site of synthesis, and localization of the luxA and luxB genes at distant chromosomal sites impaired formation of the LuxAB heterodimer. Importantly, the studies reported here show for the first time that gene location influences the interactions among MCP proteins and is critical to the higher order structure and function of bacterial MCPs. They also provide an example of how gene position can guide assembly and even determine protein function in large multi-protein complexes.
Experimental procedures
Chemicals and Reagents
Antibiotics, vitamin B12 (CN-B12), coenzyme B12 (AdoB12), NAD+, NADH, NADP+, and NADPH are from Sigma-Aldrich (St. Louis, MO). KOD DNA polymerase and restriction enzymes and T4 ligase were respectively from Novagen (Cambridge, MA) and New England Biolabs (Beverly, MA). Isopropyl-β-D-1-thiogalactopyranoside (IPTG) was from Diagnostic Chemicals Limited (Charlotteville, PEI, Canada. Choice Taq Blue Mastermix was from Denville Scientific (South Plainfield, NJ). Other chemicals were from Fisher Scientific (Pittsburgh, PA).
Bacterial strains, media, and growth conditions
The bacterial strains used are listed in Table 5. All strains are derivatives of Salmonella enterica serovar Typhimurium strain LT2, referred to as LT2. The rich medium used was Luria-Bertani/Lennox (lysogeny broth) (LB) medium (Difco, Detroit, MI). For selecting transformants TYE media was used (Chang & Cronan, 1982) where bacto-tryptone, yeast extract and agar were purchased from Difco Laboratories (Difco, Detroit, MI). The minimal medium used was no carbon-E (NCE) medium supplemented with 1 mM MgSO4, 0.3 mM each of valine, isoleucine, leucine, and threonine and 50 μM ferric citrate (Berkowitz et al., 1968, Cheng et al., 2011, Price-Carter et al., 2001).
Table 5.
Bacterial strains used in this study
| Strains | Genotype/Variant | Source |
|---|---|---|
| WT | Salmonella enterica serovar Typhimurium strain LT2 | Lab collection |
| BE293 | LT2/pKD46 | Lab collection |
| BE464 | LT2, ΔpduA::frt | Lab collection |
| BE184 | LT2, ΔpduJ | Lab collection |
| CC33 | LT2, ΔpduJ ΔpduA::frt | This Study |
| CC3 | LT2, PduAS40L | This Study |
| CC17 | LT2, PduJS39L | This Study |
| CC32 | PduAS40L-PduJS39L | This Study |
| BE287 | LT2/pLac22 | Lab collection |
| BE1351 | LT2, ΔpduA::frt/pLac22 | Lab collection |
| TA1414 | LT2, ΔpduJ/pLac22 | Lab collection |
| BE1350 | LT2, ΔpduA::frt/pLac22-pduA | Lab collection |
| CC16 | LT2, ΔpduA::frt/pLac22-pduJ | This study |
| CC10 | LT2, ΔpduJ/pLac22-pduA | This study |
| TA1412 | LT2, ΔpduJ/pLac22-pduJ | Lab collection |
| CC76 | LT2, ΔpduA::pduJM | This study |
| CC79 | LT2, ΔpduA::pduJMS39L | This study |
Construction of chromosomal mutations
Chromosomal pduA and pduJ deletions and the point mutations surrounding the pore region were constructed by recombineering as described (Chowdhury et al., 2015, Datsenko & Wanner, 2000, Sinha et al., 2014). All the mutants were confirmed by sequencing.
P22 Transduction
In order to make double mutants, the sacB-cat cassette at pduA loci was moved to a ΔpduJ mutant by transduction as previously described, using P22 HT105/1 int-210 (Schmieger, 1971). Transductants were tested for phage contamination and sensitivity by streaking on green plates against H5- int. Finally, the sacB-cat cassette was replaced by recombination with single stranded oligo having the desired mutation. Deletion mutations were confirmed by PCR followed by DNA sequencing.
Construction of clones for complementation studies
The coding sequences of pduA, and pduJ were cloned into pLac22 (Cheng et al., 2011, Warren et al., 2000) using Bgl II and Hind III restriction sites. The DNA sequences of all clones were verified. The recombinant plasmids were transformed into respective deletion mutants using electroporation (Cheng et al., 2011).
Chromosomal replacement of pduA with pduJ
To determine if chromosomal position has any effect on complementation, the chromosomal pduA gene was replaced with the pduJ gene using a recombineering method (Chowdhury et al., 2015, Datsenko & Wanner, 2000, Sinha et al., 2014). A codon-modified pduJ gene (pduJM) was designed using a codon optimization tool from Integrated DNA Technologies, (Coralville, IA). A Shine Dalgarno sequence was placed after the stop codon of pduJM to assure proper expression of downstream genes. The pduJM gene with needed flanking sequences was synthesized by Integrated DNA Technolgies as a gBlock with the sequence shown in Fig. S6. This construct was used to transform the cells where a sacB-cat cassette was inserted at pduA locus in a strain expressing the λ-red recombinase from pKD46 (Datsenko & Wanner, 2000). Positive transformants were confirmed through DNA sequencing.
Growth studies, MCP Purification and Diol dehydratase activity
Growth studies were performed at limiting CN-B12 (25 nM) as previously described using a Synergy HT Microplate reader (BioTek, Winooski, VT) (Cheng et al., 2011). Experiments were done in triplicate. Pdu MCPs were purified according to a prior protocol (Chowdhury et al., 2015, Sinha et al., 2012). Their protein content was verified by SDS PAGE. To check MCP integrity, the purified MCPs were negatively stained with uranyl acetate (2%) and visualized using a transmission electron microscope (JEOL 2100, Peabody, MA) as described (Chowdhury et al., 2015, Sinha et al., 2012). Purified MCPs were assayed for diol dehydratase using the coupled NADH-dependent alcohol dehydrogenase reaction as described (Chowdhury et al., 2015, Fan & Bobik, 2011).
Protein identification by LC-MS/MS
To verify the incorporation of PduJM into microcompartment shell, the MCP particles were purified and fractionated by SDS PAGE. Protein band of interest was excised from a SDS-polyacrylamide gel, cut into small pieces and distained with mixture of 25 mM ammonium bicarbonate and acetonitrile (ACN) (v/v 1:1) according to the protocol described earlier (Shevchenko et al., 2006). The proteins inside the gel were reduced with 1 mM DTT and cysteine residues were alkylated with iodoacetamide. Trypsin (proteomics grade, Promega) was added to the gel pieces and incubated for 16–18 h according to a published procedure (Shevchenko et al., 2006). The trypsin digested peptides were extracted and subjected to liquid chromatography mass spectrometry analysis according to the manufacturer's instructions (Thermo, Q Exactive™ Hybrid Quadrupole-Obrbitrap Mass Spectrometer). Tandem mass spectra were searched against the known protein sequence from S. enterica serovar Typhimurium LT2 using both MASCOT and Sequest HT (Thermo Proteome discoverer software from Thermo Fisher) with maximum mixed cleavage sites 2, fixed carbamidomethylation and variable methionine oxidation. The precursor mass tolerance was set to 5 ppm and fragment mass tolerance was 0.05 Da. Maximum delta Cn value was set to 0.05.
PduJ structure determination
For ease of purification and characterization a PduJ K25A mutant was constructed by site-directed mutagenesis method. The mutation greatly increased the solubility of the PduJ protein as was previously observed for the analogous mutation in PduA (K26A). The PduJ K25A and PduA K26A mutations (which are on the edges of the hexamers) reduce interactions that lead to protein sheet formation at low protein concentrations (Sinha et al., 2014). To construct this mutation, the coding sequence of full-length pduJ of S. enterica serovar Typhimurium LT2 was cloned into pET22b(+) vector (Novagen). Then, site-directed mutagenesis was performed using a QuikChange kit (Stratagene) to generate the PduJ K25A mutant. The construct was confirmed by DNA sequencing.
PduJ K25A construct was transformed in BL21 CodonPlus(DE3)-RIL E. coli cells (Stratagene), grown in LB media with ampicillin (100 μg ml−1) and chloramphenicol (34 μg/ml) at 37 °C, shaking at 225 rpm. When cell cultures reached an optical density at wavelength 600 nm (OD600) of 0.4, temperature was dropped to 30 °C; and at OD600 of 0.6, protein expression was induced with 0.5 mM isopropyl-D-1-thiogalactopyranoside (IPTG) for 4 hours. Cells were pelleted at 4,000 rpm for 15 min and resuspended (15 g cell pellet per 35 ml total) in 50 mM Tris-HCl and 200 mM NaCl, pH 9.0 (Buffer A). Resuspended cells were stored at −80°C until purification.
Before purification the cells were thawed and homogenized in the presence of lysozyme, protease inhibitor cocktail, DNase I and 1 mM MgCl2. Cells were lysed by sonication in 3 cycles, increasing amplitude from 40-60% per cycle. Cell lysate was centrifuged at 17,000 × g for 30 min and the supernatant was clarified by 0.45μm filtration. The clarified supernatant was loaded on a HisTrap column (CV=5 ml, GE Healthcare). The column was washed with 10% Buffer B (50 mM Tris-HCl, 200 mM NaCl, 500 mM Imidazole, pH 8.0) and eluted with a linear gradient (Δ90% Buffer B/45ml of elution). PduJ K25A construct was eluted between 30-50% Buffer B. Fractions containing the fairly pure and large quantities of protein migrating at 10 kDa were pooled and dialyzed into crystallization buffer (30 mM Tris. HCl, 50 mM NaCl, 1% v/v glycerol, pH 8.0) overnight at 4 °C. PduJ K25A construct was then concentrated to 100-250 mg/mL each, flash frozen with liquid N2, and stored at −80°C.
Crystallization trials were set up in 96-well plate by hanging drop method including screens from Emerald Biosystems, Hampton Research, and Qiagen. PduJ K25A (65.7 mg ml−1) crystallized in hexagonal plate crystals in multiple conditions. PduJ K25A crystals that were used for structure determination were optimized from 0.1 MES sodium salt pH 6.5, 2.0 M ammonium sulfate, 5% PEG 400 (w/v).
Before data collection, protein crystals were first cryoprotected with 35% glycerol to 65% well solution. An initial data set was collected in-house and the data were integrated, merged, and scaled using the XDS suite programs for crystallography. A molecular replacement solution was obtained with PHASER using PduA (PDB ID: 3NGK) as a search model. A higher resolution data set was later collected at the Advanced Photon Source, beamline 24-ID-C, at Argonne National Laboratories, IL. Refinement with the new data set began with rigid body refinement using coordinates from the previous molecular replacement.
Supplementary Material
Acknowledgements
The authors thank the Iowa State University microscopy and nano-imaging facility for help in electron microscopy. The authors also acknowledge the ISU DNA sequencing and synthesis facility and Protein facility for DNA sequence and LC- MS/MS analyses. The authors also thank Michael Collazo and Duilio Cascio for assistance in protein crystallization and data collection, and the synchrotron staff at APS beamline 24-ID-C. This work was supported by NIH grant R01AI081146 (TOY and TAB). Work in the Yeates lab was supported by the BER program of the DOE Office of Science. SC was supported by a CBI NIH training grant (T32-GM008496) and the by the UCLA Graduate Division. We thank M. Capel, K. Rajashankar, N. Sukumar, J. Schuermann, I. Kourinov, and F. Murphy (NECAT Beamline 24-ID-C at Advanced Photon Source, which is supported by National Center for Research Resources Grant 5P41RR015301-10 and National Institute of General Medical Sciences Grant 8 P41 GM103403-10 from the National Institutes of Health). Use of the Advanced Photon Source is supported by the Department of Energy under Contract DE-AC02-06CH11357
Footnotes
Accession Numbers
The coordinates of the refined models and the merged structure factors have been deposited in the Protein Data Bank with code 5D6V.
References
- Abdul-Rahman F, Petit E, Blanchard JL. The distribution of polyhedral bacterial microcompartments suggests frequent horizontal transfer and operon reassembly. J. Phylogen. Evolution. Biol. 2013;1 doi 10.4172/2329-9002.1000118. [Google Scholar]
- Berkowitz D, Hushon JM, Whitfield HJ, Jr., Roth J, Ames BN. Procedure for identifying nonsense mutations. J Bacteriol. 1968;96:215–220. doi: 10.1128/jb.96.1.215-220.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1, 2-propanediol degradation. J Bacteriol. 1999;181:5967–5975. doi: 10.1128/jb.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobik TA, Lehman BP, Yeates TO. Bacterial microcompartments: widespread prokaryotic organelles for isolation and optimization of metabolic pathways. Mol Microbiol. 2015;98:193–207. doi: 10.1111/mmi.13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai F, Sutter M, Bernstein SL, Kinney JN, Kerfeld CA. Engineering bacterial microcompartment shells: chimeric shell proteins and chimeric carboxysome shells. ACS Synth Biol. 2015;4:444–453. doi: 10.1021/sb500226j. [DOI] [PubMed] [Google Scholar]
- Cai F, Sutter M, Cameron JC, Stanley DN, Kinney JN, Kerfeld CA. The structure of CcmP, a tandem bacterial microcompartment domain protein from the b carboxysome, forms a subcompartment within a microcompartment. J Biol Chem. 2013;288:16055–16063. doi: 10.1074/jbc.M113.456897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA. Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell. 2013;155:1131–1140. doi: 10.1016/j.cell.2013.10.044. [DOI] [PubMed] [Google Scholar]
- Chang YY, Cronan JE., Jr. Mapping nonselectable genes of Escherichia coli by using transposon Tn10: location of a gene affecting pyruvate oxidase. J Bacteriol. 1982;151:1279–1289. doi: 10.1128/jb.151.3.1279-1289.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Liu Y, Crowley CS, Yeates TO, Bobik TA. Bacterial microcompartments: their properties and paradoxes. Bioessays. 2008;30:1084–1095. doi: 10.1002/bies.20830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Sinha S, Fan C, Liu Y, Bobik TA. Genetic analysis of the protein shell of the microcompartments involved in coenzyme B12-dependent 1,2-propanediol degradation by Salmonella. J Bacteriol. 2011;193:1385–1392. doi: 10.1128/JB.01473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury C, Chun S, Pang A, Sawaya MR, Sinha S, Yeates TO, Bobik TA. Selective molecular transport through the protein shell of a bacterial microcompartment organelle. Proc Natl Acad Sci U S A. 2015;112:2990–2995. doi: 10.1073/pnas.1423672112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury C, Sinha S, Chun S, Yeates TO, Bobik TA. Diverse bacterial microcompartment organelles. Microbiol Mol Biol Rev. 2014;78:438–468. doi: 10.1128/MMBR.00009-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cot SS, So AK, Espie GS. A multiprotein bicarbonate dehydration complex essential to carboxysome function in cyanobacteria. J Bacteriol. 2008;190:936–945. doi: 10.1128/JB.01283-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craciun S, Balskus EP. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci U S A. 2012;109:21307–21312. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley CS, Cascio D, Sawaya MR, Kopstein JS, Bobik TA, Yeates TO. Structural insight into the mechanisms of transport across the Salmonella enterica Pdu microcompartment shell. J Biol Chem. 2010;285:37838–37846. doi: 10.1074/jbc.M110.160580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley CS, Sawaya MR, Bobik TA, Yeates TO. Structure of the PduU shell protein from the Pdu microcompartment of Salmonella. Structure. 2008;16:1324–1332. doi: 10.1016/j.str.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbilgin O, McDonald KL, Kerfeld CA. Characterization of a planctomycetal organelle: a novel bacterial microcompartment for the aerobic degradation of plant saccharides. Appl Environ Microbiol. 2014;80:2193–2205. doi: 10.1128/AEM.03887-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Bobik TA. The N-terminal region of the medium subunit (PduD) packages adenosylcobalamin-dependent diol dehydratase (PduCDE) into the Pdu microcompartment. J Bacteriol. 2011;193:5623–5628. doi: 10.1128/JB.05661-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Cheng S, Liu Y, Escobar CM, Crowley CS, Jefferson RE, Yeates TO, Bobik TA. Short N-terminal sequences package proteins into bacterial microcompartments. Proc Natl Acad Sci U S A. 2010;107:7509–7514. doi: 10.1073/pnas.0913199107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Cheng S, Sinha S, Bobik TA. Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments. Proc Natl Acad Sci U S A. 2012;109:14995–15000. doi: 10.1073/pnas.1207516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havemann GD, Bobik TA. Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J Bacteriol. 2003;185:5086–5095. doi: 10.1128/JB.185.17.5086-5095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havemann GD, Sampson EM, Bobik TA. PduA is a shell protein of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar typhimurium LT2. J Bacteriol. 2002;184:1253–1261. doi: 10.1128/JB.184.5.1253-1261.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldt D, Frank S, Seyedarabi A, Ladikis D, Parsons JB, Warren MJ, Pickersgill RW. Structure of a trimeric bacterial microcompartment shell protein, EtuB, associated with ethanol utilization in Clostridium kluyveri. Biochem J. 2009;423:199–207. doi: 10.1042/BJ20090780. [DOI] [PubMed] [Google Scholar]
- Jorda J, Lopez D, Wheatley NM, Yeates TO. Using comparative genomics to uncover new kinds of protein-based metabolic organelles in bacteria. Protein Sci. 2013;22:179–195. doi: 10.1002/pro.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. Protein structures forming the shell of primitive bacterial organelles. Science. 2005;309:936–938. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- Kinney JN, Salmeen A, Cai F, Kerfeld CA. Elucidating essential role of conserved carboxysomal protein CcmN reveals common feature of bacterial microcompartment assembly. J Biol Chem. 2012;287:17729–17736. doi: 10.1074/jbc.M112.355305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, Kerfeld CA. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J Mol Biol. 2009;392:319–333. doi: 10.1016/j.jmb.2009.03.056. [DOI] [PubMed] [Google Scholar]
- Kuehl JV, Price MN, Ray J, Wetmore KM, Esquivel Z, Kazakov AE, Nguyen M, Kuehn R, Davis RW, Hazen TC, Arkin AP, Deutschbauer A. Functional genomics with a comprehensive library of transposon mutants for the sulfate-reducing bacterium Desulfovibrio alaskensis G20. MBio. 2014;5:e01041–01014. doi: 10.1128/mBio.01041-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang A, Liang M, Prentice MB, Pickersgill RW. Substrate channels revealed in the trimeric Lactobacillus reuteri bacterial microcompartment shell protein PduB. Acta Crystallogr D Biol Crystallogr. 2012;68:1642–1652. doi: 10.1107/S0907444912039315. [DOI] [PubMed] [Google Scholar]
- Pang A, Warren MJ, Pickersgill RW. Structure of PduT, a trimeric bacterial microcompartment protein with a 4Fe-4S cluster-binding site. Acta Crystallogr D Biol Crystallogr. 2011;67:91–96. doi: 10.1107/S0907444910050201. [DOI] [PubMed] [Google Scholar]
- Parsons JB, Frank S, Bhella D, Liang M, Prentice MB, Mulvihill DP, Warren MJ. Synthesis of empty bacterial microcompartments, directed organelle protein incorporation, and evidence of filament-associated organelle movement. Mol Cell. 2010;38:305–315. doi: 10.1016/j.molcel.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Penrod JT, Roth JR. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J Bacteriol. 2006;188:2865–2874. doi: 10.1128/JB.188.8.2865-2874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit E, LaTouf WG, Coppi MV, Warnick TA, Currie D, Romashko I, Deshpande S, Haas K, Alvelo-Maurosa JG, Wardman C, Schnell DJ, Leschine SB, Blanchard JL. Involvement of a bacterial microcompartment in the metabolism of fucose and rhamnose by Clostridium phytofermentans. PLoS One. 2013;8:e54337. doi: 10.1371/journal.pone.0054337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price-Carter M, Tingey J, Bobik TA, Roth JR. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar typhimurium on ethanolamine or 1,2-propanediol. J Bacteriol. 2001;183:2463–2475. doi: 10.1128/JB.183.8.2463-2475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Badger MR. Isolation and characterization of high CO2-requiring-mutants of the cyanobacterium Synechococcus PCC7942 : two phenotypes that accumulate inorganic carbon but are apparently unable to generate CO2 within the Carboxysome. Plant. Physiol. 1989;91:514–525. doi: 10.1104/pp.91.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae BD, Long BM, Badger MR, Price GD. Functions, compositions, and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol Mol Biol Rev. 2013;77:357–379. doi: 10.1128/MMBR.00061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagermann M, Ohtaki A, Nikolakakis K. Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment. Proc Natl Acad Sci U S A. 2009;106:8883–8887. doi: 10.1073/pnas.0902324106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson EM, Bobik TA. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J Bacteriol. 2008;190:2966–2971. doi: 10.1128/JB.01925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieger H. A method for detection of phage mutants with altered transducing ability. Mol Gen Genet. 1971;110:378–381. doi: 10.1007/BF00438281. [DOI] [PubMed] [Google Scholar]
- Scott KP, Martin JC, Campbell G, Mayer CD, Flint HJ. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium “Roseburia inulinivorans”. J Bacteriol. 2006;188:4340–4349. doi: 10.1128/JB.00137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf H, Fricke WF, Veith B, Bruggemann H, Liesegang H, Strittmatter A, Miethke M, Buckel W, Hinderberger J, Li F, Hagemeier C, Thauer RK, Gottschalk G. The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc Natl Acad Sci U S A. 2008;105:2128–2133. doi: 10.1073/pnas.0711093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- Shieh YW, Minguez P, Bork P, Auburger JJ, Guilbride DL, Kramer G, Bukau B. Operon structure and cotranslational subunit association direct protein assembly in bacteria. Science. 2015 doi: 10.1126/science.aac8171. [DOI] [PubMed] [Google Scholar]
- Sinha S, Cheng S, Fan C, Bobik TA. The PduM protein is a structural component of the microcompartments involved in coenzyme B12-dependent 1,2-propanediol degradation by Salmonella enterica. J Bacteriol. 2012;194:1912–1918. doi: 10.1128/JB.06529-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Cheng S, Sung YW, McNamara DE, Sawaya MR, Yeates TO, Bobik TA. Alanine scanning mutagenesis identifies an asparagine-arginine-lysine triad essential to assembly of the shell of the Pdu microcompartment. J Mol Biol. 2014;426:2328–2345. doi: 10.1016/j.jmb.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO. Atomic-level models of the bacterial carboxysome shell. Science. 2008;319:1083–1086. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Sawaya MR, Phillips M, Yeates TO. Insights from multiple structures of the shell proteins from the b-carboxysome. Protein Sci. 2009;18:108–120. doi: 10.1002/pro.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Sawaya MR, Yeates TO. Structure and mechanisms of a protein-based organelle in Escherichia coli. Science. 2010;327:81–84. doi: 10.1126/science.1179513. [DOI] [PubMed] [Google Scholar]
- Thompson MC, Cascio D, Leibly DJ, Yeates TO. An allosteric model for control of pore opening by substrate binding in the EutL microcompartment shell protein. Protein Sci. 2015;24:956–975. doi: 10.1002/pro.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren JW, Walker JR, Roth JR, Altman E. Construction and characterization of a highly regulable expression vector, pLAC11, and its multipurpose derivatives, pLAC22 and pLAC33. Plasmid. 2000;44:138–151. doi: 10.1006/plas.2000.1477. [DOI] [PubMed] [Google Scholar]
- Wheatley NM, Gidaniyan SD, Liu Y, Cascio D, Yeates TO. Bacterial microcompartment shells of diverse functional types possess pentameric vertex proteins. Protein Sci. 2013;22:660–665. doi: 10.1002/pro.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates TO, Jorda J, Bobik TA. The shells of BMC-type microcompartment organelles in bacteria. J Mol Microbiol Biotechnol. 2013;23:290–299. doi: 10.1159/000351347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.