Abstract
The patient medical record contains a wealth of information consisting of prior observations, interpretations, and interventions that need to be interpreted and applied towards decisions regarding current patient care. Given the time constraints and the large—often extraneous—amount of data available, clinicians are tasked with the challenge of performing a comprehensive review of how a disease progresses in individual patients. To facilitate this process, we demonstrate a neuro-oncology workstation that assists in structuring and visualizing medical data to promote an evidence-based approach for understanding a patient’s record. The workstation consists of three components: 1) a structuring tool that incorporates natural language processing to assist with the extraction of problems, findings, and attributes for structuring observations, events, and inferences stated within medical reports; 2) a data modeling tool that provides a comprehensive and consistent representation of concepts for the disease-specific domain; and 3) a visual workbench for visualizing, navigating, and querying the structured data to enable retrieval of relevant portions of the patient record. We discuss this workstation in the context of reviewing cases of glioblastoma multiforme patients.
General Terms: Management, Documentation, Design, Standardization
1. INTRODUCTION
The patient record is an amalgamation of a variety of reports written by different authors in multiple departments and institutions that capture descriptions of prior observations, interpretations, and interventions. An appropriate review of a patient’s medical record often requires that a physician sift through multiple clinical documents while mentally noting issues related to what the findings were, the chronology of events, spatial/temporal patterns of disease progression, the effects of interventions, and the possible causal lines of explanation of observed findings. The physician also needs to filter out those pieces of information not related to the current clinical context of care. Given the time constraints, data complexity, and data volume associated with chronic patient cases as in oncology, a complete review of a patient’s chart is in reality rarely performed. The inability to perform a thorough review may negatively impact patient care in several ways: lack of coordination among caregivers, poor integration of examination results for diagnosis, initiation of treatments before a definitive diagnosis, administration of inappropriate therapies, and/or the performance of redundant and unnecessary studies [1]. In current practice, source medical reports are delivered directly to caregivers, placing the burden on physicians to extract relevant information and to create a mental information model for each patient. This task becomes increasingly difficult for complex cases. In other investigative disciplines such as law, for example, a large amount of effort is put into systematically conditioning, procuring, and linking of data items in some logical way before critical judgments are made. In medicine, much of this process is tasked to the individual physician. However, like a lawyer who has only a limited amount of time to present his case, a physician only has a limited time to digest all the implications of various forms of medical evidence. Clearly, a new paradigm for management of patient medical data is needed.
Our solution to this challenge is to develop an integrated neuro-oncology workstation that simplifies the process of structuring patient records to improve the retrieval and presentation of this data to clinicians and to enable the creation of knowledge repositories that capture the characteristics of diseases through routine observational data. The workstation consists of three components: 1) a structuring tool that incorporates a natural language processing (NLP) toolkit; 2) a data modeling tool that captures the concepts and attributes that are important for a given domain; and 3) a visualization workbench that summarizes the structured information and facilitates exploration of patient data. The workstation is designed to enable both clinicians and knowledge workers with appropriate domain knowledge to easily structure patient records. It facilitates the comprehensive and consistent representation of concepts and their attributes (e.g., spatial, temporal, existential) for a specific disease. We describe this system in the context of structuring patient cases with an aggressive form of primary brain cancer called glioblastoma multiforme (GBM).
2. BACKGROUND
A summarization and organizational framework for medical records was first posited by Weed [2] through his notion of the Problem-Oriented Medical Record (POMR). The difficulty of adopting such a paradigm is two-fold: 1) the task of data entry is both time-consuming and manually-intensive; and 2) the representation of information is ad hoc and not standardized. Ultimately, the final presentation of a patient’s record should maximize the cognitive understanding and reasoning capabilities of a physician for a particular task while minimizing user effort and fatigue. In this section, we review some of the efforts towards addressing challenges in structuring, modeling, and visualizing information in clinical reports.
2.1.1 Structuring medical reports
Structured reporting systems have attempted to address various challenges in the areas of data entry, information content, representation, and user interface issues. They have been demonstrated in limited areas such as radiology, pathology and progress notes [3–5]. The advantages of structured reporting include: 1) consistent coverage, as a form-based system can help enforce physician reporting consistency; 2) decision support, as forms provide important reminders of possible states/interpretations for a finding; and 3) compliance, as the entered data can be immediately checked to see whether they meet regulatory/accreditation standards and institutional requirements. Health Level Seven (HL7) has established a workgroup with the mission of creating and promoting the development document templates. Despite these important benefits, widespread adoption has yet to occur in part due to: 1) reporting bias (e.g., pigeonholing a diagnosis or description into an available selection choice); 2) lack of expressibility; 3) reporting neglect (e.g., a tendency not to report unusual conditions); 4) labor-intensive and time-consuming tasks (e.g., hierarchical menus); and 5) difficulty with inputting updates (e.g., introduction of new vocabularies). One approach to overcoming these challenges has been to leverage NLP to automate the process of structuring existing medical record narratives. The main advantage of an NLP-based approach is it does not change the modus operandi for reporting. This allows the physician to dictate a report normally: users are not restricted in their use of vocabulary or grammatical style. A computer program automatically interprets the physician’s free-text descriptions and formulates the information into knowledge structures (e.g., logic-based frames). A comprehensive review of NLP systems is provided in [6].
2.1.2 Modeling and visualization
While automating the process of structuring patient records is important, the full utility of extracting and characterizing patient data is not achieved until the structured data is represented and visualized in a way that facilitates understanding of the importance and trends in the data. The goal of modeling and visualization is to tailor the presentation of the patient record for a specific medical task (e.g., monitoring of tumor burden, effect of anatomical abnormality on physiologic function) given domain knowledge provided by a data model. In this work, extracted concepts and attributes are organized around four fundamental aspects of any disease phenomenon: 1) anatomic (spatial) properties, 2) temporal trends, 3) existence, and 4) causality. Anatomic descriptions of findings are important to disease understanding. Location of a finding helps explain an observation and can provide a common reference frame for facilitating spatial inferencing related to patient outcomes [7]. Time is a basic dimension of nature; the manner in which temporal aspects related to events, object states, interventional actions, and natural processes are modeled influence how we reason causally about a phenomenon. [8] and [9] discuss past efforts for representing and visualizing temporal data in medical records, respectively. Existence characterizes the belief of whether a problem or finding is present and how it changes over time; it is a widely explored problem in the NLP resulting in algorithms such as ConText [10]. The notion of causality is inherently intertwined with clinical medicine. decisions on treatment are driven by causal considerations: symptoms are observed due to an underlying cause. While theoretical constructs have been developed to generate causal models [11], they have yet to be practically demonstrated in the medical domain. Having a curated repository of structured patient data would be an initial step towards identifying causal relationships among a large population of patients with GBM.
3. SYSTEM FRAMEWORK
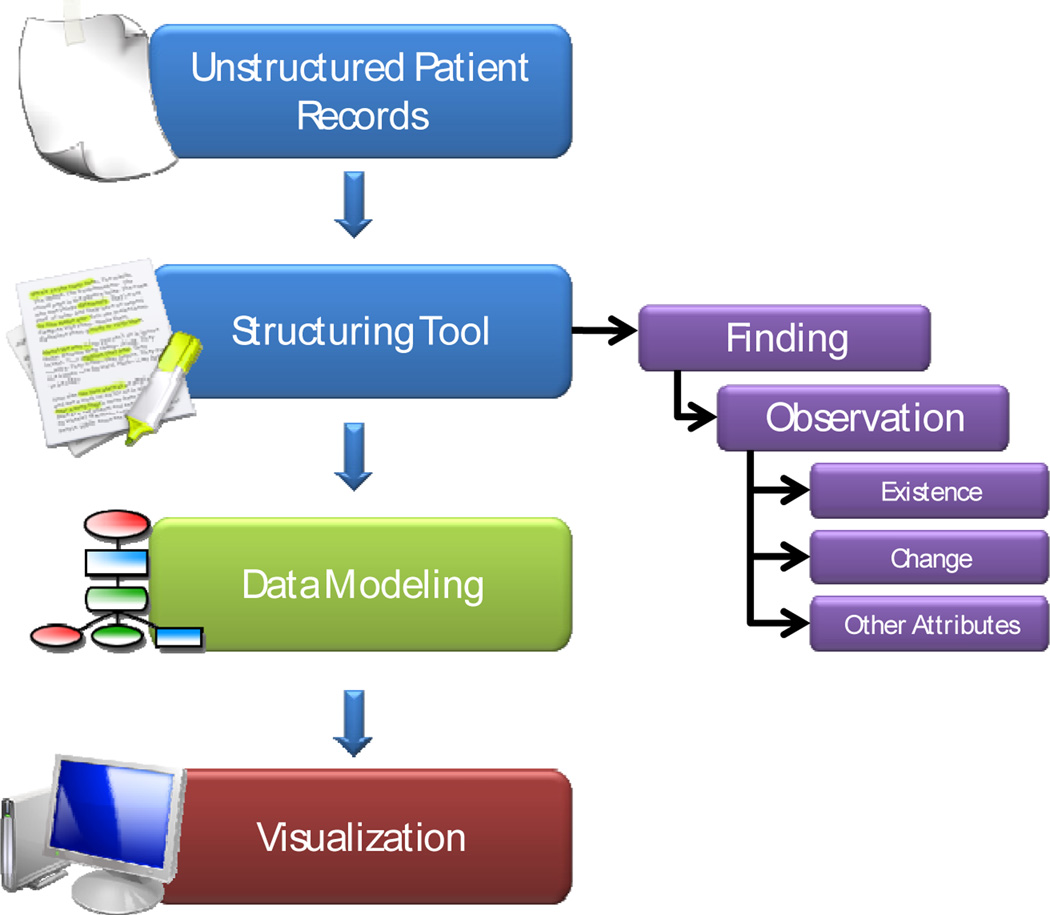
The information flow of the neuro-oncology workstation is illustrated in Fig. 1.
Figure 1.
Flow diagram of how unstructured patient records are processed in the neuro-oncology workstation.
3.1 Structuring Tool
The structuring tool guides the user who is responsible for abstracting relevant problems, findings, and attributes from the patient record to structure its contents for a given disease with as little user effort as possible. The system stores intermediate results, recommends choices, and provides useful context relevant to a specific structuring task. The process proceeds as follows:
Step 1: Once the user selects a patient case, the system provides a listing of all patient reports.
Step 2: When the user selects a report from the report list, the structuring tool automatically identifies all findings/problems and references to anatomical location mentioned within the report and displays a highlighted version of the document. We leverage an existing natural language processing (NLP) toolkit to classify a concept as being a problem, finding, or location [12]. A list of finding concepts is presented to the user. When a particular finding concept is selected, possible anatomic locations are listed. The user identifies the appropriate location for the selected finding and the spatial relationship that characterizes the location description (e.g., “in”, “adjacent to”, “extending to”, “at junction of”). As the user performs this task, a hierarchical list is dynamically generated summarizing the mentioned findings and their associated location relations within the report.
Step 3: For each instance of a finding/problem, the user then specifies whether that occurrence is the first mention of the finding/problem or has already been previously mentioned in the selected document. Findings may be assigned one of several co-reference types of relations including: 1) identical-equivalence relation; 2) part-whole or spatially encapsulated relationship (e.g., necrosis within a mass); 3) causal or associative relations (e.g., T1 hyperintensity is consistent with acute hemorrhage); 4) ellipses – null references through words like ‘appearance’ (e.g., tumor … the appearance is compatible with …); 5) general-specific references (mass effect, … focal effacement); 6) large to small scale references – references of radiologic use of ‘tumor’ (as entire collection, e.g., mass) versus pathology use of ‘tumor’ (as cell); and 7) group–to–group member relations (e.g., multiple nodules are seen, the largest is 4cm).
Step 4: For each problem/finding, the system presents the specific object frame that allows precise characterization of the important attributes and allowed values for the finding. This frame is generated using a domain-specific model created using the modeling tool described in Section 3.2.
Step 5: For each instantiated finding, the user is asked to characterize attributes related to existence of the finding. These attributes include: 1) how the finding was observed (e.g., direct observation versus inference); 2) certainty of the finding's existence (e.g., definite, appears to be, less likely); 3) relevance of the finding to the disease (e.g., significant, incidental); 4) visibility (e.g., clearly seen); 5) study quality (e.g., acceptable); 6) newness (e.g., previously seen, newly diagnosed); and 7) multiplicity (e.g., single, multiple).
Step 6: As the user works through each report in the record, the information is encoded using an extensible markup language (XML) schema to represent the information captured by the data structuring process. For a given report, this representation maintains a list of problems and all occurrences related to the problem.
Step 7: Once the user has gone through each report, the final step involves linking descriptions of finding across reports so that each individual finding can be represented with respect to time. The user first selects a document that has been previously structured. The task for the user is to link findings mentioned in prior reports to current findings. The system maintains a master problem list and presents the user with all occurrences of the problem over all previously linked studies. This information is used by the abstractor to decide if a current finding is associated (e.g., identical to) with a prior modeled findings allowing findings to be characterized over time.
Step 8: The complete patient model is then encoded in XML; this file can then be used to populate a database or drive the visualization described in Section 3.3.
3.2 Data Modeling
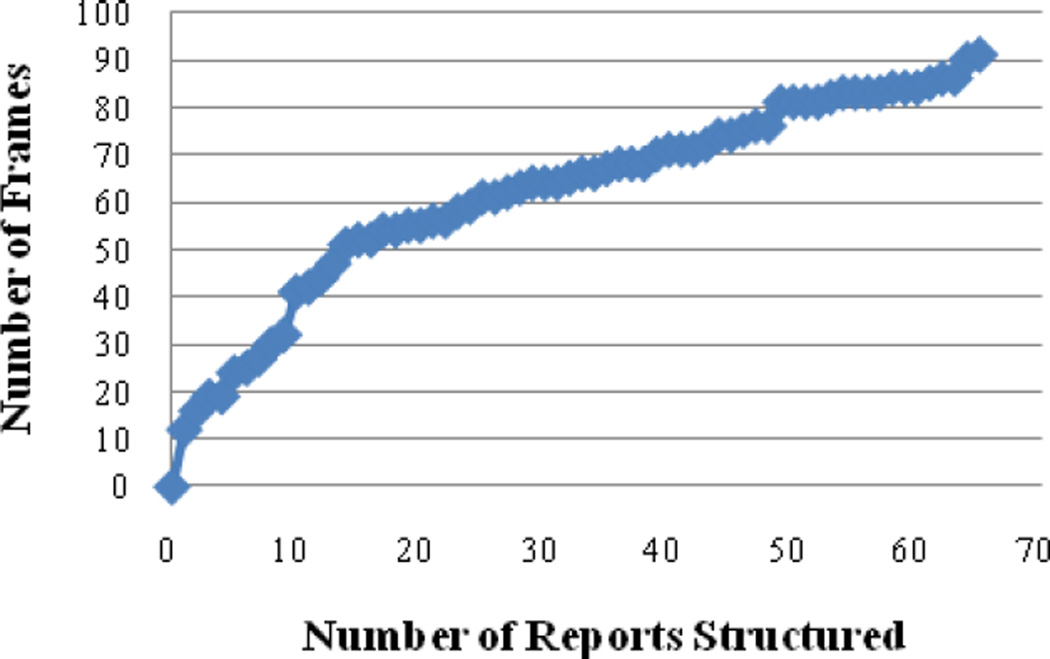
The data modeling tool provides a method for defining a domain by enumerating all possible concepts related to a disease. Given our aim to support brain cancer management, our data model contains concepts related to findings, signs and symptoms, treatments, and medical characteristics of brain tumors. Using the tool, the user can create a global model of the information space spanning a medical record and formally organizing the model to support disease-centric reasoning at the patient and population levels. The current focus of the tool is to identify and model objects, relations, attributes, values, processes that are specific to neuro-oncology. The situational ontology provides a consistent and comprehensive representation and is in a form that is useable for either disease modeling, or case visualization. The final representation is implemented as a collection of linked informational templates similar to efforts ongoing in structured reporting (e.g., HL7 Reference Information Model). Each finding type (e.g., mass, edema, hemorrhage, etc.) is modeled by a frame specifying their own particular characterizing attributes and allowed values. The majority of the frames could be expressed in terms of Boolean propositions. Some exceptions include size measurements and drug names. Fig. 2 shows a plot of the number of reports analyzed versus the number of new modeling elements. Currently, about 91 finding frames (229 properties, 813 possible values) for GBM have been defined. The average time (and standard deviation) for structuring a report with the system is 876.2 seconds (+/− 311 seconds). The average size of the reports is 2816 characters (+/− 1043). Development of the frames associated with each specific medical entity (findings, problems, prescribed drugs) proceeds using the general principles of ontology engineering (e.g., Helixz-Spindle Model). The tool is designed to allow an iterative (building/testing) and incremental approach. We have chosen a "bottom-up" approach to create the model for neuro-oncology by defining concepts from two sources: 1) individual patient cases, in which findings and attributes encountered in GBM cases are characterized and recorded; or 2) controlled vocabularies of related efforts such as the National Cancer Institute’s (NCI) Visually AcesSAble Rembrandt Images (VASARI) and cancer Biomedical Informatics Grid (caBIG) efforts. Anatomical locations of the brain are categorized according to a fixed number of brain structures defined by a neuro-oncologist.
Figure 2.
Rate frames added to the data model as additional reports are structured.
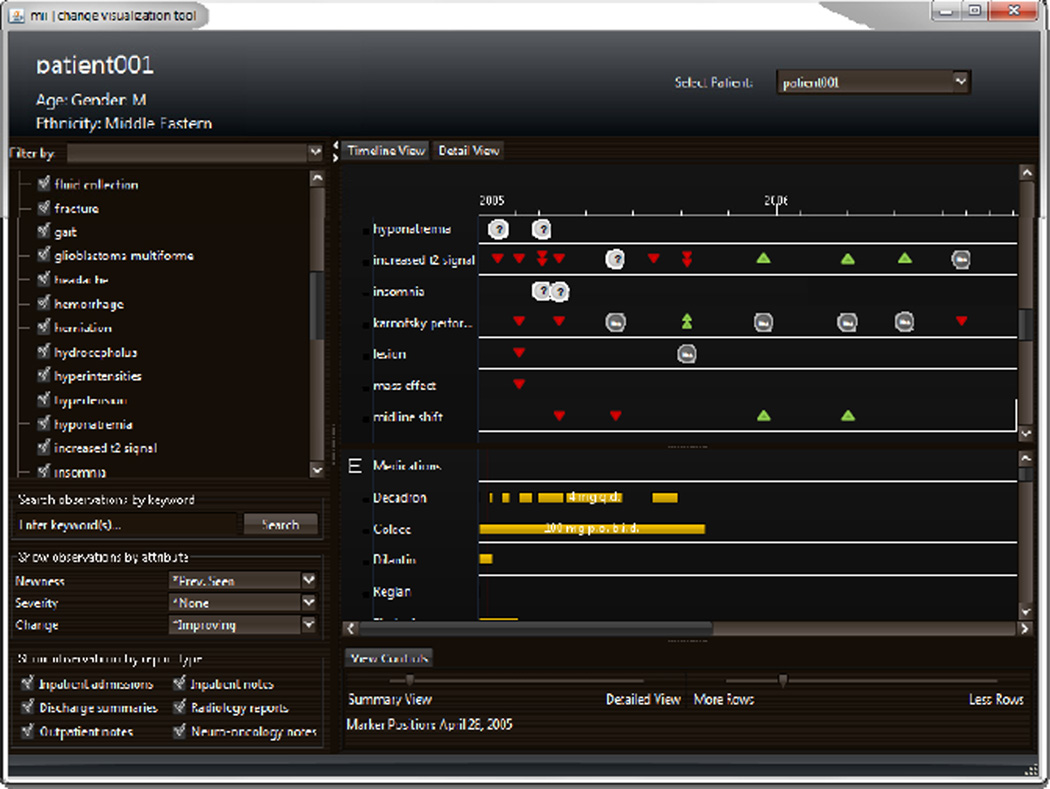
3.3 Visualization
Once a patient record has been fully structured, the information is used to generate an integrated visualization that facilitates review of and interaction with the data. We have created a workstation that visually summarizes the content of a patient record across time; a screenshot is shown in Fig. 3. The interface consists of four components: a master problem list, a query panel, a timeline, and a detail panel. The master problem list (Fig. 3, top left) serves as the primary method of navigating the patient record: users select which problems to display in the timeline. The query panel (Fig. 3, bottom left) provides additional options for customizing the display. The timeline (Fig. 3, right) visually summarizes each observation and any changes in comparison to previous observations. Additional timelines can be added to present other treatment information (e.g., medication dosages, durations), facilitating visual comparison between problems and interventions. When a user selects a specific time point for an observed problem, a detail panel appears, showing NLP-extracted information in a table with observations immediately preceding and following the selected time point. Key image slices identified from the imaging study are also displayed alongside the extracted attributes.
Figure 3.
A screen shot of the visualization that leverages the output of the structuring and data modeling tools to generate a summarization of problems, findings, and attributes documented in a record.
As part of the workstation, we have also implemented two methods to perform case-based retrieval, finding other patients cases with similar characteristics. The first method uses the latent Dirichlet allocation (LDA) statistical approach to analyze the entirety of reports for a number of patient cases and discover a topic model from the words [13]. Documents are modeled as mixtures of latent topics that indicate thematic words. Before documents are fit to the LDA model, words that do not appear in SNOMED CT are discarded to include only medically relevant concepts. To make comparisons across patients, document topic distributions are compared using the symmetric Kullback-Leibler (KL) divergence. These divergences may then be ordered to provide a ranked list of similar patients. The second method uses a Bayesian belief network (BBN) that has been parameterized using the structured patient data to generate a unique probability distribution for each case; cases are then ranked using KL divergence. We are exploring how the overall retrieval performance can be improved by weighting the LDA-based rankings with statistics from the BBN.
4. DISCUSSION
We have described the preliminary version of our neuro-oncology workstation that structures, represents, and visualizes data in a patient record. As part of future work, we intend to broaden our data model to include features characterizing other brain tumors. We have created tools that allow users to easily add frames to the neuro-oncology data model as part of the structuring process. We are improving the abilities of the underlying NLP toolkit to perform tasks such as automatically identifying and extracting findings and attributes related to brain tumors across multiple patients This feature would assist in the creation of a curated research database that would be useful for developing more robust BBN models of disease. We also intend to conduct various evaluations to gauge the efficacy of our tools. A quantitative assessment of the NLP system will be conducted to determine how well the system can automatically instantiate structured data collection forms based on our targeted neuro-oncology ontology. For clinical efficacy testing, we aim to structure 60 patient cases and ask expert physicians to answer various relevant queries from one of two systems: the proposed system (e.g., intervention) versus current workstations that exist in the clinic. Metrics to be measured include the time physicians require to answer the queries, the accuracy of their answers, and subjective opinions on interface design and usability.
Acknowledgments
The authors express gratitude to Dr. Timothy Cloughesy for access to de-identified patient cases. We would like to acknowledge Drs Hooshang Kangarloo, Denise Aberle, Suzie El-Saden, and Alex Bui for their input. We would also like to thank Lewellyn Andrada and Albert Chern for their efforts in implementing parts of this workstation. This work is supported by the National Library of Medicine grant R01-LM009961.
Contributor Information
William Hsu, Email: willhsu@mii.ucla.edu, UCLA Dept of Radiology, Los Angeles, CA.
Corey W. Arnold, Email: cwarnold@mii.ucla.edu, UCLA Dept of Radiology, Los Angeles, CA.
Ricky K. Taira, Email: rtaira@mii.ucla.edu, UCLA Dept of Radiology, Los Angeles, CA.
REFERENCE
- 1.Fletcher RH, O'Malley MS, Fletcher SW, Earp JA, Alexander JP. Measuring the continuity and coordination of medical care in a system involving multiple providers. Med Care. 1984 May;22(5):403–411. doi: 10.1097/00005650-198405000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Weed LL. Medical records that guide and teach. N Engl J Med. 1968 Mar 21;278(12):652–657. doi: 10.1056/NEJM196803212781204. [DOI] [PubMed] [Google Scholar]
- 3.Hussein R, Engelmann U, Schroeter A, Meinzer H. DICOM structured reporting. Radiographics. 2004;24(3):897–909. doi: 10.1148/rg.243035722. [DOI] [PubMed] [Google Scholar]
- 4.Kuhn K, Gaus W, Wechsler J, Janowitz P, Tudyka J, Kratzer W, Swobodnik W, Ditschuneit H. Structured reporting of medical findings: evaluation of a system in gastroenterology. Methods Inf Med. 1992;31(4):268–274. [PubMed] [Google Scholar]
- 5.Weir C, Hurdle J, Felgar M, Hoffman J, Roth B, Nebeker J. Direct text entry in electronic progress notes. Methods Inf Med. 2003;42(1):61–67. [PubMed] [Google Scholar]
- 6.Friedman C, Hripcsak G. Natural language processing and its future in medicine. Acad Med. 1999;74(8):890–895. doi: 10.1097/00001888-199908000-00012. [DOI] [PubMed] [Google Scholar]
- 7.Ogunyemi O. Methods for reasoning from geometry about anatomic structures injured by penetrating trauma. J Biomed Inform. 2006;39(4):389–400. doi: 10.1016/j.jbi.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou L, Hripcsak G. Temporal reasoning with medical data--a review with emphasis on medical natural language processing. J Biomed Inform. 2007;40(2):183–202. doi: 10.1016/j.jbi.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plaisant C, Milash B, Rose A, Widoff S, Shneiderman B. LifeLines: visualizing personal histories. New York, NY, USA, City: ACM; 1996. [Google Scholar]
- 10.Harkema H, Dowling J, Thornblade T, Chapman W. ConText: An algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform. 2009;42(5):839–851. doi: 10.1016/j.jbi.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearl J. Causality: models, reasoning, and inference. Cambridge Univ Press; 2000. [Google Scholar]
- 12.Taira R, Bashyam V, Kangarloo H. A field theoretical approach to medical natural language processing. IEEE Trans Inf Technol Biomed. 2007;11(4):364–375. doi: 10.1109/titb.2006.884368. [DOI] [PubMed] [Google Scholar]
- 13.Blei D, Ng A, Jordan M. Latent dirichlet allocation. JMLR. 2003;3:993–1022. [Google Scholar]