Abstract
Influenza B viruses (IBVs) cause annual outbreaks of respiratory illness in humans and are increasingly recognized as a major cause of influenza-associated morbidity and mortality. Studying influenza viruses requires the use of secondary methodologies to identify virus-infected cells. To this end, replication-competent influenza A viruses (IAVs) expressing easily traceable fluorescent proteins have been recently developed. In contrast, similar approaches for IBV are mostly lacking. In this report, we describe the generation and characterization of replication-competent influenza B/Brisbane/60/2008 viruses expressing fluorescent mCherry or GFP fused to the C-terminal of the viral non-structural 1 (NS1) protein. Fluorescent-expressing IBVs display similar growth kinetics and plaque phenotype to wild-type IBV, while fluorescent protein expression allows for the easy identification of virus-infected cells. Without the need of secondary approaches to monitor viral infection, fluorescent-expressing IBVs represent an ideal approach to study the biology of IBV and an excellent platform for the rapid identification and characterization of antiviral therapeutics or neutralizing antibodies using high-throughput screening approaches. Lastly, fluorescent-expressing IBVs can be combined with the recently described reporter-expressing IAVs for the identification of novel therapeutics to combat these two important human respiratory pathogens.
Keywords: Influenza B virus, mCherry, GFP, Neutralizing antibodies, Antivirals, Fluorescent-based Microneutralization, assays
1. Introduction
Influenza viruses belong to the family Orthomyxoviridae and are divided into types A, B, and C (Palese and Shaw, 2007). Despite the use of vaccines, type A and B influenza viruses (IAVs and IBVs, respectively) infect humans regularly and are responsible of yearly seasonal epidemics associated with significant public health and economic consequences (Molinari et al., 2007). It is estimated that, each year, thousands of people worldwide contract influenza and develop acute respiratory infection with significant morbidity and mortality (Luckhaupt et al., 2012; Molinari et al., 2007; Thompson et al., 2003). IAVs are further classified into different subtypes based on the antigenic major surface glycoproteins: hemagglutinin (HA; 18 subtypes) and neuraminidase (NA; 11 subtypes) (Palese and Shaw, 2007; Tong et al., 2012; Tong et al., 2013). Unlike IAVs, IBVs are not divided into antigenically distinct subtypes, although since the 1980s two lineages diverged from the ancestral influenza virus B/Lee/1940 strain and have been co-circulating in the human population (Chen and Holmes, 2008; McCullers et al., 2004). Currently, only H3N2 and H1N1 IAV subtypes and IBVs circulate in humans (Shaw and Palese, 2013; Tong et al., 2012; Tong et al., 2013). IAVs and IBVs follow a rather diffuse cyclical epidemic pattern based on prevalence, typical once every 3 years (Hite et al., 2007; Li et al., 2008; Lin et al., 2004; Olson et al., 2007). IBV epidemics tend to be less severe than H3N2 IAVs but more severe than H1N1 AIVs in adults and the elderly (Ohmit and Monto, 1995; Olson et al., 2007; Thompson et al., 2003; Van Voris et al., 1982) However, IBV infections are associated with excess morbidity and mortality in the pediatric population (Belshe, 2010; Hite et al., 2007; Li et al., 2008; Olson et al., 2007). Contrary to IAVs, which has a broad host reservoir in many avian and mammalian species, IBVs are mainly restricted to humans (Wright et al., 2007), although occasional infections of seals have been documented (Osterhaus et al., 2000).
Vaccines and antivirals are available to combat influenza viruses (Baker et al., 2015b; Burnham et al., 2013; Jackson et al., 2011a; Krammer et al., 2015; Nguyen et al., 2010; Seibert et al., 2010). Historically, influenza vaccines contain viral antigens corresponding to the prevalent H3N2 and H1N1 IAVs as well as and one lineage of influenza type B (Victoria or Yamagata) (Belshe et al., 2007; Yang, 2013). More recently and due to increasing co-circulation of both IBV lineages with significant antigenic divergence, the Advisory Committee on Immunization Practices (ACIP) recommended that influenza vaccines should be available in quadrivalent formulations (Grohskopf et al., 2014; Sun, 2012). With respect to antivirals, there are four classes of FDA-approved drugs for use against influenza infections. Rimantadine and amantadine that target the viral matrix 2 (M2) ion channel and inhibit viral entry (Hay et al., 1985) but are not effective against the M2 protein of IBVs (Beigel and Bray, 2008). Zanamivir and oseltamivir target the sialidase activity of the influenza virus NA and inhibit virus release and are effective against both IAVs and IBVs (Jackson et al., 2011b). Low clinical effectiveness of NA inhibitors against IBVs in children and the emergence of drug resistant variants during treatment has been reported (Burnham et al., 2013; Farrukee et al., 2013).
Plasmid-based reverse genetics to generate recombinant influenza viruses (Fodor et al., 1999; Martinez-Sobrido and Garcia-Sastre, 2010; Neumann et al., 1999) has significantly contributed to a better understanding of the biology of these important human respiratory pathogens (Engelhardt, 2013; Jackson et al., 2011a), for the identification and characterization of antivirals (Baker et al., 2014; Ozawa and Kawaoka, 2011; Roberts et al., 2015), and for the development of alternative influenza vaccines (Baker et al., 2015a; Martinez-Sobrido and Garcia-Sastre, 2010; Nogales et al., 2014a; Subbarao and Katz, 2004). Reverse genetics have allowed for the generation of replication-competent IAVs expressing reporter genes as novel powerful tools to track viral infections without the requirement of secondary methodologies to detect viral-infected cells (Eckert et al., 2014; Fiege and Langlois, 2015; Fukuyama et al., 2015; Kittel et al., 2004; Manicassamy et al., 2010; Nogales et al., 2014a; Pan et al., 2013; Perez et al., 2013; Reuther et al., 2015; Tran et al., 2013). In contrast, similar approaches have not been implemented to the same extend for IBV. Here we describe and characterize recombinant IBVs based on the B/Brisbane/60/2008 (Victoria lineage) strain, where fluorescent proteins (mCherry or GFP) were fused to the C-terminal end of the viral NS1 (Nogales et al., 2014a). Fluorescent-expressing IBVs have similar growth kinetics and plaque phenotype than the respective wild-type (WT) counterpart. Importantly, IBV infections were easily tracked in real-time using fluorescence microscopy and conveniently quantified using a fluorescent plate reader. By eliminating the use of secondary approaches to monitor IBV infections, these replication-competent, fluorescent-expressing IBVs represent a promising option to study the biology of IBV and an excellent platform to easily identify and evaluate antivirals or neutralizing antibodies (NAbs) using high-throughput screening (HTS) approaches, both currently needed to combat this important human respiratory pathogen.
2. Materials and methods
2.1. Cell lines
Human embryonic kidney 293T (ATCC CRL-11268) and canine Madin-Darby canine kidney (MDCK; ATCC CCL-34) cells were grown at 37 °C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), and 1% PSG (penicillin, 100 units/ml; streptomycin 100 μg/ml; lglutamine, 2 mM) (Nogales et al., 2014b).
2.2. Construction of the NS plasmids
To engineer an IBV NS segment where the C-terminal of NS1 is fused to a fluorescent mCherry or codon-optimized maxGFP (GFP; Amaxa), a recombinant NS segment was synthesized de novo (Biomatik) with appropriate restriction sites for subcloning into the ambisense plasmid pDP-2002 (Pena et al., 2013). This plasmid, named pDP-NS-2xBsmBI, contained the NS1 open reading frame (ORF), without the stop codon or splice acceptor site, and two BsmBI sites in opposite orientation followed by the porcine teschovirus-1 (PTV-1) 2A autoproteolytic cleavage site (ATNFSLLKQAGDVEENPGP) and the entire sequence of the nuclear export protein, NEP (Nogales et al., 2014a). The mCherry and GFP ORFs were amplified by PCR using oligonucleotides designed to introduce complementary BsmBI sites, and then cloned into the pDP-NS-2xBsmBI to generate the pDP-NS-mCherry and pDP-NS-GFP plasmids for IBV rescues. Oligonucleotides for the cloning of mCherry and GFP into the pDP-NS-2xBsmBI plasmid are available upon request. Plasmid constructs were confirmed by sequencing (ACGT, Inc.).
2.3. Rescue of recombinant fluorescent-expressing viruses and viral infections
Ambisense pDP-2002 plasmids were used for the rescue of WT and reporter-expressing B/Brisbane/60/2008 as previously described (Baker et al., 2014; Pena et al., 2013). Briefly, co-cultures (1:1) of 293T/MDCK cells (6-well plate format, 106 cells/well) were co-transfected in suspension with the eight ambisense pDP-2002-PB2, -PB1, -PA, -HA, -NP, -NA, -M and WT NS, NS-mCherry, or NS-GFP plasmids. Clonal WT, mCherry- or GFP-expressing IBVs were selected by plaque assay and the virus stocks were propagated in MDCK cells at 33 °C in a 5% CO2 atmosphere for 3-4 days. Influenza A/California/04/2009H1N1 (pH1N1) WT and mCherry viruses have been previously described (Baker et al., 2013; Nogales et al., 2014a). For infections, virus stocks were diluted in phosphate buffered saline (PBS) supplemented with 0.3% bovine albumin (BA) and 1% PS (PBS/BA/PS). After viral infections, cells were maintained in DMEM supplemented with 0.3% BA, 1% PSG, and 1 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Sigma) (Martinez-Sobrido and Garcia-Sastre, 2010). Virus titers were determined by standard plaque assay (plaque forming units (PFU)/ml) in MDCK cells (Nogales et al., 2014b).
2.4. Virus growth kinetics and titrations
Multicycle virus growth kinetics were performed in MDCK cells (12-well plate format, 5 × 105 cells/well) infected with the indicated viruses (triplicates) at multiplicity of infection (MOI) of 0.001. Virus titers in the tissue culture supernatants were determined by immunofocus assay (fluorescent focus-forming units, FFU/ml) (Nogales et al., 2014b) using mouse monoclonal antibodies (MAbs) against IAV (HT103) (O'Neill et al., 1998) or IBV (B017; Abcam) NP, and a fluorescein isothiocyanate (FITC)-conjugated anti-mouse secondary antibody (Dako). Mean value and standard deviation (SD) were calculated using Microsoft Excel software.
2.5. Protein gel electrophoresis and Western blots
Cell extracts from either mock or virus-infected (MOI 1.0) MDCK cells (6-well plate format, 106 cells/well) were lysed at 18 h post-infection (hpi) in passive lysis buffer (Promega) and separated by denaturing electrophoresis as previously described (Nogales et al., 2014b). Membranes were blocked for 1 h with 5% dried skim milk in PBS containing 0.1% Tween 20 (T-PBS) and incubated overnight at 4 °C with the indicated primary monoclonal or polyclonal (PAb) antibodies against IBV NP (mouse MAb B017; Abcam), mCherry (rabbit PAb; Raybiotech), IBV NS1 (rabbit PAb; kindly provided by Dr. T. Wolff) (Dauber et al., 2006) or actin (mouse MAb; Sigma). Bound primary antibodies were detected with horseradish peroxidase (HRP)-conjugated anti-mouse or anti-rabbit antibodies (GE-Healthcare). Proteins were detected by chemiluminescence (Hyglo, Dennville Scientific), according to the manufacturer's recommendations.
2.6. RT-PCR
Total RNA from mock or virus-infected (MOI 1.0) MDCK cells was collected at 18 hpi and purified using TRIzol reagent (Invitrogen) according to the manufacturer's specifications. The cDNAs were synthesized using SuperScript® II Reverse Transcriptase (Invitrogen) and an oligo-dT primer to amplify total mRNAs (mCherry and GAPDH); or specific primers for the NS and NP vRNAs. The cDNAs were used as templates for semi-quantitative PCR with primers specific for the NS and the NP vRNAs and for the mCherry and canine GAPDH mRNAs (available upon request).
2.7. Plaque assays
Confluent monolayers of MDCK cells (6-well plate format, 106 cells/well) were infected with WT or fluorescent-expressing IBVs for 1 h at room temperature, overlaid with agar and incubated at 33 °C. Three days post-infection, cells were fixed overnight with 4% paraformaldehyde (PFA), and the overlays were removed. For visualization of mCherry, PBS was added and plates were imaged with a Kodak image station (4000MM Pro molecular imaging system; Carestream Health, Inc., NY) (Nogales et al., 2014a) and Kodak molecular imaging software (v5.0.1.30). Plates were then permeabilized (0.5% Triton X-100 in PBS for 15 min at room temperature) and prepared for immunostaining as previously described (Nogales et al., 2014a) using the anti IBV NP MAb B017 and vector kits (Vectastain ABC kit and DAB HRP Substrate Kit: Vector), according to manufacturer's specifications.
2.8. Indirect immunofluorescence assays
MDCK cells were mock-infected or infected (MOI 1.0) with WT or fluorescent-expressing IBVs. At 18 hpi, cells were fixed with 4% PFA and permeabilized with 0.5% Triton X-100 in PBS for 15 min at room temperature. The staining was performed as described previously (Nogales et al., 2014a), using primary IBV NP (MAb B017; Abcam) or NS1 (PAb) (Dauber et al., 2006) antibodies, and FITC-conjugated anti-mouse (NP) or anti-rabbit (NS1) antibodies (Dako). The cell nucleus were stained with 4′,6′ -diamidino-2-phenylindole (DAPI, Research Organics). Images were captured using a fluorescence microscope (Nikon Eclipse TE2000) at X20 magnification, and pictures were processed using Adobe Photoshop CS4 (v11.0) software.
2.9. Virus neutralization and fluorescence-based microneutralization assays
Virus neutralization assays were performed with WT and mCherry IBVs and IAVs as previously described (Nogales et al., 2014a). Briefly, goat PAbs against influenza B/Hong Kong/8/1973 or pH1N1HA (BEI Resources NR-3165 and NR-15696, respectively) were heat inactivated (56 °C, 45 min) and serially diluted (2-fold) in PBS using 96-well plates (starting dilutions of 1:200). Five hundred FFUs of each virus were then added to the antibody dilutions and incubated for 1 h at room temperature. MDCK cells (96-well plate format, 5 × 104 cells/well, triplicates) were then infected with the antibody:virus mixture and incubated for 48 h at 33 °C. For WT or mCherry IAVs and IBVs, virus neutralization was determined by hemagglutination inhibition (HI) assay using tissue culture supernatants from infected cells and 0.5% turkey red blood cells for 30 min on ice (Guo et al., 2011). For the fluorescence-based microneutralization assays, cells were washed with PBS prior to red fluorescence imaging using a fluorescence microscopy or quantification using a fluorescence plate reader (DTX-880, Becton Dickenson) (Martinez-Sobrido et al., 2010). Fluorescence values of mCherry virus-infected cells in the absence of antibody (No Ab) were used to calculate 100% viral infection. Cells in the absence of viral infection (No virus) were used to calculate the fluorescence background. Triplicate wells were used to calculate the mean and SD of neutralization. The inhibitory concentration 50 (IC50) was determined by a sigmoidal dose response curve (Graphpad Prism, v4.0).
2.10. Antiviral assays
Antiviral-mediated inhibition of WT and mCherry IBVs and IAVs was evaluated as previously described (Nogales et al., 2014a). Briefly, confluent MDCK cells (2.5 × 105 cells, 24-well plate format, triplicates) were infected with the indicated viruses (MOI 0.001). After 1 h infection at room temperature, infectious media was supplemented with 2-fold serial dilutions (starting concentrations of 100 μM) of ribavirin (Sidwell et al., 1972) or amantadine (Sigma). At 48 hpi, virus titers in tissue culture supernatants were determined by FFU (Nogales et al., 2014b). To determine antiviral activity against mCherry viruses, confluent MDCK cells (96-well plate format; 5 × 104 cells/well; triplicates) were infected (MOI 0.001) and treated with the indicated compounds as previously described (Nogales et al., 2014a). At 48 hpi, cells were washed with PBS prior to red fluorescence imaging using a fluorescence microscopy or quantification using a fluorescence plate reader (DTX-880, Becton Dickenson) (Martinez-Sobrido et al., 2010). Fluorescence values of mCherry virus-infected cells in the absence of drug (No drug) were used to calculate 100% viral infection. Cells in the absence of viral infection (No virus) were used to calculate the fluorescence background. Triplicate wells were used to calculate the mean and SD of neutralization. The inhibitory concentration 50 (IC50) was determined by a sigmoidal dose response curve (Graphpad Prism, v4.0).
3. Results
3.1. Generation of a replication-competent IBV expressing the mCherry fluorescent protein (IBV-mCherry)
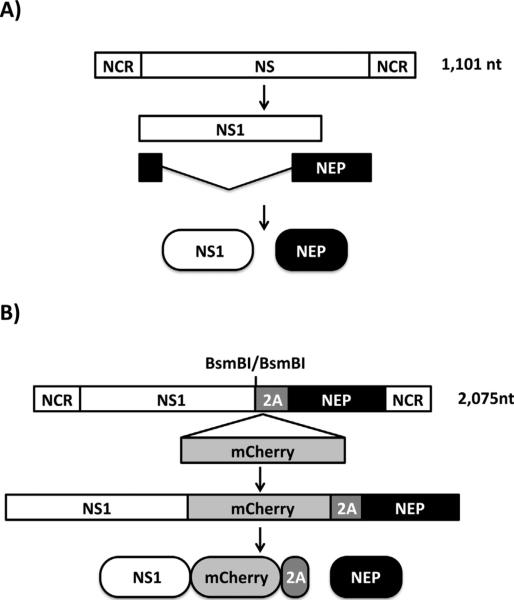
In order to engineer a replication-competent IBV expressing mCherry, segment 8 which encodes the NS1 and NEP open reading frames, was modified following a similar previously described approach (Manicassamy et al., 2010; Nogales et al., 2014a). Briefly, a modified IBV NS segment containing two BsmBI restriction sites in opposite orientation was synthesized de novo to introduce mCherry in frame with the NS1 sequence. Because the NS segment is alternatively spliced to encode NEP (Fig. 1A), two silent mutations in the splice acceptor site were introduced to avoid splicing (Hale et al., 2008; Kochs et al., 2007; Manicassamy et al., 2010; Nogales et al., 2014a). To produce NEP, the porcine teschovirus-1 (PTV-1) 2A autoproteolytic cleavage site was inserted between NS1 and NEP in order for both viral proteins to be translated individually (Fig. 1B) (Manicassamy et al., 2010, 2014a). Importantly, and to produce a full length NEP, the NS1 and NEP N-terminal overlapping first 10 amino acids were duplicated downstream of the PTV-1 2A site (Manicassamy et al., 2010; Nogales et al., 2014a). The mCherry sequence was cloned in the modified NS segment using the BsmBI restriction sites. We then use influenza B/Brisbane/60-008 plasmid-based reverse genetics (Baker et al., 2014; Pena et al., 2013) to generate a recombinant IBV expressing mCherry (IBV-mCherry) fused to the C-terminus of NS1.
Fig. 1.
Schematic representation of the WT (A) and modified (B) influenza B/Brisbane/60/2008 NS segments: IBV WT NS segment is indicated with a white box. IBV NS viral products are indicated by white (NS1) or black (NEP) boxes. NCR denotes non-coding regions. Sequences of mCherry and PTVI-2A are indicated by gray boxes. BsmBI restriction sites for cloning of fluorescent proteins fused to the C-terminal end of IBV NS1 are indicated.
3.2. Characterization of IBV-mCherry
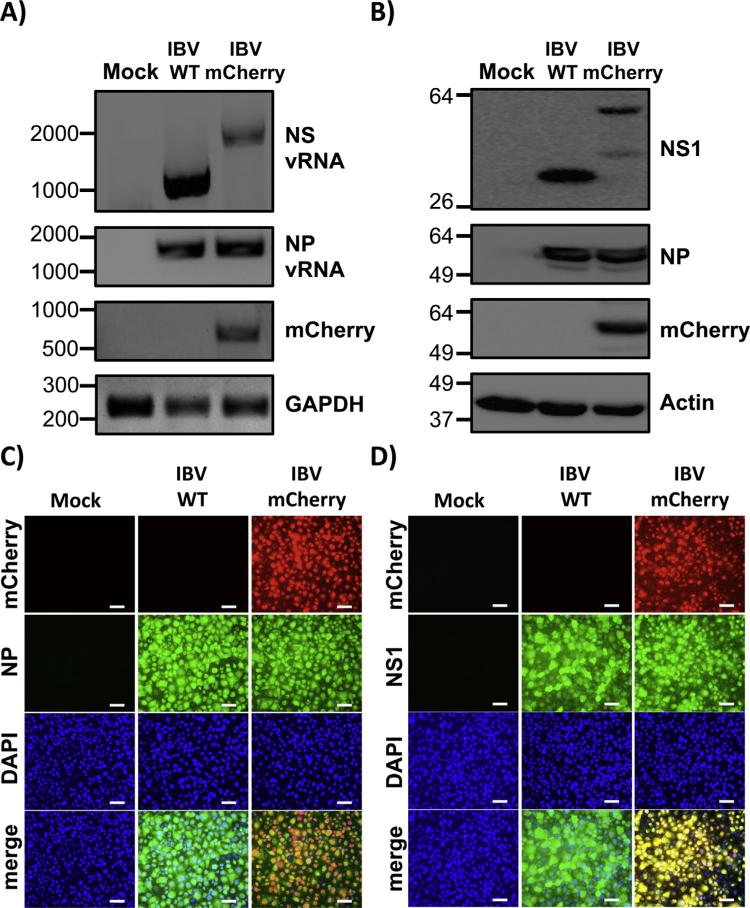
The identity of IBV-mCherry was first confirmed by RT-PCR (Fig. 2A). Total RNA was extracted from mock, IBV WT and IBV-mCherry infected (MOI 1.0) cells at 18 hpi. Expected band sizes of approximately 1100 and 2075 nts were observed, corresponding to the WT and mCherry NS vRNA segments, respectively (Fig. 2A). Primers specific for mCherry only amplified a band of the expected size (~708 nt) in IBV-mCherry infected cells (Fig. 2A). To control for viral infections and cellular transcription, viral NP vRNA and GAPDH mRNA levels, respectively, were also assessed (Fig. 2A). We next evaluated protein expression levels by Western blot (Fig. 2B). Total cell extracts from either mock, IBV WT or IBV-mCherry infected (MOI 1.0) MDCK cells were examined at 18 hpi using antibodies specific for NS1, mCherry, NP, and actin (loading control). Western blot analysis showed specific bands for NS1 or NS1-mCherry proteins with the anti-NS1 antibody (Fig. 2B). In cell extracts from IBV-mCherry infected cells, we observed an additional band of lower molecular size that might represent a NS1-mCherry degradation product. Western blot analysis with the mCherry antibody detected a specific band in cell extracts from IBV-mCherry infected cells (Fig. 2B). Notably, the levels of NP were similar between WT and mCherry IBV-infected cells (Fig. 2B).
Fig. 2.
Characterization of mCherry-expressing influenza B/Brisbane/60/08 virus (IBV-mCherry). (A) Analysis of RNA expression: MDCK cells were mock infected (Mock) or infected (MOI 1.0) with influenza B/Brisbane/60/08 WT or mCherry-expressing viruses. At 18 hpi, total RNA from infected cells was extracted. cDNA synthesis for NS or NP vRNAs was performed using NS and NP vRNA specific primers. cDNA synthesis of mRNA (mCherry and GAPDH) was performed using oligo dT. Specific primers for PCR were used to amplify NS and NP vRNAs and mCherry and GAPDH mRNAs. (B) Analysis of protein expression: MDCK cells were mock infected (Mock) or infected (MOI 1.0) with WT or mCherry-expressing IBVs as indicated above and protein expression levels of NS1, NP and mCherry were evaluated using protein specific antibodies. Actin was used as a loading control. Numbers indicate the size of molecular markers in nucleotides (A) or kDa (B) for DNA and protein products, respectively. (C–D) Protein expression by fluorescence and immunofluorescence: MDCK cells were mock infected (Mock) or infected (MOI 1.0) with WT or mCherry-expressing IBVs. At 18 hpi, cells were fixed and permeabilized, visualized for mCherry expression, and stained with IBV NP (C) or NS1 (D) antibodies. DAPI was used for nuclear staining. Merged images for mCherry, viral NP (C) or NS1 (D), and DAPI are illustrated at the bottom. Representative images (20X magnification) are included. Scale bar, 50 μm.
To assess whether IBV-mCherry could be directly visualized and used to evaluate IBV infections, confluent monolayers of MDCK cells were mock infected or infected (MOI 1.0) with WT or mCherry IBVs and examined by fluorescence (mCherry) and indirect immunofluorescence microscopy using specific antibodies for IBV NP (Fig. 2C) or NS1 (Fig. 2D) at 18 hpi. Subcellular nuclear localization of IBV NP was similar in both WT and mCherry infected cells (Fig. 2C). Likewise, IBV NS1 was similarly distributed in WT and mCherry infected MDCK cells (Fig. 2D). Importantly, only IBV-mCherry infected cells were fluorescent upon direct examination under a fluorescent microscope using a red filter (Fig. 2C and D). Altogether, these data demonstrate that MDCK cells infected with IBV-mCherry express high levels of mCherry that can be used as a valid surrogate to monitor IBV infections without the need of secondary approaches to detect viral infections.
3.3. Characterization of IBV-mCherry growth properties and plaque phenotype
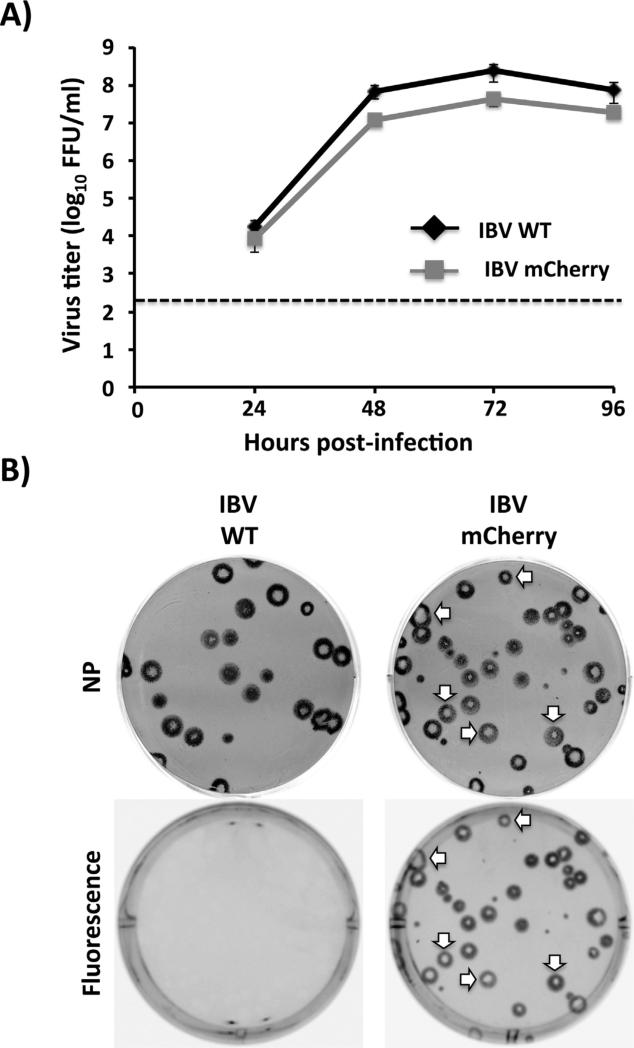
Virus kinetics of WT and mCherry IBVs were next examined in tissue culture by examining multicycle growth properties (Fig. 3A) and plaque formation (Fig. 3B). For virus growth kinetics, confluent monolayers of MDCK cells were infected (MOI 0.001) with IBV WT and IBV-mCherry and presence of virus in tissue culture supernatants were quantified at different hpi (Fig. 3A). Recombinant mCherry-expressing IBV replicates to similar levels than IBV WT, reaching maximum titers at 72 hpi. Thus, introduction of mCherry did not significantly affect IBV growth kinetics and viral replication (Fig. 3A). When we evaluated the plaque phenotype of WT and mCherry IBVs in MDCK cells, the size of infected viral foci produced by the IBV-mCherry was similar to those produced by IBV WT (Fig. 3B). Importantly, all plaques detected using the anti-NP IBV MAb expressed mCherry (Fig. 3B, white arrows), indicating that all infectious recombinant IBVs express mCherry. As expected, only IBV-mCherry formed fluorescent plaques under direct fluorescent visualization.
Fig. 3.
Growth kinetics and plaque phenotype of mCherry-expressing IBV. (A) Multicycle growth kinetics: virus titers from tissue culture supernatants of WT and mCherry IBV-infected (MOI 0.001) MDCK cells at the indicated times post-infection were analyzed by immunofocus assay (FFU/ml). Dotted line denotes the limit of detection (200 FFU/ml). Data represent the means ± SD of triplicates. (B) Plaque phenotype: Plaque sizes of WT and mCherry-expressing IBVs in MDCK cells were evaluated 3 days post-infection by immunostaining using a monoclonal antibody against IBV NP (MAb B017) or by fluorescence expression. For IBV-mCherry infections, white arrows indicate correlation between NP positive (top) and mCherry fluorescent (bottom) plaques.
3.4. Use of IBV-mCherry for the identification of neutralizing antibodies (NAb)
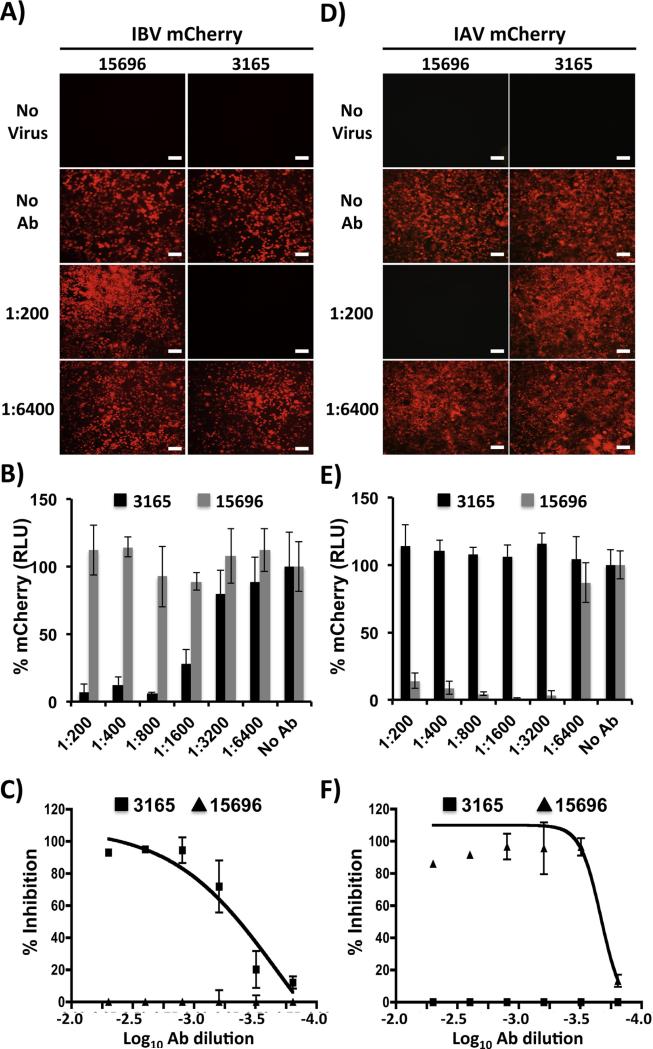
NAbs against influenza virus are important to evaluate protection efficacies of vaccine approaches (Baker et al., 2015b; Couch et al., 2013; Cox and Brokstad, 1999; Hancock et al., 2009; Martinez-Sobrido et al., 2010). However, with the exception of HA inhibition (HI) assays, approaches to assess antibody-mediated virus neutralization usually require secondary methods to detect the presence of the virus, such as cell staining, immunofluorescence assay, or ELISA (Bauman et al., 2013; Webster et al., 2002). This limitation can be sidestepped by using recombinant influenza viruses harboring easily traceable fluorescent proteins whose expression may be directly monitored (Martinez-Sobrido et al., 2010; Nogales et al., 2014a; Shaner et al., 2005). To demonstrate that mCherry-expressing IBV can be used to evaluate NAb responses, confluent monolayers of MDCK cells were infected with mCherry-expressing IBV or IAV, which were previously incubated with HA-specific NAbs for IBV (NR-3165) (Baker et al., 2014) or IAV pH1N1 (NR-15696) (Couch et al., 2013) and visualized by fluorescent expression using fluorescence microscopy (Fig. 4A and D) or quantified using a fluorescence plate reader (Fig. 4B and E), respectively. As expected, IBV-mCherry was specifically neutralized by NR-3165 but not by NR-15696 (Fig. 4A–E). In turn, IAV-mCherry was exclusively neutralized by NR-15696 but not NR-3165 (Fig. 4A–E). We then used a conventional virus neutralization (VN) assay to evaluate neutralization of WT and mCherry-expressing viruses by these influenza type-specific antibodies. Results show comparable levels of neutralization for WT and mCherry viruses (Table 1). Moreover, VN data correlated with that previously observed in the fluorescence approach, when the percentage of inhibition was calculated using sigmoidal dose responses (Fig. 4C and F and Table 2). Altogether, these results demonstrate that IBV-mCherry can be used to evaluate NAb responses with a fluorescence-based microneutralization assay, as we have recently showed for IAV mCherry (Nogales et al., 2014a).
Fig. 4.
A fluorescent-based microneutralization assay for the identification of IBV NAbs: MDCK cells were infected (MOI 0.001) with mCherry-expressing IBV (A–C) or IAV (D–F) that were pre-incubated with 2-fold serial dilutions (starting dilution of 1:200) of HA specific IAV (15,696) or IBV (3165) PAbs. At 48 hpi, virus neutralization was evaluated under a fluorescent microscope (A and D), quantified using a fluorescent microplate reader (B and E), and the percentage of inhibition calculated using sigmoidal dose response curves (C and F). Mock infected cells (No Virus) and viruses in the absence of antibodies (No Abs) were used as internal controls. Percent neutralization was normalized to infection in the absence of antibody. Data show means ± SD of the results determined for triplicates. Representative fluorescence images (10X magnification) are shown. Scale bars, 200 μM.
Table 1.
NT50 of PAbs by VN assay.
| NT50a (SD) | ||
|---|---|---|
| Virus | 15696 | 3165 |
| IBV WT | >1:200b | 533 (231) |
| IBV mCherry | >1:200b | 800 |
| IAV WT | 5333 (1847) | >1:200b |
| IAV mCherry | 5333 (1847) | >1:200b |
PAb dilution determined by serial-titration of Ab with Triplicate infections.
Highest dilution used without detectable neutralizing effect.
Table 2.
NT50 of PAbs by fluorescence-based assay.
PAb dilution determined by serial-titration of Ab with triplicate infections.
Highest dilution used without detectable neutralizing effect.
3.5. Use of IBV-mCherry for the identification of antivirals
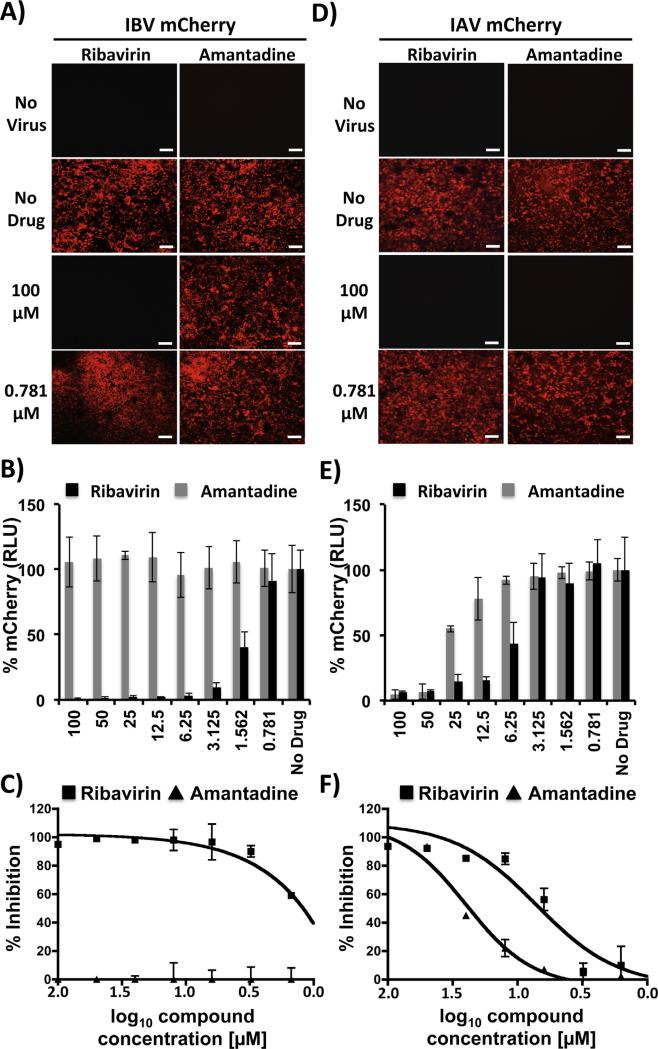
Since expression of the viral-encoded mCherry is dependent on IBV infection, mCherry can be used as a valid surrogate of IBV infection. To demonstrate that IBV-mCherry can be used for the identification of antivirals, we examined the ability of ribavirin (Cheung et al., 2014; Nguyen et al., 2010, 2009; Nogales et al., 2014a) and amantadine (Nguyen et al., 2010, 2009) to inhibit mCherry-expressing IAV and IBV (Fig. 5). Ribavirin is a nucleoside analog that has been shown to be efficient against IBV and IAV infections (Beigel and Bray, 2008). In contrast, amantadine is an inhibitor that specifically targets the M2 protein of IAVs, with no antiviral activity against IBVs (Oxford et al., 2003a). MDCK cells were cultured in 96-well plates and infected with mCherry-expressing IBV (Fig. 5A and B) and IAV (Fig. 5D and E). After infection, cells were incubated with medium containing serial 2-fold dilutions (starting concentration 100 μM) of ribavirin or amantadine. Infected MDCK cells lacking antiviral compounds were used as internal controls. At 48 hpi, mCherry expression was determined via fluorescence microscopy (Fig. 5A and D) or quantified using a fluorescent plate reader (Fig. 5B and E). As expected, mCherry-expressing IBV and IAV were inhibited by ribavirin in a dose-dependent manner (Fig. 5A–E). However, amantadine showed a dose-dependent antiviral activity against IAV-mCherry but not against IBV-mCherry (Fig. 5A–E). To demonstrate that mCherry-expressing viruses recapitulate similar antiviral profiles as those of WT viruses, we evaluated the antiviral activity of ribavirin and amantadine from tissue culture supernatants of MDCK cells infected with WT IAV and IBV. Virus release into tissue culture supernatants was determined by calculating the respective IC50 values using a classical sigmoidal dose response curve (Table 3). Regardless of the drug or virus used, IC50 values were similar, within 1–2 dilution values (Table 3), and consistent with those previously reported in the literature (Nguyen et al., 2010, 2009; Sidwell et al., 2005) and in our fluorescence-based microneutralization assay (Fig. 5C and F; and Table 4). It is important to note that IAVs WT and mCherry in the 2009 pH1N1 virus backbone have higher IC50 values for amantadine than other IAV strains due to a mutation (S31N) in the M2 channel that has been associated with drug resistance (Nguyen et al., 2010; Sidwell et al., 2005). Overall, these data demonstrate that IBV-mCherry can be used as a valid surrogate system for the rapid and easy identification of IBV inhibitors.
Fig. 5.
A fluorescent-based assay for the identification of IBV antivirals: MDCK cells were infected (MOI 0.001) with mCherry-expressing IBV (A–C) or IAV (D–F) and incubated with serial 2-fold dilutions (starting concentration 100 μM) of ribavirin or amantadine. As internal control, infected cells were not treated with antivirals (No Drug). At 48 hpi, viral inhibition was evaluated under a fluorescent microscope (A and D); quantified using a fluorescent microplate reader (B and E), and the percentage of inhibition calculated using sigmoidal dose response curves (C and F). Mock-infected cells (No virus) were used as control. Percent viral inhibition was normalized to infection in the absence of antivirals. Data show means ± SD of the results determined for triplicates. Representative fluorescence microscopy images (10X magnification) are shown. Scale bars, 200 μM.
Table 3.
IC50 of compounds by FFU titration method.
| IC50a |
||
|---|---|---|
| Virus | Ribavirin | Amantadine |
| IBV WT | 0.98 | >100b |
| IBV mCherry | 4.29 | >100b |
| IAV WT | 2.36 | 73.41 |
| IAV mCherry | 7.51 | 69.64 |
Compound concentration (μM) determined by serial-titration of drug with triplicate infections.
Highest concentration used without detectable antiviral effect.
Table 4.
IC50 of compounds by fluorescence-based assay.
Compound concentration (μM) determined by serial-titration of drug with tirplicate infections.
Highest concentration used without detectable antiviral effect.
3.6. Generation and characterization of a replication-competent IBV expressing GFP (IBV-GFP)
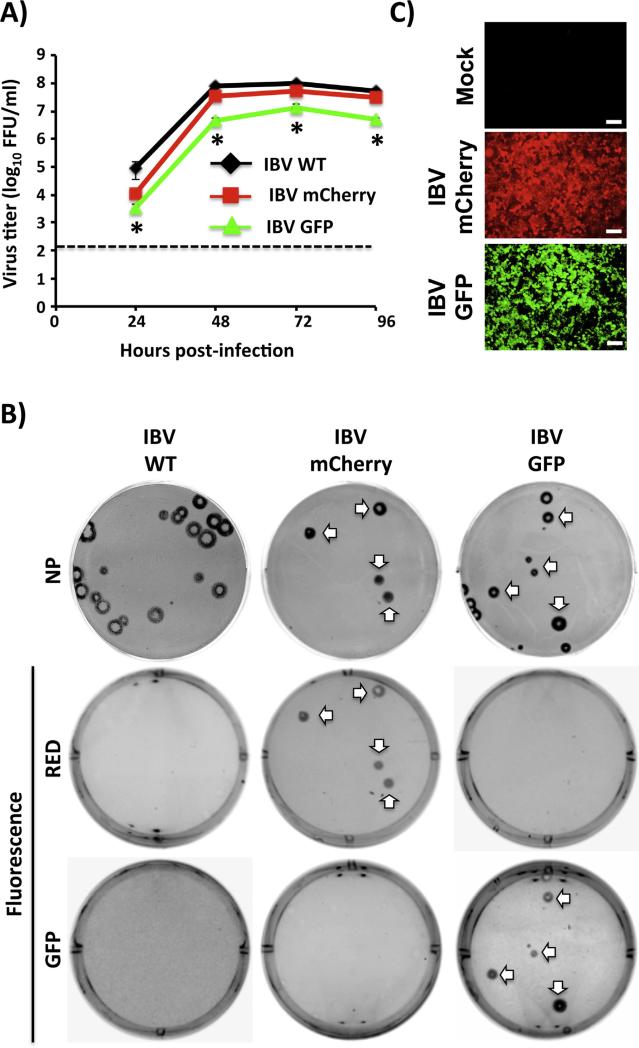
To further validate the use of this molecular approach for the generation of reporter-expressing IBVs, a recombinant IBV expressing GFP (IBV-GFP) was generated (Fig. 6). IBV-GFP showed a slightly delayed kinetics (Fig. 6A) but similar plaque size (Fig. 6B) than IBV WT or IBV-mCherry. Interestingly, IBV-GFP showed less stability than IBV-mCherry (data not shown), which resembles similar observations with IAV-GFP (Fukuyama et al., 2015; Manicassamy et al., 2010). However, we were able to obtain a stable IBV-GFP clone after 3 serial plaque purifications in MDCK cells. Importantly, MDCK cells infected with IBV-GFP express high levels of GFP (Fig. 6C) that can be used as an alternative reporter or in combination with mCherry-expressing IAV and/or IBV to evaluate virus-infected cells, NAbs and/or antivirals. These data demonstrate that replication-competent IBVs expressing different fluorescence proteins are feasible and stable, increasing the arsenal of these type of recombinant viruses that are safe and amenable to routine virus neutralization and antiviral assays as recently reported with IAVs (Fukuyama et al., 2015; Manicassamy et al., 2010).
Fig. 6.
Characterization of a GFP-expressing influenza B/Brisbane/60/08 virus (IBV-GFP). (A) Multicycle growth kinetics: Virus titers from tissue culture supernatants of MDCK cells infected (MOI 0.001) with IBV WT, mCherry, and GFP at the indicated times post-infection were analyzed by immunofocus assay (FFU/ml). Dotted line indicates the limit of detection (200 FFU/ml). Data represent the means ± SD of triplicates. *(WT vs GFP) P < 0.05 using an unpaired one-tailed Student's t test (n = 3 per time point). (B) Plaque phenotype: Plaque sizes of WT and mCherry- or GFP-expressing IBVs in MDCK cells were evaluated 3 days post-infection by immunostaining (top) using a MAb against IBV NP (B017) or by red and green fluorescence. White arrows indicate correlation between NP positive and fluorescent (mCherry or GFP) plaques. (C) Fluorescence protein expression: MDCK cells were mock infected or infected (MOI 1.0) with mCherry- or GFP-expressing IBVs. At 18 hpi, cells were assessed for GFP or mCherry expression under a fluorescent microscope using specific filters. Representative images (20X magnification) are included. Scale bar, 100 μm.
4. Discussion
Recombinant viruses expressing fluorescent proteins have been generated to monitor viral replication, including DNA (e.g., Vaccinia virus (Diallo et al., 2011; Dominguez et al., 1998) and human cytomegalovirus (Marschall et al., 2000)), and double-stranded (e.g., HIV (Daelemans et al., 2005)), positive-stranded (e.g., murine hepatitis virus (Freeman et al., 2014)), negative-stranded segmented e.g., arenaviruses (Ortiz-Riano et al., 2013), and non-segmented (e.g., Newcastle disease virus (Vigil et al., 2007) and Ebola virus (Towner et al., 2005)) RNA viruses. These recombinant systems are powerful tools to monitor viral infections in real time (Fiege and Langlois, 2015; Fukuyama et al., 2015; Manicassamy et al., 2010), to evaluate antivirals (Nogales et al., 2014a; Towner et al., 2005), and to identify NAb (Baker et al., 2015b; Nogales et al., 2014a; Rimmelzwaan et al., 2011).
With the development of reverse genetics techniques, similar approaches have been implemented for the generation and characterization of replication-competent fluorescent-expressing IAVs (Avilov et al., 2012; Baker et al., 2015b; Fiege and Langlois, 2015; Fukuyama et al., 2015; Manicassamy et al., 2010; Nogales et al., 2015, 2014a; Reuther et al., 2015; Rimmelzwaan et al., 2011). However, similar methods have not been used for the generation of recombinant fluorescent-expressing IBVs. While preparing this manuscript, Fulton et al. described the generation of replication competent IBVs expressing reporter genes from the viral polymerase PB2, PB1 or PA viral segments using the backbone of influenza B/Yamagata/16/1988 (B/Yamagata lineage) (Fulton et al., 2015). Here, we produced and characterized replication-competent reporter-expressing viruses in the backbone of the more recent influenza B/Brisbane/60/2008 (B/Victoria lineage) using a strategy similar to the one that we previously used to generate fluorescent IAV (Nogales et al., 2014a). This was achieved by fusing the mCherry ORF at the C-terminal end of the viral NS1 followed by the PTV-1 2A autoproteolytic cleavage site, and the entire viral NEP ORF (Fig. 1). PTV-1 2A mediates co-translational separation of NS1-mCherry and NEP by a mechanism termed “stop-carry on” recoding (Sharma et al., 2012), generating two functional and independent proteins. Although this approach has been successfully used previously for the generation of recombinant fluorescent-expressing IAV (Manicassamy et al., 2010; Nogales et al., 2014a), we demonstrate the feasibility of using this same methodology for the generation of IBVs encoding fluorescent proteins. Importantly, the identity of the modified IBV-mCherry NS segment was confirmed by RT-PCR (Fig. 2A) and Western blot (Fig. 2B). Furthermore, we observed good correlation of NP positive cells with mCherry expression (Fig. 2C), and co-localization of NS1 and mCherry in IBV-infected cells (Fig. 2D), demonstrating that mCherry expression can be used as a valid surrogate of IBV infection. Importantly, when multi-growth kinetics or plaque assays were performed, IBV-mCherry showed no significant attenuation as compared to its WT counterpart in vitro (Fig. 3).
Vaccination represents our best prophylactic option to combat IAV and IBV infections (Pica and Palese, 2013) and a simple and rapid assay for detecting NAbs will assist to evaluate humoral immune responses induced by traditional or novel influenza vaccine approaches (Belshe et al., 2000; He et al., 2015). Moreover, new interest has arisen in the identification of NAbs with type-specific or broadly neutralizing properties due to their therapeutic benefit in passive immunization of people expose to influenza viruses (Tan et al., 2012). NAbs against IAVs or IBVs typically bind to HA, and traditional methods to detect the presence of influenza NAbs include HI and VN assays, each of them having advantages and limitations. The HI assay provides a rapid read-out (same day), but sensitivity is limited because of the large amount of virus required to agglutinate erythrocytes (Killian, 2008). In addition, the HI assay has been shown to not be effective in the identification of broadly neutralizing antibodies (He et al., 2015; Killian, 2008). It is generally accepted that VN assays are more sensitive and reliable than HI assays (Rimmelzwaan et al., 1998). However, VN assays are more laborious, time consuming and usually require the use of secondary approaches for the identification of virus-infected cells (de Jong et al., 2003; Rowe et al., 1999). In this regard, fluorescent-expressing IBVs represent an excellent platform for the rapid, sensitive and simple identification and characterization of NAbs using a fluorescent-based microneutralization assay (Fig. 4). Importantly, with this novel microneutralization approach we obtained similar neutralization titers to those obtained using conventional microneutralization assays (Tables 1 and 2). Moreover, neutralization results using IAV- and IBV-mCherry were similar to those obtained with WT influenza viruses (Table 1).
Although vaccination is the primary method for controlling influenza, antivirals provide an additional line of defense, which is particularly important for controlling new rapidly spreading, antigenic drift and/or potentially pandemic virus (Garcia-Sastre, 2006; Oxford et al., 2003b; Seibert et al., 2010). Therapeutic options for the treatment and prevention of influenza infections include NA-targeting zanamivir and oseltamivir and M2-targeting amantadine and rimantadine inhibitors (Jackson et al., 2011b; Nguyen et al., 2010, 2009). However, NA inhibitors are the only class of antivirals approved for the prophylaxis and treatment of patients infected with IBVs (Burnham et al., 2013), although data from clinical trials suggest that oseltamivir appears less effective against IBVs than IAVs (Farrukee et al., 2013). In addition the emergence of drug-resistant variants is an increasing problem for controlling influenza infections (Nguyen et al., 2010; Seibert et al., 2010). Thus, there is an urgent medical need for the identification and characterization of novel antivirals for the treatment of influenza infections in humans, a task that will be accelerated by the development of rapid and sensitive screening assays amenable to HTS approaches (Beyleveld et al., 2013). Identification of compounds that inhibit influenza has typically relied on the use of time-consuming secondary assays to assess viral infection. Thus, alternative approaches that facilitate detection of IBVs and/or IAVs would help to identify new antivirals. Importantly, these methodologies should be compatible with HTS approaches to help simplify the screening of large compound libraries to identify hits with IBVs and/or IAVs antiviral activity. To demonstrate the implementation of fluorescent-expressing IBVs to identify and characterize antiviral drugs, we evaluated the antiviral activity of ribavirin and amantadine against both IAV and IBV (Beigel and Bray, 2008; Nguyen et al., 2010, 2009; Nogales et al., 2014a; Oxford et al., 2003a; Sidwell et al., 2005) (Fig. 5). Using our novel fluorescent-based microneutralization assay, we observed dose-dependent inhibition of IAV-mCherry by both amantadine and ribavirin, while only ribavirin had a dose-dependent inhibitory effect on IBV-mCherry. Importantly, viral inhibition by these two antivirals in our fluorescent-based microneutralization assays was similar to that obtained using WT IAV and IBV in conventional VN assays (Tables 3 and 4). Altogether, these results demonstrate the feasibility of using replication-competent fluorescent-expressing IBVs as a novel platform, amenable to HTS, for the identification and characterization of novel antivirals.
When GFP was introduced in IBV instead of mCherry, we observed high levels of GFP expression (Fig. 6), suggesting the feasibility of generating replication-competent IBVs expressing others fluorescence proteins, similar to the situation recently described for IAVs (Fukuyama et al., 2015; Manicassamy et al., 2010). However, like with IAVs, GFP-expressing IBV seems to be less stable than IBV-mCherry (data not shown) (Fukuyama et al., 2015; Manicassamy et al., 2010). Nevertheless, we have been able to generate a stable stock of IBV-GFP after three serial plaque purifications in MDCK cells. Similar to IAVs these fluorescent IBV variants (mCherry and GFP) could be used to address interesting questions in the biology of IBV, including the frequency of viral co-infections and the generation of reassortants (Baker et al., 2014). Moreover, fluorescent-expressing IBVs could be used in combination with the previously described fluorescent-expressing IAVs (Fukuyama et al., 2015; Nogales et al., 2014a) in bi-fluorescent-based microneutralization assays (Baker et al., 2015b) for the rapid and easy identification of compounds with antiviral activity against both influenza viruses. Likewise, this strategy could also be used for the detection of neutralizing antibodies with broad-antiviral activity against IAVs and IBVs.
Acknowledgments
We thank Dr. T. Wolff at the Division of Influenza and Other Respiratory Viruses at the Robert Koch-Institut (Germany) for the NS1 PAb. The polyclonal antibodies NR-3165 and NR-15696 were obtained through BEI Resources. This research was funded by the 2014 University of Rochester Research Award to LM-S.
References
- Avilov SV, Moisy D, Munier S, Schraidt O, Naffakh N, Cusack S. Replication-competent influenza A virus that encodes a split-green fluorescent protein-tagged PB2 polymerase subunit allows live-cell imaging of the virus life cycle. J. Virol. 2012;86(3):1433–1448. doi: 10.1128/JVI.05820-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SF, Guo H, Albrecht RA, Garcia-Sastre A, Topham DJ, Martinez-Sobrido L. Protection against lethal influenza with a viral mimic. J. Virol. 2013;87(15):8591–8605. doi: 10.1128/JVI.01081-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SF, Nogales A, Finch C, Tuffy KM, Domm W, Perez DR, Topham DJ, Martinez-Sobrido L. Influenza A and B virus intertypic reassortment through compatible viral packaging signals. J. Virol. 2014;88(18):10778–10791. doi: 10.1128/JVI.01440-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SF, Nogales A, Martinez-Sobrido L. Downregulating viral gene expression: codon usage bias manipulation for the generation of novel influenza A virus vaccines. Future Virol. 2015a;10(6):715–730. doi: 10.2217/fvl.15.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SF, Nogales A, Santiago FW, Topham DJ, Martinez-Sobrido L. Competitive detection of influenza neutralizing antibodies using a novel bivalent fluorescence-based microneutralization assay (BiFMA). Vaccine. 2015b;33(30):3562–3570. doi: 10.1016/j.vaccine.2015.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman JD, Patel D, Baker SF, Vijayan RS, Xiang A, Parhi AK, Martinez-Sobrido L, LaVoie EJ, Das K, Arnold E. Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors. ACS Chem. Biol. 2013;8(11):2501–2508. doi: 10.1021/cb400400j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J, Bray M. Current and future antiviral therapy of severe seasonal and avian influenza. Antiviral Res. 2008;78(1):91–102. doi: 10.1016/j.antiviral.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshe RB. The need for quadrivalent vaccine against seasonal influenza. Vaccine. 2010;28(Suppl 4):D45–D53. doi: 10.1016/j.vaccine.2010.08.028. [DOI] [PubMed] [Google Scholar]
- Belshe RB, Gruber WC, Mendelman PM, Mehta HB, Mahmood K, Reisinger K, Treanor J, Zangwill K, Hayden FG, Bernstein DI, Kotloff K, King J, Piedra PA, Block SL, Yan L, Wolff M. Correlates of immune protection induced by live, attenuated, cold-adapted, trivalent, intranasal influenza virus vaccine. J. Infect. Dis. 2000;181(3):1133–1137. doi: 10.1086/315323. [DOI] [PubMed] [Google Scholar]
- Belshe RB, Newman FK, Wilkins K, Graham IL, Babusis E, Ewell M, Frey SE. Comparative immunogenicity of trivalent influenza vaccine administered by intradermal or intramuscular route in healthy adults. Vaccine. 2007;25(37–38):6755–6763. doi: 10.1016/j.vaccine.2007.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyleveld G, White KM, Ayllon J, Shaw ML. New-generation screening assays for the detection of anti-influenza compounds targeting viral and host functions. Antiviral Res. 2013;100(1):120–132. doi: 10.1016/j.antiviral.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham AJ, Baranovich T, Govorkova EA. Neuraminidase inhibitors for influenza B virus infection: efficacy and resistance. Antiviral Res. 2013;100(2):520–534. doi: 10.1016/j.antiviral.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Holmes EC. The evolutionary dynamics of human influenza B virus. J. Mol. Evol. 2008;66(6):655–663. doi: 10.1007/s00239-008-9119-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung PP, Watson SJ, Choy KT, Fun Sia S, Wong DD, Poon LL, Kellam P, Guan Y, Malik Peiris JS, Yen HL. Generation and characterization of influenza A viruses with altered polymerase fidelity. Nat. Commun. 2014;5:4794. doi: 10.1038/ncomms5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch RB, Atmar RL, Franco LM, Quarles JM, Wells J, Arden N, Nino D, Belmont JW. Antibody correlates and predictors of immunity to naturally occurring influenza in humans and the importance of antibody to the neuraminidase. J. Infect. Dis. 2013;207(6):974–981. doi: 10.1093/infdis/jis935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RJ, Brokstad KA. The postvaccination antibody response to influenza virus proteins. APMIS. 1999;107(3):289–296. doi: 10.1111/j.1699-0463.1999.tb01556.x. [DOI] [PubMed] [Google Scholar]
- Daelemans D, Pannecouque C, Pavlakis GN, Tabarrini O, De Clercq E. A novel and efficient approach to discriminate between pre- and post-transcription HIV inhibitors. Mol. Pharmacol. 2005;67(5):1574–1580. doi: 10.1124/mol.104.010249. [DOI] [PubMed] [Google Scholar]
- Dauber B, Schneider J, Wolff T. Double-stranded RNA binding of influenza B virus nonstructural NS1 protein inhibits protein kinase R but is not essential to antagonize production of alpha/beta interferon. J. Virol. 2006;80(23):11667–11677. doi: 10.1128/JVI.01142-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong JC, Palache AM, Beyer WE, Rimmelzwaan GF, Boon AC, Osterhaus AD. Haemagglutination-inhibiting antibody to influenza virus. Dev. Biol. (Basel) 2003;115:63–73. [PubMed] [Google Scholar]
- Diallo JS, Roy D, Abdelbary H, De Silva N, Bell JC. Ex vivo infection of live tissue with oncolytic viruses. J. Vis. Exp. 2011;52:2854. doi: 10.3791/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez J, Lorenzo MM, Blasco R. Green fluorescent protein expressed by a recombinant vaccinia virus permits early detection of infected cells by flow cytometry. J. Immunol. Methods. 1998;220(1–2):115–121. doi: 10.1016/s0022-1759(98)00156-2. [DOI] [PubMed] [Google Scholar]
- Eckert N, Wrensch F, Gartner S, Palanisamy N, Goedecke U, Jager N, Pohlmann S, Winkler M. Influenza A virus encoding secreted Gaussia luciferase as useful tool to analyze viral replication and its inhibition by antiviral compounds and cellular proteins. PLoS One. 2014;9(5):e97695. doi: 10.1371/journal.pone.0097695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt OG. Many ways to make an influenza virus—review of influenza virus reverse genetics methods. Influenza Other Respir. Viruses. 2013;7(3):249–256. doi: 10.1111/j.1750-2659.2012.00392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrukee R, Mosse J, Hurt AC. Review of the clinical effectiveness of the neuraminidase inhibitors against influenza B viruses. Expert. Rev. Anti. Infect. Ther. 2013;11(11):1135–1145. doi: 10.1586/14787210.2013.842466. [DOI] [PubMed] [Google Scholar]
- Fiege JK, Langlois RA. Investigating influenza A virus infection: tools to track infection and limit tropism. J. Virol. 2015;89(12):6167–6170. doi: 10.1128/JVI.00462-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor E, Devenish L, Engelhardt OG, Palese P, Brownlee GG, Garcia-Sastre A. Rescue of influenza A virus from recombinant DNA. J. Virol. 1999;73(11):9679–9682. doi: 10.1128/jvi.73.11.9679-9682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman MC, Graham RL, Lu X, Peek CT, Denison MR. Coronavirus replicase-reporter fusions provide quantitative analysis of replication and replication complex formation. J. Virol. 2014;88(10):5319–5327. doi: 10.1128/JVI.00021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuyama S, Katsura H, Zhao D, Ozawa M, Ando T, Shoemaker JE, Ishikawa I, Yamada S, Neumann G, Watanabe S, Kitano H, Kawaoka Y. Multi-spectral fluorescent reporter influenza viruses (Color-flu) as powerful tools for in vivo studies. Nat. Commun. 2015;6:6600. doi: 10.1038/ncomms7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton BO, Palese P, Heaton NS. Replication-competent influenza b reporter viruses as tools for screening antivirals and antibodies. J. Virol. 2015;89(23):12226–12231. doi: 10.1128/JVI.02164-15. http://dx.doi.org/10.1128/JVI.02164-15, Epub 2015 Sep 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A. Antiviral response in pandemic influenza viruses. Emerg. Infect. Dis. 2006;12(1):44–47. doi: 10.3201/eid1201.051186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohskopf LA, Olsen SJ, Sokolow LZ, Bresee JS, Cox NJ, Broder KR, Walter EB. Prevention and control of seasonal influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP)—United States, 2014–15 influenza season. MMWR Morb. Mortal Wkly. Rep. 2014;63(32):691–697. [PMC free article] [PubMed] [Google Scholar]
- Guo H, Santiago F, Lambert K, Takimoto T, Topham DJ. T cell-mediated protection against lethal 2009 pandemic H1N1 influenza virus infection in a mouse model. J. Virol. 2011;85(1):448–455. doi: 10.1128/JVI.01812-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale BG, Randall RE, Ortin J, Jackson D. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 2008;89(Pt 10):2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- Hancock K, Veguilla V, Lu X, Zhong W, Butler EN, Sun H, Liu F, Dong L, DeVos JR, Gargiullo PM, Brammer TL, Cox NJ, Tumpey TM, Katz JM. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N. Engl. J. Med. 2009;361(20):1945–1952. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4(11):3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Mullarkey CE, Miller MS. Measuring the neutralization potency of influenza A virus hemagglutinin stalk/stem-binding antibodies in polyclonal preparations by microneutralization assay. Methods. 2015;(15):00191–00197. doi: 10.1016/j.ymeth.2015.04.037. http://dx.doi.org/10.1016/j.ymeth.2015.04.037 [Epub ahead of print] pii: S1046-2023. [DOI] [PubMed]
- Hite LK, Glezen WP, Demmler GJ, Munoz FM. Medically attended pediatric influenza during the resurgence of the Victoria lineage of influenza B virus. Int. J. Infect. Dis. 2007;11(1):40–47. doi: 10.1016/j.ijid.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Jackson D, Elderfield RA, Barclay WS. Molecular studies of influenza B virus in the reverse genetics era. J. Gen. Virol. 2011a;92(Pt 1):1–17. doi: 10.1099/vir.0.026187-0. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Cooper KL, Tappenden P, Rees A, Simpson EL, Read RC, Nicholson KG. Oseltamivir, zanamivir and amantadine in the prevention of influenza: a systematic review. J. Infect. 2011b;62(1):14–25. doi: 10.1016/j.jinf.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Killian ML. Hemagglutination assay for the avian influenza virus. Methods Mol. Biol. 2008;436:47–52. doi: 10.1007/978-1-59745-279-3_7. [DOI] [PubMed] [Google Scholar]
- Kittel C, Sereinig S, Ferko B, Stasakova J, Romanova J, Wolkerstorfer A, Katinger H, Egorov A. Rescue of influenza virus expressing GFP from the NS1 reading frame. Virology. 2004;324(1):67–73. doi: 10.1016/j.virol.2004.03.035. [DOI] [PubMed] [Google Scholar]
- Kochs G, Garcia-Sastre A, Martinez-Sobrido L. Multiple anti-interferon actions of the influenza A virus NS1 protein. J. Virol. 2007;81(13):7011–7021. doi: 10.1128/JVI.02581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, Palese P, Steel J. Advances in universal influenza virus vaccine design and antibody mediated therapies based on conserved regions of the hemagglutinin. Curr. Top. Microbiol. Immunol. 2015;386:301–321. doi: 10.1007/82_2014_408. [DOI] [PubMed] [Google Scholar]
- Li WC, Shih SR, Huang YC, Chen GW, Chang SC, Hsiao MJ, Tsao KC, Lin TY. Clinical and genetic characterization of severe influenza B-associated diseases during an outbreak in Taiwan. J. Clin. Virol. 2008;42(1):45–51. doi: 10.1016/j.jcv.2007.11.026. [DOI] [PubMed] [Google Scholar]
- Lin YP, Gregory V, Bennett M, Hay A. Recent changes among human influenza viruses. Virus Res. 2004;103(1–2):47–52. doi: 10.1016/j.virusres.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Luckhaupt SE, Sweeney MH, Funk R, Calvert GM, Nowell M, D'Mello T, Reingold A, Meek J, Yousey-Hindes K, Arnold KE, Ryan P, Lynfield R, Morin C, Baumbach J, Zansky S, Bennett NM, Thomas A, Schaffner W, Jones T. Influenza-associated hospitalizations by industry, 2009–10 influenza season, United States. Emerg. Infect. Dis. 2012;18(4):556–562. doi: 10.3201/eid1804.110337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, Garcia-Sastre A. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc. Natl. Acad. Sci. U. S. A. 2010;107(25):11531–11536. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall M, Freitag M, Weiler S, Sorg G, Stamminger T. Recombinant green fluorescent protein-expressing human cytomegalovirus as a tool for screening antiviral agents. Antimicrob. Agents Chemother. 2000;44(6):1588–1597. doi: 10.1128/aac.44.6.1588-1597.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Sobrido L, Cadagan R, Steel J, Basler CF, Palese P, Moran TM, Garcia-Sastre A. Hemagglutinin-pseudotyped green fluorescent protein-expressing influenza viruses for the detection of influenza virus neutralizing antibodies. J. Virology. 2010;84(4):2157–2163. doi: 10.1128/JVI.01433-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Sobrido L, Garcia-Sastre A. Generation of recombinant influenza virus from plasmid DNA. J. Vis. Exp. 2010;42 doi: 10.3791/2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers JA, Saito T, Iverson AR. Multiple genotypes of influenza B virus circulated between 1979 and 2003. J. Virol. 2004;78(23):12817–12828. doi: 10.1128/JVI.78.23.12817-12828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari NA, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, Weintraub E, Bridges CB. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine. 2007;25(27):5086–5096. doi: 10.1016/j.vaccine.2007.03.046. [DOI] [PubMed] [Google Scholar]
- Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. U. S. A. 1999;96(16):9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen JT, Hoopes JD, Le MH, Smee DF, Patick AK, Faix DJ, Blair PJ, de Jong MD, Prichard MN, Went GT. Triple combination of amantadine, ribavirin, and oseltamivir is highly active and synergistic against drug resistant influenza virus strains in vitro. PLoS One. 2010;5(2):e9332. doi: 10.1371/journal.pone.0009332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen JT, Hoopes JD, Smee DF, Prichard MN, Driebe EM, Engelthaler DM, Le MH, Keim PS, Spence RP, Went GT. Triple combination of oseltamivir, amantadine, and ribavirin displays synergistic activity against multiple influenza virus strains in vitro. Antimicrob. Agents Chemother. 2009;53(10):4115–4126. doi: 10.1128/AAC.00476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales A, Baker SF, Domm W, Martinez-Sobrido L. Development and applications of single-cycle infectious influenza A virus (sciIAV). Virus Res. 2015 doi: 10.1016/j.virusres.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales A, Baker SF, Martinez-Sobrido L. Replication-competent influenza A viruses expressing a red fluorescent protein. Virology. 2014a;476C:206–216. doi: 10.1016/j.virol.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales A, Baker SF, Ortiz-Riano E, Dewhurst S, Topham DJ, Martinez-Sobrido L. Influenza A virus attenuation by codon deoptimization of the NS gene for vaccine development. J. Virol. 2014b;88(18):10525–10540. doi: 10.1128/JVI.01565-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill RE, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17(1):288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmit SE, Monto AS. Influenza vaccine effectiveness in preventing hospitalization among the elderly during influenza type A and type B seasons. Int. J. Epidemiol. 1995;24(6):1240–1248. doi: 10.1093/ije/24.6.1240. [DOI] [PubMed] [Google Scholar]
- Olson DR, Heffernan RT, Paladini M, Konty K, Weiss D, Mostashari F. Monitoring the impact of influenza by age: emergency department fever and respiratory complaint surveillance in New York City. PLoS Med. 2007;4(8):e247. doi: 10.1371/journal.pmed.0040247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Riano E, Cheng BY, Carlos de la Torre J, Martinez-Sobrido L. Arenavirus reverse genetics for vaccine development. J. Gen. Virol. 2013;94(Pt 6):1175–1188. doi: 10.1099/vir.0.051102-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhaus AD, Rimmelzwaan GF, Martina BE, Bestebroer TM, Fouchier RA. Influenza B virus in seals. Science. 2000;288(5468):1051–1053. doi: 10.1126/science.288.5468.1051. [DOI] [PubMed] [Google Scholar]
- Oxford JS, Bossuyt S, Balasingam S, Mann A, Novelli P, Lambkin R. Treatment of epidemic and pandemic influenza with neuraminidase and M2 proton channel inhibitors. Clin. Microbiol. Infect. 2003a;9(1):1–14. doi: 10.1046/j.1469-0691.2003.00564.x. [DOI] [PubMed] [Google Scholar]
- Oxford JS, Mann A, Lambkin R. A designer drug against influenza: the NA inhibitor oseltamivir (Tamiflu). Expert Rev. Anti. Infect. Ther. 2003b;1(2):337–342. [PubMed] [Google Scholar]
- Ozawa M, Kawaoka Y. Taming influenza viruses. Virus Res. 2011;162(1–2):8–11. doi: 10.1016/j.virusres.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P, Shaw ML. Orthomyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, editors. Fields Virology. 5th edition Lippincott Williams and WIlkins; 2007. [Google Scholar]
- Pan W, Dong Z, Li F, Meng W, Feng L, Niu X, Li C, Luo Q, Li Z, Sun C, Chen L. Visualizing influenza virus infection in living mice. Nat. Commun. 2013;4 doi: 10.1038/ncomms3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena L, Sutton T, Chockalingam A, Kumar S, Angel M, Shao H, Chen H, Li W, Perez DR. Influenza viruses with rearranged genomes as live-attenuated vaccines. J. Virol. 2013;87(9):5118–5127. doi: 10.1128/JVI.02490-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez JT, García-Sastre A, Manicassamy B. Current Protocols in Microbiology. John Wiley & Sons, Inc.; 2013. Insertion of a GFP reporter gene in influenza virus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pica N, Palese P. Toward a universal influenza virus vaccine: prospects and challenges. Annu. Rev. Med. 2013;64:189–202. doi: 10.1146/annurev-med-120611-145115. [DOI] [PubMed] [Google Scholar]
- Reuther P, Gopfert K, Dudek AH, Heiner M, Herold S, Schwemmle M. Generation of a variety of stable Influenza A reporter viruses by genetic engineering of the NS gene segment. Sci. Rep. 2015;5:11346. doi: 10.1038/srep11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmelzwaan GF, Baars M, Claas EC, Osterhaus AD. Comparison of RNA hybridization, hemagglutination assay, titration of infectious virus and immunofluorescence as methods for monitoring influenza virus replication in vitro. J. Virol. Methods. 1998;74(1):57–66. doi: 10.1016/s0166-0934(98)00071-8. [DOI] [PubMed] [Google Scholar]
- Rimmelzwaan GF, Verburgh RJ, Nieuwkoop NJ, Bestebroer TM, Fouchier RA, Osterhaus AD. Use of GFP-expressing influenza viruses for the detection of influenza virus A/H5N1 neutralizing antibodies. Vaccine. 2011;29(18):3424–3430. doi: 10.1016/j.vaccine.2011.02.082. [DOI] [PubMed] [Google Scholar]
- Roberts KL, Manicassamy B, Lamb RA. Influenza A virus uses intercellular connections to spread to neighboring cells. J. Virol. 2015;89(3):1537–1549. doi: 10.1128/JVI.03306-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe T, Abernathy RA, Hu-Primmer J, Thompson WW, Lu X, Lim W, Fukuda K, Cox NJ, Katz JM. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J. Clin. Microbiol. 1999;37(4):937–943. doi: 10.1128/jcm.37.4.937-943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibert CW, Kaminski M, Philipp J, Rubbenstroth D, Albrecht RA, Schwalm F, Stertz S, Medina RA, Kochs G, Garcia-Sastre A, Staeheli P, Palese P. Oseltamivir-resistant variants of the 2009 pandemic H1N1 influenza A virus are not attenuated in the guinea pig and ferret transmission models. J. Virol. 2010;84(21):11219–11226. doi: 10.1128/JVI.01424-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2(12):905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Sharma P, Yan F, Doronina VA, Escuin-Ordinas H, Ryan MD, Brown JD. 2A peptides provide distinct solutions to driving stop-carry on translational recoding. Nucleic Acids Res. 2012;40(7):3143–3151. doi: 10.1093/nar/gkr1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw ML, Palese P. Fields Virology. 6 edition. Lippincott Williams and Wilkins; Philadelphia: 2013. Orthomyxoviridae. [Google Scholar]
- Sidwell RW, Bailey KW, Wong MH, Barnard DL, Smee DF. In vitro and in vivo influenza virus-inhibitory effects of viramidine. Antiviral Res. 2005;68(1):10–17. doi: 10.1016/j.antiviral.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Sidwell RW, Huffman JH, Khare GP, Allen LB, Witkowski JT, Robins RK. Broad-spectrum antiviral activity of Virazole: 1-beta-d-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science. 1972;177(4050):705–706. doi: 10.1126/science.177.4050.705. [DOI] [PubMed] [Google Scholar]
- Subbarao K, Katz JM. Influenza vaccines generated by reverse genetics. Curr. Top. Microbiol. Immunol. 2004;283:313–342. doi: 10.1007/978-3-662-06099-5_9. [DOI] [PubMed] [Google Scholar]
- Sun W. Approval Letter—Fluarix Quadrivalent (BL 125127/513) U.S. Food and Drug Administration (FDA); 2012. [Google Scholar]
- Tan GS, Krammer F, Eggink D, Kongchanagul A, Moran TM, Palese P. A pan-H1 anti-hemagglutinin monoclonal antibody with potent broad-spectrum efficacy in vivo. J. Virol. 2012;86(11):6179–6188. doi: 10.1128/JVI.00469-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289(2):179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. U. S. A. 2012;109(11):4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9(10):e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Paragas J, Dover JE, Gupta M, Goldsmith CS, Huggins JW, Nichol ST. Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology. 2005;332(1):20–27. doi: 10.1016/j.virol.2004.10.048. [DOI] [PubMed] [Google Scholar]
- Tran V, Moser LA, Poole DS, Mehle A. Highly sensitive real-time in vivo imaging of an influenza reporter virus reveals dynamics of replication and spread. J. Virol. 2013;87(24):13321–13329. doi: 10.1128/JVI.02381-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Voris LP, Belshe RB, Shaffer JL. Nosocomial influenza B virus infection in the elderly. Ann. Intern. Med. 1982;96(2):153–158. doi: 10.7326/0003-4819-96-2-153. [DOI] [PubMed] [Google Scholar]
- Vigil A, Park MS, Martinez O, Chua MA, Xiao S, Cros JF, Martinez-Sobrido L, Woo SL, Garcia-Sastre A. Use of reverse genetics to enhance the oncolytic properties of Newcastle disease virus. Cancer Res. 2007;67(17):8285–8292. doi: 10.1158/0008-5472.CAN-07-1025. [DOI] [PubMed] [Google Scholar]
- Webster RG, Webster RCN, Stoehr K. WHO Manual on Animal Influenza Diagnosis and Surveillance. 2002 WHO/CDS/CSR/NCS/2002.5 Rev. 1. [Google Scholar]
- Wright PF, Neumann G, Kawaoka Y. Orthomyxoviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, editors. Fields Virology. 5th edition. Lippincott Williams and WIlkins; 2007. [Google Scholar]
- Yang LP. Recombinant trivalent influenza vaccine (flublok®): a review of its use in the prevention of seasonal influenza in adults. Drugs. 2013;73(12):1357–1366. doi: 10.1007/s40265-013-0103-6. [DOI] [PubMed] [Google Scholar]