Abstract
Stiffness of biomaterial substrates plays a critical role in regulation of cell behavior. Although the effect of substrate stiffness on cell behavior has been extensively studied, molecular mechanisms of regulation rather than those involving cytoskeletal activities still remain elusive. In this study, we fabricated aligned ultrafine fibers and treated the fiber with different annealing temperatures to produce fibrous substrates with different stiffness. Human mesenchymal stem cells (hMSCs) were then cultured on these fibrous substrates. Our results showed that annealing treatment did not change the diameter of electrospun fibers but increased their polymer crystallinity and mechanical properties. The mRNA expression of RUNX2 was upregulated while the mRNA expression of scleraxis was downregulated in response to an increase in substrate stiffness, suggesting that increased stiffness favorably drives hMSCs into the osteogenic lineage. With subsequent induction of osteogenic differentiation, osteogenesis of hMSCs on stiffer substrates was increased compared to that of the cells on control substrates. Cells on stiffer substrates increasingly activated AKT and YAP and upregulated transcript expression of YAP target genes compared to those on control substrates, and inhibition of AKT led to decreased expression of YAP and RUNX2. Furthermore, macrophage migration inhibitory factor (MIF) was increasingly produced by the cell on stiffer substrates, and knocking down MIF by siRNA resulted in decreased AKT phosphorylation. Taken together, we hereby demonstrate that simply using the annealing approach can manipulate stiffness of an aligned fibrous substrate without altering the material chemistry, and substrate stiffness dictates hMSC differentiation through the MIF-mediated AKT/YAP/RUNX2 pathway.
Keywords: electrospun fiber, stiffness, mesenchymal stem cell, differentiation, macrophage migration inhibitory factor
1. Introduction
Mesenchymal stem cells (MSCs) are capable of differentiating into a variety of connective tissue cell types, such as osteoblasts, chondrocytes, adipocytes, myoblasts [1]. It has been reported that the extracellular matrix (ECM) delivers biochemical and biophysical signals to direct MSC differentiation [2-5]. Among the biophysical signals, ECM stiffness plays an important role in modulating stem cell differentiation and functions [6, 7]. It has been demonstrated that MSCs cultured on substrates with stiffness mimicking that of brain, muscle, or bone tissue preferentially become neurons, muscles, or osteoblasts, respectively [7]. Moreover, the early mesendoderm differentiation and terminal osteogenic differentiation of embryonic stem cells (ESCs) were both enhanced when ESCs were cultured on a stiff polydimethylsiloxane substrate compared to a soft one [8]. Hence, a deeper understanding of how stem cells respond to matrix stiffness is crucial for developing viable strategies to improve the efficiency of stem cell differentiation.
Recently, electrospun ultrafine fibers have attracted great interest in constructing biomimetic scaffolds for tissue engineering [9-11]. Previous studies have demonstrated that cell adhesion, proliferation, and differentiation can be regulated by the polymer chemistry and structural architecture of electrospun fibers [12-14]. However, there are few reports focusing on how the stiffness of electrospun fibers regulates MSC differentiation. Jiang et al. altered the stiffness of pullulan/dextran nanofibers by differential in situ crosslinking during electrospinning and they found stiff nanofibers promoted MSC neuronal differentiation compared to soft ones [15]. However, their approach of using different crosslinkers to change substrate stiffness also unavoidably alters chemistry of the material. It is therefore difficult to conclude if substrate stiffness, material chemistry, or both is the cause to regulation of cell response in their study. With that, it is important to have a setup capable of decoupling substrate stiffness from material chemistry for studying the effect of substrate stiffness on cell response. Recently, Baker et al. have used UV to modulate the stiffness of electrospun methacrylated dextran fibers without altering the fiber chemistry. They demonstrated that lower fiber stiffness permitted active cellular forces to recruit nearby fibers, dynamically increasing ligand density at the cell surface and promoting the formation of focal adhesions and related signaling [16].
The pro-inflammatory cytokine macrophage migration inhibitory factor (MIF) is produced by several cell types, including monocytes, macrophages, vascular smooth muscle cells, and cardiomyocytes [17-19]. MIF plays multiple roles in mediating inflammation, apoptosis, autophagy, and carbohydrate metabolism [20-23]. It has also been shown that MIF production can be modulated in response to arterial stiffness [24]. Recent studies have shown that MIF regulates AKT activity in various types of cells to affect their behavior [25-28]. Our group has previously demonstrated that endogenous MIF produced in hMSCs under hypoxia activates AKT signaling to delay the progression of cellular senescence [28]. While a previous study has shown that a substrate with higher stiffness enhances osteogenic differentiation of MSCs through activation of AKT signaling [29], it is unclear whether MIF is involved in the regulatory mechanism. Moreover, yes-associated protein (YAP) is a transcription factor that is known to play a role in stiffness-mediated cell activities [30], which suggests that YAP may be a downstream molecule of the AKT signaling pathway.
In this study, we hypothesized that stiff substrates increase the expression of MIF in hMSCs, which in turn regulates AKT/YAP signaling to direct differentiation of hMSCs. To test the hypothesis, aligned fibrous substrates were fabricated by stable jet electrospinning (SJES), and then subjected to annealing treatment to alter the substrate stiffness. In addition to determining the effect of substrate stiffness on regulation of hMSC behavior, we were particularly interested in finding out whether the regulation is mediated through the MIF-mediated AKT/YAP pathway. We aimed to identify a new mechanism, rather than cytoskeleton-mediated regulation, of how hMSC activities are modulated by substrate stiffness.
2. Materials and methods
2.1. Electrospun aligned ultrafine fibers and annealing treatment
Aligned PLLA ultrafine fibers were electrospun as described in our pervious study [31]. Briefly, poly (ethylene oxide) (PEO, Mw ~5,000,000, Sigma-Aldrich, 1 w/v%)-doped poly(L-lactic acid) (PLLA, Mw 100,000, Polysciences, 5 w/v%) in 2,2,2-trifluoroethanol (TFE, Sigma-Aldrich) was prepared and then electrospun onto a rotatory shaft at the rotating speed of 1,000 rpm to produce aligned ultrafine fibers. Electrospun aligned fibrous mats were placed in a vacuum oven for about 24 h to remove any residual organic solvent before further use. Annealing of fibers was conducted by first treating fibrous mats with the temperature at 65°C (65PLLA) or 75°C (75PLLA) under mechanical tension for 3 h and then allowing it to cool down under room temperature. Fibrous mats without annealing treatment were used as an untreated control (PLLA).
2.2. Characterization of annealing-treated aligned ultrafine fibers
Morphology of electrospun fibrous mats was analyzed by scanning electron microscopy (SEM, TM-1000, Hitachi) at an accelerating voltage of 8-10 kV. Prior to imaging, samples were sputter-coated with gold for 50 sec. The average diameter of electrospun fibers was determined by measuring fibers shown in the SEM images using the ImageJ software. For each sample, an average of 50 ultrafine fibers was counted.
X-ray diffractometer (D/Max-2550 PC, Rigaku, Japan) was used to determine the X-ray diffraction (XRD) pattern of as-electrospun PLLA fibers with Cu Kα radiation in the range of 2θ around 5°~35°. To determine the surface roughness of fibrous substrates, atomic force microscopy (AFM, Dimension FastScan Bio, Bruker, Germany) was used to scan the surface of specimens.
Mechanical properties of substrates were determined using a tensile testing machine (H5K-S, Hounsfield, UK) equipped with a 50 N load cell. Rectangular-shaped specimens (50 mm × 10 mm × 0.10-0.15 mm) were stretched at a constant cross-head speed of 10 mm/min. Five specimens were tested for each type of samples. In addition to the analysis of tensile properties, the surface stiffness of fibrous substrates was determined by measuring mechanical properties of individual fibers subjected to nanoindentation using AFM. Spherical contact tips were applied in contact mode to image and indent fibers to determine Young’s modulus based on the Hertz model.
2.3. Human MSC isolation and culture
With approval from the Institutional Review Board at the University of Wisconsin-Madison, bone marrow-derived hMSCs were harvested from femoral heads of patients undergoing total hip arthroplasty. Human MSCs were isolated following a previously described protocol. Briefly, after being curetted from the interior of femoral head and neck, whole bone marrow was mixed with Dulbecco’s modified Eagle medium (DMEM; Gibco, Carlsbad, CA, USA). A syringe with an 18-gauge needle was used to filter out bone debris from the bone marrow/DMEM mixture. The collected medium was then centrifuged at 1,000 rpm for 5 min. After removing the supernatant, the resulting cell pellet was reconstituted using 25 mL of Hank’s Balanced Salt Solution (Invitrogen, Carlsbad, CA, USA), and then slowly added into a 50-mL conical tube containing 20 mL of Ficoll solution (GE Health, Pittsburgh, PA, USA). After centrifugation at 500 g for 30 min, mononuclear cells were collected and plated in cell culture flasks with culture medium composed of low-glucose DMEM, 10% fetal bovine serum (FBS; Atlanta Biologicals, Atlanta, GA, USA) and antibiotics. The cells were maintained in an incubator at 37°C in a humidified 5% CO2 atmosphere. When reaching 70 to 80% density confluence, the cells were trypsinized using 0.05% trypsin/EDTA (Gibco) and re-plated at a seeding density of 1000 cells/cm2. Culture medium was replaced every 3 days. Cells were expanded for 2 passages prior to experiments.
2.4. Cell seeding and culture on fibrous substrates
For cell seeding, PLLA ultrafine fibrous substrates with different stiffness were sterilized by ultraviolet (UV) light irradiation for 2 h. Substrates were then hydrated with a series of decreasing ethanol concentration gradients (100%, 70%, 50%, and 25%), pure water, and Hank’s Balanced Salt Solution (HBSS) (Thermo-Fischer Scientific, Pittsburgh, PA). Six-well tissue culture plates were coated with 0.5% polyhema/ethnol solution at 37°C overnight. Human MSCs were then seeded onto fibrous substrates at a density of 5,000 cells/cm2 for cell morphology and proliferation studies. For gene expression and western blotting assays, the cells were seeded onto fibrous substrates at a density of 50,000 cells/cm2. Cell culture medium was changed every other day.
2.5. Cell morphology and proliferation assays
After 3 days of culture, the morphology of hMSCs on fibrous substrates was analyzed. Briefly, cellular substrates were fixed with 4% paraformaldehyde, washed three times with PBS, and then permeabilized with 1 mL of 0.1% (v/v) Triton X-100 (Aldrich, USA). The samples were then washed again with PBS, followed by 30-min incubation with 150 μL of rhodamine-conjugated phalloidin (1:40 dilution in PBS, Gibco, USA) in the dark at room temperature for staining actin filaments. After washed three additional times with PBS, the cells were stained for cell nuclei with 1 mg/mL 4’,6-diamidino-2-phenylindole (DAPI) (Sigma, USA) in PBS for 15 min at 37°C, and observed under a fluorescence microscope (Olympus, Shinjuku, Tokyo, Japan). To quantitatively evaluate cellular shape, ImageJ was used to delineate the contour of each cell in microscopic images. The shape index of hMSCs was assessed by the aspect ratio (AR), defined by the equation AR = L/W, where L and W are the cell length and width, respectively. The AR, ranging from 1 (a perfect circle) to ∞ (a straight line), reveals the degree of elongation of a cell. Human MSCs seeded on substrates with different stiffness were harvested at Days 1, 4 and 7. The total amount of double-stranded DNA (dsDNA) was analyzed to determine cell proliferation using the Quant-iT PicoGreen dsDNA assay (Invitrogen) following the manufacturer’s instructions.
2.6. Total RNA extraction and quantitative RT-PCR analysis
For quantitative PCR analysis, total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. Briefly, 1 mL of Trizol solution was first added to lyse hMSCs within fibrous substrates. After grinding the cellular substrates using a pestle, 200 μL of chloroform was added before RNA was precipitated with 500 μL of isopropanol. After centrifugation at 12,000 rpm for 15 min, the resulting pellet was washed three times with 1 mL of 75% ethanol and then dissolved in 20 μL of DEPC-treated water. The amount of RNA yield was determined based on the measurement of A260/A280 using the NanoDrop 1000 spectrophotometer (Thermo Scientific, Waltham, MA). First strand cDNA was reverse-transcribed using the High Capacity cDNA Reverse Transcription kit (AB Applied Biosystems, Carlsbad, CA) following the manufacturer’s instructions. Quantitative PCR analysis was performed using SYBR Green Supermix (Bio-Rad, Hercules, CA) with the primers targeting octamer binding transcription factor 3/4 (OCT3/4), NANOG, scleraxis (SCX), RUNX2, sex determining region Y-box 9 (SOX9), peroxisome proliferator-activated receptor gamma (PPARG), MIF, connective tissue growth factor (CTGF), ankyrin repeat domain 1 (ANKRD1), alkaline phosphatase (ALP), osteocalcin (OCN) and the internal control ubiquitin C (UBC). The primer sequences are listed in Table 1. The relative mRNA expression levels of a target gene were determined using the 2-ΔCt method and compared to those of the housekeeping gene UBC.
Table 1.
Primer sequences for quantitative RT-PCR analysis.
| Gene name | Accession number | Primer sequences (5’ to 3’) |
|---|---|---|
| UBC | NM_021009.4 | F:TGAAGACACTCACTGGCAAGACCA |
| R:CAGCTGCTTTCCGGCAAAGATCAA | ||
| OCT3/4 | NM_002701.4 | F:TGGAGAAGGAGAAGCTGGAGCAAA |
| R: GGCAGATGGTCGTTTGGCTGAATA | ||
| NANOG | NM_021865.2 | F:GCTGAGATGCCTCACACGGAG |
| R:TCTGTTTCTTGACCGGGACCTTGTC | ||
| SCX | NM_001090514.1 | F: ACACCCAGCCCAAACAGA |
| R: GCGGTCCTTGCTCAACTTTC | ||
| SOX9 | NM_000346.3 | F: TAAAGGCAACTCGTACCCAA |
| R: ATTCTCCATCATCCTCCACG | ||
| RUNX2 | NM_004348.3 | F: GGTTCCAGCAGGTAGATGAG |
| R: AGACACCAAACTCCACAGCC | ||
| PPARG | NM_138711.3 | F: ATGACAGCGACTTGGCAATA |
| R: GGCTTGTAGCAGGTTGTCTTG | ||
| ALP | NM_000478.3 | F:CAAAGGCTTCTTCTTGCTGG |
| R:GGTCAGAGTGTCTTCCGAGG | ||
| OCN | NM_199173.3 | F: GACTGTGACGAGTTGGCTAGA |
| R: GGAAGAGGAAAGAAGGGTGC | ||
| MIF | NM_002415.1 | F: CTCCACCTTCGCCTAAGAGC |
| R: TTCTCCCCACCAGAAGGTTG | ||
| ANKRD1 | NM_014391.2 | F: AGTAGAGGAACTGGTCACTGG |
| R: TGGGCTAGAAGTGTCTTCAGAT | ||
| CTGF | NM_001901.2 | F: AGGAGTGGGTGTGTGACGA |
| R: CCAGGCAGTTGGCTCTAATC |
Forward and reverse primers are indicated as “F” and “R”, respectively.
2.7. Protein extraction and western blotting analysis
To extract protein from hMSCs, the cells were lyzed using RIPA buffer composed of 50 mM Tris-HCl (pH 7.5), 0.25% Na-deoxycholate, 1% Nonidet P-40, 150 mM NaCl, 1 mM EDTA, and complete protease inhibitor cocktail tablets (Roche, Indianapolis, IN, USA). After centrifugation at 14,000 rpm for 10 min, the supernatant was collected. Protein concentration was measured using the BCA Protein Assay kit (Pierce, Rockford, IL, USA). A 20-μg protein sample was loaded into each lane of a 10% polyacramide gel (Bio-Rad) for electrophoresis, and the separated proteins were then transferred from the gel onto a polyvinylidene fluoride membrane (Bio-Rad). The membrane was incubated with primary antibodies against MIF (R&D Systems, Minneapolis, MN), AKT, phospho-AKT (Ser473), YAP, RUNX2 and GAPDH (Cell Signaling, Danvers, MA, USA) in a blocking solution composed of Tris-buffered saline containing 5% nonfat milk (Bio-Rad) and 0.1% Tween20 (Sigma-Aldrich) overnight at 4°C. After removing unbound antibodies, the membrane was incubated with horseradish peroxidase-linked secondary antibody in the blocking solution for 1 h at room temperature. The immuno-detected protein bands on the membrane were visualized using the SuperSignal West Pico Chemiluminescent Substrate (Pierce), and then documented by the Kodak Image Station 4000R Pro system (Kodak, Rochester, NY, USA).
2.8. Evaluation of osteogenic differentiation of hMSCs
Human MSCs seeded on PLLA and 75PLLA were first maintained in growth medium for 7 days and then induced by osteogenic medium (DMEM-LG, 10% FBS, 0.1 mM dexamethasone, 10 mM b-glycerophosphate, 50 mg/mL ascorbic acid-2-phosphate (Sigma-Aldrich), and antibiotics) for another 7 or 14 days. To evaluate osteogenesis, the expression of mRNA transcripts of bone-related markers was analyzed by quantitative RT-PCR. To quantify ALP activity, the cells were first digested in the digestion buffer containing 2% Triton X-100, 0.15 mM Tris base, 0.1 mM ZnCl2, 0.1 mM MgCl2·6H2O at pH 9.0 for 1 h at 37°C and then overnight at 4°C. ALP activity was measured as the reaction kinetics with p-nitrophenyl phosphate (Sigma-Aldrich). The results were normalized to the dsDNA content determined by the PicoGreen assay (Invitrogen).
2.9. Immunofluorescent labeling
The procedure of immunofluorescence staining was performed following our previously published method [32]. Briefly, after 14 days of osteogenic induction, cellular substrates were collected and fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 30 min. Substrates were then washed three times with PBS, followed by permeabilization with 1 mL of 0.1% (v/v) Triton X-100 (Aldrich, USA). After additional washing, samples were blocked with a buffer solution containing 1% bovine serum albumin in PBS for 30 min at room temperature and washed again with PBS. Thereafter, samples were stained with human OCN antibody (R&D Systems, Minneapolis, MN, USA) diluted to 1:200 in blocking buffer for 45 min at room temperature, followed by a 45-min incubation with anti-mouse IgG fluorescein isothiocyanate (FITC) (eBioscience, San Diego, CA, USA) at room temperature. Stained samples were then mounted with the ProLong Gold antifade reagent containing 4’, 6-diamidino-2- phenylindole (DAPI) (Life Technologies, Grand Island, NY, USA) and observed under a fluorescence microscope (Olympus, Shinjuku, Tokyo, Japan).
Immunofluorescence staining following the same procedure described above was used to detect YAP in nuclei of hMSCs cultured on fibrous substrates. Specifically, cellular substrates were incubated with a primary antibody detecting YAP (sc-271134, Santa Cruz Biotechnology, Santa Cruz, CA, USA), followed by incubation with a secondary antibody conjugated with FITC (eBioscience, San Diego, CA, USA) before being imaged by a confocal laser scanning microscope (Nikon A1RS, Japan) for analysis.
Bromodeoxyuridine (BrdU) (Roche, St. Louis, MO, USA) was used to label mitotic cells seeded on fibrous substrates. After 4 days of culture, cells on substrates were incubated with 10 μM BrdU at 37°C for 24 h and then fixed with 4% paraformaldehyde in PBS at 4°C for 1 h. After washed with PBS 3 times, cellular substrates were incubated in 1 mL of 0.1% Triton X-100 (Aldrich, USA), followed by an additional incubation with 2N HCl for 30 min at room temperature to denature the DNA. After additional wash, cellular substrates were incubated with an FITC-conjugated antibody detecting BrdU localized in cell nuclei (sc-32323, Santa Cruz Biotechnology, Santa Cruz, CA, USA).
2.10. Loss-of-function assays
To investigate whether stiff substrates regulate the osteogenic potential of hMSCs by activating the AKT signaling pathway, hMSCs cultured on 75PLLA and control PLLA were treated with 0.1 μM AKT Inhibitor IV (EMD Millipore) for 6 h before analysis.
To determine the role of MIF in the mechanism of our interest, hMSCs maintained in basal growth medium after 48 h were transfected with MIF siRNA (sc-37137; Santa Cruz Biotechnology, Santa Cruz, CA, USA) using the Lipofectamine 2000 transfection reagent (Life Technologies) following the manufacturer’s protocol. Briefly, hMSCs were transfected and incubated at 37°C for 6 h and further cultured with 1 mL of DMEM containing 20% FBS without antibiotics for additional 18 h. The transfection medium was then removed and replaced with basal growth medium to allow the cells to grow for another 24 h. Control cells were treated with scrambled siRNA (sc-37007). The cells were then collected and seeded on PLLA and 75PLLA substrates for 6 h.
2.11. Statistical analysis
All quantitative data were presented as the means ± standard deviation, as the assays were performed with samples in technical triplicate (n = 3). Statistical analysis was performed using the Origin software (OriginLab, Northampton, MA, USA). One-way ANOVA with post-hoc Tukey’s HSD Test was used to make pairwise comparisons between groups. P-values < 0.05 were considered statistically significant.
3. Results
3.1. Characterization of annealing-treated fibrous substrates
Aligned fibrous substrates produced by stable jet electrospinning and then annealing-treated, 65PLLA (Fig. 1B) and 75PLLA (Fig. 1C), exhibited the morphology similar to that of control PLLA (Fig. 1A). The average diameter of PLLA, 65PLLA or 75PLLA fibers was measured to be 2.07 ±0.38, 1.86 ±0.24, or 1.92 ±0.22 μm, respectively. To determine the impact of annealing treatment on the chemical structure of PLLA ultrafine fibers, XRD profiles of the samples were analyzed (Fig. 1D). The result of control PLLA did not show noticeable crystalline diffraction peaks, whereas that of 65PLLA or 75PLLA showed a crystalline diffraction peak at about 16.7° of the 2θ angle, which indicates the presence of the α-form homocrystal of PLLA [33]. The XRD results suggest that annealing treatment increases the crystallinity of PLLA. Specifically, the crystallinity of 65PLLA and 75PLLA were determined to be 27.21% and 28.54%, respectively, 1.35- and1.41- fold higher than that of control PLLA (20.22%). Further, the measurement of surface roughness by AFM showed that the 3 fibrous substrates shared similar roughness with 112 ± 23.3, 117 ± 37.5, and 133 ± 19.1 nm for control PLLA, 65PLLA, and 75PLLA, respectively (Fig. 1E). While similar in fiber diameter and microstructure, PLLA, 65PLLA, and 75PLLA exhibited quite different mechanical properties (Fig. 1F). The tensile modulus and strength of control PLLA are 696.90 ± 36.34 MPa and 16.83 ± 3.48 MPa, respectively, compared to those of 65PLLA 820.79 ± 32.83 MPa and 21.41 ± 1.25 MPa and those of 75PLLA 944.57 ± 29.14 MPa and 23.53 ± 2.45 MPa (Fig. 1G, H). To determine the surface stiffness of fibrous substrates, force volume images and nanoindentation force curves of AFM following the Hertz model were analyzed. The calculated Young’s modulus of an individual fiber of control PLLA, 65PLLA or 75PLLA was 77.4 ± 17.7, 729 ± 229, or 1124 ± 119 MPa, respectively (Fig. 1I). The results suggest that annealing treatment is able to increase the mechanical properties of fibrous substrates.
Figure 1.
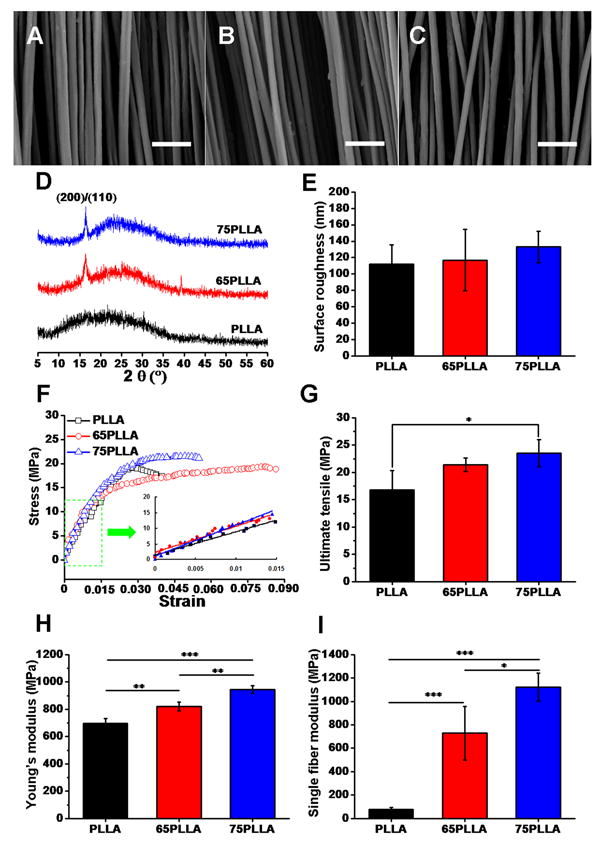
Characterization of fibrous substrates. SEM micrographs of control PLLA (A), 65PLLA (B), and 75PLLA (C). Analysis of XRD (D), surface roughness (E), tensile properties (F-H), and single fiber modulus (I) of fibrous substrates with or without annealing treatment. * p < 0.05, **p < 0.01, ***p < 0.001, n = 5, Scale bar = 10 μm.
3.2. Stiffness of fibrous substrates affected cell morphology, cytoskeleton, and proliferation
We next compared cell attachment and morphology on fibrous substrates with different stiffness. After 3 days, hMSCs on stiff substrates appeared to be more elongated than those on soft ones, and stress fibers in the culture of stiff substrates were more prominent than those in the culture of soft substrates (Fig. 2A). In addition, ARs of hMSCs on substrates were determined and the results showed that the average of ARs of cells on control PLLA was lower than that of cells on 65PLLA and 75PLLA although the difference was not significant (Fig. 2B), suggesting that the AR of hMSCs increases with substrate stiffness. The analysis of cell proliferation determined by BrdU incorporation revealed that significantly more hMSCs (86.4 ± 5.0%) stained positive for BrdU on 75PLLA than those (74.7 ± 3.2%) on 65PLLA or (41.4 ± 4.9%) on control PLLA (Fig. 2C, D), suggesting that cell proliferation increases with stiffness of fibrous substrates. Furthermore, the PicoGreen assay was also performed to measure total DNA amounts of hMSCs on different fibrous substrates at different time points. By Days 4 and 7, no significant difference in cell number between 65PLLA and 75PLLA was found but there were significantly more cells on 65PLLA and 75PLLA than on control PLLA (Fig. 2E).
Figure 2.
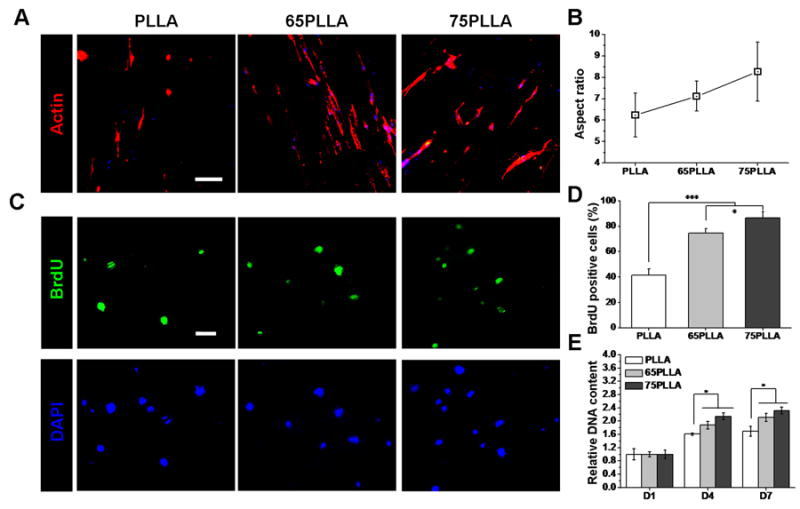
Cell morphology and proliferation regulated by stiffness of fibrous substrates. (A) Cytoskeleton of hMSCs on PLLA, 65PLLA, and 75PLLA after 3 days of culture stained by phalloidin. Scale bar = 100 μm. (B) Aspect ratios of hMSCs cultured on different fibrous substrates. (C) Proliferation of hMSCs cultured on PLLA, 65PLLA, and 75PLLA for 4 days detected by BrdU staining. Scale bar = 100 μm. (D) Quantitation of hMSC proliferation measured as the percentage of BrdU-positive cells in DAPI-positive cells. *p < 0.05, ***p < 0.001. (E) Numbers of hMSCs cultured on different fibrous substrates determined by quantification of total DNA content. *p < 0.05, n = 3.
3.3. Stiffness of fibrous substrates directed lineage-specific commitment of hMSCs
To investigate whether substrate stiffness can preferentially determine the fate of hMSCs prior to differentiation induction, the cells were cultured on substrates with different stiffness in basal growth medium and analyzed for mRNA expression using quantitative RT-PCR. Stiffer substrates significantly downregulated the expression levels of stem cell stemness markers, OCT3/4 and NANOG compared to control substrates on Day 3 (Fig. 3). In addition, the expression levels of SCX, a key transcription factor for tenogenic differentiation, in hMSCs cultured on stiffer substrates were downregulated compared to those in the cells on control substrates while the expression of RUNX2, a transcription factor essential for osteogenesis, was upregulated in hMSCs cultured on stiffer substrates compared to that in the cells on control substrates. After 21 days of culture, the expression levels of PPARG and SOX9, key transcription factors for adipogenesis and chondrogenesis, respectively were comparable among the cells on all substrates while it is notable that hMSCs on 75PLLA expressed a significantly higher level of RUNX2 than the cells on control PLLA (Fig. 3). These findings suggest that a stiff substrate downregulates the stemness property of hMSCs and directs the cells toward the osteogenic lineage.
Figure 3.
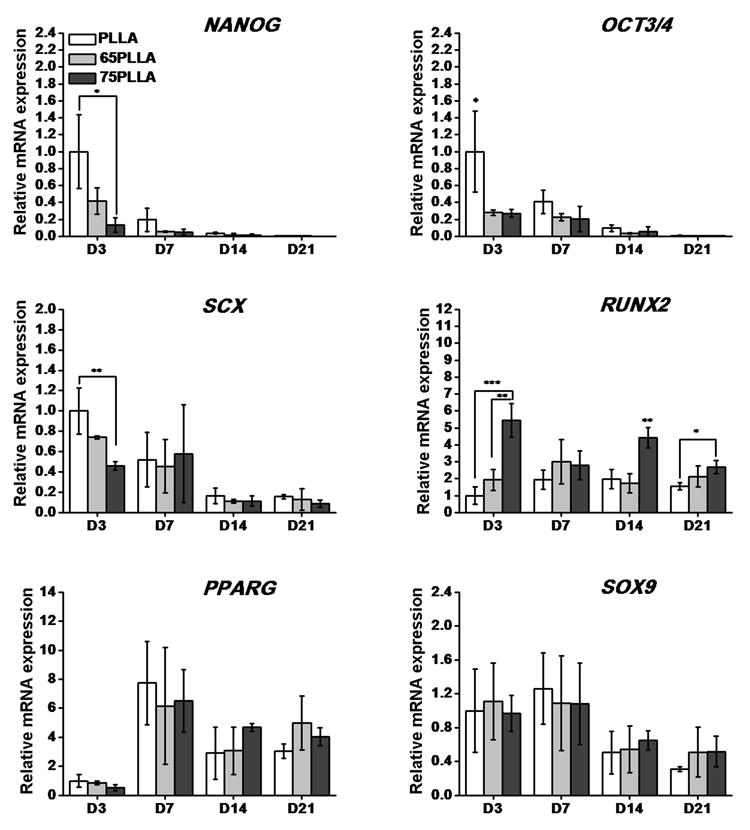
Expression levels of mRNA transcripts of stemness markers and lineage-specific transcription factors in hMSCs on different fibrous substrates prior to differentiation induction. *p < 0.05, **p < 0.01, ***p < 0.001, n = 3.
We next investigated whether the upregulated expression of RUNX2 in hMSCs stimulated by stiff substrates was able to drive enhanced osteogenesis upon differentiation induction. To this end, hMSCs were first cultured on 75PLLA and control PLLA substrates in basal growth medium for 7 days and then induced by differentiation medium for osteogenesis. After 7 and 14 days of osteogenic induction, the mRNA levels of bone-related markers, RUNX2, ALP, and OCN, in hMSCs on 75PLLA were upregulated compared to those in the cells on PLLA (Fig. 4A-C). In addition to the mRNA expression of bone-related markers, the quantitative results of ALP activity indicated that hMSCs on 75PLLA produced more ALP during 14 days of culture and significantly more on Day 7 than those on control PLLA (Fig. 4D). The analysis of immunofluorescence staining showed that a greater intensity of OCN labeling was observed in hMSCs on 75PLLA compared to that in the cells on control PLLA (Fig. 4E). Collectively, the results of mRNA and protein expression suggest that stiff 75PLLA substrates drive and enhance osteogenic differentiation of hMSCs compared to control PLLA.
Figure 4.
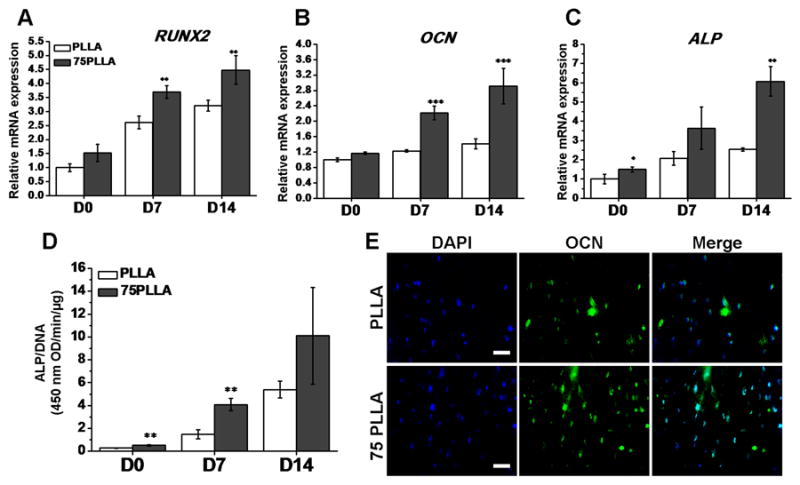
Ostegenic differentiation of hMSCs cultured on PLLA and 75PLLA. Cells were cultured in basal medium for 7 days before induced in osteogenic differentiation medium for additional 14 days. (A-C) Expression levels of mRNA transcripts of bone-associated markers in hMSCs determined by RT-PCR. (D) ALP activity in hMSCs measured and normalized by DNA content. (E) Immunofluorescence labeling of OCN (green) in hMSCs with cell nuclei stained with DAPI (blue). Scale bar = 50 μm. *p < 0.05, **p < 0.01, ***p < 0.001, n = 3.
3.4. Substrate stiffness modulated the osteogenic potential of hMSCs through a MIF-mediated mechanism
To identify a mechanism by which substrate stiffness regulates the expression level of RUNX2 of hMSCs as shown in Figure 3, we picked 75PLLA as a representative stiff substrate and PLLA as a control substrate to culture hMSCs for comparison. Given that AKT and YAP have been reported to be involved in the regulation of osteogenesis of hMSCs [29, 30], we focused on investigating the role of AKT and YAP in the mechanism regulated by substrate stiffness, which thereby directs hMSCs fates prior to differentiation induction. The results of western blotting showed that the levels of total AKT and pAKT in hMSCs on stiff substrates were higher than those in the cells on control substrates, suggesting that AKT activation is increased with substrate stiffness (Fig. 5A). Likewise, 75PLLA increased the expression level of total YAP (Fig. 5A), localization of YAP in cell nuclei (Fig. 5B, C) and the expression level of RUNX2 (Fig. 5A) in hMSCs compared to control PLLA substrates. It is noted that more YAP was localized in nuclei of hMSCs on 75PLLA than on control PLLA (indicated by arrows) (Fig. 5B), resulting in a significant increase in the intensity of nuclear YAP in hMSCs on 75PLLA compared to that in the cell on control PLLA (Fig. 5C). Interestingly, we also found that the level of MIF in hMSCs cultured on 75PLLA was greater than that in the cells on control PLLA (Fig.5A). Significantly increased levels of YAP target genes, ANKRD1 (p < 0.05) and CTGF (p < 0.001) (Fig. 5D, E), were found in hMSCs on 75PLLA compared to those on control PLLA, indicating an increase in YAP activation in the 75PLLA culture. Consistent with the result of protein expression in Figure 5A, the expression of MIF and RUNX2 mRNA transcripts in hMSCs on 75PLLA was significantly greater (p < 0.05) than that in the cell on control PLLA (Fig. 5F, G). These results together suggest that MIF, AKT, YAP and RUNX2 may be involved in the mechanism by which stiff substrates direct hMSCs toward the osteogenic lineage.
Figure 5.
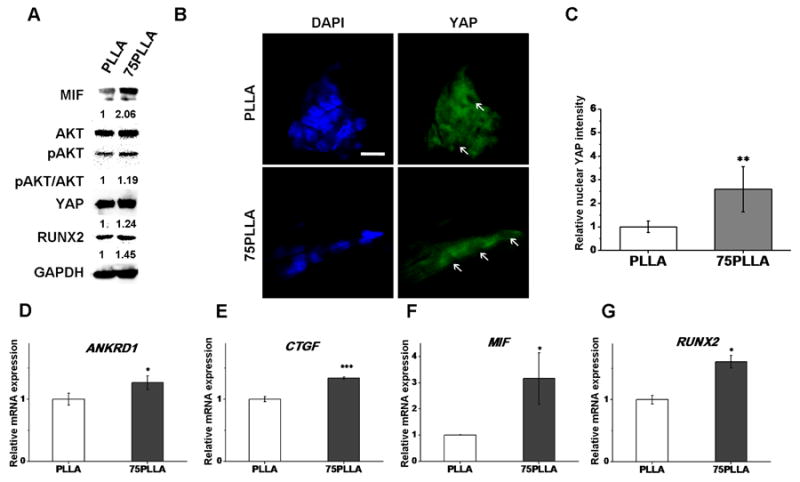
Identification of regulatory molecules involved in a substrate stiffness-induced mechanism. (A) Detection of signaling molecules and transcription factors in hMSCs cultured on PLLA and 75PLLA by western blotting of whole cell lysate. (B) Confocal laser scanning microscope images of immunofluorescent-labeled YAP (green) and DAPI-labeled nuclei (blue) in hMSCs cultured on fibrous substrates. Arrows indicate the location of cell nuclei. Scale bar = 200 μm. (C) Quantification of intensity of YAP staining in nuclei of hMSCs on PLLA and 75PLLA. (D-G) Expression levels of mRNA transcripts of YAP target genes, ANKRD1 and CTGF, MIF, and RUNX2 analyzed by quantitative RT-PCR. *p < 0.05, ***p < 0.001, n = 3.
To determine the role of AKT in the mechanism induced by substrate stiffness, we attenuated the activity of AKT by treating the cells cultured on 75PLLA with AKT inhibitor IV. Our results showed that AKT inhibitor IV was able to effectively reduce the activation of AKT without affecting the level of total AKT (Fig. 6A). The reduction of AKT activity also resulted in a decrease in the levels of YAP and RUNX2 but did not affect the level of MIF in hMSCs, suggesting that AKT regulates YAP and RUNX2 and MIF is likely to be an upstream molecule that activates AKT.
Figure 6.
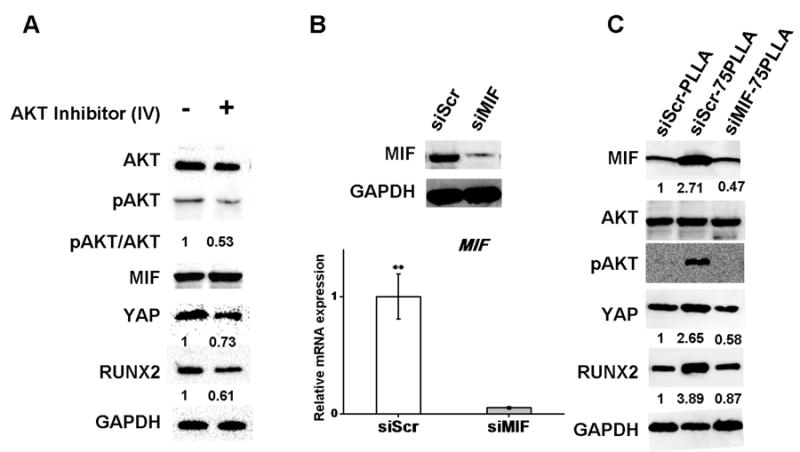
Detection of proteins and mRNA transcripts in hMSCs cultured on fibrous substrates. (A) Western blots of proteins extracted from cells on 75PLLA treated with or without AKT Inhibitor IV. (B) Protein and transcript expression of MIF from cells on 75PLLA treated with MIF siRNA or scrambled control. (C) Western blots of proteins extracted from cells on PLLA and 75PLLA treated with MIF siRNA or scrambled control.
To determine whether MIF acts through the AKT/YAP signaling pathway to regulate the expression of RUNX2, MIF siRNA (siMIF) was used to knock down MIF in hMSCs with scrambled siRNA (siSCR) as a negative control. Both of the transfected cells were then seeded on stiff and control substrates for 6 h before harvested for analysis. Both siMIF- and siSCR-transfected hMSCs displayed a similar spindle-shaped morphology (data not shown). The western blotting and mRNA expression results showed that our siRNA treatment effectively knocked down the mRNA and protein levels of MIF (Fig. 6B). Moreover, we found that knocking down the elevated level of MIF in hMSCs, induced by increased stiffness of 75PLLA, reduced the increase in activation of AKT/YAP signaling and regulation of RUNX2 expression (Fig. 6C). These results suggest that MIF is an upstream factor that regulates the AKT/YAP/RUNX2 signaling pathway to modulate the osteogenic potential of hMSCs.
4. Discussion
Studies have demonstrated that substrate stiffness plays a critical role in the regulation of stem cell-biomaterial interactions. In this study, we used electrospinning and annealing treatment to fabricate ultrafine fibrous substrates with the same microstructure and surface chemistry but distinct stiffness. Using this straightforward setup, we demonstrate that increasing substrate stiffness alone leads to elevated production of intracellular MIF, which increases the activation of AKT and YAP in hMSCs to enhance osteogenic differentiation, suggesting that the factor of substrate stiffness alone can regulate hMSC differentiation. Based on our findings, we propose a mechanism by which substrate stiffness regulates the osteogenic capacity of pre-differentiated hMSCs through the MIF-mediated AKT/YAP signaling pathway (Fig. 7).
Figure 7.
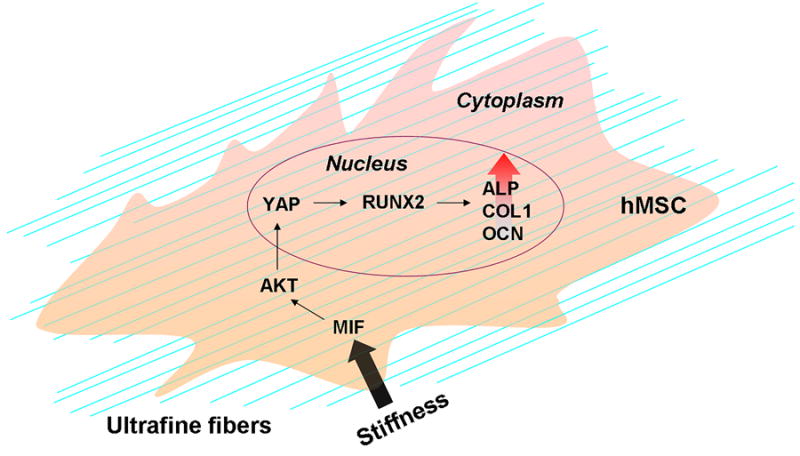
Illustration of an identified molecular mechanism elucidating how substrate stiffness directs hMSCs toward the bone-specific lineage.
Substrate stiffness is closely associated with the chemistry of a substrate, and it is challenging to keep one of these two properties unchanged while the other is altered. For instance, approaches used to alter substrate stiffness through modification of polymer compositions or crosslinking methods simultaneously introduce changes into surface chemistry of the substrates [15, 34-36]. Recently, Nam et al. have reported that they used core-shell electrospinning to produce different scaffolds that can share the same microstructure and surface chemistry but distinct mechanical properties [37], suggesting that it is possible to decouple substrate stiffness and chemistry for studying effects of substrate stiffness on cell behavior. However, their approach involving multiple procedures may not be as straightforward as our approach using annealing treatment in this study. Previous studies have demonstrated that annealing treatment can improve mechanical properties of polymeric materials by increasing the crystallinity of the polymer [38-40]. Likewise, in this study we have demonstrated that annealing treatment provides a simple approach to alter substrate stiffness without affecting substrate chemistry.
Our results show the effect of substrate stiffness on regulation of cell morphology and proliferation, in agreement with the findings of several published reports [8, 41-44]. In addition to these findings, several other studies have demonstrated that stiffness modulates osteogenesis of MSCs [7, 8, 41, 45], which is also consistent with our current finding. However, it should be noted that different from these previous studies, our study aimed to gain further understanding of how substrate stiffness regulates hMSC differentiation. In this study, we investigate effects of substrate stiffness on hMSCs with focus on the cell prior to differentiation induction rather than during differentiation. We demonstrate that stiff substrates prime the osteogenic capacity of pre-differentiated hMSCs by increasing the expression of RUNX2. It is known that RUNX2 is an early regulator of osteoblast differentiation [46], which functions as a key transcription factor to activate osteopontin and OCN through binding to the promoter region of these bone-associated molecules [47]. Our results also show that further induced by osteogenic medium, RUNX2-upregulated hMSCs on stiff substrates are increasingly differentiated into osteoblast-like cells compared to the cells on control substrates. These findings elucidate the importance of the mechanical environment on determination of the lineage-specific fate of hMSCs both before and during differentiation.
Our results show that stiffer substrates increase the amount of intracellular MIF and thereby activate AKT/YAP signaling in pre-differentiated hMSCs to upregulate the level of RUNX2 to prime their osteogenic capacity. Previous studies have shown that in addition to the AKT signaling pathway [29], other pathways involving integrin, RhoA, Smad, and/or FAK signaling molecules can be activated by substrate stiffness to regulate hMSC activities [48]. In this study, we have identified a new mechanism that involves MIF in regulation of hMSC activities by substrate stiffness. MIF is a proinflammatory cytokine capable of regulating various biological activities, such as immune response [49], neural stem cell properties [20] and cellular senescence [28, 50, 51]. Particularly, Rammos et al. have reported that MIF is associated with vascular dysfunction and its level in the body can be used as an independent indicator to predict arterial stiffness [24]. Seib et al. have also demonstrated that hMSCs cultured on rigid substrates produce more MIF than the cells on compliant substrates [2]. In his study, we have shown that MIF is involved in a regulatory mechanism by which substrates with greater stiffness direct hMSCs toward the osteogenic lineage by increasing the intracellular level of MIF in the cell. We have further demonstrated that knocking down MIF using siRNA leads to a drastic reduction in AKT phosphorylation and YAP expression, thus downregulating osteogenic differentiation of hMSCs. Although the finding of MIF regulating AKT activity in cell types other than hMSCs has been reported by other groups [25-28], to the best of our knowledge our new finding that stiff substrates direct hMSCs toward the osteogenic lineage through the MIF-mediated AKT/YAP mechanism has not been reported yet.
Orthopedic surgery for treatment of musculoskeletal disorders often causes changes in the structure and/or property of tissue, such as alteration of matrix stiffness [52, 53], which likely modifies the mechanical microenvironment of tissue. For example, heterotopic ossification commonly found in soft tissue, such as muscle or tendon, of orthopedic patients after surgery is a problem of bone being formed at an undesired tissue site. While it is unclear how heterotopic ossification is induced, one of the hypotheses to the pathogenesis is that changes in the microenvironment of tissue lead to alteration of stem cell behavior. Our study shows that the fate of lineage-specific differentiation of hMSCs is dependent on substrate stiffness with the results showing that stiff substrates direct the cells toward the osteogenic lineage while soft ones direct the cells toward the tenogenic lineage. Our findings may provide insight into one of the pathological mechanisms underlying the formation of heterotopic ossification in muscle or tendon.
5. Conclusion
In this study, we use annealing treatment to alter stiffness of fibrous substrates without changing the material chemistry, and further utilize the setup to identify a mechanism by which substrate stiffness modulates MIF to regulate the AKT/YAP signaling pathway to direct hMSC differentiation. Our findings provide insight into one of the mechanisms governing how stiffness regulates hMSC activities, which may help us enhance our understanding of interactions between hMSCs and their surrounding matrix or substrates for developing viable applications of tissue engineering and regenerative medicine.
Statement of Significance.
Stiffness of biomaterial substrates plays a critical role in regulation of cell behavior. Although the effect of substrate stiffness on cell behavior has been extensively studied, molecular mechanisms of regulation rather than those involving cytoskeletal activities still remain elusive. In this manuscript, we report our new findings that simply using the annealing approach can manipulate stiffness of an aligned fibrous substrate without altering the material chemistry, and substrate stiffness dictates human mesenchymal stem cell (hMSC) differentiation through the macrophage migration inhibitory factor-mediated AKT/YAP/RUNX2 pathway. The findings are novel and interesting because we have identified a new mechanism rather than those involving cytoskeleton activity, by which substrate stiffness regulates hMSC behavior.
Acknowledgments
This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases R01 AR064803 and partially by the National Natural Science Foundation of China (51073032, 31570969) as the Key Project of Science and Technology Commission of Shanghai Municipality (14JC1490100). We would like to thank the China Scholarship Council for the funding support (201406630033) and Donghua University for providing the Doctoral Student Innovation Fund (CUSF-DH-D-2015051) to sponsor HuiHua Yuan.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 2.Seib FP, Prewitz M, Werner C, Bornhaeuser M. Matrix elasticity regulates the secretory profile of human bone marrow-derived multipotent mesenchymal stromal cells (MSCs) Biochem Bioph Res Co. 2009;389:663–667. doi: 10.1016/j.bbrc.2009.09.051. [DOI] [PubMed] [Google Scholar]
- 3.Meyers VE, Zayzafoon M, Douglas JT, McDonald JM. RhoA and cytoskeletal disruption mediate reduced osteoblastogenesis and enhanced adipogenesis of human mesenchymal stem cells in modeled microgravity. J Bone Miner Res. 2005;20:1858–1866. doi: 10.1359/JBMR.050611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patwari P, Lee RT. Mechanical control of tissue morphogenesis. Circ Res. 2008;103:234–243. doi: 10.1161/CIRCRESAHA.108.175331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 6.Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 2009;5:17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 8.Evans ND, Minelli C, Gentleman E, LaPointe V, Patankar SN, Kallivretaki M, Chen XY, Roberts CJ, Stevens MM. Substate stiffness affects early differentiation events in embryonic stem cells. Eur Cells Mater. 2009;18:1–14. doi: 10.22203/ecm.v018a01. [DOI] [PubMed] [Google Scholar]
- 9.Mauck RL, Baker BM, Nerurkar NL, Burdick JA, Li WJ, Tuan RS, Elliott DM. Engineering on the straight and narrow: The mechanics of nanofibrous assemblies for fiber-reinforced tissue regeneration. Tissue Eng Part B. 2009;15:171–193. doi: 10.1089/ten.teb.2008.0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li WJ, Laurencin CT, Caterson EJ, Tuan RS, Ko FK. Electrospun nanofibrous structure: A novel scaffold for tissue engineering. J Biomed Mater Res. 2002;60:613–621. doi: 10.1002/jbm.10167. [DOI] [PubMed] [Google Scholar]
- 11.Sill TJ, von Recum HA. Electro spinning: Applications in drug delivery and tissue engineering. Biomaterials. 2008;29:1989–2006. doi: 10.1016/j.biomaterials.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Zhang C, Yuan HH, Liu HH, Chen X, Lu P, Zhu T, Yang L, Yin Z, Heng BC, Zhang YZ, Ouyang HW. Well-aligned chitosan-based ultrafine fibers committed teno-lineage differentiation of human induced pluripotent stem cells for Achilles tendon regeneration. Biomaterials. 2015;53:716–730. doi: 10.1016/j.biomaterials.2015.02.051. [DOI] [PubMed] [Google Scholar]
- 13.Zhang YZ, Ouyang HW, Lim CT, Ramakrishna S, Huang ZM. Electrospinning of gelatin fibers and gelatin/PCL composite fibrous scaffolds. J Biomed Mater Res B. 2005;72B:156–165. doi: 10.1002/jbm.b.30128. [DOI] [PubMed] [Google Scholar]
- 14.Yoo HS, Kim TG, Park TG. Surface-functionalized electrospun nanofibers for tissue engineering and drug delivery. Adv Drug Deliver Rev. 2009;61:1033–1042. doi: 10.1016/j.addr.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Jiang X, Nai MH, Lim CT, Le VC, Chan JKY, Chew SY. Polysaccharide nanofibers with variable compliance for directing cell fate. J Biomed Mater Res A. 2015;103:959–968. doi: 10.1002/jbm.a.35237. [DOI] [PubMed] [Google Scholar]
- 16.Baker BM, Trappmann B, Wang WY, Sakar MS, Kim IL, Shenoy VB, et al. Cell-mediated fibre recruitment drives extracellular matrix mechanosensing in engineered fibrillar microenvironments. Nat Mater. 2015:1262–1270. doi: 10.1038/nmat4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burger-Kentischer A, Goebel H, Seder R, Fraedrich G, Schaefer HE, Dimmeler S, Kleemann R, Bernhagen J, Ihling C. Expression of macrophage migration inhibitory factor in different stages of human atherosclerosis. Circulation. 2002;105:1561–1566. doi: 10.1161/01.cir.0000012942.49244.82. [DOI] [PubMed] [Google Scholar]
- 18.Calandra T, Bernhagen J, Mitchell RA, Bucala R. Macrophage is an important and previously unrecognized source of macrophage migration inhibitory factor. J Exp Med. 1994;179:1895–1902. doi: 10.1084/jem.179.6.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willis MS, Carlson DL, DiMaio JM, White MD, White DJ, Adams GA, Horton JW, Giroir BP. Macrophage migration inhibitory factor mediates late cardiac dysfunction after burn injury. Am J Physiol-Heart C. 2005;288:H795–H804. doi: 10.1152/ajpheart.00189.2004. [DOI] [PubMed] [Google Scholar]
- 20.Ohta S, Misawa A, Fukaya R, Inoue S, Kanemura Y, Okano H, Kawakami Y, Toda M. Macrophage migration inhibitory factor (MIF) promotes cell survival and proliferation of neural stem/progenitor cells. J Cell Sci. 2012;125:3210–3220. doi: 10.1242/jcs.102210. [DOI] [PubMed] [Google Scholar]
- 21.Xu X, Pacheco BD, Leng L, Bucala R, Ren J. Macrophage migration inhibitory factor plays a permissive role in the maintenance of cardiac contractile function under starvation through regulation of autophagy. Cardiovasc Res. 2013;99:412–421. doi: 10.1093/cvr/cvt116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calandra T, Bernhagen J, Metz CN, Spiegel LA, Bacher M, Donnelly T, Cerami A, Bucala R. MIF as a glucocorticoid-induced modulator of cytokine production. Nature. 1995;377:68–71. doi: 10.1038/377068a0. [DOI] [PubMed] [Google Scholar]
- 23.Benigni F, Atsumi T, Calandra T, Metz C, Echtenacher B, Peng T, Bucala R. The proinflammatory mediator macrophage migration inhibitory factor induces glucose catabolism in muscle. J Clin Invest. 2000;106:1291–1300. doi: 10.1172/JCI9900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rammos C, Hendgen-Cotta UB, Sobierajski J, Adamczyk S, Hetzel GR, Kleophas W, Dellanna F, Kelm M, Rassaf T. Macrophage migration inhibitory factor is associated with vascular dysfunction in patients with end-stage renal disease. Int J Cardiol. 2013;168:5249–5256. doi: 10.1016/j.ijcard.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 25.Denz A, Pilarsky C, Muth D, Rueckert F, Saeger HD, Gruetzmann R. Inhibition of MIF leads to cell cycle arrest and apoptosis in pancreatic cancer cells. J Surg Res. 2010;160:29–34. doi: 10.1016/j.jss.2009.03.048. [DOI] [PubMed] [Google Scholar]
- 26.Lue H, Thiele M, Franz J, Dahl E, Speckgens S, Leng L, Fingerle-Rowson G, Bucala R, Luscher B, Bernhagen J. Macrophage migration inhibitory factor (MIF) promotes cell survival by activation of the Akt pathway and role for CSN5/JAB1 in the control of autocrine MIF activity. Oncogene. 2007;26:5046–5059. doi: 10.1038/sj.onc.1210318. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz V, Lue H, Kraemer S, Korbiel J, Krohn R, Ohl K, Bucala R, Weber C, Bernhagen J. A functional heteromeric MIF receptor formed by CD74 and CXCR4. Febs Lett. 2009;583:2749–2457. doi: 10.1016/j.febslet.2009.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palumbo S, Tsai TL, Li WJ. Macrophage migration inhibitory factor regulates AKT signaling in hypoxic culture to modulate senescence of human mesenchymal stem cells. Stem Cells Dev. 2014;23:852–865. doi: 10.1089/scd.2013.0294. [DOI] [PubMed] [Google Scholar]
- 29.Xue R, Li JYS, Yeh Y, Yang L, Chien S. Effects of matrix elasticity and cell density on human mesenchymal stem cells differentiation. J Orthop Res. 2013;31:1360–1365. doi: 10.1002/jor.22374. [DOI] [PubMed] [Google Scholar]
- 30.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–U212. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 31.Yuan HH, Zhao SF, Tu HB, Li BY, Li Q, Feng B, Peng HJ, Zhang YZ. Stable jet electrospinning for easy fabrication of aligned ultrafine fibers. J Mater Chem. 2012;22:19634–19638. [Google Scholar]
- 32.Chiu CY, Tsai TL, Jr, Vanderby R, Bradica G, Lou SL, Li WJ. Ostoblastogenesis of mesenchymal stem cells in 3-D culture enhacned by low-intensity pulsed ultrasound through soluble receptor activator of nuclear factor B ligand. Ultrasound Med Biol. 2015;41:1842–1852. doi: 10.1016/j.ultrasmedbio.2015.03.017. [DOI] [PubMed] [Google Scholar]
- 33.Puiggali J, Ikada Y, Tsuji H, Cartier L, Okihara T, Lotz B. The frustrated structure of poly(L-lactide) Polymer. 2000;41:8921–8930. [Google Scholar]
- 34.Li M, Mondrinos MJ, Gandhi MR, Ko FK, Weiss AS, Lelkes PI. Electrospun protein fibers as matrices for tissue engineering. Biomaterials. 2005;26:5999–6008. doi: 10.1016/j.biomaterials.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 35.Bianco A, Di Federico E, Moscatelli I, Camaioni A, Armentano I, Campagnolo L, Dottori M, Kenny JM, Siracusa G, Gusmano G. Electrospun poly (e-caprolactone)/Ca-deficient hydroxyapatite nanohybrids: microstructure, mechanical properties and cell response by murine embryonic stem cells. Mat Sci Eng C. 2009;29:2063–2071. [Google Scholar]
- 36.Li WJ, Cooper JA, Mauck RL, Tuan RS. Fabrication and characterization of six electrospun poly (α-hydroxy ester)-based fibrous scaffolds for tissue engineering applications. Acta Biomater. 2006;2:377–385. doi: 10.1016/j.actbio.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Jin N, Johnson J, Lannutti JJ, Agarwal S. Modulation of embryonic mesenchymal progenitor cell differentiation via control over pure mechanical modulus in electrospun nanofibers. Acta Biomater. 2011;7:1516–1524. doi: 10.1016/j.actbio.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takasaki M, Ito H, Kikutani T. Development of stereocomplex crystal of polylactide in high-speed melt spinning and subsequent drawing and annealing processes. J Macromol Sci B. 2003;42:403–420. [Google Scholar]
- 39.Furuhashi Y, Kimura Y, Yoshie N, Yamane H. Higher-order structures and mechanical properties of stereocomplex-type poly (lactic acid) melt spun fibers. Polymer. 2006;47:5965–5972. [Google Scholar]
- 40.Tsuji H, Tezuka Y. Stereocomplex formation between enantiomeric poly (lactic acid) s. 12. Spherulite growth of low-molecular-weight poly (lactic acid) s from the melt. Biomacromolecules. 2004;5:1181–1186. doi: 10.1021/bm049835i. [DOI] [PubMed] [Google Scholar]
- 41.Rowlands AS, George PA, Cooper-White JJ. Directing osteogenic and myogenic differentiation of MSCs: interplay of stiffness and adhesive ligand presentation. Am J Physiol-Cell Ph. 2008;295:C1037–C44. doi: 10.1152/ajpcell.67.2008. [DOI] [PubMed] [Google Scholar]
- 42.Khatiwala CB, Peyton SR, Metzke M, Putnam AJ. The regulation of osteogenesis by ECM rigidity in MC3T3‐E1 cells requires MAPK activation. J Cell Physiol. 2007;211:661–672. doi: 10.1002/jcp.20974. [DOI] [PubMed] [Google Scholar]
- 43.Peyton SR, Raub CB, Keschrumrus VP, Putnam AJ. The use of poly (ethylene glycol) hydrogels to investigate the impact of ECM chemistry and mechanics on smooth muscle cells. Biomaterials. 2006;27:4881–4893. doi: 10.1016/j.biomaterials.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 44.Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Funaki M, Zahir N, Ming WY, Weaver V, Janmey PA. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion, Cell Motil. Cytoskel. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 45.Murphy CM, Matsiko A, Haugh MG, Gleeson JP, O’Brien FJ. Mesenchymal stem cell fate is regulated by the composition and mechanical properties of collagen–glycosaminoglycan scaffolds. J Mech Behav Biomed. 2012;11:53–62. doi: 10.1016/j.jmbbm.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 46.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 47.Sierra OL, Cheng SL, Loewy AP, Charlton-Kachigian N, Towler DA. MINT, the Msx2 interacting nuclear matrix target, enhances Runx2-dependent activation of the osteocalcin fibroblast growth factor response element. J Biol Chem. 2004;279:32913–32923. doi: 10.1074/jbc.M314098200. [DOI] [PubMed] [Google Scholar]
- 48.Lv H, Li L, Sun M, Zhang Y, Chen L, Rong Y, Li YL. Mechanism of regulation of stem cell differentiation by matrix stiffness. Pathways. 2015;1:17. doi: 10.1186/s13287-015-0083-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat Rev Immunol. 2003;3:791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia W, Xie C, Jiang M, Hou M. Improved survival of mesenchymal stem cells by macrophage migration inhibitory factor. Mol Cell Biochem. 2015;404:11–24. doi: 10.1007/s11010-015-2361-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia W, Zhang F, Xie C, Jiang M, Hou M. Macrophage migration inhibitory factor confers resistance to senescence through CD74-dependent AMPK-FOXO3a signaling in mesenchymal stem cells. Stem Cell Res Ther. 2015;6:82. doi: 10.1186/s13287-015-0076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hollander JL. Environment and musculoskeletal diseases, Arch. Environ Health. 1963;6:527–536. doi: 10.1080/00039896.1963.10663436. [DOI] [PubMed] [Google Scholar]
- 53.Virgilio KM, Martin KS, Peirce SM, Blemker SS. Multiscale models of skeletal muscle reveal the complex effects of muscular dystrophy on tissue mechanics and damage susceptibility. Interface focus. 2015;5 doi: 10.1098/rsfs.2014.0080. 0140080. [DOI] [PMC free article] [PubMed] [Google Scholar]


