Abstract
Modeling Amyotrophic Lateral Sclerosis (ALS) with human induced pluripotent stem cells (iPSCs) aims to reenact embryogenesis, maturation, and aging of spinal motor neurons (spMNs) in vitro. As the maturity of spMNs grown in vitro compared to spMNs in vivo remains largely unaddressed, it is unclear to what extent this in vitro system captures critical aspects of spMN development and molecular signatures associated with ALS. Here, we compared transcriptomes among iPSC-derived spMNs, fetal, and adult spinal tissues. This approach produced a maturation scale revealing that iPSC-derived spMNs were more similar to fetal spinal tissue than to adult spMNs. Additionally, we resolved gene networks and pathways associated with spMN maturation and aging. These networks enriched for pathogenic familial ALS genetic variants and were disrupted in sporadic ALS spMNs. Altogether, our findings suggest that developing strategies to further mature and age iPSC-derived spMNs will provide more effective iPSC models of ALS pathology.
Introduction
Amyotrophic Lateral Sclerosis (ALS) is a devastating neurodegenerative disorder, which typically presents in adulthood, has no effective treatments, and involves paralysis and death within 3-5 years of diagnosis [1]. While animal models of ALS have captured molecular and physiological aspects of disease onset and progression, genetic differences between humans and animals limit the interpretation and relevance of phenotypic results. A powerful, complementary in vitro system to animal models of ALS is patient-derived induced pluripotent stem cells (iPSCs) [2, 3]. iPSCs from ALS patients possess the advantage of harboring the patient's complex genetic makeup that contributes to their disease. In disease modeling with iPSCs, the goal is to recapitulate in a dish the embryonic development, maturation, and aging of cell types involved with ALS pathology. Spinal MNs (spMNs) are one of the primary cell types that die in ALS. While their embryonic development is understood to enable their differentiation in vitro from pluripotent cells [4, 5], the fidelity of in vitro to in vivo spMNs remains unresolved. Thus, further optimizing spMN differentiation protocols from iPSCs could enhance the monitoring of disease onset and reveal molecular mechanisms to therapeutically target [6].
spMN progression from the embryonic to the mature and aged state, the point at which they degenerate in ALS, occurs over decades. It therefore remains unclear whether iPSC-derived MNs (iMNs), which are produced in vitro in several weeks, can recapitulate the decades of complex in vivo events leading to MN degeneration [7]. Notably, a systems level comparison has not been conducted between iMNs and adult spMNs, the in vivo counterparts that iMNs attempt to model. We thus performed a direct and global transcriptional comparison of spMNs and iMNs to examine the expression kinetics of gene networks as cells progress from pluripotency to matured and aging adult states. These analyses indicated that iMNs are more similar to fetal rather than adult spinal tissue and revealed a sequential activation and repression of molecular pathways in spinal motor tissue through stages of embryonic development, maturation, and aging. Strikingly, these networks enriched for genetic variants associated with MN disease, revealing that maturation- and age-related pathways may play roles in disease initiation. Finally, these maturation- and aging-associated gene networks are dysregulated in familial and sporadic ALS. Collectively, these findings suggest that strategies to mature and age iPSC-derived spMNs may provide more relevant iPSC models of ALS.
Results
iPSC-derived MNs resemble fetal rather than adult MNs
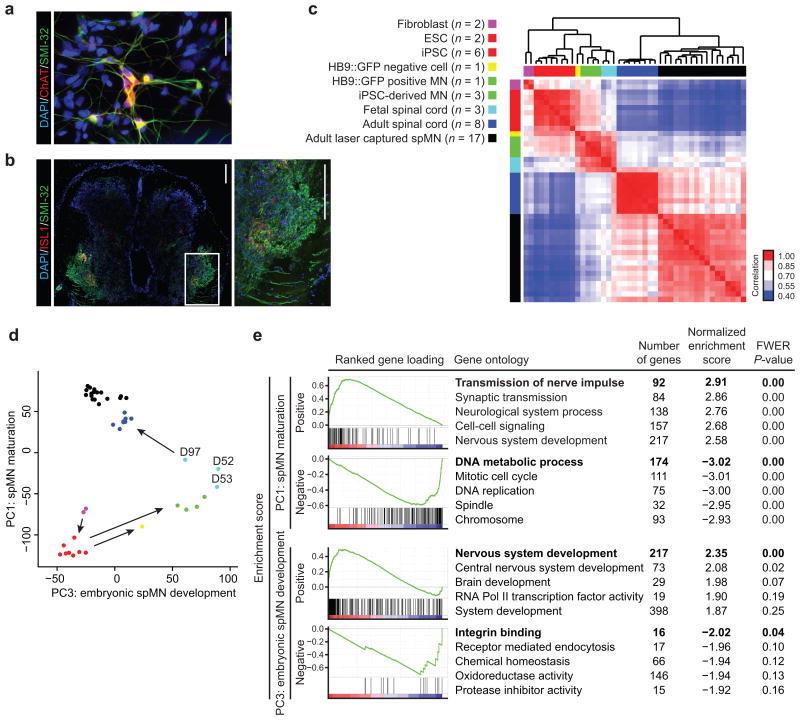
We compared expression profiles among fibroblasts, iPSCs, fetal spinal cord, whole adult spinal cord and laser captured MNs from the spinal cords of control and ALS patients, which included those with mutations in SOD1 or CHMP2B [8, 9] (Supplementary Table 1a). Selecting profiles from the same microarray platform reduced the likelihood of confounding batch effects. An established 7 week in vitro protocol was used to generate cultures with 33 – 45% choline acetyl transferase (ChAT)- and SMI-32-double positive differentiated iMNs [10] (Fig. 1a). We also obtained expression data for MN cultures from human embryonic stem cells containing an HB9::GFP motor neuron reporter, which were sorted into GFP-positive and negative fractions [11]. To represent immature in vivo spinal tissue during embryonic development, we generated expression data from human fetal spinal cords. ISL1 and SMI-32 immunostaining revealed MNs in the ventral horn (Fig. 1b). After normalizing the expression for 10,605 genes in all samples (n = 43, Supplementary Table 2a), hierarchical clustering and Pearson correlation revealed that iMNs were more similar to fetal rather than adult spinal cord tissue or iPSCs from which they were differentiated (Fig. 1c). Principal component analysis (PCA) revealed that PC1 distinguished between pluripotent cells and adult spMNs (Supplementary Fig. 1a). PC2 distinguished non-laser captured adult spinal tissues from all other samples, possibly due to strong gene expression contributed by heterogeneous adult spinal tissue that were absent in other samples. Notably, PC3 distinguished iMNs and fetal spinal cords from all other samples. When considering only PC1 and PC3 (Fig. 1d), an intuitive progression of MN development can be visualized. Regression analysis of linear models for sample traits against PC coordinates also revealed that tissue type best explained PC1 and PC3 (Supplementary Fig. 1b and Supplementary Table 1b).
Figure 1.
iPSC-derived MNs resemble fetal rather than adult MNs. (a) Immunostaining of iMNs used in expression profiling. ChAT (red) and SMI-32 (green) positive cells indicate the presence of MNs differentiated from iPSCs. Nuclear DAPI stains are shown in blue. n = 3 independent experiments and differentiation efficiency is quantified by ChAT- and SMI-32-double positive motor neurons. Scale bar = 4.75 μm. (b) Immunofluorescence staining of a 63-day-old fetal spinal cord. Inset depicts ISL1 (red) and SMI-32 (green) positive cells in the motor horn with ventrally projecting processes, indicating the presence of MNs at this developmental stage. Nuclear DAPI stains are shown in blue. Scale bars = 200 μm. (c) Unsupervised hierarchical clustering of 10,605 mRNA transcripts in n = 43 samples. Heatmap indicates Pearson's correlation between pairwise sample comparisons and dendrogram indicates average linkage distance between samples. The color legend for tissue types is indicated to the left and refers to d as well. (d) Principal component analysis of 10,605 mRNA transcripts in n = 43 samples. Sample coordinates along Principal components (PC) 1 and 3 are shown. The percentages of total variance are 14.61% and 7.01% for PC1 and PC3, respectively. Sample colors refer to c. Arrows depict the progression of fibroblasts (magenta) reprogrammed to iPSCs (red), the subsequent differentiation of iPSCs into either HB9::GFP negative cells (yellow) or HB9::GFP positive MNs (green), which project towards fetal spinal cords (cyan). Arrows also depict the progression of fetal spinal cords towards adult spinal tissues (blue and black). The days post conception of fetal spinal cord donors are indicated next to the cyan data points. (e) Gene set enrichment analysis of ranked gene loadings from PC1 and PC3 for gene ontology (GO) terms. “Positive” and “Negative” categories indicate GO terms enriched among genes whose loadings contribute most to the respective positive or negative direction of each principal component. Enriched GO terms for each category are listed along with family-wise error rate (FWER) corrected P-values. Enrichment plots are shown for bolded GO terms. For additional information, see Supplementary Fig. 1 (for all PCs and linear model statistics), Supplementary Tables 1a (for sample meta data), 2a (for linear expression values), and 2b (for full list of significantly enriched gene sets and P-values).
Next, molecular pathways associated with PC1 and PC3 were examined. Gene set enrichment analysis (GSEA) [12] was performed on the ranked gene loadings for PC1 and PC3 to detect gene sets that concordantly place each sample along each component (Fig. 1e and Supplementary Table 2b). Pathways related to synaptic transmission were enriched among positive gene loadings along PC1, supporting that MN maturation is characterized in part by changes in electrophysiology [13]. Conversely, pathways related to cell cycle and DNA repair were enriched among negative gene loadings along PC1. Furthermore, pathways related to nervous system development were enriched among positive gene loadings along PC3. Interestingly, genes associated with integrin binding, which regulate neuronal migration during development [14], were enriched among negative gene loadings along PC3. Together, these data account for differences in global expression as well as specific pathways across all samples, further supporting that iMNs are molecularly restricted to a fetal-like state. By summarizing the diverse GO terms enriched along their PC gene loadings for this expression data set and for simplification, PC1 is best described as spMN maturation, and PC3 is best described as embryonic spMN development.
Network analysis resolves development, maturation, and aging
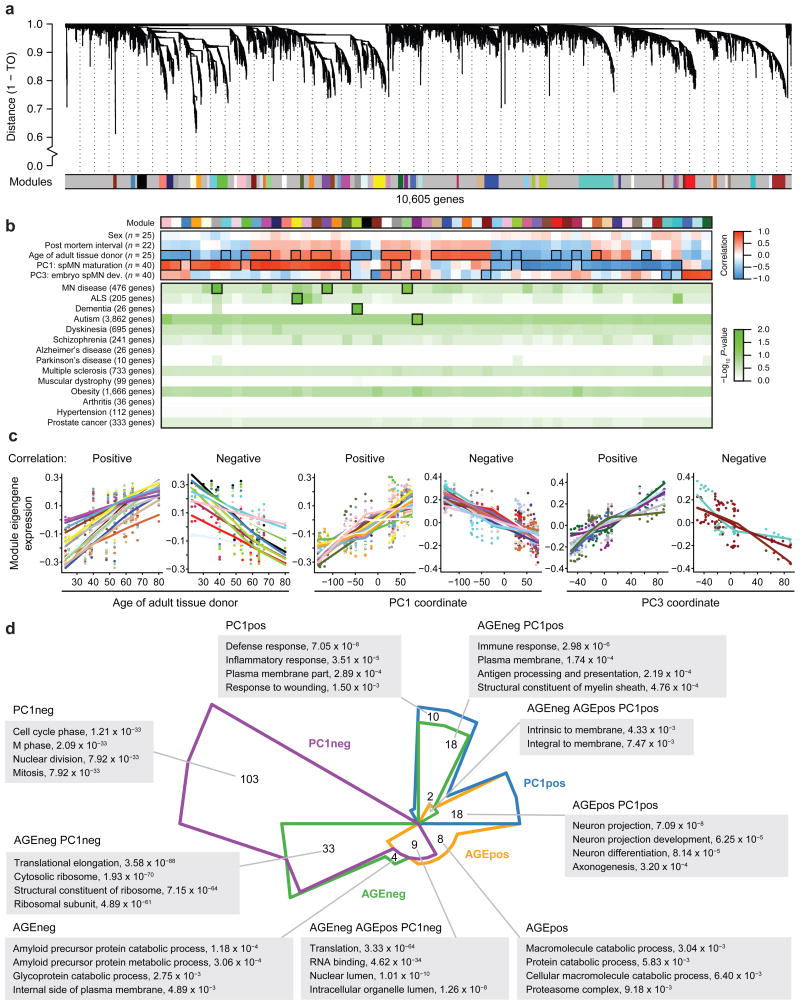
Gene co-expression network analysis is a useful method to link tightly co-expressed gene modules to phenotypic traits [15]. Using weighted gene co-expression network analysis (WGCNA), 10,605 genes across 40 samples were hierarchically clustered based on topological overlap (Fig. 2a). This analysis identified 55 modules ranging in size from 30 to 511 genes with a median size of 66 genes (Supplementary Table 2c). Altogether, 4,711 genes were assigned to modules (Fig. 2a, colored bars) and 5,894 genes were not classified into any modules (Fig. 2a, grey bars).
Figure 2.
Network analysis resolves co-expression modules. (a) WGCNA clustered 10,605 genes across pluripotent cells, iMNs, fetal spinal tissues, and adult spinal tissues based on similar network topology (n = 40 samples). Height metric on dendrogram indicates topological overlap (TO) distance between genes. A dynamic tree-cutting algorithm grouped tightly networked genes, illustrated as low hanging branches, into 55 modules represented by arbitrary colors directly below the branches. Genes falling onto the predominant light grey color are not classified into any module. (b) Upper and middle panel: Heatmap indicates Pearson's correlation of module eigengenes with 5 sample traits. n = number of samples for which there is data for the indicated sample trait, and thus used in the correlation. Outlined panels indicate correlations with a Bonferroni-corrected P-value < 0.01, and these modules were kept for subsequent GO analysis. Lower panel: Gene variants associated with diseases in the ClinVar database were tested against each of the 55 modules for enrichment. n = number of genes with variants associated with the indicated disease, represented on the human microarray platform, and thus used in the enrichment analysis. Green heatmap indicates the Benjamini-Hochberg corrected negative log10 P-value from each hypergeometric test. Corrected P-values < 0.05 are called significant and outlined in black. This data is also shown in Fig. 5b. (c) For each module eigengene that significantly correlates (Positive) or anti-correlates (Negative) with age, PC1, or PC3 (Bonferroni-corrected P-value < 0.01), the module eigengene expression values are graphed in a scatter plot against either sample age or PC coordinate. Locally weighted scatterplot smoothing lines are graphed for each module. (d) Chow-Ruskey diagram illustrating the number of overlapping and distinct GO terms (Bonferroni-corrected P-value < 0.05) enriched in modules identified as significantly correlated or anti-correlated to age (AGEpos or AGEneg, respectively) or spMN maturation (PC1pos or PC1neg, respectively). Representative pathways are listed in grey boxes extending from diagram, along with the lowest Bonferroni-corrected P-values across all modules. For additional information, see Supplementary Fig. 2 (for additional WGCNA and Chow-Ruskey diagrams), Supplementary Tables 2a (for linear expression values), 2c (for module assignments and properties), 2d, 2e (for gene set enrichments and P-values), and 2j (for module-trait correlations, P-values, and Clinvar enrichment P-values).
The module eigengene represents the expression of all genes classified into that module. When each eigengene expression was correlated against a sample trait using Pearson's method (Supplementary Table 1a), only the age of the spinal tissue donor significantly correlated positively or negatively to a subset of eigengenes (Fig. 2b, Age of adult tissue donor, outlined panels). In addition to sample traits, each sample analyzed by PCA was assigned a coordinate along each PC. When each eigengene expression was correlated against PC1, the component that describes spMN maturation, or against PC3, the component that describes embryonic spMN development, subsets of eigengenes were significantly correlated to each PC in an independent manner (Fig. 2b, PC1 and PC3, outlined panels). This analysis thus identified gene co-expression networks that strongly associate with age, spMN maturation, and embryonic spMN development (Fig. 2c).
While neuron maturation and aging are physiological features likely involved in ALS and other late onset, neurodegenerative diseases [7, 16], their distinction has been difficult to define. While regression analysis highlighted tissue type as the main covariate that correlated with PC1, the age of tissue donor slightly, but nevertheless significantly correlated with PC1 as well (Supplementary Fig. 1b). Interestingly, correlation performed on the 55 modules revealed that some modules significantly correlated with spMN maturation but not age and vice versa (Fig. 2b, Age of adult tissue donor and PC1, outlined panels). Additionally, some modules correlated with both spMN maturation and age in the same direction, while other modules correlated with both in opposite directions. WGCNA was thus able to finely resolve gene co-expression networks associated with the processes of neuron maturation and aging.
ALS genetic variants disrupt spMN maturation and aging genes
To test if any of the 55 modules identified by WGCNA were associated with age-related diseases, enrichment analyses were performed using the ClinVar database [17]. Strikingly, pathogenic genetic variants associated with the search terms “motor neuron” were enriched in the dark grey, purple, and lightgreen modules, and those associated with “Amyotrophic Lateral Sclerosis” were enriched in the yellow module (Fig. 2b, MN disease and ALS, outlined panels). Notably, these four modules were significantly correlated with spMN maturation, age, or both (Fig. 2b, Age of adult tissue donor and PC1, outlined panels). Interestingly, variants associated with dementia were enriched in the greenyellow module, which significantly anti-correlated with age, and variants associated with autism were enriched in the darkmagenta module, which significantly correlated with embryonic development. Enrichment for MN disease and ALS variants in these modules were specific, as there was no significant enrichment for genetic variants associated with other age-related disease like Alzheimer's disease, hypertension, or prostate cancer (Fig. 2b, lower panels). Modules identified using WGCNA on a data set where all ALS patient samples were removed yielded similar results (Supplementary Fig. 2a), suggesting that the age- and spMN maturation-correlated modules are indeed modifiers of ALS and MN disease presentation. We subsequently analyzed modules built with the expression data set that included the ALS samples so as to 1) increase the sample size of spMNs, thereby increasing the robustness of the detected modules and 2) provide some representation of co-expression networks as they occur in spMNs from ALS patients. We note that mutations in CHMP2B are of uncertain pathogenicity in ALS, given that there is insufficient evidence of segregation with disease in familial pedigrees and the presence of a rare, candidate polymorphism in non-ALS populations [18, 19].
Network analysis resolves spMN maturation and aging pathways
To gain a biological understanding of network-to-sample trait relationships, the age- or spMN maturation-associated modules were tested for enrichment of genes belonging to gene ontology (GO) terms. Each of the 55 modules was grouped into four classes: significant positive (PC1pos) or negative (PC1neg) correlation with spMN maturation, and positive (AGEpos) or negative (AGEneg) correlation with age (Supplementary Table 2d). Performing a four-way intersection, this revealed numbers of overlapping and distinct GO terms enriched in each class (Fig. 2d, Supplementary Fig. 2b, and 2c).
Pathways involved in cell cycle and mitosis were exclusively enriched in PC1neg (Fig. 2d), consistent with GSEA results (Fig. 1e). Neuron projection modules positively correlated with both PC1 and age, suggesting that axonogenesis is a continual process throughout spMN maturation and aging. Translation and ribosomal pathways were enriched in both PC1neg and AGEneg (Fig. 2d), indicating that co-regulated gene networks associated with protein production decline in cells during spMN maturation and aging. Conversely, proteasome-associated genes increased with age, suggesting a role for protein degradation and catabolism in older cells. Notably, amyloid precursor processing genes were enriched in AGEneg, indicating a progressively declining ability to prevent glycoprotein aggregates in older cells. Interestingly, immune response and myelin sheath genes were enriched in PC1pos and AGEneg (Fig. 2d), suggesting that these gene networks increase in expression during spMN maturation, but decrease in expression during aging. Altogether, these analyses effectively resolve biological processes that are associated with spMN maturation, aging, or both traits. Furthermore, pathway enrichment through WGCNA recovered additional ontologies not detected using PCA with GSEA alone (Supplementary Table 2e), indicating that WGCNA is a more sensitive approach to discover pathways of interest.
Key markers assess spMN maturation and embryonic development
Having identified expression networks that significantly associated with the processes of spMN maturation and embryonic spMN development, we investigated if the gene properties within those networks could identify key markers of both processes. A reduced list of key markers would enable a comparison among samples that were expression profiled on different microarray platforms and provide a robust indicator of the embryonic developmental and maturation status of pluripotent cell-derived MNs. To this end, we scored each of the 10,605 genes along three properties with respect to PC1 and PC3 that reflect spMN maturation and embryonic spMN development, respectively (Fig. 1d). These properties were 1) preferential gene loading, 2) gene significance, and 3) intramodule gene membership. The preferential gene loading was the net contribution of each gene to one particular PC and minimal contribution to all other PCs. The gene significance is the Pearson's correlation between the gene expression value and a sample trait, as in this instance, spMN maturation or embryonic development. The intramodule membership is the correlation between the gene expression value and the eigengene of the module to which it is assigned.
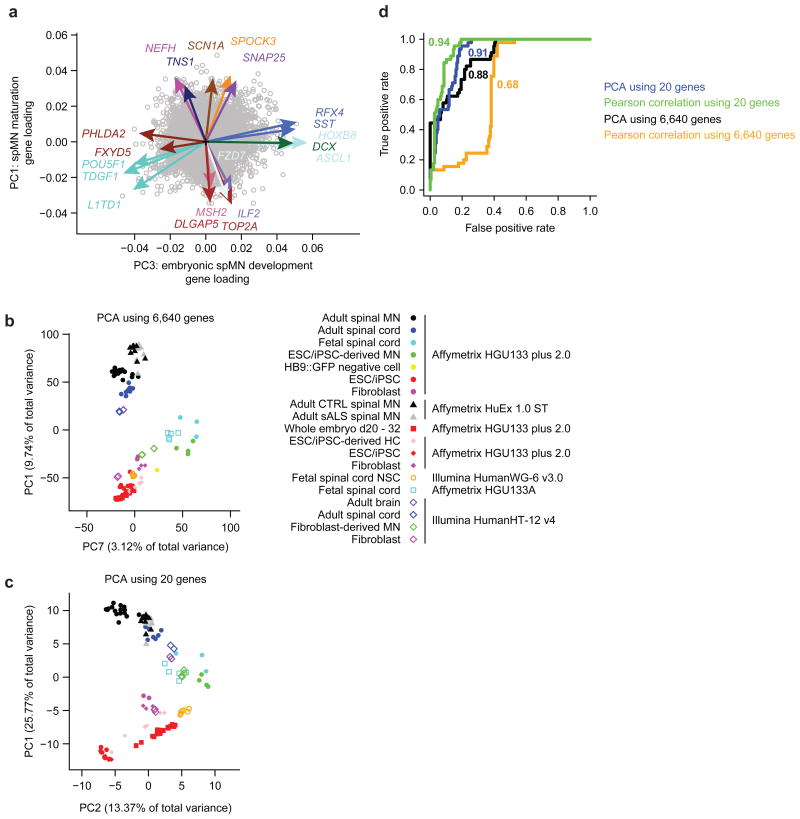
After generating gene scores for each of these properties, genes were partitioned into four classes: PC1pos, PC1neg, PC3pos, and PC3neg, based on the signed value of their gene loadings in PC1 or PC3, and ranked by a score summarizing the three properties (Supplementary Tables 2c and 2e – h). From each of these ranked lists, five genes were selected based on the best gene scores, as well as for diverse representation of module colors and prior knowledge of biological relevance to spMN maturation or embryonic spMN development. Together, this totaled 20 genes that best predicted the sample clustering along the axes of PC1 and PC3 (Fig. 3a), and RT-qPCR validated microarray expression patterns (Supplementary Fig. 3a and Supplementary Table 3a).
Figure 3.
Principal component and network analyses reveal key spMN maturation and embryonic development markers. (a) Gene loadings for 20 genes selected to best represent spMN maturation and embryonic spMN development. Colored arrows and gene labels depict gene loadings and module assignments for the 20 genes along PC1 and PC3. Open grey circles represent gene loadings for 10,585 genes not selected for the panel. PCA was performed on n = 43 samples depicted in Fig. 1d. (b) Principal component analysis performed on 6,640 quantile normalized gene expression values across 120 samples. Samples are plotted by their coordinates along PC1 and PC7. Sample legend is shown on the right. Colors of data points indicate similar sample types, and shapes of data points indicate the study from which the data were obtained. Microarray platforms are also indicated. (c) As in b, except PCA was performed using 20 genes depicted in a. Samples are plotted by their coordinates along PC1 and PC2. Sample legend is the same as for b. (d) Receiver-operator characteristic analysis performed on four methods classifying n = 77 samples in the validation data set as pluripotent stem cells, fetal-like cells, or adult spinal cord cells. Classifications were based on sample correlation to the median expression values of target cell types in the training data set using 6,640 genes (red) or 20 genes (green) or based on sample coordinates along the spMN maturation or embryonic development principal components using 6,640 genes (black) or 20 genes (blue). The area under the curve is shown next to each like-colored curve, and summarizes the overall performance of each classification method. For additional information, see Supplementary Fig. 3 (validation data), Supplementary Tables 1a (for sample meta data), 2c (for module assignments and properties), 2f – i (for gene scoring properties), 3a (for qPCR primer sequences), and 3b (for normalized linear expression values used in validation analyses).
To test the robustness of this marker panel to assay spMN maturation and embryonic spMN development, expression data were downloaded from additional studies as validation data sets utilizing a variety of microarray platforms (Supplementary Table 1a). Expectedly, expression values for 6,640 overlapping genes from different platforms and studies exhibited varied distribution patterns (Supplementary Fig. 3b, colored curves). Quantile normalization conformed all values to a single distribution pattern (Supplementary Fig. 3b, black curve and Supplementary Table 3b).
Performing PCA on all 6,640 quantile normalized genes produced PCs that resembled the training data set (Figs. 1d, 3b, and Supplementary Fig. 3c). Specifically, PC1 again best described spMN maturation and PC7 in this instance best described embryonic spMN development. Notably, PC3 captures variation due to differences between Illumina and Affymetrix microarray platforms, despite quantile normalization minimizing these differences (Supplementary Figs. 3b and c). Strikingly, PCA performed using the 20 key genes reflected spMN maturation in PC1 and embryonic spMN development in PC2 (Fig. 3c and Supplementary Fig. 3d). Importantly, variations arising from different microarray platforms were not noticeably captured. A receiver-operating characteristic analysis on the validation data set demonstrated that the 20 genes outperformed 6,640 genes with either PCA or Pearson correlation, yielding a higher area under the curve (Fig. 3d and Supplementary Figs. 3e – g). These observations support the efficacy and universal adaptability of combining PCA and WGCNA to identify key markers of spMN maturation and embryonic spMN development.
mtSOD1 distinctively affects spMNs and iMNs networks
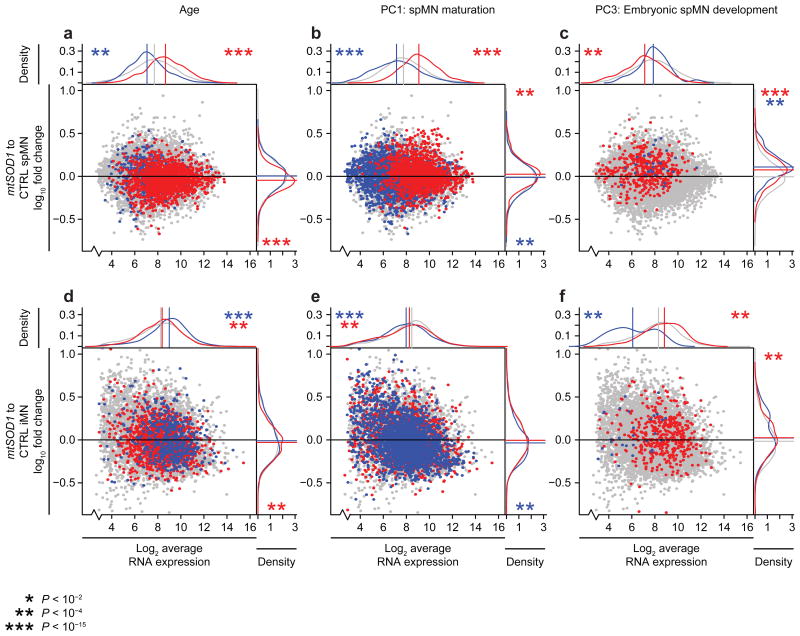
The expression of modules associated with age, spMN maturation, and embryonic spMN development were next investigated in spMN and iMN samples comparing a familial form of ALS caused by mutant SOD1 (mtSOD1) to controls (Fig. 4, Supplementary Tables 1a, 4a, and 4b) [9, 20]. mtSOD1 significantly affected several module classes. Notably, modules correlated to age were downregulated by mtSOD1 in spMNs (Fig. 4a). These age-associated modules in mtSOD1 spMNs were dysregulated in the opposite manner in which they are endogenously expressed during aging. This suggests that the aging process enacts homeostatic gene expression programs that may protect spMNs, which otherwise undergo cell death due to misexpression of these transcriptional programs in the mtSOD1 condition. Interestingly, all of the embryonic spMN development-associated modules were significantly upregulated in mtSOD1 spMNs (Fig. 4c), indicating that some embryonic pathways are also affected in mtSOD1-induced ALS.
Figure 4.
Gene expression networks are distinctively affected by familial ALS in spMNs and iMNs. Scatter and density plots depicting expression levels and mtSOD1-induced fold changes of age-, spMN maturation-, or embryonic spMN development-associated module genes as they were defined by WGCNA performed on the expression data set lacking ALS spMNs (Supplementary Fig. 2a). This was done to prevent modules from being defined by mtSOD1-induced gene expression changes in spMNs. Grey data points indicate no membership in module categories, red data points indicate membership in correlated modules, and blue data points indicate membership in anti-correlated modules. Grey data points are plotted behind red and blue data points in all panels. Red data points are plotted in front of blue data points in a and b. Blue data points are plotted in front of red data points in c – f. 8,830 overlapping genes represented in both spMN and iMN data sets are shown in each plot. X-axis indicates the log2 average RNA expression values among all mtSOD1 and control (CTRL) samples. Y-axis indicates the log10 fold change in average RNA expression when comparing mtSOD1 to control samples, and the scale has cut off some outlier data points in order to visualize distribution shifts. The density plots account for all data points in the expression set, and the straight lines mark the median value of each distribution. Asterisks indicate the Kolmogorov-Smirnov two-sided P-value for like-colored categories tested against the grey distribution, and its location indicates the direction of the shifted distributions. (a – c) mtSOD1 (n = 3) versus control (n = 3) spMNs [9]. (d – f) mtSOD1 (n = 2) versus control (n= 3) iMNs [20]. For additional information, see Supplementary Tables 2c (for module assignments and properties), 4a (for linear expression values and module assignments), and 4b (for Kolmogorov-Smirnov test statistics and P-values).
Mapping the expression of these trait-associated modules in an independent data set for mtSOD1 and control iMNs (Supplementary Tables 1a, 4a, and 4b) [20] revealed patterns that were similar to spMNs in some aspects, but distinct in others. When comparing module expression levels between mtSOD1 and control iMNs, the modules correlated to age, though not as highly expressed as in spMNs, were significantly downregulated (Figs. 4a and 4d). These observations suggest that age-associated gene expression networks affected by mtSOD1 ALS in spMNs may be recapitulated to some extent in iMNs. Yet, the expression of modules correlated to spMN maturation was not significantly different in iMNs (Figs. 4b and 4e). These observations suggest that iMNs do not faithfully recapitulate the expression profile of mature spMNs affected by mtSOD1. Lastly, modules that were correlated or anti-correlated to embryonic spMN development were respectively highly or lowly expressed overall in iMNs (Fig. 1f). Thus, the expression pattern of these embryonic spMN development-associated modules in an independent expression data set supports the idea that iMNs are more similar to fetal spinal tissue than adult spMNs.
Sporadic ALS dysregulates spMN maturation and age modules
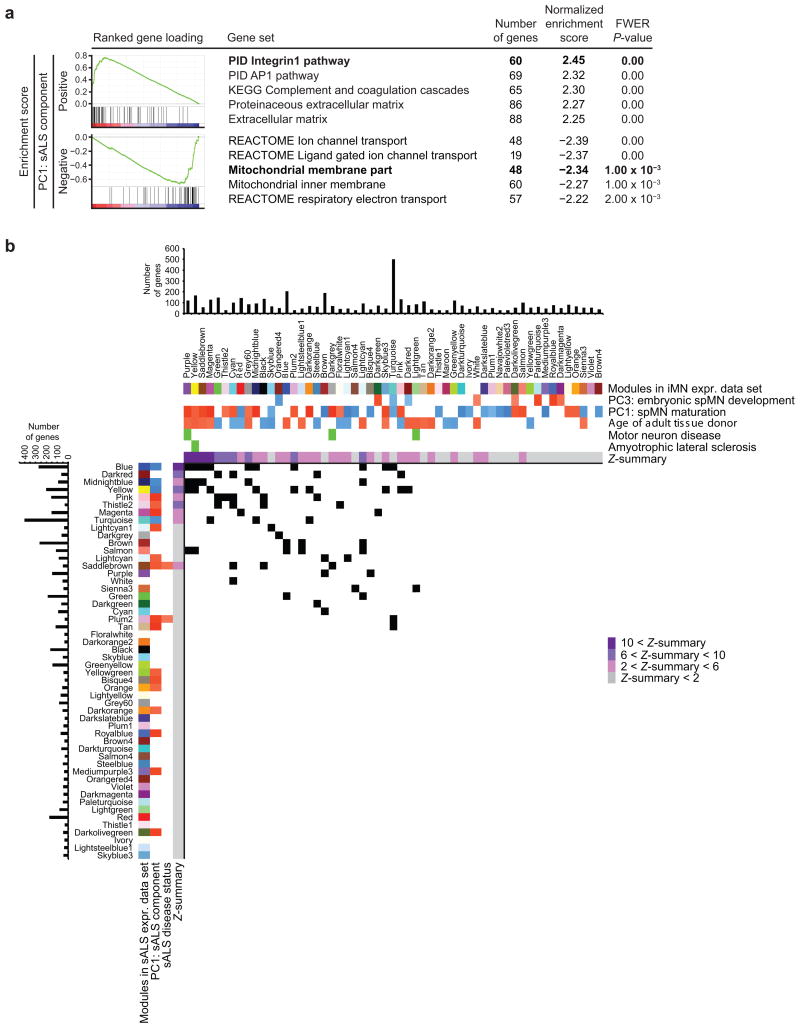
WGCNA with the composite data set analyzed thus far (hereinafter referred to as iMN expression data set) (Supplementary Table 2a) did not identify modules significantly correlated to ALS conditions (data not shown). This was likely due to an insufficiently large representation of samples associated with ALS in the data set, perhaps also compounded by distinct expression changes induced by the two genotypes of ALS represented: mtSOD1 and mtCHMP2B. Thus, a separate analysis was performed to identify gene networks that significantly associate with the ALS condition in spMNs. An independent transcriptomic data set of 15,614 genes that specifically focused on comparing expression between sporadic ALS (sALS) and control spMNs [21] was analyzed (n = 22, age range = 47 – 81 years, median = 73 years) (Supplementary Tables 1a and 5a). This data set, also used in the gene reduction validation analysis (Fig. 3b), similarly compared the transcriptional profiles of laser captured spMNs between 12 sALS patients and 10 control subjects. However, this data set was profiled on a different microarray platform than was used for the composite iMN expression data set. Therefore, PCA and WGCNA were separately performed on this expression data set (hereinafter referred to as the sALS expression data set) in order to independently validate co-expression networks and observe if their expression is affected between sALS and control conditions. PCA and linear regression analysis revealed that PC1 was best distinguished by sALS and control spMNs (Supplementary Figs. 4a, 4b, and Supplementary Table 1c), therefore this PC was best described as the sALS component.
GSEA was performed on the ranked gene loadings for this component to explore pathways and gene ontologies enriched among genes that were dysregulated in sALS spMNs (Fig. 5a and Supplementary Table 5b). Interestingly, pathways related to extracellular matrix were enriched among genes that were upregulated in sALS, consistent with previously described gene expression changes [21]. Conversely, pathways related to mitochondrial ion and electron transport were enriched among genes that were downregulated in sALS. These observations were not previously described for this data set [21], highlighting the sensitivity of GSEA using PCA gene loadings to detect enriched pathways without thresholding on an arbitrarily assigned, fold change cut-off.
Figure 5.
spMN maturation and age modules are dysregulated in sporadic ALS. (a) Gene set enrichment analysis of 15,614 ranked gene loadings from PC1 for pathways and GO terms. “Positive” and “Negative” categories indicate gene sets enriched among genes whose loadings contribute most to the respective positive or negative direction of the sALS component (PC1 in a PCA performed on n = 22 samples). Enriched gene sets for each category are listed along with family-wise error rate (FWER) corrected P-values. Enrichment plots are shown for bolded gene sets. (b) For each of the 52 sALS modules, a hypergeometric test was performed to detect enrichment for genes from each of the 55 iMN modules. Upper panel: iMN modules are displayed along with the sample traits with which they are significantly associated, as identified and also shown in Fig. 2b. Enrichment for ClinVar pathogenic variants in motor neuron disease or ALS is also shown. The Z-summary value for each iMN module measures the extent of module preservation in the sALS data set. For the likelihood of module preservation, Z-summary > 10 indicates strong evidence; 10 > Z-summary > 2 indicates moderate to weak evidence, and 2 > Z-summary indicates no evidence. Bar graphs above indicate the number of genes assigned to each iMN module that were also represented by probe sets on the Affymetrix Human Exon 1.0 ST Array. Left panel: sALS modules are displayed along with the sample traits with which they are significantly correlated or anti-correlated, as identified in Supplementary Fig. 4d. The Z-summary value for each sALS module measures the extent of module preservation in the iMN data set. Bar graphs to the left indicate the number of genes assigned to each sALS module that were also represented by probe sets on the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array. A matrix of P-values from hypergeometric tests performed for each iMN and sALS module overlap were corrected by the Benjamini-Hochberg method, and subsequent P-values < 0.05 are marked as a black square panels and illustrated in the matrix diagram. For additional information, see Supplementary Figs. 4 (for all PCs and linear model statistics, WGCNA), 5 (for module preservation statistics and intramodule membership of genetic variants), 6 (for Chow-Ruskey diagrams), Supplementary Tables 2c (for iMN module assignments and properties), 5a (for linear expression values of sALS data set), 5c (for sALS module assignments and properties), 5d (for module preservation statistics), 5b, 5e, 5f (for full lists of significantly enriched gene sets and P-values), and 5g (for hypergeometric test P-values).
Next, gene co-expression networks were built using only expression data from the sALS expression data set. WGCNA on this data set identified 52 modules ranging in size from 31 to 401 genes with a median size of 51 genes (Supplementary Table 5c). Altogether 4,444 genes were assigned to modules (Supplementary Fig. 4c, colored bars) and 11,170 genes were not classified into any modules (Supplementary Fig. 4c, grey bars). Each eigengene was then correlated using Pearson's method against sex, sALS disease status, site of ALS onset, age of tissue donor, disease course, and post mortem interval (Supplementary Table 1a). Additionally, each eigengene was correlated against the sALS component. Among these sample traits, only sALS disease status and the sALS component significantly correlated or anti-correlated to a subset of modules (Supplementary Fig. 4d, outlined panels).
Given this observation, the possibility that these gene modules were also involved with spMN maturation and age was hypothesized. Therefore, the extents of overlap were examined between modules defined in the iMN and sALS expression data sets, hereinafter referred to as iMN and sALS modules, respectively. To this end, each sALS module was systematically tested for enrichment of each iMN module (Fig. 5b and Supplementary Table 5g). This analysis demonstrated that some iMN modules significantly overlapped with one or more sALS modules, and vice versa.
In addition to cross-tabulating module assignments between the two data sets, the stability of networks defined in each data set was examined using module preservation Z-statistics [22]. These measures indicated the likelihood that the network structures of iMN modules also occurred in the sALS data set by random chance. Consistent with the overlap analysis, the most significantly overlapping modules were also those whose network structures were most significantly preserved across data sets (Fig. 5b, Supplementary Fig. 5, and Supplementary Table 5d).
Despite an imperfect one-to-one module correspondence, overlapping iMN and sALS modules tended to significantly correlate or anti-correlate to sample traits in a consistent way. For instance, iMN modules that significantly correlated with age and spMN maturation tended to have strong overlap with sALS modules that significantly anti-correlated to the sALS component. Conversely, iMN modules that significantly anti-correlated with age and spMN maturation tended to have strong overlap with sALS modules that significantly correlated to the sALS component. Additionally, three of the four modules that enriched for genetic variants associated with MN disease or ALS significantly overlapped with at least one sALS module that in turn significantly associated with the sALS component or sALS disease status. These observations thus support that networks involved in spMN maturation and age are also affected in sALS.
To gain a biological understanding of these module-to-trait relationships, enrichment analysis was performed on each significantly associated module for GO terms. Modules were then classified into two groups: those that significantly correlated with the sALS component positively (sALSpos) or negatively (sALSneg) (Supplementary Table 5e). Similar to the comparisons in the iMN data set, pathway enrichment using WGCNA in the sALS data set recovered additional ontologies not detected using PCA with GSEA alone (Supplementary Table 5f). Performing a four-way intersection analysis with either AGEpos and AGEneg, PC1pos and PC1neg, or PC3pos and PC3neg (Fig. 2d, Supplementary Figs. 2b, and c), these analyses revealed numbers of overlapping and distinct GO terms enriched in each of the different groups (Supplementary Fig. 6). Interestingly, pathways such as RNA processing and translational elongation, which are enriched within AGEneg and PC1neg, are also enriched within sALSpos (Supplementary Figs. 6a and 6b). Notably, the immune response pathway, which is enriched within PC1pos and AGEneg, is also enriched in sALSpos. Conversely, pathways such as neuron differentiation and cell projection, which are enriched within AGEpos and PC1pos, are also enriched within sALSneg. To a lesser extent, some transcriptional pathways, which are enriched within PC3pos, are also enriched within sALSpos (Supplementary Fig. 6c).
spMN maturation, aging, and sALS converge on hub genes
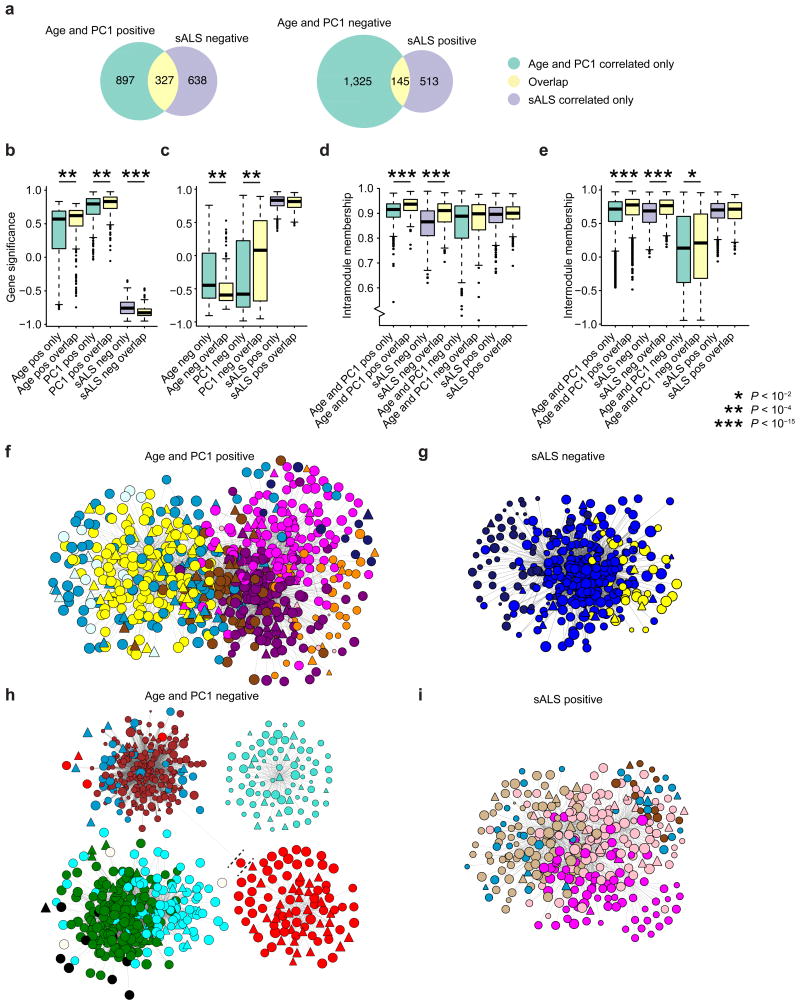
An alternative approach was taken to explore the network properties of genes affected by maturation, aging, and ALS. Four module classes were defined: modules that 1) increased or 2) decreased expression with aging and maturation (AGEpos and PC1pos or AGEneg and PC1neg, respectively), modules that 3) increase or 4) decrease with sALS (sALSpos or sALSneg, respectively). Consistent with overlap comparisons performed on a module-by-module basis (Fig. 5b), this comparison of combined modules demonstrates that age and spMN maturation modules had extensive overlap with sALS modules in opposite directions (Fig. 6a). Having identified which genes within the module classes are commonly affected (hereinafter referred to as overlaps), the gene expression networks in both the iMN and sALS expression data sets were observed for any properties that distinguished overlaps from genes that were uniquely found in either of the module classes (hereinafter referred to as non-overlaps).
Figure 6.
Genes associated with spMN maturation, aging, and sALS tend to be hub genes. (a) Overlap analysis of genes that are represented in WGCNA modules independently built in either the iMN expression data set or in the sALS expression data set. Modules built from the iMN data set are classified as Age positive or negative based on whether their module eigengene significantly correlates or anti-correlates, respectively, with the age of tissue donor from the iMN data set. Modules built from the iMN data set are classified as PC1 positive or negative based on whether their module eigengene significantly correlates or anti-correlates, respectively, with PC1 in the PCA performed on the iMN data set. To be included into the Age and PC1 positive group, genes must be classified as either Age positive, PC1 positive, or both. Additionally, their modules must have significant overlap, based on Fig. 5b, with at least one sALS module that correlates with the sALS component. These criteria are likewise for the Age and PC1 negative group which overlaps with sALS positive modules. Modules built from the sALS data set are similarly classified based on their relationship to the PCA performed on the sALS data set. Venn diagrams indicate the number of genes assigned to each class. (b) Boxplots of gene significance for the overlapping and non-overlapping genes from the comparison depicted in a, left. Asterisks indicate P-values determined by the Wilcoxon rank-sum test. For the “Age pos only” and “Age pos overlap” genes, the y-axis indicates gene significance values against age in the iMN expression data set. For the “PC1 pos only” and “PC1 pos overlap” genes, the y-axis indicates gene significance values against PC1 in the iMN expression data set. For the “sALS neg only” and “sALS neg overlap” genes, the y-axis indicates gene significance values against PC1 in the sALS expression data set. In each boxplot, the center line is the median, the lower and upper limits of the box are respectively the first and third quartile, and the lower and upper whiskers extend to either the respective minimum or maximum values of the distribution, or up to 1.5 times the interquartile range. Outliers are plotted as open circles. (c) Similar presentation as in b, except applied to genes in a, right. For the “Age neg only” and “Age neg overlap” genes, the y-axis indicates gene significance values against age in the iMN expression data set. For the “PC1 neg only” and “PC1 neg overlap” genes, the y-axis indicates gene significance values against PC1 in the iMN expression data set. For the “sALS pos only” and “sALS pos overlap” genes, the y-axis indicates gene significance values against PC1 in the sALS expression data set. (d) Boxplots of intramodule membership for the genes in a. (e) Boxplots of intermodule membership for the genes in a. (f) Cytoscape network maps for iMN gene modules significantly correlated to age and spMN maturation (PC1) in the iMN expression data set. The top 1% of gene-to-gene connections is shown (627 nodes and 3,197 edges). Node colors indicate module assignments. Circular nodes represent genes correlated with age and spMN maturation in the iMN expression data set, and also anti-correlated with the sALS component in the sALS expression data set. Triangular nodes represent genes detected in the iMN modules that correlate with age and spMN maturation, but are not detected in sALS modules that anti-correlate with the sALS component. The relative sizes of all nodes reflect their gene significance towards age in the iMN expression data set. The 99th percentile of edge weights were filtered for display and plotted using the prefuse force layout method. (g) As in f, except for sALS gene modules significantly anti-correlated to the sALS component in the sALS expression data set (323 nodes and 2,886 edges). Larger nodes in this instance have gene significance values closer to -1 and are therefore more anti-correlated to the sALS component. (h) As in f, except for iMN gene modules significantly anti-correlated to age and spMN maturation (PC1) in the iMN expression data set (669 nodes and 6,175 edges). Larger nodes have gene significance values closer to -1 and are therefore more anti-correlated to age. Since there are three distinct, contiguous network clusters with no edges connecting them, they were rotated and repositioned relative to each other so that node shapes are more visible. Therefore, node-to-node spatial relationships within, but not across, contiguous network clusters are accurate. Edges that intersect with dashed lines were shortened and repositioned in order to scale the diagram for visibility. No other modifications to the layout were made. (i) As in g, except for sALS gene modules significantly correlated to the sALS component in the sALS expression data set (312 nodes and 878 edges). Larger nodes have greater gene significance towards the sALS component. For additional information, see Supplementary Tables 2c (for iMN module assignments and properties), 2d (for full lists of significantly enriched GO terms and P-values), 2e (for comparison of gene set enrichments), 5c (for sALS module assignments and properties), 5e (for full lists of significantly enriched GO terms and P-values), 5f (for comparison of gene set enrichments), and 6 (for Wilcoxon rank-sum test statistics).
When comparing the network property of gene significance, overlaps had more biologically significant roles than non-overlaps. This is particularly the case for genes classified as Age and PC1 positive and sALS negative (Fig. 6b and Supplementary Table 6) but not the case for genes classified as Age and PC1 negative and sALS positive (Fig. 6c and Supplementary Table 6). Additionally, overlaps tended to have higher intramodule membership within Age and PC1 positive modules as well as within sALS negative modules (Fig. 6d and Supplementary Table 6). Furthermore, pathogenic genetic variants associated with ALS tend to have higher intramodule membership than genes not affected by ALS variants (Supplementary Fig. 5e). Together, these observations further underscore vulnerability of cellular systems to disruptions targeting network hub genes in both the familial and sporadic ALS conditions. Lastly, intermodule membership for each gene measures its expression correlation to the eigengenes of other modules that significantly correlate or anti-correlate to the same sample trait [15]. This metric can be used to identify hub genes that connect multiple modules that act on distinct but coordinated pathways. This comparison showed that overlaps tended to have higher intermodule membership within Age and PC1 positive modules as well as within sALS negative modules (Fig. 6e and Supplementary Table 6), suggesting that they are functionally conserved as intermodule hub genes.
These gene network properties can be simultaneously visualized as network maps (Figs. 6f – i). Within Age and PC1 positive modules, a network map filtered for the strongest 1% of gene-to-gene connections comprised of multiple interconnected modules (Fig. 6f). This network contained far more overlaps (circular nodes) than non-overlaps (triangular nodes), and overlaps tended to have greater gene significance with respect to age (relative node size) and also tended to have greater intramodule membership (central positions within like-colored clusters). Non-overlaps demonstrated a lower degree of intramodule membership compared to overlaps, as they occupied more peripheral positions within like-colored clusters (Fig. 6f). The same trends were observed in sALS negative modules (Fig. 6g), Age and PC1 negative modules (Fig. 6h), and sALS positive modules (Fig. 6i). Altogether, these data highlight the importance of hub genes within networks associated with MN maturation and aging, evidenced by the preservation of their network structures within the strongest connections detected in the sALS data set. Furthermore, the high gene significance measures of these hub genes suggest they are the most responsive genes to perturbations caused by sALS.
Discussion
Many iPSC differentiated tissues represent an immature or fetal stage of development [23, 24]. This is presumably due to resetting the epigenetic state to that of an embryo during iPSC production, which can occur with samples even over 90-years old [25]. Our study wholly demonstrates that iPSC-derived MNs are more similar to their fetal in vivo counterparts than adult MNs based on their transcriptome.
Through an unbiased, genome-wide approach, we identified network hub genes described as markers or functional drivers of neuronal development or maturation. For example, DCX, NEFH, and SNAP25 have been shown as histological markers of early, intermediate, and late neuronal maturation, respectively [26]. Additionally, loss of function mutations in SCN1A are implicated in a failure of interneurons to develop mature action potentials in Dravet syndrome [27], and ASCL1 is the key pioneering transcription factor driving direct fibroblast conversion to neurons [28], which subsequently demonstrate slower maturation kinetics [29]. Using only 20 markers, we reduced the number of genes tested and thereby circumvented microarray platform-specific biases that confound accurate comparison of maturation states across samples. This technique will be extremely helpful to develop methods to efficiently mature spMNs in culture, and can be applied to other iPSC-derived cell types such as upper MN models of ALS [30], given the transcriptomic data available for in vivo brain tissues at different developmental stages [31, 32].
Using MNs to model neurological disease may require their aging in the dish. Attempts to simulate aspects of aging in iPSC-derived neurons include prolonging time in culture or introducing agents of cellular stress by chemical or genetic means [33-37]. Notably, these strategies can concomitantly bring about age-related molecular features along with phenotypes associated with neurodegeneration. However, our data indicate that molecular processes of neuronal aging and neuronal maturation are distinct, substantiating a perspective held by others [38]. Therefore, it is unclear whether these techniques accelerated iPSC progression from the embryonic to a mature neuronal state, or simply induced isolated aging pathways in immature cells. Since cellular age is collectively a multi-faceted, syndromic condition [7], it is most effectively assayed in a systems-wide manner. Thus, the global gene expression and network analysis we have employed is well-suited to assay cellular maturity and age and can provide a comprehensive method to assess iPSC-derived tissue models under stress-induced conditions. A caveat to acknowledge is that the adult spinal cord samples used in our composite data set are younger than the adult laser captured MNs. We therefore recognize that some genes in the age-associated modules may actually be more associated with different sample types. Because a large number of in vivo spinal cords profiled on the same microarray platform along with detailed clinical data were difficult to obtain, we combined all available expression profiles in order to achieve more robust statistical power. Therefore, our data set provides a resource to generate hypotheses; however, the burden will be on follow-up investigations to eliminate such confounding factors.
Studies using direct neuronal reprogramming from aged adult fibroblasts have shown that the matured epigenetic state may be maintained [39]. While MNs have been directly converted from embryonic and fetal fibroblasts [40, 41], it would be interesting to see if similar techniques performed on aged fibroblasts maintain a more mature state through the transition process. Nevertheless, we provide data and methods that future studies can apply in addressing these questions.
There are increasing genetic variants reported to be associated with several human diseases in the ClinVar database [17]. Strikingly, some modules correlated with maturation or age (or both) enriched for variants pathogenically linked to ALS or MN disease. This lends strong evidence for maturation- and age-associated networks and pathways acting as causative effectors of late onset diseases. Future characterization of novel genetic variants classified as risk factors may explain how they act collectively to modify the penetrance or onset time of disease in individual patients [42].
The mitochondrial free radical theory of aging posits that as cells age, their electron transport chain stability declines, and accumulating reactive oxygen species oxidatively damage nuclear and mitochondrial DNA [43]. Our analysis revealed that respiratory chain components decreased expression during spMN maturation, consistent with observations in ALS patient blood [44]. Additionally, DNA repair pathways decreased expression as spMNs mature, suggesting a reduced ability to mitigate oxidative DNA damage. Our analysis also revealed that sALS further downregulated mitochondrial respiratory chain genes. Thus, the combination of spMN maturation and sALS can exacerbate a condition in which spMNs are vulnerable to oxidative damage. Notably, these mitochondrial components were reported to be downregulated in mtSOD1 iMNs [20], indicating that despite being in a fetal context, some key pathways can already be affected.
Interestingly, a dynamic expression pattern of gene networks associated with antigen presentation and immune response was highlighted. Whereas these processes increased expression as spMNs mature, they decreased expression as spMNs age. This model is consistent with the role of microglia in pruning synaptic connections during neuronal maturation and their hyperactivity in late-onset diseases [45]. The immune response and complement activation pathways were also upregulated in sALS, similar to observations in spinal cords of C9orf72-deficient ALS mouse models [46]. In summary, sALS antagonizes the endogenous expression pattern of these immune activation pathways and that of protein translation and degradation. These observations therefore support that the expression kinetics for these pathways serve a homeostatic, protective role in aging spMNs that can be derailed by ALS, resulting in neurodegeneration.
Our analysis also provided another dimension to the role of genes within maturation and aging expression networks. The scale-free architectures of natural systems are robustly tolerant against perturbations to the majority of its nodes, but at the cost of being vulnerable to disruptions targeting hub nodes [47, 48]. Our observation that ALS preferentially disrupts hub genes within maturation and aging expression networks underscores its devastation to critical cellular systems. Understanding which central genes are the most vulnerable to ALS will guide effective therapies aimed at rescuing the function of these hub genes.
The stem cell modeling and ALS communities can apply the data presented here towards their own research in several ways. This resource includes a variety of transcriptomic data tables composed from and normalized across multiple studies; an atlas of gene networks that can be examined for common or distinct roles in maturation, aging, or ALS in other tissues; enriched gene sets and pathways among those gene networks; and a panel of marker genes that can accurately indicate spMN maturity, either by RT-qPCR or microarrays. Importantly, this study describes an analytical framework for cross-study transcriptomic comparisons, and this methodology can be broadly applied to other models of disease.
Altogether, our findings support a strong interaction between gene networks and pathways associated with spMN maturation, aging, and affected in ALS. This suggests that reenacting the endogenous spMN maturation and aging pathways in iMNs can sensitize them to ALS-induced dysfunction. Nevertheless, it is possible that achieving a mature and aged state in iMNs is superfluous to effective ALS modeling. Disease-relevant phenotypes have been gleaned from immature iMNs, including RNA foci, protein inclusion bodies, altered electrophysiology, nuclear pore deficits, and cell death [10, 13, 20, 34, 49, 50]. While some of these phenotypes have spurred attempts at therapeutic strategies in ALS patients, it remains to be seen whether they are indeed relevant to disease etiology in adults. For instance, if ALS patients treated with retigabine, an antiepileptic drug that reduces hyperexcitability in mtSOD1 iMNs, demonstrate a positive response to the treatment, this would validate the efficacy of current iMN models in predicting events in adult spMNs. Otherwise, maturation and aging pathways should be considered as modifiers of disease presentation, and our present findings lay the groundwork for future efforts to achieve a higher fidelity model of the molecular and pathological events of ALS as they occur in vivo.
Online Methods
Tissue culture and processing
iMN differentiation was performed as previously described [10]. Briefly, mTeSR1 medium was removed from confluent iPSC cultures and replaced with Iscove's modified Dulbecco's medium supplemented with 2% B27-vitamin A and 1% N2 (neural induction medium) for six days. The cells were then Accutase-treated to single cell suspension, and centrifuged in 384-well PCR plates in the presence of Matrigel and the neural induction medium (now using Neurobasal medium) that was further supplemented with 0.1μM all-trans retinoic acid. After suspension culture for nine days, 1μM purmorphamine was added to the medium and aggregates were cultured for another eight days. Thereafter, dissociated aggregates were plated onto poly-ornithine/laminin-coated coverslips and cultured in Dulbecco's modified Eagle's medium (DMEM)/F12 supplemented with 2% B27, 0.1μM all-trans retinoic acid, 1μM purmorphamine, 1μM dibutyryl cyclic adenosine monophosphate, 200 ng/ml ascorbic acid, 10 ng/ml brain-derived neurotrophic factor, and 10 ng/ml glial cell line-derived neurotrophic factor for 7 weeks. HB9::GFP positive MNs and HB9::GFP negative cell samples were processed directly from their published study [11]. Specifically, the samples analyzed in this study were differentiated using 200 ng/ml SHH as a ventralizing factor.
Fetal tissue was obtained from the Birth Defects Research Laboratory at the University of Washington under their approved IRB, consent, and privacy guidelines. All protocols were performed in accordance with the Institutional Review Board's guidelines at the Cedars-Sinai Medical Center under the auspice IRB-SCRO Protocol Pro00021505. Upon receipt, tissue samples were renamed D52, D53, D63, or D97 to reflect their estimated gestational stage. Samples arrived as fully or partially intact spinal columns, and spinal columns were opened prior to shipment. Vertebrae were removed and were partitioned into cervical, thoracic, and lumbar sections. Since only spinal columns were received, the exact anatomical reference for each somite could not be accurately determined, therefore the labeling of cervical, thoracic, and lumbar sections were estimated.
RNA extraction and processing
Total RNA was isolated from all frozen spinal sections using the RNeasy kit (QIAGEN) with on column DNase digestion. For each fetal spinal cord, equal amounts of total RNA from each section were pooled for expression profiling. RNA from iMNs and ESC-derived MNs were respectively obtained directly from samples previously reported [10, 11]. Prior to expression profiling, all RNA samples were run through RNeasy kit columns with on column DNase digestion to produce similarly sized products as the fetal spinal cord samples. RT-qPCR was performed using the Reverse Transcription System (Promega A3500), SYBR Select Master Mix (Life Technologies 4472908), and the BIO-RAD CFX384 Real-Time System. Primer sequences are listed in Supplementary Table 3a. Triplicate Ct values for each gene primer were normalized against averaged triplicate GAPDH Ct values and referenced against one transcript of that gene set at 40 Ct. RNA expression profiling was performed on the Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays at the UCLA microarray core facility. A list of all expression data sets used (either generated by or downloaded) in this study is given in Supplementary Table 1a.
Immunofluorescence
For D63, spinal cord was isolated as before and fixed in 4% paraformaldehyde for 48 hours. 25μm sections were taken using cryostat (Leica) at -20°C and directly mounted on glass slides (Fisher Scientific). Lumbar tissue section was blocked in PBS containing 5% normal donkey serum (Sigma) and 0.25% Triton-X for 1.5 hours. Primary antibody solution containing mouse anti-SMI32 (Covance SMI-32P-100) and goat anti-Islet-1 (R&D AF1387) were incubated overnight at 4°C. Donkey anti-mouse Alexa-flour 488 and donkey anti-goat 594 secondary antibodies (Life Technologies A21202 and A21289) were incubate for one hour at room temperature. Samples were mounted in Fluoromount-G (SouthernBiotech) and acquired at 10x using automated stitching on a Leica DM 6000 microscope. iMNs were fixed in 4% paraformaldehyde, rinsed with PBS, and blocked in 5% donkey serum and 0.2% Triton-X. Primary antibody solution containing goat anti-ChAT (Millipore AB144P) and mouse anti-SMI32 (Covance SMI-32P-100) were incubated, rinsed in PBS, and incubated in species-specific Alexa-fluor secondary antibodies. The immunostaining shown in Fig. 1a was observed in all iMN lines 00i, 14i, and 83i, which were derived from three individual patients. The immunostaining shown in Fig. 1b was observed in at least 20 sections of the days 52, 53, 63, and 97 fetal spinal cords.
Expression data pre-processing
Previously published mRNA microarray expression data from human fibroblasts (n = 2), embryonic stem cells (ESCs) (n = 2), and iPSCs (n = 3) were chosen to represent cell types relevant to human somatic cell reprogramming [51, 52]. To represent mature in vivo whole spinal cord, we obtained previously published mRNA microarray expression data from adult spinal cords (n = 8, age range = 23 – 53 years, median = 36.5 years) [53] and for mature in vivo spMNs data from laser capture micro-dissected spMNs from familial ALS and control patients [8, 9]. Additionally, expression data for spMN and oculomotor neurons from non-ALS patients [54] were included. These 17 in vivo laser-captured MN specimens came from individuals with an age range from 40 to 80 years (median age, 63 years).
All CEL files considered for use in this study were submitted to ArrayAnalysis.org to inspect RNA and microarray hybridization quality, and samples that failed to meet the recommended standards were removed from further analysis. Affymetrix GeneChip Human Genome U133 Plus 2.0 Array CEL files downloaded from GEO as well as produced in this study were normalized together with Robust Multichip Analysis (rma) using the affy package in Bioconductor R. The accession number for the microarray data produced in this study is GEO: GSE75701. Non-informative probesets were then filtered out using the pvac package in R [55]. This is an unbiased, intuitive method of filtering that is more objective than setting an arbitrary probeset fluorescence intensity cut-off that penalizes lowly expressed transcripts that are nonetheless important. A probeset can be considered non-informative due to being lowly expressed. In addition, a highly expressed probeset can also be considered non-informative if the probes constituting the probeset are highly discordant. “As all probes in a probeset are designed to target the same transcript or a transcript cluster, these probes should largely perform concordantly when gene expression is measured.” [55]. Probeset discordance may arise due to the post mortem collection of RNA from human tissue, where heat or exonuclease activity can degrade RNA at either the 5′ or 3′ ends of the transcript. Inclusion of these probes would adversely affect the interpretation of PCA, thereby justifying a method to remove these probesets. pvac is the proportion of variation accounted for by the first principal component. It measures consistency among probes belonging to the same probeset. The higher the pvac score, the more concordance is observed among probes within a probeset. The pvac package in R generates an empirical distribution of pvac score for all probesets in an Affymetrix expression set. It also generates an empirical distribution of pvac scores for all probesets called absent by the MAS5 command on the affy package. These probesets are postulated to have low concordance among their probes, therefore their distribution of pvac scores serves as the threshold for filtering concordant and informative probesets. The application of pvac filtering on our data set serves two purposes. It can remove probesets that are highly expressed but discordant, ruling out inaccurately quantified transcripts that have possibly been degraded in post mortem samples. It can also remove lowly expressed probesets that have a comparable degree of discordance as those probesets deemed to be absent by the MAS5 algorithm. Conversely, lowly expressed probesets can also be retained if their probes have a sufficiently high pvac score. These probesets may have otherwise been discarded if an arbitrary intensity threshold was applied to the expression set.
Filtered probesets were then annotated to their HGNC symbol using the Affymetrix annotation file for the GeneChip Human Genome U133 Plus 2.0 Array Release 35, summarized to the gene level by taking the probeset with the maximum expression value to represent the maximal transcriptional activity associated with that gene, and the resulting 10,605 gene expression values were quantile normalized on the linear scale using the normalize.quantiles function in the preprocessCore package. The processed Affymetrix Human Genome U133 Plus 2.0 Array expression values for 10,605 genes are listed in Supplementary Table 2a. For RNAseq data from mtSOD1 and control iMNs [20], normalized counts for each sample as they were provided through GEO accession series GSE54409 were compiled into an expression table for 14,422 Ensembl identifiers, re-annotated to HGNC symbols based on gene symbols selected through Ensembl BioMart, and summarized to the gene level by taking the transcripts with the maximum expression value to represent the maximal transcriptional activity associated with each gene. This list of genes was intersected with the list of 10,605 genes used for the mtSOD1 and control spMN samples [9] to produce the expression table for 8,830 genes listed in Supplementary Table 4.
For the sALS data set [21], CEL files downloaded from GEO accession series GSE18920 and normalized with Robust Multichip Analysis (rma) using the oligo package in Bioconductor R with the pd.huex.1.0.st.v2 array library. Expression values for transcript clusters were re-annotated to Affymetrix Human Genome U133 Plus 2.0 Array probesets, summarized to the gene level by taking the probeset with the maximum expression value to represent the maximal transcriptional activity associated with that gene, and the resulting gene expression values were quantile normalized on the linear scale using the preprocessCore package. This produced the expression table for 15,614 genes listed in Supplementary Table 5a. Power analysis was not calculated prior to determine a justified sample size. Composite data sets were created to compare as many fetal and adult spinal tissues as possible, limited by available data generated on a common microarray platform to minimize batch effects. For network modeling using WGCNA, data sets with similar ratios of sample types, such as pluripotent, fetal, and adult cells or control to sporadic ALS tissue were composed in order to provide equal representation of the relevant cellular states. There was no randomization of the data assigned to classes, and there was no blinding when performing data collection and analysis of the experiments. For analysis comparing mtSOD1 to control spMNs in Fig. 4a – c, three of the five control spMNs were selected for analysis based on the highest Pearson correlation among all five samples to match the sample size of mtSOD1 spMNs (n = 3).
Gene expression analysis
Pearson correlations, statistical tests, and multiple testing corrections were performed in R. Unsupervised hierarchical clustering was performed in Cluster 3.0 and heat maps were visualized using Java Treeview. Principal component analysis (PCA) was performed in Cluster 3.0 or R. The signed values for principal component coordinates of samples and gene loadings were reversed as necessary to maintain consistency across analyses. PCA plots, as well a scatter, density, and box plots were visualized in R with the basic R plotting tools. To generate univariate linear models, the lm function was used in R. Principal component coordinates, or log2 transformed principal component coordinates were used as the dependent variable in order to satisfy the assumption of normally distributed standardized residuals. Sample traits were used as independent variables. P-values determined from the F-statistic were Bonferroni-corrected based on the number of principal components tested against the number of sample traits. Venn and Chow-Ruskey diagrams were generated using the Vennerable package in R. Gene Set Enrichment Analysis (GSEA) [12] was performed on pre-ranked lists generated using PCA gene loadings with 1,000 permutations of the gene sets to generate a null distribution. Gene ontology (GO) enrichment was performed using DAVID, and genes represented on the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array (Supplementary Table 2d) or Affymetrix Human Exon 1.0 ST Array (Supplementary Table 5e) were used as background. Adjusted P-value or FDR q-value thresholds for enriched gene sets and GO terms are indicated for each figure in their respective legends.
WGCNA was performed using its package in R [15]. The two fibroblast samples as well as the HB9::GFP negative sample were removed before network building. To create networks and modules from gene expression data sets, a similarity matrix is calculated for all pairwise genes xi and xj using Pearson's correlation and then transformed into an adjacency matrix using a signed network power function, aij = (0.5 +0.5cor(xixj))^β. The resulting relationships between each pairwise set of genes are known as their connection strength. This connection strength can be increased if two genes have a similar profile of connection strengths with all other genes, or it can be decreased if the two genes have a very dissimilar profile of connection strengths with all other genes. This is regarded as their topological overlap measure (TOM). Genes are then hierarchically clustered using average linkage with their TOM distances, and the resulting dendrogram is used to classify genes into modules (low hanging branches on the dendrogram) by specifying the branch height to cut as well as the minimal number of genes to include into a module. Key parameters used for both the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array (iMN) and Human Exon 1.0 ST Array (sALS) expression data sets were as follows: 30 and 21, respectively, were chosen as the soft threshold powers (β) to transform each of the similarity matrices into adjacency matrices, yielding networks with scale-free topology model fits that were greater than 0.8, a value satisfying the proposed scale-free topology criterion [56]. The one step network construction and module detection command blockwiseModules was used, with power = 30 or 21, maxBlockSize = 10,606 (iMN data set) or 15,615 (sALS data set), deepSplit = 3, pamStage = FALSE, TOMType = “signed”, networkType = “signed”, minModuleSize = 30 (this sets the minimum number of genes needed to form a module to 30 genes), reassignThreshold = 0, mergeCutHeight = 0, numericLabels = TRUE, pamRespectsDendro = FALSE, saveTOMs = TRUE, saveTOMFileBase = “mnTOM”, and verbose = 3. In the data set excluding the familial ALS spMNs, 10,605 genes were used for WGCNA, the soft threshold power was set to 30, and the mergeCutHeight was set to 2.5. This produced the results shown in Supplementary Fig. 2a. The eigengene expression of the resulting modules, which is the first principal component of all expression values of genes assigned to that module, were then correlated to the samples traits or principal component coordinates using Pearson's method, the P-values of each correlation were determined based on the degrees of freedom for each test, (sample size minus 2), and P-values were Bonferroni-corrected based on the number of modules tested (55 for the iMN data set, 52 for the sALS data set, and 15 for the iMN data set lacking familial ALS spMNs). The gene module assignments and correlations to sample traits are listed in Supplementary Tables 2c (iMN) and 5c (sALS). Module-trait correlations with Bonferroni-corrected P-values < 0.01 were called significant and kept for GO term enrichment analysis through DAVID shown in Supplementary Tables 2d (iMN) and 5e (sALS). Non-linear associations between a) module eigengenes and sample traits or b) principal component coordinates and sample traits were tested using 1) Spearman's method for age and post mortem interval, 2) the Kruskal-Wallis test for the categorical variables tissue type, sex, study, disease status, site of ALS onset, and sample processing, and 3) Cox-regression for ALS disease duration. These tests (subjected to multiple testing corrections) revealed no significant associations that were not shown to be significant using linear models. One exception was that by Spearman's but not Pearson's correlation, the paleturquoise module anti-correlates with age. This observation did not impact the main interpretations, therefore we based our discussions on results from the linear correlations.
We note that the gene sets and pathways enriched by using principal component gene loadings and GSEA serve as confirmatory indicators of sensible pathways describing the differences between samples, while the pathways enriched by using DAVID on modules serve as more discovery and hypothesis generators. WGCNA is more sensitive than PCA, because WGCNA accounts for subtler, but consistent gene-to-gene co-expression relationships that need not have larger gene loadings per principal component to be detected. These larger gene loadings are rather rewarded in GSEA at the cost of detecting significantly correlated gene networks with lower gene loadings in embryonic spMN development or maturation that nonetheless may also be physiologically important.
To identify the 20 key marker genes, fibroblasts were removed from the iMN data set, and PCA was applied to the resulting “training data set” of 10,605 genes in 41 samples to produce 41 principal components. We considered three properties of each gene to select for the most effective markers of spMN maturation and embryonic development. These properties were 1) gene significance, 2) intramodule membership, and 3) preferential gene loading. As an example, HOXB8 has a gene significance value of 0.78 along embryonic spMN development (PC3). However, the gene with the highest gene significance along embryonic spMN development is CXXC4 (0.92). Among all the genes classified into the category PC3pos, CXXC4 has the highest gene significance score (0.92/0.92 = 1.00), and HOXB8 has a gene significance score of 0.78/0.92 = 0.85. A gene with high intramodule membership best represents the expression of all genes within the module to which it is assigned. HOXB8 belongs to the paleturquoise module and has an intramodule membership of 0.94. However, CAMK2N1, which belongs to the darkolivegreen module, has an intramodule membership of 0.97, which is the highest intramodule membership value among all the genes classified into the category PC3pos. Therefore, HOXB8 has an intramodule membership score of 0.94/0.97 = 0.97. Among all 10,605 genes in the iMN data set, HOXB8 has the highest gene loading along PC3 (0.06). However, HOXB8 can just as well have high gene loadings in the other 40 principal components. Therefore to quantify to what extent HOXB8 preferentially contributes to PC3, the average gene loading for HOXB8 in the other 40 principal components is subtracted from its gene loading in PC3. Before performing this operation, the gene loadings of HOXB8 in all principal components were squared to convert them into positive values. These values needed to be scaled by the percentage contribution of each principal component to the total variance. Dividing each eigenvalue by the sum of all 41 eigenvalues and multiplying by 100 produced the percentage contribution of each eigenvalue to the total variance, and the squared gene loading for each component was multiplied by this percentage contribution. Because principal components 35, 36, etc. have lower eigenvalues, a large gene loading for HOXB8 in these components are less detrimental to the preferential gene loading score. As observed in Supplementary Table 2i, HOXB8 has the highest preferential PC3pos gene loading score, because it has 1) the highest gene loading in PC3, and 2) it has relatively low gene loadings in all other principal components. Normalizing the scores by the maximum value in each category for each of the gene properties evenly weights their contribution in determining the best marker genes for each of the four categories PC1pos, PC1neg, PC3pos, and PC3neg. The total gene scores were summed from all three properties and ranked from largest to smallest. These values are listed in Supplementary Tables 2f – i. The ROCR package was used to calculate and plot the Receiver Operator Characteristics and Area Under the Curve analyses. Predictions were based on correct classification of pluripotent stem cells, fetal-like cells, and adult spinal cord cells. Tests were performed using either 1) the Pearson correlation between the expression values in the test data set and the median expression values among those cell types in the training data set, or 2) the negative threshold below PC1 coordinate for pluripotent stem cells, positive threshold above PC1 coordinate for adult spinal cord cells, and positive threshold above PC7 coordinate (6,640 genes) or PC2 coordinate (20 genes) for fetal-like cells. PC coordinates were generated by performing PCA with 6,640 or 20 genes on all samples from both the training and test data sets, but the prediction table used to generate the ROC curve and AUC values are without the training data set.
Hypergeometric tests for module enrichment were performed using custom scripts applying the dhyper function in R, and the resulting hypergeometric P-values were corrected with the Benjamini-Hochberg method. For ClinVar enrichment in the iMN modules, 22,608 genes represented on the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array were used as the background number of genes. For iMN module enrichment in the sALS modules, 21,279 genes represented on both the Affymetrix GeneChip Human Genome U133 Plus 2.0 and Human Exon 1.0 ST Arrays were used as the background number of genes.
Module preservation was performed using the WGCNA package in R [22]. 8,715 overlapping genes were represented in both the iMN and sALS expression data sets and were thus used in the preservation analysis. The expression values for these 8,715 genes were loaded as two data tables into one multiExpr object: one data table of expression values from the iMN data set, and the other data table of expression values from the sALS data set. Two lists of module color assignments were also loaded into a multiColor object: one listing the module color assignments for the 8,715 genes when WGCNA was performed using 10,605 genes in the iMN data set, and the other listing the module color assignment for the 8,715 genes when WGCNA was performed using 15,614 genes in the sALS data set. The command modulePreservation was called on the multiExpr and multiColor objects, first by using the iMN data set as the reference and the sALS data set as the test, and then vice versa. 200 permutations were used to randomly assign genes to module colors in the test data. This generates a distribution of preservation statistics under the null hypothesis that no modules are preserved across the reference and test data sets. Z-statistics are defined for preservation statistic such as module density and module connectivity. The Z-summary summarizes all individual Z-statistics. Since Z-summary is heavily influenced by module size, the median rank measure for each module is more appropriate when comparing the relative preservation among differently sized modules.
Cytoscape 3.0 was used to visualize network topology for each of the four classes of human networks depicted in Figs. 6f – 6i. For each class, the 99th percentile of edge weights were filtered followed by filtering for source nodes with at least 30 edges with target nodes. Edge and node attributes, as well as graphical layout constraints are described in the figure legends.
Data availability
The data that support the findings of this study are available in the Supplementary Figures, Supplementary Tables, and Statistics Reporting Checklist. The microarray data generated in this study are available at the Gene Expression Omnibus under the accession code GSE75701. The programming scripts written for the analysis performed in this study are compiled into a text document and available https://github.com/ritchieho/NN-RS55118B_R_scripts.git.
A Supplementary Methods Checklist is available.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the following: B. Shelley, L. Garcia, and L. Ornelas for assistance with experiments and reagent organization; B. Berman and D. Rushton for statistical and programming advice; B. Berman, V. Mattis, and S. Svendsen for critical reading and comments on the manuscript. This work was supported by the following grants: NIH/NINDS (U54NS091046-01) (C.N.S) and the ALS Association (R.H., C.N.S.). The Project ALS Foundation supported work done by H.W. and M.W.A.
Footnotes
Accession Codes: The microarray data generated in this study are available at the Gene Expression Omnibus under the accession code GSE75701.
Competing Financial Interests: The authors declare no competing financial interests.
Author Contributions: Conceptualization: R.H. and C.N.S.; Methodology: R.H.; Software: R.H.; Formal Analysis: R.H.; Investigation: R.H., S.S., G.G., M.W.A., J.G.O., A.S., D.S., and C.N.S; Resources: H.W., R.H.B., D.S., and C.N.S.; Data Curation: R.H.; Writing – Original Draft: R.H. and C.N.S.; Writing – Review & Editing: R.H., S.S., M.W.A, and C.N.S.; Visualization: R.H.; Supervision: C.N.S.; Funding Acquisition: C.N.S.
References
- 1.Hardiman O, van den Berg LH, Kiernan MC. Clinical diagnosis and management of amyotrophic lateral sclerosis. Nat Rev Neurol. 2011;7(11):639–49. doi: 10.1038/nrneurol.2011.153. [DOI] [PubMed] [Google Scholar]
- 2.Dimos JT, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321(5893):1218–21. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 3.Sterneckert JL, Reinhardt P, Scholer HR. Investigating human disease using stem cell models. Nat Rev Genet. 2014;15(9):625–39. doi: 10.1038/nrg3764. [DOI] [PubMed] [Google Scholar]
- 4.Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1(1):20–9. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 5.Wichterle H, et al. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110(3):385–97. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 6.Sances S, et al. Modeling ALS with motor neurons derived from human induced pluripotent stem cells. Nat Neurosci. 2016;16(4):542–53. doi: 10.1038/nn.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arbab M, Baars S, Geijsen N. Modeling motor neuron disease: the matter of time. Trends Neurosci. 2014;37(11):642–52. doi: 10.1016/j.tins.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Cox LE, et al. Mutations in CHMP2B in lower motor neuron predominant amyotrophic lateral sclerosis (ALS) PLoS One. 2010;5(3):e9872. doi: 10.1371/journal.pone.0009872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirby J, et al. Phosphatase and tensin homologue/protein kinase B pathway linked to motor neuron survival in human superoxide dismutase 1-related amyotrophic lateral sclerosis. Brain. 2011;134(Pt 2):506–17. doi: 10.1093/brain/awq345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sareen D, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5(208):208–149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amoroso MW, et al. Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J Neurosci. 2013;33(2):574–86. doi: 10.1523/JNEUROSCI.0906-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devlin AC, et al. Human iPSC-derived motoneurons harbouring TARDBP or C9ORF72 ALS mutations are dysfunctional despite maintaining viability. Nat Commun. 2015;6:5999. doi: 10.1038/ncomms6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wojcik-Stanaszek L, Gregor A, Zalewska T. Regulation of neurogenesis by extracellular matrix and integrins. Acta Neurobiol Exp (Wars) 2011;71(1):103–12. doi: 10.55782/ane-2011-1827. [DOI] [PubMed] [Google Scholar]
- 15.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das MM, Svendsen CN. Astrocytes show reduced support of motor neurons with aging that is accelerated in a rodent model of ALS. Neurobiol Aging. 2015;36(2):1130–9. doi: 10.1016/j.neurobiolaging.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 17.Landrum MJ, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42(Database issue):D980–5. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ince PG, et al. Molecular pathology and genetic advances in amyotrophic lateral sclerosis: an emerging molecular pathway and the significance of glial pathology. Acta Neuropathol. 2011;122(6):657–71. doi: 10.1007/s00401-011-0913-0. [DOI] [PubMed] [Google Scholar]
- 19.Schymick JC, Talbot K, Traynor BJ. Genetics of sporadic amyotrophic lateral sclerosis. Hum Mol Genet. 2007;16 Spec No. 2:R233–42. doi: 10.1093/hmg/ddm215. [DOI] [PubMed] [Google Scholar]
- 20.Kiskinis E, et al. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell. 2014;14(6):781–95. doi: 10.1016/j.stem.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabin SJ, et al. Sporadic ALS has compartment-specific aberrant exon splicing and altered cell-matrix adhesion biology. Hum Mol Genet. 2010;19(2):313–28. doi: 10.1093/hmg/ddp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langfelder P, et al. Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7(1):e1001057. doi: 10.1371/journal.pcbi.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdelalim EM, Emara MM. Advances and challenges in the differentiation of pluripotent stem cells into pancreatic beta cells. World J Stem Cells. 2015;7(1):174–81. doi: 10.4252/wjsc.v7.i1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Batalov I, Feinberg AW. Differentiation of Cardiomyocytes from Human Pluripotent Stem Cells Using Monolayer Culture. Biomark Insights. 2015;10(Suppl 1):71–6. doi: 10.4137/BMI.S20050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lapasset L, et al. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev. 2011;25(21):2248–53. doi: 10.1101/gad.173922.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarnat HB. Clinical neuropathology practice guide 5-2013: markers of neuronal maturation. Clin Neuropathol. 2013;32(5):340–69. doi: 10.5414/NP300638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bender AC, et al. SCN1A mutations in Dravet syndrome: impact of interneuron dysfunction on neural networks and cognitive outcome. Epilepsy Behav. 2012;23(3):177–86. doi: 10.1016/j.yebeh.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wapinski OL, et al. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell. 2013;155(3):621–35. doi: 10.1016/j.cell.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chanda S, et al. Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Reports. 2014;3(2):282–96. doi: 10.1016/j.stemcr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc. 2012;7(10):1836–46. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- 31.Hawrylycz M, et al. Canonical genetic signatures of the adult human brain. Nat Neurosci. 2015;18(12):1832–1844. doi: 10.1038/nn.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478(7370):483–9. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Almeida S, et al. Induced pluripotent stem cell models of progranulin-deficient frontotemporal dementia uncover specific reversible neuronal defects. Cell Rep. 2012;2(4):789–98. doi: 10.1016/j.celrep.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bilican B, et al. Mutant induced pluripotent stem cell lines recapitulate aspects of TDP-43 proteinopathies and reveal cell-specific vulnerability. Proc Natl Acad Sci U S A. 2012;109(15):5803–8. doi: 10.1073/pnas.1202922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller JD, et al. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell. 2013;13(6):691–705. doi: 10.1016/j.stem.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen HN, et al. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011;8(3):267–80. doi: 10.1016/j.stem.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi Y, et al. A human stem cell model of early Alzheimer's disease pathology in Down syndrome. Sci Transl Med. 2012;4(124):124–29. doi: 10.1126/scitranslmed.3003771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Studer L, Vera E, Cornacchia D. Programming and Reprogramming Cellular Age in the Era of Induced Pluripotency. Cell Stem Cell. 2015;16(6):591–600. doi: 10.1016/j.stem.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mertens J, et al. Directly Reprogrammed Human Neurons Retain Aging-Associated Transcriptomic Signatures and Reveal Age-Related Nucleocytoplasmic Defects. Cell Stem Cell. 2015 doi: 10.1016/j.stem.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu ML, et al. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat Commun. 2013;4:2183. doi: 10.1038/ncomms3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Son EY, et al. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9(3):205–18. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cady J, et al. Amyotrophic lateral sclerosis onset is influenced by the burden of rare variants in known amyotrophic lateral sclerosis genes. Ann Neurol. 2015;77(1):100–13. doi: 10.1002/ana.24306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kennedy SR, Loeb LA, Herr AJ. Somatic mutations in aging, cancer and neurodegeneration. Mech Ageing Dev. 2012;133(4):118–26. doi: 10.1016/j.mad.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin J, et al. Specific electron transport chain abnormalities in amyotrophic lateral sclerosis. J Neurol. 2009;256(5):774–82. doi: 10.1007/s00415-009-5015-8. [DOI] [PubMed] [Google Scholar]
- 45.Chung WS, et al. Do glia drive synaptic and cognitive impairment in disease? Nat Neurosci. 2015;18(11):1539–45. doi: 10.1038/nn.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Rourke JG, et al. C9orf72 is required for proper macrophage and microglial function in mice. Science. 2016;351(6279):1324–9. doi: 10.1126/science.aaf1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406(6794):378–82. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 48.Langfelder P, Mischel PS, Horvath S. When is hub gene selection better than standard meta-analysis? PLoS One. 2013;8(4):e61505. doi: 10.1371/journal.pone.0061505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wainger BJ, et al. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 2014;7(1):1–11. doi: 10.1016/j.celrep.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang K, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525(7567):56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chin MH, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5(1):111–23. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maherali N, et al. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3(3):340–5. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roth RB, et al. Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics. 2006;7(2):67–80. doi: 10.1007/s10048-006-0032-6. [DOI] [PubMed] [Google Scholar]
- 54.Brockington A, et al. Unravelling the enigma of selective vulnerability in neurodegeneration: motor neurons resistant to degeneration in ALS show distinct gene expression characteristics and decreased susceptibility to excitotoxicity. Acta Neuropathol. 2013;125(1):95–109. doi: 10.1007/s00401-012-1058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu J, et al. Principal component analysis-based filtering improves detection for Affymetrix gene expression arrays. Nucleic Acids Res. 2011;39(13):e86. doi: 10.1093/nar/gkr241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available in the Supplementary Figures, Supplementary Tables, and Statistics Reporting Checklist. The microarray data generated in this study are available at the Gene Expression Omnibus under the accession code GSE75701. The programming scripts written for the analysis performed in this study are compiled into a text document and available https://github.com/ritchieho/NN-RS55118B_R_scripts.git.
A Supplementary Methods Checklist is available.