Abstract
Bipolar disorder (BD) is a multifactorial illness thought to result from an interaction between genetic susceptibility and environmental stimuli. Epigenetic mechanisms, including DNA methylation, can modulate gene expression in response to the environment, and therefore might account for part of the heritability reported for BD. This paper aims to review evidence of the potential role of DNA methylation in the pathophysiology and treatment of BD. In summary, several studies suggest that alterations in DNA methylation may play an important role in the dysregulation of gene expression in BD, and some actually suggest their potential use as biomarkers to improve diagnosis, prognosis, and assessment of response to treatment. This is also supported by reports of alterations in the levels of DNA methyltransferases in patients and in the mechanism of action of classical mood stabilizers. In this sense, targeting specific alterations in DNA methylation represents exciting new treatment possibilities for BD, and the ‘plastic’ characteristic of DNA methylation accounts for a promising possibility of restoring environment-induced modifications in patients.
Keywords: bipolar disorder, DNA methylation, epigenetics, mood stabilizers, DNA methyltransferase, mood disorders
1. Introduction
Complex psychiatric disorders, including mood disorders such as major depressive disorder (MDD) and bipolar disorder (BD), have been shown to run in families and present considerable heritability (Dunn et al., 2015; Kerner, 2015). This is based on observations that first-degree relatives of patients with mood disorders have a higher risk to develop psychiatric disorders than the general population (Potash and DePaulo, 2000; Sullivan et al., 2000), and that monozygotic twins have higher concordance rates than dizygotic twins for these phenotypes (Rice et al., 2002; Smoller and Finn, 2003).
However, most of the studies that aimed at defining the genetic basis of these disorders have failed to accurately detect the same magnitude of heritability in the form of common genetic variants (‘heritability gap’), which suggests that additional mechanisms are operative. Thus, the current model for BD views the disease as the result of the interaction between genetic susceptibility and environmental stimuli (Kerner, 2015). Accordingly, several environmental events have been reported to be associated with a higher incidence of BD, including childhood trauma and chronic stress (Brietzke et al., 2012). These external stimuli are thought to interact with a susceptible genotype in the pathogenesis of BD, which has been empirically shown by a few studies. For instance, patients who carry the Met allele at the brain-derived neurotrophic factor (BDNF) Val66Met polymorphism have been shown to be more likely to develop depressive episodes following stressful life events than Val allele homozygous (Hosang et al., 2010). In this same vein, a recent report has shown that pathogen exposure interacts with a single nucleotide polymorphism in the toll-like receptor 2 (TLR2) gene in the modulation of the risk of BD (Oliveira et al., 2016), and the same polymorphism has also been suggested to amplify the negative effects of childhood sexual abuse on age at onset of BD (Oliveira et al., 2015). Finally, interactions between early trauma and polymorphisms in or near genes coding for calcium channel activity-related proteins have also been suggested to have a potential effect on the development and manifestation of BD (Anand et al., 2015).
Such interactions suggest the action of so-called ‘epigenetic mechanisms’, which can modulate gene expression in response to the environment, and therefore might partly underlie the multifactorial heritability of BD. These mechanisms include the actions of microRNAs, long noncoding RNAs, and covalent modifications of histones and DNA, forming either repressive or permissive chromatin, thereby modulating gene expression (Breiling and Lyko, 2015; Kouzarides, 2007; Roundtree and He, 2015). DNA methylation is the most stable form of epigenetic alteration, and several studies suggested a critical role for this mechanism in BD (Li et al., 2015; Nestler et al., 2015). This paper aims to survey the literature to shed light on the potential role of DNA methylation in the pathophysiology and treatment of BD.
2. DNA methylation
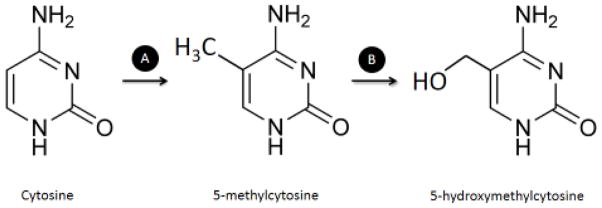
DNA methylation in higher eukaryotes is the addition of a methyl (CH3) group to the carbon-5 of cytosines in the DNA sequence, generating a modified nucleotide called 5-methylcytosine (5mC) (Figure 1) (Breiling and Lyko, 2015). Formation of 5mC has been classically involved in gene silencing and repressive chromatin (heterochromatin) (Schubeler, 2015), but recent evidence suggests additional functions for DNA methylation (Reddington et al., 2013). In vertebrates, DNA methylation occurs throughout the entire genome at cytosines of CpG dinucleotides. Even though these dinucleotides are underrepresented in the whole genome, they are frequently enriched around promoter regions, in so-called CpG islands (CGI) (Lo and Weksberg, 2014).
Figure 1.
Catalysis of DNA methylation and hydroxylation. A) Cytosine at CpG dinucleotides can be methylated at carbon-5 by DNMTs, which use SAM as methyl-carrier, generating a modified nucleotide called 5-methylcytosine (5mC). B) Possibly as first step of an active demethylation process, 5mC can be oxidized by TET enzymes to 5-hydroxymethylcytosines (5hmC), whose exact function is still unknown. DNMT – DNA methyltransferase; TET – ten-eleven translocation.
While CpG dinucleotides are commonly methylated across the genome, CGI regions are typically maintained in an unmethylated form (Lo and Weksberg, 2014). The higher CpG density of CGIs facilitates the DNA methylation-dependent control of promoter activity, in which methylation has been consistently linked to gene repression. In fact, methylation is responsible for several forms of epigenetic repression, such as imprinting, X chromosome inactivation, and silencing of repetitive DNA (Schubeler, 2015). Mechanistically, it can repress the activation of a promoter by either (a) sterically inhibiting the binding of transcription factors necessary for RNA polymerase II recruitment and initiation of transcription, or by (b) actively recruiting repressor proteins such as methyl CpG binding protein 2 (MeCP2) and histone deacetylases (HDAC), which will ultimately lead to the formation of heterochromatin (Breiling and Lyko, 2015; Klose and Bird, 2006). In fact, several proteins recognize and bind to methylated CpGs due to the presence of specific DNA-binding domains in their structures, such as the methyl-binding domain (MBD). At present, eleven proteins containing MBDs have been identified, of which the MeCP2 was the first described (Du et al., 2015). On the contrary, unmethylated CpGs are recognized by Cys-X-X-Cys (CXXC)-type zinc finger domains (where XX represent two any other amino acids), which are found in several proteins with functions related to DNA or chromatin modification (Frauer et al., 2011; Schubeler, 2015). These domains can target and recruit other proteins, such as the CXXC finger protein 1 (CFP1) or the histone demethylase KDM2A and KDM2B, to help maintain the unmethylated state of a particular cytosine (Schubeler, 2015).
Methylated cytosines are also prone to spontaneous deamination, yielding thymines and thus generating a C to T transition (Cortazar et al., 2011). The CG to TG transition is by far the most frequent single nucleotide polymorphism in the mammalian genomes; this is explained by the fact that deamination of unmethylated cytosines yields uracils, which can easily be detected by the repair machinery, while thymines are bona fide DNA nucleobases and repair of the mismatch is expected to have a 50% chance of leaving the thymine. This is also consistent with the higher density of CpGs in the typically unmethylated CGIs.
Of note, the role of DNA methylation in the cell is not confined to gene repression. Recent reports have suggested specific roles for DNA methylation in alternative splicing (Lev Maor et al., 2015) and transcription elongation (Wen and Tang, 2014), among other lesser understood mechanisms (Reddington et al., 2013; Wen and Tang, 2014). Moreover, in some cases DNA methylation has been shown to actually enhance gene transcription (Bockmuhl et al., 2015). Specifically, methylation at the CpG island shore of the nuclear receptor subfamily 3, group C, member 1 (Nr3c1) gene, which encodes the glucocorticoid receptor, has been shown to disrupt the binding of a repressive protein, hindering its ability to recruit other repressive proteins that would otherwise induce post-translational modifications of histones and ultimately lead to a repressive chromatin (Bockmuhl et al., 2015). Such a mechanism is opposite to the known canonical repressive role of DNA methylation, and suggests that its effects are dependent on the location of the CpG. Accordingly, gene body methylation has also been recently shown to increase gene expression by a series of potential mechanisms, including the blocking of initiation of intragenic promoters, interfering with the activity of repetitive DNAs within the transcription unit (Maunakea et al., 2010), or forming an ordered structure within the nucleosome that might increase the rate of transcription either by elongation or splicing (Yang et al., 2014).
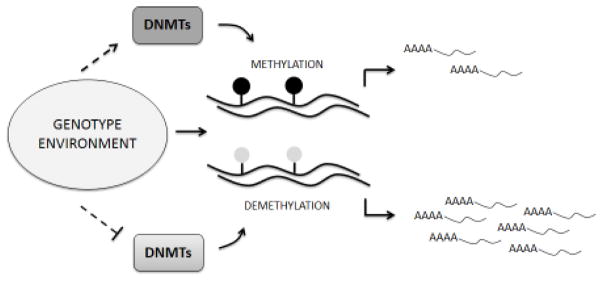
A specific group of enzymes called DNA methyltransferases (DNMTs) is responsible for catalyzing methylation of DNA and is known to be constitutively expressed (Lo and Weksberg, 2014). According to their roles and specificities they have been divided into (a) maintenance DNMTs, such as DNMT1, and (b) de novo DNMTs, which include DNMT3A, DNMT3B and DNMT3L (Sadakierska-Chudy et al., 2015). By recognizing hemimethylated DNA strands, DNMT1 faithfully maintains the pattern of DNA methylation across cell divisions, even though some patterns can be lost passively through imperfect maintenance (Schubeler, 2015). In contrast, active demethylation has been suggested to be initiated by the ten-eleven translocation (TET) family of proteins, including TET1, TET2, and TET3, by converting 5mC into 5-hydroxymethylcytosine (5hmC) (Figure 1) (Sadakierska-Chudy et al., 2015). Further oxidations catalyzed by TET enzymes leads to the formation of 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), which can then be efficiently removed by thymine-DNA glycosylase (TDG) (Schubeler, 2015). The orchestrated action of DNMTs, TETs and TDG is responsible for establishing and maintaining specific DNA methylation patterns in different cell types (Figure 2). Of note, particular roles for the oxidized forms of 5mC are only starting to be understood. The levels of 5hmC are very high in the central nervous system, which led to the hypothesis that hydroxyl methylation might play a role in the epigenetic control of neuronal function (Tognini et al., 2015). In addition, 5hmC is quite abundant in gene bodies and promoter regions of active genes, and evidence shows that MeCP2 can bind to it with an affinity comparable to 5mC (Spruijt et al., 2013). Of note, several other proteins have been shown to bind to 5hmC, 5fC and 5caC, including DNA glycosylases and DNA repair proteins, and each of them seems to recruit a distinct and dynamic set of proteins (Spruijt et al., 2013).
Figure 2.
Typical dynamics of DNA methylation. DNA methylation patterns are determined by a combination of genetic background and environmental exposure. By means of modulating the activity of DNMTs, CpG dinucleotides will undergo methylation or (passive) demethylation of promoters, gene bodies and/or gene regulatory regions. Hypermethylation of CGIs in promoter regions have been traditionally associated with decreased gene expression, whereas their hypomethylation has been linked to increased gene expression. DNMT – DNA methyltransferase; CGI – CpG island.
Apart from the studies of basic mechanisms, the translational analysis of DNA methylation in pathological conditions, such as BD, can be of great value by producing biomarkers of state, prognosis and treatment response, in addition to advancing our understanding of pathophysiological mechanisms. Moreover, the ready availability of genome-wide techniques has sparked intense research efforts to delineate 5mC patterns in health and disease. A particular challenge for these analyses is the cellular heterogeneity (different from genetic sequence, DNA methylation is cell specific and will only directly inform about the tissue being used in the analysis). The next sections will discuss findings reported for alterations in DNA methylation and DNMTs in BD, followed by a review of the modulation of these mechanisms induced by mood stabilizers.
3. DNA methylation in BD
BD affects around 1% of the world’s population and represents one of the leading causes of disability (Grande et al., 2016). It is a lifelong episodic illness with a variable course that is characterized by the occurrence of at least one manic episode (BD I) or one hypomanic and one major depressive episode (BD II). Specifically, manic or hypomanic episodes are states of elevated mood and increased motor drive that impair social or occupational functioning, and a comprehensive biological, social, and psychological approach is required for its treatment and investigation (Grande et al., 2016). Of note, no biomarker has yet been approved for its diagnosis or prognosis (Carvalho et al., 2016; Frey et al., 2013). Nevertheless, alterations in DNA methylation patterns in patients with BD have been extensively investigated for the past years, being considered a promising marker that could integrate both genotype and environmental effects. Genome-wide methylation as well as methylation of several specific candidate genes known to be linked to BD, such as brain-derived neurotrophic factor (BDNF) and the serotonin receptors, are discussed below and are summarized in Table 1.
Table 1.
Alterations in DNA methylation in bipolar disorder
| Gene | Cell/tissue | Method | Main findings | Reference |
|---|---|---|---|---|
| 5-HTR1A | Leukocytes | HRM | Hypermethylation of 5HTR1A gene. | Carrard et al., 2011 |
| 5-HTR2A | Saliva | MSP and bisulfite sequencing | Hypomethylation of the 5-HTR2A gene at the T102C polymorphic region. | Ghadirivasfi et al., 2011 |
| 5-HTR2A | Postmortem frontal lobe | MSP | Hypermethylation of the 5-HTR2A promoter region around −1438A/G polymorphic region. Hypomethylation of the T102C region in patients. | Abdolmaleki et al., 2011 |
| 5-HT3AR | Peripheral blood | Bisulfite pyrosequencing | Methylation of two CpGs mediated the effects of childhood maltreatment on the severity of the disorder in adulthood. | Perroud et al., 2016 |
| 5-HTT | Postmortem frontal lobe | MSP | Tendency of a hypermethylation of 5HTT in antipsychotic-free BD patients. | Abdolmaleki et al., 2014 |
| BDNF | Peripheral blood mononuclear cells | Bisulfite sequencing | Hypermethylation of BDNF in BD II compared to BD I. Hypermethylation in depressed patients compared with manic/mixed patients. Lower methylation levels in patients on lithium and valproate. | Dell’Osso et al., 2014 |
| BDNF | Peripheral blood mononuclear cells | MSP | BDNF exon I methylation was increased in MDD subjects compared to BD patients and controls. Increased methylation associated with antidepressant use. | Carlbert et al., 2014 |
| BDNF | Venous blood | MassARRAY platform | Different methylation degree between BD and controls for 11 of 36 CpG units. | Strauss et al., 2013 |
| BDNF | Peripheral blood mononuclear cells | MSP | Increased BDNF methylation in BD II (but not type I). Negative correlation between methylation and gene expression. | D’Addario et al., 2012 |
| Chromosome X | Buccal mucosa and peripheral leukocytes | Restriction enzymes analysis | Discordant twins for BD are more discordant for the methylation of chromosome X, especially compared to concordant twins for BD. | Rosa et al., 2008 |
| DTNBP1 | Saliva | Bisulfite sequencing | Higher DTNBP1 promoter methylation in BD patients with psychotic depression compared to other BD patients. | Abdolmaleky et al., 2015 |
| FAM63B | Whole peripheral blood | iPLEX assay | Hypomethylation of two CpG sites in the exon 9 of the FAM63B gene. | Starnawska et al., 2016 |
| FKBP5 | Peripheral blood mononuclear cells | Bisulfite pyrosequencing | Increased FKBP5 methylation. | Fries et al., 2014 |
| GAD1 regulatory network genes | Post-mortem hippocampus | Bisulfite sequencing | Diagnosis- and circuit-specific methylation changes at a subset of GAD1 regulatory network genes. | Ruzicka et al., 2015 |
| Genome-wide methylation | Post-mortem brain region BA9 | MeDIP-seq | 16,599 differentially methylated regions in BD compared to controls (6,836 hypermethylated and 9,763 hypomethylated). | Zhao et al., 2015 |
| Genome-wide methylation; CYP11A1 | Blood | Illumina HumanMethylation450 BeadChip and pyrosequencing | Hypomethylation at the CYP11A1 gene in manic patients. Association of this methylation with inflammatory markers. | Sabunciyan et al., 2015 |
| Genome-wide methylation | Blood | Illumina HumanMethylation450 BeadChip | Altered methylation patterns in BD patients in use of quetiapine and valproic acid after adjusting for drug-related changes on cell type composition. | Houtepen et al., 2016 |
| Genome-wide methylation | Blood | Illumina HumanMethylation450 BeadChip | Altered methylation in carriers of a haplotype linked to BD and MDD, including genes related to neurodevelopment and ion channel activity (such as FANCI). | Walker et al., 2016 |
| Genome-wide methylation | Blood | MeDIP-Seq | Thousands of differentially methylation regions located preferentially in promoters 3′-UTRs and 5′-UTRs of genes. | Li et al., 2015 |
| Genome-wide methylation | Frontal cortex and anterior cingulate | MeDIP-seq | Several differentially methylated regions in BD compared to controls. Different distributions of methylation across the genome between the two brain regions. | Xiao et al., 2014 |
| Genome-wide methylation | Cerebellum | Illumina Infinium HumanMethylation27 | CpG gene pairs were found to significantly correlate with the differential expression and methylation of PIK3R1, BTN3A3, NHLH1, and SLC16A7 genes. | Chen et al., 2014 |
| Genome-wide methylation | Peripheral blood and postmortem brain | Illumina Infinium HumanMethylation27 | Hypomethylation of the promoter region of the ST6GALNAC1 gene in affected twins with psychosis and in brains from patients. | Dempster et al., 2011 |
| Genome-wide methylation | Postmortem frontal cortex and germline | CpG-island microarrays | Specific differences in methylation between males and females. No difference between controls and patients in the methylome of germlines. | Mill et al., 2008 |
| Global methylation | Whole blood | ELISA-based assay | Lower 5hmC levels in BD patients, but no difference in global 5mC levels compared to controls. | Soeiro-de-Souza et al., 2013 |
| Global methylation | Leukocyte | Luminometric Methylation Assay | Methylation in BD patients stable on medication was significantly influenced by insulin resistance, second-generation antipsychotic use and smoking. | Burghardt, 2015 |
| Global methylation | Transformed lymphoblasts | ELISA | Decreased methylation in BD subjects and their relatives compared to controls | Huzayyin et al., 2014 |
| Global methylation | Leukocytes | Cytosine-extension assay | No diferences between patients and controls. | Bromberg et al., 2009 |
| Global methylation, COX-2, BDNF, debrin-like protein | Postmortem frontal cortex | MSP and ELISA (for global methylation) | Global hypermethylation; COX-2 hypomethylation; BDNF hypermethylation; debrin-like protein gene hypermethylation. | Rao et al., 2012 |
| HCG9 | Postmortem prefrontal cortex | Bisulfite pyrosequencing | Significant differences between patients and controls detected in CpG modifications. | Pal et al., 2016 |
| HCG9 | Postmortem brain tissues, peripheral blood cells, and sperm | Bisulfite pyrosequencing | Low HCG9 methylation in patients. | Kaminsky et al., 2012 |
| KCNQ3 | Postmortem prefrontal cortex | Bisulfite pyrosequencing | Lower methylation of the KCNQ3 exon 11 in BD. | Kaminsky et al., 2015 |
| MB-COMT | Saliva | MSP | Hypomethylation of the MB-COMT promoter. | Nohesara et al., 2011 |
| MB-COMT | Postmortem frontal lobe | MSP and bisulfite sequencing | Hypomethylation of the MB-COMT promoter. | Abdolmaleky et al., 2006 |
| RELN | Postmortem forebrain | Restriction enzymes analysis and PCR | No correlation between RELN methylation and age (which was shown in controls). | Tamura et al., 2007 |
| RGS4 | Dorsolateral prefrontal cortex | Bisulfite sequencing | No alteration in the methylation status of CpG islands of the RGS4 gene. | Ding et al., 2016 |
| SLC6A4 | Lymphoblastoic cell lines and postmortem prefrontal cortex | Bisulfite sequencing | Hypermethylation of the SLC6A4 gene. | Sugawara et al., 2011 |
| Upstream regions of SMS and PPIEL | Transformed lymphoblastoid cells | MS-RDA | Aberrant methylation in upstream regions of the SMS and PPIEL genes. | Kuratomi et al., 2008 |
5mC – 5-methylcytosine; 5hmC – 5-hydroxymethylcytosine; 5-HT3AR – serotonin receptor 3A; 5HTR1A – serotonin receptor 1A; 5HTR2A – serotonin receptor 2A; 5-HTT – serotonin transporter; BD – bipolar disorder; BDNF – brain-derived neurotrophic factor; BTN3A3 - butyrophilin, subfamily 3, member A3; COX-2 - cyclooxygenase-2; CYP11A1 - Cytochrome P450, Family 11, Subfamily A, Polypeptide 1; DTNBP1 - dystrobrevin binding protein 1; ELISA – enzyme-linked immunosorbent assay; FAM63B - family with sequence similarity 63, member B; FANCI - Fanconi anemia, complementation group I; FKBP5 – FK506-binding protein 5; GAD1 - glutamate decarboxylase 1; HCG9 - HLA Complex Group 9; HRM – high-resolution melting; KCNQ3 - potassium channel, voltage gated KQT-like subfamily Q, member 3; MB-COMT – membrane-bound catechol-O-methyltransferase; MDD – major depressive disorder; MeDIP - Methylated DNA immunoprecipitation; MSP – methylation-specific PCR; MS-RDA – methylation-sensitive representational difference analysis; NHLH1 - nescient helix loop helix 1; PIK3R1 - phosphoinositide-3-kinase, regulatory subunit 1; PPIEL - peptidylprolyl isomerase E-like; RGS4 - regulator of G protein signaling 4; RELN - reelin; SLC16A7 - solute carrier family 1 (neurotransmitter transporter, serotonin) member 7; SLC6A4 - solute carrier family 6 (neurotransmitter transporter, serotonin) member 4; SMS – spermine synthase.
3.1 BDNF
BDNF is one of the most extensively studied genes in BD, and its involvement with BD has been suggested by several studies (Wu et al., 2014). In particular, increased methylation of the BDNF promoter region (exon 1) was found in peripheral blood mononuclear cells from BD II patients, in which BDNF gene expression levels were also significantly downregulated when compared with controls (D’Addario et al., 2012; Dell’Osso et al., 2014). The authors also found higher levels of methylation at the BDNF gene in BD subjects on pharmacological treatment with mood stabilizers in combination with antidepressants (predominantly BD II) compared with those exclusively on mood-stabilizing agents (predominantly BD I). In another study, the degree of methylation differed between BD participants and controls for 11 of 36 CpG units analyzed in the promoters 3 and 5 of the BDNF gene (Strauss et al., 2013). Several of the significantly different CpGs overlapped with or were immediately adjacent to transcription factor binding sites in promoter 5, suggesting a possible role for DNA methylation in the modulation of gene expression in the BDNF gene. Interestingly, a recent study found a high correlation for BDNF methylation between post-mortem human peripheral and brain tissues, suggesting that peripheral DNA methylation of this gene can be used as a proxy for brain-specific alterations (Stenz et al., 2015). Accordingly, BDNF has been shown to be hypermethylated in post-mortem frontal cortex of patients compared to controls (Rao et al., 2012a), as well. Of note, one study found no differences in BDNF exon 1 methylation between BD patients and controls, which suggests that this marker is sample-specific and might not be generalizable to all populations (Carlberg et al., 2014). Nevertheless, DNA methylation alterations might be underlying the differences in BDNF expression that seem to play an important role in BD; thus targeting BDNF methylation might be beneficial for BD treatment.
As a limitation, it is important to note that BDNF expression is highly dependent on the promoter being modulated (Pruunsild et al., 2007), as well as on genetic polymorphisms that can interfere with its processing and function (Egan et al., 2003; Hong et al., 2011). Based on this, the study of BDNF methylation in BD will need to systematically take those features into account before we actually have an established picture of how it acts in determining clinical features in different populations. Moreover, even though most of the studies have focused on BDNF (which makes it look like the top ranked gene being modulated by methylation in BD), it is noteworthy that no genome-wide study found it as one of the top differentially methylated genes in BD, showing that the overemphasis on this gene is coming from candidate gene studies.
3.2 Serotonergic system
DNA methylation in genes related to the serotonergic system have also been consistently shown to be altered in BD. Particularly, DNA methylation in the promoter regions of the serotonin transporter and receptor genes, including the serotonin transporter (5-HTT) and the serotonin receptor 1A (5-HTR1A), were shown to be increased in leukocytes and post-mortem brain samples of BD patients, possibly explaining the decrease of their expression reported in patients (Abdolmaleky et al., 2014; Carrard et al., 2011). Interestingly, many studies have suggested that alterations in methylation levels may be stratified by specific DNA polymorphisms. For instance, the promoter region of the serotonin receptor 2A (5-HTR2A) gene was found to be hypermethylated at the −1438A/G polymorphic site, but hypomethylated at the T102C polymorphic site compared to controls (Ghadirivasfi et al., 2011). The T102C SNP is in perfect linkage disequilibrium with the −1438 SNP and has a role in gene expression (Polesskaya et al., 2006). Likewise, hypermethylation of the solute carrier family 6 (neurotransmitter transporter, serotonin) member 4 (SLC6A4) gene, which codes for the serotonin transporter, was significantly correlated with lower mRNA expression levels only in individuals with the two short alleles (S/S, known as risk genotype) of the gene (Sugawara et al., 2011). For the serotonin receptor 3A (5-HT3AR) gene, the methylation status has been shown to mediate the relationship between childhood maltreatment and clinical severity outcomes in adulthood of patients with BD and other psychiatric illnesses (Perroud et al., 2016). In summary, the findings related to serotonergic genes seem to be consistent and even correlate with important clinical parameters in patients. Along with BDNF gene, this system has been the most studied one in regards to methylation in BD, which might be attributed to the previously reported alterations in expression in patients and the known relevance of serotonergic receptors and transporters in BD. The stratification of some of the analyses by genotype adds a more complex level of interpretation to the analysis of DNA methylation, and suggests this approach to be taken into account in the study of other loci, as well.
3.3 GAD1
Methylation analyses of genes beyond the neurotrophic and serotonergic system include, for instance, the glutamate decarboxylase 1 (GAD1). Abnormal expression of this gene has been reported in schizophrenia and BD, and there are diagnosis- and circuit-specific DNA methylation changes at a subset of GAD1 regulatory network genes in the hippocampus in these disorders (Ruzicka et al., 2015). These genes have been shown to participate in chromatin regulation and cell cycle control, supporting the concept that the established GABAergic dysfunction in these disorders is related to disruption of GABAergic interneuron physiology at specific circuit locations within the human hippocampus.
3.4 HCG9
Studies in two brain tissue cohorts have reported lower DNA methylation in BD patients compared with controls at an extended HLA complex group 9 (HCG9) region (Kaminsky et al., 2012; Pal et al., 2016), which seems to depend on age and DNA sequence variation. Particularly, a hypomethylation of this gene has also been shown to occur in white blood cells and in the germline, suggesting a potential use of this alteration as a biomarker of disease (Kaminsky et al., 2012). The authors of the latter study have argued that the mildly predictive value found for this marker may be accounted for the cellular heterogeneity in the white blood cells and brain tissue DNA, which seems to be an issue in most of the DNA methylation studies. Interestingly, given that a hypomethylation of HCG9 was also found in the sperm of BD patients, it is possible to speculate that this methylation pattern is transmitted to the offspring and may account for the familial risk of BD shown by this population (Kaminsky et al., 2012).
3.5 MB-COMT
Alterations have also been reported at the gene coding for the membrane-bound catechol-O-methyltransferase (MB-COMT), which is involved in the degradation of synaptic dopamine in the brain and is one of the most investigated genes in psychiatry (Abdolmaleky et al., 2006; Nohesara et al., 2011). Particularly in the postmortem frontal lobe, a hypomethylation of its promoter in patients was also accompanied by a reduction in the transcript levels (Abdolmaleky et al., 2006), which could lead to an increase in the rate of dopamine degradation and thereby to a hypodopaminergic state in the frontal lobe. Of particular relevance, a later study by the same group also showed that the MB-COMT hypomethylation can be detected in DNA derived from saliva, suggesting it as a potential epigenetic biomarker of disease (Nohesara et al., 2011).
3.6 FKBP5
A study by Fries et al. (2015) has assessed the patterns of methylation at the FK-506 binding protein 5 (FKBP5) gene in patients with BD, unaffected siblings, and healthy controls as a means to explore mechanisms associated with the glucocorticoid receptor resistance found in patients. FKBP5 is a negative modulator of the glucocorticoid receptor, and the methylation of specific intronic regions of its gene has been shown to modulate the receptor activity by interfering with the FKBP5 ultra-short feedback loop (Fries et al., 2015; Klengel et al., 2013). Patients showed an increased methylation at introns 7 and 2 of FKBP5 gene, and negative correlations between the number of previous manic episodes and the methylation status at two CpG sites were also reported. These epigenetic alterations might be partly responsible for the glucocorticoid receptor resistance presented by patients, ultimately leading to an impaired hypothalamus-pituitary-adrenal axis activity and stress resilience.
3.7 RELN
Reelin (RELN) is an extracellular matrix protease responsible for normal lamination of the brain during embryogenesis, and it has been shown to be involved in cell signaling and synaptic plasticity in adults (Fatemi, 2011). A significant reduction of the expression of RELN has been consistently reported in postmortem brain samples from BD patients, which supports its connection to the illness (Fatemi et al., 2000; Guidotti et al., 2000; Ovadia and Shifman, 2011). Interestingly, RELN has been shown to be epigenetically regulated by DNA methylation (Guidotti et al., 2016), and the down-regulation of RELN expression in neurons from BD patients has been associated with an overexpression of DNMT1 and DNMT3a (Veldic et al., 2007), suggesting that a hypermethylation of the gene might be responsible for its decreased expression. One study has examined methylation levels of the RELN gene in postmortem forebrains from patients as a means to further explore the regulation of RELN expression in BD (Tamura et al., 2007). Even though their results found no differences in the average levels of DNA methylation among groups, patients with BD did not show a correlation between age and RELN methylation levels, which was seen in healthy individuals. These results have not been replicated yet, but suggest that an epigenetic aberration from the normal DNA methylation control of RELN may confer susceptibility for the disorder (Tamura et al., 2007).
3.8 DTNBP1
The dystrobrevin binding protein 1 (DTNBP1), also known as dysbindin, has been suggested to be involved in glutamatergic neurotransmission by influencing exocytotic glutamate release (Domschke et al., 2011). Due to the role of glutamate in BD and psychosis, several studies have been assessing the potential role of genetic variations in the DTNBP1 gene in these disorders (Corvin et al., 2008; Joo et al., 2007; Yun et al., 2008). Accordingly, a hypermethylation of CpG sites upstream of this gene has been found in the brain of female BD patients (Mill et al., 2008), whereas a hypomethylation of its promoter has been shown in patients, especially in those who were under drug treatment (Abdolmaleky et al., 2015). Of note, methylation status was significantly lower in non-psychotic patients compared to psychotic BD or schizophrenic patients, and a correlation was found between the extent of antipsychotic drug use and DTNBP1 expression in the brains of BD patients (Abdolmaleky et al., 2015). This suggests that the overall reduction in methylation seen in BD patients might be due to drug treatment effects. The same study also found a hypermethylation of DTNBP1 promoter in BD with psychotic depression compared to other BD patients, suggesting it as a marker of psychotic phenotype. Interestingly, even though it hasn’t been assessed in the saliva of BD patients, DTNBP1 methylation might also represent a biomarker of illness (in this case, of psychosis, since a hypermethylation was reported in saliva from schizophrenic patients) (Abdolmaleky et al., 2015).
3.9 KCNQ3
Previous studies have hypothesized that the dysregulated neuronal hyperexcitability in BD might be accounted for altered function of specific voltage-gated potassium channels called M-channels. These channels are heterodimers comprised of the Kv7.2 and Kv7.3 proteins, which are encoded by the potassium channel, voltage gated KQT-like subfamily Q, members 2 and 3 (KCNQ2 and KCNQ3) genes, respectively (Kaminsky et al., 2015). In a series of experiments Kaminsky et al. (2015) demonstrated that postmortem prefrontal cortex of patients with BD present significantly lower methylation of CpGs located in the exon 11 of KCNQ3 gene, which was significantly correlated with mRNA levels. Interestingly, the authors were also able to show that mood stabilizers increase the methylation of this region in rats. By doing so, it was hypothesized that these drugs might restore the ion channel dysfunction and channelopathy that is seen in BD (Kaminsky et al., 2015).
3.10 PPIEL
After a comprehensive scan of DNA methylation in lymphoblastoid cells, the putatitve promoter region of the peptidylprolyl isomerase E-like (PPIEL) was shown to be globally hypomethylated in a patient with BD compared to his unaffected co-twin (Kuratomi et al., 2008). The methylation status was significantly correlated with PPIEL expression, and the hypomethylation of PPIEL was also replicated in an independent sample of patients with BD (Kuratomi et al., 2008). Even though the role of the PPIEL protein is still unknown, it is believed that it might be involved in specific neuronal function due to its peptidyl-prolyl cis-trans isomerase (PPI) domain. Future studies are now required to validate this finding and to identify the relevance of this alteration in BD.
3.11 FAM63B
The family with sequence similarity 63, member B (FAM63B) gene was found to be one of the top ranked loci identified in an epigenome-wide association study (EWAS) with schizophrenic patients (Aberg et al., 2014). Because of the high degree of comorbidity found between schizophrenia and BD, the methylation status of FAM63B has been recently assessed in whole blood from patients with BD (Starnawska et al., 2016), and the results suggest that patients present a lower methylation at this gene compared to controls. As was the case for several of the findings reviewed here, variations in cell composition might be contributing to the variation in FAM63B methylation. In addition, even though the biological function of this gene is still not know, it is suggested to play a role in circadian clock (Starnawska et al., 2016), which is interesting considering its known link with psychiatric disorders (Karatsoreos, 2014).
3.12 RGS4
A recent study has looked for the effects of genotype and methylation status of the regulator of G protein signaling 4 (RGS4) gene in postmortem dorsolateral prefrontal cortex from patients with BD, schizophrenia, and healthy controls (Ding et al., 2016), following up on previous evidence of reduced expression of RGS4 in the brain of schizophrenic patients (Mirnics et al., 2001). Of note, a trend toward lower RGS4-2 mRNA expression has been reported in the brain of BD patients (Ding and Hegde, 2009), as well as a decrease in the mRNA ratio of RGS4-2 to RGS4-3 and RGS4-2 to RGS4-5 compared to controls (which are different splice variants of the gene). Contrary to the initial hypothesis, no evidence of hypermethylation of RGS4 was found in patients compared to controls, suggesting that DNA methylation may not play a role in the decrease of RGS4 expression in the brain of BD patients.
3.13 Global methylation
Global methylation patterns have also been assessed in bipolar patients, and were shown to be significantly influenced by insulin resistance, second-generation antipsychotics use, and smoking (Burghardt et al., 2015). In an earlier study, no differences between bipolar patients and controls were found in leukocytes (Bromberg et al., 2009). However, a marked global hypermethylation has been reported in postmortem frontal cortex from affected individuals compared to controls (Rao et al., 2012a), while two other studies have reported a decrease in global methylation in transformed lymphoblasts (Huzayyin et al., 2014) and in whole blood from BD subjects (Soeiro-de-Souza et al., 2013). These discrepancies might be accounted for not only by the different populations studied, but also by the different methods used, which have been shown to vary significantly (Lisanti et al., 2013): three studies have assessed global methylation by ELISA (Huzayyin et al., 2014; Rao et al., 2012b; Soeiro-de-Souza et al., 2013), while others have used luminometric methylation assay (Burghardt et al., 2015) or cytokine-extension assay (Bromberg et al., 2009). Of note, the overall clinical relevance of alterations in global methylation still need to be determined.
3.14 Genome-wide methylation studies
Genome-wide methylation differences have also been analyzed in blood (Dempster et al., 2011; Houtepen et al., 2016; Li et al., 2015; Sabunciyan et al., 2015; Walker et al., 2016) and in different post-mortem brain regions, such as the BA9 (Zhao et al., 2015), frontal cortex (Mill et al., 2008; Xiao et al., 2014), anterior cingulate (Xiao et al., 2014), and cerebellum (Chen et al., 2014) from BD patients and controls.
Six out of the 9 published genome-wide methylation studies in BD have been performed in blood (with one paper analyzing both blood and postmortem brain (Dempster et al., 2011)), and they suggest that several differentially methylated regions (DMRs) can be found in the periphery of BD patients. Moreover, four studies have analyzed genome-wide methylation changes in postmortem brain tissues, and they suggest that differences between groups (for example, cases and controls) strongly depend on the brain region (Xiao et al., 2014). As in the case in the blood analysis, the altered cellular heterogeneity between distinct regions might be playing a role in the methylome differences. Interestingly, in the frontal cortex and anterior cingulate, only a few differentially methylated regions overlapped with promoters, whereas a greater proportion occurred in introns and intergenic regions (Xiao et al., 2014). A similar pattern was found in another study, where a high percent of DMRs was found in introns and repeat elements (Zhao et al., 2015) in the brain region BA9. In this particular study, hypermethylated DMRs significantly overlapped with long intergenic non-coding RNAs (lincRNAs), while hypomethylated DMRs significantly overlapped with introns, as well as with short and long interspersed nuclear elements. Interestingly, many of the intronic DMRs overlapped with microRNAs, suggesting a role for them in DMR-induced gene expression changes in the BA9 region of patients of BD (Zhao et al., 2015). By using different datasets, a highly significant correlation between methylation and gene expression has been reported in the cerebellum, as well (Chen et al., 2014). Collectively, these studies suggest that alterations in DNA methylation may play an important role in the dysregulation of gene expression in BD (either directly or indirectly by microRNAs or lincRNAS), and a few studies have actually suggested their potential to be used as biomarkers to improve diagnosis and to better understand the pathophysiology of this illness.
So far the inconsistency between studies and lack of replicated findings warrant more investigation of the topic. Specifically, the inconsistencies are coming not only from the different tissues analyzed (blood, prefrontal cortex, anterior cingulate, cerebellum, and sperm), but also due to methodological variations in the studies: three studies have used methylated DNA immunoprecipitation (MeDIP-seq) followed by next generation sequencing (Li et al., 2015; Xiao et al., 2014; Zhao et al., 2015), two studies have used the Illumina Infinium HumanMethylation27 beadchip (Chen et al., 2014; Dempster et al., 2011), three studies have used the Illumina Infinium HumanMethylation450 beadchip (Houtepen et al., 2016; Sabunciyan et al., 2015; Walker et al., 2016), and one used CpG-island microarrays (Mill et al., 2008) for the assessment of methylation changes. Of note, even though MeDIP-seq and microarray-based analyses have been shown to present a good positive correlation, MeDIP-seq allows a wider interrogation of methylation regions of the genome, including thousands of non-RefSeq genes and repetitive elements that are not assessed in microarrays (Clark et al., 2012). Moreover, different studies use distinct software algorithms in the data analysis, even when analyzing data from a similar method. That includes, for instance, different preprocessing methods (quality control and data filtering), distinct stringency levels for the identification of DMRs (from false discovery rate < 0.1 following multiple testing correction (Walker et al., 2016) to p < 0.05 without adjustment (Sabunciyan et al., 2015) for the analysis of the HumanMethylation450 chip, for example), and controlling or not for differences in cellular composition.
4. The role of DNMTs in BD
Based on several studies reporting alterations in DNA methylation patterns in BD patients, it is reasonable to assume that DNMTs might be altered and may even be taken as future targets for the treatment of this disorder. However, so far only a few studies have been performed to examine the role of these enzymes – particularly of DNMT1 – in BD (Table 2), and when taken together their results are still inconclusive.
Table 2.
Alterations in DNMT expression reported in BD
| Cell/tissue | Findings | References |
|---|---|---|
| Post-mortem prefrontal cortex (BA9) and cerebellar tissue | Increased binding of DNMT1 to GABAergic and glutamatergic promoters in the cortex but not in the cerebellum in BD and schizophrenia. Increased binding of DNMT1 positively correlated with increased binding of MBD2. | Dong et al, 2015 |
| Post-mortem prefrontal cortex | Increase in DNMT1 mRNA-positive neurons in psychotic patients (including BD). | Guidotti et al., 2013 |
| Post-mortem brain | Co-variations of DNMT1 and DNMT3B with GABRB2 expression in BD samples. | Zhao et al., 2012 |
| Peripheral blood mononuclear cells | Reduction in the mRNA expression of DNMT1 in bipolar depression, but not in euthimia. | Higuchi et al., 2011 |
| BA9 and GABAergic neurons | Increased S-adenosyl methionine and DNMT1 mRNA expression in patients with BD and schizophrenia. | Guidotti et al., 2007 |
| BA9 and GABAergic neurons | Increased DNMT1 mRNA and protein levels in patients with BD and psychosis. | Veldic et al., 2005 |
| GABAergic caudate nucleus and putamen neurons | No increase of reelin and GAD67 mRNA expression after DNMT1 overexpression in neurons from patients with BD (which occurs in cells from schizophrenic patients). | Veldic et al., 2007 |
BA9 - brodmann area 9; BD - bipolar disorder; DNMT - DNA methyltransferase; GABRB2 - gamma-aminobutyric acid (GABA) A receptor, beta 2; GAD67 - glutamic acid decarboxylase 67; MBD2 - methyl-CpG binding domain protein 2; MDD – major depressive disorder.
The methyl donor S-adenosyl methionine (SAM) was shown to be increased by about two-fold in the prefrontal cortex in schizophrenia and BD, which was also associated with an overexpression of DNMT1 mRNA in Brodmann’s area 9 GABAergic neurons (Guidotti et al., 2007). This increase of DNMT1 expression was also reported by other studies (Dong et al., 2015; Guidotti et al., 2013; Veldic et al., 2005; Veldic et al., 2007). In addition, Dong et al. (2015) has shown that higher expression of DNMT1 and TET1 enzymes was associated with an increase in the DNMT1 binding to the promoters of GAD1, RELN and BDNF, as well as with downregulation of these genes in the brains of schizophrenia and BD patients (Dong et al., 2015). These data are consistent with the hypothesis that the down-regulation of specific GABAergic and glutamatergic genes in schizophrenia and BD patients may be mediated, at least in part, by a brain region- and promoter-specific DNMT1 action.
A role for DNMT1 in the treatment of BD has been further evidenced by the inhibitory effect of at least some antidepressants in DNMT1 enzymatic activity (Gassen et al., 2015a; Zimmermann et al., 2012). Of note, this inhibitory action appears to depend on FKBP51, a regulator of the glucocorticoid receptor (Wochnik et al., 2005) which has been implicated in antidepressant response (Binder et al., 2004; Gassen et al., 2015b; Gassen et al., 2014).
Of particular interest, the expression of DNMT1 seems to be mood state-dependent, since the levels of DNMT1 mRNA have been shown to be decreased in the depressive but not in the euthymic state of MDD and BD (Higuchi et al., 2011). Altogether, these few studies support the hypothesis that DNMTs, particularly DNMT1, may play a role in BD, possibly being taken as targets for future novel treatments. Indeed, as reviewed in the next section, currently available medications for the treatment of these disorders are able to alter DNA methylation, and it is likely that they accomplish this by directly or indirectly modulating DNMT expression or activity.
5. The effects of mood stabilizers on DNA methylation
Pharmacological treatment of BD typically includes mood stabilizers (e.g. lithium, valproate, lamotrigine, and carbamazepine), antidepressants and atypical antipsychotics (Jann, 2014). In this section we will focus on mood stabilizers, which are the main class of medications used in BD. Although their mechanisms of action have been studied by different groups, most of the information currently available focuses on the acute and subchronic effects of these drugs. In other words, the current model for evaluating a drug’s effect focuses on synaptic signaling, receptor/transporter regulation, intracellular signaling and posttranslational modification, while its effects on gene expression and neuroplasticity appear less investigated. While the acute effects are important to decipher the initial events, it is also important to evaluate the long-term molecular events that parallel symptoms relieve. Long-term changes in DNA methylation are attractive read-outs for potential long-term effects of medications’ effects. In this section we will attempt to structure the current understanding of DNA methylation and the use of mood stabilizers in BD (Table 3).
Table 3.
Effects of mood stabilizers on DNA methylation
| Drug | Gene | Disease/model | Method | Main effects | Reference |
|---|---|---|---|---|---|
| Lithium | 5mC and 5hmC | Rat primary cortical neurons | Immunocytochemis try | Prevention of rotenone-induced increase in DNA methylation. | Scola et al., 2014 |
| Lithium | BDNF | PBMCs from BD patients | MSP | Reduction of BDNF methylation. | D’Addario et al., 2012 |
| Lithium | BDNF | PBMCs from BD patients | MSP | Reduction of BDNF methylation. | Dell’Osso et al., 2014 |
| Lithium | Bdnf | Rat hippocampal neurons | MSP | Hypomethylation of Bdnf exon IV promoter. | Dwivedi and Zhang, 2014 |
| Lithium | Genome-wide methylation | SK-N-SH cells | Infinium HumanMethylation 27 | Hypermethylation of 345 genes and hypomethylation of 138 genes. | Asai et al., 2013 |
| Lithium | Global methylation | Lymphoblasts from BD patients | ELISA | No effect on global methylation in patients (it remained decreased compared to controls after treatment). Rescue of methylation levels in relatives. | Huzayyin et al., 2014 |
| Lithium | Igf2, Igf2r, H19 | Mouse ESCs | Bisulfite sequencing | Hypomethylation of imprinted control regions via Dnmr3a2. | Popkie et al., 2010 |
| Valproate | BDNF | PBMCs from BD patients | MSP | Reduction of BDNF methylation. | D’Addario et al., 2012 |
| Valproate | BDNF | PBMCs from BD patients | MSP | Reduction of BDNF methylation. | Dell’Osso et al., 2014 |
| Valproate | Genome-wide methylation | SK-N-SH cells | Infinium HumanMethylation 27 | Hypermethylation of 64 genes and hypomethylation of 36 genes. | Asai et al., 2013 |
| Valproate | Global methylation | Primary rat astreocyte | LUMA | Global hypomethylation. | Perisic et al., 2010 |
| Valproate | Glt-1 | Primary rat cortical astrocyte | Bisulfite sequencing | Demethylation of Glt-1 promoter. | Perisic et al., 2010 |
| Valproate | p21 | Mouse hippocampal cells | PCR and restriction enzymes analysis | Demethylation of the distal region of p21. | Aizawa and Yamamuro, 2015 |
| Valproate | RELN | Mouse striatum (frontal cortex) | 5-methyl CpG ChIP | Decrease of RELN methylation after treatment to increase DNMT activity. | Dong et al., 2008 |
| Carbamazepine | Genome-wide methylation | SK-N-SH cells | Infinium HumanMethylation 27 | Hypermethylation of 64 genes and hypomethylation of 14 genes. | Asai et al., 2013 |
| Lamotrigine | Global methylation | Primary rat cortical astrocyte | LUMA | No changes in DNA methylation. | Perisic et al., |
BD – bipolar disorder; Igf2 - insulin-like growth factor 2; Igf2r - insulin-like growth factor 2 receptor; HTR2A – serotonin receptor 2A; BDNF – brain-derived neurotrophic factor; 5mC – 5-methylcytosine; 5-hmC – 5-hydroxymethylcytosine; SLC6A4 - solute carrier family 6 (neurotransmitter transporter, serotonin) member 4; RELN - reelin; Glt-1 - glutamate type I transporter; LUMA - luminometric methylation analysis; MSP – methylation-specific PCR; PBMC – peripheral blood mononuclear cells; ChIP – chromatin immunoprecipitation.
Lithium is the most commonly prescribed mood stabilizer and the foundation of treatment of BD (Jann, 2014), yet the specific mechanisms by which it exerts its effects are incompletely understood. It is known that lithium increases inhibitory (GABA) neurotransmission and decreases excitatory (dopamine and glutamate) neurotransmission (Malhi et al., 2013), but the long-term effects of lithium on gene expression are only recently being evaluated. It is believed that lithium plays a role in inositol and glycogen synthase kinase-3β (GSK-3β) signaling leading to the downregulation of Dnmt3a2, thereby inducing a reduction in DNA methylation and increasing gene expression at specific loci (Lee et al., 2015). Indeed, lithium treatment resulted in hypomethylation of Igf2 (which encodes a protein involved in cell proliferation, differentiation and survival), Igf2r, and H19 (regulation of cell proliferation) in mouse embryonic stem cells (Popkie et al., 2010), as well as of the BDNF gene in peripheral blood mononuclear cells (PBMCs) from patients (D’Addario et al., 2012; Dell’Osso et al., 2014) and in rat hippocampal neurons (Dwivedi and Zhang, 2014). In addition, global 5mC levels were reduced by lithium in transformed lymphoblasts from relatives from BD patients (Huzayyin et al., 2014). Interestingly, lithium did not show any effect on global methylation in BD patients, which remained decreased when compared to controls even after treatment (Huzayyin et al., 2014).
Valproate is an anticonvulsant typically prescribed for epilepsy that has been shown to be effective in controlling impulsive behavior, as is observed in BD patients. As seen with lithium, the exact mechanisms of action of valproate are still unknown but they have been linked to the blockade of voltage-dependent sodium channels, as well as potentiation of inhibitory GABAergic transmission (Lee et al., 2015; Phiel et al., 2001). In addition, valproate has been shown to be a potent inhibitor of HDACs (Phiel et al., 2001), which are believed to have a direct influence on DNA methylation (Dobosy and Selker, 2001). In line, Milutinovic et al. found that valproate induced histone acetylation and activated DNA demethylation in the same gene systems in HEK293 cells, suggesting that valproate might trigger demethylation of genes through histone acetylation (Milutinovic et al., 2007). Indeed, in human PBMCs, treatment with valproate led to a significant reduction of ~24% in BDNF promoter methylation (D’Addario et al., 2012; Dell’Osso et al., 2014), whereas it was also shown to decrease the methylation of p21 (Aizawa and Yamamuro, 2015), RELN (Dong et al., 2008), and glutamate type I transporter (GLT-1) genes (Perisic et al., 2010), as well as induce a significantly altered methylation signature in BD patients (Houtepen et al., 2016).
Regarding other mood stabilizers, carbamazapine has been shown to induce hypermethylation of 64 genes and hypomethylation of 14 genes in a neuroblastoma cell line (Asai et al., 2013), while lamotrigine induced no changes in global DNA methylation in another study (Perisic et al., 2010). Altogether, these findings suggest that the effects of mood stabilizers on DNA methylation are drug-specific and still warrant substantial analysis in different samples, even though common effects on the methylation of a specific group of genes have been reported (Asai et al., 2013). Similar effects have also been suggested to take place in the mechanism of action of antidepressants (Menke and Binder, 2014) and antipsychotics (Guidotti and Grayson, 2014; Houtepen et al., 2016), which further corroborates the hypothesis that the reversal of symptoms induced by BD treatment involves the modulation of DNA methylation and thereby alteration of gene expression.
6. Perspectives
In summary, the findings reviewed in this paper indicate the occurrence of alterations in DNA methylation in patients with BD compared to healthy controls. Some studies take the analysis further and also suggest a role for this mechanism in determining specific clinical features, response to treatment, and prognosis. Some genes, such as BDNF and those encoding serotonin transporters, have been extensively analyzed in BD, which is not surprising considering that previous studies had already shown alterations in their expression. In fact, most of the studies have analyzed DNA methylation as a potential modulator of gene expression, which is true for most cases but might not be the case in others. The nuances and possibilities of DNA methylation, as discussed earlier, are still beginning to be understood.
6.1 The clinical utility of DNA methylation
The enthusiasm about the potential benefits of more detailed information on methylation profiles is leading to the initiation of more extended clinical trials with BD patients. Moreover, as DNA methylation appears to be determined by genetic polymorphisms as well, this information should also be taken into account in studies assessing methylation patterns. In other words, it is reasonable to assume that the combination of genotype and DNA methylation determines the ability to respond to a particular medication, as well as other clinical features. By understanding the effects of such alterations in patients, we might be able to identify distinct groups of patients according to methylation markers and appropriately treat them in a more tailored and targeted way. Of note, from the evidence gathered in this review, no methylation marker identified so far seems to have enough specificity and sensitivity to be taken as a clinically useful marker of diagnosis or treatment prediction of BD at this point, and no such approach is currently being used in clinical routines. However, this review strongly suggests that DNA methylation plays an important role in the pathophysiology of BD, and it is likely that specific markers will be clinically useful in the near future. For that to occur, however, clinical studies need to start adding DNA methylation as one of their outcomes, and longitudinal studies are warranted. This is particularly relevant when considering the potential role of DNA methylation in the risk of BD, which is supported by several theoretical frameworks but not by strong empirical evidence.
6.2 Peripheral methylation as a proxy of the brain
Of note, most of the studies have analyzed DNA methylation in peripheral tissues of patients with BD. There are in fact some reports showing concordances in methylation patterns between blood and brain, suggesting the utility of peripheral blood in human epigenetic studies (Davies et al., 2012). Specifically, Horvarth et al. was able to find a correlation of around r = 0.9 across the 2 tissues (Horvath et al., 2012), and DNA methylation changes in ten genes identified in the brain of schizophrenic and BD patients have also been confirmed in peripheral blood samples (Xiao et al., 2014). In addition, methylation in peripheral blood has been correlated with brain volume in healthy individuals and schizophrenia patients (Liu et al., 2015), and it has also been correlated with symptoms of major psychiatric disorders, including schizophrenia, depression and BD (Liu et al., 2014; Sabunciyan et al., 2015).
However, the studies on this topic are still controversial, and some findings indicate that most DNA methylation markers in peripheral blood do not reliably predict brain DNA methylation status (Walton et al., 2016). Only a specific subset of peripheral data was shown to proxy methylation status of brain tissue, which suggests that studies on peripheral methylation should ideally restrict their analysis to these markers (Walton et al., 2016). In addition to peripheral blood, similar patterns of methylation have been observed in saliva and post-mortem tissue in two studies (Ghadirivasfi et al., 2011; Nohesara et al., 2011). Interestingly, saliva and blood methylation have been shown to present a positive correlation overall, although DNA methylation in saliva seems to be more similar to patterns of specific brain regions than methylation in blood (Smith et al., 2015). Taking these limitations into account and being aware that results from peripheral tissues should be interpreted with caution, we believe that the assessment of peripheral methylation can be valuable for psychiatric epigenetics, particularly for BD, mostly for allowing its use in longitudinal studies in living subjects and for potentially providing biomarkers of illness.
6.3 Candidate gene vs. genome-wide methylation studies
So far, the majority of published studies have chosen to analyze the methylation of specific genes rather than assess methylation in a genome-wide fashion. Today, psychiatric consortiums with significantly large sample sizes are being formed, allowing for genome-wide studies with later validation and replication for the identification of biologically significant DNA methylation changes between patients and controls. Different BD populations have been analyzed by these techniques, but the results between them are still not consistent or concordant enough. This might be due to the lack of control for genotype, differences in the tissue being analyzed, sample heterogeneity, as well as still insufficient sample sizes. The variability between studies has also been pointed out by a recent systematic review (Teroganova et al., 2016) of findings in peripheral tissues (blood and saliva), especially in regards to the different methods used for the assessment of DNA methylation, DNA extraction, and lack of control for potential effects of clinical variables (such as diet, exercise, smoking, and ethnicity).
6.4 Non-canonical roles of DNA methylation
Several studies have been reshaping the traditional view of DNA methylation as simply a repressor of gene expression and are unraveling novel functions and features of this epigenetic alteration (Kulis et al., 2013; Reddington et al., 2013), including the regulation of alternative intragenic promoters, regulation of intragenic non-coding RNAs and transposable elements, and RNA processing (reviewed by Kulis et al., 2013). By doing so, the role of DNA methylation in BD might eventually become clearer and ultimately help tackle specific pathways for its treatment. For instance, the recently suggested effects of DNA methylation on alternative splicing mechanisms likely contribute to the splicing alterations reported in BD patients, where a dysfunctional gene splicing is being considered as a pathophysiological hallmark of the disorder (Glatt et al., 2009; Glatt et al., 2011). In addition, correlations of DNA methylation markers with gene expression and transcripts profiles, as well as with dynamic cell functions, will lead to more groundbreaking and certainly more relevant results to be followed-up. Of note, even though analyses in cancers have revealed that differences in DNA methylation only result in gene expression changes in a low percentage of genes (Simmer et al., 2012), this does not seem to be the consensus in neuronal cells, where strong correlations are seen (especially with methylation loci around the transcription start site and gene bodies) (Kozlenkov et al., 2016). These discrepancies suggest that the extent to which DNA methylation modulates gene expression is also dependent on the tissue being analyzed, in addition to other still unknown factors that might link the association between these two events.
Of particular relevance, DNA methylation is now beginning to be seen as a dynamic process, as well. A growing body of evidence suggests that it can be modified by neuronal activity and thus play a key role in dynamic processes, such as learning and memory formation (Lister and Mukamel, 2015). This is especially important when considering the highly dynamic and phasic nature of the course of BD, and suggests that some methylation markers might parallel changes in mood, as well.
6.5 DNA methylation in the risk of BD
Even though the hypothesis that methylation has functional effects on the risk to develop BD is supported by preliminary findings in patients and preclinical models, only recently have researchers started to actually measure the so-called ‘non genetic intergenerational transmission’ of DNA methylation patterns over generations (Weber-Stadlbauer et al., 2016; Yehuda et al., 2015), which will in fact demonstrate whether the risk shown by populations at familial risk of BD can be modulated by such alterations. Specifically, part of the ‘heritability gap’ might be certainly accounted for by environmental events, but recent preclinical studies with the ‘cross-fostering’ approach (that eliminates the ‘environment factor’) are also suggesting that stress-related DNA methylation markers can be transmitted by non-genetic intergenerational transmission, as well (Klengel et al., 2016a, 2016b). In this sense, it is possible that such markers may account for part of the familial risk of BD proposed by family and twin studies (when in combination with multiple genetic markers of small effect and the effects of environmental triggers), although this still needs to be empirically assessed. Of note, there are no published studies (neither cross-sectional nor longitudinal) on DNA methylation in populations at high risk of BD, including offspring and siblings, even though preliminary findings suggest that they might share altered DNA methylation patterns compared to healthy controls (Fries et al., 2016).
6.6 Targeting DNA methylation in BD
The use of methylation inhibitors is promising in regards to the possibility of permanently reversing several alterations in DNA methylation that have been consistently shown in patients. In fact, preclinical studies in animal models of relevance for BD show promising results (Sales et al., 2011; Sales and Joca, 2016), and studies have shown that targeting DNMT is a crucial part of the mechanism of action of antidepressants and the resolution of depressive symptoms (Gassen et al., 2015a; Zimmermann et al., 2012). In this sense, several DNMT inhibitors have been identified and are being tested in different medical conditions, especially cancers (Erdmann et al., 2015). Of note, it is known that DNMTs are targeted to specific sequences and that adaptor proteins are required for it (Szyf, 2011), suggesting that targeting those proteins might also be relevant in a methylation-modifying therapy.
At present, however, the lack of specificity of these inhibitors is still a major issue. Because methylation is known to determine and maintain the different cell phenotypes within an organism, the potential carcinogenic property of these drugs needs to be considered. In fact, a screening for methylation alterations has actually been proposed for the assessment of carcinogenicity of drugs in addition to the tradition mutagenesis screening (Szyf, 2011). Moreover, because methylation has been shown to play a role in learning and long-term memory storage, one could also argue that a generalized demethylation could affect those individual characteristics, as well. In summary, the rationale for using such methylation inhibitors, albeit very promising in light of the full range of alterations reviewed in this article, may still depend on our ability to target methylation at specific loci rather than simply inhibiting non-specific enzymes.
7. Conclusions
In conclusion, the literature reviewed here strongly indicates an important role for DNA methylation in the pathophysiology and treatment of BD. However, its potential in the risk of developing the disorder still needs to be investigated. Of particular interest, targeting specific alterations in DNA methylation represents exciting new treatment possibilities for BD, and the ‘plastic’ characteristic of DNA methylation accounts for a promising possibility of restoring environment-induced modifications in patients.
Highlights.
DNA methylation is a stable epigenetic modification that can alter gene expression
DNA methylation might partly account for BD heritability
Several genome-wide and gene-specific methylation patterns have been found in BD
Bipolar patients have been shown to present alterations in DNMTs, particularly DNMT1
Mood stabilizers have been shown to alter DNA methylation in several genes
Acknowledgments
The Translational Psychiatry Program (USA) is funded by the Department of Psychiatry and Behavioral Sciences, McGovern Medical School, The University of Texas Health Science Center at Houston (UTHealth).
Laboratory of Neurosciences (Brazil) is one of the centers of the National Institute for Molecular Medicine (INCT-MM) and one of the members of the Center of Excellence in Applied Neurosciences of Santa Catarina (NENASC). Its research is supported by grants from CNPq (JQ), FAPESC (JQ); Instituto Cérebro e Mente (JQ) and UNESC (JQ). JQ is a 1A CNPq Research Fellow.
This study was also supported in part by grants from the Pat Rutherford, Jr. Endowed Chair in Psychiatry (JCS), John S. Dunn Foundation from United States (JCS), and NIMH (R01MH085667).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdolmaleky HM, Cheng KH, Faraone SV, Wilcox M, Glatt SJ, Gao F, Smith CL, Shafa R, Aeali B, Carnevale J, Pan H, Papageorgis P, Ponte JF, Sivaraman V, Tsuang MT, Thiagalingam S. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Human molecular genetics. 2006;15:3132–3145. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdolmaleky HM, Nohesara S, Ghadirivasfi M, Lambert AW, Ahmadkhaniha H, Ozturk S, Wong CK, Shafa R, Mostafavi A, Thiagalingam S. DNA hypermethylation of serotonin transporter gene promoter in drug naive patients with schizophrenia. Schizophrenia research. 2014;152:373–380. doi: 10.1016/j.schres.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdolmaleky HM, Pajouhanfar S, Faghankhani M, Joghataei MT, Mostafavi A, Thiagalingam S. Antipsychotic drugs attenuate aberrant DNA methylation of DTNBP1 (dysbindin) promoter in saliva and post-mortem brain of patients with schizophrenia and Psychotic bipolar disorder. American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2015;168:687–696. doi: 10.1002/ajmg.b.32361. [DOI] [PubMed] [Google Scholar]
- Aberg KA, McClay JL, Nerella S, Clark S, Kumar G, Chen W, Khachane AN, Xie L, Hudson A, Gao G, Harada A, Hultman CM, Sullivan PF, Magnusson PK, van den Oord EJ. Methylome-wide association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA psychiatry. 2014;71:255–264. doi: 10.1001/jamapsychiatry.2013.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa S, Yamamuro Y. Valproate administration to mice increases hippocampal p21 expression by altering genomic DNA methylation. Neuroreport. 2015;26:915–920. doi: 10.1097/WNR.0000000000000448. [DOI] [PubMed] [Google Scholar]
- Anand A, Koller DL, Lawson WB, Gershon ES, Nurnberger JI. Genetic and childhood trauma interaction effect on age of onset in bipolar disorder: An exploratory analysis. Journal of affective disorders. 2015;179:1–5. doi: 10.1016/j.jad.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Bundo M, Sugawara H, Sunaga F, Ueda J, Tanaka G, Ishigooka J, Kasai K, Kato T, Iwamoto K. Effect of mood stabilizers on DNA methylation in human neuroblastoma cells. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum (CINP) 2013;16:2285–2294. doi: 10.1017/S1461145713000710. [DOI] [PubMed] [Google Scholar]
- Binder EB, Salyakina D, Lichtner P, Wochnik GM, Ising M, Putz B, Papiol S, Seaman S, Lucae S, Kohli MA, Nickel T, Kunzel HE, Fuchs B, Majer M, Pfennig A, Kern N, Brunner J, Modell S, Baghai T, Deiml T, Zill P, Bondy B, Rupprecht R, Messer T, Kohnlein O, Dabitz H, Bruckl T, Muller N, Pfister H, Lieb R, Mueller JC, Lohmussaar E, Strom TM, Bettecken T, Meitinger T, Uhr M, Rein T, Holsboer F, Muller-Myhsok B. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nature genetics. 2004;36:1319–1325. doi: 10.1038/ng1479. [DOI] [PubMed] [Google Scholar]
- Bockmuhl Y, Patchev AV, Madejska A, Hoffmann A, Sousa JC, Sousa N, Holsboer F, Almeida OF, Spengler D. Methylation at the CpG island shore region upregulates Nr3c1 promoter activity after early-life stress. Epigenetics. 2015;10:247–257. doi: 10.1080/15592294.2015.1017199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiling A, Lyko F. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenetics & chromatin. 2015;8:24. doi: 10.1186/s13072-015-0016-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brietzke E, Kauer Sant’anna M, Jackowski A, Grassi-Oliveira R, Bucker J, Zugman A, Mansur RB, Bressan RA. Impact of childhood stress on psychopathology. Revista brasileira de psiquiatria (Sao Paulo, Brazil : 1999) 2012;34:480–488. doi: 10.1016/j.rbp.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Bromberg A, Bersudsky Y, Levine J, Agam G. Global leukocyte DNA methylation is not altered in euthymic bipolar patients. Journal of affective disorders. 2009;118:234–239. doi: 10.1016/j.jad.2009.01.031. [DOI] [PubMed] [Google Scholar]
- Burghardt KJ, Goodrich JM, Dolinoy DC, Ellingrod VL. DNA methylation, insulin resistance and second-generation antipsychotics in bipolar disorder. Epigenomics. 2015;7:343–352. doi: 10.2217/epi.15.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlberg L, Scheibelreiter J, Hassler MR, Schloegelhofer M, Schmoeger M, Ludwig B, Kasper S, Aschauer H, Egger G, Schosser A. Brain-derived neurotrophic factor (BDNF)-epigenetic regulation in unipolar and bipolar affective disorder. Journal of affective disorders. 2014;168:399–406. doi: 10.1016/j.jad.2014.07.022. [DOI] [PubMed] [Google Scholar]
- Carrard A, Salzmann A, Malafosse A, Karege F. Increased DNA methylation status of the serotonin receptor 5HTR1A gene promoter in schizophrenia and bipolar disorder. Journal of affective disorders. 2011;132:450–453. doi: 10.1016/j.jad.2011.03.018. [DOI] [PubMed] [Google Scholar]
- Carvalho AF, Kohler CA, Fernandes BS, Quevedo J, Miskowiak KW, Brunoni AR, Machado-Vieira R, Maes M, Vieta E, Berk M. Bias in emerging biomarkers for bipolar disorder. Psychological medicine. 2016:1–11. doi: 10.1017/S0033291716000957. [DOI] [PubMed] [Google Scholar]
- Chen C, Zhang C, Cheng L, Reilly JL, Bishop JR, Sweeney JA, Chen HY, Gershon ES, Liu C. Correlation between DNA methylation and gene expression in the brains of patients with bipolar disorder and schizophrenia. Bipolar disorders. 2014;16:790–799. doi: 10.1111/bdi.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark C, Palta P, Joyce CJ, Scott C, Grundberg E, Deloukas P, Palotie A, Coffey AJ. A comparison of the whole genome approach of MeDIP-seq to the targeted approach of the Infinium HumanMethylation450 BeadChip((R)) for methylome profiling. PloS one. 2012;7:e50233. doi: 10.1371/journal.pone.0050233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortazar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, Steinacher R, Jiricny J, Bird A, Schar P. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- Corvin A, Donohoe G, Nangle JM, Schwaiger S, Morris D, Gill M. A dysbindin risk haplotype associated with less severe manic-type symptoms in psychosis. Neuroscience letters. 2008;431:146–149. doi: 10.1016/j.neulet.2007.11.031. [DOI] [PubMed] [Google Scholar]
- D’Addario C, Dell’Osso B, Palazzo MC, Benatti B, Lietti L, Cattaneo E, Galimberti D, Fenoglio C, Cortini F, Scarpini E, Arosio B, Di Francesco A, Di Benedetto M, Romualdi P, Candeletti S, Mari D, Bergamaschini L, Bresolin N, Maccarrone M, Altamura AC. Selective DNA methylation of BDNF promoter in bipolar disorder: differences among patients with BDI and BDII. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2012;37:1647–1655. doi: 10.1038/npp.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, Coarfa C, Harris RA, Milosavljevic A, Troakes C, Al-Sarraj S, Dobson R, Schalkwyk LC, Mill J. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome biology. 2012;13:R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Osso B, D’Addario C, Carlotta Palazzo M, Benatti B, Camuri G, Galimberti D, Fenoglio C, Scarpini E, Di Francesco A, Maccarrone M, Altamura AC. Epigenetic modulation of BDNF gene: differences in DNA methylation between unipolar and bipolar patients. Journal of affective disorders. 2014;166:330–333. doi: 10.1016/j.jad.2014.05.020. [DOI] [PubMed] [Google Scholar]
- Dempster EL, Pidsley R, Schalkwyk LC, Owens S, Georgiades A, Kane F, Kalidindi S, Picchioni M, Kravariti E, Toulopoulou T, Murray RM, Mill J. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Human molecular genetics. 2011;20:4786–4796. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Hegde AN. Expression of RGS4 splice variants in dorsolateral prefrontal cortex of schizophrenic and bipolar disorder patients. Biological psychiatry. 2009;65:541–545. doi: 10.1016/j.biopsych.2008.10.026. [DOI] [PubMed] [Google Scholar]
- Ding L, Styblo M, Drobna Z, Hegde AN. Expression of the Longest RGS4 Splice Variant in the Prefrontal Cortex Is Associated with Single Nucleotide Polymorphisms in Schizophrenia Patients. Frontiers in psychiatry. 2016;7:26. doi: 10.3389/fpsyt.2016.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobosy JR, Selker EU. Emerging connections between DNA methylation and histone acetylation. Cellular and molecular life sciences : CMLS. 2001;58:721–727. doi: 10.1007/PL00000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domschke K, Lawford B, Young R, Voisey J, Morris CP, Roehrs T, Hohoff C, Birosova E, Arolt V, Baune BT. Dysbindin (DTNBP1)--a role in psychotic depression? Journal of psychiatric research. 2011;45:588–595. doi: 10.1016/j.jpsychires.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Dong E, Nelson M, Grayson DR, Costa E, Guidotti A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13614–13619. doi: 10.1073/pnas.0805493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Ruzicka WB, Grayson DR, Guidotti A. DNA-methyltransferase1 (DNMT1) binding to CpG rich GABAergic and BDNF promoters is increased in the brain of schizophrenia and bipolar disorder patients. Schizophrenia research. 2015;167:35–41. doi: 10.1016/j.schres.2014.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Luu PL, Stirzaker C, Clark SJ. Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics. 2015;7:1051–1073. doi: 10.2217/epi.15.39. [DOI] [PubMed] [Google Scholar]
- Dunn EC, Brown RC, Dai Y, Rosand J, Nugent NR, Amstadter AB, Smoller JW. Genetic determinants of depression: recent findings and future directions. Harvard review of psychiatry. 2015;23:1–18. doi: 10.1097/HRP.0000000000000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi T, Zhang H. Lithium-induced neuroprotection is associated with epigenetic modification of specific BDNF gene promoter and altered expression of apoptotic-regulatory proteins. Frontiers in neuroscience. 2014;8:457. doi: 10.3389/fnins.2014.00457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, Lu B, Weinberger DR. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Erdmann A, Halby L, Fahy J, Arimondo PB. Targeting DNA methylation with small molecules: what’s next? Journal of medicinal chemistry. 2015;58:2569–2583. doi: 10.1021/jm500843d. [DOI] [PubMed] [Google Scholar]
- Fatemi SH. Reelin, a marker of stress resilience in depression and psychosis. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2011;36:2371–2372. doi: 10.1038/npp.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Earle JA, McMenomy T. Reduction in Reelin immunoreactivity in hippocampus of subjects with schizophrenia, bipolar disorder and major depression. Molecular psychiatry. 2000;5:654–663. 571. doi: 10.1038/sj.mp.4000783. [DOI] [PubMed] [Google Scholar]
- Frauer C, Rottach A, Meilinger D, Bultmann S, Fellinger K, Hasenoder S, Wang M, Qin W, Soding J, Spada F, Leonhardt H. Different binding properties and function of CXXC zinc finger domains in Dnmt1 and Tet1. PloS one. 2011;6:e16627. doi: 10.1371/journal.pone.0016627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey BN, Andreazza AC, Houenou J, Jamain S, Goldstein BI, Frye MA, Leboyer M, Berk M, Malhi GS, Lopez-Jaramillo C, Taylor VH, Dodd S, Frangou S, Hall GB, Fernandes BS, Kauer-Sant’Anna M, Yatham LN, Kapczinski F, Young LT. Biomarkers in bipolar disorder: a positional paper from the International Society for Bipolar Disorders Biomarkers Task Force. The Australian and New Zealand journal of psychiatry. 2013;47:321–332. doi: 10.1177/0004867413478217. [DOI] [PubMed] [Google Scholar]
- Fries GR, Gassen NC, Schmidt U, Rein T. The FKBP51-Glucocorticoid Receptor Balance in Stress-Related Mental Disorders. Current molecular pharmacology. 2015;9:126–140. doi: 10.2174/1874467208666150519114435. [DOI] [PubMed] [Google Scholar]
- Fries GR, Quevedo J, Soares JC, Walss-Bass C. Genome-wide Expression and Methylation Analyses in Pediatric Bipolar Disorder and High-risk Offspring: A Preliminary Study. Biological Psychiatr. 2016;79:229S. [Google Scholar]
- Gassen NC, Fries GR, Zannas AS, Hartmann J, Zschocke J, Hafner K, Carrillo-Roa T, Steinbacher J, Preissinger SN, Hoeijmakers L, Knop M, Weber F, Kloiber S, Lucae S, Chrousos GP, Carell T, Ising M, Binder EB, Schmidt MV, Ruegg J, Rein T. Chaperoning epigenetics: FKBP51 decreases the activity of DNMT1 and mediates epigenetic effects of the antidepressant paroxetine. Science signaling. 2015a;8:ra119. doi: 10.1126/scisignal.aac7695. [DOI] [PubMed] [Google Scholar]
- Gassen NC, Hartmann J, Zannas AS, Kretzschmar A, Zschocke J, Maccarrone G, Hafner K, Zellner A, Kollmannsberger LK, Wagner KV, Mehta D, Kloiber S, Turck CW, Lucae S, Chrousos GP, Holsboer F, Binder EB, Ising M, Schmidt MV, Rein T. FKBP51 inhibits GSK3beta and augments the effects of distinct psychotropic medications. Molecular psychiatry. 2015b;21:277–289. doi: 10.1038/mp.2015.38. [DOI] [PubMed] [Google Scholar]
- Gassen NC, Hartmann J, Zschocke J, Stepan J, Hafner K, Zellner A, Kirmeier T, Kollmannsberger L, Wagner KV, Dedic N, Balsevich G, Deussing JM, Kloiber S, Lucae S, Holsboer F, Eder M, Uhr M, Ising M, Schmidt MV, Rein T. Association of FKBP51 with priming of autophagy pathways and mediation of antidepressant treatment response: evidence in cells, mice, and humans. PLoS medicine. 2014;11:e1001755. doi: 10.1371/journal.pmed.1001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadirivasfi M, Nohesara S, Ahmadkhaniha HR, Eskandari MR, Mostafavi S, Thiagalingam S, Abdolmaleky HM. Hypomethylation of the serotonin receptor type-2A Gene (HTR2A) at T102C polymorphic site in DNA derived from the saliva of patients with schizophrenia and bipolar disorder. American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2011;156b:536–545. doi: 10.1002/ajmg.b.31192. [DOI] [PubMed] [Google Scholar]
- Glatt SJ, Chandler SD, Bousman CA, Chana G, Lucero GR, Tatro E, May T, Lohr JB, Kremen WS, Everall IP, Tsuang MT. Alternatively Spliced Genes as Biomarkers for Schizophrenia, Bipolar Disorder and Psychosis: A Blood-Based Spliceome-Profiling Exploratory Study. Current pharmacogenomics and personalized medicine. 2009;7:164–188. doi: 10.2174/1875692110907030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatt SJ, Cohen OS, Faraone SV, Tsuang MT. Dysfunctional gene splicing as a potential contributor to neuropsychiatric disorders. American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2011;156b:382–392. doi: 10.1002/ajmg.b.31181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande I, Berk M, Birmaher B, Vieta E. Bipolar disorder. Lancet (London, England) 2016;387:1561–1572. doi: 10.1016/S0140-6736(15)00241-X. [DOI] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Di-Giorgi-Gerevini V, Dwivedi Y, Grayson DR, Impagnatiello F, Pandey G, Pesold C, Sharma R, Uzunov D, Costa E. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Archives of general psychiatry. 2000;57:1061–1069. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- Guidotti A, Dong E, Gavin DP, Veldic M, Zhao W, Bhaumik DK, Pandey SC, Grayson DR. DNA methylation/demethylation network expression in psychotic patients with a history of alcohol abuse. Alcoholism, clinical and experimental research. 2013;37:417–424. doi: 10.1111/j.1530-0277.2012.01947.x. [DOI] [PubMed] [Google Scholar]
- Guidotti A, Grayson DR. DNA methylation and demethylation as targets for antipsychotic therapy. Dialogues in clinical neuroscience. 2014;16:419–429. doi: 10.31887/DCNS.2014.16.3/aguidotti. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Grayson DR, Caruncho HJ. Epigenetic RELN Dysfunction in Schizophrenia and Related Neuropsychiatric Disorders. Frontiers in cellular neuroscience. 2016;10:89. doi: 10.3389/fncel.2016.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Ruzicka W, Grayson DR, Veldic M, Pinna G, Davis JM, Costa E. S-adenosyl methionine and DNA methyltransferase-1 mRNA overexpression in psychosis. Neuroreport. 2007;18:57–60. doi: 10.1097/WNR.0b013e32800fefd7. [DOI] [PubMed] [Google Scholar]
- Higuchi F, Uchida S, Yamagata H, Otsuki K, Hobara T, Abe N, Shibata T, Watanabe Y. State-dependent changes in the expression of DNA methyltransferases in mood disorder patients. Journal of psychiatric research. 2011;45:1295–1300. doi: 10.1016/j.jpsychires.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Hong CJ, Liou YJ, Tsai SJ. Effects of BDNF polymorphisms on brain function and behavior in health and disease. Brain research bulletin. 2011;86:287–297. doi: 10.1016/j.brainresbull.2011.08.019. [DOI] [PubMed] [Google Scholar]
- Horvath S, Zhang Y, Langfelder P, Kahn RS, Boks MP, van Eijk K, van den Berg LH, Ophoff RA. Aging effects on DNA methylation modules in human brain and blood tissue. Genome biology. 2012;13:R97. doi: 10.1186/gb-2012-13-10-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosang GM, Uher R, Keers R, Cohen-Woods S, Craig I, Korszun A, Perry J, Tozzi F, Muglia P, McGuffin P, Farmer AE. Stressful life events and the brain-derived neurotrophic factor gene in bipolar disorder. Journal of affective disorders. 2010;125:345–349. doi: 10.1016/j.jad.2010.01.071. [DOI] [PubMed] [Google Scholar]
- Houtepen LC, van Bergen AH, Vinkers CH, Boks MP. DNA methylation signatures of mood stabilizers and antipsychotics in bipolar disorder. Epigenomics. 2016;8:197–208. doi: 10.2217/epi.15.98. [DOI] [PubMed] [Google Scholar]
- Huzayyin AA, Andreazza AC, Turecki G, Cruceanu C, Rouleau GA, Alda M, Young LT. Decreased global methylation in patients with bipolar disorder who respond to lithium. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum (CINP) 2014;17:561–569. doi: 10.1017/S1461145713001569. [DOI] [PubMed] [Google Scholar]
- Jann MW. Diagnosis and treatment of bipolar disorders in adults: a review of the evidence on pharmacologic treatments. American health & drug benefits. 2014;7:489–499. [PMC free article] [PubMed] [Google Scholar]
- Joo EJ, Lee KY, Jeong SH, Chang JS, Ahn YM, Koo YJ, Kim YS. Dysbindin gene variants are associated with bipolar I disorder in a Korean population. Neuroscience letters. 2007;418:272–275. doi: 10.1016/j.neulet.2007.03.037. [DOI] [PubMed] [Google Scholar]
- Kaminsky Z, Jones I, Verma R, Saleh L, Trivedi H, Guintivano J, Akman R, Zandi P, Lee RS, Potash JB. DNA methylation and expression of KCNQ3 in bipolar disorder. Bipolar disorders. 2015;17:150–159. doi: 10.1111/bdi.12230. [DOI] [PubMed] [Google Scholar]
- Kaminsky Z, Tochigi M, Jia P, Pal M, Mill J, Kwan A, Ioshikhes I, Vincent JB, Kennedy JL, Strauss J, Pai S, Wang SC, Petronis A. A multi-tissue analysis identifies HLA complex group 9 gene methylation differences in bipolar disorder. Molecular psychiatry. 2012;17:728–740. doi: 10.1038/mp.2011.64. [DOI] [PubMed] [Google Scholar]
- Karatsoreos IN. Links between Circadian Rhythms and Psychiatric Disease. Frontiers in behavioral neuroscience. 2014;8:162. doi: 10.3389/fnbeh.2014.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner B. Toward a Deeper Understanding of the Genetics of Bipolar Disorder. Frontiers in psychiatry. 2015;6:105. doi: 10.3389/fpsyt.2015.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Dias BG, Ressler KJ. Models of Intergenerational and Transgenerational Transmission of Risk for Psychopathology in Mice. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2016a;41:219–231. doi: 10.1038/npp.2015.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Howell B, Guzman D, Morin E, Binder EB, Ressler KJ, Sanchez M. Intergenerational Effects of Infant Maltreatment on Epigenetic and Functional Outcomes in Rhesus Monkeys. Biological Psychiatry. 2016b;79:143S. [Google Scholar]
- Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, Pace TW, Mercer KB, Mayberg HS, Bradley B, Nemeroff CB, Holsboer F, Heim CM, Ressler KJ, Rein T, Binder EB. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nature neuroscience. 2013;16:33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends in biochemical sciences. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kozlenkov A, Wang M, Roussos P, Rudchenko S, Barbu M, Bibikova M, Klotzle B, Dwork AJ, Zhang B, Hurd YL, Koonin EV, Wegner M, Dracheva S. Substantial DNA methylation differences between two major neuronal subtypes in human brain. Nucleic acids research. 2016;44:2593–2612. doi: 10.1093/nar/gkv1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulis M, Queiros AC, Beekman R, Martin-Subero JI. Intragenic DNA methylation in transcriptional regulation, normal differentiation and cancer. Biochimica et biophysica acta. 2013;1829:1161–1174. doi: 10.1016/j.bbagrm.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Kuratomi G, Iwamoto K, Bundo M, Kusumi I, Kato N, Iwata N, Ozaki N, Kato T. Aberrant DNA methylation associated with bipolar disorder identified from discordant monozygotic twins. Molecular psychiatry. 2008;13:429–441. doi: 10.1038/sj.mp.4002001. [DOI] [PubMed] [Google Scholar]
- Lee RS, Pirooznia M, Guintivano J, Ly M, Ewald ER, Tamashiro KL, Gould TD, Moran TH, Potash JB. Search for common targets of lithium and valproic acid identifies novel epigenetic effects of lithium on the rat leptin receptor gene. Translational psychiatry. 2015;5:e600. doi: 10.1038/tp.2015.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev Maor G, Yearim A, Ast G. The alternative role of DNA methylation in splicing regulation. Trends in genetics : TIG. 2015;31:274–280. doi: 10.1016/j.tig.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Li Y, Camarillo C, Xu J, Arana TB, Xiao Y, Zhao Z, Chen H, Ramirez M, Zavala J, Escamilla MA, Armas R, Mendoza R, Ontiveros A, Nicolini H, Magana AA, Rubin LP, Li X, Xu C. Genome-wide methylome analyses reveal novel epigenetic regulation patterns in schizophrenia and bipolar disorder. BioMed research international. 2015;2015:201587. doi: 10.1155/2015/201587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisanti S, Omar WA, Tomaszewski B, De Prins S, Jacobs G, Koppen G, Mathers JC, Langie SA. Comparison of methods for quantification of global DNA methylation in human cells and tissues. PloS one. 2013;8:e79044. doi: 10.1371/journal.pone.0079044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA. Turning over DNA methylation in the mind. Frontiers in neuroscience. 2015;9:252. doi: 10.3389/fnins.2015.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Chen J, Ehrlich S, Walton E, White T, Perrone-Bizzozero N, Bustillo J, Turner JA, Calhoun VD. Methylation patterns in whole blood correlate with symptoms in schizophrenia patients. Schizophrenia bulletin. 2014;40:769–776. doi: 10.1093/schbul/sbt080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Siyahhan Julnes P, Chen J, Ehrlich S, Walton E, Calhoun VD. The association of DNA methylation and brain volume in healthy individuals and schizophrenia patients. Schizophrenia research. 2015;169:447–452. doi: 10.1016/j.schres.2015.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo R, Weksberg R. Biological and biochemical modulation of DNA methylation. Epigenomics. 2014;6:593–602. doi: 10.2217/epi.14.49. [DOI] [PubMed] [Google Scholar]
- Malhi GS, Tanious M, Das P, Coulston CM, Berk M. Potential mechanisms of action of lithium in bipolar disorder. Current understanding. CNS drugs. 2013;27:135–153. doi: 10.1007/s40263-013-0039-0. [DOI] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJ, Haussler D, Marra MA, Hirst M, Wang T, Costello JF. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke A, Binder EB. Epigenetic alterations in depression and antidepressant treatment. Dialogues in clinical neuroscience. 2014;16:395–404. doi: 10.31887/DCNS.2014.16.3/amenke. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, Jia P, Assadzadeh A, Flanagan J, Schumacher A, Wang SC, Petronis A. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. American journal of human genetics. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milutinovic S, D’Alessio AC, Detich N, Szyf M. Valproate induces widespread epigenetic reprogramming which involves demethylation of specific genes. Carcinogenesis. 2007;28:560–571. doi: 10.1093/carcin/bgl167. [DOI] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Stanwood GD, Lewis DA, Levitt P. Disease-specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Molecular psychiatry. 2001;6:293–301. doi: 10.1038/sj.mp.4000866. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Pena CJ, Kundakovic M, Mitchell A, Akbarian S. Epigenetic Basis of Mental Illness. The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry. 2015 doi: 10.1177/1073858415608147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohesara S, Ghadirivasfi M, Mostafavi S, Eskandari MR, Ahmadkhaniha H, Thiagalingam S, Abdolmaleky HM. DNA hypomethylation of MB-COMT promoter in the DNA derived from saliva in schizophrenia and bipolar disorder. Journal of psychiatric research. 2011;45:1432–1438. doi: 10.1016/j.jpsychires.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Oliveira J, Etain B, Lajnef M, Hamdani N, Bennabi M, Bengoufa D, Sundaresh A, Chaabane AB, Bellivier F, Henry C, Kahn JP, Charron D, Krishnamoorthy R, Leboyer M, Tamouza R. Combined effect of TLR2 gene polymorphism and early life stress on the age at onset of bipolar disorders. PloS one. 2015;10:e0119702. doi: 10.1371/journal.pone.0119702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira J, Kazma R, Le Floch E, Bennabi M, Hamdani N, Bengoufa D, Dahoun M, Manier C, Bellivier F, Krishnamoorthy R, Deleuze JF, Yolken R, Leboyer M, Tamouza R. Toxoplasma gondii exposure may modulate the influence of TLR2 genetic variation on bipolar disorder: a gene-environment interaction study. International journal of bipolar disorders. 2016;4:11. doi: 10.1186/s40345-016-0052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovadia G, Shifman S. The genetic variation of RELN expression in schizophrenia and bipolar disorder. PloS one. 2011;6:e19955. doi: 10.1371/journal.pone.0019955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M, Ebrahimi S, Oh G, Khare T, Zhang A, Kaminsky ZA, Wang SC, Petronis A. High Precision DNA Modification Analysis of HCG9 in Major Psychosis. Schizophrenia bulletin. 2016;42:170–177. doi: 10.1093/schbul/sbv079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perisic T, Zimmermann N, Kirmeier T, Asmus M, Tuorto F, Uhr M, Holsboer F, Rein T, Zschocke J. Valproate and amitriptyline exert common and divergent influences on global and gene promoter-specific chromatin modifications in rat primary astrocytes. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2010;35:792–805. doi: 10.1038/npp.2009.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud N, Zewdie S, Stenz L, Adouan W, Bavamian S, Prada P, Nicastro R, Hasler R, Nallet A, Piguet C, Paoloni-Giacobino A, Aubry JM, Dayer A. Methylation of serotonin receptor 3A in ADHD, borderline personality, and bipolar disorders: link with severity of the disorders and childhood maltreatment. Depression and anxiety. 2016;33:45–55. doi: 10.1002/da.22406. [DOI] [PubMed] [Google Scholar]
- Phiel CJ, Zhang F, Huang EY, Guenther MG, Lazar MA, Klein PS. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. The Journal of biological chemistry. 2001;276:36734–36741. doi: 10.1074/jbc.M101287200. [DOI] [PubMed] [Google Scholar]
- Polesskaya OO, Aston C, Sokolov BP. Allele C-specific methylation of the 5-HT2A receptor gene: evidence for correlation with its expression and expression of DNA methylase DNMT1. Journal of neuroscience research. 2006;83:362–373. doi: 10.1002/jnr.20732. [DOI] [PubMed] [Google Scholar]
- Popkie AP, Zeidner LC, Albrecht AM, D’Ippolito A, Eckardt S, Newsom DE, Groden J, Doble BW, Aronow B, McLaughlin KJ, White P, Phiel CJ. Phosphatidylinositol 3-kinase (PI3K) signaling via glycogen synthase kinase-3 (Gsk-3) regulates DNA methylation of imprinted loci. The Journal of biological chemistry. 2010;285:41337–41347. doi: 10.1074/jbc.M110.170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potash JB, DePaulo JR., Jr Searching high and low: a review of the genetics of bipolar disorder. Bipolar disorders. 2000;2:8–26. doi: 10.1034/j.1399-5618.2000.020103.x. [DOI] [PubMed] [Google Scholar]
- Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics. 2007;90:397–406. doi: 10.1016/j.ygeno.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao JS, Keleshian VL, Klein S, Rapoport SI. Epigenetic modifications in frontal cortex from Alzheimer’s disease and bipolar disorder patients. Translational psychiatry. 2012a;2:e132. doi: 10.1038/tp.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao JS, Kellom M, Reese EA, Rapoport SI, Kim HW. Dysregulated glutamate and dopamine transporters in postmortem frontal cortex from bipolar and schizophrenic patients. Journal of affective disorders. 2012b;136:63–71. doi: 10.1016/j.jad.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Reddington JP, Pennings S, Meehan RR. Non-canonical functions of the DNA methylome in gene regulation. The Biochemical journal. 2013;451:13–23. doi: 10.1042/BJ20121585. [DOI] [PubMed] [Google Scholar]
- Rice F, Harold G, Thapar A. The genetic aetiology of childhood depression: a review. Journal of child psychology and psychiatry, and allied disciplines. 2002;43:65–79. doi: 10.1111/1469-7610.00004. [DOI] [PubMed] [Google Scholar]
- Roundtree IA, He C. RNA epigenetics-chemical messages for posttranscriptional gene regulation. Current opinion in chemical biology. 2015;30:46–51. doi: 10.1016/j.cbpa.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka WB, Subburaju S, Benes FM. Circuit- and Diagnosis-Specific DNA Methylation Changes at gamma-Aminobutyric Acid-Related Genes in Postmortem Human Hippocampus in Schizophrenia and Bipolar Disorder. JAMA psychiatry. 2015;72:541–551. doi: 10.1001/jamapsychiatry.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabunciyan S, Maher B, Bahn S, Dickerson F, Yolken RH. Association of DNA Methylation with Acute Mania and Inflammatory Markers. PloS one. 2015;10:e0132001. doi: 10.1371/journal.pone.0132001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadakierska-Chudy A, Kostrzewa RM, Filip M. A comprehensive view of the epigenetic landscape part I: DNA methylation, passive and active DNA demethylation pathways and histone variants. Neurotoxicity research. 2015;27:84–97. doi: 10.1007/s12640-014-9497-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sales AJ, Biojone C, Terceti MS, Guimaraes FS, Gomes MV, Joca SR. Antidepressant-like effect induced by systemic and intra-hippocampal administration of DNA methylation inhibitors. British journal of pharmacology. 2011;164:1711–1721. doi: 10.1111/j.1476-5381.2011.01489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sales AJ, Joca SR. Effects of DNA methylation inhibitors and conventional antidepressants on mice behaviour and brain DNA methylation levels. Acta neuropsychiatrica. 2016;28:11–22. doi: 10.1017/neu.2015.40. [DOI] [PubMed] [Google Scholar]
- Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- Simmer F, Brinkman AB, Assenov Y, Matarese F, Kaan A, Sabatino L, Villanueva A, Huertas D, Esteller M, Lengauer T, Bock C, Colantuoni V, Altucci L, Stunnenberg HG. Comparative genome-wide DNA methylation analysis of colorectal tumor and matched normal tissues. Epigenetics. 2012;7:1355–1367. doi: 10.4161/epi.22562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AK, Kilaru V, Klengel T, Mercer KB, Bradley B, Conneely KN, Ressler KJ, Binder EB. DNA extracted from saliva for methylation studies of psychiatric traits: evidence tissue specificity and relatedness to brain. American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2015;168b:36–44. doi: 10.1002/ajmg.b.32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller JW, Finn CT. Family, twin, and adoption studies of bipolar disorder. American journal of medical genetics. Part C, Seminars in medical genetics. 2003;123c:48–58. doi: 10.1002/ajmg.c.20013. [DOI] [PubMed] [Google Scholar]
- Soeiro-de-Souza MG, Andreazza AC, Carvalho AF, Machado-Vieira R, Young LT, Moreno RA. Number of manic episodes is associated with elevated DNA oxidation in bipolar I disorder. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum (CINP) 2013;16:1505–1512. doi: 10.1017/S1461145713000047. [DOI] [PubMed] [Google Scholar]
- Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, Eberl HC, Mensinga A, Brinkman AB, Lephikov K, Muller U, Walter J, Boelens R, van Ingen H, Leonhardt H, Carell T, Vermeulen M. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Starnawska A, Demontis D, McQuillin A, O’Brien NL, Staunstrup NH, Mors O, Nielsen AL, Borglum AD, Nyegaard M. Hypomethylation of FAM63B in bipolar disorder patients. Clinical epigenetics. 2016;8:52. doi: 10.1186/s13148-016-0221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenz L, Zewdie S, Laforge-Escarra T, Prados J, La Harpe R, Dayer A, Paoloni-Giacobino A, Perroud N, Aubry JM. BDNF promoter I methylation correlates between postmortem human peripheral and brain tissues. Neuroscience research. 2015;91:1–7. doi: 10.1016/j.neures.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Strauss JS, Khare T, De Luca V, Jeremian R, Kennedy JL, Vincent JB, Petronis A. Quantitative leukocyte BDNF promoter methylation analysis in bipolar disorder. International journal of bipolar disorders. 2013;1:28. doi: 10.1186/2194-7511-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara H, Iwamoto K, Bundo M, Ueda J, Miyauchi T, Komori A, Kazuno A, Adati N, Kusumi I, Okazaki Y, Ishigooka J, Kojima T, Kato T. Hypermethylation of serotonin transporter gene in bipolar disorder detected by epigenome analysis of discordant monozygotic twins. Translational psychiatry. 2011;1:e24. doi: 10.1038/tp.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. The American journal of psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- Szyf M. The implications of DNA methylation for toxicology: toward toxicomethylomics, the toxicology of DNA methylation. Toxicological sciences : an official journal of the Society of Toxicology. 2011;120:235–255. doi: 10.1093/toxsci/kfr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Kunugi H, Ohashi J, Hohjoh H. Epigenetic aberration of the human REELIN gene in psychiatric disorders. Molecular psychiatry. 2007;12:519, 593–600. doi: 10.1038/sj.mp.4002014. [DOI] [PubMed] [Google Scholar]
- Teroganova N, Girshkin L, Suter CM, Green MJ. DNA methylation in peripheral tissue of schizophrenia and bipolar disorder: a systematic review. BMC genetics. 2016;17:27. doi: 10.1186/s12863-016-0332-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognini P, Napoli D, Pizzorusso T. Dynamic DNA methylation in the brain: a new epigenetic mark for experience-dependent plasticity. Frontiers in cellular neuroscience. 2015;9:331. doi: 10.3389/fncel.2015.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldic M, Guidotti A, Maloku E, Davis JM, Costa E. In psychosis, cortical interneurons overexpress DNA-methyltransferase 1. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2152–2157. doi: 10.1073/pnas.0409665102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldic M, Kadriu B, Maloku E, Agis-Balboa RC, Guidotti A, Davis JM, Costa E. Epigenetic mechanisms expressed in basal ganglia GABAergic neurons differentiate schizophrenia from bipolar disorder. Schizophrenia research. 2007;91:51–61. doi: 10.1016/j.schres.2006.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RM, Christoforou AN, McCartney DL, Morris SW, Kennedy NA, Morten P, Anderson SM, Torrance HS, Macdonald A, Sussmann JE, Whalley HC, Blackwood DH, McIntosh AM, Porteous DJ, Evans KL. DNA methylation in a Scottish family multiply affected by bipolar disorder and major depressive disorder. Clinical epigenetics. 2016;8:5. doi: 10.1186/s13148-016-0171-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton E, Hass J, Liu J, Roffman JL, Bernardoni F, Roessner V, Kirsch M, Schackert G, Calhoun V, Ehrlich S. Correspondence of DNA Methylation Between Blood and Brain Tissue and Its Application to Schizophrenia Research. Schizophrenia bulletin. 2016;42:406–414. doi: 10.1093/schbul/sbv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Stadlbauer U, Richetto J, Labouesse MA, Bohacek J, Mansuy IM, Meyer U. Transgenerational transmission and modification of pathological traits induced by prenatal immune activation. Molecular psychiatry. 2016 doi: 10.1038/mp.2016.41. In press. [DOI] [PubMed] [Google Scholar]
- Wen L, Tang F. Genomic distribution and possible functions of DNA hydroxymethylation in the brain. Genomics. 2014;104:341–346. doi: 10.1016/j.ygeno.2014.08.020. [DOI] [PubMed] [Google Scholar]
- Wochnik GM, Ruegg J, Abel GA, Schmidt U, Holsboer F, Rein T. FK506-binding proteins 51 and 52 differentially regulate dynein interaction and nuclear translocation of the glucocorticoid receptor in mammalian cells. The Journal of biological chemistry. 2005;280:4609–4616. doi: 10.1074/jbc.M407498200. [DOI] [PubMed] [Google Scholar]
- Wu R, Fan J, Zhao J, Calabrese JR, Gao K. The relationship between neurotrophins and bipolar disorder. Expert review of neurotherapeutics. 2014;14:51–65. doi: 10.1586/14737175.2014.863709. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Camarillo C, Ping Y, Arana TB, Zhao H, Thompson PM, Xu C, Su BB, Fan H, Ordonez J, Wang L, Mao C, Zhang Y, Cruz D, Escamilla MA, Li X, Xu C. The DNA methylome and transcriptome of different brain regions in schizophrenia and bipolar disorder. PloS one. 2014;9:e95875. doi: 10.1371/journal.pone.0095875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R, Daskalakis NP, Bierer LM, Bader HN, Klengel T, Holsboer F, Binder EB. Holocaust Exposure Induced Intergenerational Effects on FKBP5 Methylation. Biological psychiatry. 2015 doi: 10.1016/j.biopsych.2015.08.005. In press. [DOI] [PubMed] [Google Scholar]
- Yun DH, Pae CU, Drago A, Mandelli L, De Ronchi D, Patkar AA, Paik IH, Serretti A, Kim JJ. Effect of the dysbindin gene on antimanic agents in patients with bipolar I disorder. Psychiatry investigation. 2008;5:102–105. doi: 10.4306/pi.2008.5.2.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Xu J, Pang L, Zhang Y, Fan H, Liu L, Liu T, Yu F, Zhang G, Lan Y, Bai J, Li X, Xiao Y. Genome-wide DNA methylome reveals the dysfunction of intronic microRNAs in major psychosis. BMC medical genomics. 2015;8:62. doi: 10.1186/s12920-015-0139-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann N, Zschocke J, Perisic T, Yu S, Holsboer F, Rein T. Antidepressants inhibit DNA methyltransferase 1 through reducing G9a levels. The Biochemical journal. 2012;448:93–102. doi: 10.1042/BJ20120674. [DOI] [PubMed] [Google Scholar]