Abstract
Macrophages mount complex responses to pathogens. Though several key signaling pathways have been identified, it remains unclear how they work together to provide specificity. In a recent paper, Gottschalk et al. report that differential dose response behavior of NFκB and MAPK pathways allows for dose-specific gene expression programs.
How immune activation is controlled continues to be an important research question; a well-functioning immune system, or its dysregulation, is a key determinant of human health and disease. Immediate recognition of pathogen-derived substances or antigens by innate and adaptive immune receptor molecules has been studied in atomic detail to provide some clarity on the basis for molecular specificity and the distinction between self and non-self. However, our understanding of the specificity of the signaling pathways that are activated by these receptor-ligand interactions lags far behind.
Progress has been made in identifying the molecular components of prominent signaling pathways such as those controlling the activities of transcription factors NFκB, interferon regulatory factors (IRF), or kinases of the c-Jun N-terminal kinases (JNK), or MAPK/ERK families. Thus, recent studies have been able to focus on how the signaling characteristics of each pathway are generated. Experimental approaches that allow for single cell resolution, temporal sequence, and true quantitation are revealing new, emergent properties of pathways which in turn determine their biological function. However, by and large these studies have been focused on the functioning of a single signaling pathway at a time.
The biological response involves the coordinated functioning of several pathways, and combinations of pathways are known to be activated by the ligands of pathogen-sensing receptors or immune receptors. Also, immune response genes whose expression are upregulated by them, are thought to integrate combinations of transcription factors (Figure 1).
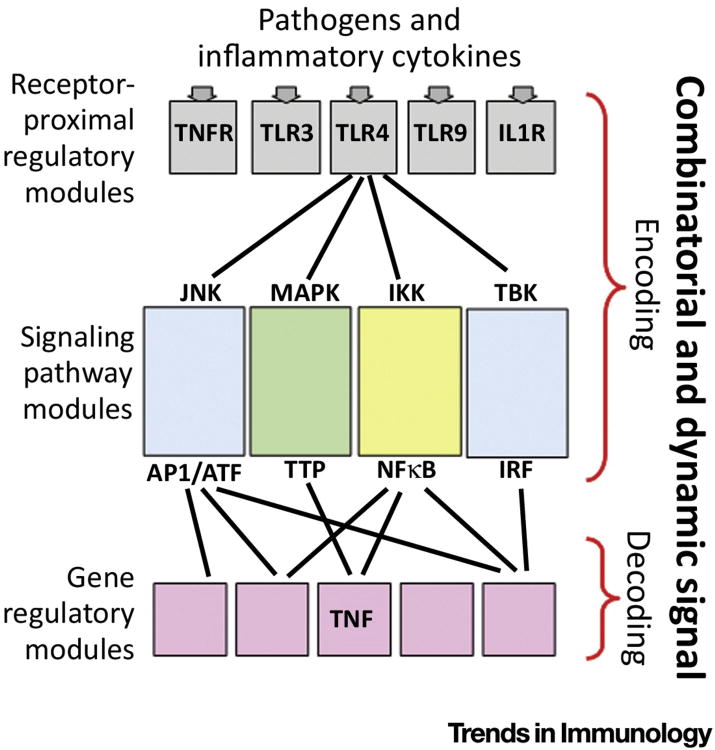
Figure 1. Encoding and Decoding of Inflammatory Signals.
Schematic of the immune response signaling network, emphasizing a modular structure. In this view, receptors that sense the presence of pathogen-derived substances or inflammatory cytokines engage receptor-proximal regulatory modules that trigger the activation of several signaling pathways. These pathways regulate the activities of transcription factors or other regulators that combine in gene regulatory modules to control immune response gene expression. For example, endotoxin binding to TLR4 is known to trigger the activation at least four kinases; two downstream effectors are known to control TNF protein expression [5]. Gottschalk et al. show that the endotoxin dose-response behavior of MAPK and NFκB are quite distinct, supporting the view that cells encode the presence of pathogens in both combinatorial and dynamic intra-cellular signaling events, allowing gene regulatory modules to decode these to produce gene expression that is both ligand- and dose-appropriate. TNFR: Tumor necrosis factor receptor, TLR: Toll-like receptor, JNK: c-Jun N-terminal kinase, MAPK: mitogen-activated protein kinase, IKK: IκB kinase, TBK: TANK binding kinase, AP-1: activator protein 1, ATF: activating transcription factor, TTP: tristetraprolin, IRF: interferon regulatory factor.
The hypothesis of a combinatorial signaling code posits that regulatory modules encode information about the environment in combinations of intra-cellular signals (such as kinase activities), and effector-associated regulatory modules (such as gene regulatory modules) decode combinations of intra-cellular signals to provide a response (Figure 1). However, diagrams of immune response networks show that two prominent pathways, NFκB and MAPK, emanate from virtually all immune activation receptors, whether they are pathogen, cytokine or antigen sensors [1].
A recently published paper, Gottschalk et al, shows that these two prominent signaling pathways of NFκB and MAPK in fact have differential dose-response behaviors; thus the control of dose response behavior allows cells to express different sets of genes at different doses of the same ligand [2]. Specifically, the authors found that tumor necrosis factor (TNF) protein production in individual cells as assayed by flow cytometry was highly thresholded, a phenomenon also described as ultra-sensitivity, despite the fact that NFκB-responsive transcriptomes could be observed even at subthreshold concentrations. As TNF production is not only dependent on NFκB-driven transcription, but also MAPK-driven mRNA splicing, mRNA half-life stabilization, and protein processing, the authors hypothesized that the MAPK signaling pathway may have a higher threshold than the NFκB pathway. Indeed, probing IκBα degradation and MAPK/p38 or ERK phosphorylation showed differential dose responses, with the dose response of TNF production resembling that of Erk or p38 phosphorylation more than that of IκBα degradation. Interestingly, thresholded dose response behavior in the expression of other genes was generally correlated with MAPK dependence, and while gene expression of macrophages obtained from human donors may differ in some aspects the thresholded MAPK dose response was generally conserved.
The present paper emphasizes the importance of understanding the molecular mechanisms that function together in producing even simple emergent systems properties such as dose-response behavior. Within the Toll-like receptor (TLR) pathway, recent studies have begun to describe how signaling behavior may be “encoded” and “decoded”. As multiple molecular mechanisms function together and must be considered quantitatively, mathematical modeling is a hallmark of such mechanistic studies. Although this study does not provide extensive timecourse information, dose response behaviors are the result of kinetic molecular mechanisms that also produce dynamic behavior. For example, one recent study explored the mechanisms by which TLR “signal encoding” is thresholded and identified the formation of an oligomeric signalosome, the Myddosome, as a source of this ultra-sensitivity [3]; a previous study showed substantial ultra-sensitivity in the MAPK cascade itself [4], which is not present in the NFκB signaling module.
On the other hand, “signal decoding” may be mediated by both nuclear and cytoplasmic mechanisms. Specifically, in the case of TNF, control of mRNA processing, half-life, translation, and protein-processing and secretion, were shown to be controlled by MAPK [5]. Consistent with the present study, the key role of MAPK in TNF production renders NFκB activity a poor predictor of TNF production at a single cell level [6]. However, these studies do not shed light on the differential dose response behavior of MAPK and NFκB pathways, and further quantitative molecular mechanistic studies particularly in the MAPK pathways are required to produce well-founded math model research tools. When such experimental and modeling frameworks are established, future studies may investigate whether the dose response behavior might be stimulus-specific such that different ligands might produce different gene expression responses, particularly at non-saturating concentrations.
The present paper challenges future studies to integrate the combinatorial and dynamic signaling codes [7] to derive a mechanistic understanding of how combinations of dynamic signaling events are encoded by signaling pathways and decoded by receptors. A first framework for TNF [5], provides one example (though lacking single cell resolution) revealing that TNF's autocrine functions are largely restricted to pathogen ligands that do not engage the TIR-domain-containing adapter inducing interferon-β (TRIF) signaling pathway. It will be interesting and important to discover how combinatorial engagement of distinct signaling pathways, each with specific dynamic regulation [8], provides for specificity. Such analyses, at the single cell level, also allow employing information theoretic techniques to characterize the performance of signaling networks. Considering the NFκB pathway alone, the information-carrying capacity in response to TLR4 appears to be more limited than anticipated due to substantial cell-to-cell heterogeneity [9,10]. Initial analysis of MAPK signaling at a single time point in response to TNF indicated little improvement [9]. However, within the framework of both combinatorial and dynamical coding, information carrying capacities of pathogen responsive signaling may be much greater. Thus, with the advent of live cell experimental probes that allow measuring activities of multiple pathways simultaneously, future work may address the mutual information benefit of a combinatorial code within a noisy cellular environment.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Oda K, Kitano H. A comprehensive map of the toll-like receptor signaling network. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100057. 2006 0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottschalk RA, Martins AJ, Angermann BR, Dutta B, Ng CE, Uderhardt S, Tsang JS, Fraser ID, Meier-Schellersheim M, Germain RN. Distinct NF-kappaB and MAPK Activation Thresholds Uncouple Steady-State Microbe Sensing from Anti-pathogen Inflammatory Responses. Cell Syst. 2016;2:378–390. doi: 10.1016/j.cels.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng Z, Taylor B, Ourthiague DR, Hoffmann A. Distinct single-cell signaling characteristics are conferred by the MyD88 and TRIF pathways during TLR4 activation. Sci Signal. 2015;8:ra69. doi: 10.1126/scisignal.aaa5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang CY, Ferrell JE., Jr Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc Natl Acad Sci U S A. 1996;93:10078–10083. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldwell AB, Cheng Z, Vargas JD, Birnbaum HA, Hoffmann A. Network dynamics determine the autocrine and paracrine signaling functions of TNF. Genes Dev. 2014;28:2120–2133. doi: 10.1101/gad.244749.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Junkin M, Kaestli AJ, Cheng Z, Jordi C, Albayrak C, Hoffmann A, Tay S. High-Content Quantification of Single-Cell Immune Dynamics. Cell Rep. 2016;15:411–422. doi: 10.1016/j.celrep.2016.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behar M, Hoffmann A. Tunable signal processing through a kinase control cycle: the IKK signaling node. Biophys J. 2013;105:231–241. doi: 10.1016/j.bpj.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Werner SL, Barken D, Hoffmann A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 2005;309:1857–1861. doi: 10.1126/science.1113319. [DOI] [PubMed] [Google Scholar]
- 9.Cheong R, Rhee A, Wang CJ, Nemenman I, Levchenko A. Information transduction capacity of noisy biochemical signaling networks. Science. 2011;334:354–358. doi: 10.1126/science.1204553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selimkhanov J, Taylor B, Yao J, Pilko A, Albeck J, Hoffmann A, Tsimring L, Wollman R. Systems biology. Accurate information transmission through dynamic biochemical signaling networks. Science. 2014;346:1370–1373. doi: 10.1126/science.1254933. [DOI] [PMC free article] [PubMed] [Google Scholar]