Abstract
Objective
To develop and validate a novel decision tree-based clinical algorithm to differentiate Kawasaki disease (KD) from other pediatric febrile illnesses that share common clinical characteristics.
Study design
Using clinical and laboratory data from 801 subjects with acute KD (533 for development, and 268 for validation) and 479 febrile control subjects (318 for development, and 161 for validation), we developed a step-wise KD diagnostic algorithm combining our previously developed linear-discriminant-analysis (LDA)-based model with a newly developed, tree-based algorithm.
Results
The primary model (LDA) stratified the 1,280 subjects into FC (276), indeterminate (247), and KD (757) subgroups. The subsequent model (decision trees) further classified the indeterminate group into FC (103) and KD (58) subgroups, leaving only 29 of 801 (3.6%) KD and 57 of 479 (11.9%) FC subjects indeterminate. The 2-step algorithm had a sensitivity of 96.0% and a specificity of 78.5% and correctly classified all KD subjects who later developed coronary artery aneurysms.
Conclusion
The addition of a decision tree step increased sensitivity and specificity in KD/FC classification over our previously described LDA model. A multicenter trial is needed to prospectively determine its utility as a point of care diagnostic test for KD.
Keywords: KD diagnosis, LDA, Random forest, Incomplete KD, 2-step algorithm
More effective methods for the early diagnosis of acute Kawasaki disease (KD) are required to permit timely IVIG administration and prevention of adverse outcomes. The classic KD diagnostic criteria adopted by the American Heart Association (AHA) include fever plus at least four of five principal clinical signs (Figure 1)(1). These guidelines, although widely adopted by clinicians, occasionally fail to differentiate KD from other pediatric rash/fever illnesses(2). Moreover, despite supplementary laboratory criteria to aid in the diagnosis of KD patients who manifest only 2 or 3 clinical signs, these incomplete cases may still be missed by clinicians(1). Missing the diagnosis can lead to delayed treatment, thus increasing the risk of developing coronary artery lesions(3–5).
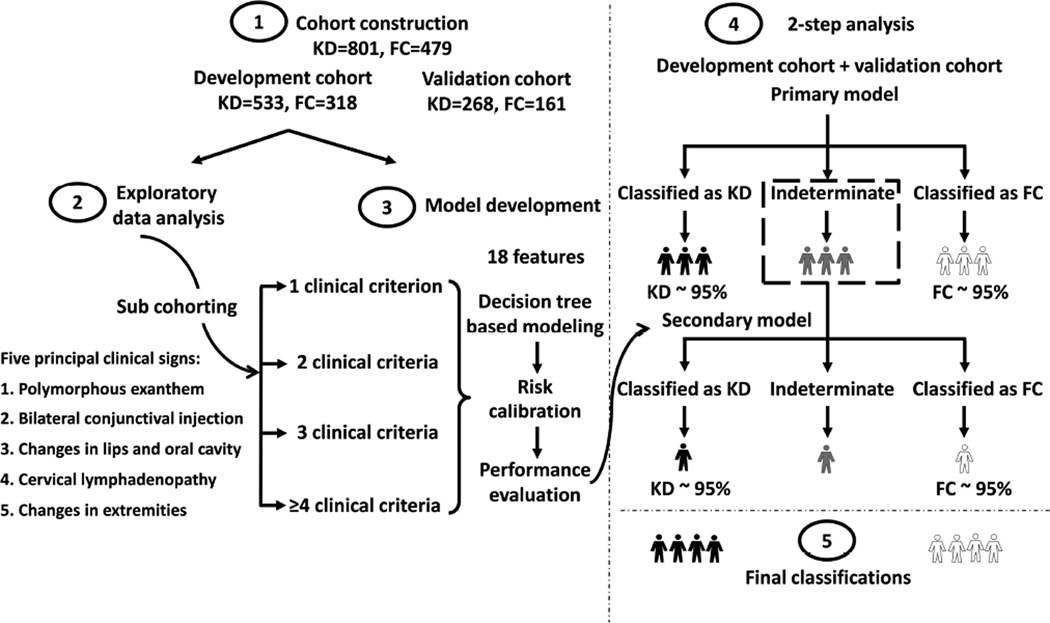
Figure 1.
Workflow to create a 2-step statistical algorithm for distinguishing KD and FC subjects. LDA- and decision-tree-based models developed based on clinical and laboratory test variables were applied in sequence to construct a 2-step algorithm, partitioning the subjects into 3 diagnostic classifications (FC, KD, and indeterminate). PPV and NPV of 95% were achieved at each step.
We previously applied statistical learning using clinical and laboratory test variables, and developed a linear-discriminant-analysis (LDA)-based scoring system to differentiate KD from febrile controls (FCs)(6) with a sensitivity of 92–94% and a specificity of 88–89%. However, 20–30% of subjects in either the KD or FC group remained unclassified, and the algorithm performance on KD subjects with incomplete clinical criteria was not investigated(6, 7).
In this study, we tested the hypothesis that applying separate tree-based algorithms following the LDA algorithm would improve the classification accuracy in differentiating KD from FC subjects. This novel integrated algorithm was validated with an independent subject cohort.
Methods
Subjects with KD and febrile controls meeting inclusion criteria were identified from the database maintained at the UCSD KD Research Center. Complete demographic and clinical data were collected prospectively on all KD and FC subjects. A total of 1,280 subjects (801 KD and 479 FC) were included in this study (Figure 2; available at www.jpeds.com). KD subjects in this study were: a) patients with fever (≥38.0 °C rectally or orally) for no more than 10 days plus at least four of the five principal clinical criteria, b) patients meeting fewer criteria but with coronary artery abnormalities (Z-score≥2.5 for left anterior descending (LAD) and/or right coronary arteries (RCA)) documented by echocardiogram, and c) patients meeting fewer than four criteria but meeting the American Heart Association (AHA) criteria for incomplete KD by laboratory criteria(1). A concomitant viral infection by RT-PCR did not disqualify the patient as a KD subject. Every subject was evaluated clinically by one of two expert KD clinicians and the final assignment of a KD diagnosis was based on the opinion of these two experts. FC subjects were recruited from the Emergency Department (ED) at Rady Children’s Hospital San Diego. All FC subjects had unexplained fever, at least one of the five principal clinical criteria for KD, and had laboratory tests performed including those commonly ordered for evaluation of KD, which included a complete blood count with manual differential, erythrocyte sedimentation rate (ESR), and levels of C-reactive protein (CRP), alanine aminotransferase (ALT), and gamma glutamyl transferase (GGT). All patients referred to the ED for evaluation of possible KD (approximately 50% of the FC cohort) were offered enrollment as FC subjects in our study. We enrolled the remaining FCs from children in the ED presenting with fever and at least one of the clinical signs of KD, and excluded patients who had an obvious respiratory or gastrointestinal infection because KD would be unlikely to present in this manner. The final diagnoses of the FCs were determined by chart review by two expert clinicians (JCB and JK) from prospectively collected clinical and laboratory data and from review of microbiologic and serologic results and subsequent clinical encounters. Only 3.8% of the FCs (18 of 479) had echocardiography to evaluate for possible KD.
Signed consent or assent forms were obtained from the parents of all subjects and from all subjects >6 years of age. The study was approved by the Institutional Review Boards of the University of California San Diego (UCSD) and Stanford University.
For each subject, we collected the 18 clinical and laboratory test variables retained in the final model of the LDA-based algorithm(6). Clinical data included six clinical signs associated with KD: illness days (temperature≥38.0 °C); cervical lymph node of at least 1.5 cm; rash; conjunctival injection; extremity changes including red, swollen, or peeling hands or feet; and oropharyngeal changes including red pharynx, red, fissured lips, or strawberry tongue. Laboratory test data (obtained prior to administration of IVIG for KD subjects) included total white blood cell count (WBC), percentages of monocytes, lymphocytes, eosinophils, neutrophils, and immature neutrophils (bands), platelet count, hemoglobin concentration normalized for age (ZHgb), CRP, GGT, ALT, and ESR. For test results that exceeded the upper or lower limit of the test, we used the numeric value for the limit. Subgroups of KD subjects were defined as having either normal coronary arteries (RCA and LAD Z-score always<2.5), transiently dilated coronary arteries (RCA and/or LAD Z-score≥2.5 and resolving within 8 weeks of KD onset), or aneurysms (Z ≥ 5.0 or dilated segment 1.5 times the internal diameter of the adjacent segment). We performed a multivariate analysis on the clinical and laboratory test variables for KD and FC discrimination in our total dataset. Panels combining 18 clinical and laboratory test variables were evaluated and the resulting odds ratios, p-values and variable effects on the final model were calculated.
Of the 1,280 subjects (801 KD and 479 FC), 489 (261 KD and 228 FC) were from the development cohort in our previous study(6) and remained in this study’s development cohort. The remaining 791 subjects were assigned into 2 cohorts while maintaining the same ratio of KD and FC subjects across cohorts. Of the entire cohort of 1,280 subjects, 228 of 801 KD subjects and 287 of 479 FC subjects had at least one missing value for the laboratory variables. Missing values were imputed among KD and FC subjects, respectively, using a method of weighted K-nearest neighbors (Appendix 2; available at www.jpeds.com)(8). There were 533 subjects with KD and 318 FCs for model development, and 268 subjects with KD and 161 FCs for model validation. The study design is outlined in Figure 1. A 2-step algorithm was developed using the 6 clinical and 12 laboratory test variables to stratify the subjects into three subgroups: FC, indeterminate, and KD. 95% PPV and NPV for KD and FC classification were targeted at each step.
Primary model
The previously developed KD algorithm was developed using an LDA method, with days of fever, 5 principal clinical criteria, and 12 laboratory test variables as input variables. The output of the algorithm was a unique score describing the probability of KD diagnosis for each subject(6). Two cutoffs were set to stratify these subjects into 3 classification subgroups: FC, indeterminate, and KD(6), allowing 95% accuracy in both KD and FC subgroups.
After applying the LDA model, 9.6% (51 of 533) of KD and 33.0% of (105 of 318) FC subjects in the development cohort remained indeterminate. The proportions of subjects with indeterminate scores, however, differed among the 4 sub-cohorts based on the number of principal clinical criteria manifested by each subject. The LDA model performed less well for subjects with fewer clinical criteria, yielding indeterminate scores for 28.4% (29 of 102) of KD and 43.9% (72 of 164) of FC subjects who manifested only 2 or 3 clinical criteria. Therefore, an additional model was developed to improve the adjudication of indeterminate subjects based on the number of clinical criteria present.
Secondary model
To improve the classification of subjects in the indeterminate group from the first analysis, we used 2-step data mining methods to combine the advantages of multiple models to achieve better predictive accuracy than is possible with any individual model(9). Random forest models constructed by a set of decision trees were developed(10, 11). Subjects were divided into 4 sub-cohorts based on the number of KD criteria that they manifested (Figure 1). Separate models were then developed for each sub-cohort. Specifically, subjects in the development cohort were further randomly partitioned into two sub-cohorts (Sub-cohort I and Sub-cohort II). A ‘forest’ of 300 binary ‘trees’ was constructed using randomly selected samples and variables (clinical and laboratory test variables) of Sub-cohort I. At each node, ‘trees’ were split by choosing a split variable value producing the maximum node separation. ‘Trees’ were constructed until each of the terminal nodes reached a sample size of 1. Final decisions were reached by averaging the decisions of each ‘tree’. The derived algorithm was then calibrated with Sub-cohort II by setting two thresholds that stratified all the subjects into 3 classification subgroups (FC, indeterminate, and KD), allowing 95% PPV and NPV. The performance of the algorithm was tested on the validation cohort. The modeling details appear in Appendix 3 (available at www.jpeds.com).
Performance analyses
Performance of the 2-step model was demonstrated by sensitivity, specificity, PPV, and NPV. Classification of incomplete KD subjects and subjects developing coronary artery abnormalities was analyzed. Indeterminate subjects were analyzed to explore the model limitations. Performance of models derived with reduced numbers of input variables (missing data) was tested to explore its robustness in KD/FC classification.
Results
The demographic and clinical details of development and validation cohorts are presented in Table I. Asian patients were over-represented among KD subjects and underrepresented among FCs, compared with the San Diego population at large (12%). FCs had a clinically determined or culture-proven etiology for their febrile illnesses (Table II; available at www.jpeds.com). Viral diagnosis was established by viral culture, direct fluorescent antibody testing, or polymerase chain reaction assays. Viral syndrome was defined as a febrile illness that resolved without specific treatment and for which no specific pathogens could be identified.
Table 1.
Demographic characteristics of study cohorts
| Characteristics | Development cohort | Validation cohort | ||||
|---|---|---|---|---|---|---|
| KD (n=533) | FC (n=318) | P | KD (n=268) | FC (n=161) | P | |
| Age, months, median (IQRa) |
29.8 (15.8, 52.0) |
30.7 (15.4, 61.8) |
0.22b | 30.4 (16.8, 52.6) |
45.0 (18.9, 79.1) |
<0.001b |
| Males, n (%) | 337 (63.2) | 191 (60.1) | 0.38c | 157 (58.6) | 98 (61) | 0.69c |
| Race/ethnicity, n (%) | <0.001c | 0.03c | ||||
| African American | 22 (4.1) | 8 (2.5) | 11 (4.1) | 3 (2) | ||
| Native American | 2 (0.4) | 0 (0) | 1 (0.4) | 0 (0) | ||
| Asian | 91 (17.1) | 26 (8.2) | 45 (16.8) | 11 (7) | ||
| Caucasian | 120 (22.5) | 83 (26.1) | 72 (26.9) | 45 (28) | ||
| Hispanic | 175 (32.8) | 124 (39.0) | 84 (31.3) | 59 (37) | ||
| Mixed | 109 (20.5) | 60 (18.9) | 45 (16.8) | 32 (20) | ||
| Other/unknown | 14 (2.6) | 17 (5.3) | 10 (3.7) | 11 (7) | ||
Interquartile range
Rank sum test
Fisher’s exact test
Multivariate analysis and 2-step analyses of KD and FC
We compared KD and FC subjects using Fisher exact tests for categorical variables, and odds ratios and likelihood ratio tests for continuous variables (Tables III and IV; available at www.jpeds.com). Each sub-cohort had different statistically significant clinical variables in the univariate analysis and independent predictors in the multivariate analysis (Tables III and IV), supporting the need to develop models for each sub-cohort separately. The impacts of each variable to the classification decision in secondary models were measured by the percent increase of model mean square error due to the permutation of the variable values (Table V; available at www.jpeds.com).
By applying the previously derived primary LDA-based model and the score cutoffs that achieved 95% PPV and NPV(11), 90.0% (721 of 801) of KD subjects and 57.4% (275 of 479) of FC subjects were correctly classified; 0.1% (1 of 801) of KD subjects and 7.5% (36 of 479) of FC subjects were erroneously classified, and 9.9% (79 of 801) of KD subjects and 35.1% (168 of 479) of FC subjects were left indeterminate.
The secondary random forest models, applied to 4 sub-cohorts of remaining indeterminate subjects, correctly classified 60.8% (48 of 79) of KD subjects and 60.1% (101 of 168) of FC subjects. The secondary models erroneously classified 2.5% (2 of 79) of KD subjects and 6.0% (10 of 168) of FC subjects, and 36.7% (29 of 79) of KD and 33.9% (57 of 168) of FCs remained indeterminate.
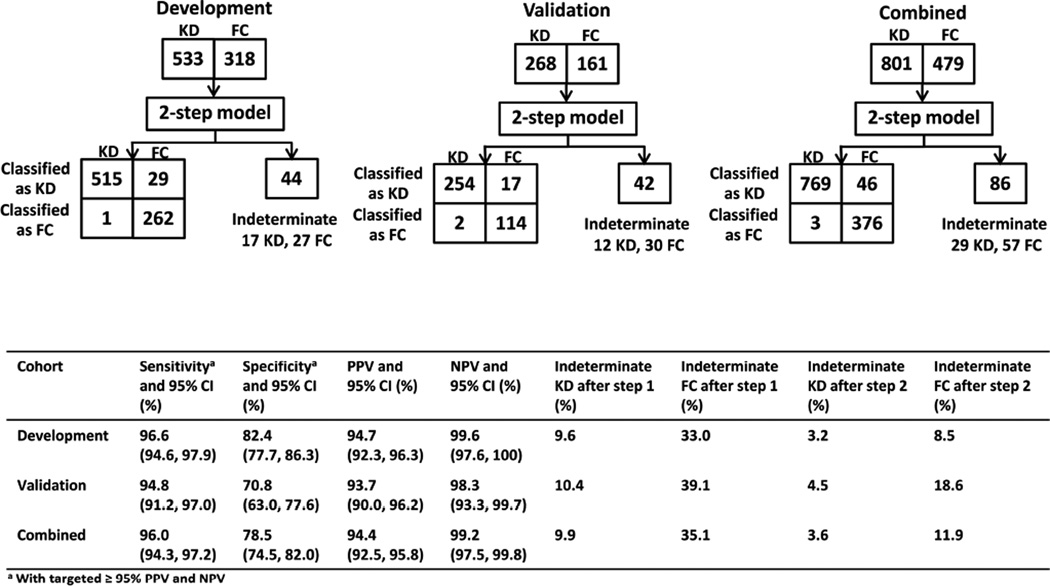
The 2-step algorithm correctly classified 96.0% (769 of 801) of KD subjects and 78.5% (376 of 479) of FC subjects (Figure 3), with targeted ≥ 95% PPV and NPV. Only 3.6% (29 of 801) and 11.9% (57 of 479) of KD and FC subjects remained indeterminate, whereas 9.9% of KD subjects and 35.1% of FC subjects were left indeterminate by the original LDA model.
Figure 3.
Diagnostic performance of the 2-step algorithm applied to the development and validation cohorts. Top: classification of subjects. Bottom: sensitivity, specificity, PPV, NPV, and proportions of subjects with indeterminate scores.
We compared the ability of the 2-step algorithm in terms of sensitivity, specificity, PPV, and NPV to the use of the AHA guidelines for KD diagnosis in the absence of echocardiography (Figure 4; available at www.jpeds.com). Results showed that the algorithm had a sensitivity of 96.0% versus AHA guidelines of only 72.2%. AHA guidelines had a higher specificity of 93.5% versus our specificity of 78.5%. However, when it came to PPV and NPV, the AHA guidelines and the 2-step algorithm had the same PPV around 95%, and our NPV was 99.2% whereas AHA guidelines had a NPV of 66.8%. Thus, use of the algorithm was better at picking up more patients with KD, and having a better NPV for patients without KD.
Algorithm performance in sub-cohorts stratified by age, illness day, and CRP
The diagnosis of KD in young infants can be particularly challenging. This algorithm performed well (Table VI; available at www.jpeds.com) among subjects ≤ 6 months of age (n=92; 69 KD and 23 FC subjects). The PPV and NPV for these infants were both 100%. The sensitivity was 97.1% and specificity was 87.0%. Only 2 (3%) KD subjects and 3 (13%) FC subjects were indeterminate. Among subjects > 6 months of age (n=1188; 732 KD and 456 FC subjects), the PPV, NPV, sensitivity, and specificity were slightly lower (93.9%, 99.2%, 95.9%, and 78.1%, respectively). The indeterminate frequency of these older KD and FC subjects were 3.7% (27 of 732) and 11.8% (54 of 456). The distribution of correctly classified, erroneously classified, and indeterminate subjects did not differ significantly among the two age groups (P = 0.12 by Chi-squared test). Importantly, the algorithm performed well in different age groups including the most vulnerable age group: patients under 6 months of age.
To determine the effect of duration of illness on algorithm performance, we divided the subjects into four sub-cohorts based on illness day (≤ 3 days [n=251]; 4–5 days [n=435]; 6–7 days [n=390]; 8–10 days [n=204]) and analyzed the algorithm’s performance for each sub-cohort (Table VI). PPVs, NPVs, and sensitivities remained similar (< 7% variation) among these sub-cohorts. The specificity levels decreased monotonically with illness duration from 85.7% the group of ≤ 3 days of illness to 61.2% the sub-cohort of 8–10 days of illness. The distribution of correctly classified, erroneously classified, and indeterminate subjects did not differ significantly among different sub-cohort of illness days (P = 0.27 by Chi-squared test).
In our study there were 242 of 479 (50.5%) FC subjects who had CRP values of at least 3.0 mg/dL. Of these FCs, the algorithm correctly classified 72.7% (176 of 242) of the subjects, erroneously classified 12.8% (31 of 242) of the subjects, and left 14.5% (35 of 242) of the subjects indeterminate. Of these 242 subjects, 61 subjects fulfilled the criteria for incomplete KD based on AHA guidelines. This algorithm correctly identified 42.6% (26 of 61) as FCs and left 41.0% (25 of 61) as indeterminate subjects requiring further evaluation. For the 237 FC subjects who had CRP values less than 3.0 mg/dL, the algorithm classified 84.4% (200) correctly as FC, 6.3% (15) erroneously as KD, and 9.3% (22) as indeterminate. Such results demonstrate the utility of the algorithm as a classification tool for front-line clinicians to evaluate suspected KD when echocardiography is not readily available.
Algorithm performance for subjects with incomplete KD
Of 801 KD subjects, 646 had complete KD, 155 met AHA criteria for incomplete KD with 62 showing coronary changes (57 had transiently dilated coronary arteries and 5 had aneurysms) on the initial echocardiogram. For the 93 incomplete KD subjects with normal echocardiograms, the algorithm classified 80.6% (75) correctly as KD, 1.1% (1) erroneously as FC, and 18.3% (17) as indeterminate. Compared with the original LDA model (26.9% indeterminate), the 2-step model improved the correct adjudication of incomplete KD by almost one-third.
Classification of KD with coronary artery abnormalities
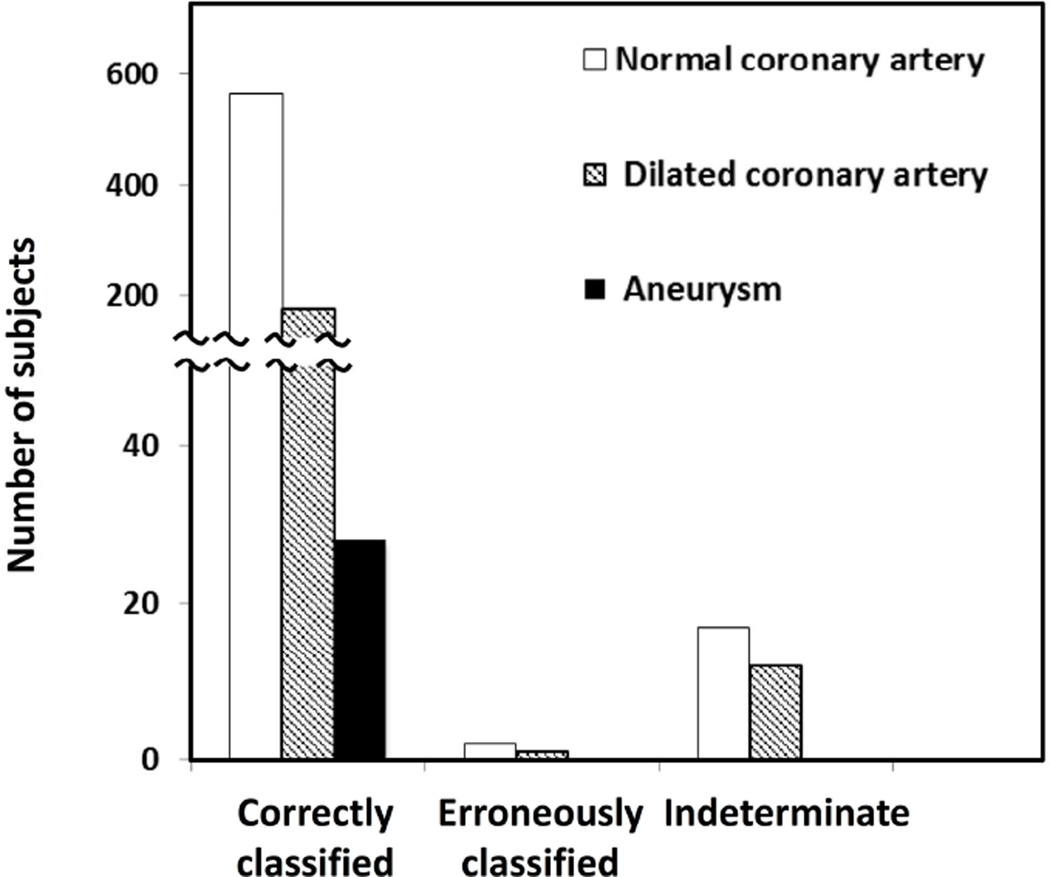
Because the prompt diagnosis of the subset of KD subjects who developed coronary artery aneurysms is of paramount importance, the model’s performance was separately evaluated for subjects in regard to coronary artery status (Figure 5). Of the 32 KD subjects who were erroneously classified or indeterminate, 26 had normal and 6 had transiently dilated coronary arteries (with worst Z-scores ranging from 2.66 to 4.35 for the LAD and/or RCA). Thus, the algorithm correctly classified all 28 subjects who developed aneurysms based on baseline clinical criteria and laboratory test results before the echo was done. The distribution of these 28 subjects over the three sub-cohorts was shown in Figure 6 (available at www.jpeds.com). Five of these 28 subjects manifested 2 or 3 criteria and were diagnosed by echocardiography. In addition, 57 subjects who manifested 2 or 3 criteria had dilation of the coronary arteries (Z score >2.5) and were diagnosed on this basis. The decision support tool correctly identified 81% (50 of 62) of subjects with incomplete KD diagnosed by echocardiographic criteria. Thus, the decision support tool could be used on the initial examination to improve the diagnosis of KD.
Figure 5.
Performance of the algorithm according to coronary artery status of subjects.
Erroneously classified and indeterminate subjects
Clinical and laboratory test variables were analyzed to profile the subjects with erroneous or indeterminate classification by the model. Of the 32 erroneously classified or indeterminate KD subjects, 30 manifested 3 or fewer KD principal criteria. Thus, the majority exhibited incomplete clinical characteristics at the time that the algorithm was applied. On the other hand, 70 of 103 erroneously classified or indeterminate FC subjects manifested 3 or more KD principal criteria. Distributions of the 12 laboratory test variables were compared among the correctly classified, erroneously classified, and indeterminate KD and FC subgroups (Figure 7; available at www.jpeds.com). The KD subjects erroneously classified as FC had laboratory test values comparable with those of correctly classified FCs. Conversely, the laboratory test values of indeterminate KD/FC subjects were intermediate to those of the correctly classified KD and FC subjects. Adenovirus is well-known to mimic many of the clinical and laboratory features of KD(12). For the 28 FC subjects with adenovirus infection documented either by culture, direct fluorescent antibody testing, or PCR in the validation cohort, the algorithm classified 57% (16) correctly as FC, 18% (5) erroneously as KD, and 25% (7) as indeterminate. Such clinical and laboratory test result patterns likely explain the misclassification by the algorithm.
Impact of variable reduction in algorithm performance
Because patients typically have incomplete data early in their evaluations, we studied the effect of eliminating variables, beginning with the least-weighted (Figure 8; available at www.jpeds.com). The frequency of certain classifications decreased with the reduction in variable number from 18 to 3. A 9-variable algorithm including 6 clinical variables (5 KD principal criteria plus illness days) and 3 laboratory variables (ZHgb, eosinophil percentage, and WBC count) had an 80% classification certainty rate for KD and FC subjects and 42% for incomplete KD subjects. In our study cohort, there were 228 of 801 KD subjects and 287 of 479 FC subjects having missing laboratory values. The impact of these subjects on algorithm performance was also explored (Appendix 2 and Table VII; Table VII available at www.jpeds.com). Missing laboratory data did not affect the algorithm performance and both the negative and positive predictive value was preserved (Table VII).
Discussion
The sequential use of a primary LDA-derived algorithm to perform initial classification and secondary decision-tree-based algorithms applied in parallel to sub-cohorts of indeterminate cases resulted in improved classification certainty in differentiating KD from clinically similar febrile illnesses. The diagnosis of patients with incomplete KD criteria is also challenging. The algorithm correctly classified 80.6% of subjects with incomplete KD who fulfilled AHA laboratory criteria. In contrast to the AHA algorithm that requires an echocardiogram as part of the evaluation, this algorithm is intended for use at the point of care in settings where echocardiography would not be readily available. The algorithm correctly classified 80.6% of KD subjects who manifested three or fewer KD principal criteria and were diagnosed by echocardiography. Furthermore, the algorithm correctly classified all 28 KD subjects who went on to develop the most severe complication, coronary artery aneurysms.
There are both strengths and limitations to our study. We enrolled well-characterized, phenotypically similar control subjects, of whom approximately one-half were referred to our ED specifically for evaluation of possible KD. Thus, we used development and validation cohorts that mirror the patient population for which a classification algorithm would be most useful. In addition, the algorithm used widely available laboratory tests coupled with easily observable clinical signs and can be adapted as a computer- or smart phone-based tool. Nonetheless, 3.6% and 11.9% of KD and FC subjects, respectively, remained indeterminate. The algorithm performed less well when ≤3 clinical criteria were present. Although subject age did not adversely affect algorithm performance, illness day did have an impact with a greater proportion of FC correctly identified early in the course of the illness. The algorithm had a higher sensitivity but lower specificity for subjects having 8 to 10 days of fever compared with those having 3 or fewer days of fever. The natural evolution of laboratory values in acute KD is for the inflammatory markers to diminish with time. Thus, by 8 to 10 days of fever, many of the key components of the algorithm were already starting to normalize, thus making some of the KD subjects look more like the FC(13). The goal of KD management is to treat with IVIG as soon as the diagnosis can be established. Thus, the fact that the algorithm performed well discriminating KD patients from FC in the early phase of their illness makes the algorithm more valuable as a tool to ensure timely diagnosis and treatment. Integration into this algorithm of additional biomarkers that better differentiate KD and FCs could help to improve its performance. Incorporating nuanced clinical data such as limbal sparing of conjunctival injection or perineal accentuation of rash could also result in better diagnostic performance. Those data were not captured for this study, however, as the intention was to computationally capture the differences between patients to provide support for the more inexperienced practitioner. In the absence of a diagnostic test for KD diagnosis, there is always a possibility that subjects were erroneously classified. Thus, the best diagnostic tool will only be as good as expert clinicians until the etiology of KD is discovered and specific diagnostic tests can be devised. Before this algorithm can be widely adopted, it must be evaluated as a clinical device by the Food and Drug Administration, which will require prospective testing in larger cohorts from different medical centers where the "gold standard", including the use of echocardiography on FC subjects, is established by other experts. The detailed algorithm will be made available to interested investigators upon request.
Supplementary Material
Acknowledgments
Supported by the American Heart Association (to H.C. and X.L.), Stanford University Spark Program (H.C. and X.L.), the David Gordon Louis Daniel Foundation (to J.B.), the Mario Batali Foundation (J.B.), the National Institutes of Health, National Heart, Lung, Blood Institute (HL69413 [to JCB]), the Hartwell Foundation (to A.T.), and the Harold Amos Medical Faculty Development Program/Robert Wood Johnson Foundation (to A.T.).
We thank our colleagues at the Stanford University Pediatric Proteomics Group for critical discussions, and the Stanford University IT group for Linux cluster support and assistance in data analysis and software development. We also thank Joan Pancheri, RN, BSN, for data collection and assistance with patient enrollment.
Abbreviations
- AHA
American Heart Association
- ALT
alanine aminotransferase
- CRP
C-reactive protein
- ED
Emergency Department
- ESR
erythrocyte sedimentation rate
- GGT
gamma-glutamyl transferase
- ZHgb
hemoglobin concentration normalized for age
- IVIG
intravenous immunoglobulin
- KD
Kawasaki disease
- LAD
left anterior descending
- LDA
linear discriminant analysis
- RCA
right coronary artery
- WBC
white blood cell count
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- 1.Newburger JW, Takahashi M, Gerber MA, Gewitz MH, Tani LY, Burns JC, et al. Diagnosis, treatment, and long-term management of Kawasaki disease: a statement for health professionals from the Committee on Rheumatic Fever, Endocarditis and Kawasaki Disease, Council on Cardiovascular Disease in the Young, American Heart Association. Circulation. 2004;110:2747–2771. doi: 10.1161/01.CIR.0000145143.19711.78. [DOI] [PubMed] [Google Scholar]
- 2.Yellen ES, Gauvreau K, Takahashi M, Burns JC, Shulman S, Baker AL, et al. Performance of 2004 American Heart Association recommendations for treatment of Kawasaki disease. Pediatrics. 2010;125:e234–e241. doi: 10.1542/peds.2009-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rowley AH, Gonzalez-Crussi F, Gidding SS, Duffy CE, Shulman ST. Incomplete Kawasaki disease with coronary artery involvement. The Journal of pediatrics. 1987;110:409–413. doi: 10.1016/s0022-3476(87)80503-6. [DOI] [PubMed] [Google Scholar]
- 4.Manlhiot C, Christie E, McCrindle BW, Rosenberg H, Chahal N, Yeung RS. Complete and incomplete Kawasaki disease: two sides of the same coin. European journal of pediatrics. 2012;171:657–662. doi: 10.1007/s00431-011-1631-2. [DOI] [PubMed] [Google Scholar]
- 5.Ha KS, Jang G, Lee J, Lee K, Hong Y, Son C, et al. Incomplete clinical manifestation as a risk factor for coronary artery abnormalities in Kawasaki disease: a meta-analysis. European journal of pediatrics. 2013;172:343–349. doi: 10.1007/s00431-012-1891-5. [DOI] [PubMed] [Google Scholar]
- 6.Ling XB, Kanegaye JT, Ji J, Peng S, Sato Y, Tremoulet A, et al. Point-of-care differentiation of Kawasaki disease from other febrile illnesses. The Journal of pediatrics. 2013;162:183–188. e3. doi: 10.1016/j.jpeds.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling XB, Lau K, Kanegaye JT, Pan Z, Peng S, Ji J, et al. A diagnostic algorithm combining clinical and molecular data distinguishes Kawasaki disease from other febrile illnesses. BMC Med. 2011;9:130. doi: 10.1186/1741-7015-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 9.Oza NC, editor. 2006. Ensemble data mining methods. [Google Scholar]
- 10.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 11.Breiman L. Bagging predictors. Machine Learning. 1996;24:123–140. [Google Scholar]
- 12.Jaggi P, Kajon AE, Mejias A, Ramilo O, Leber A. Human adenovirus infection in Kawasaki disease: a confounding bystander? Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2013;56:58–64. doi: 10.1093/cid/cis807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tremoulet AH, Jain S, Chandrasekar D, Sun X, Sato Y, Burns JC. Evolution of laboratory values in patients with Kawasaki disease. Pediatr Infect Dis J. 2011;30:1022–1026. doi: 10.1097/INF.0b013e31822d4f56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.