Abstract
Even though MHC class Ia and many Ib molecules have similarities in structure, MHC class Ib molecules tend to have more specialized functions, which include the presentation of non-peptidic antigens to non-classical T cells. Likewise, non-classical T cells also have unique characteristics, including an innate-like phenotype in naïve animals and rapid effector functions. In this review, we discuss the role of MAIT and NKT cells during infection, but also the contribution of less studied MHC class Ib-restricted T cells such as Qa-1-, Qa-2-, and M3-restricted T cells. We focus on describing the types of antigens presented to non-classical T cells, their response and cytokine profile following infection, as well as the overall impact of these T cells to the immune system.
Keywords: Non-classical T cells, Infectious diseases, MHC class Ib, NKT & MAIT cells
A. Introduction
The innate and adaptive branches of the immune system are not mutually exclusive, and there is a growing interest in innate-like T cells, many of which are restricted by non-classical MHC molecules. Cytotoxic CD8+ T cells are restricted by MHC class I molecules and categorized into two groups: classical MHC class Ia and non-classical MHC class Ib. MHC class Ia molecules are highly polymorphic and encoded in the Mhc locus by H-2K, H-2D, and H-2L in mice and HLA-A, HLA-B, and HLA-C in humans. The MHC class Ib family has evolved more diverse, and specialized, functions than their classical counterparts. These molecules are predominantly found in the H2-Q, H2-T, and H2-M regions in mice and HLA-E, HLA-F, and HLA-G in humans within the Mhc locus. Many are capable of presenting antigens, while others are incapable of antigen binding or participate in responses outside of the immune system. For example, murine M1 and M10 bind to V2R G protein-coupled receptors and play a role in pheromone detection (Loconto et al. 2003), while ZAG in humans and mice binds fatty acids and polyethylene glycol, contributing to lipid metabolism (Delker et al. 2004; Hirai et al. 1998). MHC class Ib molecules are also well known to interact with NK cell receptors (Braud et al. 1998; Lee et al. 1998b; Vance et al. 1999; Vance et al. 1998). Some have proposed to classify MHC class Ib molecules according to their age, such as ‘Young,’ ‘Middle-aged,’ and ‘Old’ (Rodgers and Cook 2005). For example, ‘Old’ genes diverged during early vertebrate evolution and many members of this subset fall outside the Mhc gene locus in humans and rodents, such as CD1, MR1, and HFE (Rodgers and Cook 2005). Generally, non-classical MHC molecules present a more diverse array of antigens, e.g. CD1 presents glycolipid antigens (Beckman et al. 1994), MR1 presents Vitamin B metabolites (Kjer-Nielsen et al. 2012), and M3 presents formylated peptides (Smith et al. 1994) (Table 1). These molecules also tend to have more restricted tissue localization, lower expression at the cell surface, limited polymorphism, and shorter cytoplasmic tails (Stroynowski and Lindahl 1994). In this review, we will discuss MHC class Ib-restricted T cell responses in humans and mice in the context of infection. We will focus on 1) the antigens (or lack thereof) presented by this family of molecules, 2) the cytokine profile of MHC class Ib-restricted T cells, and 3) the overall contribution of non-classical T cells to the immune response (Table 2).
Table 1.
MHC class Ib molecules that participate in TCR mediated responses
| Mouse | Human | Mhc locus | TCR-interaeting ligand |
|---|---|---|---|
| CD1d | CD1a, CD1b, CD1c, CD1d, CD1e | No | Lipids |
| MR1 | MR1 | No | Vitamin B derivatives |
| HFE | HFE | No | Transferrin receptors |
| H2-M3 | - | Yes | N-formylated peptides |
| Qa-1 | HLA-E | Yes | Peptides |
| TL | - | Yes | None |
| T10/T22 | - | Yes | None |
| Qa-2 | - | Yes | Peptides |
| - | HLA-G | Yes | Peptides |
Table 2.
The antimicrobial response of MHC class Ib-restricted T cells in mice and humans
| T cell | Positive Selection | Recognized Antigens | PIZF | Cytokine Prolife |
|---|---|---|---|---|
| Group 1 CD 1-restricted T cells | HC | Mtb lipids | Yes | IFN-γ, TNF-α, IL-6, IL-13 |
| Type I NKT cells | HC | α-Galcer, microbial lipids | Yes | IFN-γ, TNF-α, IL-4, IL-13, IL-17 etc. |
| Type II NKT cells | HC | Sulfatide, glycolipids and phospholipids | Yes | IFN-γ, TNF-α, IL-17 |
| MAIT cells | HC | Bacterial and fungal riboflavin metabolites | Yes | IFN-γ, TNF-α, IL-2, IL-17 |
| HFE-restricted T cells | ND | N/A | No | IL-6, IL-10, hepcidin |
| M3-restricted T cells | HC or TEC | Bacterial and fungal N-formylatcd peptides | No | IFN-γ , TNF-α |
| Qa-1/HLA-E-restricted T cells | HC or TEC | CMV- and GroEL-derived peptides | ND | IFN-γ, TNF-α, IL-4, IL-5, IL-10, IL-13 |
| TL-restricted T cells | ND | N/A | ND | IFN-γ |
| Q9-restricted T cells | ND | MPyV-derived peptide | ND | IFN-γ, TNF-α |
| HLA-G-restricted T cells | ND | HCMV-derived peptide | ND | ND |
HC: hematopoietic cell; TEC: thymic epithelial cell; ND; no data
B. Positive selection of non-classically restricted CD8+ T cells
Non-classical CD8+ T cells often have an innate-like phenotype, which includes increased expression of CD44 and decreased CD62L expression (Jay et al. 2008; Kurepa et al. 2003). It has been proposed that this results from unusual positive selection. For example, the conditions that T10- and T22-restricted γδ T cells undergo positive selection affect their effector phenotype. Cells that develop in the presence of T22 are able to produce IFN-γ, whereas antigen-naïve T10- and T22-reactive γδ T cells during development produce IL-17 (Jensen et al. 2008). Thymic epithelial cells (TECs) are essential for the positive selection of conventional T cells (Anderson et al. 1994). On the other hand, invariant natural killer T (iNKT) cells are selected by CD4+CD8+ double positive (DP) cortical thymocytes that present CD1d (Bendelac 1995). Similarly to iNKT cells, MAIT cells are also selected by hematopoietic cells (HCs) (Treiner et al. 2003), which are DP thymocytes expressing MR1 (Seach et al. 2013). MHC class Ib-restricted CD8+ T cells that are specific for Listeria monocytogenes antigens are also selected for by HCs, whereas MHC class Ia-restricted T cells are inadequately selected (Urdahl et al. 2002). Interestingly, it was recently shown that M3-restricted T cells could be selected by TECs or HCs, but that the selecting cell type played a role in their phenotype (Chiu et al. 1999b; Cho et al. 2011). Cells selected by HCs acquired enhanced effector functions (Cho et al. 2011). Similarly, using transgenic mice possessing a TCR specific for a Qa-1- presented insulin-derived peptide, it was determined that either TECs or HCs selected Qa-1-restricted CD8+ T cells (Sullivan et al. 2002). However, in contrast to M3’s role during positive selection, there was no observable difference in phenotype between these two differentially selected populations (Sullivan et al. 2002). Altogether, these findings demonstrate that individual MHC class Ib-restricted T cell populations have unique requirements for positive selection.
C. MHC class Ib-restricted CD8+ T cells and their participation during infection
The CD1-restricted family of T cells
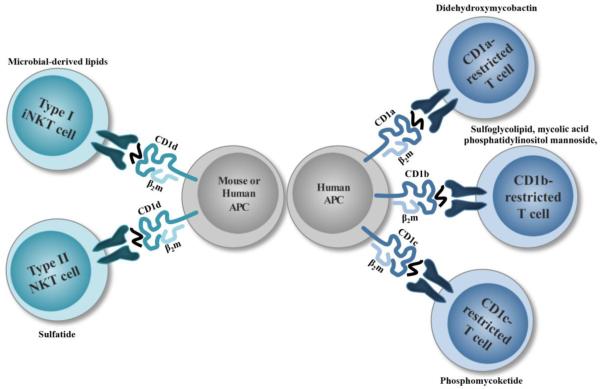
The CD1 locus is not linked to the Mhc locus. CD1 presents self and foreign lipid antigens to multiple CD1-restricted T cell populations, rather than peptides (Figure 1) (Beckman et al. 1994). Five isoforms of human CD1 are expressed, which are organized into three groups: Group 1 CD1 (CD1a, b, c), Group 2 CD1 (CD1d), and Group 3 CD1 (CD1e). T cells are able to directly recognize lipids presented by all CD1 isoforms except CD1e, which is thought to aid CD1b in ligand processing and presentation (de la Salle et al. 2005). Unlike MHC class Ia molecules, the transporter associated with antigen processing (TAP) is not required for CD1 antigen loading (Brutkiewicz et al. 1995; Hanau et al. 1994). CD1d is the only isoform expressed in rodents. There are two types of Group 2 CD1-restricted T cells, type I iNKT cells and type II NKT cells, which were initially classified based on TCR diversity (Cardell et al. 1995). iNKT cells participate during a variety of infectious diseases and can be activated in a TCR-dependent or TCR-independent manner by responding to environmental cytokines, like IL-12 and IL-18, rather than ligand stimulation (Leite-De-Moraes et al. 1999). This is evident following murine cytomegalovirus (MCMV) infection, where iNKT cells become activated in an IL-12-dependent manner, and is partially contingent on type I interferons (Holzapfel et al. 2014; Tyznik et al. 2014; Wesley et al. 2008). On the other hand, type II NKT cell activation mainly occurs in a TCR-dependent manner to self-glycolipids of self-phospholipids, whose antigen repertoire can be either exclusive or promiscuous (Jahng et al. 2004; Tatituri et al. 2013). Type II NKT cells appear to have opposing roles, capable of participating in protective responses or promoting pathology. However, the information about this subset has remained limited because type II NKT cells cannot be labeled as a single population with CD1d tetramers like iNKT cells. In addition, early studies to determine the functions of type II NKT cells using Jα18−/− mice may need to be revisited, due to an impaired TCR repertoire of the original mice (Bedel et al. 2012).
Figure 1. The family of CD1-restricted αβ T cells.
Group 1 CD1-specific T cells are depicted with examples of Mtb glycolipid antigens.
iNKT cells respond to CD1d-presented ligands derived from a number of bacteria and even protozoa, including: bacteroides fragilis-derived sphingolipid α-galactosylceramide (An et al. 2014; Wieland Brown et al. 2013); borrelia burgdorferi-derived galactosyl diacylglycerol (Kinjo et al. 2006); helicobacter pylori-derived cholesteryl α-glucoside (Ito et al. 2013); Sphingomonas bacteria-derived α-linked galacturonic acid (Kinjo et al. 2005); Streptococus pneumoniae-derived glycolipids containing diacylglycerol (Kinjo et al. 2011); and lipopeptidophosphoglycan derivatives from the protozoan entamoeba histolytica (Lotter et al. 2009). Human and murine iNKT cells also respond to M. tuberculosis (Mtb) phosphatidylinositol mannoside (PIM) ligands (Fischer et al. 2004). However, iNKT cells are dispensable during mycobacterial infection, as illustrated using CD1d−/− animals (Behar et al. 1999). In contrast, iNKT cells are physiologically relevant for clearance and protection against other pathogenic microorganisms. iNKT cells produce IFN-γ in response to B. burgdorferi infection in vivo and both CD1d-deficient animals (Kumar et al. 2000) and Jα18−/−BALB/c mice were more susceptible to infection, developing chronic joint inflammation and arthritis (Tupin et al. 2008). iNKT cell participation is also implied during E. histolytica infection, which causes increased amebic liver abscesses in the absence of iNKT cells in Jα18−/− mice (Lotter et al. 2006) and CD1d−/− mice, (Lotter et al. 2009). Jα18−/− animals have also been reported to be susceptible to Streptococcus infections (Kawakami et al. 2003). Predictably, due to expression of CD1d ligands the CD1d-restricted response was shown to be necessary for protection against S. pneumoniae and Group B Streptococcus through IFN-γ and IL-17 production in the lung (Kinjo et al. 2011). This CD1d restricted response was validated using Nur77GFP transgenic mice, which upregulate GFP following TCR engagement (Holzapfel et al. 2014). Unexpectedly however, although it was shown that iNKT cells respond to S. typhimurium infection in a CD1d restricted manner (Brigl et al. 2003), iNKT cells produced IFN-γ without TCR engagement (Holzapfel et al. 2014).
In contrast to type I and II NKT cells, investigations into Group 1 CD1-restricted T cell responses have mainly focused on mycobacterial infection. CD1a, CD1b, and CD1c molecules present different types of glycolipids, owing to structural differences in their antigen-binding grooves (Gadola et al. 2002; Scharf et al. 2010; Zajonc et al. 2005). Circulating CD1-restricted T cells are observed in patients previously infected with Mtb or immunized with Mycobacterium bovis bacillus Calmette-Guerin (BCG) (Kawashima et al. 2003; Ulrichs et al. 2003). These Mycobacterium-specific T cells are capable of producing IFN-γ ex vivo and recognize M. bovis BCG-infected cells (Kawashima et al. 2003). Interestingly, there is also a small population of CD1b-restricted germline-encoded, mycolyl lipid-reactive (GEM) T cells present in uninfected patients (Van Rhijn et al. 2013). To counteract the lack of an animal model to study Group 1 CD1 molecules in vivo, human group 1 CD1 transgenic (hCD1Tg) mice were generated, which express all Group 1 CD1 isoforms (Felio et al. 2009). Mtb infection and immunization of hCD1Tg mice are both capable of inducing a CD1-restricted T cell response, characteristic of classical T cells; this includes a slow primary response to immunization and rapid secondary response (Felio et al. 2009). Importantly, in hCD1Tg mice expressing a mycolic acid-specific TCR transgene, immune protection against Mtb was observed (Zhao et al. 2015). A second group investigated the CD1 repertoire in a humanized mouse model using NSG mice engrafted with human fetal thymus and fetal liver, as well as CD34+ hematopoietic cells (Lockridge et al. 2011). CD1a, CD1b, CD1c, and CD1d were all expressed in these animals, and Group 1 CD1-restricted T cells were present (Lockridge et al. 2011), though their response following Mtb infection still remains to be seen. To date, Group 1 CD1 molecules have been shown to present eight Mycobacterium-derived ligands, the majority of which are loaded in CD1b (Siddiqui et al. 2015). However, a role for Group 1-restricted T cell populations during other infections has not been reported.
MR1-restricted mucosal associated invariant T (MAIT) cells
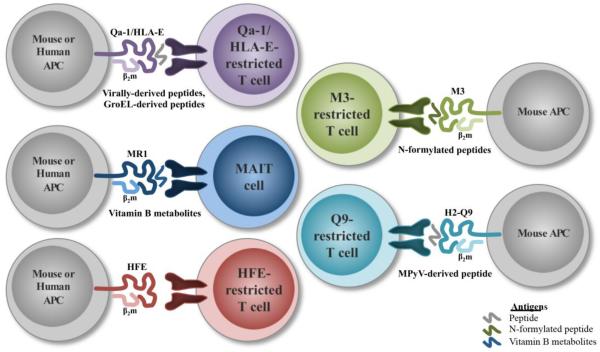
MAIT cells were first described in 1993 (Porcelli et al. 1993), but it was not until six years later that they were recognized as a distinct population (Figure 2) (Tilloy et al. 1999). MAIT cells are unique innate-like T cells found at mucosal sites and in the circulation of humans and mammals. MAIT cells are restricted by MR1 (MHC-related protein 1) (Treiner et al. 2003), which is encoded outside the Mhc gene locus. There is 90% sequence homology between MR1 in humans and mice (Riegert et al. 1998). Wild-type mice generally have low frequencies of MAIT cells (Rahimpour et al. 2015), but they are more abundant in humans and make up approximately 1-4% of circulating T cells (Martin et al. 2009). The antimicrobial role of MAIT cells was first alluded to based on their absence in germ-free (GF) mice (Treiner et al. 2003), however they can successfully expand in GF mice following inoculation with a single bacterial species, i.e. Bacteroides thetaiotaomicron, Bifidobacterium animalis, Enterobacter cloacae, or Lactobacillus casei (Le Bourhis et al. 2010). Two groups then found that MAIT cells are able to respond to a number of bacterial and fungal species in vitro using infected PMDCs or BMDCs, but not to viruses, e.g. Lactobacillus acidophilus, Mtb, Pseudomonas aeroginosa, Salmonella typhimurium, Staphylococcus aureus, Saccharomyces cerevisiae, and Candida albicans (Gold et al. 2010; Le Bourhis et al. 2010). A major breakthrough in this field was the discovery that MR1 presents Vitamin B metabolites, such as Vitamin B2 (riboflavin) and Vitamin B9 (folic acid) derivatives (Kjer-Nielsen et al. 2012). MR1 is able to bind and stabilize the unstable intermediates of the riboflavin biosynthesis pathway for presentation (Corbett et al. 2014). Interestingly, these ligands can be activating or non-activating in nature by presenting riboflavin or folic acid derivatives, respectively (Kjer-Nielsen et al. 2012).
Figure 2. Examples of non-classical αβ T cell populations during microbial infection.
MAIT cells are now thought to be involved in the early control of a number of bacterial pathogens. Following up on the observation that β2m−/− mice are more vulnerable to Klebsiella pneumonia than wild-type mice (Cogen and Moore 2009), Georgel et al. determined that MR1−/− mice also had increased susceptibility compared to MR1-sufficient animals (Georgel et al. 2011). MAIT cells also robustly expand in the lungs of mice infected with the live vaccine strain (LVS) of Francisella tularensis in an MR1- and IL-12p40-dependent manner (Meierovics et al. 2013). Their expansion inversely correlated with bacterial burden, and was accompanied by the production of IFN-γ, TNF-α, and IL-17A (Meierovics et al. 2013). Interestingly, MAIT cells were also observed to contribute during chronic infection of F. tularensis LVS, even when classical CD4+ and CD8+ T cells were recruited, but were insufficient for bacterial clearance alone (Meierovics et al. 2013). MR1−/− mice also have increased bacterial burden on Day 10 following M. bovis BCG infection, compared to wild-type mice (Chua et al. 2012). This disparity was no longer observed at later time points (Day 30), illustrating the importance of MAIT cells during early immunological control (Chua et al. 2012). Interestingly, this was suggested to be an MR1-independent, but IL-12-dependent response (Chua et al. 2012). In contrast, it appears that human MAIT cells require MR1 for appropriate IFN-γ production in response to Mtb infected APCs (Gold et al. 2010; Le Bourhis et al. 2010). Many studies have observed decreased numbers of circulating MAIT cells in tuberculosis (TB) patients (Gold et al. 2010; Le Bourhis et al. 2010). The remaining MAIT cell population in patients with active TB produced significantly more IFN-γ and TNF-α in response to BCG and decreased cytokine production following E. coli infection (Jiang et al. 2014). This suggests that MAIT cells are enriched in this environment to respond to Mtb (Jiang et al. 2014). In support of this concept, it was shown that the heterogeneity of the MAIT cell TCR repertoire might allow for pathogen specificity (Eckle et al. 2014; Gold et al. 2014). Cell lines expressing MAIT cell TCRs also become activated in an MR1-dependent manner following S. enterica serovar Typhimurium infection (Reantragoon et al. 2012). Additionally, human MAIT cells produce IFN-γ, TNF-α, and IL-2 following incubation with either S. typhimurium supernatant or the synthetic riboflavin derivative 6-hydroxymethyl-8-D-ribityllumazine (rRL-6-CH2OH) in the presence of MR1-expressing APCs (Reantragoon et al. 2013). Overall however, the functional role of MAIT cells is more ambiguous in humans than mice.
The MAIT cell field is relatively young, but has recently burgeoned due to the development of MR1 tetramers (Reantragoon et al. 2013). Nevertheless, a number of lingering questions remain. For instance, the role of MAIT cells during bacterial infection has been relatively well documented, however the in vivo role of MR1- restricted T cells has not been well defined for yeast. There is also the potential that MR1 could bind and present additional ligands to Vitamin B derivatives. Finally, it appears that MAIT cell activation can occur in a TCR-independent manner, similarly to iNKT cells, irrespective of bacterial/fungal riboflavin metabolism (Chua et al. 2012; Meierovics et al. 2013; Ussher et al. 2014). This opens up potential avenues to study MAIT cell responses during viral infections and autoimmune disorders, such as HIV (Fernandez et al. 2015; Leeansyah et al. 2013), multiple sclerosis (Treiner and Liblau 2015), inflammatory bowel disease (Treiner 2015), and Celiac disease (Dunne et al. 2013).
HFE-specific CD8+ T cells
HFE, or human hemochromatosis protein, was first discovered due to its association with hereditary hemochromatosis (HH) patients, a genetic disorder that results in iron overload (Feder et al. 1996). The HFE heavy chain forms a noncovalent bond with an associated β2m light chain (Feder et al. 1996; Feder et al. 1997), similarly to many other MHC class Ib molecules. However, in one common HFE mutation seen in HH patients, the C282Y mutation, the ability to bind β2m is disrupted. This prevents HFE expression at the cell surface by perturbing a critical disulfide bridge in the α3 domain (Feder et al. 1997). HFE-deficient and β2m-deficient mice both recapitulate the HH phenotype (Santos et al. 1996; Zhou et al. 1998). Even though HFE is structurally similar to other MHC class I molecules, its peptide-binding groove does not support antigen binding (similarly to TL, see below). This is due to the α1 and α2 domains being in closer proximity, since the α1 helix has a 4Å translocation towards the α2 domain this results in a narrower groove (Lebron et al. 1998). Rather than antigen binding, HFE is predominantly known for associating with the transferrin receptors to regulate iron homeostasis (Goswami and Andrews 2006; Lebron et al. 1998; Parkkila et al. 1997). However, there is evidence that suggests a potential immunological role for HFE as well. For example, the iron overload phenotype is even more pronounced in mice that are deficient for both β2m and RAG1, compared to β2m−/− animals (Santos et al. 2000). HFE-deficient animals on a RAG1 background also have increased iron overload, compared to HFE−/− animals (Miranda et al. 2004).
A role for CD8+ T cells was initially proposed because many HH patients have unusually small CD8+ T cell populations in circulation (Macedo et al. 2010; Porto et al. 1994). However, the exact nature of this deficiency is unclear, as it could be an indirect result of iron overload or a direct result of HFE regulating CD8+ T cells (Costa et al. 2015; Reuben et al. 2014). Interestingly, HFE is thought to impede activation via its α1 and α2 helixes by influencing MHC class I antigen processing and presentation (Reuben et al. 2014). Rohrlich et al. also showed that HFE influences the TCR repertoire, as evidenced by a decreased number of Vα6 TCRs in HFE-deficient mice (Rohrlich et al. 2005). Importantly, a subset of CD8+ T cells directly recognize HFE via their TCR and produce IL-6, IL-10, and hepcidin (Boucherma et al. 2012; Rohrlich et al. 2005). Overall, although HFE is not capable of binding and presenting antigens, there is evidence for an immunological role of HFE. Additional studies will be necessary to investigate the functions of HFE-reactive CD8+ T cells, as well as the immune system’s role in iron metabolism.
TL-restricted CD8+ T cells
Thymus leukemia antigen (TL) was first discovered during the development of spontaneous or radiation induced leukemia (Old and Boyse 1963) and subsequently mapped to the Mhc locus (Boyse et al. 1964). TL is encoded by the H2-T3 and H2-T18 genes in mice and is considered an ancient MHC class Ib gene that diverged over 100 million years ago (Davis et al. 2002). There is no human homologue for TL, but it has been suggested that HLA-G is a functional homologue (Attinger et al. 2005; Huang et al. 2011). TL is expressed on intestinal epithelial cells (IELs) (Hershberg et al. 1990; Wu et al. 1991) and immature thymocytes of certain mouse strains (Chen et al. 1985). Following activation, T cells and APCs also express TL (Cook and Landolfi 1983; Madakamutil et al. 2004). T cells can also “snatch” TL from intestinal epithelial cells to present it on their cell surface (Pardigon et al. 2006). The expression of TL at the cell surface is dependent on β2m (Yokoyama et al. 1982), but not on peptide binding (Weber et al. 2002). Even though TL exhibits approximately 70% identity with MHC class Ia molecules, its peptide-binding groove is closed due to the α1 helix being 7Å closer to the α2 helix (Liu et al. 2003). TL cell surface expression is also TAP-independent, which distinguishes it from classical MHC class Ia molecules (Holcombe et al. 1995; Rodgers et al. 1995). TL binds the CD8αα homodimer with higher affinity than to CD8αβ (Leishman et al. 2001; Tsujimura et al. 2001). This is a result of three exposed amino acids in the α3 helix of TL (Attinger et al. 2005). In contrast, MHC class Ia molecules have comparable affinities to CD8αβ and CD8αα (Kern et al. 1999). Unlike CD8αβ heterodimers (Bosselut et al. 1999), the CD8αα homodimer does not act as a co-receptor, rather it inhibits activation by acting as a co-repressor (Cheroutre and Lambolez 2008). CD8αβ T cells that co-express CD8αα are abundant in the intestinal mucosa. However, TL is not required for the formation of CD8+ T cell memory (Williams and Bevan 2005). It has also been shown that TL is important for controlling IEL function, for example inhibiting IEL proliferation (Olivares-Villagomez et al. 2008). Both αβ (Morita et al. 1994) and γδ TCRs (Tsujimura et al. 1996) can recognize TL. These TL-specific cytotoxic CD8+ T cells recognize the α1 and α2 domains of TL with CD8αα helping to stabilize the TL/TCR interaction (Tsujimura et al. 2003) in a TAP-independent mechanism (Tsujimura et al. 2000). Perhaps not surprisingly, due to its closed binding groove, the role of TL-restricted T cells during infectious disease clearance is limited.
M3-specific CD8+ T cells
In contrast to MHC class Ia molecules and many MHC class Ib molecules, M3 binds N-formylated peptides (Shawar et al. 1993; Smith et al. 1994). Thus, M3 is able to present peptides of prokaryotic or mitochondrial origin (Wang et al. 1991). There is also evidence that M3 can bind non-formylated peptides (Byers and Fischer Lindahl 1998), however M3 binds N-formylated peptides with much higher affinity than unformylated ones (Smith et al. 1994). The crystal structure of M3 showed that the specificity for N-formylated peptides was a result of alterations in its peptide-binding groove (Wang et al. 1995). There are very few endogenous N-formylated peptides available, which results in low M3 expression at the cell surface (Levitt et al. 2001), and sequesters M3 in the endoplasmic reticulum. TAP is required for M3 stabilization in the ER, while tapasin is necessary for intracellular peptide loading (Chun et al. 2001a). Dependency on TAP further differentiates M3 from other MHC class Ib molecules like CD1 and TL (Brutkiewicz et al. 1995; Hanau et al. 1994; Holcombe et al. 1995; Rodgers et al. 1995). However, both TAP-dependent and -independent presentation have been observed for L. monocytogenes peptides (Rolph and Kaufmann 2000). Exogenous antigens are capable of inducing M3 expression at the cell surface, unlike lowered temperatures (Chiu et al. 1999a), which occurs with MHC class Ia molecules (Ljunggren et al. 1990).
M3-restricted CD8+ T cells specific to a number of intracellular pathogens have been characterized, e.g. Mtb, L. monocytogenes, Chlamydia pneumonia, and S. enterica serovar Typhimurium (Chun et al. 2001b; Gulden et al. 1996; Lenz et al. 1996; Princiotta et al. 1998; Tvinnereim and Wizel 2007; Ugrinovic et al. 2005). The most well studied M3-restricted CD8+ T cells are specific to L. monocytogenes. Using MHC class Ia-deficient (KbDb−/−) mice, it has been shown that M3-restricted CD8+ T cells are sufficient to protect against L. monocytogenes infection (D'Orazio et al. 2003; Seaman et al. 1999). In this context, M3 presents three L. monocytogenes-derived peptides: Attm (f-MIVTLF) (Princiotta et al. 1998), Fr38 (f-MIVIL) (Gulden et al. 1996), and LemA (f-MIGWII) (Lenz et al. 1996). However, M3-restricted CD8+ T cells are not required for protection against L. monocytogenes (D'Orazio et al. 2006). Nevertheless, using M3-deficient animals it was determined that the M3-restricted and MHC class Ia-restricted immune responses are not redundant (Xu et al. 2006). The significance of M3-restricted memory CD8+ T cells during L. monocytogenes infection is more ambiguous. In contrast to primary infection, after secondary infection M3-restricted CD8+ T cells do not significantly expand (Kerksiek et al. 1999). It has been suggested that rather than contributing to a memory phenotype during a secondary infection, M3-restricted CD8+ T cells are already in a memory state in naïve animals, due to interactions with cross-reactive antigens from commensal bacteria (Lenz and Bevan 1997). Alternatively, it has been proposed that M3-restricted memory CD8+ T cells are constrained in the presence of MHC class Ia-restricted memory cells (Hamilton et al. 2004). M3 also appears to contribute to the immune response against MTB infection. Chun et al. showed that the Mtb genome contained a number of N-formylated peptides capable of binding to M3, and that M3-restricted CD8+ T cells can recognize a number of these peptides in mice (Chun et al. 2001b). Immunization of mice with dendritic cells pulsed with an N-formylated Mtb peptide are also able to elicit an H2-M3-mediated response (Doi et al. 2007). However, although MHC class Ib-restricted T cells accumulate in the lung, they provide minimal protection against Mtb infection (Urdahl et al. 2003). Further investigations are necessary to determine whether N-formylated peptides from other bacterial species elicit an M3-specific response.
CD8+ T cells-restricted by Qa-2 and HLA-G
The H2-Q6, -Q7, -Q8, and -Q9 genes in mice encode Qa-2. It is thought that the Qa-2 region resulted from a series of gene pair duplications, for example H2-Q7 and H2-Q9 are nearly identical, with over 99% homology (Devlin et al. 1985). Originally, Qa-2 was thought to have a restricted peptide repertoire (Rotzschke et al. 1993), however it is capable of binding a diverse array of endogenous and foreign peptides (Joyce et al. 1994; Tabaczewski et al. 1997). The crystal structure of Q9 revealed that it associates with β2m, and the peptide-binding groove is more hydrophobic and shallower than classical MHC molecules, which could play a role in its promiscuous peptide repertoire (He et al. 2001). Qa-2 is unique among MHC molecules for being anchored to the cell membrane by a glycophosphatidyl inositol (GPI) linker (Stroynowski et al. 1987), which is necessary for T cell activation (Robinson et al. 1989). In addition, there are soluble and membrane-linked forms of Qa-2, both of which require TAP (Tabaczewski and Stroynowski 1994), that arise because of alternative splicing or cleavage post-translation (Tabaczewski et al. 1994). However, the α3 domain of Qa-2 is unable to effectively interact with CD8 to appropriately activate cytotoxic T cells (Teitell et al. 1993).
There is evidence that Qa-2 participates in resistance to the murine parasite Taenia crassiceps. This was based on the observation that BALB/cAnN mice (Qa-2null) are susceptible to T. crassiceps, while BALB/cJ mice (Qa-2+) are resistant (Fragoso et al. 1996). In support of these findings, it was later shown that Qa-2 transgenic mice have increased clearance of the parasite (Fragoso et al. 1998). In addition, Q9-specific CD8+ T cells have been extensively characterized during the response to mouse polyoma virus (MPyV). Q9-restricted T cells from KbDb−/− mice are able to control MPyV infection, impede tumor formation, and recognize a nonameric peptide derived from the virus’s VP2 capsid protein (termed VP2.139) (Swanson et al. 2008). This population is present in wild-type mice as well. However, Q9-restricted T cells are somewhat different from classical CD8+ T cells because they form an inflationary population during persistent infection for approximately 12 weeks (Swanson et al. 2008). Further characterization showed that immunization with a VP2.139 peptide in mice carrying different MHC haplotypes could generate an MPyV-specific non-classical CD8+ T cell response (Hofstetter et al. 2013). However, the authors were not able to determine whether this population provided enhanced control during MPyV infection (Hofstetter et al. 2013).
Importantly, human HLA-G is proposed to be the functional homologue of Qa-2 (Comiskey et al. 2003). The expression of HLA-G is primarily limited to placental tissues such as cytotrophoblasts (Kovats et al. 1990), whereas classical MHC molecules are believed to be poorly expressed (Hunt et al. 1987). There are four membrane-bound isoforms of HLA-G, HLA-G1-G4, and three soluble isoforms, HLA-G5-G7, which occur via alternative splicing (Fujii et al. 1994; Ishitani and Geraghty 1992; Kirszenbaum et al. 1994; Paul et al. 2000). Secreted and membrane-bound forms can both present endogenous nonameric peptides (Diehl et al. 1996; Lee et al. 1995), however truncated isoforms do not associate with β2m (Morales et al. 2007). HLA-G has a number of immunomodulatory effects (Amiot et al. 2014; Guleria and Sayegh 2007), presumably mediated via interaction with the inhibitory receptors ILT2, ILT4, and KIR2DL4 (CD158d) (LeMaoult et al. 2005; Rajagopalan and Long 2012). HLA-G may be critical for immune tolerance during pregnancy to protect the fetus from rejection by maternal effector cells (Rouas-Freiss et al. 1997). In mice, Qa-2 is thought to participate during embryonic cleavage division and survival following preimplantation (McElhinny et al. 2000; Warner et al. 1987). Similarly to Qa-2, HLA-G molecules are capable of invoking a cytotoxic T cell response that is specific to HLA-G. This was first illustrated using HLA-G transgenic mice and skin graft experiments (Horuzsko et al. 1997; Schmidt et al. 1997). In a follow up experiment utilizing HLA-G tetramers for the predominant human cytomegalovirus (HCMV) peptide pp65, some HCMV-specific CD8+ T cells restricted by HLA-G were observed (Lenfant et al. 2003). However, the relevance of these HLA-G-restricted T cells, and their presence in humans, remains to be seen.
Qa-1 and HLA-E-restricted CD8+ T cells
Before HLA-E was known to present antigenic peptides to HLA-E-restricted T cell populations, it was first shown to be a ligand for the CD94/NKG2 family of NK cell receptors (NKG2A, NKG2B, and NKG2C) (Braud et al. 1998; Lee et al. 1998b). Qa-1 in mice (encoded by H2-T23) can similarly bind to CD94/NKG2 receptors (Vance et al. 1999; Vance et al. 1998). Expression of HLA-E at the cell surface is dependent on TAP (Lee et al. 1998a) and loading of nonamer peptides derived from the leader sequences of HLA-A, HLA-B, HLA-C, or HLA-G molecules (Braud et al. 1997; Lee et al. 1998a). Likewise, Qa-1 also binds signal sequence-derived peptides (AMAPRTLLL) from MHC class Ia molecules in a TAP-dependent manner (Aldrich et al. 1994). The primary ligands for Qa-1 and HLA-E are denoted Qdm, or Qa-1 determinant modifier, however other peptides can be loaded under different circumstances. For instance, the self-peptide FL9 (FYAEATPML) was recently identified in ERAAP-deficient mice (Nagarajan et al. 2012). These are not the only similarities between Qa-1 and HLA-E, which are considered functional homologues that arose by convergent evolution (Yeager et al. 1997). Both have low cell surface expression and broad tissue distribution, limited polymorphism, and structural homology (Zeng et al. 2012). This also includes similarities within their peptide-binding groove, such as unique substitutions at positions 143 and 147 (Connolly et al. 1993). HLA-E, for example, is the least polymorphic of the human non-classical MHC molecules. Caucasians only have two alleles, designated HLA-E*0101 and HLA-E*0103, which differ from each other at one amino acid position (Geraghty et al. 1992; Grimsley et al. 2002).
In addition to the role of HLA-E and Qa-1 during innate immunity, these MHC class Ib molecules can also present microbial antigens to CD8+ T cells. HLA-E and Qa-1 bind peptides from bacteria, such as S. typhimurium, S. enterica serovar Typhi, L. monocytogenes, and Mtb (Bouwer et al. 1997; Caccamo et al. 2015; Lo et al. 2000; Salerno-Goncalves et al. 2004; van Meijgaarden et al. 2015). Interestingly, in the absence of Qdm, a peptide derived from heat shock protein 60 (Hsp60) is primarily loaded into Qa-1 (GMKFDRGYI) (Davies et al. 2003). Hsp60 is well conserved in prokaryotes, whose homologue is GroEL in bacteria. In agreement with these observations, it was found that Qa-1 presents a S. typhimurium-derived peptide from GroEL (GMQFDRGYL) to CD8+ cytotoxic T cells (Lo et al. 2000). These GroEL-specific CD8+ T cells were also cross-reactive with Hsp60 and lyse stressed macrophages (Lo et al. 2000). Similarly, HLA-E presentation of GroEL-derived antigens from S. enterica caused targeted cell lysis of infected cells, as well as IFN-γ production (Salerno-Goncalves et al. 2004). HLA-E also participates during Mtb infection. This was first proposed when it was determined that the predominant CD8+ T cell response in latently infected patients was MHC class Ia and CD1 independent (Heinzel et al. 2002; Lewinsohn et al. 2000). The recognition of Mtb--derived peptides presented by HLA-E (Caccamo et al. 2015; van Meijgaarden et al. 2015) differs from the classically restricted CD8+ T cell response. HLA-E-restricted T cells appear to acquire a Th2 phenotype, producing TNF-α, IL-4, IL-5, IL-10, and IL-13, but have poor cytotoxicity in response to stimulation (Caccamo et al. 2015; van Meijgaarden et al. 2015). HLA-E and Qa-1 are also capable of presenting virally derived peptides. For example, HLA-E-reactive T cells to Epstein-Barr virus recognize a peptide from its BZLF-1 protein (SQAPLPCVL) (Garcia et al. 2002; Jorgensen et al. 2012). A peptide derived from a Hepatitis C virus (HCV) core protein (YLLPRRGPRL) was also shown to bind HLA-E, stabilize its cell surface expression, and protect cells from NK cell lysis (Nattermann et al. 2005). Additionally, 40% of an HCV infected cohort was determined to have an HLA-E-reactive T cell response, resulting in IFN-γ production, which was not observed from healthy control samples (Schulte et al. 2009). Interestingly, there was an increased incidence of HLA-E-specific CD8+ T cells to HCV in patients with the HLA-E*0101 allele, compared to the HLA-E*0103 allele (Schulte et al. 2009).
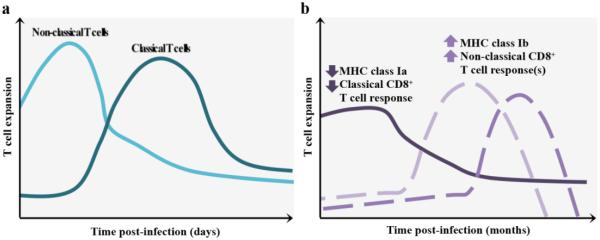
Perhaps the most intriguing HLA-E-restricted response occurs during HCMV infection. HCMV employs a variety of mechanisms to downregulate the expression of conventional HLA molecules at the cell surface and avoid the classical CD8+ T cell response. One immunoevasive mechanism HCMV employs is to inhibit ERAP1 function, the human homolog of murine ERAAP, through miR-US4-1 (Kim et al. 2011). This miRNA obstructs cytotoxic CD8+ T cells from lysing infected cells by inhibiting HCMV antigen derivation (Kim et al. 2011). HCMV also provides its own peptide, derived from the signal sequence of its glycoprotein UL40 (gpUL40) to load in HLA-E molecules, increasing cell surface expression independently of TAP (Tomasec et al. 2000; Ulbrecht et al. 2000). The UL40 leader sequence from the AD169 and Toledo HCMV strains are both able to provide peptides (VMAPRTLIL and VMAPRTLVL, respectively) that bind HLA-E. This is thought to be a mechanism for escaping the NK cell response because UL40 deletion mutants are unable to evade NK cells through CD94/NKG2A inhibition (Wang et al. 2002). However, HLA-E-restricted CD8+ T cells can also recognize these gpUL40-derived peptides via their TCR (Pietra et al. 2003). HLA-E-restricted HCMV-specific T cells have an effector memory phenotype, can kill HCMV infected target cells, and produce IFN-γ in response to contact with UL40 leader peptides (Mazzarino et al. 2005). Interestingly, the gpUL40 leader sequence of AD169 and Toledo HCMV strains is identical to certain HLA-A and HLA-Cw leader peptide alleles, possibly leading to a non-classical T cell evasion mechanism (Pietra et al. 2010; Pietra et al. 2003). In addition to increased expression of HLA-E, HCMV also disrupts ERAP1 function (Kim et al. 2011). As mentioned previously, Qa-1-restricted cells kill ERAAP-deficient cells in naï ve mice (Nagarajan et al. 2012). Therefore, it would be interesting to determine whether HLA-E-restricted CD8+ T cells play a similar role during HCMV infection. Overall, the role of Qa-1/HLA-E-restricted T cells may only be revealed when the MHC class I and/or conventional CD8+ T cell response is failing (Figure 3, see proposed model below).
Figure 3. Proposed roles of non-classical CD8+ αβ T cell populations in humans and mice, compared to conventional CD8+ T cells.
A) MHC class Ib-restricted CD8+ T cells, e.g. iNKT cells and MAIT cells, have a faster effector response following infection, compared to MHC class Ia-restricted T cells. B) During chronic infection, microorganisms can employ immunoevasion mechanisms to dampen the classical T cell response and/or NK cell response. MHC class Ib-specific responses could represent a backup system that responds following MHC class Ib upregulation, for example HLA-E.
T cell responses restricted by unidentified MHC class Ib molecules and other non-classical T cell responses
Certain non-classical T cell responses are a result of, as of yet, undetermined MHC class Ib molecules. For example, non-classically restricted CD8+ T cells from KbDb−/− mice are sufficient to control chronic γ-herpesvirus 68 (Braaten et al. 2006). While the exact restriction of this population is currently unknown, it is dependent on β2m and CD1d is dispensable (Braaten et al. 2006). In addition, MHC class Ib-restricted T cells respond following lymphocytic choriomeningitis virus (LCMV) infection in both KbDb−/− and KbDbCIITA−/− mice, which also lack MHC class II molecules, however they were inadequate to fully clear the virus (Chen et al. 2011). Non-classical T cells are also present in a wide range of species. For instance, in the amphibian Xenopus laevis, iVα6 T cells are restricted by the MHC class Ib molecule XNC10 and resemble iNKT cells found mice and humans (Edholm et al. 2013). XNC10-restricted T cells were essential for an appropriate antiviral response, and successful viral clearance, following infection with frog virus 3 (Edholm et al. 2015). Studies such as these illustrate the importance of non-classical MHC class Ib-restricted T cells, and their biological relevance, in a wide range of species.
Concluding remarks and proposed models
Non-classical T cells are unique in many ways – strategic localization at barrier sites, recognition of a wide array of unique microbial pathogens, and rapid effector responses. These features led to the hypothesis that the primary function of non-classical T cells such as iNKT and MAIT cells is to rapidly respond to infections. We propose that another, non-mutually exclusive, function for these cells may be revealed during chronic infection when classical T cell and NK cell responses are impaired (Figure 3). This has been recently documented in the case of Qa-1/HLA-E-restricted T cells, which exploit pathogen immunoevasion adaptations (Hansen et al. 2016). Together with the low polymorphism of MHC class Ib molecules, the unique characteristics of MHC class Ib-restricted T cells render them attractive targets for vaccine development, especially when the immune system is compromised.
Acknowledgements
We would like to thank Timothy Erick for critical reading of the manuscript. This work was supported by National Institutes of Health Research Grant RO1 AI46709 and AAI Careers in Immunology Fellowship (L.B.) and National Institutes of Health Fellowship F31 AI124556 (C. K. A.)
References
- Aldrich CJ, DeCloux A, Woods AS, Cotter RJ, Soloski MJ, Forman J. Identification of a Tap-dependent leader peptide recognized by alloreactive T cells specific for a class Ib antigen. Cell. 1994;79:649–658. doi: 10.1016/0092-8674(94)90550-9. [DOI] [PubMed] [Google Scholar]
- Amiot L, Vu N, Samson M. Immunomodulatory properties of HLA-G in infectious diseases. J Immunol Res. 2014;2014:298569. doi: 10.1155/2014/298569. doi:10.1155/2014/298569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. doi:10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson G, Owen JJ, Moore NC, Jenkinson EJ. Thymic epithelial cells provide unique signals for positive selection of CD4+CD8+ thymocytes in vitro. The Journal of experimental medicine. 1994;179:2027–2031. doi: 10.1084/jem.179.6.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attinger A, et al. Molecular basis for the high affinity interaction between the thymic leukemia antigen and the CD8alphaalpha molecule. Journal of immunology (Baltimore, Md : 1950) 2005;174:3501–3507. doi: 10.4049/jimmunol.174.6.3501. [DOI] [PubMed] [Google Scholar]
- Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells Nature. 1994;372:691–694. doi: 10.1038/372691a0. doi:10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- Bedel R, Matsuda JL, Brigl M, White J, Kappler J, Marrack P, Gapin L. Lower TCR repertoire diversity in Traj18-deficient mice. Nature immunology. 2012;13:705–706. doi: 10.1038/ni.2347. doi:10.1038/ni.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar SM, Dascher CC, Grusby MJ, Wang CR, Brenner MB. Susceptibility of mice deficient in CD1D or TAP1 to infection with Mycobacterium tuberculosis. The Journal of experimental medicine. 1999;189:1973–1980. doi: 10.1084/jem.189.12.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. The Journal of experimental medicine. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosselut R, Zhang W, Ashe JM, Kopacz JL, Samelson LE, Singer A. Association of the adaptor molecule LAT with CD4 and CD8 coreceptors identifies a new coreceptor function in T cell receptor signal transduction. The Journal of experimental medicine. 1999;190:1517–1526. doi: 10.1084/jem.190.10.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucherma R, et al. Loss of central and peripheral CD8+ T-cell tolerance to HFE in mouse models of human familial hemochromatosis. European journal of immunology. 2012;42:851–862. doi: 10.1002/eji.201141664. doi:10.1002/eji.201141664. [DOI] [PubMed] [Google Scholar]
- Bouwer HG, Seaman MS, Forman J, Hinrichs DJ. MHC class Ib-restricted cells contribute to antilisterial immunity: evidence for Qa-1b as a key restricting element for Listeria-specific CTLs. Journal of immunology (Baltimore, Md : 1950) 1997;159:2795–2801. [PubMed] [Google Scholar]
- Boyse EA, Old LJ, Luell S. Genetic Determination of the Tl (Thymusleukaemia) Antigen in the Mouse. Nature. 1964;201:779. doi: 10.1038/201779a0. [DOI] [PubMed] [Google Scholar]
- Braaten DC, McClellan JS, Messaoudi I, Tibbetts SA, McClellan KB, Nikolich-Zugich J, Virgin HW. Effective control of chronic gamma-herpesvirus infection by unconventional MHC Class Ia-independent CD8. T cells PLoS pathogens. 2006;2:e37. doi: 10.1371/journal.ppat.0020037. doi:10.1371/journal.ppat.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braud V, Jones EY, McMichael A. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. European journal of immunology. 1997;27:1164–1169. doi: 10.1002/eji.1830270517. doi:10.1002/eji.1830270517. [DOI] [PubMed] [Google Scholar]
- Braud VM, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. doi:10.1038/35869. [DOI] [PubMed] [Google Scholar]
- Brigl M, Bry L, Kent SC, Gumperz JE, Brenner MB. Mechanism of CD1d-restricted natural killer. T cell activation during microbial infection Nature immunology. 2003;4:1230–1237. doi: 10.1038/ni1002. doi:10.1038/ni1002. [DOI] [PubMed] [Google Scholar]
- Brutkiewicz RR, Bennink JR, Yewdell JW, Bendelac A. TAP-independent, beta 2-microglobulin-dependent surface expression of functional mouse CD1.1. The Journal of experimental medicine. 1995;182:1913–1919. doi: 10.1084/jem.182.6.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers DE, Fischer Lindahl K. H2-M3 presents a nonformylated viral epitope to CTLs generated in vitro. Journal of immunology (Baltimore, Md : 1950) 1998;161:90–96. [PubMed] [Google Scholar]
- Caccamo N, et al. Human CD8 T lymphocytes recognize Mycobacterium tuberculosis antigens presented by HLA-E during active tuberculosis and express type 2 cytokines. European journal of immunology. 2015;45:1069–1081. doi: 10.1002/eji.201445193. doi:10.1002/eji.201445193. [DOI] [PubMed] [Google Scholar]
- Cardell S, Tangri S, Chan S, Kronenberg M, Benoist C, Mathis D. CD1-restricted CD4+ T cells in major histocompatibility complex class II-deficient mice. The Journal of experimental medicine. 1995;182:993–1004. doi: 10.1084/jem.182.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Jay DC, Fairbanks JD, He X, Jensen PE. An MHC class Ib-restricted CD8+ T cell response to lymphocytic choriomeningitis virus. Journal of immunology (Baltimore, Md : 1950) 2011;187:6463–6472. doi: 10.4049/jimmunol.1101171. doi:10.4049/jimmunol.1101171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YT, Obata Y, Stockert E, Old LJ. Thymus-leukemia (TL) antigens of the mouse. Analysis of TL mRNA and TL cDNA TL+ and TL- strains. The Journal of experimental medicine. 1985;162:1134–1148. doi: 10.1084/jem.162.4.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheroutre H, Lambolez F. Doubting the TCR coreceptor function of CD8alphaalpha. Immunity. 2008;28:149–159. doi: 10.1016/j.immuni.2008.01.005. doi:10.1016/j.immuni.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Chiu NM, Chun T, Fay M, Mandal M, Wang CR. The majority of H2-M3 is retained intracellularly in a peptide-receptive state and traffics to the cell surface in the presence of N-formylated peptides. The Journal of experimental medicine. 1999a;190:423–434. doi: 10.1084/jem.190.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu NM, Wang B, Kerksiek KM, Kurlander R, Pamer EG, Wang CR. The selection of M3-restricted T cells is dependent on M3 expression and presentation of N-formylated peptides in the thymus. The Journal of experimental medicine. 1999b;190:1869–1878. doi: 10.1084/jem.190.12.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Bediako Y, Xu H, Choi HJ, Wang CR. Positive selecting cell type determines the phenotype of MHC class Ib-restricted CD8+ T cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13241–13246. doi: 10.1073/pnas.1105118108. doi:10.1073/pnas.1105118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua WJ, Truscott SM, Eickhoff CS, Blazevic A, Hoft DF, Hansen TH. Polyclonal mucosa-associated invariant T cells have unique innate functions in bacterial infection. Infect Immun. 2012;80:3256–3267. doi: 10.1128/IAI.00279-12. doi:10.1128/IAI.00279-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun T, Grandea AG, 3rd, Lybarger L, Forman J, Van Kaer L, Wang CR. Functional roles of TAP and tapasin in the assembly of M3-N-formylated peptide complexes. Journal of immunology (Baltimore, Md : 1950) 2001a;167:1507–1514. doi: 10.4049/jimmunol.167.3.1507. [DOI] [PubMed] [Google Scholar]
- Chun T, Serbina NV, Nolt D, Wang B, Chiu NM, Flynn JL, Wang CR. Induction of M3-restricted cytotoxic T lymphocyte responses by N-formylated peptides derived from Mycobacterium tuberculosis. The Journal of experimental medicine. 2001b;193:1213–1220. doi: 10.1084/jem.193.10.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogen AL, Moore TA. Beta2-microglobulin-dependent bacterial clearance and survival during murine Klebsiella pneumoniae bacteremia. Infect Immun. 2009;77:360–366. doi: 10.1128/IAI.00909-08. doi:10.1128/IAI.00909-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comiskey M, Goldstein CY, De Fazio SR, Mammolenti M, Newmark JA, Warner CM. Evidence that HLA-G is the functional homolog of mouse Qa-2, the Ped gene product. Hum Immunol. 2003;64:999–1004. doi: 10.1016/j.humimm.2003.08.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly DJ, et al. A cDNA clone encoding the mouse Qa-1a histocompatibility antigen and proposed structure of the putative peptide binding site. Journal of immunology (Baltimore, Md : 1950) 1993;151:6089–6098. [PubMed] [Google Scholar]
- Cook RG, Landolfi NF. Expression of the thymus leukemia antigen by activated peripheral T lymphocytes. The Journal of experimental medicine. 1983;158:1012–1017. doi: 10.1084/jem.158.3.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett AJ, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509:361–365. doi: 10.1038/nature13160. doi:10.1038/nature13160. [DOI] [PubMed] [Google Scholar]
- Costa M, et al. Lymphocyte gene expression signatures from patients and mouse models of hereditary hemochromatosis reveal a function of HFE as a negative regulator of CD8+ T-lymphocyte activation and differentiation in vivo. PloS one. 2015;10:e0124246. doi: 10.1371/journal.pone.0124246. doi:10.1371/journal.pone.0124246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Orazio SE, Halme DG, Ploegh HL, Starnbach MN. Class Ia MHC-deficient BALB/c mice generate CD8+ T cell-mediated protective immunity against Listeria monocytogenes infection. Journal of immunology (Baltimore, Md : 1950) 2003;171:291–298. doi: 10.4049/jimmunol.171.1.291. [DOI] [PubMed] [Google Scholar]
- D'Orazio SE, Shaw CA, Starnbach MN. H2-M3-restricted CD8+ T cells are not required for MHC class Ib-restricted immunity against Listeria monocytogenes. The Journal of experimental medicine. 2006;203:383–391. doi: 10.1084/jem.20052256. doi:10.1084/jem.20052256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies A, et al. A peptide from heat shock protein 60 is the dominant peptide bound to Qa-1 in the absence of the MHC class Ia leader sequence peptide Qdm. Journal of immunology (Baltimore, Md : 1950) 2003;170:5027–5033. doi: 10.4049/jimmunol.170.10.5027. [DOI] [PubMed] [Google Scholar]
- Davis BK, Cook RG, Rich RR, Rodgers JR. Hyperconservation of the putative antigen recognition site of the MHC class I-b molecule TL in the subfamily Murinae: evidence that thymus leukemia antigen is an ancient mammalian gene. Journal of immunology (Baltimore, Md : 1950) 2002;169:6890–6899. doi: 10.4049/jimmunol.169.12.6890. [DOI] [PubMed] [Google Scholar]
- de la Salle H, et al. Assistance of microbial glycolipid antigen processing by CD1e. Science. 2005;310:1321–1324. doi: 10.1126/science.1115301. doi:10.1126/science.1115301. [DOI] [PubMed] [Google Scholar]
- Delker SL, West AP, Jr., McDermott L, Kennedy MW, Bjorkman PJ. Crystallographic studies of ligand binding by Zn-alpha2-glycoprotein. J Struct Biol. 2004;148:205–213. doi: 10.1016/j.jsb.2004.04.009. doi:10.1016/j.jsb.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Devlin JJ, Weiss EH, Paulson M, Flavell RA. Duplicated gene pairs and alleles of class I genes in the Qa2 region of the murine major histocompatibility complex: a comparison. The EMBO journal. 1985;4:3203–3207. doi: 10.1002/j.1460-2075.1985.tb04066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl M, Munz C, Keilholz W, Stevanovic S, Holmes N, Loke YW, Rammensee HG. Nonclassical HLA-G molecules are classical peptide presenters. Curr Biol. 1996;6:305–314. doi: 10.1016/s0960-9822(02)00481-5. [DOI] [PubMed] [Google Scholar]
- Doi T, Yamada H, Yajima T, Wajjwalku W, Hara T, Yoshikai Y. H2-M3-restricted CD8+ T cells induced by peptide-pulsed dendritic cells confer protection against Mycobacterium tuberculosis. Journal of immunology (Baltimore, Md : 1950) 2007;178:3806–3813. doi: 10.4049/jimmunol.178.6.3806. [DOI] [PubMed] [Google Scholar]
- Dunne MR, Elliott L, Hussey S, Mahmud N, Kelly J, Doherty DG, Feighery CF. Persistent changes in circulating and intestinal gammadelta T cell subsets, invariant natural killer T cells and mucosal-associated invariant T cells in children and adults with coeliac disease. PloS one. 2013;8:e76008. doi: 10.1371/journal.pone.0076008. doi:10.1371/journal.pone.0076008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckle SB, et al. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. The Journal of experimental medicine. 2014;211:1585–1600. doi: 10.1084/jem.20140484. doi:10.1084/jem.20140484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edholm ES, et al. Nonclassical MHC class I-dependent invariant T cells are evolutionarily conserved and prominent from early development in amphibians. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:14342–14347. doi: 10.1073/pnas.1309840110. doi:10.1073/pnas.1309840110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edholm ES, Grayfer L, De Jesus Andino F, Robert J. Nonclassical MHC-Restricted Invariant Valpha6 T Cells Are Critical for Efficient Early Innate Antiviral Immunity in the Amphibian Xenopus laevis. Journal of immunology (Baltimore, Md : 1950) 2015;195:576–586. doi: 10.4049/jimmunol.1500458. doi:10.4049/jimmunol.1500458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JN, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. doi:10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- Feder JN, et al. The hemochromatosis founder mutation in HLA-H disrupts beta2-microglobulin interaction and cell surface expression. The Journal of biological chemistry. 1997;272:14025–14028. doi: 10.1074/jbc.272.22.14025. [DOI] [PubMed] [Google Scholar]
- Felio K, et al. CD1-restricted adaptive immune responses to Mycobacteria in human group 1 CD1 transgenic mice. The Journal of experimental medicine. 2009;206:2497–2509. doi: 10.1084/jem.20090898. doi:10.1084/jem.20090898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez CS, Amarasena T, Kelleher AD, Rossjohn J, McCluskey J, Godfrey DI, Kent SJ. MAIT cells are depleted early but retain functional cytokine expression in HIV infection Immunol. Cell Biol. 2015;93:177–188. doi: 10.1038/icb.2014.91. doi:10.1038/icb.2014.91. [DOI] [PubMed] [Google Scholar]
- Fischer K, et al. Mycobacterial phosphatidylinositol mannoside is a natural antigen for CD1d-restricted T cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10685–10690. doi: 10.1073/pnas.0403787101. doi:10.1073/pnas.0403787101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso G, Lamoyi E, Mellor A, Lomeli C, Govezensky T, Sciutto E. Genetic control of susceptibility to Taenia crassiceps cysticercosis. Parasitology. 1996;112:119–124. doi: 10.1017/s003118200006515x. Pt 1. [DOI] [PubMed] [Google Scholar]
- Fragoso G, Lamoyi E, Mellor A, Lomeli C, Hernandez M, Sciutto E. Increased resistance to Taenia crassiceps murine cysticercosis in Qa-2 transgenic mice. Infect Immun. 1998;66:760–764. doi: 10.1128/iai.66.2.760-764.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T, Ishitani A, Geraghty DE. A soluble form of the HLA-G antigen is encoded by a messenger ribonucleic acid containing intron 4. Journal of immunology (Baltimore, Md : 1950) 1994;153:5516–5524. [PubMed] [Google Scholar]
- Gadola SD, et al. Structure of human CD1b with bound ligands at 2.3 A, a maze for alkyl chains. Nature immunology. 2002;3:721–726. doi: 10.1038/ni821. doi:10.1038/ni821. [DOI] [PubMed] [Google Scholar]
- Garcia P, et al. Human T cell receptor-mediated recognition of HLA-E. European journal of immunology. 2002;32:936–944. doi: 10.1002/1521-4141(200204)32:4<936::AID-IMMU936>3.0.CO;2-M. doi:10.1002/1521-4141(200204)32:4<936::AID-IMMU936>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Georgel P, Radosavljevic M, Macquin C, Bahram S. The non-conventional MHC class I MR1 molecule controls infection by Klebsiella pneumoniae in mice. Mol Immunol. 2011;48:769–775. doi: 10.1016/j.molimm.2010.12.002. doi:10.1016/j.molimm.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Geraghty DE, Stockschleader M, Ishitani A, Hansen JA. Polymorphism at the HLA-E locus predates most HLA-A and -B polymorphism. Hum Immunol. 1992;33:174–184. doi: 10.1016/0198-8859(92)90069-y. [DOI] [PubMed] [Google Scholar]
- Gold MC, et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 2010;8:e1000407. doi: 10.1371/journal.pbio.1000407. doi:10.1371/journal.pbio.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold MC, et al. MR1-restricted MAIT cells display ligand discrimination and pathogen selectivity through distinct T cell receptor usage. The Journal of experimental medicine. 2014;211:1601–1610. doi: 10.1084/jem.20140507. doi:10.1084/jem.20140507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami T, Andrews NC. Hereditary hemochromatosis protein, HFE, interaction with transferrin receptor 2 suggests a molecular mechanism for mammalian iron sensing. The Journal of biological chemistry. 2006;281:28494–28498. doi: 10.1074/jbc.C600197200. doi:10.1074/jbc.C600197200. [DOI] [PubMed] [Google Scholar]
- Grimsley C, et al. Definitive high resolution typing of HLA-E allelic polymorphisms: Identifying potential errors in existing allele data. Tissue Antigens. 2002;60:206–212. doi: 10.1034/j.1399-0039.2002.600302.x. [DOI] [PubMed] [Google Scholar]
- Gulden PH, et al. A Listeria monocytogenes pentapeptide is presented to cytolytic T lymphocytes by the H2-M3 MHC class Ib molecule. Immunity. 1996;5:73–79. doi: 10.1016/s1074-7613(00)80311-8. [DOI] [PubMed] [Google Scholar]
- Guleria I, Sayegh MH. Maternal acceptance of the fetus: true human tolerance. Journal of immunology (Baltimore, Md : 1950) 2007;178:3345–3351. doi: 10.4049/jimmunol.178.6.3345. [DOI] [PubMed] [Google Scholar]
- Hamilton SE, Porter BB, Messingham KA, Badovinac VP, Harty JT. MHC class Ia-restricted memory T cells inhibit expansion of a nonprotective MHC class Ib (H2-M3)-restricted memory response. Nature immunology. 2004;5:159–168. doi: 10.1038/ni1026. doi:10.1038/ni1026. [DOI] [PubMed] [Google Scholar]
- Hanau D, et al. CD1 expression is not affected by human peptide transporter deficiency. Hum Immunol. 1994;41:61–68. doi: 10.1016/0198-8859(94)90086-8. [DOI] [PubMed] [Google Scholar]
- Hansen SG, et al. Broadly targeted CD8(+) T cell responses restricted by major histocompatibility complex. E Science. 2016;351:714–720. doi: 10.1126/science.aac9475. doi:10.1126/science.aac9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Tabaczewski P, Ho J, Stroynowski I, Garcia KC. Promiscuous antigen presentation by the nonclassical MHC Ib Qa-2 is enabled by a shallow, hydrophobic groove and self-stabilized peptide conformation. Structure. 2001;9:1213–1224. doi: 10.1016/s0969-2126(01)00689-x. [DOI] [PubMed] [Google Scholar]
- Heinzel AS, et al. HLA-E-dependent presentation of Mtb-derived antigen to human CD8+ T cells. The Journal of experimental medicine. 2002;196:1473–1481. doi: 10.1084/jem.20020609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Eghtesady P, Sydora B, Brorson K, Cheroutre H, Modlin R, Kronenberg M. Expression of the thymus leukemia antigen in mouse intestinal epithelium. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:9727–9731. doi: 10.1073/pnas.87.24.9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai K, Hussey HJ, Barber MD, Price SA, Tisdale MJ. Biological evaluation of a lipid-mobilizing factor isolated from the urine of cancer patients. Cancer Res. 1998;58:2359–2365. [PubMed] [Google Scholar]
- Hofstetter AR, Evavold BD, Lukacher AE. Peptide immunization elicits polyomavirus-specific MHC class ib-restricted CD8 T cells in MHC class ia allogeneic mice. Viral Immunol. 2013;26:109–113. doi: 10.1089/vim.2012.0052. doi:10.1089/vim.2012.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcombe HR, Castano AR, Cheroutre H, Teitell M, Maher JK, Peterson PA, Kronenberg M. Nonclassical behavior of the thymus leukemia antigen: peptide transporter-independent expression of a nonclassical class I molecule. The Journal of experimental medicine. 1995;181:1433–1443. doi: 10.1084/jem.181.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzapfel KL, Tyznik AJ, Kronenberg M, Hogquist KA. Antigen-dependent versus -independent activation of invariant NKT cells during infection. Journal of immunology (Baltimore, Md : 1950) 2014;192:5490–5498. doi: 10.4049/jimmunol.1400722. doi:10.4049/jimmunol.1400722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horuzsko A, Antoniou J, Tomlinson P, Portik-Dobos V, Mellor AL. HLA-G functions as a restriction element and a transplantation antigen in mice. International immunology. 1997;9:645–653. doi: 10.1093/intimm/9.5.645. [DOI] [PubMed] [Google Scholar]
- Huang Y, et al. Mucosal memory CD8(+) T cells are selected in the periphery by an MHC class I molecule. Nature immunology. 2011;12:1086–1095. doi: 10.1038/ni.2106. doi:10.1038/ni.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt JS, Andrews GK, Wood GW. Normal trophoblasts resist induction of class I HLA. Journal of immunology (Baltimore, Md : 1950) 1987;138:2481–2487. [PubMed] [Google Scholar]
- Ishitani A, Geraghty DE. Alternative splicing of HLA-G transcripts yields proteins with primary structures resembling both class I and class II antigens. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:3947–3951. doi: 10.1073/pnas.89.9.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, et al. Helicobacter pylori cholesteryl alpha-glucosides contribute to its pathogenicity and immune response by natural killer T cells. PloS one. 2013;8:e78191. doi: 10.1371/journal.pone.0078191. doi:10.1371/journal.pone.0078191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahng A, Maricic I, Aguilera C, Cardell S, Halder RC, Kumar V. Prevention of autoimmunity by targeting a distinct, noninvariant CD1d-reactive T cell population reactive to sulfatide. The Journal of experimental medicine. 2004;199:947–957. doi: 10.1084/jem.20031389. doi:10.1084/jem.20031389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay DC, Reed-Loisel LM, Jensen PE. Polyclonal MHC Ib-restricted CD8+ T cells undergo homeostatic expansion in the absence of conventional MHC-restricted T cells. Journal of immunology (Baltimore, Md : 1950) 2008;180:2805–2814. doi: 10.4049/jimmunol.180.5.2805. [DOI] [PubMed] [Google Scholar]
- Jensen KD, et al. Thymic selection determines gammadelta T cell effector fate: antigen-naive cells make interleukin-17 and antigen-experienced cells make interferon gamma. Immunity. 2008;29:90–100. doi: 10.1016/j.immuni.2008.04.022. doi:10.1016/j.immuni.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, et al. Mucosal-associated invariant T-cell function is modulated by programmed death-1 signaling in patients with active tuberculosis. Am J Respir Crit Care Med. 2014;190:329–339. doi: 10.1164/rccm.201401-0106OC. doi:10.1164/rccm.201401-0106OC. [DOI] [PubMed] [Google Scholar]
- Jorgensen PB, Livbjerg AH, Hansen HJ, Petersen T, Hollsberg P. Epstein-Barr virus peptide presented by HLA-E is predominantly recognized by CD8(bright) cells in multiple sclerosis patients. PloS one. 2012;7:e46120. doi: 10.1371/journal.pone.0046120. doi:10.1371/journal.pone.0046120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce S, Tabaczewski P, Angeletti RH, Nathenson SG, Stroynowski I. A nonpolymorphic major histocompatibility complex class Ib molecule binds a large array of diverse self-peptides. The Journal of experimental medicine. 1994;179:579–588. doi: 10.1084/jem.179.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, et al. Critical role of Valpha14+ natural killer T cells in the innate phase of host protection against Streptococcus pneumoniae infection. European journal of immunology. 2003;33:3322–3330. doi: 10.1002/eji.200324254. doi:10.1002/eji.200324254. [DOI] [PubMed] [Google Scholar]
- Kawashima T, et al. Cutting edge: major CD8 T cell response to live bacillus Calmette-Guerin is mediated by CD1 molecules. Journal of immunology (Baltimore, Md : 1950) 2003;170:5345–5348. doi: 10.4049/jimmunol.170.11.5345. [DOI] [PubMed] [Google Scholar]
- Kerksiek KM, Busch DH, Pilip IM, Allen SE, Pamer EG. H2-M3-restricted T cells in bacterial infection: rapid primary but diminished memory responses. The Journal of experimental medicine. 1999;190:195–204. doi: 10.1084/jem.190.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern P, Hussey RE, Spoerl R, Reinherz EL, Chang HC. Expression, purification, and functional analysis of murine ectodomain fragments of CD8alphaalpha and CD8alphabeta dimers. The Journal of biological chemistry. 1999;274:27237–27243. doi: 10.1074/jbc.274.38.27237. [DOI] [PubMed] [Google Scholar]
- Kim S, et al. Human cytomegalovirus microRNA miR-US4-1 inhibits CD8(+) T cell responses by targeting the aminopeptidase ERAP1. Nature immunology. 2011;12:984–991. doi: 10.1038/ni.2097. doi:10.1038/ni.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo Y, et al. Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nature immunology. 2011;12:966–974. doi: 10.1038/ni.2096. doi:10.1038/ni.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo Y, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nature immunology. 2006;7:978–986. doi: 10.1038/ni1380. doi:10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- Kinjo Y, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–525. doi: 10.1038/nature03407. doi:10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- Kirszenbaum M, Moreau P, Gluckman E, Dausset J, Carosella E. An alternatively spliced form of HLA-G mRNA in human trophoblasts and evidence for the presence of HLA-G transcript in adult lymphocytes. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:4209–4213. doi: 10.1073/pnas.91.10.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer-Nielsen L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012;491:717–723. doi: 10.1038/nature11605. doi:10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science. 1990;248:220–223. doi: 10.1126/science.2326636. [DOI] [PubMed] [Google Scholar]
- Kumar H, Belperron A, Barthold SW, Bockenstedt LK. Cutting edge: CD1d deficiency impairs murine host defense against the spirochete, Borrelia burgdorferi. Journal of immunology (Baltimore, Md : 1950) 2000;165:4797–4801. doi: 10.4049/jimmunol.165.9.4797. [DOI] [PubMed] [Google Scholar]
- Kurepa Z, Su J, Forman J. Memory phenotype of CD8+ T cells in MHC class Ia-deficient mice. Journal of immunology (Baltimore, Md : 1950) 2003;170:5414–5420. doi: 10.4049/jimmunol.170.11.5414. [DOI] [PubMed] [Google Scholar]
- Le Bourhis L, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nature immunology. 2010;11:701–708. doi: 10.1038/ni.1890. doi:10.1038/ni.1890. [DOI] [PubMed] [Google Scholar]
- Lebron JA, et al. Crystal structure of the hemochromatosis protein HFE and characterization of its interaction with transferrin receptor. Cell. 1998;93:111–123. doi: 10.1016/s0092-8674(00)81151-4. [DOI] [PubMed] [Google Scholar]
- Lee N, Goodlett DR, Ishitani A, Marquardt H, Geraghty DE. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. Journal of immunology (Baltimore, Md : 1950) 1998a;160:4951–4960. [PubMed] [Google Scholar]
- Lee N, Llano M, Carretero M, Ishitani A, Navarro F, Lopez-Botet M, Geraghty DE. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proceedings of the National Academy of Sciences of the United States of America. 1998b;95:5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Malacko AR, Ishitani A, Chen MC, Bajorath J, Marquardt H, Geraghty DE. The membrane-bound and soluble forms of HLA-G bind identical sets of endogenous peptides but differ with respect to TAP association. Immunity. 1995;3:591–600. doi: 10.1016/1074-7613(95)90130-2. [DOI] [PubMed] [Google Scholar]
- Leeansyah E, et al. Activation, exhaustion, and persistent decline of the antimicrobial MR1-restricted MAIT-cell population in chronic HIV-1 infection. Blood. 2013;121:1124–1135. doi: 10.1182/blood-2012-07-445429. doi:10.1182/blood-2012-07-445429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leishman AJ, et al. T cell responses modulated through interaction between CD8alphaalpha and the nonclassical MHC class I molecule, TL. Science. 2001;294:1936–1939. doi: 10.1126/science.1063564. doi:10.1126/science.1063564. [DOI] [PubMed] [Google Scholar]
- Leite-De-Moraes MC, et al. A distinct IL-18-induced pathway to fully activate NK T lymphocytes independently from TCR engagement. Journal of immunology (Baltimore, Md : 1950) 1999;163:5871–5876. [PubMed] [Google Scholar]
- LeMaoult J, Zafaranloo K, Le Danff C, Carosella ED. HLA-G up-regulates ILT2, ILT3, ILT4, and KIR2DL4 in antigen presenting cells, NK cells, and T cells. FASEB J. 2005;19:662–664. doi: 10.1096/fj.04-1617fje. doi:10.1096/fj.04-1617fje. [DOI] [PubMed] [Google Scholar]
- Lenfant F, Pizzato N, Liang S, Davrinche C, Le Bouteiller P, Horuzsko A. Induction of HLA-G-restricted human cytomegalovirus pp65 (UL83)-specific cytotoxic T lymphocytes in HLA-G transgenic mice. The Journal of general virology. 2003;84:307–317. doi: 10.1099/vir.0.18735-0. doi:10.1099/vir.0.18735-0. [DOI] [PubMed] [Google Scholar]
- Lenz LL, Bevan MJ. CTL responses to H2-M3-restricted Listeria epitopes. Immunological reviews. 1997;158:115–121. doi: 10.1111/j.1600-065x.1997.tb00997.x. [DOI] [PubMed] [Google Scholar]
- Lenz LL, Dere B, Bevan MJ. Identification of an H2-M3-restricted Listeria epitope: implications for antigen presentation by M3. Immunity. 1996;5:63–72. doi: 10.1016/s1074-7613(00)80310-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt JM, Howell DD, Rodgers JR, Rich RR. Exogenous peptides enter the endoplasmic reticulum of TAP-deficient cells and induce the maturation of nascent MHC class I molecules. European journal of immunology. 2001;31:1181–1190. doi: 10.1002/1521-4141(200104)31:4<1181::aid-immu1181>3.0.co;2-j. doi:10.1002/1521-4141(200104)31:4<1181::AID-IMMU1181>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Lewinsohn DM, Briden AL, Reed SG, Grabstein KH, Alderson MR. Mycobacterium tuberculosis-reactive CD8+ T lymphocytes: the relative contribution of classical versus nonclassical HLA restriction. Journal of immunology (Baltimore, Md : 1950) 2000;165:925–930. doi: 10.4049/jimmunol.165.2.925. [DOI] [PubMed] [Google Scholar]
- Liu Y, et al. The crystal structure of a TL/CD8alphaalpha complex at 2.1 A resolution: implications for modulation of T cell activation and memory. Immunity. 2003;18:205–215. doi: 10.1016/s1074-7613(03)00027-x. [DOI] [PubMed] [Google Scholar]
- Ljunggren HG, et al. Empty MHC class I molecules come out in the cold. Nature. 1990;346:476–480. doi: 10.1038/346476a0. doi:10.1038/346476a0. [DOI] [PubMed] [Google Scholar]
- Lo WF, Woods AS, DeCloux A, Cotter RJ, Metcalf ES, Soloski MJ. Molecular mimicry mediated by MHC class Ib molecules after infection with gram-negative pathogens. Nat Med. 2000;6:215–218. doi: 10.1038/72329. doi:10.1038/72329. [DOI] [PubMed] [Google Scholar]
- Lockridge JL, et al. Analysis of the CD1 antigen presenting system in humanized SCID mice. PloS one. 2011;6:e21701. doi: 10.1371/journal.pone.0021701. doi:10.1371/journal.pone.0021701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loconto J, et al. Functional expression of murine V2R pheromone receptors involves selective association with the M10 and M1 families of MHC class Ib molecules. Cell. 2003;112:607–618. doi: 10.1016/s0092-8674(03)00153-3. [DOI] [PubMed] [Google Scholar]
- Lotter H, et al. Natural killer T cells activated by a lipopeptidophosphoglycan from Entamoeba histolytica are critically important to control amebic liver abscess. PLoS pathogens. 2009;5:e1000434. doi: 10.1371/journal.ppat.1000434. doi:10.1371/journal.ppat.1000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotter H, Jacobs T, Gaworski I, Tannich E. Sexual dimorphism in the control of amebic liver abscess in a mouse model of disease. Infect Immun. 2006;74:118–124. doi: 10.1128/IAI.74.1.118-124.2006. doi:10.1128/IAI.74.1.118-124.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo MF, Porto G, Costa M, Vieira CP, Rocha B, Cruz E. Low numbers of CD8+ T lymphocytes in hereditary haemochromatosis are explained by a decrease of the most mature CD8+ effector memory T cells Clin Exp. Immunol. 2010;159:363–371. doi: 10.1111/j.1365-2249.2009.04066.x. doi:10.1111/j.1365-2249.2009.04066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madakamutil LT, et al. CD8alphaalpha-mediated survival and differentiation of CD8 memory T cell precursors. Science. 2004;304:590–593. doi: 10.1126/science.1092316. doi:10.1126/science.1092316. [DOI] [PubMed] [Google Scholar]
- Martin E, et al. Stepwise development of MAIT cells in mouse and human. PLoS Biol. 2009;7:e54. doi: 10.1371/journal.pbio.1000054. doi:10.1371/journal.pbio.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzarino P, et al. Identification of effector-memory CMV-specific T lymphocytes that kill CMV-infected target cells in an HLA-E-restricted fashion. European journal of immunology. 2005;35:3240–3247. doi: 10.1002/eji.200535343. doi:10.1002/eji.200535343. [DOI] [PubMed] [Google Scholar]
- McElhinny AS, Exley GE, Warner CM. Painting Qa-2 onto Ped slow preimplantatiom embryos increases the rate of cleavage. Am J Reprod Immunol. 2000;44:52–58. doi: 10.1111/j.8755-8920.2000.440108.x. doi:DOI 10.1111/j.8755-8920.2000.440108.x. [DOI] [PubMed] [Google Scholar]
- Meierovics A, Yankelevich WJ, Cowley SC. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E3119–3128. doi: 10.1073/pnas.1302799110. doi:10.1073/pnas.1302799110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda CJ, Makui H, Andrews NC, Santos MM. Contributions of beta2-microglobulin-dependent molecules and lymphocytes to iron regulation: insights from HfeRag1(−/−) and beta2mRag1(−/−) double knock-out mice. Blood. 2004;103:2847–2849. doi: 10.1182/blood-2003-09-3300. doi:10.1182/blood-2003-09-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales PJ, Pace JL, Platt JS, Langat DK, Hunt JS. Synthesis of beta(2)-microglobulin-free, disulphide-linked HLA-G5 homodimers in human placental villous cytotrophoblast cells. Immunology. 2007;122:179–188. doi: 10.1111/j.1365-2567.2007.02623.x. doi:10.1111/j.1365-2567.2007.02623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita A, et al. TL antigen as a transplantation antigen recognized by TL-restricted cytotoxic T cells. The Journal of experimental medicine. 1994;179:777–784. doi: 10.1084/jem.179.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan NA, Gonzalez F, Shastri N. Nonclassical MHC class Ib-restricted cytotoxic T cells monitor antigen processing in the endoplasmic reticulum. Nature immunology. 2012;13:579–586. doi: 10.1038/ni.2282. doi:10.1038/ni.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nattermann J, et al. The HLA-A2 restricted T cell epitope HCV core 35-44 stabilizes HLA-E expression and inhibits cytolysis mediated by natural killer cells. The American journal of pathology. 2005;166:443–453. doi: 10.1016/S0002-9440(10)62267-5. doi:10.1016/S0002-9440(10)62267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Old LJ, Boyse EA. Antigenic Properties of Experimental Leukemias. I. Serological Studies in Vitro with Spontaneous and Radiation-Induced Leukemias. J Natl Cancer Inst. 1963;31:977–995. [PubMed] [Google Scholar]
- Olivares-Villagomez D, Mendez-Fernandez YV, Parekh VV, Lalani S, Vincent TL, Cheroutre H, Van Kaer L. Thymus leukemia antigen controls intraepithelial lymphocyte function and inflammatory bowel disease. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17931–17936. doi: 10.1073/pnas.0808242105. doi:10.1073/pnas.0808242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardigon N, et al. CD8 alpha alpha-mediated intraepithelial lymphocyte snatching of thymic leukemia MHC class Ib molecules in vitro and in vivo. Journal of immunology (Baltimore, Md : 1950) 2006;177:1590–1598. doi: 10.4049/jimmunol.177.3.1590. [DOI] [PubMed] [Google Scholar]
- Parkkila S, et al. Association of the transferrin receptor in human placenta with HFE, the protein defective in hereditary hemochromatosis. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:13198–13202. doi: 10.1073/pnas.94.24.13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul P, et al. Identification of HLA-G7 as a new splice variant of the HLA-G mRNA and expression of soluble HLA-G5, -G6, and -G7 transcripts in human transfected cells. Hum Immunol. 2000;61:1138–1149. doi: 10.1016/s0198-8859(00)00197-x. [DOI] [PubMed] [Google Scholar]
- Pietra G, Romagnani C, Manzini C, Moretta L, Mingari MC. The emerging role of HLA-E-restricted CD8+ T lymphocytes in the adaptive immune response to pathogens and tumors. Journal of biomedicine & biotechnology. 2010;2010:907092. doi: 10.1155/2010/907092. doi:10.1155/2010/907092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietra G, et al. HLA-E-restricted recognition of cytomegalovirus-derived peptides by human CD8+ cytolytic T lymphocytes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:10896–10901. doi: 10.1073/pnas.1834449100. doi:10.1073/pnas.1834449100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8- alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. The Journal of experimental medicine. 1993;178:1–16. doi: 10.1084/jem.178.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porto G, Reimao R, Goncalves C, Vicente C, Justica B, de Sousa M. Haemochromatosis as a window into the study of the immunological system: a novel correlation between CD8+ lymphocytes and iron overload. Eur J Haematol. 1994;52:283–290. doi: 10.1111/j.1600-0609.1994.tb00097.x. [DOI] [PubMed] [Google Scholar]
- Princiotta MF, Lenz LL, Bevan MJ, Staerz UD. H2-M3 restricted presentation of a Listeria-derived leader peptide. The Journal of experimental medicine. 1998;187:1711–1719. doi: 10.1084/jem.187.10.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimpour A, et al. Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. The Journal of experimental medicine. 2015;212:1095–1108. doi: 10.1084/jem.20142110. doi:10.1084/jem.20142110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S, Long EO. KIR2DL4 (CD158d): An activation receptor for HLA-G. Frontiers in immunology. 2012;3:258. doi: 10.3389/fimmu.2012.00258. doi:10.3389/fimmu.2012.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reantragoon R, et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. The Journal of experimental medicine. 2013;210:2305–2320. doi: 10.1084/jem.20130958. doi:10.1084/jem.20130958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reantragoon R, et al. Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. The Journal of experimental medicine. 2012;209:761–774. doi: 10.1084/jem.20112095. doi:10.1084/jem.20112095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuben A, Phenix M, Santos MM, Lapointe R. The WT hemochromatosis protein HFE inhibits CD8(+) T-lymphocyte activation. European journal of immunology. 2014;44:1604–1614. doi: 10.1002/eji.201343955. doi:10.1002/eji.201343955. [DOI] [PubMed] [Google Scholar]
- Riegert P, Wanner V, Bahram S. Genomics, isoforms, expression, and phylogeny of the MHC class I-related MR1 gene. Journal of immunology (Baltimore, Md : 1950) 1998;161:4066–4077. [PubMed] [Google Scholar]
- Robinson PJ, Millrain M, Antoniou J, Simpson E, Mellor AL. A glycophospholipid anchor is required for Qa-2-mediated T cell activation. Nature. 1989;342:85–87. doi: 10.1038/342085a0. doi:10.1038/342085a0. [DOI] [PubMed] [Google Scholar]
- Rodgers JR, Cook RG. MHC class Ib molecules bridge innate and acquired immunity. Nature reviews Immunology. 2005;5:459–471. doi: 10.1038/nri1635. doi:10.1038/nri1635. [DOI] [PubMed] [Google Scholar]
- Rodgers JR, Mehta V, Cook RG. Surface expression of beta 2-microglobulin-associated thymus-leukemia antigen is independent of TAP2. European journal of immunology. 1995;25:1001–1007. doi: 10.1002/eji.1830250421. doi:10.1002/eji.1830250421. [DOI] [PubMed] [Google Scholar]
- Rohrlich PS, et al. Direct recognition by alphabeta cytolytic T cells of Hfe, a MHC class Ib molecule without antigen-presenting function. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12855–12860. doi: 10.1073/pnas.0502309102. doi:10.1073/pnas.0502309102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolph MS, Kaufmann SH. Partially TAP-independent protection against Listeria monocytogenes by H2-M3-restricted CD8+ T cells. Journal of immunology (Baltimore, Md : 1950) 2000;165:4575–4580. doi: 10.4049/jimmunol.165.8.4575. [DOI] [PubMed] [Google Scholar]
- Rotzschke O, Falk K, Stevanovic S, Grahovac B, Soloski MJ, Jung G, Rammensee HG. Qa-2 molecules are peptide receptors of higher stringency than ordinary class I molecules. Nature. 1993;361:642–644. doi: 10.1038/361642a0. doi:10.1038/361642a0. [DOI] [PubMed] [Google Scholar]
- Rouas-Freiss N, Goncalves RM, Menier C, Dausset J, Carosella ED. Direct evidence to support the role of HLA-G in protecting the fetus from maternal uterine natural killer cytolysis. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:11520–11525. doi: 10.1073/pnas.94.21.11520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salerno-Goncalves R, Fernandez-Vina M, Lewinsohn DM, Sztein MB. Identification of a human HLA-E-restricted CD8+ T cell subset in volunteers immunized with Salmonella enterica serovar Typhi strain Ty21a typhoid vaccine. Journal of immunology (Baltimore, Md : 1950) 2004;173:5852–5862. doi: 10.4049/jimmunol.173.9.5852. [DOI] [PubMed] [Google Scholar]
- Santos M, Schilham MW, Rademakers LH, Marx JJ, de Sousa M, Clevers H. Defective iron homeostasis in beta 2-microglobulin knockout mice recapitulates hereditary hemochromatosis in man. The Journal of experimental medicine. 1996;184:1975–1985. doi: 10.1084/jem.184.5.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos MM, de Sousa M, Rademakers LH, Clevers H, Marx JJ, Schilham MW. Iron overload and heart fibrosis in mice deficient for both beta2-microglobulin and Rag1. The American journal of pathology. 2000;157:1883–1892. doi: 10.1016/s0002-9440(10)64827-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf L, et al. The 2.5 A structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity. 2010;33:853–862. doi: 10.1016/j.immuni.2010.11.026. doi:10.1016/j.immuni.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CM, Garrett E, Orr HT. Cytotoxic T lymphocyte recognition of HLA-G in mice. Hum Immunol. 1997;55:127–139. doi: 10.1016/s0198-8859(97)00097-9. [DOI] [PubMed] [Google Scholar]
- Schulte D, et al. The HLA-E(R)/HLA-E(R) genotype affects the natural course of hepatitis C virus (HCV) infection and is associated with HLA-E-restricted recognition of an HCV-derived peptide by interferon-gamma-secreting human CD8(+) T cells. The Journal of infectious diseases. 2009;200:1397–1401. doi: 10.1086/605889. doi:10.1086/605889. [DOI] [PubMed] [Google Scholar]
- Seach N, et al. Double-positive thymocytes select mucosal-associated invariant T cells. Journal of immunology (Baltimore, Md : 1950) 2013;191:6002–6009. doi: 10.4049/jimmunol.1301212. doi:10.4049/jimmunol.1301212. [DOI] [PubMed] [Google Scholar]
- Seaman MS, Perarnau B, Lindahl KF, Lemonnier FA, Forman J. Response to Listeria monocytogenes in mice lacking MHC class Ia molecules. Journal of immunology (Baltimore, Md : 1950) 1999;162:5429–5436. [PubMed] [Google Scholar]
- Shawar SM, Vyas JM, Shen E, Rodgers JR, Rich RR. Differential amino-terminal anchors for peptide binding to H-2M3a or H-2Kb and H-2Db. Journal of immunology (Baltimore, Md : 1950) 1993;151:201–210. [PubMed] [Google Scholar]
- Siddiqui S, Visvabharathy L, Wang CR. Role of Group 1 CD1-Restricted T Cells in Infectious Disease. Frontiers in immunology. 2015;6:337. doi: 10.3389/fimmu.2015.00337. doi:10.3389/fimmu.2015.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GP, Dabhi VM, Pamer EG, Lindahl KF. Peptide presentation by the MHC class Ib molecule, H2-M3. International immunology. 1994;6:1917–1926. doi: 10.1093/intimm/6.12.1917. [DOI] [PubMed] [Google Scholar]
- Stroynowski I, Lindahl KF. Antigen presentation by non-classical class I molecules. Current opinion in immunology. 1994;6:38–44. doi: 10.1016/0952-7915(94)90031-0. [DOI] [PubMed] [Google Scholar]
- Stroynowski I, Soloski M, Low MG, Hood L. A single gene encodes soluble and membrane-bound forms of the major histocompatibility Qa-2 antigen: anchoring of the product by a phospholipid tail. Cell. 1987;50:759–768. doi: 10.1016/0092-8674(87)90334-5. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Kraj P, Weber DA, Ignatowicz L, Jensen PE. Positive selection of a Qa-1-restricted T cell receptor with specificity for insulin. Immunity. 2002;17:95–105. doi: 10.1016/s1074-7613(02)00343-6. [DOI] [PubMed] [Google Scholar]
- Swanson PA, 2nd, Pack CD, Hadley A, Wang CR, Stroynowski I, Jensen PE, Lukacher AE. An MHC class Ib-restricted CD8 T cell response confers antiviral immunity. The Journal of experimental medicine. 2008;205:1647–1657. doi: 10.1084/jem.20080570. doi:10.1084/jem.20080570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabaczewski P, Chiang E, Henson M, Stroynowski I. Alternative peptide binding motifs of Qa-2 class Ib molecules define rules for binding of self and nonself peptides. Journal of immunology (Baltimore, Md : 1950) 1997;159:2771–2781. [PubMed] [Google Scholar]
- Tabaczewski P, Shirwan H, Lewis K, Stroynowski I. Alternative splicing of class Ib major histocompatibility complex transcripts in vivo leads to the expression of soluble Qa-2 molecules in murine blood. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:1883–1887. doi: 10.1073/pnas.91.5.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabaczewski P, Stroynowski I. Expression of secreted and glycosylphosphatidylinositol-bound Qa-2 molecules is dependent on functional TAP-2 peptide transporter. Journal of immunology (Baltimore, Md : 1950) 1994;152:5268–5274. [PubMed] [Google Scholar]
- Tatituri RV, et al. Recognition of microbial and mammalian phospholipid antigens by NKT cells with diverse TCRs. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1827–1832. doi: 10.1073/pnas.1220601110. doi:10.1073/pnas.1220601110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitell M, Holcombe H, Cheroutre H, Aldrich CJ, Stroynowski I, Forman J, Kronenberg M. The alpha 3 domain of the Qa-2 molecule is defective for CD8 binding and cytotoxic T lymphocyte activation The. Journal of experimental medicine. 1993;178:2139–2145. doi: 10.1084/jem.178.6.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilloy F, et al. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. The Journal of experimental medicine. 1999;189:1907–1921. doi: 10.1084/jem.189.12.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasec P, et al. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science. 2000;287:1031. doi: 10.1126/science.287.5455.1031. [DOI] [PubMed] [Google Scholar]
- Treiner E. Mucosal-associated invariant T cells in inflammatory bowel diseases: bystanders, defenders, or offenders? Frontiers in immunology. 2015;6:27. doi: 10.3389/fimmu.2015.00027. doi:10.3389/fimmu.2015.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiner E, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003;422:164–169. doi: 10.1038/nature01433. doi:10.1038/nature01433. [DOI] [PubMed] [Google Scholar]
- Treiner E, Liblau RS. Mucosal-Associated Invariant T Cells in Multiple Sclerosis: The Jury is Still Out. Frontiers in immunology. 2015;6:503. doi: 10.3389/fimmu.2015.00503. doi:10.3389/fimmu.2015.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimura K, Obata Y, Iwase S, Matsudaira Y, Ozeki S, Takahashi T. The epitope detected by cytotoxic T lymphocytes against thymus leukemia (TL) antigen is TAP independent. International immunology. 2000;12:1217–1225. doi: 10.1093/intimm/12.9.1217. [DOI] [PubMed] [Google Scholar]
- Tsujimura K, et al. Thymus leukemia antigen (TL)-specific cytotoxic T lymphocytes recognize the alpha1/alpha2 domain of TL free from antigenic peptides. International immunology. 2003;15:1319–1326. doi: 10.1093/intimm/dxg131. [DOI] [PubMed] [Google Scholar]
- Tsujimura K, Obata Y, Matsudaira Y, Ozeki S, Yoshikawa K, Saga S, Takahashi T. The binding of thymus leukemia (TL) antigen tetramers to normal intestinal intraepithelial lymphocytes and thymocytes. Journal of immunology (Baltimore, Md : 1950) 2001;167:759–764. doi: 10.4049/jimmunol.167.2.759. [DOI] [PubMed] [Google Scholar]
- Tsujimura K, Takahashi T, Morita A, Hasegawa-Nishiwaki H, Iwase S, Obata Y. Positive selection of gamma delta CTL by TL antigen expressed in the thymus. The Journal of experimental medicine. 1996;184:2175–2184. doi: 10.1084/jem.184.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tupin E, et al. NKT cells prevent chronic joint inflammation after infection with Borrelia burgdorferi. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19863–19868. doi: 10.1073/pnas.0810519105. doi:10.1073/pnas.0810519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvinnereim A, Wizel B. CD8+ T cell protective immunity against Chlamydia pneumoniae includes an H2-M3-restricted response that is largely CD4+ T cell-independent. Journal of immunology (Baltimore, Md : 1950) 2007;179:3947–3957. doi: 10.4049/jimmunol.179.6.3947. [DOI] [PubMed] [Google Scholar]
- Tyznik AJ, Verma S, Wang Q, Kronenberg M, Benedict CA. Distinct requirements for activation of NKT and NK cells during viral infection. Journal of immunology (Baltimore, Md : 1950) 2014;192:3676–3685. doi: 10.4049/jimmunol.1300837. doi:10.4049/jimmunol.1300837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugrinovic S, Brooks CG, Robson J, Blacklaws BA, Hormaeche CE, Robinson JH. H2-M3 major histocompatibility complex class Ib-restricted CD8 T cells induced by Salmonella enterica serovar Typhimurium infection recognize proteins released by Salmonella serovar Typhimurium. Infect Immun. 2005;73:8002–8008. doi: 10.1128/IAI.73.12.8002-8008.2005. doi:10.1128/IAI.73.12.8002-8008.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrecht M, Martinozzi S, Grzeschik M, Hengel H, Ellwart JW, Pla M, Weiss EH. Cutting edge: the human cytomegalovirus UL40 gene product contains a ligand for HLA-E and prevents NK cell-mediated lysis. Journal of immunology (Baltimore, Md : 1950) 2000;164:5019–5022. doi: 10.4049/jimmunol.164.10.5019. [DOI] [PubMed] [Google Scholar]
- Ulrichs T, Moody DB, Grant E, Kaufmann SH, Porcelli SA. T-cell responses to CD1-presented lipid antigens in humans with Mycobacterium tuberculosis infection. Infect Immun. 2003;71:3076–3087. doi: 10.1128/IAI.71.6.3076-3087.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdahl KB, Liggitt D, Bevan MJ. CD8+ T cells accumulate in the lungs of Mycobacterium tuberculosis-infected Kb−/−Db−/− mice, but provide minimal protection. Journal of immunology (Baltimore, Md : 1950) 2003;170:1987–1994. doi: 10.4049/jimmunol.170.4.1987. [DOI] [PubMed] [Google Scholar]
- Urdahl KB, Sun JC, Bevan MJ. Positive selection of MHC class Ib-restricted CD8(+) T cells on hematopoietic cells. Nature immunology. 2002;3:772–779. doi: 10.1038/ni814. doi:10.1038/ni814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ussher JE, et al. CD161++ CD8+ T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. European journal of immunology. 2014;44:195–203. doi: 10.1002/eji.201343509. doi:10.1002/eji.201343509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meijgaarden KE, Haks MC, Caccamo N, Dieli F, Ottenhoff TH, Joosten SA. Human CD8+ T-cells recognizing peptides from Mycobacterium tuberculosis (Mtb) presented by HLA-E have an unorthodox Th2-like, multifunctional, Mtb inhibitory phenotype and represent a novel human T-cell subset. PLoS pathogens. 2015;11:e1004671. doi: 10.1371/journal.ppat.1004671. doi:10.1371/journal.ppat.1004671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rhijn I, et al. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nature immunology. 2013;14:706–713. doi: 10.1038/ni.2630. doi:10.1038/ni.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance RE, Jamieson AM, Raulet DH. Recognition of the class Ib molecule Qa-1(b) by putative activating receptors CD94/NKG2C and CD94/NKG2E on mouse natural killer cells. The Journal of experimental medicine. 1999;190:1801–1812. doi: 10.1084/jem.190.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance RE, Kraft JR, Altman JD, Jensen PE, Raulet DH. Mouse CD94/NKG2A is a natural killer cell receptor for the nonclassical major histocompatibility complex (MHC) class I molecule Qa-1(b) The Journal of experimental medicine. 1998;188:1841–1848. doi: 10.1084/jem.188.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CR, Castano AR, Peterson PA, Slaughter C, Lindahl KF, Deisenhofer J. Nonclassical binding of formylated peptide in crystal structure of the MHC class Ib molecule H2-M3. Cell. 1995;82:655–664. doi: 10.1016/0092-8674(95)90037-3. [DOI] [PubMed] [Google Scholar]
- Wang CR, Loveland BE, Lindahl KF. H-2M3 encodes the MHC class I molecule presenting the maternally transmitted antigen of the mouse. Cell. 1991;66:335–345. doi: 10.1016/0092-8674(91)90623-7. [DOI] [PubMed] [Google Scholar]
- Wang EC, et al. UL40-mediated NK evasion during productive infection with human cytomegalovirus. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:7570–7575. doi: 10.1073/pnas.112680099. doi:10.1073/pnas.112680099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner CM, Gollnick SO, Flaherty L, Goldbard SB. Analysis of Qa-2 antigen expression by preimplantation mouse embryos: possible relationship to the preimplantation-embryo-development (Ped) gene product. Biol Reprod. 1987;36:611–616. doi: 10.1095/biolreprod36.3.611. [DOI] [PubMed] [Google Scholar]
- Weber DA, et al. Peptide-independent folding and CD8 alpha alpha binding by the nonclassical class I molecule, thymic leukemia antigen. Journal of immunology (Baltimore, Md : 1950) 2002;169:5708–5714. doi: 10.4049/jimmunol.169.10.5708. [DOI] [PubMed] [Google Scholar]
- Wesley JD, Tessmer MS, Chaukos D, Brossay L. NK cell-like behavior of Valpha14i NK T cells during MCMV infection. PLoS pathogens. 2008;4:e1000106. doi: 10.1371/journal.ppat.1000106. doi:10.1371/journal.ppat.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland Brown LC, et al. Production of alpha-galactosylceramide by a prominent member of the human gut microbiota. PLoS Biol. 2013;11:e1001610. doi: 10.1371/journal.pbio.1001610. doi:10.1371/journal.pbio.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MA, Bevan MJ. Cutting edge: a single MHC class Ia is sufficient for CD8 memory T cell differentiation. Journal of immunology (Baltimore, Md : 1950) 2005;175:2066–2069. doi: 10.4049/jimmunol.175.4.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, van Kaer L, Itohara S, Tonegawa S. Highly restricted expression of the thymus leukemia antigens on intestinal epithelial cells. The Journal of experimental medicine. 1991;174:213–218. doi: 10.1084/jem.174.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Chun T, Choi HJ, Wang B, Wang CR. Impaired response to Listeria in H2-M3-deficient mice reveals a nonredundant role of MHC class Ib-specific T cells in host defense. The Journal of experimental medicine. 2006;203:449–459. doi: 10.1084/jem.20051866. doi:10.1084/jem.20051866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager M, Kumar S, Hughes AL. Sequence convergence in the peptide-binding region of primate and rodent MHC class Ib molecules. Mol Biol Evol. 1997;14:1035–1041. doi: 10.1093/oxfordjournals.molbev.a025709. [DOI] [PubMed] [Google Scholar]
- Yokoyama K, Stockert E, Old LJ, Nathenson SG. Structural evidence that the small subunit found associated with the TL antigen is beta 2-microglobulin. Immunogenetics. 1982;15:543–549. doi: 10.1007/BF00347048. [DOI] [PubMed] [Google Scholar]
- Zajonc DM, et al. Molecular mechanism of lipopeptide presentation by CD1a. Immunity. 2005;22:209–219. doi: 10.1016/j.immuni.2004.12.009. doi:10.1016/j.immuni.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Zeng L, et al. A structural basis for antigen presentation by the MHC class Ib molecule, Qa-1b. Journal of immunology (Baltimore, Md : 1950) 2012;188:302–310. doi: 10.4049/jimmunol.1102379. doi:10.4049/jimmunol.1102379. [DOI] [PubMed] [Google Scholar]
- Zhao J, Siddiqui S, Shang S, Bian Y, Bagchi S, He Y, Wang CR. Mycolic acid-specific T cells protect against Mycobacterium tuberculosis infection in a humanized transgenic mouse model. Elife. 2015;4 doi: 10.7554/eLife.08525. doi:10.7554/eLife.08525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XY, et al. HFE gene knockout produces mouse model of hereditary hemochromatosis. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:2492–2497. doi: 10.1073/pnas.95.5.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]