Abstract
Long Interspersed Nucleotide Element 1 (LINE-1) retrotransposons are heavily methylated and are the most abundant transposable elements in mammalian genomes. Here, we investigated the differential DNA methylation within the LINE-1 under normal conditions and in response to environmentally relevant doses of sparsely and densely ionizing radiation. We demonstrate that DNA methylation of LINE-1 elements in the lungs of C57BL6 mice is dependent on their evolutionary age, where the elder age of the element is associated with the lower extent of DNA methylation. Exposure to 5-aza-2′-deoxycytidine and methionine-deficient diet affected DNA methylation of selective LINE-1 elements in an age- and promoter type-dependent manner. Exposure to densely IR, but not sparsely IR, resulted in DNA hypermethylation of older LINE-1 elements, while the DNA methylation of evolutionary younger elements remained mostly unchanged. We also demonstrate that exposure to densely IR increased mRNA and protein levels of LINE-1 via the loss of the histone H3K9 dimethylation and an increase in the H3K4 trimethylation at the LINE-1 5′-untranslated region, independently of DNA methylation. Our findings suggest that DNA methylation is important for regulation of LINE-1 expression under normal conditions, but histone modifications may dictate the transcriptional activity of LINE-1 in response to exposure to densely IR.
Keywords: epigenetics, high-LET radiation, histone methylation, ionizing radiation, transposable elements
1. Introduction
The terrestrial environment and various diagnostic and treatment modalities are primarily characterized by sparsely ionizing radiation (IR), while densely IR is prevalent in the space environment. Recently, densely IR is being increasingly utilized in the clinic, due to the possibility of delivering a higher dose to a confined volume, thus potentially increasing the effects on the tumor tissue while decreasing the normal tissue toxicity (Durante, 2014; Girdhani et al., 2013).
Aside from the recognized genotoxic potential (Girdhani et al., 2013; Goodhead et al., 1993; Prise et al., 1994), exposure to densely IR is also characterized by epigenetic alterations, including aberrant global and TEs-associated DNA methylation (Aypar et al., 2011; Jangiam et al., 2015; Lima et al., 2014; Miousse et al., 2014; Nzabarushimana et al., 2014). DNA methylation is a fundamental epigenetic mechanism that controls the proper expression of genetic information in a tissue-, cell type- and sex-dependent manner and plays a key role in the silencing of transposable elements (TEs) (Jones, 2012). Long Interspersed Nucleotide Elements 1 (LINE-1) constitute the dominant category of TEs comprising ~20% of mammalian genomes (Miousse et al., 2015). By nature, these TEs are autonomous retrotransposons that propagate through the genome via a “copy-paste” mechanism. Aberrant methylation and, associated with it, the activation and retrotransposition of LINE-1 elements have been reported in numerous pathological states, including human cancers (Helman et al., 2014; Lee et al., 2012; Miousse and Koturbash, 2015; Rodić et al., 2014). Furthermore, hypomethylation of the LINE-1 5′-untranslated region (UTR) is thought to alter the expression of nearby genes, and hypomethylation of LINE-1 inserts in the promoters or introns of host genes can also alter expression of the latter (Hur et al., 2014; Wolff et al., 2010).
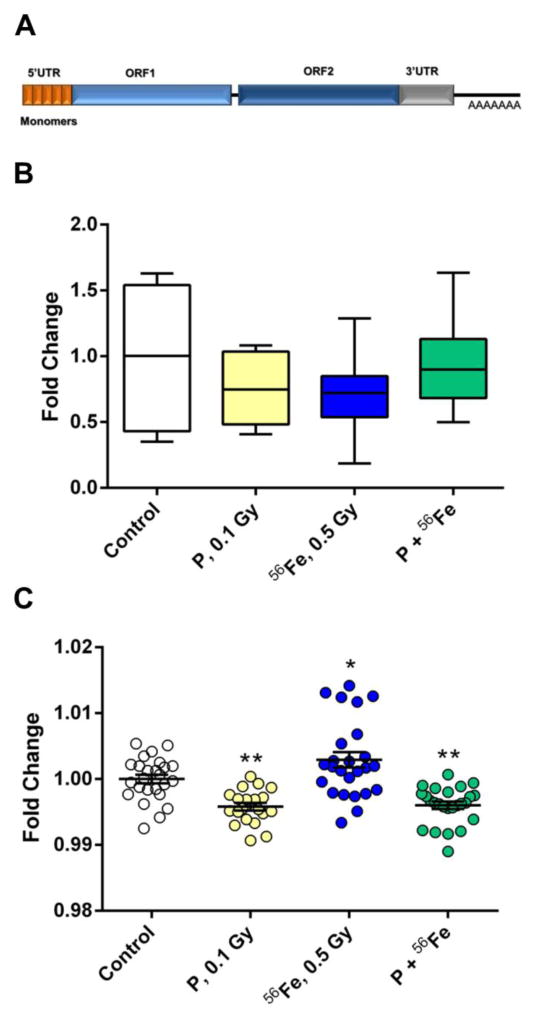
The full-length LINE-1 elements, aside from a ~900 bp long 5′-UTR that serves as an internal promoter, also contain a bicistronic open reading frame that encodes two proteins - ORF1p (a 40 kDa trimeric protein that possesses RNA binding and nucleic acid chaperone activity) and ORF2p (a 150 kDa protein that encodes an endonuclease, reverse transcriptase, and zinc finger-like protein and is responsible for the actual LINE-1 retrotransposition) and a 3′-UTR with a poly(A) tail (Cost et al., 2002; Feng et al., 1996) (Fig. 1A). At the same time, evolutionary patterns of LINE-1, characterized by the continuous replacement of extinct families by more recently evolved families, predetermines substantial differences between these elements (Khan et al., 2006). These differences primarily stem from the 5′-UTR ends sequence and structure, while the ORF sequences remain highly homologous between the families. The recent advances in computational biology have allowed the classification of murine LINE-1 families according to each family’s unique 5′-UTR sequence and evolutionary age (Sookdeo et al., 2013).
Figure 1.
Effects of exposure to densely IR on LINE-1 DNA methylation. (A) LINE-1 schematic representation. (B) Analysis of LINE-1 ORF2 DNA methylation. Data are presented as ± SEM. (C) Analysis of LINE-1 ORF1 DNA methylation. Cumulative data from five CpG sites presented as changes in the percentage of methylated CpG sites ± SEM. Asterisks (***) denotes significant (P<0.001) difference from control (one-way ANOVA). P – protons; 56Fe – heavy iron ions; P+56Fe – consequent exposure to protons and heavy iron ions.
Both sparsely and densely IR are capable of causing pulmonary fibrosis and lung cancer in humans and in experimental models (Maddams et al., 2011; Munley et al., 2011; Yarnold and Brotons, 2010). The role of epigenetic alterations, including aberrant methylation and expression of LINE-1 elements in the development and promotion of radiation-induced fibrosis and lung cancer is becoming increasingly recognized (Ikeda et al., 2013; Saito et al., 2010; Weigel et al., 2015). In our previous studies, we reported the long-term dose-dependent genomic and LINE-1 ORF1-associated DNA hypermethylation in the mouse lung 5 months after exposure to heavy iron ions (56Fe) (Nzabarushimana et al., 2014), but failed to identify changes in LINE-1 ORF1 methylation four weeks after exposure to either protons or 56Fe exposure, or as a result of the sequential exposure to both (Nzabarushimana et al., 2015). In this study, we aimed to investigate whether or not exposure to environmentally relevant low absorbed mean doses of densely IR would result in evolutionary- and family-dependent alterations in the DNA methylation status of LINE-1 in the mouse lung.
2. Methods
2.1. Animals and radiation exposures
All animals in the study were eight-week-old male C57BL/6J mice purchased from Jackson Laboratory (Bar Harbor, ME, USA). For the proton and 56Fe exposure, animals were shipped to Brookhaven National Laboratories (BNL) in Upton, NY, USA. After a one-week acclimation period, the mice were either sham irradiated or received whole-body irradiation [protons 0.1 Gy, 150 MeV/n; 56Fe 0.5 Gy, 600 MeV/n; or combined sequential exposure to 0.1 Gy of protons (Day 1) and 0.5 Gy of 56Fe (Day 2); n = 16 mice/group] at the dose rate of 0.5 Gy/min. The dose of protons was chosen as likely to occur during a solar particle event (SPE) (Wu et al., 2009). The energy of 150 MeV is commonly used in a therapeutic setting and also represents energy near the maximum abundance of protons expected in most SPEs (NCRP., 2006). The acute dose of 56Fe was selected to reflect the estimated cumulative dose to which astronauts could be potentially exposed during deep space exploration due to the galactic cosmic rays and is the lowest dose previously shown to cause cell loss in in vivo settings during deep space exploration due to the galactic cosmic rays (Rola et al., 2008). Sequential irradiation with protons before iron ions reflects the likely exposure of cells in the space environment where daily traversals by protons are accompanied by infrequent traversals by heavy ions (Ramadan et al., 2016). At the selected energy of 600 MeV/n, thorough penetration of the animals with a relatively flat Bragg peak entrance region is expected. Dosimetry was performed by the NASA Space Radiation Laboratory physics dosimetry group at BNL to ensure the quality of exposure. For each exposure, animals were individually placed into clear Lucite cubes (3 in × 1½ in × 1½ in) with breathing holes. Sham irradiated mice served as controls and were placed into the same enclosures and for the same amount of time, since previous studies reported no effect of sham irradiation on molecular end-points (Miousse et al., 2014). One week after irradiation, the mice were shipped to Oregon Health and Science University (OHSU) where they underwent behavioral testing reported elsewhere (Raber et al., 2015). Food (PicoLab Rodent Diet 20, no. 5053; PMI Nutrition International, St. Louis, MO, USA) and water were provided ad libitum.
To compare the response to exposure between densely IR and sparsely IR, animals were housed at the University of Arkansas for Medical Sciences (UAMS). After a one-week acclimation period, the mice were randomly divided into three groups (n=6/group): sham or whole-body irradiation to 0.1 or 1 Gy of 137Cs (dose rate 1.21 Gy/min) Dose of 0.1 Gy was chosen given the previously reported relative biological effectiveness (RBE) values for protons vs gamma rays to be ~ 1 (ranging from 0.7 to 1.6) (Paganetti et al., 2002) Dose of 1 Gy was selected given the previously reported RBE values for 56Fe when compared to gamma-rays in the range of 0.9–3.3 (Brooks et al., 2001) (Brooks et al., 2001; Paganetti et al., 2002). Irradiation was performed with a J. L. Shepherd Mark I [model 25 137Cs irradiator (J. L. Shepherd & Associates, San Fernando, CA, USA)]. Un-anesthetized mice were placed in cylindrical, well-ventilated Plexiglas chambers (J. L. Shepherd & Associates) divided into four 90° “pie slice” compartments by vertical dividers made of T-6061 aluminum with a gold anodized coating. Chambers were placed on a turntable rotating at 5 rpm in the position furthest away from the radiation source.
All animals were killed by cervical dislocation four weeks after irradiation; lungs were excised and immediately frozen in liquid nitrogen. All procedures were approved by the Institutional Animal Care and Use Committee at UAMS, OHSU and BNL.
2.2. Animals and diet administration
After a one-week acclimation period at UAMS, the mice were randomly divided into two groups (n=5/group): animals fed a methionine/choline adequate diet (TD.140520) and animals fed a methionine/choline deficient diet (TD.90262) (Supplemental Table S1) (Harlan Teklad, Madison, WI, USA). The diets and water were provided ad libitum. Animals were killed by cervical dislocation after eight weeks of diet administration. Lungs were excised and immediately frozen in liquid nitrogen. All procedures were approved by the UAMS Institutional Animal Care and Use Committee.
2.3. Cell culture and treatment with 5-Aza-2′-deoxycytidine
RAW 264.7 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were grown in DMEM with 10% FBS and Pen/Strep (all - Life Technologies, Carlsbad, CA, USA), and were maintained in a humidified incubator at 37°C, 5% CO2. After allowing cells to adhere (24 h), they were treated with 2 μM of 5-Aza-2′-deoxycytidine (Sigma-Aldrich, St. Louis, MO, USA) for 96 h. Cells were then lysed and the DNA was extracted using TRIzol (Life Technologies) extraction according to manufacturer’s instructions.
2.4. Nucleic acids extraction
RNA and DNA were extracted simultaneously from flash-frozen tissue using the AllPrep DNA/RNA extraction kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. DNA concentrations and integrity were analyzed by the Nanodrop 2000 (Thermo Scientific, Waltham, MA, USA) and 1% agarose gel.
2.5. Analysis of LINE-1 ORF2 DNA methylation by McrBC-methylation sensitive quantitative real-time PCR (MS qRT PCR)
Analysis was carried out as follows: 500 ng of total genomic DNA were digested overnight using 1 U pf McrBC in 1X NEB buffer, 200 μg/mL BSA, and 1 mM GTP (New England Biolabs, Ipswich, MA, USA) at 37°C. One μg of digested DNA was then analyzed via qRT-PCR using SYBR Select (Life Technologies) and a ViiA 7 Real-Time PCR System (Applied Biosystems, Forrest City, CA, USA). Assays are listed in Supplemental Table S2. The threshold cycle (Ct) values were converted into the absolute amount of input DNA using the absolute standard curve method and further normalized towards rDNA readings.
2.6. Analysis of LINE-1 ORF1 DNA methylation by pyrosequencing
Bisulfite conversion was performed on 1 μg of DNA using the EZ DNA Methylation™ Kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s protocol. Pyrosequencing analysis was performed using the PyroMark Q96 system (Qiagen) on five CpG sites within the murine LINE-1 ORF1. Percentage of the LINE-1 ORF1 DNA methylation is the percentage of methylated cytosines over the sum of methylated and un-methylated cytosines.
2.7. LINE-1 families’ consensus sequences
LINE-1 families’ consensus sequences were obtained from the Genetic Information Research Institute (GIRI) Database: http://www.girinst.org/ (Bao et al., 2015; Jurka et al., 2005).
2.8. Construction of LINE-1 phylogenetic tree
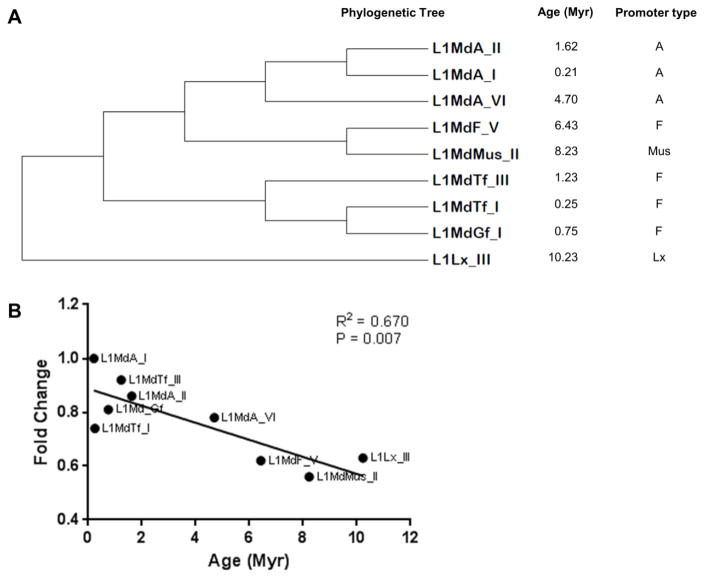
The evolutionary history was inferred by using the Maximum Likelihood method based on the Hasegawa-Kishino-Yano model (Hasegawa et al., 1985). The original tree was obtained by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood approach, and then selecting the topology with superior log likelihood value. The bootstrap consensus tree inferred from 1000 replicates was taken to represent the evolutionary history of the murine LINE-1 families analyzed. Evolutionary analyses were conducted in MEGA 6.06 software (Tamura et al., 2013). The analysis involved nine 5′-UTR sequences of the investigated LINE-1 families. Sequences were aligned using MUSCLE 3.8.31 software with default settings (Edgar, 2004) (Fig. 2).
Figure 2.
Evolutionary age of LINE-1 elements. (A) Phylogenetic tree, promoter type and evolutionary age of the investigated murine LINE-1 elements. (B) Correlation between the extent of DNA methylation and the evolutionary age of LINE-1 elements in the normal mouse lung. Data normalized to the values of the youngest LINE-1 family, L1MdA_I. Myr – million years.
2.9. Analysis of DNA methylation within the 5′-UTR of selective LINE-1 elements
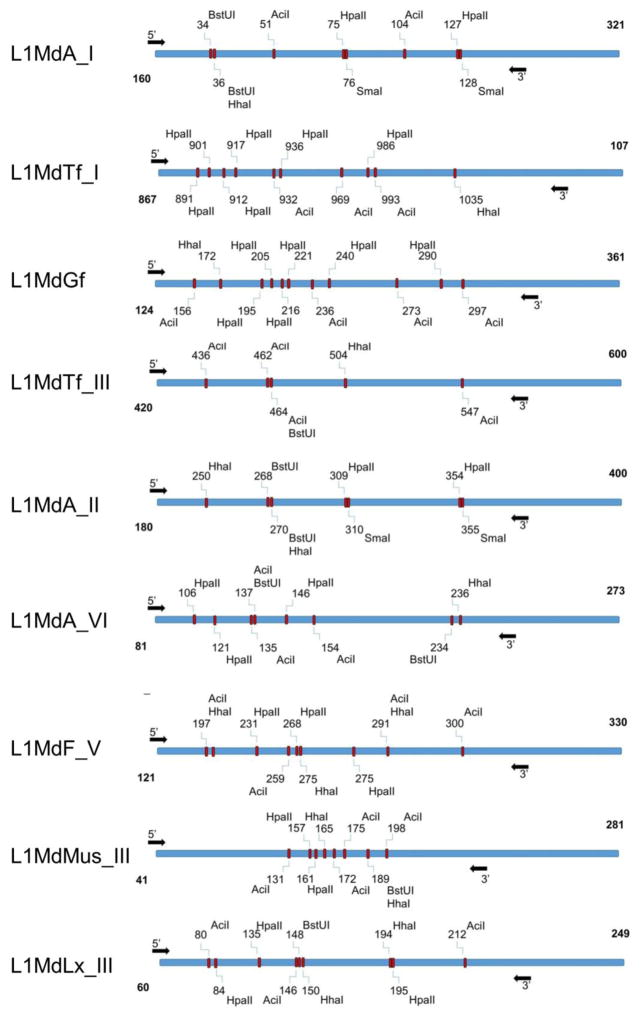
First, the 5′-UTRs of nine LINE-1 elements were analyzed using NEBcutter® (http://nc2.neb.com/NEBcutter2/). Five most frequent CpG sites that can be cleaved by the methylation-sensitive restriction enzymes (AciI, BstUI, HhaI, HpaII, and SmaI) were identified (Fig. 3). Analysis of the DNA methylation was performed as follows: one μg of genomic DNA was digested with 1 U of SmaI in 1X CutSmart buffer at 25°C for 2 h. This was followed by a 16 h digestion at 37°C in the presence of 1 U of the HpaII, HhaI, and AciI in 1X CutSmart buffer. The digestion was finalized by adding 0.5 U of BstUI in 1X CutSmart buffer for 4 h at 60°C (New England Biolabs). Digested DNA was then analyzed by qRT-PCR on a ViiA 7 Real-Time PCR System (Applied Biosystems). DNA samples not digested with the restriction enzyme mix served as a positive control, while samples lacking the specific primers for DNA amplification and/or DNA template served as negative controls. The Ct was defined as the fractional cycle number that passes the fixed threshold. The Ct values were converted into the absolute amount of input DNA using the absolute standard curve method and further normalized towards readings from the respective to each LINE-1 element ORF1 region that lacks CpG sites. Assays for determination of 5′-UTR LINE-1 DNA methylation are provided in Supplementary Table S2.
Figure 3.
Schematic representation of the 5′-UTRs of nine LINE-1 elements with methylation-sensitive restriction sites. Arrows define the amplification loci.
2.10. Analysis of LINE-1 5′-UTR by Methylated DNA Immunoprecipitation (MeDIP) Assay
Five μg of genomic DNA was randomly sheared to an average length of 0.2–1.0 kb by sonication and divided into immunoprecipitated and input portions. DNA from the immunoprecipitated portions was incubated overnight at 4°C with a monoclonal antibody (5 μg) against 5-methylcytosine (Abcam, Cambridge, MA, USA), followed by overnight incubation with Pan-mouse IgG Dynal magnetic beads (Invitrogen, Carlsbad, CA, USA) at 4°C. The methylated DNA/antibody complexes were then digested with proteinase K, and enriched DNA was recovered by phenol-chloroform extraction followed by ethanol precipitation. Purified DNA from immunoprecipitated and input DNA samples was analyzed by qPCR on a ViiA 7 Real-Time PCR System (Applied Biosystems) as described above. The relative changes in the extent of LINE1 methylation were determined by measuring the amount of DNA in immunoprecipitated DNA after normalization to the input DNA.
2.11. Identification of L1MdA_II evolutionary insertional mutagenesis events
The targets of L1MdA_II insertional mutagenesis were identified using the UCSC genome browser: https://genome.ucsc.edu/
2.12. Gene expression analysis using quantitative reverse transcription PCR
cDNA was synthesized using random primers and a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s protocol. The levels of gene transcripts were determined by qRT-PCR using TaqMan Gene Expression Assays (Supplemental Table S2) (Life Technologies). Each plate contained one experimental gene and a housekeeping gene. The Ct for each sample was determined from the linear region of the amplification plot. The ΔΔCt were calculated using each exposed group means relative to control group means. The fold change data were calculated from the ΔΔCt values. All qRT-PCR reactions were conducted in triplicate and repeated twice.
2.13. Analysis of the LINE-1 protein levels by Western Blot
Total cellular proteins were extracted using RIPA buffer. A total of 2 μg of total lysate was loaded on a 12% SDS-PAGE gel and transferred on a PVDF membrane. The membrane was blotted with primary antibodies against LINE-1 ORF1p (clone M-300, Santa Cruz Biotechnology, Dallas, TX, USA) and a goat anti-rabbit HRP secondary antibody (Millipore, Billerica, MA, USA). Chemoluminescence was detected with the SuperSignal West Pico Kit (Thermo Scientific, Grand Island, NY, USA). The loading control was produced by staining the same membrane with Ponceau solution.
2.14. Chromatin Immunoprecipitation (ChIP)
Frozen murine lung tissue was minced and cross-linked in 1% formaldehyde for 10 min, after which the reaction was quenched with 125 mM glycine. The tissues were then homogenized to a single cell suspension and then sonicated for 30 min at 4°C. M280 sheep anti-rabbit IgG Dynabeads (Life Technologies) were labeled with appropriate antibodies [anti-histone H3 # ab1791, anti-histone H3 (dimethyl K9) # ab1220, or anti-histone H3 (trimethyl K4) # ab12209 (Abcam)] overnight at 4°C. The sonicated sample slurry was then added to the labeled beads and incubated for 4 h at 4°C. After reversal of cross-linking, the immunoprecipitated DNA was chemically precipitated via phenol/chloroform/isoamyl alcohol and ethanol extracted. One ng of DNA for each sample was then analyzed by qRT-PCR using SYBR Select (Life Technologies). Values were normalized to the internal control, L1Lx_I ORF1 using the ΔΔCt method, and further normalized to average values from precipitation with anti-histone H3.
2.15. Statistical analysis
All data are presented as mean ± standard error of mean(s). Statistically significant differences for each treatment compared to the control (at α=95%) were assessed using one-way ANOVAs followed by Dunnett’s or Tukey’s posthoc tests. Statistical analyses were performed using GraphPad Prism 6 (GraphPad Software Inc. LaJolla, CA, USA).
3. Results
3.1. Lack of radiation-induced changes in DNA methylation of ORF1 and ORF2
First, the DNA methylation status of the LINE-1 ORF2 in response to exposure to protons, 56Fe or sequential exposure to protons and 56Fe was assessed in the murine lung using McrBC-qPCR. No significant differences were detected between the control and exposed mice four weeks after irradiation (Fig. 1B).
Next, pyrosequencing – the most sensitive approach to evaluate locus-specific DNA methylation – was utilized, in order to confirm these findings. The loss in LINE-1 ORF1 DNA methylation after exposure to protons (−0.46%, P<0.001) and sequential exposure to protons and 56Fe (−0.45%, P<0.001) was identified (Fig. 1C). No significant changes in the DNA methylation of LINE-1 ORF1 in response to 56Fe exposure were detected.
3.2. Influence of the LINE-1 evolutionary age and 5′-UTR structure on its DNA methylation status
We hypothesized that although no substantial differences in the DNA methylation of LINE-1 within ORF1 and ORF2 were detected, such differences could be identified within the 5′-UTR of LINE-1 elements that belong to particular families. Furthermore, murine LINE-1 ORF1 and ORF2 contain substantially lower number of CpG sites in comparison with the 5′-UTR, suggesting that the 5′-UTR may serve as a better substrate in the analysis of LINE-1 DNA methylation. Therefore, next we addressed the 5′-UTR methylation status of nine LINE-1 elements that belong to distinct families based on their promoter types - A, Tf, Gf, Mus, and Lx – and which also substantially differ by their evolutionary age (from 0.21 Myr old LINE-1 LMdA_1 to 10.23 Myr old LINE-1 L1Lx_III) (Fig. 2). In the normal mouse lung, the 5′-UTRs of evolutionary elder elements were significantly hypomethylated in comparison with the elements that evolved later (R2=0.670, P=0.007; Fig. 2B). No promoter type-specific differences in DNA methylation of LINE-1 elements were detected.
3.3. Demethylating agent 5-Aza-2′-deoxycytidine – selectively affects younger LINE-1 elements in vitro
Next, we hypothesized that exogenous stressors may differentially affect the methylation status of 5′-UTRs that belong to distinct LINE-1 families. In order to test this hypothesis, RAW 264.7 murine macrophages were treated with 5-Aza-2′-deoxycytidine – a potent DNA demethylating agent (Yang et al., 2010). Indeed, loss of DNA methylation occurred primarily within the 5′-UTR of evolutionary younger LINE-1 elements, while the methylation status of elder elements was affected to a lesser extent (R2=0.375, P=0.0009; Supplemental Fig. S1A).
3.4. Administration of a methionine-deficient diet results in DNA hypomethylation of LINE-1 elements that belong to the A-type promoter in vivo
To explore the selective LINE-1 demethylation in in vivo conditions, we fed C57BL/6J mice a methionine/choline-deficient diet (MDD) for 2 months. Previous studies reported the loss of global and LINE-1 DNA methylation as a result of administration of MDD (Shivapurkar et al., 1986; Wilson et al., 1984). Here, we found that a methionine-deficient diet resulted in a significant loss of DNA methylation within the 5′-UTR of two A-type LINE-1 elements: L1MdA_I (0.21 Myr) and L1MdA_VI (4.7 Myr) (−1.52-fold, P<0.05 and −1.78-fold, P<0.001, respectively) (Supplemental Fig. S1B). At the same time, the LINE-1 elements that carry other types of promoter – Tf and F were characterized by either no changes in DNA methylation, as observed in L1MdTf_III (1.23 Myr), or even DNA hypermethylation, although insignificant, observed in the L1MdF_V (6.43 Myr) elements (1.46-fold, P<0.1).
3.5. Exposure to low mean absorbed doses of densely but not sparsely ionizing radiation affects DNA methylation of selective LINE-1 elements in vivo
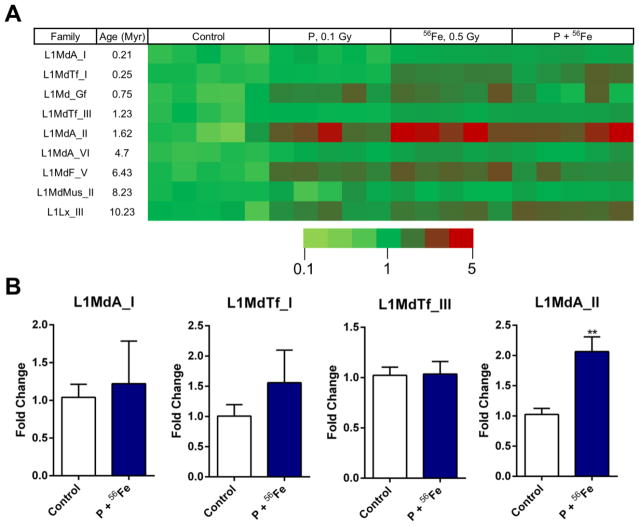
Taking into account these findings, we hypothesized that exposure to densely IR may also lead to alterations in DNA methylation within the 5′-UTRs of selective LINE-1 elements. Therefore, next we addressed the methylation status of the abovementioned nine families of LINE-1 elements after exposure to either protons, 56Fe, or sequential exposure to protons and 56Fe. All three regimens of exposure have led to substantial hypermethylation of L1MdA_II (1.62 Myr) LINE-1 element (3.37-fold increase, P<0.01; 4.60-fold increase, P<0.001; 3.95-fold increase, P<0.01 after exposure to protons, 56Fe, or sequential exposure to protons and 56Fe, respectively) and two other, evolutionary old, LINE-1 families: L1MdF_V (6.43 Myr) (2.24–2.93-fold increase range; P<0.05) and L1Lx_III (10.2 Myr) (1.79–2.46-fold increase range; P<0.05) (Fig. 4A; Supplemental Table S3). Of smaller magnitude, but still significant DNA hypermethylation was observed in the 5′-UTR of evolutionary younger L1MdTf_I element (2.17- and 2.18- fold increase after exposure to 56Fe and sequential exposure to protons and 56Fe, respectively; P<0.05). No statistically significant differences were detected in the youngest, L1MdA_I (0.21 Myr) element. These findings were confirmed with the MeDIP analysis of selective LINE-1 5′-UTRs, where a 2.11-fold increase in DNA methylation was detected in the 5′-UTR of L1MdA_II element, along with an absence of significant changes in the 5′-UTRs of L1MdA_I and L1MdTf_III elements (Fig. 4B).
Figure 4.
Effects of exposure to densely IR on DNA methylation in LINE-1 5′-UTR. (A) Exposure to protons, heavy iron ions (56Fe) alone or in combination results in DNA hypermethylation within the 5′-UTRs of selective LINE-1 elements. Fold change for each sample is represented by a color change on the heat map. (B) DNA methylation within the 5′-UTR of selective LINE-1 elements as measured by MeDIP. Asterisks (**) denotes significant (P<0.01) difference from control.
In order to investigate whether the exposure to low doses of sparsely IR is capable of causing similar persistent changes in the lung DNA methylation of LINE-1 elements, we exposed mice to 0.1 and 1 Gy of 137Cs. First, we compared the DNA methylation status of LINE-1 elements between the two cohorts of sham-irradiated mice. No significant differences were observed (data not shown). Next, we addressed the methylation status of nine LINE-1 families in the control and exposed mice four weeks after 137Cs irradiation. No substantial changes in the 5′-UTR DNA methylation were detected in any of the nine tested LINE-1 elements (Supplemental Fig. S2).
3.6. DNA hypermethylation of L1MdA_II LINE-1 elements does not lead to altered expression of genes that evolutionary acquired their insertions
Because the DNA methylation status of individual LINE-1 elements may affect the transcription levels of the genes that evolutionary acquired their insertions (Hur et al., 2014; Wolff et al., 2010), we next used the USCS genome browser to identify genes carrying the functional 5′-UTR unit of the L1MdA_II LINE-1 element. Among the identified genes, the most substantial changes were detected in the Cntnap gene, where the sequential exposure to protons and 56Fe resulted in a 2.2-fold increase in its mRNA levels (P<0.05) (Fig. 5). No substantial changes were detected in the expression of other genes, suggesting that the changes in the DNA methylation status of L1MdA_II LINE-1 do not necessarily lead to changes in the expression of genes linked to this element.
Figure 5.
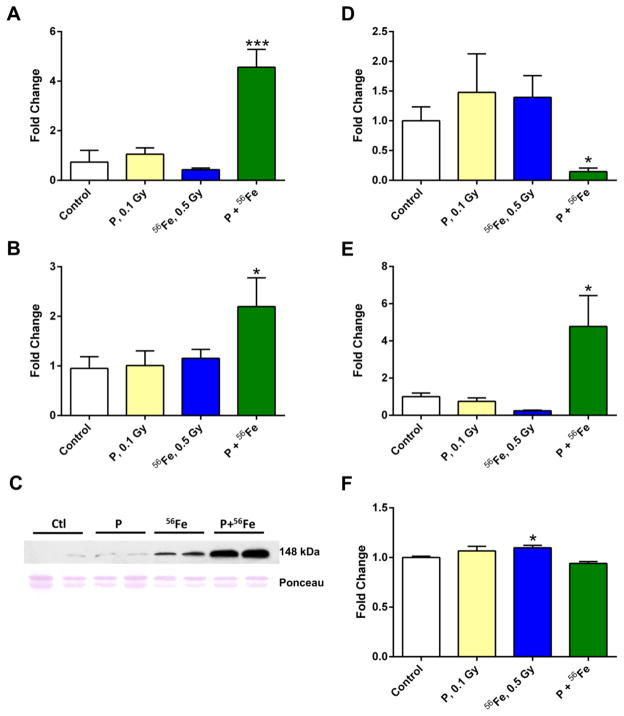
Analysis of mRNA levels of genes that evolutionary acquired L1MdA_II element insertions. The differential expression of genes was determined by quantitative RT-PCR. The fold change data were calculated from the ΔΔCt values. Asterisks (*) denotes significant (P<0.05), difference from control (n=5). All qRT-PCR reactions were repeated twice. P – protons; 56Fe – heavy iron ions; P+56Fe – consequent exposure to protons and heavy iron ions.
3.7. Sequential exposure to protons and 56Fe but not to single exposure to either protons or
56Fe results in reactivation of LINE-1 elements Methylation of DNA is considered as a key mechanism that prevents the transcriptional activation of LINE-1. Taking this into consideration, next we addressed the mRNA levels of LINE-1 in the lungs of control and exposed mice. Sequential exposure to protons and 56Fe resulted in a substantial increase in LINE-1 ORF1 mRNA levels (4.3-fold, P<0.001, Fig. 6A). Similar increases after sequential exposure to protons and 56Fe were observed in mRNA levels of LINE-1 ORF2 (2.2-fold, P<0.05, Fig. 6B). These findings were further confirmed by Western blot analysis, indicating elevated cytoplasmic levels of ORF1p (Fig. 6C). At the same time, exposure to either protons or 56Fe alone did not affect significantly the expression of either of the ORFs. Similarly, exposure to either 0.1 or 1 Gy of 137Cs did not lead to significant alterations in the expression of LINE-1 four weeks after irradiation (data not shown).
Figure 6.
Exposure to densely IR results in LINE-1 reactivation via histone modifications. (A-B) Analysis of LINE-1 ORF1 (A) and ORF2 (B) mRNA levels by reverse transcription qRT-PCR. (C) Western immunoblot analysis of the LINE-1 ORF1p levels in the mouse lung. P – protons; 56Fe – heavy iron ions; P+56Fe – consequent exposure to protons and heavy iron ions (n=4). (D, E) Analysis of the histone H3 Lysine 9 (H3K9) dimethylation (D) and histone H3 Lysine 4 (H3K4) trimethylation (E) at the L1MdA-I 5′-UTR by ChIP analysis. Samples were normalized to L1Lx_I ORF1. Data are presented as ± SEM (one-way ANOVA). (F) Analysis of the Ehmt2/G9a histone methyltransferase mRNA levels using qRT-PCR. Asterisks (*) denotes significant (P<0.05), and (***) denotes significant (P<0.001) difference from control. P – protons; 56Fe – heavy iron ions; P+56Fe – consequent exposure to protons and heavy iron ions.
3.8. Histone H3 modifications play a key role in the post-exposure regulation of LINE-1 expression
Given that the observed changes in DNA methylation cannot explain the reactivation of LINE-1, we examined whether this effect was mediated by histone modifications. Recent studies demonstrated the role of histone H3 lysine 9 dimethylation (H3K9me2) as a key silencing heterochromatic mark critical for the regulation of LINE-1 expression (Martens et al., 2005; Rangasamy, 2013). Therefore, using the ChIP-qPCR approach, we next addressed the status of H3K9me2 at the 5′-UTR of L1MdA_I, since only evolutionarily young LINE-1 elements are capable of reactivation and retrotransposition.
Levels of H3K9me2 were significantly decreased after sequential exposure to protons and 56Fe (5.2-fold, P<0.05), while some non-significant increases were observed in the samples exposed to protons or 56Fe alone (Fig. 6D). These changes were also paralleled by the decreased mRNA levels of Ehmt2/G9a, a histone methyltransferase involved in the trimethylation of the H3K9me2 histone mark, which is also implicated in the regulation of LINE-1 (Di Giacomo et al., 2014) (Fig. 6F). Furthermore, a significant and substantial increase was observed in the levels of histone H3 lysine 4 trimethylation (H3K4me3), a euchromatic mark associated with transcriptional activation in the lung tissue after consecutive exposure to protons and 56Fe, but not to either protons or 56Fe alone (4.6-fold, P<0.05; Fig. 6E). In addition, we examined these histone modifications in evolutionarily older LINE-1 families, L1MdA_VI and L1MdF_V, but were not able to detect any significant changes (data not shown).
4. Discussion
IR is a potent genotoxic stressors that is also capable of causing substantial and persistent epigenetic alterations both in vitro and in vivo. Studies indicate that exposures to higher doses of IR are usually associated with the loss of global and repetitive elements-associated DNA methylation (Koturbash et al., 2007, 2005). At the same time, analyses of the effects of exposure to low doses of IR on DNA methylation are more challenging.
In our previous study, we reported a lack of repetitive elements-associated changes in DNA methylation in mice four weeks after exposure to low absorbed mean doses of protons, 56Fe or sequential exposure to protons and 56Fe (Nzabarushimana et al., 2015). Analysis of LINE-1 DNA methylation in that study, similar to the vast majority of other studies, was performed on the LINE-1 ORF1. In this study, we addressed the levels of LINE-1 ORF2 using MS qRT-PCR and LINE-1 ORF1 using the pyrosequencing approach. No significant changes in DNA methylation were detected within ORF2. At the same time, exposure to protons alone or in combination with 56Fe resulted in DNA hypomethylation within the LINE-1 ORF1, although the loss of DNA methylation was very small.
A recent study indicated that certain differences exist in the DNA methylation of LINE-1 elements in human embryonic stem cells based on their evolutionary age (Guo et al., 2014). Such differences may also predetermine the extent of LINE-1 DNA methylation changes in response to environmental stressors. Indeed, studies have reported differential alterations in DNA methylation of two LINE-1 elements in response to IR and particulate matter exposure (Byun et al., 2013; Lima et al., 2014).
Taking these findings into account, we first sought to investigate whether particular differences exist between nine LINE-1 elements in the murine lung that belong to different families and are distinct by their evolutionary age. Congruent to the findings in human embryonic cells, the degree of LINE-1 elements DNA methylation within the 5′-UTR strongly correlated with their evolutionary age. The elder elements were significantly less methylated, when compared to evolutionary younger elements. This finding may be explained by the fact that evolutionary older elements acquired a higher number of mutations in comparison with the younger elements. Indeed, 5-methylcytosine is prone to spontaneous deamination resulting in the loss of the methyl group due to the conversion into thymine. Another possible explanation is that only young LINE-1 elements are capable of retrotransposition and, thus, DNA methylation of 5′-UTR is necessary only in these elements, but not in inactivated and truncated older elements.
Treatment of RAW-264.7 murine macrophages with a potent DNA demethylation agent, 5-Aza-2′-deoxycytidine, has led, in agreement with previous studies (Kulis and Esteller, 2010; Yang et al., 2010), to the loss of DNA methylation in LINE-1 elements. This hypomethylation, at the same time, was also dependent on the evolutionary age of the elements, where the younger elements experienced a more substantial loss of DNA methylation in comparison to the older elements. This is probably due to the higher DNA demethylation capacity of the younger elements as dictated by their substantially higher basal levels of DNA methylation.
Validation of the in vitro findings in the in vivo model by administration of the MDD, which leads to the loss of global DNA methylation in the murine and rat models, showed promoter type-dependent changes in LINE-1 DNA methylation, rather than evolutionary age-dependent. Feeding mice MDD for two months resulted in the loss of DNA methylation from the LINE-1 elements that belong to the A-type promoter, while the elements that contain Tf and V promoters were either not affected or were even characterized by some degree of DNA hypermethylation. These differences between the in vitro and in vivo studies could be possibly explained by the principally different mechanisms of DNA demethylation exerted by 5-Aza-2′-deoxycytidine and MDD. While the former primarily targets the DNA methyltransferases (Song et al., 2011), MDD results in decreased levels of S-adenosylmethionine (SAM), which serves as a major donor of methyl groups needed for DNA methylation (Cavuoto and Fenech, 2012).
Exposures to low absorbed mean doses of protons, 56Fe or sequential exposure to protons and 56Fe have led to DNA hypermethylation within the 5′-UTR of several LINE-1 elements. The most substantial changes were observed in L1MdA_II, an element that carries the A-type promoter and is 1.62 Myr old, reaching a 4.6-fold (P<0.05) increase in DNA methylation after exposure to 56Fe alone. Some minor increases in DNA methylation were observed in the 5′-UTRs of L1MdTf_I and L1MdGf elements (0.25 and 0.75 Myr, respectively) and no changes were detected in the L1MdA_I elements (0.21 Myr), which are evolutionary the youngest elements and that are still active in the mouse genome. These findings may be explained by the fact that, given our abovementioned observations, young elements are heavily methylated in comparison with the older elements and, thus, have a lower capacity for the acquisition of new methyl groups within their CpG sites.
The overall trend towards global DNA hypermethylation, given the genomic abundance of LINE-1 elements, is in good agreement with previous findings which reported global and LINE-1-associated DNA hypermethylation after exposure to various types of densely IR (Jangiam et al., 2015; Lima et al., 2014; Miousse et al., 2014; Nzabarushimana et al., 2014). At the same time, exposure to low doses of sparsely IR (137Cs) did not affect substantially the methylation of individual LINE-1 elements in the lung tissue and confirmed the findings from an earlier study reporting lack of changes in DNA methylation in the murine lung after total body irradiation with x-rays (Pogribny et al., 2004).
Changes in the LINE-1 DNA methylation may significantly affect the expression of genes-carriers of LINE-1 insertions. For instance, hypomethylation of the LINE-1 insertion within the MET oncogene was associated with its aberrant transcription in prostate and pancreatic cancer and strongly correlated with the tumor metastatic potential (Hur et al., 2014; Wolff et al., 2010). The observed DNA hypermethylation of the 5′-UTR of a specific LINE-1 family, in this study, did not lead to substantial changes in the expression of candidate genes. However, it has to be taken into consideration that these changes in DNA methylation were present in the LINE-1 element which is considered to be inactive in the mouse genome. Furthermore, aberrant gene expression reported in these studies was associated with the hypomethylated LINE-1 status, while LINE-1 hypermethylation was observed in our study.
Of particular interest are the unexpected (despite the strong DNA hypermethylation response of older elements and lack of changes within the youngest element) and dramatic increases in the mRNA levels of both LINE-1 ORFs – ORF1 and ORF2 – and ORF-1 protein levels. These findings suggest that other epigenetic mechanisms, such as histone modifications, may be responsible for the IR-induced reactivation of LINE-1 elements. Indeed, our data indicate that consequent exposure to protons and 56Fe, but not to any of those IR types alone, results in a substantial loss of H3K9me2 and increase in H3K4me3 within the 5′-UTR of the youngest LINE-1 element L1MdA_I. Our finding confirms the previous suggestion that DNA methylation presumably serves as an additional “lock” to reinforce the silencing of repetitive elements induced by repressive histone marks (Jones, 2012), and has little influence on the regulation of their expression under genotoxic stress conditions.
The increase in LINE-1 expression may result in its retrotransposition and the latter has been documented as a consequence of exposure to IR (Banaz-Yaşar et al., 2012). Furthermore, increased LINE-1 expression and retrotransposition are reported in numerous human diseases, including cancer, and may play a driving role in carcinogenesis (Rodić and Burns, 2013). Increased LINE-1 expression and retrotransposition events have been recently reported in human lung cancer (Lee et al., 2012; Rodić et al., 2014). This knowledge is of particular importance, since lung cancer has been considered as the largest potential cancer risk from space travel (Shay et al., 2011) and is the most frequent radiation treatment-induced secondary malignancy (Maddams et al., 2011). Furthermore, a very recent study demonstrated the increase in lung tumors in mice after exposure to 0.4 Gy of 56Fe (Christofidou-Solomidou et al., 2015). Given that the used doses of densely IR and exposure settings correspond to the real space environment, the results of this study have to be taken into consideration in regards to the potential health effects associated with deep space exploration.
In conclusions, the data presented in this study demonstrates that DNA methylation of murine LINE-1 elements is dependent on their evolutionary age, where the elder age of the element is associated with the lower extent of DNA methylation. Furthermore, we demonstrate that exogenous factors, such as 5-Aza-2′-deoxycytidine and modulation over the methionine dietary patterns, affect the DNA methylation of selective LINE-1 elements in vitro and in vivo. We also show that exposure to environmentally relevant low mean absorbed doses of densely IR, but not sparsely IR, result in DNA hypermethylation of primarily older LINE-1 elements in the murine lung, while the DNA methylation of evolutionary younger elements remain mostly unchanged. Consequent exposure to low mean absorbed doses of protons and 56Fe has led to substantial increases in mRNA and protein levels of LINE-1. Reactivation of LINE-1 was independent of DNA methylation patterns, and was associated with the loss of the heterochromatic mark H3K9 dimethylation and an increase in the euchromatic mark H3K4 trimethylation at the 5′-UTR of L1MdA_I, the evolutionary youngest LINE-1 element. These findings suggests that DNA methylation is a key player in the regulation of LINE-1 expression under normal conditions, but has limited input on the expression of LINE-1 in response to exposure to densely IR, where the histone modifications play a critical role. Furthermore, these radiation-induced changes are persistent by nature and can be identified at least four weeks after exposure to doses and settings relevant to the space environment.
Supplementary Material
Highlights.
DNA methylation of LINE-1 elements is dependent on their evolutionary age
Densely ionizing radiation affects DNA methylation of selective LINE-1 elements
Radiation-induced reactivation of LINE-1 is DNA methylation-independent
Histone modifications dictate the transcriptional activity of LINE-1
Acknowledgments
The authors would like to thank Dr. Christy Bekkevold and Erica Nicholson for the excellent animal care at the UAMS Animal Facility. The work was supported by National Institute of Health Center of Biological Research Excellence [grant number 1P20GM109005], National Institute of Health [grant number R21ES025268]; National Aeronautics and Space Administration [grant numbers NNX10AD59G, NNJ12ZSA001N]; Arkansas Space Grant Consortium through National Aeronautics and Space Administration [grant number NNX13AB29A]; National Space Biomedical Research Institute through the National Aeronautics and Space Administration NCC 9-58 [grant number RE03701]; NIH/UAMS Clinical and Translational Science Award [grant numbers UL1TR000039, KL2TR000063], and the Arkansas Biosciences Institute. The authors declare no conflict of interests.
Abbreviations
- Gy
Gray
- IR
Ionizing Radiation
- LET
Linear Energy Transfer
- LINE-1
Long Interspersed Nucleotide Element 1
- MDD
Methionine-Deficient Diet
- MAD
Methionine-Adequate Diet
- MDD
Methionine-Deficient Diet
- MeV
Megaelectron Volt
- Myr
Million Years
- ORF
Open Reading Frame
- qRT-PCR
quantitative Real Time Polymerase Chain Reaction
- SPE
Solar Particle Events
- TE
Transposable Elements
- UTR
Untranslated Region
Footnotes
The work was supported by National Institute of Health Center of Biological Research Excellence [grant number 1P20GM109005], National Institute of Health [grant number R21ES025268]; National Aeronautics and Space Administration [grant numbers NNX10AD59G, NNJ12ZSA001N]; Arkansas Space Grant Consortium through National Aeronautics and Space Administration [grant number NNX13AB29A]; National Space Biomedical Research Institute through the National Aeronautics and Space Administration NCC 9-58 [grant number RE03701]; NIH/UAMS Clinical and Translational Science Award [grant numbers UL1TR000039, KL2TR000063], and the Arkansas Biosciences Institute. All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Arkansas for Medical Sciences (UAMS), Oregon Health and Sciences University (OHSU) and Brookhaven National Laboratories (BNL).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aypar U, Morgan WF, Baulch JE. Radiation-induced epigenetic alterations after low and high LET irradiations. Mutat Res. 2011;707:24–33. doi: 10.1016/j.mrfmmm.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Banaz-Yaşar F, Gedik N, Karahan S, Diaz-Carballo D, Bongartz BM, Ergün S. LINE-1 retrotransposition events regulate gene expression after X-ray irradiation. DNA Cell Biol. 2012;31:1458–1467. doi: 10.1089/dna.2012.1676. [DOI] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:1. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks A, Bao S, Rithidech K, Couch LA, Braby LA. Relative effectiveness of HZE iron-56 particles for the induction of cytogenetic damage in vivo. Radiat Res. 2001;155:353–359. doi: 10.1667/0033-7587(2001)155[0353:reohip]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Byun H-M, Motta V, Panni T, Bertazzi PA, Apostoli P, Hou L, Baccarelli AA. Evolutionary age of repetitive element subfamilies and sensitivity of DNA methylation to airborne pollutants. Part Fibre Toxicol. 2013;10(1) doi: 10.1186/1743-8977-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavuoto P, Fenech MF. A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension. Cancer Treat Rev. 2012;38:726–736. doi: 10.1016/j.ctrv.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Christofidou-Solomidou M, Pietrofesa RA, Arguiri E, Schweitzer KS, Berdyshev EV, McCarthy M, Corbitt A, Alwood JS, Yu Y, Globus RK. Space radiation-associated lung injury in a murine model. Am J Physiol-Lung Cell Mol Physiol. 2015;308:L416–L428. doi: 10.1152/ajplung.00260.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giacomo M, Comazzetto S, Sampath SC, Sampath SC, O’Carroll D. G9a co-suppresses LINE1 elements in spermatogonia. Epigenetics Chromatin. 2014;7:1. doi: 10.1186/1756-8935-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durante M. New challenges in high-energy particle radiobiology. Br J Radiol. 2014;87 doi: 10.1259/bjr.20130626. 20130626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:1. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Girdhani S, Sachs R, Hlatky L. Biological effects of proton radiation: what we know and don’t know. Radiat Res. 2013;179:257–272. doi: 10.1667/RR2839.1. [DOI] [PubMed] [Google Scholar]
- Goodhead D, Thacker J, Cox R. Effects of radiations of different qualities on cells: molecular mechanisms of damage and repair. Int J Radiat Biol. 1993;63:543–556. doi: 10.1080/09553009314450721. [DOI] [PubMed] [Google Scholar]
- Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, Yan J, Ren X, Lin S, Li J. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–610. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Helman E, Lawrence MS, Stewart C, Sougnez C, Getz G, Meyerson M. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 2014;24:1053–1063. doi: 10.1101/gr.163659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur K, Cejas P, Feliu J, Moreno-Rubio J, Burgos E, Boland CR, Goel A. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut. 2014;63:635–646. doi: 10.1136/gutjnl-2012-304219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Shiraishi K, Eguchi A, Shibata H, Yoshimoto K, Mori T, Baba Y, Baba H, Suzuki M. Long interspersed nucleotide element 1 hypomethylation is associated with poor prognosis of lung adenocarcinoma. Ann Thorac Surg. 2013;96:1790–1794. doi: 10.1016/j.athoracsur.2013.06.035. [DOI] [PubMed] [Google Scholar]
- Jangiam W, Tungjai M, Rithidech KN. Induction of chronic oxidative stress, chronic inflammation and aberrant patterns of DNA methylation in the liver of titanium-exposed CBA/CaJ mice. Int J Radiat Biol. 2015;91:389–398. doi: 10.3109/09553002.2015.1001882. [DOI] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koturbash I, Boyko A, Rodriguez-Juarez R, McDonald RJ, Tryndyak VP, Kovalchuk I, Pogribny IP, Kovalchuk O. Role of epigenetic effectors in maintenance of the long-term persistent bystander effect in spleen in vivo. Carcinogenesis. 2007;28:1831–1838. doi: 10.1093/carcin/bgm053. [DOI] [PubMed] [Google Scholar]
- Koturbash I, Pogribny I, Kovalchuk O. Stable loss of global DNA methylation in the radiation-target tissue--a possible mechanism contributing to radiation carcinogenesis? Biochem Biophys Res Commun. 2005;337:526–533. doi: 10.1016/j.bbrc.2005.09.084. [DOI] [PubMed] [Google Scholar]
- Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, Lohr JG, Harris CC, Ding L, Wilson RK. Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima F, Ding D, Goetz W, Yang AJ, Baulch JE. High LET 56Fe ion irradiation induces tissue-specific changes in DNA methylation in the mouse. Environ Mol Mutagen. 2014;55:266–277. doi: 10.1002/em.21832. [DOI] [PubMed] [Google Scholar]
- Maddams J, Parkin DM, Darby SC. The cancer burden in the United Kingdom in 2007 due to radiotherapy. Int J Cancer. 2011;129:2885–2893. doi: 10.1002/ijc.26240. [DOI] [PubMed] [Google Scholar]
- Martens JH, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse IR, Chalbot M-CG, Lumen A, Ferguson A, Kavouras IG, Koturbash I. Response of transposable elements to environmental stressors. Mutat Res Rev Mutat Res. 2015;765:19–39. doi: 10.1016/j.mrrev.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse IR, Koturbash I. The Fine LINE: Methylation Drawing the Cancer Landscape. BiomedRes Int. 2015;2015:131547. doi: 10.1155/2015/131547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse IR, Shao L, Chang J, Feng W, Wang Y, Allen AR, Turner J, Stewart B, Raber J, Zhou D, Koturbash I. Exposure to low-dose (56)Fe-ion radiation induces long-term epigenetic alterations in mouse bone marrow hematopoietic progenitor and stem cells. Radiat Res. 2014;182:92–101. doi: 10.1667/RR13580.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munley MT, Moore JE, Walb MC, Isom SP, Olson JD, Zora JG, Kock ND, Wheeler KT, Miller MS. Cancer-prone mice expressing the Ki-ras G12C gene show increased lung carcinogenesis after CT screening exposures. Radiat Res. 2011;176:842–848. doi: 10.1667/rr2649.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCRP. NCRP Report No 153. 2006. Information needed to make radiation protection recommendations for space missions beyond low-earth orbit. [Google Scholar]
- Nzabarushimana E, Miousse IR, Shao L, Chang J, Allen AR, Turner J, Stewart B, Raber J, Koturbash I. Long-term epigenetic effects of exposure to low doses of 56Fe in the mouse lung. J Radiat Res (Tokyo) 2014;55:823–828. doi: 10.1093/jrr/rru010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nzabarushimana E, Prior S, Miousse IR, Pathak R, Allen AR, Latendresse J, Olsen RHJ, Raber J, Hauer-Jensen M, Nelson GA, Koturbash I. Combined exposure to protons and (56)Fe leads to overexpression of Il13 and reactivation of repetitive elements in the mouse lung. Life Sci Space Res. 2015;7:1–8. doi: 10.1016/j.lssr.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paganetti H, Niemierko A, Ancukiewicz M, Gerweck LE, Goitein M, Loeffler JS, Suit HD. Relative biological effectiveness (RBE) values for proton beam therapy. Int J Radiat Oncol Biol Phys. 2002;53:407–421. doi: 10.1016/s0360-3016(02)02754-2. [DOI] [PubMed] [Google Scholar]
- Pogribny I, Raiche J, Slovack M, Kovalchuk O. Dose-dependence, sex-and tissue-specificity, and persistence of radiation-induced genomic DNA methylation changes. Biochem Biophys Res Commun. 2004;320:1253–1261. doi: 10.1016/j.bbrc.2004.06.081. [DOI] [PubMed] [Google Scholar]
- Prise K, Folkard M, Newman H, Michael B. Effect of radiation quality on lesion complexity in cellular DNA. Int J Radiat Biol. 1994;66:537–542. doi: 10.1080/09553009414551581. [DOI] [PubMed] [Google Scholar]
- Raber J, Allen AR, Sharma S, Allen B, Rosi S, Olsen RH, Davis MJ, Eiwaz M, Fike JR, Nelson GA. Effects of Proton and Combined Proton and 56Fe Radiation on the Hippocampus. Radiat Res. 2015;185:20–30. doi: 10.1667/RR14222.1. [DOI] [PubMed] [Google Scholar]
- Ramadan SS, Sridharan V, Koturbash I, Miousse IR, Hauer-Jensen M, Nelson GA, Boerma M. A priming dose of protons alters the early cardiac cellular and molecular response to (56)Fe irradiation. Life Sci Space Res. 2016;8:8–13. doi: 10.1016/j.lssr.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangasamy D. Distinctive patterns of epigenetic marks are associated with promoter regions of mouse LINE-1 and LTR retrotransposons. Mob DNA. 2013;4:1. doi: 10.1186/1759-8753-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodić N, Burns KH. Long Interspersed Element–1 (LINE-1): Passenger or Driver in Human Neoplasms? PLoS Genet. 2013;9:e1003402. doi: 10.1371/journal.pgen.1003402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodić N, Sharma R, Sharma R, Zampella J, Dai L, Taylor MS, Hruban RH, Iacobuzio-Donahue CA, Maitra A, Torbenson MS. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am J Pathol. 2014;184:1280–1286. doi: 10.1016/j.ajpath.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rola R, Fishman K, Baure J, Rosi S, Lamborn KR, Obenaus A, Nelson GA, Fike JR. Hippocampal neurogenesis and neuroinflammation after cranial irradiation with 56Fe particles. Radiat Res. 2008;169:626–632. doi: 10.1667/RR1263.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Kawakami K, Matsumoto I, Oda M, Watanabe G, Minamoto T. Long interspersed nuclear element 1 hypomethylation is a marker of poor prognosis in stage IA non–small cell lung cancer. Clin Cancer Res. 2010;16:2418–2426. doi: 10.1158/1078-0432.CCR-09-2819. [DOI] [PubMed] [Google Scholar]
- Shay JW, Cucinotta FA, Sulzman FM, Coleman CN, Minna JD. From mice and men to earth and space: joint NASA–NCI workshop on lung cancer risk resulting from space and terrestrial radiation. Cancer Res. 2011;71:6926–6929. doi: 10.1158/0008-5472.CAN-11-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivapurkar N, Wilson MJ, Hoover KL, Mikol YB, Creasia D, Poirier LA. Hepatic DNA methylation and liver tumor formation in male C3H mice fed methionine-and choline-deficient diets. J Natl Cancer Inst. 1986;77:213–217. [PubMed] [Google Scholar]
- Song SH, Han SW, Bang YJ. Epigenetic-Based Therapies in Cancer : Progress to Date. Drugs. 2011;71:2391–2403. doi: 10.2165/11596690-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Sookdeo A, Hepp CM, McClure MA, Boissinot S. Revisiting the evolution of mouse LINE-1 in the genomic era. Mob DNA. 2013;4:1. doi: 10.1186/1759-8753-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013:mst197. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel C, Schmezer P, Plass C, Popanda O. Epigenetics in radiation-induced fibrosis. Oncogene. 2015;34:2145–2155. doi: 10.1038/onc.2014.145. [DOI] [PubMed] [Google Scholar]
- Wilson MJ, Shivapurkar N, Poirier LA. Hypomethylation of hepatic nuclear DNA in rats fed with a carcinogenic methyl-deficient diet. Biochem J. 1984;218:987–990. doi: 10.1042/bj2180987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff EM, Byun HM, Han HF, Sharma S, Nichols PW, Siegmund KD, Yang AS, Jones PA, Liang G. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet. 2010;6:e1000917. doi: 10.1371/journal.pgen.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Huff JL, Casey R, Kim M-H, Cucinotta FA. Risk of Acute Radiation Syndromes Due to Solar Particle Events. In: McPhee JC, Charles JB, editors. Human Health and Performance Risks of Space Exploration Missions: Evidence Reviewed by the NASA Human Research Program. Government Printing Office; 2009. pp. 171–190. [Google Scholar]
- Yang X, Lay F, Han H, Jones PA. Targeting DNA methylation for epigenetic therapy. Trends Pharmacol Sci. 2010;31:536–546. doi: 10.1016/j.tips.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnold J, Brotons M-CV. Pathogenetic mechanisms in radiation fibrosis. Radiother Oncol. 2010;97:149–161. doi: 10.1016/j.radonc.2010.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.