Summary
Cyclic di-AMP (c-di-AMP) is an emerging second messenger in bacteria. It has been shown to play important roles in bacterial fitness and virulence. However, transduction of c-di-AMP signaling in bacteria and the role of c-di-AMP in biofilm formation is not well understood. The level of c-di-AMP is modulated by activity of di-adenylyl cyclase that produces c-di-AMP and phosphodiesterase (PDE) that degrades c-di-AMP. In this study, we determined that increased c-di-AMP levels by deletion of the pdeA gene coding for a PDE promoted biofilm formation in Streptococcus mutans. Deletion of pdeA up-regulated expression of gtfB, the gene coding for a major glucan producing enzyme. Inactivation of gtfB blocked the increased biofilm by the pdeA mutant. Two c-di-AMP binding proteins including CabPA (SMU_1562) and CabPB (SMU_1708) were identified. Interestingly, only CabPA deficiency inhibited both the increased biofilm formation and the up-regulated expression of GtfB observed in the pdeA mutant. In addition, CabPA but not CabPB interacted with VicR, a known transcriptional factor that regulates expression of gtfB, suggesting that a signaling link between CabPA and GtfB through VicR. Increased biofilm by the pdeA deficiency also enhanced bacterial colonization of Drosophila in vivo. Taken together, our studies reveal a new role of c-di-AMP in mediating biofilm formation through a CabPA/VicR/GtfB signaling network in S. mutans.
Keywords: Streptococcus mutans, biofilms, c-di-AMP
Introduction
Signaling molecules are crucial for bacterial physiology and pathogenesis (Kalia et al., 2013, Antunes et al., 2010, Minvielle et al., 2013). The best known and well characterized bacterial signaling molecules are autoinducers, which mediate the quorum sensing systems, such as competence stimulating peptide in Gram-positive bacteria and homoserine lactones in Gram-negative bacteria (Senadheera & Cvitkovitch, 2008, Cvitkovitch, 2001, Kim et al., 2013, Antunes et al., 2010). Nucleotide signaling molecules such as cyclic AMP (cAMP) and (p)ppGpp have been demonstrated in mediating carbon metabolism and stringent response (Gomelsky, 2011, Kalia et al., 2013). Recently, cyclic di-GMP (c-di-GMP), a cyclic dinucleotide, has been recognized as a key signaling molecule involved in regulating bacterial biofilm formation and virulence (Hengge, 2009, Gomelsky, 2011, Römling et al., 2013, Nunes-Alves, 2014).
Cyclic di-AMP (c-di-AMP) is a newly discovered cyclic dinucleotide (Corrigan & Grundling, 2013). c-di-AMP is an essential signaling molecule in a wide variety of bacteria (Zhang et al., 2012, Kamegaya et al., 2011, Woodward et al., 2010, Witte et al., 2008, Corrigan et al., 2011, Bai et al., 2013). It is synthesized from two molecules of ATP by di-adenylyl cyclase (DAC) enzymes and degraded to pApA or AMP by distinct c-di-AMP phosphodiesterase enzymes (PDE) (Bai et al., 2013, Rao et al., 2010). DAC enzymes have been identified in a large number of bacteria (Woodward et al., 2010, Kamegaya et al., 2011, Bejerano-Sagie et al., 2006, Barker et al., 2013, Corrigan et al., 2011). These enzymes contain a conserved DisA_N domain (recently renamed as DAC) (Kamegaya et al., 2011, Woodward et al., 2010, Bejerano-Sagie et al., 2006, Barker et al., 2013, Corrigan et al., 2011). Most bacteria, including Streptococcus mutans, only possess one DAC domain containing protein (Mehne et al., 2013). The first c-di-AMP degrading phosphodiesterase enzyme named GdpP was discovered in Bacillus subtilis (Rao et al., 2010). GdpP contains a PAS sensory domain, a highly variable GGDEF domain, a DHH domain, and a DHH-associated DHHA1 domain. The C-terminal DHH–DHHA1 region is essential for PDE activity (Corrigan et al., 2011). A new type of c-di-AMP phosphodiesterase, PgpH, which contains an HD domain, was recently found in Listeria monocytogenes (Huynh et al., 2015), but no PgpH homolog was found in S. mutans. S. mutans has only one protein (PdeA) that contains GGDEF and DHH-DHHA domains. c-di-AMP plays important roles in regulating diverse cellular pathways. It modulates fatty acid synthesis and transport in Mycobacterium smegmatis (Zhang et al., 2013), and alters bacterial cell size and regulates cell envelope stress and biofilm formation in Staphylococcus aureus (Corrigan et al., 2011). Moreover, changes in c-di-AMP levels influence potassium uptake, peptidoglycan homeostasis, acid tolerance, antibiotic resistance, and bacterial virulence (Corrigan et al., 2011, Rao et al., 2010, Bai et al., 2013, Luo & Helmann, 2012). Increased c-di-AMP levels affect growth and promote biofilm formation in Streptococcus suis (Du et al., 2014). However, factors connecting c-di-AMP and biofilm formation are unknown.
S. mutans is one of the primary etiologic agents of dental caries. Formation of biofilm is a key virulence feature of this pathogen. S. mutans synthesizes extracellular polysaccharides (EPS) to promote biofilm formation using distinct glucosyltransferases (GTFs) (Koo et al., 2010). The EPS, including both water-insoluble and water-soluble glucans, mediates the initial adherence of S. mutans and other oral bacteria to the tooth surface and facilitates the formation of mature dental plaque (Rölla, 1989, Koo et al., 2009). GtfB, a major enzyme responsible for production of water-insoluble glucans, is critical for biofilm formation and virulence of S. mutans (Yamashita et al., 1993). GtfB deficiency in S. mutans inhibits biofilm formation and reduces bacterial virulence (Koo et al., 2002, Koo et al., 2010). However, it is unknown what signal modulates expression of gtfB. Understanding the underlying regulatory mechanisms of biofilm formation by the new bacterial second messenger will help to uncover new targets that are amenable to development of therapeutics to prevent and treat dental caries.
In this study, we showed that c-di-AMP mediated S. mutans biofilm formation. Increased c-di-AMP levels concurrently promoted biofilm formation via up-regulating the expression of gtfB. The enhanced biofilm formation and increased GtfB were simultaneously blunted by deletion of either a c-di-AMP binding protein (CabPA) or GtfB. Further we found that CabPA interacted with VicR, a response regulator of the VicRK two-component system, that modulates biofilm formation (Smith & Spatafora, 2012, Senadheera et al., 2012). Together, our studies demonstrate that c-di-AMP controls S. mutans biofilm formation by regulation of gtfB expression through the binding of CabPA to VicR. This study defines a new signaling network that links c-di-AMP signaling and biofilm formation.
Results
PdeA modulates cellular c-di-AMP levels in S. mutans
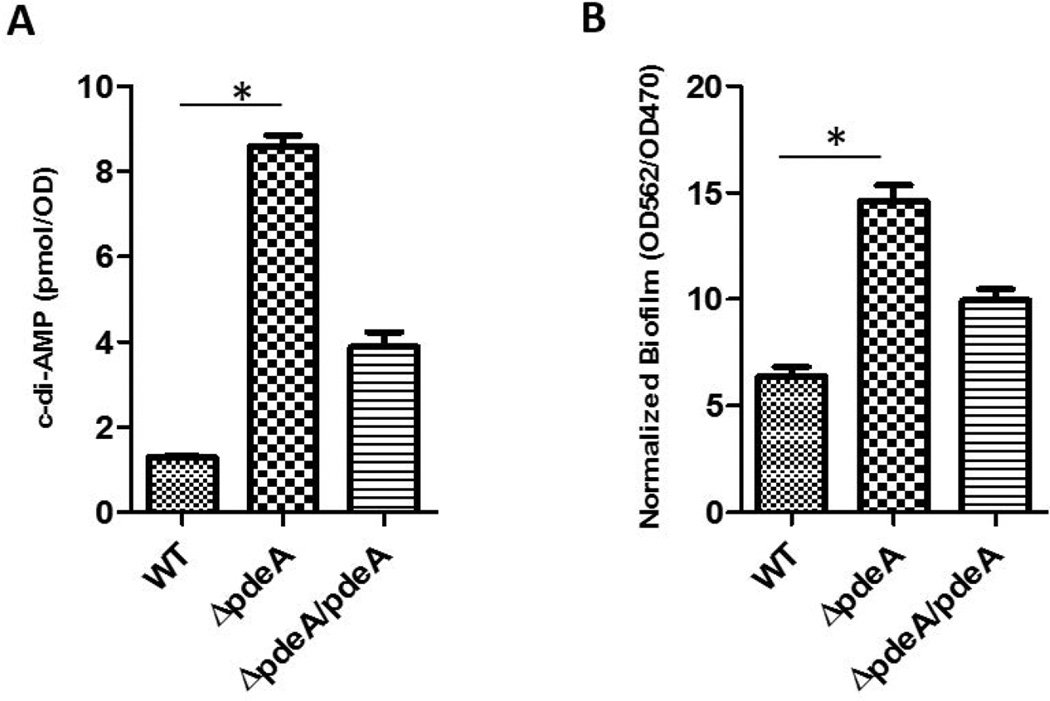
c-di-AMP PDEs have been identified in diverse bacteria. S. mutans PdeA (SMU_2140c) is a homolog of GdpP that possesses a DHH-DHHA1 domain (FIG. S1). To determine whether PdeA possesses c-di-AMP PDE activity, we generated an in-frame deletion of pdeA and measured the amount of intracellular c-di-AMP produced by the mutant. The amount of intracellular c-di-AMP was increased approximately 6-fold in the pdeA deficient mutant when compared to that in the wild type. Complementation of the mutant with the full-length pdeA gene partially restored the intracellular c-di-AMP levels (FIG. 1A). These data suggest that PdeA functions as a c-di-AMP PDE in S. mutans.
FIG 1. The PdeA deficiency increased c-di-AMP levels and promoted biofilm formation of S. mutans.
(A) Detection of bacterial c-di-AMP levels. Cell lysates of wild type, the pdeA mutant and the complemented strain were used to determine intracellular c-di-AMP levels by competitive ELISA. Data were normalized by bacterial growth OD470 of the cultures. (B) Biofilm formation by the pdeA mutant. Biofilm mass of wild type, the pdeA mutant and the complemented strain were determined using crystal violet staining assay and normalized to bacterial growth by OD562/OD470. Means of three independent experiments are shown. Error bars denote the SEM. *, P < 0.01 of Student’s t test using Prism 5.0.
The PdeA deficiency promotes biofilm formation of S. mutans
To investigate how the PdeA deficiency affects the ability of S. mutans to form biofilms, we performed biofilm assays first using a crystal violet staining method. The pdeA mutant produced significantly more biomass than the wild type bacteria, although the pdeA mutant exhibited a decreased growth (FIG. S2). The complemented strain partially restored the wild type phenotype (FIG. 1B). Similar phenotypes were observed when biofilm formation, and production of c-di-AMP of the pdeA mutant were monitored at different time points (8–24 hours) (FIG. S3).
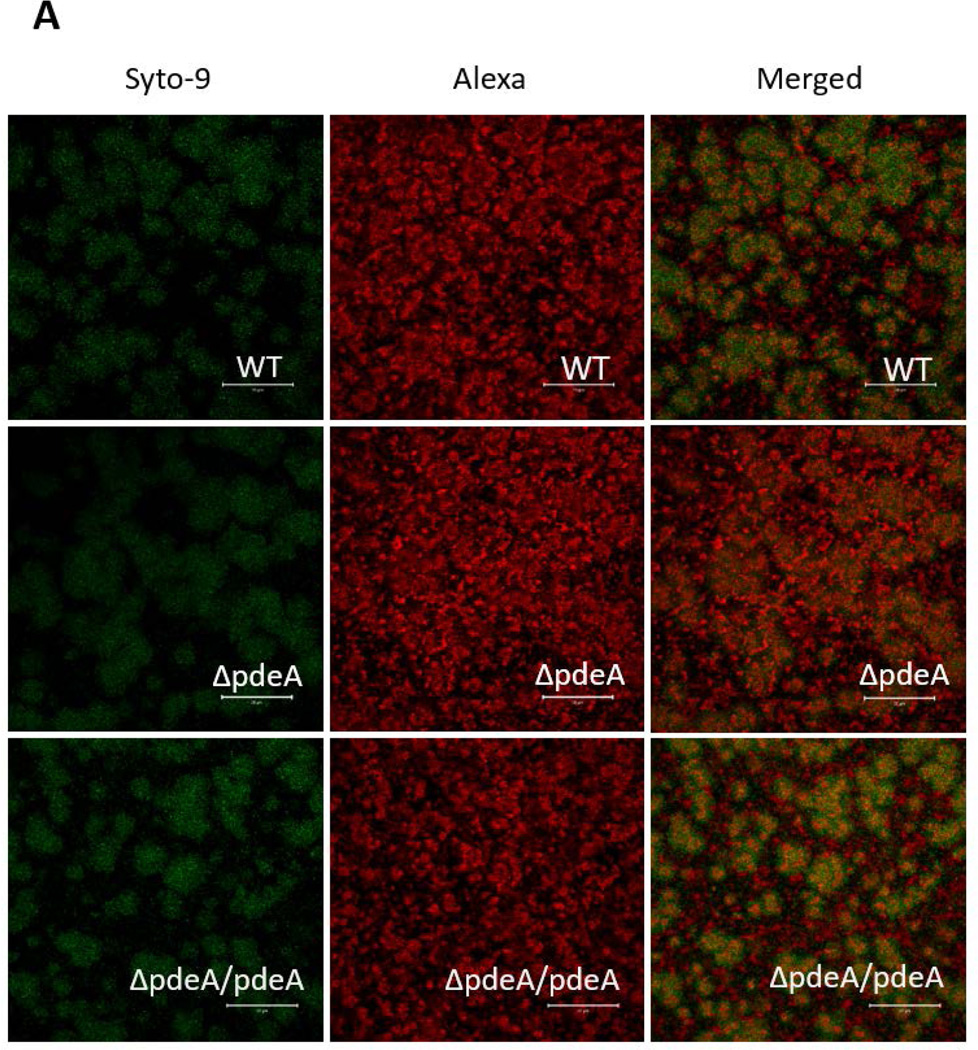
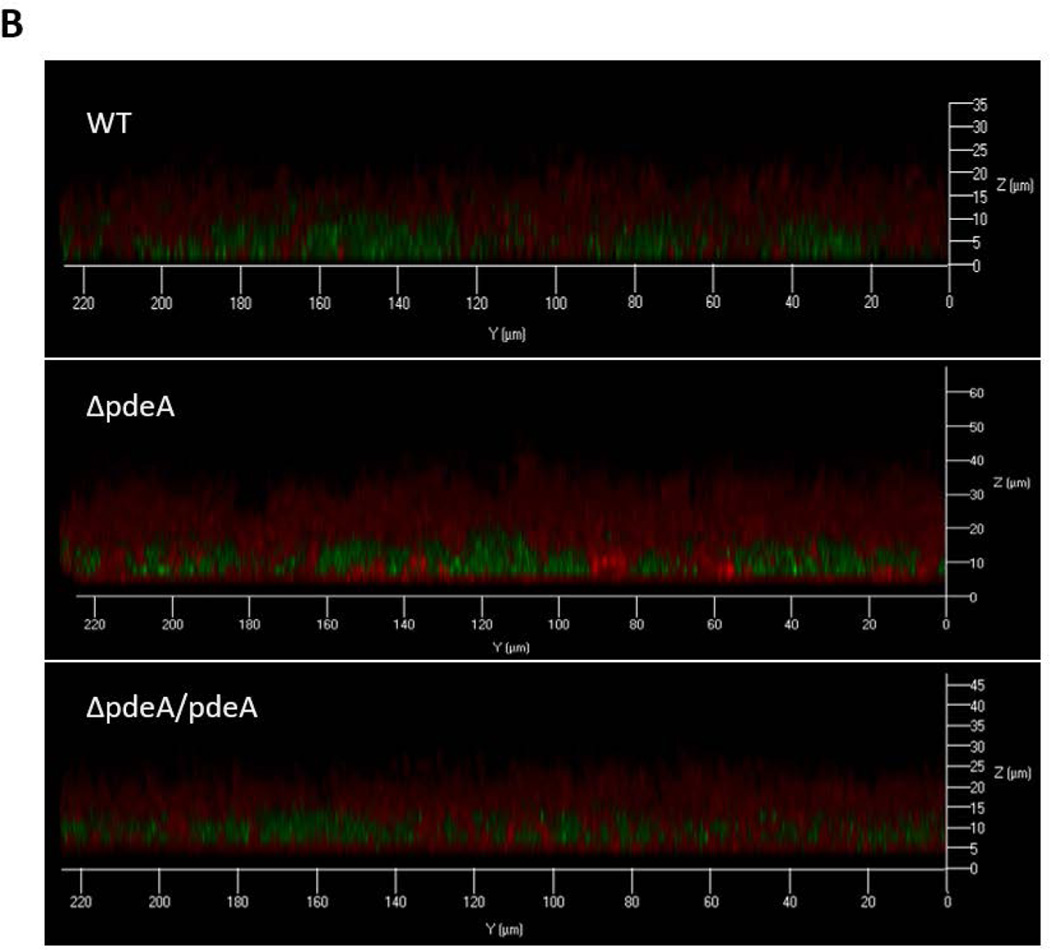
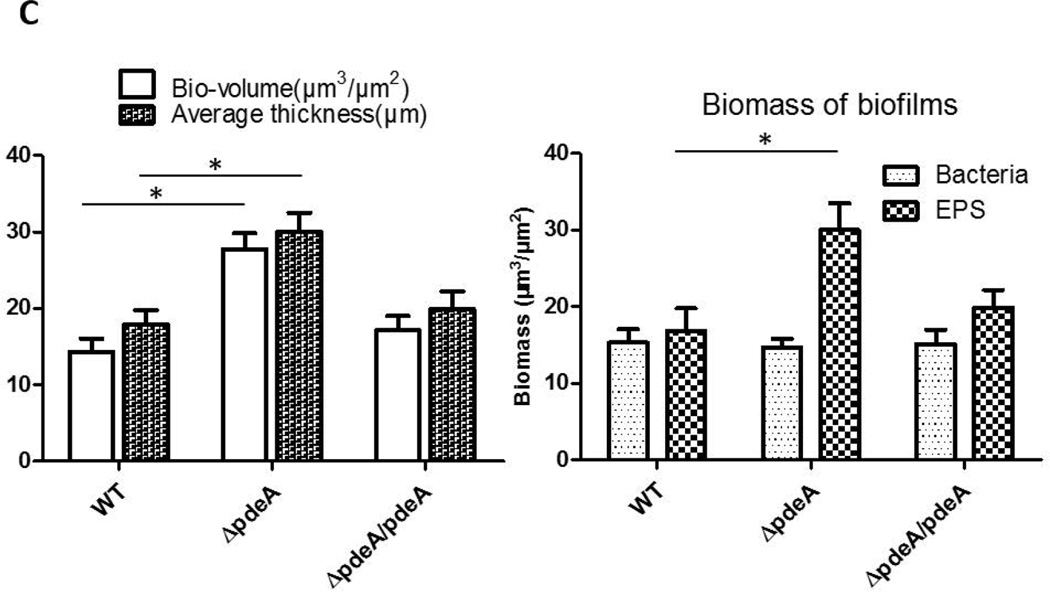
The effect of the PdeA deficiency on biofilm structure of S. mutans was also evident as assessed by glucan labeling and confocal laser scanning microscopy (CLSM). The wild type S. mutans formed typical biofilm clusters, while the pdeA mutant showed slightly altered distribution of cells and matrix, and markedly enhanced production of EPS when probed by the labeled dextran (FIG. 2A) (Xiao et al., 2012). Quantification of biofilm images (FIG. 2A) by the COMSTAT program and vertical sectioned biofilm images (FIG. 2B) also revealed that the pdeA mutant formed much thicker biofilm, produced more EPS in comparison with the wild type bacteria (FIG. 2C). Interestingly the pdeA mutant retained the bacterial level while produced more EPS (FIG. 2C, the right panel). Complementation of the pdeA mutant partially restored these phenotypes, demonstrating the role of PdeA in regulating biofilm formation. We also examined a recombinant strain that overexpressed DAC (the enzyme produces c-di-AMP) and found overexpressed DAC increased biofilm formation when compared to wild type (FIG. 3D). The PdeA deficiency and overexpression of DAC both increased c-di-AMP levels (FIG. 1A&S5) and promoted sucrose-dependent biofilm formation. Taken together, these data imply that c-di-AMP positively regulates biofilm formation.
FIG 2. PdeA deficiency-mediated biofilm formation analyzed by confocal laser scanning microscopy (CLSM).
(A) Representative images of 16 hours biofilms of each S. mutans strain analyzed using CLSM. The left panels represent bacteria stained by SYTO-9 (green), the middle panels represent EPS stained by dextran-conjugated Alexa Red, and the right panels represent merged images. Representative images from three independent experiments are shown. 50 µm scale bar was showed in each image. (B) Images of vertical sections of biofilms analyzed by CLSM, using CLSM 5 Image Browser software. (C) COMSTAT analysis of bio-volume and depths of biofilm depicted in (B). Total biomass and thickness were analyzed (Left panel); bacteria and EPS biomass were analyzed separately (Right panel). Data shown are the means with standard deviations of results from 6 representative images of each group taken from two difference days by the same person. *, P < 0.05 of Student’s t test using Prism 5.0.
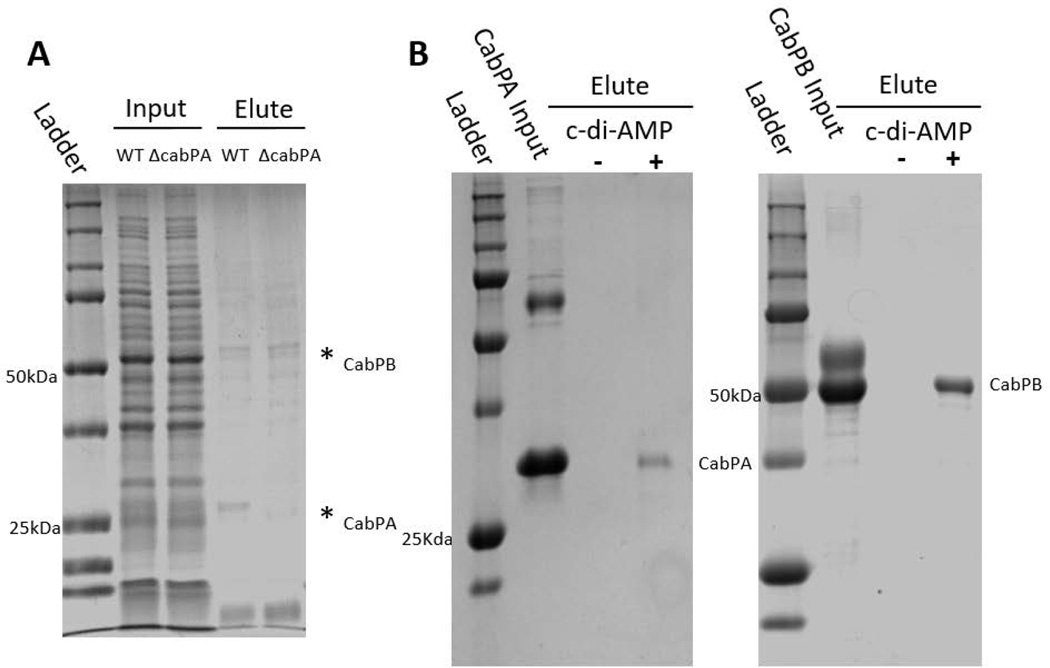
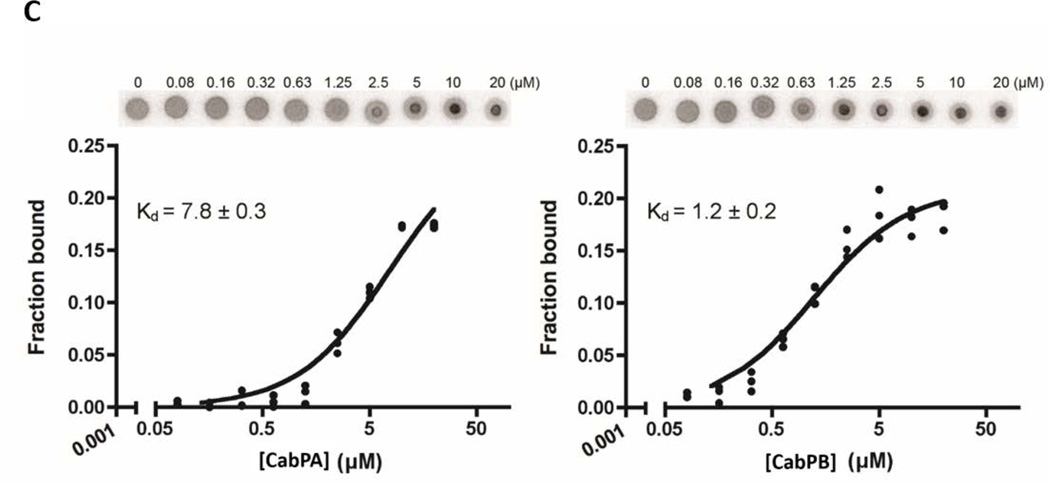
FIG 3. Identification of c-di-AMP binding proteins.
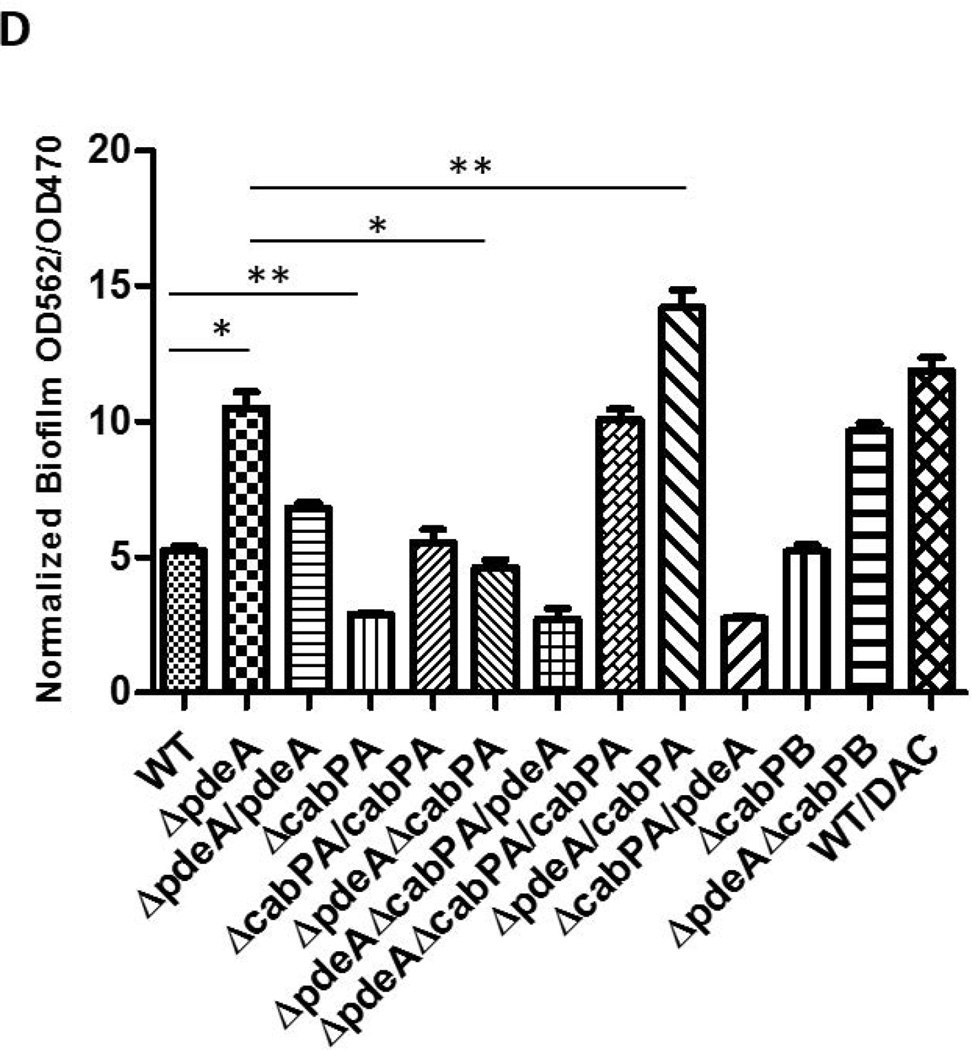
(A) Affinity purification of c-di-AMP binding proteins of S. mutans. Cell lysates prepared from the cabPA mutant and wild type strain of S. mutans were allowed to pass through a 2’-AHC-c-di-AMP agarose column. Proteins eluted from the column after three washes were subject to SDS-PAGE analysis and staining. Two protein bands (*) evident in the wild type elution were identified as CabPA and CabPB by LC/MS/MS. (B) Recombinant CabPA and CabPB proteins bind to c-di-AMP. Purified recombinant CabPA (Left panel) and CabPB (Right panel) were used in in vitro pull-down assays with c-di-AMP-coupled (+) or uncoupled (−) beads. BSA was included in the inputs with recombinant CabPA and CabPB as a negative control. Proteins bound to the beads were subject to SDS-PAGE analysis and protein staining. (C) Binding affinity of CabPA and CabPB to c-di-AMP measured by DRaCALA. CabPA had a lower affinity (in µM) binding to c-di-AMP when compared to CabPB. (D) Effects of deletion or overexpression of cabPA on biofilm formation by the pdeA mutant. Biofilm formation of wild type and different mutant variants and complements were determined using crystal violet staining assay. Means of three independent experiments are shown. Error bars denote the SEM. *, P < 0.01; **, P < 0.05 of Student’s t test using Prism 5.0.
c-di-AMP binding protein CabPA is crucial for the biofilm formation
To investigate how c-di-AMP regulates biofilm formation in S. mutans, we performed an affinity purification to identify proteins that bind c-di-AMP by using 2’-AHC-c-diAMP-agarose resin (Biolog Life Sciences Institute) (Bai et al., 2014). Two proteins were evident in the eluted fraction and identified by LC/MS/MS as CabPA with a molecular mass of ~25 kDa, and CabPB with a molecular mass of ~55 kDa (FIG. 3A). Both CabPA and CabPB contain TrkA_C and TrkA_N domains (FIG. S1), and the TrkA domains were found bound peripherally to the inner side of cytoplasmic membrane of the constitutive potassium-uptake systems (Schlosser et al., 1993), the TrkA_C domain containing proteins have been identified as c-di-AMP receptors (Corrigan et al., 2013). To further verify that both CabPA and CabPB are c-di-AMP binding proteins, we performed an in vitro pull-down assay using biotinylated c-di-AMP coupled beads. Both CabPA and CabPB were readily pulled down by c-di-AMP-coupled beads (FIG. 3B). Additionally, a differential radial capillary action of ligand assay (DRaCALA) was performed to verify the bindings. CabPA had lower affinity for c-di-AMP (Kd = 7.8±0.3 µM), when compared to CabPB (Kd= 1.2±0.2 µM), which is consistent with the observation that less CabPA was co-purified than CabPB by the pull-down assays (FIG. 3B&3C).
To determine whether CabPA and CabPB are responsible for the increased biofilm formation seen in the pdeA mutant, we mutated cabPA and cabPB, respectively, in the pdeA deficient background, and then examined their biofilms. cabPA deficiency in the pdeA mutant background inhibited the enhanced biofilm formation, and complementation of the pdeA-cabPA double mutant with cabPA restored the enhanced biofilm formation. In contrast, inactivation of cabPB in the pdeA mutant background did not alter the increased biofilm formation (FIG. 3D&S4). To determine whether these effects are due to changes in the c-di-AMP levels, we measured the c-di-AMP levels of all mutants and the DAC overexpressing strain, and found that enhanced biofilm formation is linked to increased c-di-AMP levels (FIG. S5). To determine regulation of CabPA by c-di-AMP produced from the pdeA mutant in the biofilm formation, we performed epistasis experiments. We first attempted to restore biofilm defects of cabPA, pdeA, and cabPA-pdeA by provision of cabPA and pdeA on a plasmid (FIG. 3D). Overexpression of cabPA restored the biofilm phenotype of the cabPA mutant, and significantly increased biofilm formation of the pdeA mutant, suggesting increased c-di-AMP coupled with overexpressed CabPA promoted biofilm formation. On the other hand, the pdeA gene restored the biofilm phenotype of the pdeA mutant, but failed to alter biofilm phenotype of the cabPA mutant (Fig. 3D). Together, these data suggest that CabPA is a key c-di-AMP receptor that is required for the c-di-AMP-mediated biofilm formation.
c-di-AMP regulates GtfB-dependent biofilm formation
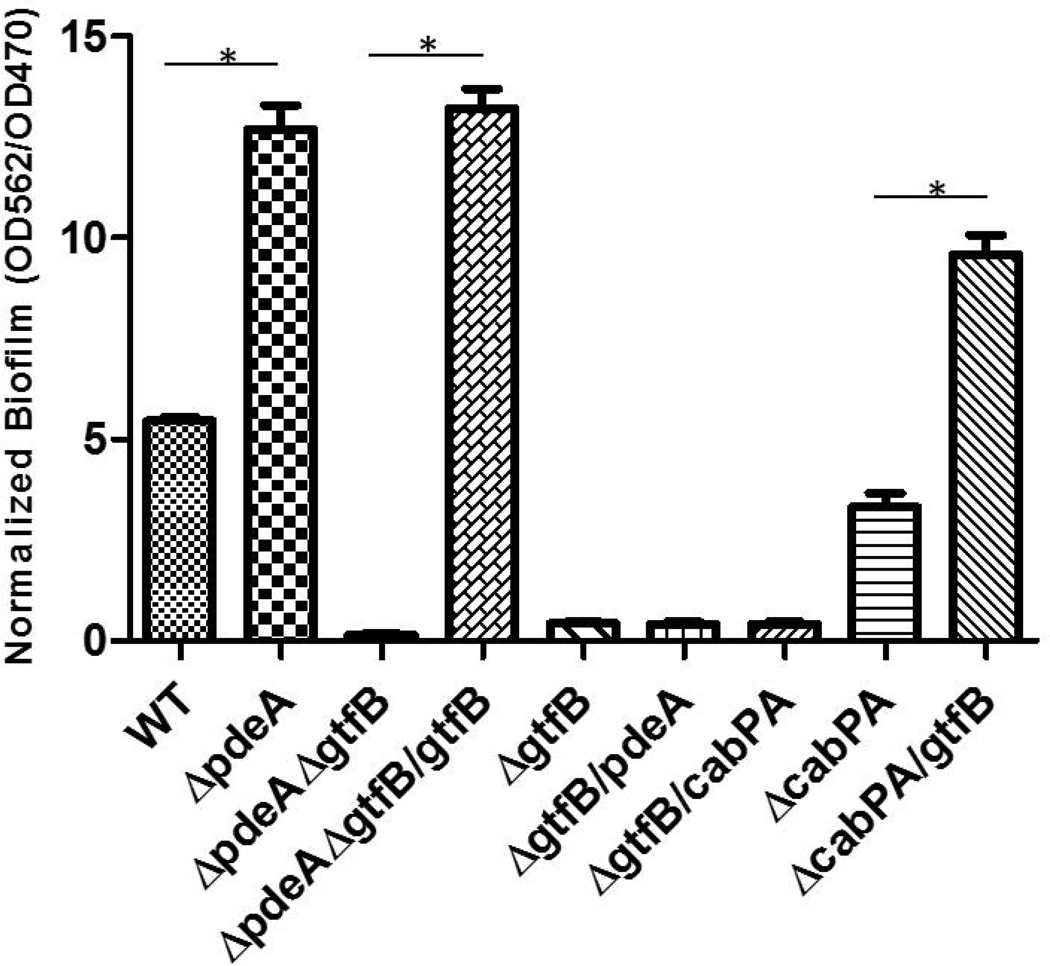
To investigate how c-di-AMP mediates biofilm formation in S. mutans, we attempted to isolate genes involved in c-di-AMP-mediated biofilm by a transposon mutagenesis scheme. A transposon library on the pdeA mutant background was generated using a Himar 1 Mariner mini-transposon system (van Opijnen & Camilli, 2010), and validated as described (Klein et al., 2012). Over 2000 individual transposon mutants were screened to isolate the mutants that would have reduced c-di-AMP-mediated biofilm formation. Two transposon mutants that exhibited lower biofilm biomass were identified and confirmed. Sequencing analysis of the transposon mutants revealed the transposon element inserted into gtfB (FIG. 4) and smu_30 (data not shown), respectively. gtfB encodes a glucosyltransferase that is critical to biofilm formation in S. mutans while smu_30 codes for a hypothetical protein with no known function in the biofilm formation. Complementation of the pdeA-gtfB double mutant with gtfB restored the enhanced biofilm formation (FIG. 4). Overexpression of gtfB in the cabPA mutant significantly increased biofilm formation, while overexpression of either pdeA or cabPA in the gtfB mutant didn’t restore biofilm formation, suggesting that GtfB is a downstream factor.
FIG 4. GtfB is crucial for c-di-AMP-mediated biofilm formation.
Biofilm deficient mutant pdeA-gtfB was identified by screening of a transposon mutant library on the pdeA mutant background. Cross-complementation of various mutants by GtfB, CabpA and PdeA was carried out to determine their effects on biofilm formation. Biofilm formation of wild type, the pdeA mutant, the identified insertional GtfB mutant and the complemented strain were determined by OD562/OD470 using a crystal violet staining assay. Means of three independent experiments are shown. Error bars denote the SEM. *, P < 0.01 of Student’s t test using Prism 5.0.
c-di-AMP regulates Gtf-dependent EPS production
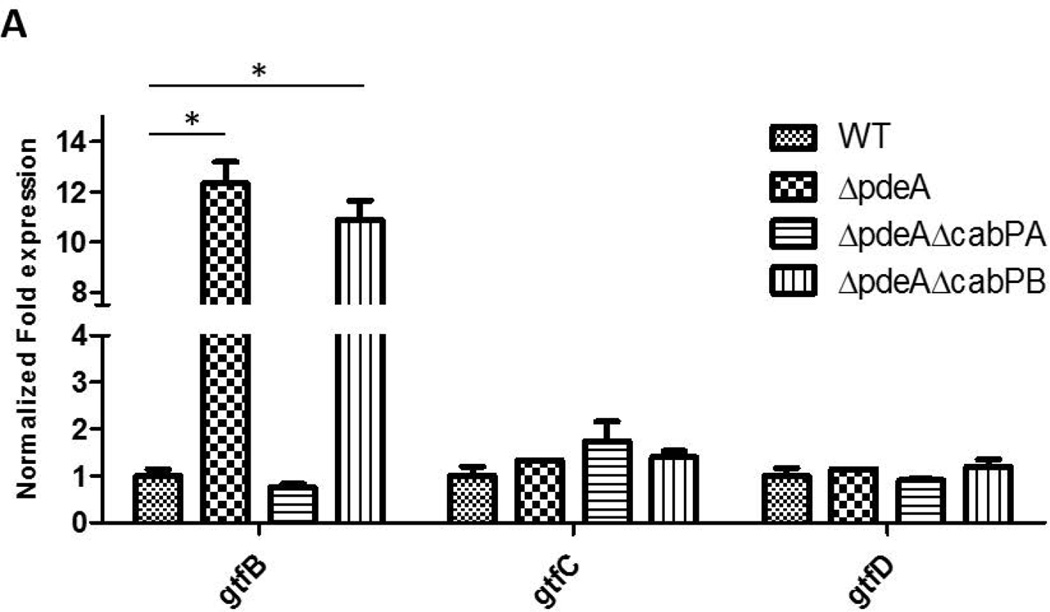
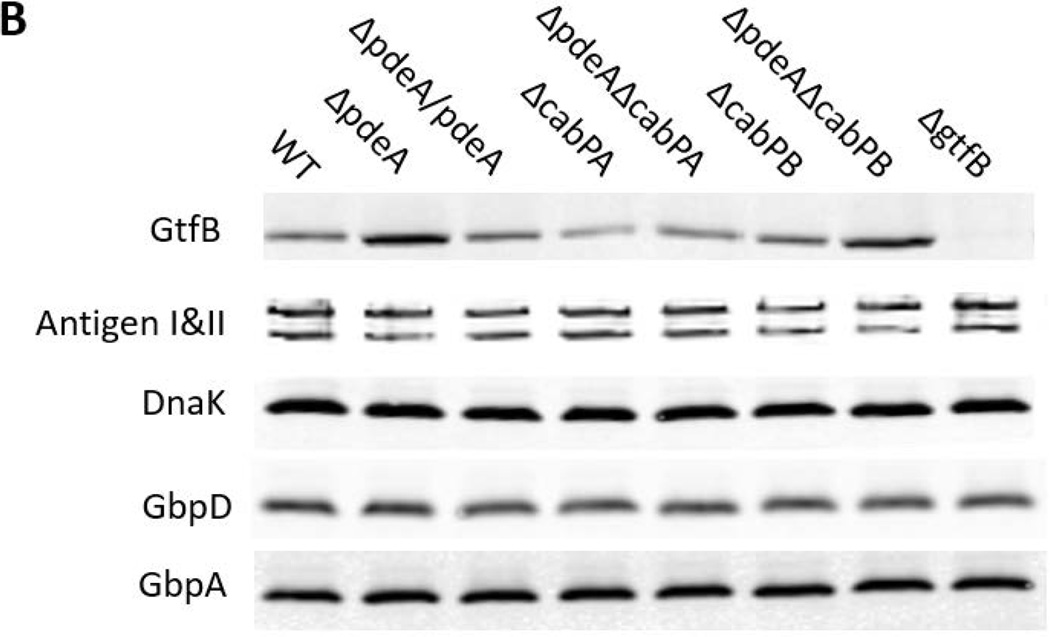
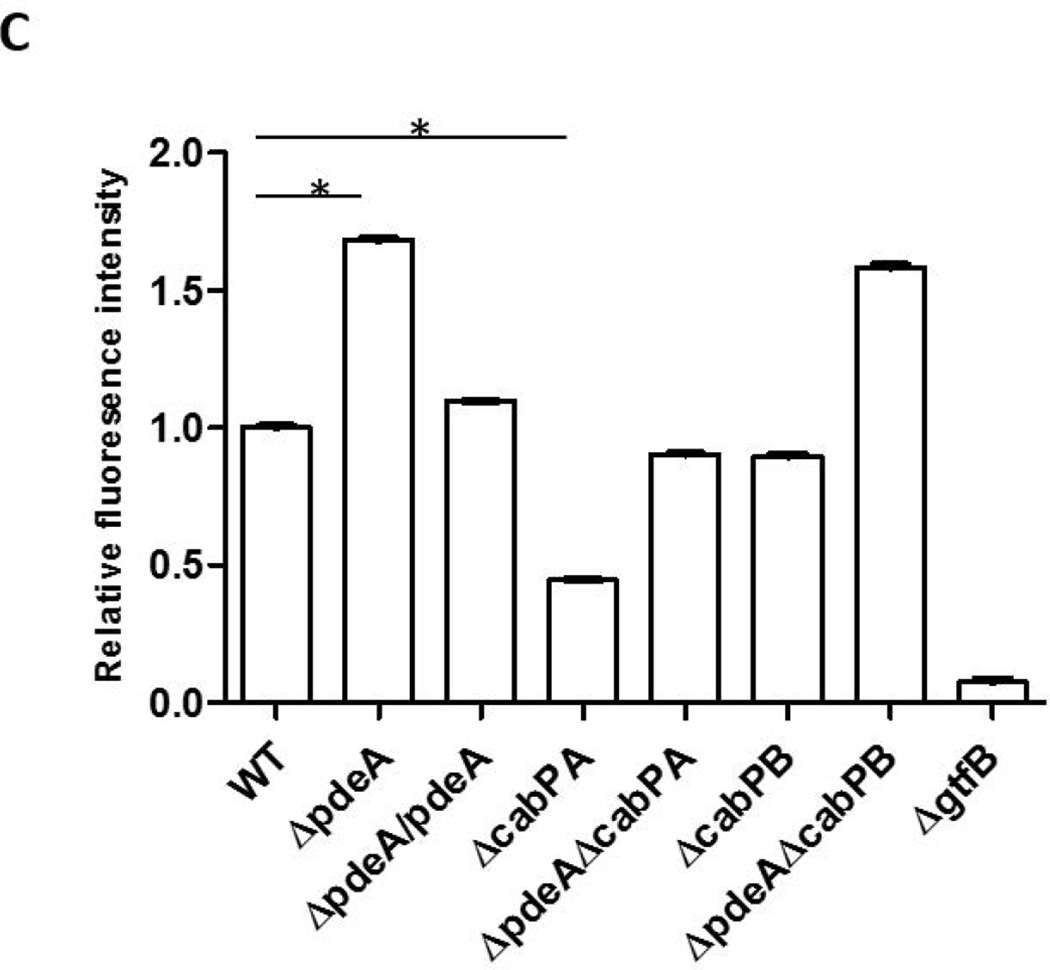
Since GtfB deficiency blunted increased biofilm formation in the pdeA mutant (FIG. 4), EPS production was enhanced in the pdeA mutant (FIG. 2A & B) and EPS is primarily synthesized by three glucosyltransferases GtfB, GtfC and GtfD, we examined how the PdeA deficiency affected expression of gtfB, gtfC and gtfD by real-time RT-PCR. Interestingly, the expression of gtfB in the pdeA mutant was up-regulated by 12-fold compared to the wild type; however there was no significant difference in expression of gtfC and gtfD (FIG. 5A). Furthermore, inactivation of cabPA but not cabPB abolished the increased expression of gtfB by the pdeA mutant (FIG. 5A). We subsequently examined the effect on the GtfB protein level using Western blot analysis. Consistently, the amount of GtfB was significantly increased in the pdeA mutant and restored to the wild type level when cabPA but not cabPB was deleted in the pdeA mutant (FIG. 5B&5C). To investigate whether c-di-AMP affects secreted GtfB, we examined GtfB in culture supernatants and found that secreted GtfB was altered similarly (FIG. S6). These findings are consistent with the changes in the biofilm formation by various mutants (FIG. 3D&S4). Other biofilm associated or glucan binding related surface proteins such as antigen I/II, GbpA and GbpD were not significantly affected (FIG. 5B), suggesting selective impact of c-di-AMP on GtfB through CabPA. Together, these data indicate that c-di-AMP, the binding receptor CabPA, and the production of GtfB are tightly coordinated with the biofilm formation in S. mutans, which constitutes a major c-di-AMP-mediated biofilm pathway. Interestingly, we found the pdeA-cabPA double mutant had more GtfB than the cabPA single mutant and produced more biofilm biomass (FIG. 5B, 5C&3D), indicating that CabPA-independent biofilm formation pathways also exist in response to elevated c-di-AMP (FIG. 8).
FIG 5. CabPA is critical for the regulation of GtfB.
(A) Quantitative analysis of expression of glucosyltransferase GtfBCD genes. Expression of gtfB, gtfC and gtfD in the indicated mutants was determined by qRT-PCR using RNA prepared from log-phase grown bacteria. Expression of mRNA was calibrated using 16S rRNA. Values represent the means of three different experiments with standard deviations. * = P < 0.01 of Student’s t test using Prism 5.0. (B) Western blotting analysis of GtfB and other S. mutans proteins. The amount of GtfB, AntigenI/II, GbpA, and GbpD in different S. mutans mutants was examined by Western blotting analysis of cell lysates of 6-h S. mutans cultures. Dnak was set as a loading control. (C) Quantification of Western blotting analysis of GtfB. Values represent the means of three experiments with standard deviations of relative fluorescence intensity of each band analyzed by using ImageJ. * = P < 0.01 of Student’s t test using Prism 5.0.
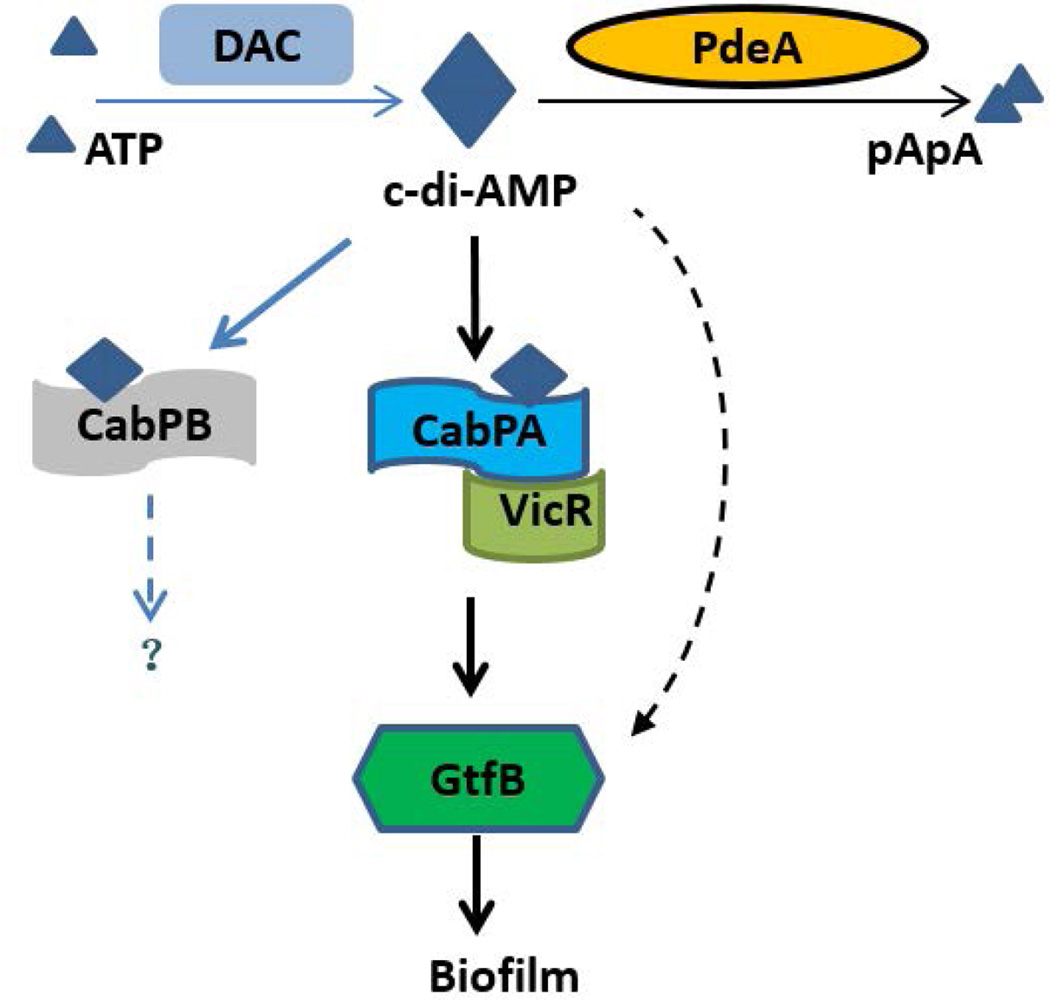
FIG 8. Working model of c-di-AMP-mediated biofilm formation.
Elevated c-di-AMP binds to its receptor CabPA, and CabPA in turn interacts with VicR, a response regulator that modulates expression of gtfB, thereby promoting biofilm formation. Two homologous c-di-AMP receptors CabPA and CabPB were identified, only CabPA was found to bind to VicR, thereby increasing expression of gtfB and promoting biofilm formation. CabPB is not involved in biofilm formation, the precise function of CabPB is unknown. Alternative pathways may exist that engage c-di-AMP receptors other than CabPA and CabPB to regulate GtfB-dependent biofilm formation. Solid lines represent well-supported findings, and broken lines represent hypothetical or unknown functions.
Binding of CabPA to VicR, a known response regulator that mediates expression of gtfB
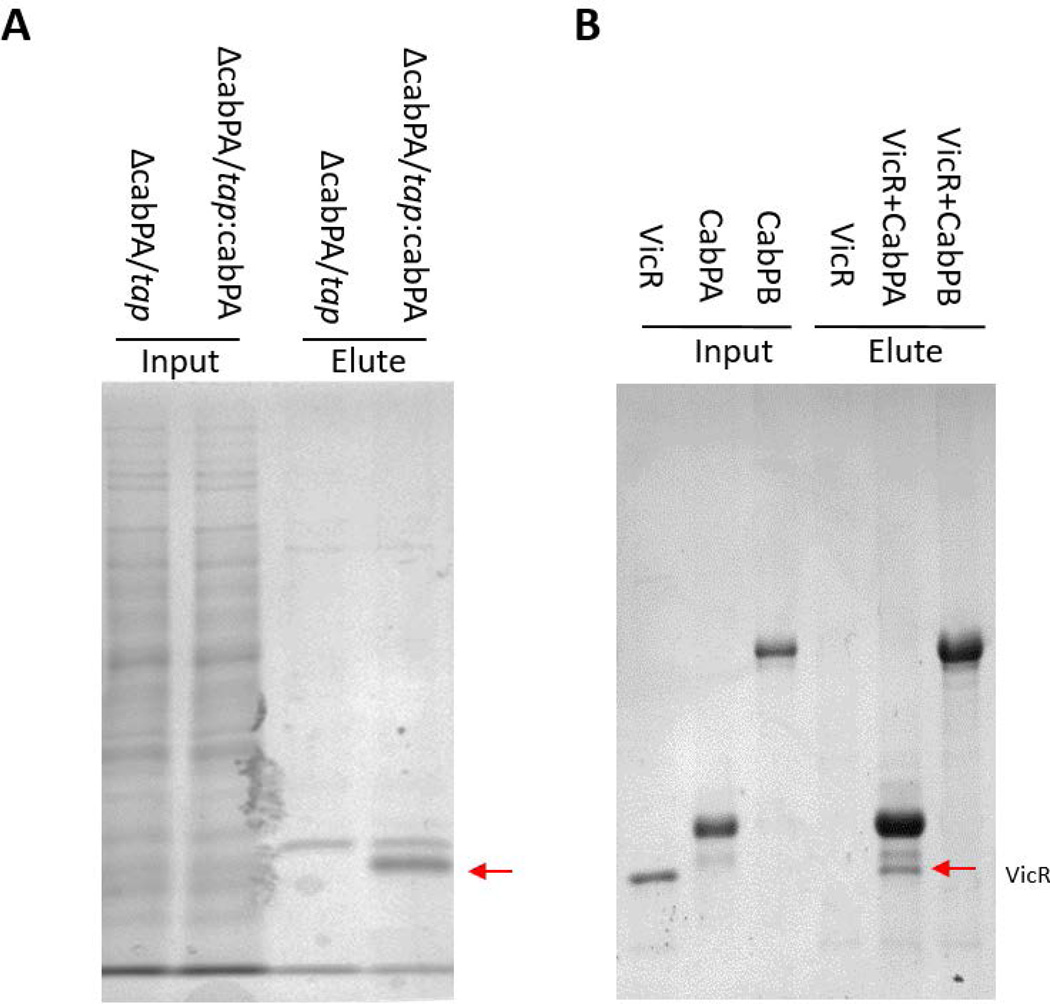
The above studies suggest that CabPA may regulate expression of gtfB. To determine how CabPA regulates GtfB. We used TAP (Tandem Affinity Purification) method to identify proteins that associate with CabPA in vivo in S. mutans. We found a ~25kDa protein was specifically purified with CabPA (FIG. 6A), and this band is evident by protein staining. It was determined to be VicR by LC/MS/MS. VicR is a response regulator of a known S. mutans two-component system. To confirm this we probed the purified proteins with VicR antiserum, the western blot demonstrated that the protein associated with CabPA is VicR. When other biofilm relevant proteins GbpA and D (Glucan-binding proteins) and BrpA (Biofilm regulatory protein, a putative transcriptional factor and important for biofilm formation), were probed by their antibodies, GbpC non-selectively bound to TAP purified proteins independent of CabPA, while BrpA was not detectable (FIG. S7), demonstrating the specific association of VicR with CabPA. To determine whether CabPA directly interacts with VicR, we performed a pull-down assay using recombinant CabPA, CabPB and VicR, and confirmed that VicR directly interacted with CabPA not CabPB in vitro (FIG. 6B). Therefore, both in vivo and in vitro data demonstrated that CabPA binds VicR. Overall, our results suggest that c-di-AMP mediates GtfB expression and biofilm formation through the interaction between CabPA and VicR.
FIG 6. CabPA, not CabPB, interacted with VicR.
(A) Coomassie blue staining SDS-PAGE of TAP purification. Both input and elute samples were analyzed. A band at 25 kDa was evident in tap:cbpA but not the tap control lane (arrow indicated) and identified as VicR by LC/MS/MS. (B) CabPA not CabPB bound to VicR as determined by a pull-down assay. Recombinant VicR could be pulled down by recombinant CabPA, not CabPB.
PdeA deficiency promotes colonization of Drosophila by S. mtuans
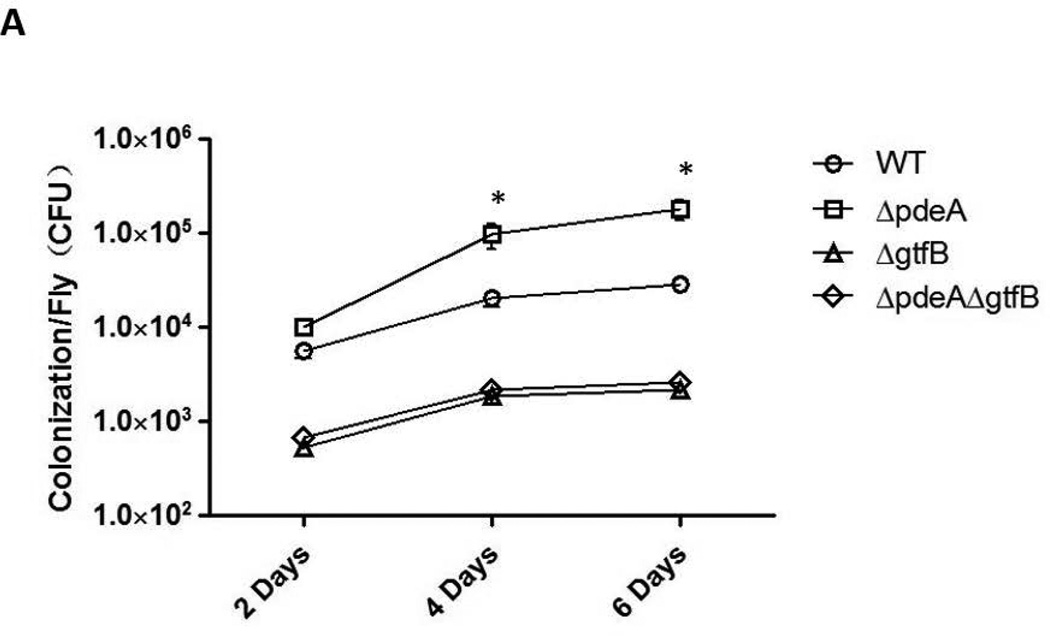
To determine if c-di-AMP impacts S. mutans colonization in vivo, we employed a widely used Drosophila colonization model (Mulcahy et al., 2011, Chugani et al., 2001). Flies were infected with mid-log phase bacterial cells of wild type S. mutans, gtfB and pdeA mutants. After 2 days of infection, there was no difference in colonization between wild type and the pdeA mutant, while the gtfB mutant, a known biofilm defective strain, showed significant reduction in colonization. However after 4 days and 6 days of infections, the difference emerged. Wild type S. mutans colonized in Drosophila at a considerable level of 3.8 × 104 per fly on day 6 post-infection while GtfB deficiency reduced colonization to 4.0 × 103 per fly, about a 10-fold decrease compared to that of the wild type. However, PdeA deficient mutant exhibited significantly increased bacterial colonization to 2.5 × 105 per fly, approximately a six-fold increase over the wild type bacteria (FIG. 7A). To visually show the colonization of Drosophila by S. mutans, we fed flies with green fluorescent protein (gfp)-tagged strains and examined effects of various mutations on bacterial colonization as described (Dionne et al., 2003). On day 6 post-infection, significantly higher fluorescence intensity was observed in guts of flies fed with the PdeA/gfp strain when compared to those fed with WT/gfp. Deletion of gtfB from the PdeA/gfp strain rendered the infected flies exhibit the minimal fluorescence similar to the flies infected with GtfB/gtf (FIG. 7B). These data again demonstrated that increased c-di-AMP promotes colonization of Drosophila by S. mutans in vivo, and the enhanced colonization by increased c-di-AMP was attenuated by the GtfB deficiency.
FIG 7. Colonization of Drosophila.
(A) Quantification of bacterial colonization. Colony forming units/fly at 2, 4 and 6 days after infected with S. mutans wild type, PdeA, GtfB and PdeA-GtfB double mutants were enumerated. * = P < 0.01 of Student’s t test using Prism 5.0. (B) Fluorescent detection of S. mutans colonized Drosophila. Representative images of Drosophila colonized by gfp-tagged S. mutans variants on 6 days were analyzed by Nikon elipse 90i microscope, equipped with an Epi-fluorescence and NIS elements AR imaging system.
Discussion
Biofilm formation is an important virulence determinant of many bacterial pathogens including S. mutans. As an emerging bacterial second messenger, c-di-AMP plays an important role in bacterial fitness and virulence (Woodward et al., 2010, Corrigan et al., 2011, Oppenheimer-Shaanan et al., 2011, Bai et al., 2014, Ye et al., 2014, Skrnjug et al., 2014, Yang et al., 2014). However, its role in bacterial biofilm formation is not well defined. In this study, we determined that c-di-AMP regulates biofilm formation of the oral pathogen S. mutans. Our studies revealed a new signaling network that integrates c-di-AMP signaling and biofilm formation of S. mutans (FIG. 8).
c-di-AMP signaling is essential for bacterial viability in many Gram-positive bacteria (Corrigan et al., 2011, Bai et al., 2012, Ye et al., 2014, Bai et al., 2013, Luo & Helmann, 2012). It participates in an array of activities, including cell wall biosynthesis, potassium homeostasis, DNA damage response, and bacterial host interactions. For instance, the increase in cross-linked peptidoglycan subunits in S. aureus gdpP mutant strengthens the peptidoglycan integrity and renders the bacteria more resistant to antibiotics. c-di-AMP has been implicated in the regulation of biofilm formation of S. aureus (Corrigan et al., 2011); however, whether it is directly involved in biofilm formation is unknown. Our studies reveal that regulation of GtfB, a major biofilm matrix building protein, by c-di-AMP and its receptor CabPA via binding VicR is the key step. Glucan binding proteins A, C and D have been implicated in the biofilm formation (Lynch et al., 2013); however, there was no apparent effect of increased c-di-AMP on these three proteins and other closely related glucosyltransferase genes gtfC and gtfD in S. mutans, suggesting the key and selective action through GtfB. Two c-di-AMP receptors from S. mutans, CabPA and CabPB, were identified by using c-di-AMP affinity purification scheme. TrkA_C domain is known c-di-AMP binding region (Corrigan et al., 2013). The homologous proteins have been shown to mediate potassium homeostasis in S. aureus (Corrigan et al., 2013) and S. pneumoniae (Bai et al., 2014). However, whether they are involved in regulating biofilm formation is unknown. Interestingly, although CabPB possesses two TrkA_C domains and exhibited higher affinity for c-di-AMP, inactivation of cabPB did not alter the increased biofilm formation in the pdeA mutant. In contrast, inactivation of CabPA blunted the increased biofilm in the pdeA mutant, again indicating the selective action by CabPA in modulating formation of biofilms. Consistently, inactivation of cabPA, not cabPB, down-regulated expression of GtfB. These observations provide convincing evidence that CabPA not CabPB regulates biofilm formation of S. mutans. Complementation using cabPA not pdeA restored the biofilm defect of a cabPA-pdeA double mutant and overexpression of cabPA in the pdeA mutant significantly increased biofilm formation (FIG. 3D), further indicating higher concentrations of c-di-AMP can interact with overexpressed CabPA to enhance biofilm formation. Furthermore, our studies indicate the receptor CabPA interacted with VicR, an essential response regulator that has been shown to mediate expression of gtfB (Senadheera et al., 2005). These data uncovered a missing link between c-di-AMP signaling and GtfB-mediated biofilm formation.
It is not clear whether potassium homeostasis plays a direct role in biofilm formation. Studies from B. subtilis hint a potential association (Lundberg et al., 2013), albeit the underlying mechanism is not known. In addition, up-regulation of a putative potassium uptake system has been associated with sucrose-dependent biofilm formation in S. mutans (Klein et al., 2010). Further studies are needed to examine association between potassium homeostasis and the VicRK system on the regulation of biofilm formation.
Intriguingly the pdeA-cabPA double mutant produced slightly more GtfB than the cabPA single mutant, also formed slightly more biofilms, which may indicate that cabPA-independent biofilm pathway exists in response to increased c-di-AMP.
Like c-di-GMP, a well characterized bacterial second messenger that regulates biofilm formation, c-di-AMP promotes biofilm formation by up-regulating production of glucans, a key component of the biofilm matrix. In c-di-GMP mediated biofilm formation, both extracellular matrix components and biofilm associated adhesins are up-regulated (Hengge, 2009). In contrast, c-di-AMP-mediated biofilm enhancement promoted production of glucans, but not other key biofilm associated adhesins such as antigen I/II (Munro et al., 1993), GbpA, and GbpD (Banas & Vickerman, 2003), suggesting that distinct regulation of biofilm formation by c-di-AMP in S. mutans. Like c-di-GMP, c-di-AMP has been implicated in modulating bacterial response to a variety of environmental stresses such as antibiotics exposure, acid, and oxidative stress (Romling, 2008, Gomelsky, 2011, Corrigan & Grundling, 2013). Unlike c-di-GMP, c-di-AMP signaling is essential for bacterial viability in many species (Mehne et al., 2013, Dengler et al., 2013, Yamamoto et al., 2012, Bai et al., 2013). Biofilm phenotypes mediated by c-di-AMP may represent a new activity independent of this essential function.
In conclusion, we determined a c-di-AMP-mediated biofilm signaling network, in which the c-di-AMP receptor CabPA controls c-di-AMP-mediated expression of a key biofilm matrix regulator, GtfB, and biofilm formation via the binding to VicR. Alternative biofilm pathways may exist to complement this major pathway. Future studies are warranted to dissect other biofilm pathways and receptors involved and c-di-AMP-mediated stress responses, which would provide a more complete picture for c-di-AMP-mediated biological activities in S. mutans.
Experimental procedures
Bacterial strains and culture conditions
S. mutans UA159, Escherichia coli Top 10 and BL-21 were used in this study. S. mutans strains were grown statically at 37°C in biofilm medium (BM) (Netuschil et al., 1996, Loo et al., 2000), Todd-Hewitt broth (THB) or on TH agar plates under an aerobic atmosphere with 5% CO2. E. coli strains were grown in Luria-Bertani (LB) broth or on LB agar plates. Appropriate antibiotics were used to select transformants on media as needed.
Construction and complementation of mutants
Mutant strains were derived from S. mutans UA159 using PCR ligation mutagenesis with the insertion of a nonpolar kanamycin resistance cassette to replace targeting genes (Ahn et al., 2006, Xie et al., 2011). Briefly, primers (Listed in Table 2) were used to amplify the regions flanking pdeA, cabPA, and cabPB, corresponding PCR products were ligated to kanamycin or erythromycin resistance cassette by fusion PCR, and the obtained PCR products were transformed into competent S. mutans. Transformants were selected on THB agar with kanamycin (1 mg ml−1) or erythromycin (10 µg ml−1). PCR and DNA sequencing methods were used to verify whether a correct mutation was introduced. The complete open reading frame of pdeA was amplified and cloned into the E. coli-Streptococcus shuttle vector pVPT (Zhou et al., 2008) to generate the corresponding complementation plasmid pVPT-pdeA. The plasmid was then transformed into the pdeA mutant strain to generate a complemented strain.
Table 2.
Primers used in this study
| Primers | Sequence (5’-3’) | Description |
|---|---|---|
| pdeAupF | TATGATATTGTCCTTCCTCATC | PdeA deletion |
| pdeAupR | TATTCTCATTTTAGCCATTATCTAAACCTCTTAAGCCCTA | PdeA deletion |
| pdeAdnF | CTGGATGAATTGTTTTAGGATGAAAGTTATTTTTTTAGCA | PdeA deletion |
| pdeAdnR | CGAAATCCACTACGATTACT | PdeA deletion |
| pedAcheckF | ATGTGGTTCATTCGATTAAA | Check PdeA mutant |
| pdeAcheckR | GAACTTCTAAATTTTCATAATTGAC | Check PdeA mutant |
| pdeAF | ATGAAAAGATTTCGTTTTGCCAC | Amplify PdeA |
| pdeAR | TTAAGCTTCCTCGTTTCCTG | Amplify PdeA |
| kanF | ATGGCTAAAATGAGAATA | Amplify kanamycin |
| kanR | CTAAAACAATTCATCCAG | Amplify kanamycin |
| ermF | GGAAGGAGTGATTACATGAA | Amplify rythromycin |
| ermR | GAAGCTGTCAGTAGTATACC | Amplify rythromycin |
| cabPAupF | GCCTTATCAGATGAAGTTGTC | CabPA deletion |
| cabPAupR | TTCATGTAATCACTCCTTCCAGCTACCCTCCTTTTGATAAC | CabPA deletion |
| cabPAdnF | GGTATACTACTGACAGCTTCGGAGGAAGAAGCTATGAAAAT | CabPA deletion |
| cabPAdnR | CTACTATCGAGCTCTTCAACTG | CabPA deletion |
| cabPAcheckF | AAGATATGTCAGATGATGACTTGA | Check cabPA mutant |
| cabPAcheckR | AGCTGGACTTTTCTAATTCTTTC | Check cabPA mutant |
| cabPBupF | CCACAGCATAAGTATGACTAA | CabPB deletion |
| cabPBupR | TTCATGTAATCACTCCTTCC GATTTACCTCCGAATTATAA | CabPB deletion |
| cabPBdnF | GGTATACTACTGACAGCTTCGCTATTATGAATAAAAGTATGG | CabPB deletion |
| cabPBdnR | TAAACTGAGAAAAGAGCGGCC | CabPB deletion |
| cabPBcheckF | TTTTTGTAATACTTTTAACGCATAT | Check cabPB mutant |
| cabPBcheckR | ACCAAGAAACATCAGTAGAAGC | Check cabPB mutant |
| cabPAF | GTCAGGATCCG ATGAAAATTATTATTGTCGGATGTGG | CabPA expression |
| cabPAR | GCCGGAGCTC TCAGAAAATAAAGCGTTCTCTC | CabPA expression |
| cabPBF | GTCAGGATCCG ATGAAAATTATTGTCGTTGGTGG | CabPB expression |
| cabPBR | GCCGGTCGAC TTACCTCTTTAATAAATCATAAA | CabPB expression |
| Q16sRNAF | AGCGTTGTCCGGATTTATTG | qRT-PCR |
| Q16sRNAR | CTACGCATTTCACCGCTACA | qRT-PCR |
| QgtfBF | CACTATCGGCGGTTACGAAT | qRT-PCR |
| QgtfBR | CAATTTGGAGCAAGTCAGCA | qRT-PCR |
| QgtfCF | GATGCTGCAAACTTCGAACA | qRT-PCR |
| QgtfCR | TATTGACGCTGCGTTTCTTG | qRT-PCR |
| QgtfDF | TTGACGGTGTTCGTGTTGAT | qRT-PCR |
| QgtfDR | AAAGCGATAGGCGCAGTTTA | qRT-PCR |
| cabPAtapF | GCCGGTCGACATGAAAATTATTATTGTCGGATGTGG | TAP construct |
| cabPAtapR | GCGCGGTACCTCAGAAAATAAAGCGTTCTCTC | TAP construct |
Detection of c-di-AMP
Bacteria strains were grown in 5 ml THB for 16 h, and the growth was determined at an optical density at 470 nm (OD470). The bacteria were harvested by centrifugation at 8,000 rpm for 3 min. The pellets were suspended in 0.5 ml 50 mM Tris-HCl (pH 8.0), sonicated for 20 s, and followed by heating for 10 min at 95°C. Bacterial debris was removed and the supernatant was collected and used to measure c-di-AMP levels by an enzyme-linked immunosorbent assay (ELISA) as described (Bai et al., 2013). The amount of c-di-AMP was normalized by OD470 representing of the bacterial growth.
Biofilm formation assay
S. mutans biofilms were grown in either THB containing 1% sucrose or a chemically defined biofilm medium (BM) containing 1% sucrose (Loo et al., 2000). Overnight cultures were subcultured into fresh THB, grown to an OD470 of 0.6 and diluted 1:100 and subsequently aliquoted in a 96 well microtiter plate and grown at 5% CO2 at 37 °C under static conditions (Li et al., 2013). Biofilm samples were collected after 16 h and stained with crystal violet. Crystal violet staining at OD562 was used to monitor biofilm formation as described previously (Wu et al., 2007). Due to the different growth rates of wild type and mutants, the biofilms were normalized by bacterial growth using OD562/OD470. Each assay was carried out with duplicate samples and replicated three times.
Confocal laser scanning microscopy (CLSM) analysis
Bacteria strains were grown in BM with 1µM dextran-conjugated alexa (Molecular Probes, Invitrogen) to label EPS on glass coverslips placed in wells of a sterile 6-well cell culture plate (Corning Costar Corp.) under 5% CO2 at 37°C for 16 h. The biofilm samples were gently washed with PBS three times to remove unattached cells, dried for 5 min, and then stained with SYTO 9 (Molecular Probes, Invitrogen) (Decker et al., 2014). The stained samples were then examined by CLSM (LSM 710; Zeiss) with a 63× oil immersion objective. Images were obtained from serial optical sections and captured at 488nm. At least three independent experiments were performed by the same person on different days, and image stacks were acquired randomly. Thickness and bio-volume of biofilm were quantified by the program COMSTAT (Heydorn et al., 2000, Kreth et al., 2004).
Generation and screening of transposon mutant library
Genomic DNA was isolated from the pdeA mutant using Gentra Puregene Yeast/Bact. Kit (Qiagen). The smupdeA transposon library was generated as described by Tim et al (van Opijnen & Camilli, 2010). Briefly, 1µg of the mutant genomic DNA was incubated in the presence of purified HimarC9 transposase and 1µg of plasmid magellan6 as a donor of the Himar1 Mariner transposon with spectinomycin resistance. After repair of the resulting transposition products with T4 DNA polymerase and E. coli DNA ligase , 1 µg mutagenized DNA was used for transformation by using natural transformation for S. mutans (Liu et al., 2011). The randomness of transposon library was determined by sequencing 10 colonies randomly picked from the library. To screen the library for mutants that have biofilm defects, we performed a crystal violet assay to measure biofilm formation of 2000 individual insertion mutants. Mutants that had significant lower biofilm formation than the pdeA mutant were identified, transposon insertion was mapped as described (van Opijnen & Camilli, 2010).
Affinity purification of c-di-AMP binding proteins
Thirty milliliters of an overnight S. mutans culture were harvested and suspended in 1 mL 10 mM Tris·HCl (pH 7.5), 50 mM NaCl buffer containing EDTA-free complete protease inhibitor (Roche). Purification of c-di-AMP binding proteins from S. mutans cell lysates was carried out by using the previously described method (Bai et al., 2014). For in vitro affinity pull-down assay, forty microliters streptavidin dynabeads (Invitrogen) coupled with 2.4 µM biotinylated c-di-AMP (Biolog Life Sciences Institute) were incubated with 5 µM of purified recombinant CabPA or CabPB in 1.5 mL 10% (vol/vol) glycerol, 1 mM MgCl2, 5 mM Tris (pH 7.5), 250 mM NaCl, 0.5 mM DTT, and 4 mM EDTA containing 50 µg mL−1 BSA for 30 min at room temperature. Samples were washed four times with the same buffer lacking BSA and suspended in 50 µL protein sample buffer. Samples were boiled for 5 min to release the protein binding to c-di-AMP beads, centrifuged to remove beads, and the supernatant samples were analyzed using SDS-PAGE (Corrigan et al., 2013).
Tandem affinity purification
pVPT-tap-CabPA was constructed by PCR amplification of cabPA from S. mutans UA159 chromosomal DNA using primers CabPA-SalI-F/CabPA-KpnI-R (Table 2), digestion with SalI and KpnI, and ligation into pVPT-tap. The tandem affinity purification (TAP) methods provide a tool that allows rapid purification under native conditions of complexes (Puig et al., 2001, Rohila et al., 2004). Tandem affinity purification was performed as described (Puig et al., 2001, Wu & Wu, 2011) with modification. In brief, 500 ml of overnight S. mutans cultures were harvested and frozen at −80 °C. The pellets were thawed by adding 30 ml NETN (100 mM NaCl, 20 mM Tris-HCl pH 8.0, 0.5 mM EDTA, 0.5% (v/v) NP-40) and lysed by sonication (10 min, 5 s on, 10 s off). The lysates were then centrifuged (20 min, 10000 rpm, 4°C). The supernatants were transferred into another tubes and centrifuged again. The purification was carried out as described except using NETN buffer.
Mass Spectrometry
The amples were subjected to 10% SDS-PAGE analysis and Comassie blue staining. Potein bands were excised from the gels and digested with trypsin. The digested peptide fragments were analyzed for protein identification by liquid chromatography-tandem mass spectrometry (LC/MS/MS) as described previously (Bu et al., 2008).
Differential Radial Capillary Action of Ligand Assay (DRaCALA)
DRaCALA is a method based on the ability of dry nitrocellulose to separate the free ligand from bound protein–ligand complexes, which allows detection of specific interactions between nucleotides and their cognate binding proteins (Roelofs et al., 2011, Corrigan et al., 2013). Briefly, [α-32P] ATP (MP Biomedicals) was converted to 32P-labeled c-di-AMP by using recombinant M. tuberculosis DisA protein and purified using a thin layer chromatography (TLC) (Bai et al., 2014). The reaction (10 µl) for DRaCALA assay was prepared by mixing 1 µL 32P-labeled c-di-AMP and appropriate concentrations of recombinant CabPA or CabPB in binding buffer (40 mM Tris, 100 mM NaCl, 20 mM MgCl2 [pH 7.5]). Reaction was incubated at room temperature for 10 min, followed by spotting 5 µL onto a nitrocellulose membrane (GE healthcare). After air dry for 10 min, the membrane was exposed on a phosphor screen for 3 h. The radioactivity was detected using a Storm 860 PhosphorImager (Molecular Dynamics). Dissociation constant was analyzed as previous reported (Roelofs et al., 2011).
Expression and purification of CabPA and CabPB
Recombinant CabPA and CabPB constructs were created by amplifying the open reading frames of cabPA and cabPB from S. mutans genomic DNA and ligating into the pET-28(b) vector, each introducing a C-terminal 6×His tag using primers listed in Table 2. The recombinant plasmids were transformed into E. coli BL-21(DE3), and induction of protein expression was analyzed by SDS-PAGE. One liter of bacterial culture for each induced recombinant strain was pelleted by centrifugation, resuspended in 30 mL lysis buffer (20 mM Tris pH 8.0, 500 mM NaCl), and lysed by sonication (10 min, 5 s on, 10 s off). Cell debris was pelleted by centrifugation for 60 min followed by passing through a HisTrap column (Ni-affinity column). The proteins were further purified using a 16/60 Superdex 75 gel-filtration column (GE Healthcare) to near homogeneity and were maintained in a solution containing 20 mM Tris–HCl (pH 8.0) and 100 mM NaCl (Zhu et al., 2013).
Transcriptional analysis by qRT-PCR
Total RNA was extracted from the same numbers of harvested bacterial cells from the wild type control and mutants groups. All harvested cells were digested by N-acetylmuramidase (mutanolysin; Sigma-Aldrich) at 20 µg mL−1 and lysozyme at 10 µg mL−1 at 37°C for 30 min. The samples were then extracted with RNeasy Mini Kit (Qiagen) and further digested by RNase-free DNase (Promega) to remove trace amounts of contaminated DNA. The isolated RNA was reverse transcribed into cDNA using the iScript cDNA synthesis kit (Bio-rad). cDNA samples were then quantified by real-time PCR using the iQ SYBR green supermix kit (Bio-Rad) (Liu et al., 2011). Primers used for qRT-PCR are listed in Table 2.
Western blotting analysis
The same cell numbers of each bacterial samples were spin down and digested by 100 µL of mutanolysin and lysozyme mixture in lysis buffer (100 mM NaCl, 20 mM Tris-HCl [pH 8.0], 0.5 mM EDTA, 0.5% [vol/vol] NP-40) for 10 min at room temperature (Liu et al., 2011). Cell lysates were harvested by centrifugation at 13,000 rpm for 5 min after the removal of cell debris and used as protein extracts. Cell supernatants were harvested and precipitated with 30% ethanol at −80°C for 15 minutes, supernatant proteins were harvested by centrifugation at 13,000 rpm for 15 min at 4°C. Protein samples were then dissolved into 5× SDS loading buffer and subjected to SDS-PAGE analysis, followed by Western blotting using GtfB, antigen I&II, GbpA, GbpD and DnaK antiserum. Western blotting was carried out using procedures described previously (Liu et al., 2011). Fluorescence intensity was quantified by the method of Gassmann et al using Image J 1.48v (Windows version of NIH Image, http://rsb.info.nih.gov/nih-image/) (Gassmann et al., 2009, Tan & Ng, 2008).
Colonization of Drosophila
Drosophila were maintained routinely on medium containing corn meal, agar, and sucrose. Infections were performed as described (Chugani et al., 2001, Mulcahy et al., 2011). Mid-log phase THB cultures of S. mutans were spun down and resuspended in 5% sucrose. Cultures were adjusted to an OD600=25 (2.5×1010 CFU per ml) in sucrose. The resuspended cells (100 µl) were spotted onto a sterile filter (Whatman) that was placed on the surface of 5 ml of solidified 5% sucrose agar in a plastic vial (VWR). The vials were allowed to dry at room temperature for 30 minutes prior to addition of Drosophila. Male Canton S flies (1–3 days old) were treated with antibiotics for 2 days and starved for 3 hours prior to add to vials (10–14 flies per vial). The vials were capped with cotton, inverted over a tray with water for humidity, and incubated at 25°C. Flies were anaesthetized by carbon dioxide throughout the sorting and transferring process. To determine the number of viable bacterial cells associated with individual flies, single flies were ground with tips in an Eppendorf tube with 100 µl of THB, and serial dilutions of the homogenate were spread on THB agar plates. To visualize the colonization of Drosophila by S. mutans, we fed flies with gfp-tagged (green fluorescent protein) strains and examined effects of various mutations on bacterial colonization as described (Dionne et al., 2003). The colonization of flies by gpf-tagged strains were analyzed by Nikon elipse 90i microscope, equipped with an Epi-fluorescence and NIS elements AR imaging system. Representative images were presented.
Supplementary Material
Table 1.
Bacterial strains and plasmids used in this study
| Strains | Description | Source |
|---|---|---|
| S.mutans UA159 | Parent strain | (Liu et al., 2011) |
| ΔpdeA | PdeA deletion; Kanr | This study |
| ΔpdeA /pdeA | PdeA complementation; Kanr; Ermr | This study |
| ΔcabPA | CabPA deletion; Ermr | This study |
| ΔcabPB | CabPB deletion; Ermr | This study |
| ΔpdeAΔcabPA | PdeA and CabPA double mutant; Kanr; Ermr | This study |
| ΔpdeAΔcabPB | PdeA and CabPB double mutant; Kanr; Ermr | This study |
| ΔpdeAΔgtfB | GtfB insertion mutant on PdeA; Kanr; Specr | This study |
| ΔgtfB | GtfB deletion; Kanr | This study |
| ΔcabPA/tap | CabPA deletion with tap; Kanr; Ermr | This study |
| ΔcabPA/tap:cabPA | Recombinant CabPA with tap; Kanr; Ermr | This study |
| ΔpdeAΔgtfB/gtfB | Complementation of PdeA-GtfB using GtfB; Ermr ;Kanr; Specr |
This study |
| WT/DAC | Overexpressed DAC on wild type S. mutans | This study |
| S. mutans/gfp | gfp-tagged S. mutans UA159; Kanr | (Liu et al., 2011) |
| ΔpdeA/gfp | gfp-tagged PdeA deletion; Kanr; Ermr | This study |
| ΔpdeAΔgtfB/gfp | gfp-tagged PdeA-GtfB double mutant; Kanr; Ermr |
This study |
| ΔgtfB/gfp | gfp-tagged GtfB deletion; Kanr; Ermr | This study |
|
E.coli pET28b–CabPA |
CabPA expression; Kanr | This study |
|
E.coli pET28b–CabPB |
CabPB expression; Kanr | This study |
|
E.coli pET28a–sumo-VicR |
VicR purification; Kanr | Wu Lab |
| E. coli Top 10 | Cloning strain | Invitrogen |
| E. coli BL-21 | Protein production strain | Invitrogen |
| Plasmids | ||
| pVPT | E. coli-Streptococci shuttle vector; Ermr | (Zhou et al., 2008) |
| pVPT-pdeA | pVPT carrying PdeA ORF; Ermr | This study |
| pVPT-tap | pVPT carrying tap tag; Ermr | Wu Lab |
| pVPT-tap-cabPA | pVPT carrying tap tag and CabPA; Ermr | This study |
| pET28b(+) | Expression plasmid; Kanr | Novagen |
| pET28a–sumo | Expression plasmid; Kanr | Wu Lab |
| pET28b–cabPA | pET28b(+) carrying cabPA ORF; Kanr | This study |
| pET28b–cabPB | pET28b(+) carrying cabPB ORF; Kanr | This study |
Acknowledgments
We thank Dr. Jeff Banas from University of Iowa for providing us with GbpA and GbpD antibodies. This work was supported by NIH/NIDCR R01DE022350 (HW) and American Heart Association grant 12SDG12080067 (GB).
References
- Ahn S-J, Wen ZT, Burne RA. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect Immun. 2006;74:1631–1642. doi: 10.1128/IAI.74.3.1631-1642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes LCM, Ferreira RB, Buckner MM, Finlay BB. Quorum sensing in bacterial virulence. Microbiology. 2010;156:2271–2282. doi: 10.1099/mic.0.038794-0. [DOI] [PubMed] [Google Scholar]
- Bai Y, Yang J, Eisele LE, Underwood AJ, Koestler BJ, Waters CM, Metzger DW, Bai G. Two DHH Subfamily 1 Proteins in Streptococcus pneumoniae Possess Cyclic Di-AMP Phosphodiesterase Activity and Affect Bacterial Growth and Virulence. Journal of Bacteriology. 2013;195:5123–5132. doi: 10.1128/JB.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Yang J, Zarrella TM, Zhang Y, Metzger DW, Bai G. Cyclic di-AMP impairs potassium uptake mediated by a cyclic di-AMP binding protein in Streptococcus pneumoniae. J Bacteriol. 2014;196:614–623. doi: 10.1128/JB.01041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Yang J, Zhou X, Ding X, Eisele LE, Bai G. Mycobacterium tuberculosis Rv3586 (DacA) is a diadenylate cyclase that converts ATP or ADP into c-di-AMP. PloS one. 2012;7:e35206. doi: 10.1371/journal.pone.0035206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banas J, Vickerman M. Glucan-binding proteins of the oral streptococci. Critical Reviews in Oral Biology & Medicine. 2003;14:89–99. doi: 10.1177/154411130301400203. [DOI] [PubMed] [Google Scholar]
- Barker JR, Koestler BJ, Carpenter VK, Burdette DL, Waters CM, Vance RE, Valdivia RH. STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during Chlamydia trachomatis infection. mBio. 2013;4:e00018–e00013. doi: 10.1128/mBio.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano-Sagie M, Oppenheimer-Shaanan Y, Berlatzky I, Rouvinski A, Meyerovich M, Ben-Yehuda S. A Checkpoint Protein That Scans the Chromosome for Damage at the Start of Sporulation in Bacillus subtilis. Cell. 2006;125:679–690. doi: 10.1016/j.cell.2006.03.039. [DOI] [PubMed] [Google Scholar]
- Bu S, Li Y, Zhou M, Azadin P, Zeng M, Fives-Taylor P, Wu H. Interaction between two putative glycosyltransferases is required for glycosylation of a serine-rich streptococcal adhesin. J Bacteriol. 2008;190:1256–1266. doi: 10.1128/JB.01078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugani SA, Whiteley M, Lee KM, D’Argenio D, Manoil C, Greenberg E. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences. 2001;98:2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Abbott JC, Burhenne H, Kaever V, Grundling A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011;7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Campeotto I, Jeganathan T, Roelofs KG, Lee VT, Gründling A. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci U S A. 2013;110:9084–9089. doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Grundling A. Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol. 2013;11:513–524. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- Cvitkovitch DG. Genetic competence and transformation in oral streptococci. Crit Rev Oral Biol Med. 2001;12:217–243. doi: 10.1177/10454411010120030201. [DOI] [PubMed] [Google Scholar]
- Decker E-M, Klein C, Schwindt D, von Ohle C. Metabolic activity of Streptococcus mutans biofilms and gene expression during exposure to xylitol and sucrose. International journal of oral science. 2014;6:195–204. doi: 10.1038/ijos.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler V, McCallum N, Kiefer P, Christen P, Patrignani A, Vorholt JA, Berger-Bachi B, Senn MM. Mutation in the C-di-AMP cyclase dacA affects fitness and resistance of methicillin resistant Staphylococcus aureus. PloS one. 2013;8:e73512. doi: 10.1371/journal.pone.0073512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne MS, Ghori N, Schneider DS. Drosophila melanogaster is a genetically tractable model host for Mycobacterium marinum. Infect Immun. 2003;71:3540–3550. doi: 10.1128/IAI.71.6.3540-3550.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du B, Ji W, An H, Shi Y, Huang Q, Cheng Y, Fu Q, Wang H, Yan Y, Sun J. Functional analysis of c-di-AMP phosphodiesterase, GdpP, in Streptococcus suis serotype 2. Microbiol Res. 2014 doi: 10.1016/j.micres.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Gassmann M, Grenacher B, Rohde B, Vogel J. Quantifying Western blots: pitfalls of densitometry. Electrophoresis. 2009;30:1845–1855. doi: 10.1002/elps.200800720. [DOI] [PubMed] [Google Scholar]
- Gomelsky M. cAMP, c-di-GMP, c-di-AMP and now cGMP: bacteria use them all! Mol Microbiol. 2011;79:562–565. doi: 10.1111/j.1365-2958.2010.07514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersbøll BK, Molin S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- Huynh TN, Luo S, Pensinger D, Sauer J-D, Tong L, Woodward JJ. An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence. Proceedings of the National Academy of Sciences. 2015;112:E747–E756. doi: 10.1073/pnas.1416485112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia D, Merey G, Nakayama S, Zheng Y, Zhou J, Luo Y, Guo M, Roembke BT, Sintim HO. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem Soc Rev. 2013;42:305–341. doi: 10.1039/c2cs35206k. [DOI] [PubMed] [Google Scholar]
- Kamegaya T, Kuroda K, Hayakawa Y. Identification of a Streptococcus pyogenes SF370 gene involved in production of c-di-AMP. Nagoya J Med Sci. 2011;73:49–57. [PMC free article] [PubMed] [Google Scholar]
- Kim JN, Stanhope MJ, Burne RA. Core-gene-encoded peptide regulating virulence-associated traits in Streptococcus mutans. J Bacteriol. 2013;195:2912–2920. doi: 10.1128/JB.00189-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BA, Tenorio EL, Lazinski DW, Camilli A, Duncan MJ, Hu LT. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC genomics. 2012;13:578. doi: 10.1186/1471-2164-13-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein MI, DeBaz L, Agidi S, Lee H, Xie G, Lin AH-M, Hamaker BR, Lemos JA, Koo H. Dynamics of Streptococcus mutans transcriptome in response to starch and sucrose during biofilm development. PloS one. 2010;5:e13478. doi: 10.1371/journal.pone.0013478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Rosalen PL, Cury JA, Park YK, Bowen WH. Effects of compounds found in propolis on Streptococcus mutans growth and on glucosyltransferase activity. Antimicrobial agents and chemotherapy. 2002;46:1302–1309. doi: 10.1128/AAC.46.5.1302-1309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Xiao J, Klein M, Jeon J. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. Journal of Bacteriology. 2010;192:3024–3032. doi: 10.1128/JB.01649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Xiao J, Klein MI. Extracellular polysaccharides matrix—an often forgotten virulence factor in oral biofilm research. International journal of oral science. 2009;1:229. doi: 10.4248/IJOS.09086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Hagerman E, Tam K, Merritt J, Wong D, Wu B, Myung N, Shi W, Qi F. Quantitative analyses of Streptococcus mutans biofilms with quartz crystal microbalance, microjet impingement and confocal microscopy. Biofilms. 2004;1:277–284. doi: 10.1017/S1479050504001516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M-Y, Huang R-J, Zhou X-D, Gregory RL. Role of sortase in Streptococcus mutans under the effect of nicotine. International journal of oral science. 2013;5:206–211. doi: 10.1038/ijos.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Worthington RJ, Melander C, Wu H. A new small molecule specifically inhibits the cariogenic bacterium Streptococcus mutans in multispecies biofilms. Antimicrobial agents and chemotherapy. 2011;55:2679–2687. doi: 10.1128/AAC.01496-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo C, Corliss D, Ganeshkumar N. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. Journal of Bacteriology. 2000;182:1374–1382. doi: 10.1128/jb.182.5.1374-1382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg ME, Becker EC, Choe S. MstX and a putative potassium channel facilitate biofilm formation in Bacillus subtilis. PloS one. 2013;8:e60993. doi: 10.1371/journal.pone.0060993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Helmann JD. A sigmaD-dependent antisense transcript modulates expression of the cyclic-di-AMP hydrolase GdpP in Bacillus subtilis. Microbiology. 2012;158:2732–2741. doi: 10.1099/mic.0.062174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch DJ, Michalek SM, Zhu M, Drake D, Qian F, Banas JA. Cariogenicity of Streptococcus mutans glucan-binding protein deletion mutants. Oral Health Dent Manag. 2013;12:191–199. [PMC free article] [PubMed] [Google Scholar]
- Mehne FM, Gunka K, Eilers H, Herzberg C, Kaever V, Stulke J. Cyclic di-AMP homeostasis in bacillus subtilis: both lack and high level accumulation of the nucleotide are detrimental for cell growth. J Biol Chem. 2013;288:2004–2017. doi: 10.1074/jbc.M112.395491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle MJ, Eguren K, Melander C. Highly Active Modulators of Indole Signaling Alter Pathogenic Behaviors in Gram-Negative and Gram-Positive Bacteria. Chemistry-A European Journal. 2013;19:17595–17602. doi: 10.1002/chem.201303510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy H, Sibley CD, Surette MG, Lewenza S. Drosophila melanogaster as an animal model for the study of Pseudomonas aeruginosa biofilm infections in vivo. PLoS Pathogens. 2011;7:e1002299. doi: 10.1371/journal.ppat.1002299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro GH, Evans P, Todryk S, Buckett P, Kelly CG, Lehner T. A protein fragment of streptococcal cell surface antigen I/II which prevents adhesion of Streptococcus mutans. Infect Immun. 1993;61:4590–4598. doi: 10.1128/iai.61.11.4590-4598.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netuschil L, Vohrer K, Riethe P, Kasloff Z, Brecx M. Antibacterial effects of amalgams on mutans streptococci in an in vitro biofilm test procedure. Acta stomatologica Belgica. 1996;93:73–78. [PubMed] [Google Scholar]
- Nunes-Alves C. Biofilms: Targeting (p)ppGpp disrupts biofilms. Nat Rev Microbiol. 2014;12:461. [Google Scholar]
- Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 2011;12:594–601. doi: 10.1038/embor.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Séraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Rölla G. Why is sucrose so cariogenic? The role of glucosyltransferase and polysaccharides. Scand J Dent Res. 1989;97:115–119. doi: 10.1111/j.1600-0722.1989.tb01439.x. [DOI] [PubMed] [Google Scholar]
- Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F, See RY, Zhang D, Toh DC, Ji Q, Liang ZX. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J Biol Chem. 2010;285:473–482. doi: 10.1074/jbc.M109.040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofs KG, Wang J, Sintim HO, Lee VT. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc Natl Acad Sci U S A. 2011;108:15528–15533. doi: 10.1073/pnas.1018949108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohila JS, Chen M, Cerny R, Fromm ME. Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. The Plant Journal. 2004;38:172–181. doi: 10.1111/j.1365-313X.2004.02031.x. [DOI] [PubMed] [Google Scholar]
- Romling U. Great times for small molecules: c-di-AMP, a second messenger candidate in Bacteria and Archaea. Sci Signal. 2008;1:e39. doi: 10.1126/scisignal.133pe39. [DOI] [PubMed] [Google Scholar]
- Schlosser A, Hamann A, Bossemeyer D, Schneider E, Bakker EP. NAD+ binding to the Escherichia coli K+-uptake protein TrkA and sequence similarity between TrkA and domains of a family of dehydrogenases suggest a role for NAD+ in bacterial transport. Molecular Microbiology. 1993;9:533–543. doi: 10.1111/j.1365-2958.1993.tb01714.x. [DOI] [PubMed] [Google Scholar]
- Senadheera D, Cvitkovitch DG. Quorum sensing and biofilm formation by Streptococcus mutans. Adv Exp Med Biol. 2008;631:178–188. doi: 10.1007/978-0-387-78885-2_12. [DOI] [PubMed] [Google Scholar]
- Senadheera DB, Cordova M, Ayala EA, de Paz LC, Singh K, Downey JS, Svensäter G, Goodman SD, Cvitkovitch D. Regulation of bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans. Journal of Bacteriology. 2012;194:1307–1316. doi: 10.1128/JB.06071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senadheera MD, Guggenheim B, Spatafora GA, Huang Y-CC, Choi J, Hung DCI, Treglown JS, Goodman SD, Ellen RP, Cvitkovitch DG. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J Bacteriol. 2005;187:4064–4076. doi: 10.1128/JB.187.12.4064-4076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrnjug I, Rueckert C, Libanova R, Lienenklaus S, Weiss S, Guzmán CA. The Mucosal Adjuvant Cyclic di-AMP Exerts Immune Stimulatory Effects on Dendritic Cells and Macrophages. PloS one. 2014;9:e95728. doi: 10.1371/journal.pone.0095728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Spatafora G. Gene Regulation in S. mutans Complex Control in a Complex Environment. Journal of dental research. 2012;91:133–141. doi: 10.1177/0022034511415415. [DOI] [PubMed] [Google Scholar]
- Tan HY, Ng TW. Accurate step wedge calibration for densitometry of electrophoresis gels. Optics Communications. 2008;281:3013–3017. [Google Scholar]
- van Opijnen T, Camilli A. Genome-Wide Fitness and Genetic Interactions Determined by Tn-seq, a High-Throughput Massively Parallel Sequencing Method for Microorganisms. Current protocols in microbiology. 2010:1E. 3.1–1E. 3.16. doi: 10.1002/9780471729259.mc01e03s36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte G, Hartung S, Buttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zeng M, Fives-Taylor P. The glycan moieties and the N-terminal polypeptide backbone of a fimbria-associated adhesin, Fap1, play distinct roles in the biofilm development of Streptococcus parasanguinis. Infect Immun. 2007;75:2181–2188. doi: 10.1128/IAI.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Wu H. A molecular chaperone mediates a two-protein enzyme complex and glycosylation of serine-rich streptococcal adhesins. Journal of Biological Chemistry. 2011;286:34923–34931. doi: 10.1074/jbc.M111.239350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Klein MI, Falsetta ML, Lu B, Delahunty CM, Yates JR, Heydorn A, Koo H. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 2012;8:e1002623. doi: 10.1371/journal.ppat.1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Okinaga T, Qi F, Zhang Z, Merritt J. Cloning-independent and counterselectable markerless mutagenesis system in Streptococcus mutans. Appl Environ Microbiol. 2011;77:8025–8033. doi: 10.1128/AEM.06362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Hara H, Tsuchiya K, Sakai S, Fang R, Matsuura M, Nomura T, Sato F, Mitsuyama M, Kawamura I. Listeria monocytogenes strain-specific impairment of the TetR regulator underlies the drastic increase in cyclic di-AMP secretion and beta interferon-inducing ability. Infect Immun. 2012;80:2323–2332. doi: 10.1128/IAI.06162-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y, Bowen WH, Burne RA, Kuramitsu HK. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect Immun. 1993;61:3811–3817. doi: 10.1128/iai.61.9.3811-3817.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Bai Y, Zhang Y, Gabrielle VD, Jin L, Bai G. Deletion of the cyclic di-AMP phosphodiesterase gene (cnpB) in Mycobacterium tuberculosis leads to reduced virulence in a mouse model of infection. Mol Microbiol. 2014;93:65–79. doi: 10.1111/mmi.12641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M, Zhang J-J, Fang X, Lawlis GB, Troxell B, Zhou Y, Gomelsky M, Lou Y, Yang XF. DhhP, a Cyclic di-AMP Phosphodiesterase of Borrelia burgdorferi, Is Essential for Cell Growth and Virulence. Infect Immun. 2014;82:1840–1849. doi: 10.1128/IAI.00030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li W, He Z-G. DarR, a TetR-like transcriptional factor, is a cyclic di-AMP-responsive repressor in Mycobacterium smegmatis. J Biol Chem. 2013;288:3085–3096. doi: 10.1074/jbc.M112.428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li W, He ZG. DarR, a TetR-like Transcriptional Factor, Is a Cyclic Di-AMP-responsive Repressor in Mycobacterium smegmatis. Journal of Biological Chemistry. 2012;288:3085–3096. doi: 10.1074/jbc.M112.428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Fives-Taylor P, Wu H. The utility of affinity-tags for detection of a streptococcal protein from a variety of streptococcal species. J Microbiol Methods. 2008;72:249–256. doi: 10.1016/j.mimet.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Wu R, Zhang H, Wu H. Structural and biochemical analysis of a bacterial glycosyltransferase. Methods Mol Biol. 2013;1022:29–39. doi: 10.1007/978-1-62703-465-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.