Abstract
The endoplasmic reticulum is the key organelle which controls protein folding, lipid biogenesis, and calcium (Ca2+) homeostasis. Cd exposure in Saccharomyces cerevisiae activated the unfolded protein response and was confirmed by the increased Kar2p expression. Cd exposure in wild-type (WT) cells increased PC levels and the PC biosynthetic genes. Deletion of the two phospholipid methyltransferases CHO2 and OPI3 modulated PC, TAG levels and the lipid droplets with cadmium exposure. Interestingly, we noticed an increase in the calcium levels upon Cd exposure in the mutant cells. This study concluded that Cd interrupted calcium homeostasis-induced lipid dysregulation leading to ER stress.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-016-0714-4) contains supplementary material, which is available to authorized users.
Keywords: Cadmium, Phosphatidylcholine, Lipid droplets, Calcium and ER stress
Introduction
The endoplasmic reticulum (ER) plays a crucial role in protein synthesis, folding, glycosylation, secretion, and lipid synthesis (Görlach et al. 2006). ER stress, induced by dithiothreitol (DTT) and tunicamycin, led to lipid overload in Saccharomyces cerevisiae and is linked to calcium homeostasis (Gardarin et al. 2010). The toxicants or chemicals induce ER stress to alter the ER calcium, ER lipid composition, reactive oxygen species (ROS), and misfolded/unfolded proteins (Biogioli et al. 2008; Fu et al. 2011). Cadmium (Cd) is a toxic metal that induces ER stress, and also competes with the essential elements such as calcium, iron, zinc, and manganese, altering their intracellular ion homeostasis (Gardarin et al. 2010). Several studies indicated that exposure to Cd increased Ca2+ level, but the mechanism remains poorly understood (Beyersmann and Hechtenberg 1997). The Cd interacts with Ca2+ transport in intracellular stores, such as interference in hepatic Ca2+ sequestration in the microsomes (Zhang et al. 1990), or inhibition of sarcoplasmic reticulum Ca2+-TPase (SERCA) (Hechtenberg and Beyersmann 1991). An alternative theory of ER stress postulates a key role for the downregulation of SERCA2 as a result of ER luminal Ca2+ depletion (Kharroubi et al. 2004).
In mammals, SERCA a type II P-type ATPase maintains ER calcium homeostasis (Brini and Carafoli 2009). In yeast cells, ER calcium signaling mechanisms are regulated by the Cod1/Spf1 ATPase (Cronin et al. 2002) and Pmr1 Ca2+-ATPase (Antebi and Fink 1992). The Pmr1 Ca2+-ATPase functions together with the calcium sequestration in the ER/Golgi complex in addition to the transportation of calcium and manganese to the secretory pathway compartments (Sorin et al. 1997; Antebi and Fink 1992). Interestingly, the ER-associated proteins are involved in maintaining ER calcium homeostasis. The molecular chaperones such as calreticulin, GRP94 or BiP, and folding enzymes (protein disulfide isomerases [PDI]) contribute to Ca2+ buffering in the ER lumen (Prins and Michalak 2011).
The lipotoxicity due to an enhanced ratio of phosphatidylcholine and phosphatidylethanolamine (PC/PE) in the ER, impairs SERCA2 and stimulates ER stress (Fu et al. 2011) and indicates the link between lipid and calcium. The absence of SERCA in Saccharomyces cerevisiae led to the discovery of another P-type ATPase, Cod1/Spf1p, that controls the Hmg2p degradation through calcium in the ER (Cronin et al. 2002). Together, these studies indicate an intimate relationship between the calcium and lipid homeostasis in the ER. We recently studied the accumulation of triacylglycerol (TAG) and lipid droplets (LDs) in yeast Saccharomyces cerevisiae, and the association of Cd stress resulted in lipid accumulation and Zn alteration (Rajakumar et al. 2016). In our lab, we also reported that Cd-induced ER stress resulted in phospholipid accumulation (Muthukumar et al. 2011). Expression of phospholipid biosynthetic enzymes get altered in response to a variety of stress conditions (Carman and Han 2009; Iwanyshyn et al. 2004) which in turn modifies the composition of phospholipids and exert tolerance against organic solvents (Ghosh et al. 2008). PC is the most abundant phospholipid in cellular membranes (Zinser et al. 1991) and crucial for the formation of the phospholipid bilayer. PC is a precursor for diverse signaling molecules (Sheikhnejad and Srivastava 1986). In ER, the two PE methyltransferases PEMT (Cho2) and PLMT (Opi3) execute a three-step methylation reaction of PE. Cho2 catalyzes the first methylation step; however, Opi3 is capable of promoting all three steps, although the last two steps with higher proficiency than the first one (Greenberg et al. 1983; Kodaki and Yamashita 1989; Summers et al. 1988; Gaynor et al. 1991; Preitschopf et al. 1993). The yeast cells respond to Cho2p or Opi3p deficiency by broadly varying the transcript levels (Thibault et al. 2012). The PC homeostasis is interconnected with neutral lipid metabolism. In most of the species like yeast and plants, PC is a major acyl donor for triacylglycerol (TAG) synthesis via the activation of lecithin cholesterol acyltransferase (LRO1) (Dahlqvist et al. 2000). The prominence of PC to health and disease was already proven by earlier studies (Farber et al. 2000; Grothe et al. 2015; Lee et al. 2014). However, the metabolism under cellular stress remains unclear. In this study, the loss of either Cho2 or Opi3 during cadmium exposure affected the calcium homeostasis in the ER and mitochondria.
Materials and methods
Chemical and reagents
Yeast extract, peptone, and bacteriological agar were purchased from Difco. Thin-layer silica gels 60 plates were purchased from Merck. BODIPY 493/503 was purchased from Invitrogen. Trizol, PMSF, and all other chemicals were purchased from Sigma unless specifically mentioned. The cDNA synthesizing kit was obtained from Bio-Rad. All solvents were purchased from Merck, and lipid standards were obtained from Avanti Polar Lipids (Alabaster, AL).
Strains and growth conditions
Saccharomyces cerevisiae strains used in the study were BY4741 [MATa his3∆1 leu2∆0 met15∆0 ura3∆0], cho2∆, and opi3∆. These strains were the kind gift from Prof. Ram Rajasekharan, CSIR-CFTRI, Mysore, INDIA. Strains were grown aerobically up to early stationary phase at 30 °C in YPD medium (1 % yeast extract, 2 % bacto-peptone, and 2 % dextrose). The yeast cells were precultured in 5 ml of YPD medium for 12 h, and then the cells were harvested and transferred to 25 ml of YPD media with or without 50-μM Cd so that the final absorption (A600) of 0.1 was achieved. Following these, cells were agitated at 180 rpm at 30 °C for 12 h.
Cell viability and growth
To study the sensitivity against cadmium, wild-type and mutant strains were diluted (4-fold) and spotted on YPD plates containing 2 % agar supplemented with 0 to 100 μM of Cd and incubated for 2 days at 30 °C. Sensitivity was found to be 50 μM and so for the further experimental purposes, the cells were grown in a liquid medium with/without 50-μM Cd.
Extraction and separation of lipids
Yeast lipids were extracted using the method described by Bligh and Dyer (1959). Briefly, to the cell pellet, 2:1 ratio of chloroform and methanol were added, followed by vortexing. To this, an equal volume of acidified water (2 % phosphoric acid) was added and vigorously vortexed. The phospholipids were separated by TLC; solvents for the first dimension were chloroform/methanol/ammonia (65:35:5, v/v); solvents for the second dimension were chloroform/methanol/acetone/acetic acid/water (50:10:20:15:5, v/v). The neutral lipids were separated by using petroleum ether: diethyl ether: acetic acid (70:30:1, v/v) as the solvent system. Individual lipids were located by comparing the Rf values of the unknown with the Rf values of the standard. For the neutral lipids, the spots were visualized by post-chromatographic staining after dipping TLC plates into a solution containing 0.8-g MnCl2 × 4H2O, 120-ml water, 120-ml methanol, and 9-ml concentrated sulphuric acid and charring at 105 °C for 30 min and quantified by densitometry scanner.
Phosphorous assay of phospholipids
Spots of phospholipid samples were scraped from TLC plate and transferred to glass tubes, and 0.65-ml perchloric acid was added and the tubes placed in a heating block until the yellow color disappears. After cooling, 3.3 ml of water, 0.5 ml of 2.5 % molybdate solution, and 0.5 ml of 10 % ascorbic acid solution were added to the tubes and mixed vigorously. The tubes were then kept in a boiling water bath for 5 min and cooled to RT. The samples (including the standards) were read at A800 nm. Digestion was not needed for the standards (1 to 5 μg P/tube). The standards were diluted with water (3.3 ml) and 0.65-ml perchloric acid. The tubes containing the silica gel were centrifuged before take reading as it will interfere with the absorbance of phosphate ( Siakotos et al. 1966).
Construction of protein-protein interaction network
The protein-protein interaction network of lipid proliferative pathways was constructed and analyzed using network biology approach. The protein interactions of Opi3 and Spf1 proteins were searched and retrieved from STRING database (Szklarczyk et al. 2015). The STRING database contains known and predicted protein-protein interactions derived from high throughput experiments, co-expression, co-occurrence, and gene fusion methods. The protein-protein interactions experimentally confirmed were retrieved by giving Opi3 and Spf1 proteins as seed proteins. The interactions only above the confidence score cut-off of 0.60 were chosen.
RNA isolation and real-time PCR
The wild and mutant cells were grown in YPD medium with and without Cd at 30 °C until mid-log phase. RNA was isolated using the RNA assay kit from Qiagen as described by the manufacturer instructions. After DNaseI digestion, complementary DNA (cDNA) was constructed using the high-capacity cDNA reverse transcription kit with 1× RT buffer, 1× random primer, 4-mM dNTP mix, 50-U/ml reverse transcriptase, and 1-μg total RNA. The primers were designed using the Primer ExpressR Software 3.0 (Applied Biosystems), and the primer sequences are listed in Table S1. Relative mRNA levels were determined by quantitative real-time RT-PCR using SYBR Green real-time PCR master mix (Applied Biosystems) by following the manufacturer’s instructions. PCR was initiated with denaturation at 94 °C for 5 min, followed by 40 cycles at 95 °C for 15 s, 60 °C for 60 s, and 95 °C for 15 s, and a final extension at 72 °C for 5 min. Amplification was measured using an AB 7500 instrument (Applied Biosystems). Samples were evaluated using the ΔΔCt method described (Livak and Schmittgen 2001). The mRNA expressions were analyzed in triplicate, and the results were analyzed using relative quantification. ACT1 was used as endogenous control for normalization.
Microscopical analysis
Yeast cells were grown with or without 50-μM Cd in YPD liquid culture. For 3, 3′-dihexyloxacarbocyanine iodide (DiOC6) (Sigma) staining, cells were harvested and washed with a TE buffer. After washing, the pellet was resuspended in the TE buffer. DiOC6 was resuspended in 1-mg/ml ethanol (as DiOC6 stock solution). One microliter of DiOC6 stock solution was added to 1 ml of the yeast cell suspension. If the cell density was A600 > 1.0 at harvesting, the cell suspension was diluted with the TE buffer to an A600 of ∼1.0. Cells stained with DiOC6 were analyzed immediately ( Block-Alper et al. 2002). For Nile Red staining, yeast cells in stationary phase were washed and resuspended in phosphate-buffered saline (PBS). The cells were stained with Nile Red solution (0.0005 % in PBS, diluted from a 0.01 % stock solution) for 15 min at room temperature in the dark. The cells were then washed six times with PBS to remove the surplus dye. Cells were resuspended in 50 μl of PBS and observed with a confocal microscope directly after staining (Kimura et al. 2004). Mitochondrial damage was studied in opi3∆ and cho2∆ cells with Aconitase GFP plasmid, gifted by Prof. Markus Ralser, University of Cambridge, UK. Transformants were diluted from an overnight culture to an A600 = 0.1 in SC medium lacking uracil and were grown till mid-exponential phase. The final experiment was done by YPD media.
Image acquisition
Samples were fixed with 0.5 % (wt/vol) agarose on microscope slides. Fluorescence microscopic images were recorded on an LSM 710 Confocal microscope (Zeiss) equipped with a 100/1.40 oil objective and an AxioCam MRM camera (Zeiss) at room temperature.
Western blotting
Yeast cell lysates were prepared by breaking the cells with striled glass beads in a lysis buffer (50-mM Tris–HCl pH 7.4, 150-mM NaCl, 5-mM EDTA, and 1-mM PMSF) and the protease inhibitor cocktail. Total protein (50 μg) was subjected to SDS-PAGE. To visualize SDS-PAGE, gels were stained using a Coomassie brilliant blue staining. For immunoblot analyses, proteins were transferred to nitrocellulose membrane at 120 V for 1 h. Blots were decorated using anti-kar2p and anti-pgk1p antibodies. The resulting membrane was incubated with a 1:5000 dilution of primary antibody for 2 h, followed by a 1:2500 dilution of ALP-conjugated goat anti-rabbit IgG for 1 h. The membrane was washed again and detected with BCIP/NBT substrate system. Band intensity was quantified using the ImageJ software.
Determination of intracellular metals
Intracellular calcium and cadmium levels were determined in the wild-type and mutant strains by inductively coupled plasma atomic emission spectrometer (ICP-OES) on a Perkin Elmer 3300 (SAIF-IIT, Chennai). The cells were grown on YPD medium, and A600 = 1 of cells were collected by centrifugation, washed twice with 10-ml ice cold 1X TE buffer and then resuspended in nitric acid and heated at 200 °C for 4 h. After complete digestion, 1 ml was diluted to 10 ml in metal-free water and subjected to intracellular metal analysis (Doner and Edge 2004).
Statistics
The data analysis was done using the tools of PRISM 5.6. Each experiment was done thrice, and the data were presented as mean ± SD. Statistical analysis was carried out by analysis of variance (Oneway-ANOVA) test. The statistical probability of *p < 0.05, **p < 0.01, and ***p < 0.001 was considered to be significant.
Results
Transcriptional alteration of PC biosynthesis during cadmium exposure
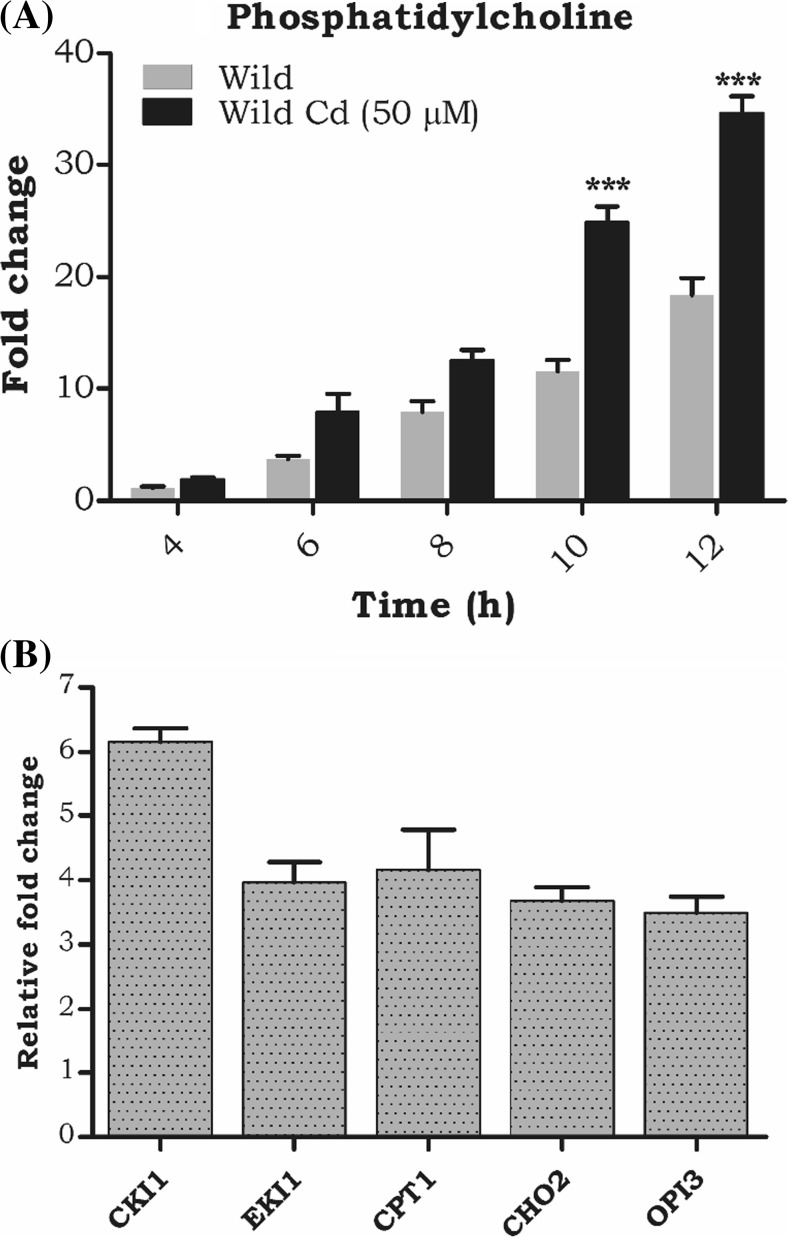
Earlier studies in our laboratory reported that cadmium exposure significantly increased the phospholipids in wild-type cells. We also analyzed the PC profiling at different time intervals (4, 6, 8, 10, and 12 h) during cadmium exposure. At 10 and 12 h, PC was increased up to 15- and 25-fold (Fig. 1A) compared to wild-type control cells. Moreover, to explore, how cadmium affects the lipid transcriptional regulation in S. cerevisiae, the quantitative PCR analysis of the PC biosynthetic genes CKI1, EK11, CPT1, CHO2, and OPI3 genes were quantified, and the expression of all the genes was significantly upregulated (Fig. 1B). Further, we studied the impact of Cd on both the Kennedy and the de novo (methylation) CDP-DAG pathway mutants. Our results show Kennedy pathway mutants (cki1∆, cpt1∆, and pct1∆) behaved similarly to the wild-type cells, whereas the de novo pathway mutants (cho2∆ and opi3∆) were more susceptible to Cd stress (Fig. S1) and so we chose the de novo pathway mutants (cho2∆ and opi3∆) for further studies.
Fig. 1.
Cd toxicity alters phosphatidylcholine (PC) synthesis. a A time-dependent study into PC accumulation during the growth phase (4–12 h), with or without 50-μM Cd. Equal amounts (A600 = 20OD) of cells were harvested at different time intervals. The lipids were extracted and separated on a silica TLC using solvent system b quantitative (qPCR) analysis of PC biosynthetic genes (CKI1, EKI1, CPT1, CHO2, and OPI3). Total RNA was isolated and converted into cDNA using 2 μg of total RNA. A1:20 dilutions of cDNA was used for the quantitative expression analysis. Actin was used as an endogenous control. Samples were analyzed in triplicate, and the expression of these genes in wild-type control cells was set as one (data not shown in the graph) and was compared with the mutants and Cd-treated cells
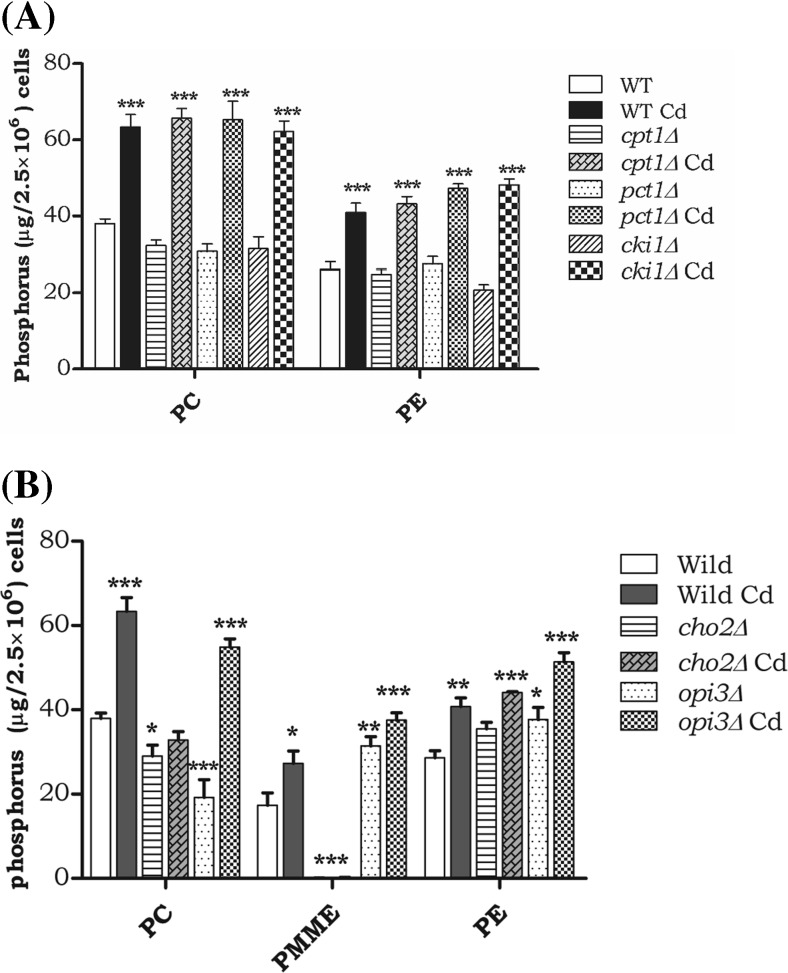
Exposure tocadmium modulates PC levels in Kennedy and methylation pathway
Wild-type cells during 12 h of Cd exposure depicted a 50 % increase in PC, and the Kennedy pathway mutants (cki1∆, cpt1∆, and pct1∆) also showed a palpable elevation in the accumulation of PC-like WT Cd-treated cells, as well as a similar trend, was observed in PE levels (Fig. 2A). The cho2∆ did not show PC alteration in the presence of Cd. But in opi3∆, the PC was reduced and was quite obvious. Upon Cd exposure, the PC was significantly increased in opi3∆. The intermediates of PC synthesis, phosphatidyl monomethyl ethanolamine (PMME) also accrued in opi3∆. Accumulation of PE was observed in both the cho2 and opi3 mutant strains and was further elevated during Cd exposure (Fig. 2B). Finally, PC alteration was apparent in methylation pathway mutants cho2∆ and opi3∆; this might have an impact in the unfolded protein response mechanism.
Fig. 2.
Defect of PC synthesis modulates the PC production upon Cd stress. The wild-type, cho2∆, and opi3∆ cells were grown up to mid-log phase on YPD medium; with or without 50-μM cadmium. An equal number of cells were taken, and lipids were extracted and resolved by two-dimensional silica TLC. The pattern of major membrane phospholipids PC and PE, and also quantified intermediate form of PC, like PMME. a The level of PC and PE in Kennedy pathway mutants (cki1∆, cpt1∆, and pct1∆) and b methylation pathway mutants ( cho2∆ and opi3) phospholipid was quantified and presented as phosphorus (μg/2.5 × 106 cells). WT control cells were compared with other groups with a statistical significance of *p < 0.05, **p < 0.01, and *** p < 0.001
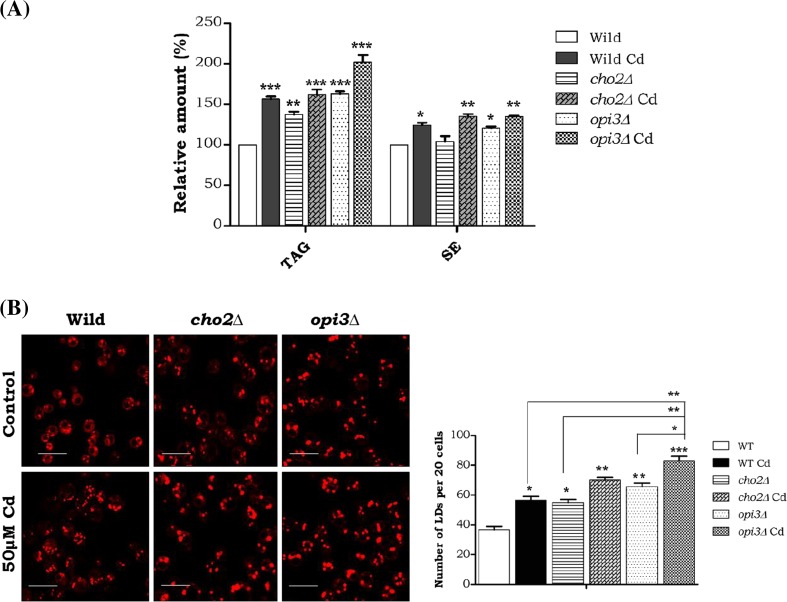
Defects of cho2 and opi3 cause TAG and LD accumulation
Both the cho2 and opi3 mutant strains showed a marked increase in TAG level (100 %), and Cd exposure elevated it further. The SE level was increased with Cd exposure in the wild-type and also the mutants (Fig. 3A). Consequently, we studied the LD formations and our result demonstrated increased number and size of LDs with cadmium exposure in all the strains, the especially Opi3 mutant showed significantly increase in LDs both in its numbers and size. (Fig. 3B) These results demonstrate that the methylation pathway PC synthesis are essential for both membrane and storage lipid homeostasis during Cd stress.
Fig. 3.
Impairment of PC synthesis leads storage lipid accumulation under Cd stress. The wild-type, cho2∆, and opi3 cells were grown up to the mid-log phase on YPD medium with or without 50-mM cadmium. An equal amount of cells were taken for lipid extraction. Lipids were resolved on silica TLC and quantified. a TAG triacylglycerol, SE sterol esters. Samples were analyzed in triplicate and compared to the control. WT control cells were compared with other groups with a statistical significance of *p < 0.05, **p < 0.01, and *** p < 0.001. b The cells were stained with Nile red (1 μg/ml) and viewed under the fluorescent microscope. The bar in the figures indicates the scale of 5.0 μm
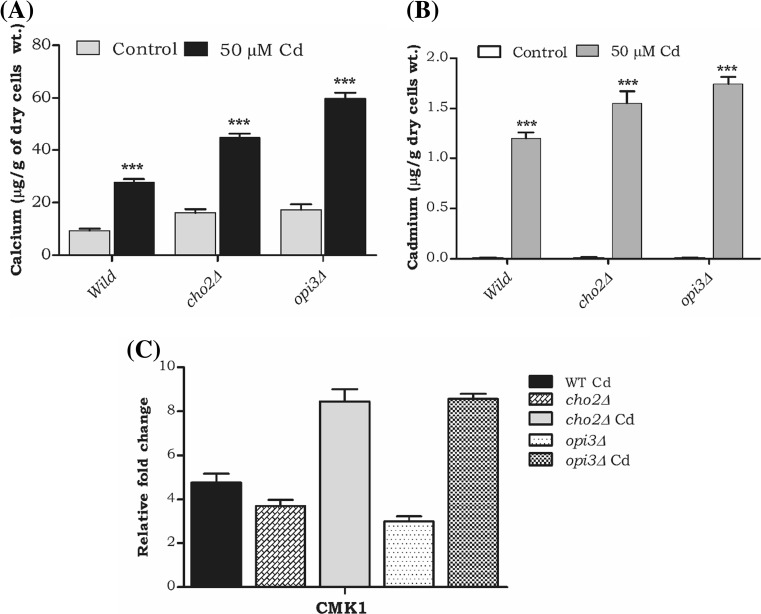
Cadmium alters intracellular calcium in the PC deletion mutants
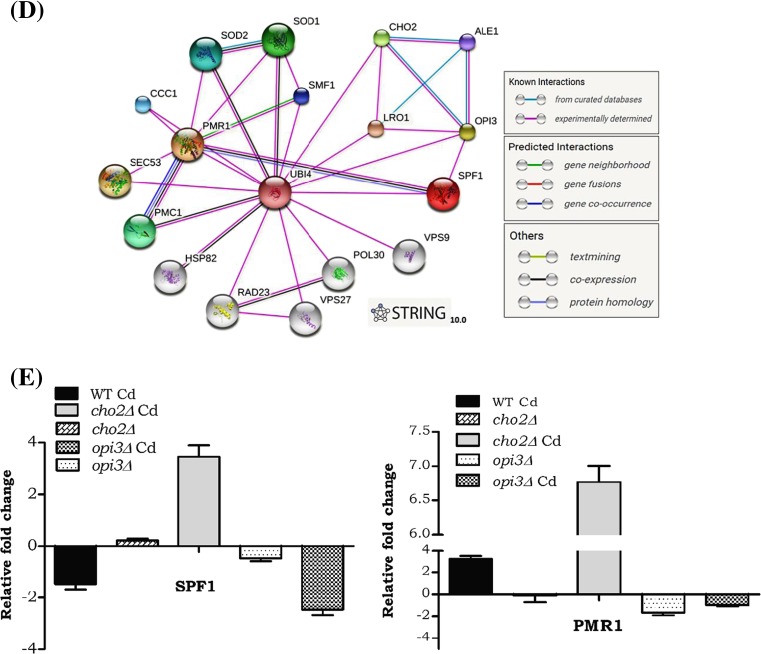
Earlier studies in our lab reported alteration of Zn during cadmium exposure and in the current study, we focused on the cadmium interference with the biologically essential mineral such as calcium. Hence, during Cd exposure, we quantified the intracellular calcium levels in PC mutants by ICP-OES. We noticed the remarkable elevation of calcium (2-, 1.5-, and 2.5-fold) in wild-type, cho2Δ and opi3Δ Cd-treated cells (Fig. 4A). Subsequently, we analyzed transcriptional alteration of calmodulin kinase 1 (CMK1). Our results showed significant upregulation of the CMK1 in cho2Δ and opi3Δ during Cd exposure (Fig. 4C). In SGD (Saccharomyces Genome database and BIOGRIDs), the gene interaction map displays an interaction between ER calcium ATPase SPF1 and OPI3 and both are localized in ER (Fig. 4D). Also, some experimental evidence suggests the negative genetic interaction between opi3 and spf1 with a high confident score (0.66) (Table 1). We also studied the gene expression of SPF1 and PMR1 in cho2Δ and opi3Δ strains during Cd exposure. Interestingly, the SPF1 and PMR1 were downregulated in opi3Δ cells and upregulated in cho2Δ cells during Cd exposure (Fig. 4E). These results suggest that in S. cerevisiae during Cd exposure, the PC levels and calcium levels were altered and so had an impact on their Ca2+-ATPasein ER.
Fig. 4.
A defect of methylation pathway PC leads to Ca2+ imbalance. a Analysis of intracellular calcium by ICP-MS. Wild and mutant strains were grown on nutrient rich (YPD) medium with or without Cd, after harvested and digested with Conc HNO3. Error bars indicate SD from two independent experiments (**p value <0.001 and*p value <0.01). b The level of intracellular cadmium and c quantitative (qPCR) analysis of calcium-calmodulin kinase1(CMK1). d Interaction mapping of phospholipid methyl transferase gene (OPI3) and ER calcium ATPase (SPF1) by using STRING 10.0. e Quantitative (qPCR) analysis of ER calcium ATPase SPF1 and PMR1. Samples were analyzed in triplicate, and the expression of these genes in wild-type control cells was set as one (data not shown in the graph) and was compared with the mutants and Cd-treated cells
Table 1.
Protein-protein interactions observed between Ca2+ homeostasis (Spf1), lipid (Cho2, Lro1, Ale1), and ubiquitin metabolic processes (Ubi4). Protein interaction summary of Opi3
| Node1 | Node2 | Node1 accession | Node2 accession | Node1 annotation | Node2 annotation | Score |
|---|---|---|---|---|---|---|
| OPI3 | ALE1 | YJR073C | YOR175C | Phospholipid methyltransferase | Broad specificity lysophospholipid acyltransferase | 0.978 |
| OPI3 | CHO2 | YJR073C | YGR157W | Phospholipid methyltransferase | Phosphatidylethanolamine methyltransferase (PEMT) | 0.999 |
| OPI3 | LRO1 | YJR073C | YNR008W | Phospholipid methyltransferase | Acyltransferase that catalyzes diacylglycerol esterification | 0.958 |
| OPI3 | SPF1 | YJR073C | YEL031W | Phospholipid methyltransferase | P-type ATPase, ion transporter of the ER membrane; ER function and Ca2+ homeostasis | 0.665 |
| OPI3 | UBI4 | YJR073C | YLL039C | Phospholipid methyltransferase | Ubiquitin, essential for the cellular stress response | 0.79 |
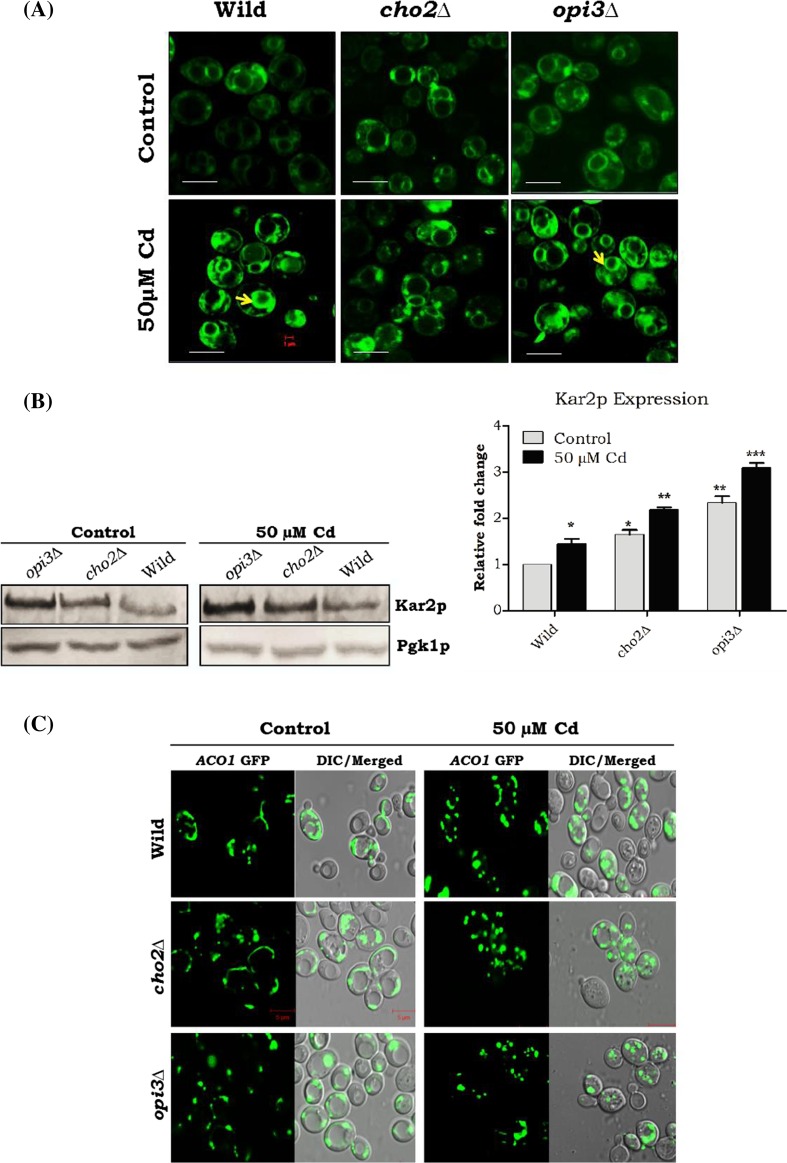
Cadmium toxicity induced membrane aberrancy, ER stress, and mitochondrial damage in PC mutants
The abundance of intracellular calcium induced ER stress and may have an impact on membrane proliferation in Saccharomyces cerevisiae. DiOC6 the membrane specific dye quantifies the intercellular membrane content of yeast cells, and DiOC6 was used to analyze the membrane aberrancy in Cd-treated cells. Our data showed bright rings arising from the ER and nuclear membranes in the wild-type and opi3∆ strains upon Cd exposure. Only faint images were observed in the cadmium-unexposed cells and cho2∆ (−/+ cadmium) strains (Fig. 5A). We also tested ER stress by measuring Kar2p expression (ER chaperone overexpressed during UPR activation). The cho2∆ and opi3∆ cells were increased Kar2p expression that was further enhanced by Cd exposure (Fig. 5B). Furthermore, calcium alteration in ER will have an impact on the mitochondria, and so we analyzed the mitochondrial damage in the cho2∆ and opi3∆ genes during Cd stress by using ACO1-pGFP (aconitase) that worked as stress sensor for the protection of mtDNA. Our micrographs showed tubular mitochondria in wild-type and cho2∆ control strains, but opi3∆ strains showed fragmented mitochondria. On the other hand, Cd-treated cells displayed total mitochondrial fragmentation (Fig. 5C).
Fig. 5.
Defects in PC methylation in inducing membrane proliferation, ER stress, and mitochondrial damage. Wild-type and mutant cells were grown up to mid-log phase on YPD medium with or without Cd at 30 °C. a Membrane proliferation was assessed by the incorporation of the lipophilic dye DiOC6 and viewed under the fluorescence microscope. The bar in the figures indicates the scale of 1.0 μm. b Cells were lysed, and 50 μg of protein was loaded and separated by 10 % SDS-PAGE followed by immunoblotting with kar2 antiserum. Band intensity was quantified by the ImageJ software and expressed as a ratio with pgk1. c The ACO1-pGFP was analyzed for mitochondria morphology. Strains wild-type and cho2∆ contain typical tubular mitochondria. Opi3∆ caused mitochondrial fragmentation gradually increased; Aco1p showed 100 % mitochondrial network fragmentation in all Cd-treated mutant yeast cells
Discussion
The alteration of intracellular calcium and UPR activation are reported in several models of metabolic disease and lipid disequilibrium (Fu et al. 2011; Ozcan et al. 2004). ER calcium-dependent proteins have an important role in the maintenance of cellular integrity. Disruption of the calcium homeostasis in the ER is a potent trigger of ER stress and apoptosis (Groenendyk et al. 2010 and Schroder and Kaufman 2005). Recent studies revealed that lipid remodeling majorly contributes to adaptive stress response and acclimatization of ER stress but the mechanistic link between lipid aberrancy and Ca2+ alteration inducing ER stress is not yet elucidated.
In yeast, several lipid metabolic defects activate the UPR, suggesting a compensatory role when these pathways are disrupted (Jonikas et al. 2009 and Thibault et al. 2012). Our earlier reports have demonstrated that Cd exposure is induced ER stress (Muthukumar et al. 2011). In our current study, the phospholipids mainly PC and PE were increased in wild-type cells upon Cd exposure (Fig. 1A). The Kennedy (CKI1, EKI1, and CPT1) and the methylation pathway (CHO2 and OPI3) genes were upregulated during Cd exposure in the wild-type cells (Fig. 1B). It is also backed up by the DMSO treatment, facilitating the anomaly in gene expression of the phospholipid synthesis pathway, the relative quantities of phospholipids in yeast (Murata et al. 2003). These results are consistent and support our previous findings (Muthukumar et al. 2011 and Rajakumar et al. 2016). The de novo PC synthesis was affected in cho2Δ and opi3Δ because the cellular membranes have a high PC turnover due to a high rate of synthesis and breakdown (Jackowski 1996). The cho2Δ cell growth was greatly inhibited than opi3Δ and wild-type cells (Fig. S1) upon cadmium exposure. In contrary, the Kennedy pathway mutants (cki1∆, pct1∆, and cpt1∆) showed more viability and also the significant increase in PC compared to the de novo pathway mutants (Fig. S1 and Fig. 2A). These might contribute to the upregulation of phospholipid methyl transferases (CHO2 and OPI3) during chemical stress (Murata et al. 2003). Intriguingly, cho2Δ reduced PC when compared to opi3Δ cells during Cd exposure (Figs. 3A and 5B) and also reduced Kar2p expression in cho2Δ than that in opi3Δ cells. It is in close grip with earlier studies; the reduction of PC suggests that Opi3p might partly compensate for cho2Δ (Kanipes and Henry 1997; Preitschopf et al. 1993). The opi3Δ showed more PC content during Cd exposure.
Our work extended to an integration of PC metabolic pathway and calcium homeostasis. The elevation of intracellular calcium leads to alteration of phospholipids, especially PC (Moore et al. 1984), and also the silencing of PEMT, accelerated the SERCA activity and reduced the ER stress (Jacobs et al. 2010). In yeast, the Biological General Repository for Interaction Datasets (BioGRIDs) showed a negative genetic interaction of OPI3 with SPF1 and PMR1 by high throughput screenings (Fig. 4D) (Schuldiner et al. 2005; Hoppins et al. 2011; Jonikas et al. 2009 and Surma et al. 2013). A problem that has yet to be resolved is why PC is regulated in response to ER calcium governance. We noticed an elevation of calcium in cho2Δ than opi3Δ cells during Cd exposure (Fig. 4A). Also, the calcium calmodulin kinase-encoding gene CMK1 expression was highly elevated, whereas SPF1 expression was reduced in opi3Δ when compared to WT Cd-treated cells (Fig. 4C and E). This result might suggest that cytosolic Ca2+ was increased, and ER calcium reduced. Inhibition of CaMKII (calcium calmodulin kinase) reduced the ER stress, mitochondrial integrity, and membrane potential in murine cells (Bracken et al. 2016). Previous studies also revealed that the ratio of phosphatidylcholine and phosphatidylethanolamine in membrane resulted in ER stress and Ca2+ imbalance in mice hepatocytes (Fu et al. 2011). Moreover, we observed higher expression of Kar2p level (ER chaperone) in opi3Δ control and Cd-treated strains and might be due to IRE1-mediated ER stress activation (Fig. 5B). These findings also support that activation of IRE1 in Opi3 deletion is likely to cause membrane or lipid-related aberrations (Promlek et al. 2011).
The lipophilic green fluorescent dye DiOC6 was used to identify the intercellular membrane abnormalities or membrane proliferation in yeast cells (Koning et al. 1996; Murata et al. 2003). Our microscopy results showed the aberrant membrane structures in Cd-treated WT and opi3 mutant strains (Fig. 5A). This result also proves the ER stress triggers the UPR and is not constantly accompanied by accumulation of unfolded proteins in the ER, but is also activated by the aberrant lipid or membrane content (Promlek et al. 2011). Together, these results demonstrate the relationship between the UPR and lipid homeostasis. Also, we studied the mitochondrial dysfunctions during impairment of PC with Cd stress. ER and mitochondria interact both physiologically and functionally, and the association is referred as mitochondrial-associated membranes (Vance 1990). The main facets of this interaction are essential for calcium signaling, lipid transport, energy metabolism, and cell survival (Malhotra and Kaufman 2011: Stone and Vance 2000). In humans, congenital muscular dystrophy patients showed defects in the PC synthesis and associated mitochondrial abnormality, directing a role of PC in mitochondrial function (Mitsuhashi et al. 2011). Therefore, we studied mitochondrial morphology by using ACO1-pGFP in wild and mutant cells. The yeast mitochondrial Aco1p is a TCA cycle enzyme that converts citrate into isocitrate, and it is essential for mtDNA stability (Shadel 2005). Our microscopical data depicted tubular mitochondria and were observed in WT and cho2∆ control strains, but fragmented mitochondria was observed in opi3∆ and Cd-treated WT strains (Fig. 5C). Altogether, impairment of methylated PC synthesis specifically affects the function and stability of TOM and SAM complexes of mitochondria in yeast (Schuler et al. 2015). This data supports that PEMT pathway is also essential for mitochondrial function during ER stress and impairment of calcium. The defect of Pmr1p and Spf1p, implicated in protein glycosylation, influenced the conditions of ER stress stimulates LD formation in Saccharomyces cerevisiae (Fei et al. 2009; Cohen et al. 2013). Besides, the role of LD biogenesis and synthesis of TAG is a sign of cellular stress, and it was proposed that alterations in PC homeostasis might be the source of TAG (Al-Saffar et al. 2002 and Iorio et al. 2003). Our data showed increased number of LDs and accumulation TAG in cho2Δ and opi3Δ cells during Cd exposure (Fig. 3). Also, we reported that exposure to cadmium in yeast accumulates TAG and LDs (Rajakumar et al. 2016).
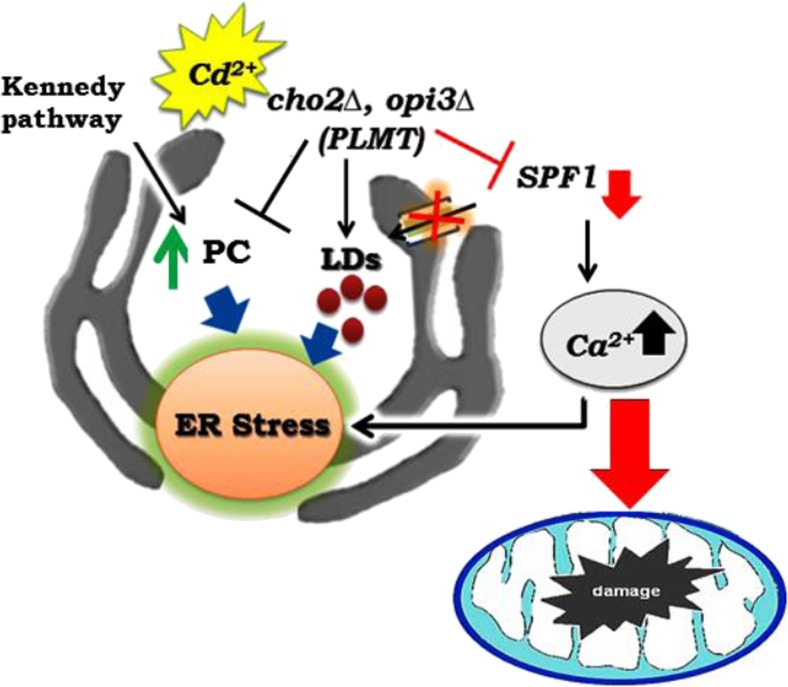
Finally, we can conclude that phospholipid has an important role in calcium homeostasis and ER stress during the Cd exposure (Fig. 6). The phospholipid methyltransferase encoding gene OPI3 and phosphatidylcholine are essential for Cd tolerance and are required for intracellular membrane maintenance and mitochondrial morphology.
Fig. 6.
A diagrammatic representation of phospholipid methylation and cadmium exposure alters calcium levels, induces ER stress mitochondrial dysfunction, and disrupts lipid homeostasis in Saccharomyces cerevisiae. (Cd cadmium, Ca calcium, PC Phosphatidylcholine, CHO2-phosphatidylethanolamine methyltransferase, OPI3-phospholipid methyltransferase, SPF1-ER Ca2+ ATPase, PMR1-ER & Golgi Ca2+ ATPase, LDs lipid droplets)
Electronic supplementary material
The Kennedy (cki1∆,cpt1∆, and pct1∆) and the methylation pathway (cho2∆, opi3∆) mutant cells were grown up to mid-log phase on YPD medium, harvested and washed twice in sterile water. Fourfold serial dilutions were spotted on to YPD agar plate with 50 μM Cd and incubated at 30 °C for 2 days (DOCX 530 kb)
(DOCX 28 kb)
Acknowledgments
Infrastructure facility by DST-FIST, Biochemistry Department and Confocal microscopy facility by DST-PURSE at Bharathidasan University is gratefully acknowledged. SR is supported by a fellowship from the Rajiv Gandhi National Fellowship Scheme from UGC, INDIA. We thank Prof. Ram Rajasekharan, CSIR-CFTRI, Mysore, INDIA, for providing mutant strains for this work. We also acknowledge Sophisticated Analytical Instruments Facility (SAIF), IIT-M, Chennai, India for helping us to conduct the ICP-OES study.
References
- Al-Saffar NM, Titley JC, Robertson D, Clarke PA, Jackson LE, Leach MO, Ronen SM. Apoptosis is associated with triacylglycerol accumulationin Jurkat T-cells. Br J Cancer. 2002;86:963–970. doi: 10.1038/sj.bjc.6600188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi A, Fink GR. The yeast Ca(2+)-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol Biol Cell. 1992;3(6):633–654. doi: 10.1091/mbc.3.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyersmann D, Hechtenberg S. Cadmium, gene regulation, and cellular signalling in mammalian cells. Toxicol Appl Pharmacol. 1997;144(2):247–261. doi: 10.1006/taap.1997.8125. [DOI] [PubMed] [Google Scholar]
- Biogioli M, Pifferi S, Ragghianti M, Rizzuto R, Pinton P (2008) Endoplasmic reticulum stress and alteration in calcium homeostasis are involved in cadmium-induced apoptosis. Cell Calcium 43:184–195 [DOI] [PubMed]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Block-Alper L, Webster P, Zhou X, Supeková L, Wong WH, Schultz PG, Meyer DI. IN02, a positive regulator of lipid biosynthesis, is essential for the formation of inducible membranes in yeast. Mol Biol Cell. 2002;1:40–51. doi: 10.1091/mbc.01-07-0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken C, Beauverger P, Duclos O, Russo RJ, Rogers KA, Husson H, Natoli TA, Ledbetter SR, Janiak P, Ibraghimov-Beskrovnaya O, Bukanov NO. CaMKII as a pathological mediator of ER stress, oxidative stress and mitochondrial dysfunction in a murine model of Nephronophthisis. Am J Phys Renal Phys. 2016 doi: 10.1152/ajprenal.00426.2015. [DOI] [PubMed] [Google Scholar]
- Brini M, Carafoli E. Calcium pumps in health and disease. Physiol Rev. 2009;89:1341–1378. doi: 10.1152/physrev.00032.2008. [DOI] [PubMed] [Google Scholar]
- Carman GM, Han GS. Regulation of phospholipid synthesis in yeast. J Lipid Res. 2009;50(Suppl):S69–S73. doi: 10.1194/jlr.R800043-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin SR, Rao R, Hampton RY. Cod1p/Spf1p is a P-type ATPase involved in ER function and Ca2+ homeostasis. J Cell Biol. 2002;157:1017–1028. doi: 10.1083/jcb.200203052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y, Megyeri M, Chen OC, Condomitti G, Riezman I, Loizides-Mangold U, Abdul-Sada A, Rimon N, Riezman H, Platt FM, Futerman AH, Schuldiner M. The yeast p5 type ATPase, spf1, regulates manganese transport into the endoplasmic reticulum. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0085519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlqvist A, Stahl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S. Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci U S A. 2000;97(12):6487–6492. doi: 10.1073/pnas.120067297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doner G, Edge A. Evaluation of digestion procedures of the determination of iron and zinc in biscuits by flame atomic absorption spectrometry. Anal Chim Acta. 2004;520:217–222. doi: 10.1016/j.aca.2004.05.069. [DOI] [Google Scholar]
- Fu S, Yang L, Li P, Hofmann O, Dicker L, Hide W, Lin X, Watkins SM, Ivanov AR, Hotamisligil GS. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473:528–531. doi: 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardarin A, Chédin S, Lagniel G, Aude JC, Godat E, Catty P, Labarre J. Endoplasmic reticulum is a major target of cadmium toxicity in yeast. Mol Microbiol. 2010;76(4):1034–1048. doi: 10.1111/j.1365-2958.2010.07166.x. [DOI] [PubMed] [Google Scholar]
- Gaynor PM, Gill T, Toutenhoofd S, Summers EF, McGraw P, Homann MJ, Henry SA, Carman GM. Regulation of phosphatidylethanolamine methyltransferase and phospholipid methyltransferase by phospholipid precursors in Saccharomyces cerevisiae. Biochim Biophys Acta. 1991;1090(3):326–332. doi: 10.1016/0167-4781(91)90197-T. [DOI] [PubMed] [Google Scholar]
- Grothe J, Riethmüller J, Tschürtz SM, Raith M, Pynn CJ, Stoll D, Bernhard W. Plasma phosphatidylcholine alterations in cystic fibrosis patients: impaired metabolism and correlation with lung function and inflammation. Cell Physiol Biochem. 2015;35(4):1437–1453. doi: 10.1159/000373964. [DOI] [PubMed] [Google Scholar]
- Ghosh AK, Ramakrishnan G, Rajasekharan R. YLR099C (ICT1) encodes a soluble acyl-CoA-dependent lysophosphatidic acid acyltransferase responsible for enhanced phospholipid synthesis on organic solvent stress in Saccharomyces cerevisiae. J Biol Chem. 2008;283(15):9768–9775. doi: 10.1074/jbc.M708418200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlach A, Klappa P, Kietzmann T. The endoplasmic reticulum: folding, calcium homeostasis, signaling, and redox control. Antioxid Redox Signal. 2006;8(9–10):1391–1418. doi: 10.1089/ars.2006.8.1391. [DOI] [PubMed] [Google Scholar]
- Greenberg ML, Klig LS, Letts VA, Loewy BS, Henry SA. Yeast mutant defective in phosphatidylcholine synthesis. J Bacteriol. 1983;153(2):791–799. doi: 10.1128/jb.153.2.791-799.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenendyk J, Sreenivasaiah PK, Kim do H, Agellon LB, Michalak M. Biology of endoplasmic reticulum stress in the heart. Circ Res. 2010;107(10):1185–1197. doi: 10.1161/CIRCRESAHA.110.227033. [DOI] [PubMed] [Google Scholar]
- Hechtenberg S, Beyersmann D. Inhibition of sarcoplasmic reticulum Ca(2+)-ATPase activity by cadmium, lead and mercury. Enzyme. 1991;45(3):109–115. doi: 10.1159/000468875. [DOI] [PubMed] [Google Scholar]
- Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, Devay RM, Lackner LL, Westermann B, Schuldiner M, Weissman JS, Nunnari J. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J Cell Biol. 2011;195(2):323–340. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Wang H, Fu X, Bielby C, Yang H. Conditions of endoplasmic reticulum stress stimulate lipid droplet formation in Saccharomyces cerevisiae. Biochem J. 2009;424(1):61–67. doi: 10.1042/BJ20090785. [DOI] [PubMed] [Google Scholar]
- Farber SA, Slack BE, Blusztajn JK. Acceleration of phosphatidylcholine synthesis and breakdown by inhibitors of mitochondrial function in neuronal cells: a model of the membrane defect of Alzheimer’s disease. FASEB J. 2000;14(14):2198–2206. doi: 10.1096/fj.99-0853. [DOI] [PubMed] [Google Scholar]
- Iorio E, Di Vito M, Spadaro F, Ramoni C, Lococo E, Carnevale R, Lenti L, Strom R, Podo F. Triacsin C inhibits the formation of 1 H NMRvisible mobile lipids and lipid bodies in HuT 78 apoptotic cells. Biochim Biophys Acta. 2003;1634:1–14. doi: 10.1016/j.bbalip.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Iwanyshyn WM, Han GS, Carman GM. Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc. J Biol Chem. 2004;279(21):21976–21983. doi: 10.1074/jbc.M402047200. [DOI] [PubMed] [Google Scholar]
- Jackowski S. Cell cycle regulation of membrane phospholipid metabolism. J Biol Chem. 1996;271(34):20219–20222. doi: 10.1074/jbc.271.34.20219. [DOI] [PubMed] [Google Scholar]
- Jacobs RL, Zhao Y, Koonen DP, Sletten T, Su B, Lingrell S, Cao G, Peake DA, Kuo MS, Proctor SD, Kennedy BP, Dyck JR, Vance DE (2010) Impaired de nova choline synthesis explains why phosphatidylethanolamine N-methyltranferase-defiecient mice are protected from diet-induced obesity. J Biol Chem 285(29):22403 [DOI] [PMC free article] [PubMed]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, Schuldiner M. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323(5922):1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanipes MI, Henry SA. The phospholipid methyltransferases in yeast. Biochim Biophys Acta. 1997;1348:134–141. doi: 10.1016/S0005-2760(97)00121-5. [DOI] [PubMed] [Google Scholar]
- Kharroubi I, Ladrière L, Cardozo AK, Dogusan Z, Cnop M, Eizirik DL. Free fatty acids and cytokines induce pancreatic beta-cell apoptosis by different mechanisms: role of nuclear factor-kappaB and endoplasmic reticulum stress. Endocrinology. 2004;145(11):5087–5096. doi: 10.1210/en.2004-0478. [DOI] [PubMed] [Google Scholar]
- Kimura K, Yamaoka M, Kamisaka Y. Rapid estimation of lipids in oleaginous fungi and yeasts using Nile red fluorescence. J Microbiol Methods. 2004;56(3):331–338. doi: 10.1016/j.mimet.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Kodaki T, Yamashita S. Characterization of the methyltransferases in the yeast phosphatidylethanolamine methylation pathway by selective gene disruption. Eur J Biochem. 1989;185(2):243–251. doi: 10.1111/j.1432-1033.1989.tb15109.x. [DOI] [PubMed] [Google Scholar]
- Koning AJ, Roberts CJ, Wright RL. Different subcellular localization of Saccharomyces cerevisiae HMG-CoA reductase isozymes at elevated levels corresponds to distinct endoplasmic reticulum membrane proliferations. Mol Biol Cell. 1996;5:769–789. doi: 10.1091/mbc.7.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Nam Y, Chung YH, Kim HR, Park ES, Chung SJ, Kim JH, Sohn UD, Kim HC, Oh KW, Jeong JH. Beneficial effects of phosphatidylcholine on high-fat diet-induced obesity, hyperlipidemia and fatty liver in mice. Life Sci. 2014;118(1):7–14. doi: 10.1016/j.lfs.2014.09.027. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Malhotra JD, Kaufman RJ. ER stress and its functional link to mitochondria: role in cell survival and death. Cold Spring Harb Perspect Biol. 2011;3:a004424. doi: 10.1101/cshperspect.a004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi S, Ohkuma A, Talim B, Karahashi M, Koumura T, Aoyama C, Kurihara M, Quinlivan R, Sewry C, Mitsuhashi H, Goto K, Koksal B, Kale G, Ikeda K, Taguchi R, Noguchi S, Hayashi YK, Nonaka I, Sher RB, Sugimoto H, Nakagawa Y, Cox GA, Topaloglu H, Nishino I. A congenital muscular dystrophy with mitochondrial structural abnormalities caused by defective de novo phosphatidylcholine biosynthesis. Am J Hum Genet. 2011;88:845–851. doi: 10.1016/j.ajhg.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, Johannsson A, Hesketh RT, Smith GA, Metcalfe JC. Calcium signals and phospholipid methylation in eukaryotic cells. Biochem J. 1984;221:675–684. doi: 10.1042/bj2210675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, Watanabe T, Sato M, Momose Y, Nakahara T, Oka S, Iwahashi H. Dimethyl sulfoxide exposure facilitates phospholipid biosynthesis and cellular membrane proliferation in yeast cells. J Biol Chem. 2003;278(35):33185–33193. doi: 10.1074/jbc.M300450200. [DOI] [PubMed] [Google Scholar]
- Muthukumar K, Rajakumar S, Sarkar MN, Nachiappan V. Glutathione peroxidase3 of Saccharomyces cerevisiae protects phospholipids during cadmium-induced oxidative stress. Antonie Van Leeuwenhoek. 2011;99:761–771. doi: 10.1007/s10482-011-9550-9. [DOI] [PubMed] [Google Scholar]
- Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Görgün C, Glimcher LH, Hotamisligil GS. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306(5695):457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- Preitschopf W, Lückl H, Summers E, Henry SA, Paltauf F, Kohlwein SD. Molecular cloning of the yeast OPI3 gene as a high copy number suppressor of the cho2 mutation. Curr Genet. 1993;23:95–101. doi: 10.1007/BF00352006. [DOI] [PubMed] [Google Scholar]
- Prins D, Michalak M (2011) Organellar calcium buffers. Cold Spring Harb Perspect Biol 3(3) [DOI] [PMC free article] [PubMed]
- Promlek T, Ishiwata-Kimata Y, Shido M, Sakuramoto M, Kohno K, Kimata Y. Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol Biol Cell. 2011;22:3520–3532. doi: 10.1091/mbc.E11-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumar S, Ravi C, Nachiappan V. Defect of zinc transporter ZRT1 ameliorates cadmium induced lipid accumulation in Saccharomyces cerevisiae. Metallomics. 2016;8(4):453–460. doi: 10.1039/C6MT00005C. [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, Weissman JS, Krogan NJ. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123(3):507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Schuler MH, Di Bartolomeo F, Böttinger L, Horvath SE, Wenz LS, Daum G, Becker T. Phosphatidylcholine affects the role of the sorting and assembly machinery in the biogenesis of mitochondrial β-barrel proteinsJ. Biol Chem. 2015;290(44):26523–26532. doi: 10.1074/jbc.M115.687921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel GS. Mitochondrial DNA, aconitase ‘wraps’ it up. Trends Biochem Sci. 2005;30:294–296. doi: 10.1016/j.tibs.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Sheikhnejad RG, Srivastava PN. Isolation and properties of a phosphatidylcholine-specific phospholipase C from bull seminal plasma. J Biol Chem. 1986;261(16):7544–7549. [PubMed] [Google Scholar]
- Siakotos AN, Rouser G, Fleischer S. Phospholipid composition of human, bovine and frog myelin isolated on a large scale from brain and spinal cord. Lipids. 1966;1:85–86. doi: 10.1007/BF02668129. [DOI] [PubMed] [Google Scholar]
- Sorin A, Rosas G, Rao R. PMR1, a Ca2 + −ATPase in yeast Golgi, has properties distinct from sarco/endoplasmic reticulum and plasma membrane calcium pumps. J Biol Chem. 1997;272(15):9895–9901. doi: 10.1074/jbc.272.15.9895. [DOI] [PubMed] [Google Scholar]
- Stone SJ, Vance JE. Phosphatidylserine synthase-1 and −2 are localized to mitochondria-associated membranes. J Biol Chem. 2000;275:34534–34540. doi: 10.1074/jbc.M002865200. [DOI] [PubMed] [Google Scholar]
- Summers EF, Letts VA, McGraw P, Henry SA. Saccharomyces cerevisiae cho2 mutants are deficient in phospholipid methylation and cross-pathway regulation of inositol synthesis. Genetics. 1988;120(4):909–922. doi: 10.1093/genetics/120.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surma MA, Klose C, Peng D, Shales M, Mrejen C, Stefanko A, Braberg H, Gordon DE, Vorkel D, Ejsing CS, Farese R, Jr, Simons K, Krogan NJ, Ernst R. A lipid E-MAP identifies Ubx2 as a critical regulator of lipid saturation and lipid bilayer stress. Mol Cell. 2013;51(4):519–530. doi: 10.1016/j.molcel.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault G, Shui G, Kim W, McAlister GC, Ismail N, Gygi SP, Wenk MR, Ng DT. The membrane stress response buffers lethal effects of lipid disequilibrium by reprogramming the protein homeostasis network. Mol Cell. 2012;48(1):16–27. doi: 10.1016/j.molcel.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance JE. Phospholipid synthesis in a membrane fraction associated with mitochondria. J Biol Chem. 1990;265:7248–7256. [PubMed] [Google Scholar]
- Zhang GH, Yamaguchi M, Kimura S, Higham S, Kraus-Friedmann N. Effects of heavy metal on rat liver microsomal Ca2(+)-ATPase and Ca2+ sequestering. Relation to SH groups. J Biol Chem. 1990;265(4):2184–2189. [PubMed] [Google Scholar]
- Zinser E, Sperka-Gottlieb CD, Fasch EV, Kohlwein SD, Paltauf F, Daum G. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J Bacteriol. 1991;173(6):2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Kennedy (cki1∆,cpt1∆, and pct1∆) and the methylation pathway (cho2∆, opi3∆) mutant cells were grown up to mid-log phase on YPD medium, harvested and washed twice in sterile water. Fourfold serial dilutions were spotted on to YPD agar plate with 50 μM Cd and incubated at 30 °C for 2 days (DOCX 530 kb)
(DOCX 28 kb)