Abstract
Plants are constantly challenged by environmental stresses, including drought and high salinity. Improvement of drought and osmotic stress tolerance without yield decrease has been a great challenge in crop improvement. The Arabidopsis ENHANCED DROUGHT TOLERANCE1/HOMEODOMAIN GLABROUS11 (AtEDT1/HDG11), a protein of the class IV HD-Zip family, has been demonstrated to significantly improve drought tolerance in Arabidopsis, rice, and pepper. Here, we report that AtEDT1/HDG11 confers drought and osmotic stress tolerance in the Chinese kale. AtEDT1/HDG11-overexpression lines exhibit auxin-overproduction phenotypes, such as long hypocotyls, tall stems, more root hairs, and a larger root system architecture. Compared with the untransformed control, transgenic lines have significantly reduced stomatal density. In the leaves of transgenic Chinese kale plants, proline (Pro) content and reactive oxygen species-scavenging enzyme activity was significantly increased after drought and osmotic stress, particularly compared to wild kale. More importantly, AtEDT1/HDG11-overexpression leads to abscisic acid (ABA) hypersensitivity, resulting in ABA inhibitor germination and induced stomatal closure. Consistent with observed phenotypes, the expression levels of auxin, ABA, and stress-related genes were also altered under both normal and/or stress conditions. Further analysis showed that AtEDT1/HDG11, as a transcription factor, can target the auxin biosynthesis gene YUCC6 and ABA response genes ABI3 and ABI5. Collectively, our results provide a new insight into the role of AtEDT1/HDG11 in enhancing abiotic stress resistance through auxin- and ABA-mediated signaling response in Chinese kale.
Keywords: AtEDT1/HDG11, drought, osmotic stress, abscisic acid, auxin, signaling response
Introduction
Drought and osmotic stress can seriously limit plant growth and productivity. Plants have developed multiple strategies to cope with drought and osmotic stress. These normally involve a mixture of stress avoidance and tolerance adaptations, which produce a range of changes at the morphological, physiological, cellular, and molecular levels (Shinozaki and Yamaguchi-Shinozaki, 2007; Ashraf, 2010). It is well known that the most efficient and important mechanisms for plants to cope with water deficit stress include maximization of water uptake through the development of deep and extensive root systems and/or minimization of water loss by stomatal closure and reduction of stomatal density (Yu et al., 2008, 2013).
Drought causes dehydration when root uptake of water from soil is insufficient to meet the transpirational requirements of plants (Jeong et al., 2010; Yoo et al., 2010). It has been reported that drought-resistant plants usually have deeper and more highly branched root systems than drought-sensitive plants. Ease of access to water and nutrients is a factor potentially limiting plant growth (Price and Tomos, 1997; Kondo et al., 2000; Yu et al., 2008; Uga et al., 2013). Literature has documented that overexpression of AtCKX, AtNAC1, OsDRO1, OsNAC5, and OsNAC10 enhanced root depth and/or thickness and led to significantly improved drought tolerance (Xie et al., 2000; Jeong et al., 2010, 2013; Uga et al., 2013).
It is well known that the majority of plant transpiration occurs through stomatal pores (Yoo et al., 2010). Thus, water loss during transpiration in the stomata is a key determinant of drought tolerance (Yu et al., 2008). Plant transpiration rates can be modulated by stomatal density and/or stomatal movements (Masle et al., 2005; Yoo et al., 2010). A pair of specialized guard cells wield control over the size of stomatal apertures, which are influenced by many factors, such as illumination, water supply, CO2, abscisic acid (ABA), reactive oxygen species (ROS), and calcium and potassium ions (Berger and Altmann, 2000; Von Groll et al., 2002). Stomatal density is also variable and is set according to the environmental factors prevailing during leaf development. In addition to these exogenous factors, stomatal density is also subject to genetic control (Berger and Altmann, 2000; Masle et al., 2005; Shpak et al., 2005). Recently, a number of genes controlling stomatal development have been identified. It has been reported that overexpression of AtERECTA, AtGTL1, AtSDD1, and OsSIK1 significantly enhanced drought tolerance in plants and was also associated with reduction of stomatal density (Masle et al., 2005; Ouyang et al., 2010; Yoo et al., 2010).
The homeodomain-leucine zipper (HD-Zip) superfamily of transcription factors is unique to the plant kingdom. They can be classified into four subfamilies, according to a set of distinctive features that include DNA-binding specificities, gene structures, additional common motifs and physiological functions (Ariel et al., 2007). Some HD-Zip proteins participate in organ and vascular development or meristem maintenance, while others mediate the action of hormones or are involved in responses to environmental conditions (Ariel et al., 2007; Agalou et al., 2008). In recent years, many efforts were undertaken to elucidate the functions of HD-Zip genes, and found a subset of genes is regulated by drought (Agalou et al., 2008). These include HD-Zip family I gene families Athb-7 and Athb-12 from Arabidopsis and Oshox22 from rice (Söderman et al., 1996; Olsson et al., 2004; Ré et al., 2014; Zhang et al., 2012). The Arabidopsis HOMEODOMAIN GLABROUS 11 (HDG11) encodes a protein of the class IV HD-Zip family, which has two known conserved domains, the homeobox domain and the START domain, and was first identified as a negative regulator of trichome branching development (Nakamura et al., 2006; Khosla et al., 2014). AtEDT1/HDG11 was identified as improving drought tolerance in the Arabidopsis enhanced drought tolerance1 (edt1) mutant through T-DNA inserted in the 5′-untranslated region of HDG11, resulting in the overexpression of HDG11, reduced stomatal density, and an improved root system (Yu et al., 2008). Additionally, ectopic expression of AtEDT1/HDG11 has been demonstrated to significantly improve drought tolerance in rice, cotton, and pepper, without yield penalty, indicating that it is a promising candidate gene for crop stress tolerance development (Yu et al., 2013, 2016; Zhu et al., 2015). However, the underlying molecular mechanisms are still not well-studied.
In this work, Arabidopsis Enhanced Drought Tolerance1/HOMEODOMAIN GLABROUS 11 (AtEDT1/HDG11) was observed to be overexpressed in Chinese kale. Our results demonstrate that the overexpression of AtEDT1/HDG11 in Chinese kale is not only responsible for its enhanced drought and osmotic tolerance, but also significantly improves biomass under both normal and stress conditions. AtEDT1/HDG11-overexpression lines also alter the expression of ABA and auxin biosynthesis, and/or the expression of response genes. These findings may provide a new insight into the mechanism of abiotic stress tolerance in Chinese kale, and represent an important genetic engineering approach for the improvement of stress tolerance in crops.
Materials and Methods
Construction and Generation of Overexpression Plants
The full-length cDNA of AtEDT1/HDG11 was isolated from the PCB2004-HDG11 plasmid vector (Yu et al., 2008) via the PCR specific primers AtHDG11-FOR and AtHDG11-REV (Supplementary Table 1). The sequence-confirmed fragment was cloned into pCAMBIA1301-Cry2Aa2-PIM. The construct was transformed into the Chinese kale (Brassica oleracea var. alboglabra) inbred line 25 using Agrobacterium tumefaciens-mediated transformation. The PCR and qRT-PCR analysis indicated that 12 T0 independent transgenic plants were generated; the Southern blot analysis indicated that most transgenic lines were harboring one copy. The positive T2 transformants were prescreened for drought resistance in the greenhouse and more than 80% of transgenic lines showed improved drought tolerance during the seedling stage. Several drought-tolerant lines (i.e., OE-1, OE-2, OE-3) were selected for further analysis (Supplementary Figure 1).
Identification of Morphological Characterization of Transgenic Chinese Kale
For morphological characterization of plants, transgenic and wild-type (untransformed) plants were grown under standard or stress conditions. Leaves of the same age and same relative position were sampled from transgenic and wild-type plants during the seedling stage. The area was measured using the portable leaf area meter (CI-203 Handheld Laser Area Meter, USA). During the reproductive stage, the plant inflorescence, pedicel, siliques, and epidermal cells between the first and second leaf length were measured. Subsequently, siliques and seeds were counted.
Morphological Characterization of Transgenic Plant Roots
For measurement of root elongation, the wild-type and transgenic seeds were germinated and then grown on an MS medium. The 7-day-old seedlings were used for hypocotyl length and root length measurement. To better observe the root system, transgenic and wild-type plants were grown in a nutrient solution. The number of roots was counted and the root length was measured. For measurement of roots in the soil, wild-type and transgenic seeds were transplanted to pots with the substrate (Tref BIO, Norway), and grown in a greenhouse under standard growth conditions for further research. For data analysis, the 8-week-old transgenic and wild-type plants were carefully removed from their pots. Subsequently, the soil was carefully separated without damaging the roots, and the root biomass was measured as fresh weight.
Drought Tolerance Assay of AtEDT1/HDG11 Transgenic Chinese Kale
For drought tolerance tests of plants in the seedling stage, 35-day-old wild-type and transgenic seedlings were subjected to drought treatment in greenhouse by withholding water for 5 days (severe drought stress), re-watering for recovery for 4 days and then calculating the survival rate of seedlings. To evaluate the maximum photochemical efficiency of PSII under moderate drought stress (water withheld for three days), Fv/Fm values were measured by the Opti-Science OS-30p (Opti-Sciences, USA). After treatment with drought stress plants leaves were harvested and frozen in liquid nitrogen for a chlorophyll, proline and H2O2 contents and superoxide dismutase (SOD) activity assay.
To measure stomatal aperture, 35-day-old seedlings underwent a 2-day drought treatment, and the imprint method was then used for stomatal measurement as previously described (Zhu et al., 2015). To measure water loss rates, roots were detached from the transgenic and wild-type plants, respectively. Plant shoots were weighed immediately after detachment, placed on a plate on a laboratory bench, and were then weighed at designated time intervals (0, 0.8, 1.6, 2.4, 3.2, 4.0, 4.8, 5.6, 6.4, and 7.2 h). The proportion of fresh weight loss was expressed using the initial weight of the plants. For data analysis, 10 plants were used for treatment.
To test Chinese kale drought tolerance at the flowering stage, the transgenic and wild-type plants were subjected to a water deficit of withheld watering for 3–5 days (3 days for the early stage and 5 days for the late stage). The plants were re-watered while drought stress symptoms (e.g., leaf wilting) were clearly observable in the transgenic lines, and then plants were left to recover for 15 days. Repeated drought treatment was sustained until the siliques and seeds were obtained and counted.
Osmotic Tolerance Assay of AtEDT1/HDG11 Transgenic Chinese Kale
For the osmotic tolerance (PEG and salt) test at the seedling stage, 40-day-old transgenic and wild-type plants underwent treatment with 25% PEG and 250mM NaCl, respectively. Osmotic tolerance treatment was sustained for 30 days and after a re-watering recovery for 7 days, the survival rate was calculated and the biomass of surviving plants was weighted. For further analysis of osmotic tolerance, transgenic plants were subjected to an osmotic stress treatment for 10 days, and leaves of similar developmental stages from stress-treated plants or normal control plants were sampled for proline and H2O2 contents and SOD activity measurement.
Quantification of Free IAA Contents
Free IAA from the indicated tissues was extracted as described by Pan et al. (2010). Free IAA contents were measured using a PhytodetekTM IAA Test kit (Agdia, Arizona, CA, USA) according to the manufacturer’s instructions.
Chlorophyll Content Measurement
To measure the chlorophyll content of transgenic plants and wild-type plants after exposure to 3 days of drought treatments, a 0.5 cm2 disk was cut from the middle of the leaf blade, and chlorophyll content was calculated and expressed as mg/g FW (Loukehaich et al., 2012).
Measurement of Proline Content
Leaves of similar developmental stages from stress-treated plants or normal control plants were used for proline content measurement. Proline was assayed as described by Bates et al. (1973).
Determination of SOD Activity
For measurement of the SOD activities, 0.1 g of leaves was sampled and homogenized in an ice-cold mortar using a 50 mM sodium phosphate buffer (pH 7.8) containing 1 % polyvinylpyrrolidone and 10 mmol L-1β-mercaptoethanol. Subsequently, centrifuged at 12,000 × g for 15 min at 4°C, the supernatant was used for the determination of SOD. Total SOD activity was measured by the SOD assay kit according to the manufacturer’s instructions (Nanjing Jiancheng Bioengineering Institute).
Measurement of H2O2 Content
Frozen tissues were ground in liquid nitrogen. Ground tissues (50 mg) were soaked in a hydrogen peroxide assay kit lysis buffer (Beyotime). The extracts were clarified by centrifugation at 1,2000 × g for 15 min at 4°C. H2O2 concentration using the hydrogen peroxide assay kit according to the manufacturer’s instructions.
Seeds Germination Test
For the germination assays, more than 40 seeds were placed on one MS agar medium containing different concentrations of ABA. To break the dormancy, seeds were incubated at 4°C for 2 days in the dark before germination and were subsequently grown in a growth chamber at room temperature. Seed germination was followed for 7 days. Seeds were counted as germinated when the radicles had emerged by 1 mm. The germination rate was calculated as a percentage of the total number of seeds planted. For data analysis the experiment was repeated at least three times.
Stomatal Density and Aperture Measurements
To measure the stomatal density, fully expanded leaves in close proximity to one another were detached. Subsequently, the leaf surface imprint method was used, as previously described, to evaluate cells and stomatal density (Zhu et al., 2015). For statistical analysis of stomatal density, proximate leaves were sampled, and five plants were sampled for the wild-type and transgenic plants, respectively.
Fully expanded leaves of the same relative position and age were detached from the transgenic and the wild-type plants. The leaves were then placed in solutions containing 20 mM KCl, 5 mM MES-KOH (pH 6.15), and 1 mM CaCl2 and exposed to light at a photon fluency rate of 150 μM m-2sec-1 for 2 h for the stomata to fully open. Subsequently, ABA was added to the solution at 1–100 μM to assay the stomata closing. The inner edges of guard cells, the height of which was between 16 and 26 μm, were focused on by an optical microscope (Carl Zeiss; Germany), and the apertures of usually 30 to 50 stomata were measured.
Measurements of Photosynthetic Rate, Transpiration Rate, and Water Use Efficiency
Photosynthesis (P) and transpiration (T) rates of transgenic and wild-type seedlings were measured using the portable photosynthesis system Li-6400XT (LI-COR, USA). This was done in the morning (9 to 11 AM), on the same plants mentioned above, before stomata observation. All of the photosynthetic measurements were taken at a constant air flow rate of 500 μmol s-1. The concentration of CO2 was 400 μmol mol-1 and delivered using the system’s CO2 injector (Li-Cor 6400-01). The chamber temperature was maintained at 26 ± 2°C, and the photosynthetic photon flux density at 1,200 μmol m-2 s-1. Three measurements were made for each plant, and five plants were used for both the wild-type and the transgenic plants. WUE was defined as P/T ratio and were derived from the measured P and T.
Real-Time Reverse Transcription (RT)-PCR Analysis
Total RNA was prepared from tissues indicated in the figures by the TRIzol (Life, USA), and 1 μg of RNA from each sample was used for the reverse transcription reaction by the Prime ScriptTM RT reagent kit with gDNA eraser (Takara, Japan). Quantitative real-time PCR analysis was performed on a Light Cycler 480 Real-Time PCR System according to the manufacturer’s instructions (Roche, Switzerland). The qPCR program included an initial denaturation step at 94°C for 8 min, followed by 40 cycles of 10 s at 94°C, 15 s at 56°C, and 30 s at 72°C. As an internal control, the tubulin8 transcript was used to quantify the relative transcript level of each target gene in each sample. The gene accession numbers and sequences of all primers used for qPCR analysis are as described (Supplementary Table 1). The values represent the mean of three biological replicates.
Promoter Analysis of Stress-Responsive and Phenotype-Related Genes
Promoter sequences at about 2 kb length upstream of the ATG start codon were obtained from BRAD (Cheng et al., 2011) with specific primers (Supplementary Table 1), and cis-elements in promoters found in the PLACE1 and Plant-CARE databases2.
Transient Transactivation Assay
Promoters of four genes ABI3, ABI5, EXPA5, and YUCCA6 were isolated from Chinese kale and cloned into the binary vector pEGFP. The effector construct 35S-AtEDT1/HDG11 was also used in the transformation. An AtEDT1/HDG11 N-terminal deletion without the HD domain was constructed in the same vector pEGFP as for the effector construct and used as a negative control. These reporter constructs were co-transformed into onion cells with either the 35S-AtEDT1/HDG11 construct as effector or a modified 35S-AtEDT1/HDG11 N-terminal deletion without the HD domain as a negative effector control (Yu et al., 2008). The transformed onion cells were observed with a confocal microscope (Carl Zeiss; Germany) using an excitation wavelength of 488 nm. GFP fluorescence intensity was quantified using ImageJ (Abràmoff et al., 2004).
Statistical Analysis
Statistically significant differences (P < 0.05 or P < 0.01) were computed based on the Student’s t-tests. Data are the means ± SD of least three independent repeat experiments.
Results
Morphological Characterization of Transgenic Chinese Kale
It this study, we generated AtEDT1/HDG11-overexpressing Chinese kale plants and the phenotype of Chinese kale was characterized at different developmental stages. During the seedling stage the transgenic plants showed auxin-overproduction phenotypes such as hypocotyl and primary root length, root hairs, and lateral numbers were significantly increased (Figures 1A,B,F–H; Supplementary Figure 2). A larger root system was also observed in transgenic plants during late periods of vegetative development (Supplementary Figures 3A,B). As a consequence, the fresh weight of 8-week-old Chinese kale was significantly improved in AtEDT1/HDG11-overexpressing lines (Supplementary Figure 3C). However, the root-to-shoot biomass ratio was almost unaltered, owing to the increased root biomass, which was nearly equivalent to the shoot biomass (data not shown).
FIGURE 1.
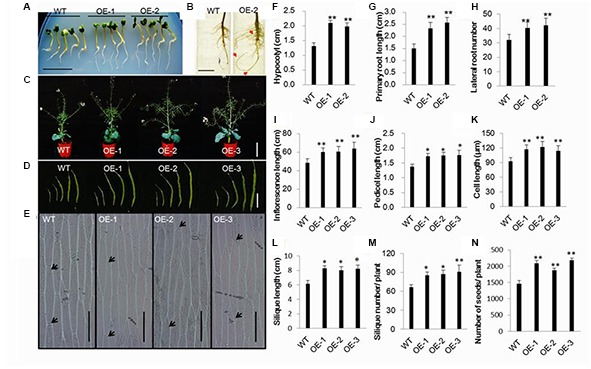
Morphological characterization of transgenic Chinese kale plants. (A) The primary root and hypocotyl of 7-day-old transgenic seedlings was longer than that of the wild-type (untransformed, WT) seedlings of the same age on the MS medium. Bar = 1.5 cm. (B) Root systems of 3-week-old transgenic and wild-type plants grown in a nutrient solution. Bar = 3cm. (C) Wild-type and transgenic plants in the flower stage. Bar = 30cm. (D) Transgenic and wild-type plants siliques 6, 10, 20, 30, and 50 day post-anthesis. Bar = 4cm. (E) Epidermal cells of the stems between the first and second leaf. (F,G) Hypocotyl and primary root length of 7-day-old seedlings. Values are mean ± SD (n = 10 plants). (H) Number of roots of 3-week-old transgenic and wild-type plants Values are mean ± SD (n = 10 plants). (I–L) Inflorescence, pedicel, epidermal cell and silique lengths of transgenic and wild-type plants. Values are mean ± SD. For analysis of inflorescence length 10 plants were used, respectively. Fifty-day-old siliques were used for silique and pedicel length measurement. Five siliques from the base of the main inflorescence of 10 plants. Epidermal cells of the stems between the first and second cauline leaves were examined. Measurements of five stems, 10 epidermal cells from each stem. (M) Silique number of transgenic and wild-type plants. Values are mean ± SD (n = 10 plants). (N) Seed numbers in transgenic and wild-type plants. Values are mean ± SD (n = 10 plants). ∗P < 0.05 and ∗∗P < 0.01 asterisks indicate Student’s t-test significant differences.
During the reproductive phase, the plant height and main inflorescence length of transgenic lines were significantly improved in comparison to the wild-type plants (Figures 1C,I), and the increased elongation was mainly caused by increased cell length and width (Figures 1E,K; Supplementary Figure 4). Accordingly, transgenic plants had more siliques than wild-type plants (Figure 1M). In addition, compared to the wild-type control, pedicel and silique lengths were also significantly improved in transgenic plants (Figures 1J,L). The siliques are blunt in the wild-type plants compared to the transgenic lines (Supplementary Figure 5). Furthermore, the number of seeds for transgenic lines was higher than in the wild-type plant (Figure 1N).
AtEDT1/HDG11 Enhanced Drought Tolerance of Transgenic Chinese Kale
Drought tolerance during the seedling stage is important for Chinese kale biomass establishment, so we carefully tested the drought tolerance in transgenic lines at the seedling stage. Under drought stress conditions, leaf rolling was significantly delayed in transgenic plants compared to wild-type plants (Figure 2A; Supplementary Figure 1). After 5 days of drought treatment and subsequent recovery for 4 days, 100% of the transgenic plants survived, when only 40% of the wild-type plants survived (Figure 2B). The majority of plant transpiration occurs via the stomatal pores, so we observed the stomata under drought conditions. After drought stress, the stomatal apertures of transgenic lines were smaller than those of the wild-type control (Figures 2C,D). Accordingly, 76% (46 of 60) of the wild-type stomata were open in contrast to only 28.3% (17 of 60) and 35% (21 of 60) in the transgenic lines OE-1 and OE-2, respectively. As a result, water loss in the transgenic plants was much slower than in the wild-type (Figure 2E).
FIGURE 2.
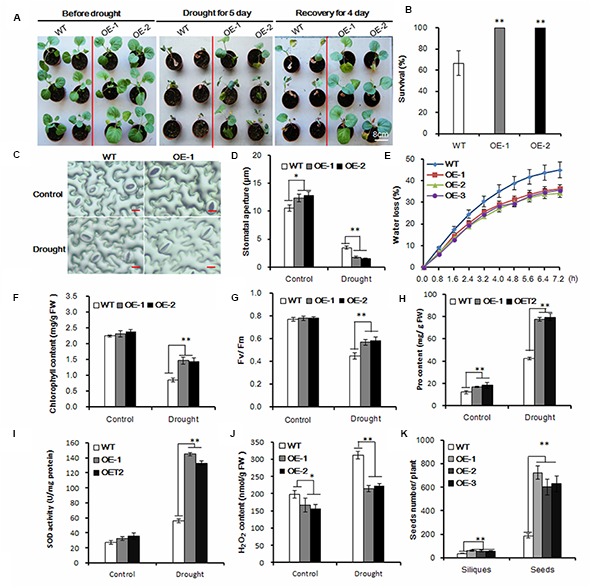
Enhanced drought tolerance in transgenic Chinese kale. (A) Drought treatment of 35-day-old transgenic plants in a greenhouse for 5 days and then recovery for 5 days. Bar = 8 cm. (B) Survival rate after 5-day drought treatment and 4 days of recovery. Values are mean ± SD (n = 50, ∗∗P < 0.01). (C) Stomatal observation of the transgenic Chinese kale lines. Thirty-five-day-old plants were treated with drought stress for 2 days. The tests were repeated three times, and typical results are presented. (D) Stomatal apertures of the transgenic Chinese kale lines as shown in (C). Values are mean ± SD (n = 50 stomata). (E) Comparison of water loss rate between transgenic and wild-type plants. Water loss was measured at the indicated time points and expressed as the percentage of the initial fresh weight (FW). Values are mean ± SD (n = 10 plants). (F–J) Chlorophyll content, chlorophyll fluorescence (Fv/Fm), Pro content, SOD activity, H2O2 content of transgenic and wild-type plants after 3-day water deficit treatment. Data represents mean ± SD (∗p < 0.05, ∗∗p < 0.01). (K) Siliques and seeds of the transgenic and wild-type plants under drought stress conditions. Values are mean ± SD (n = 10 plants). Asterisks indicate Student’s t-test significant differences
To investigate the drought tolerance of transgenic Chinese kale, the chlorophyll content was measured. The chlorophyll content was about 72 and 69% higher in the transgenic lines OE-1 and OE-2, respectively, than in the wild-type plants under drought conditions (Figure 2F). Accordingly, the Fv/Fm levels were significantly higher in transgenic plants than in the wild-type plants under drought conditions (Figure 2G). Meanwhile, proline (Pro), a common compatible osmolyte in higher plants, was measured before and after 3 days of drought stress. The Pro content of the transgenic plants was higher than that of the control under normal conditions, and under drought stress increased more significantly in the transgenic plants compared with the wild-type controls (Figure 2H). Moreover, the activity of ROS-scavenging enzymes, such as SOD, was significantly increased in the AtHDG11 transgenic plants compared with wild-type plants after stress treatments (Figure 2I), consistent with low H2O2 level detected in transgenic lines (Figure 2J). Therefore, our data indicate that the transgenic plants are better protected from oxidative damage during drought and salt stress.
Furthermore, we compared the drought tolerance performance of transgenic and the wild-type Chinese kale at the reproductive stage. Compared to the untransformed plants, the AtEDT1/HDG11-overexpressing lines have more siliques and seeds (Figure 2K). These results demonstrate that AtED1/HDG11 can significantly increase the drought tolerance of Chinese kale plants, likely at different developmental stages.
Osmotic Tolerance Was Improved in AtEDT1/HDG11-Overxpressing Chinese Kale
Drought stress was usually accompanied by high osmotic stress. To test whether the AtEDT1/HDG11-overexpressing plants were more osmotic-tolerant, we carried out PEG and salt tolerance tests in the greenhouse. For osmotic stress treatment, the 40-day-old seedlings were irrigated with 500 ML 25% PEG6000 or 250 mM NaCl solution for each pot every 3 days, respectively. After treatment with PEG6000 or NaCl for 30 days, the wild-type plants withered with many leaves dropping and dying, indicating severe osmotic stress in the plants. However, the AtEDT1/HDG11-overexpressing plants under the same conditions still showed relatively normal growth phenotypes (Figures 3A,B). After watering for recovery of 7 days, the survival rate of transgenic plants was the highest among all the plants treated with PEG6000 and NaCl (Figure 3C), respectively. Moreover, the Fv/Fm levels were significantly higher in transgenic plants than in the wild-type plants under osmotic conditions (Figure 3D). Similar to the results of drought stress, after treatment with PEG and NaCl transformed lines also showed higher levels of Pro content and SOD activity (Figures 3E,F), while H2O2 content was decreased (Figure 3G).
FIGURE 3.
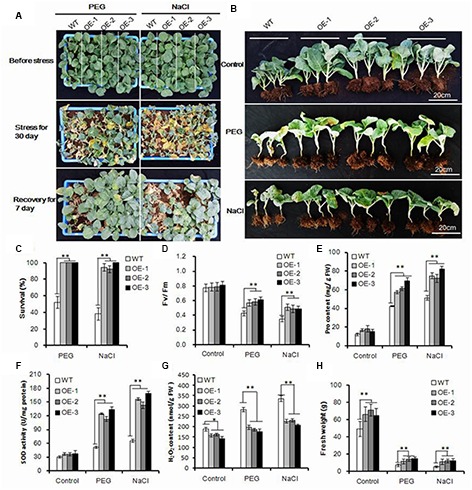
Enhanced osmotic and salt tolerance in transgenic Chinese kale. (A) Forty-day-old transgenic and wild-type plant treatment with 25% PEG 6000 and 250 mM NaCl for 30 days and recovery of 7 days. (B) Transgenic plants with or without stress treatment after recovery for 7 days. (C) Survival rate after 30-day stress treatment and 7 days of recovery. Values are mean ± SD (n = 50, ∗∗P < 0.01). (D–G) Chlorophyll fluorescence (Fv/Fm), Pro content, SOD activity, and H2O2 content of transgenic plants subjected to stress treatment for 10 days. Data represents mean ± SD of three independent experiments (∗p < 0.05, ∗∗p < 0.01). (H) Fresh weight of transgenic and wild-type plants with or without stress treatment. Values are mean ± SD (n = 10 plants, ∗∗p < 0.01). Asterisks indicate Student’s t-test significant differences.
More importantly, the transgenic Chinese kale showed significantly improved fresh weight both under normal and osmotic stress conditions, (Figure 3H). Compared with normal conditions, the biomass of HDG11 overexpressing lines decreased approximately 80%, while 85% decreased in wild-type lines. Taken together, our results show that the AtEDT1/HDG11-overexpressing Chinese kale has significantly improved osmotic tolerance.
Enlarged Leaf Area and Stomatal Size with Reduced Stomatal Density but Increased Water Use Efficiency in Transgenic Chinese Kale
Under normal conditions, transgenic plants have exhibited a larger leaf area (Supplementary Figures 6A–C). It is well known that the majority of plant transpiration occurs through stomatal pores, and plant transpiration rates can be modulated by stomatal density and/or stomatal movements. Because the water loss was significantly delayed in transgenic plants, and to uncover the underlying mechanism, the leaf surface imprint method was used to evaluate the formation of cells and stomata (Supplementary Figure 7). The average cell density of the transgenic lines OE-1 and OE-2 was reduced by 34.7 and 39.2% compared with that of the wild-type, respectively (Figure 4A). Accordingly, the stomatal density was reduced by 27.9 and 39.9%, respectively (Figure 4B). While the stomatal density was decreased, the stomatal size was increased in the AtEDT1/HDG11-overexpressing lines (Figure 4C; Supplementary Figure 7). Compared with wild-type controls the average stomatal length in transgenic lines OE-1 and OE-2 was increased by 24.5 and 21.4%, respectively (Figure 4C). Correspondingly, the average stomatal widths of transgenic lines OE-1 and OE-2 were increased by 18.2 and 19.7%, respectively (Figure 4C).
FIGURE 4.
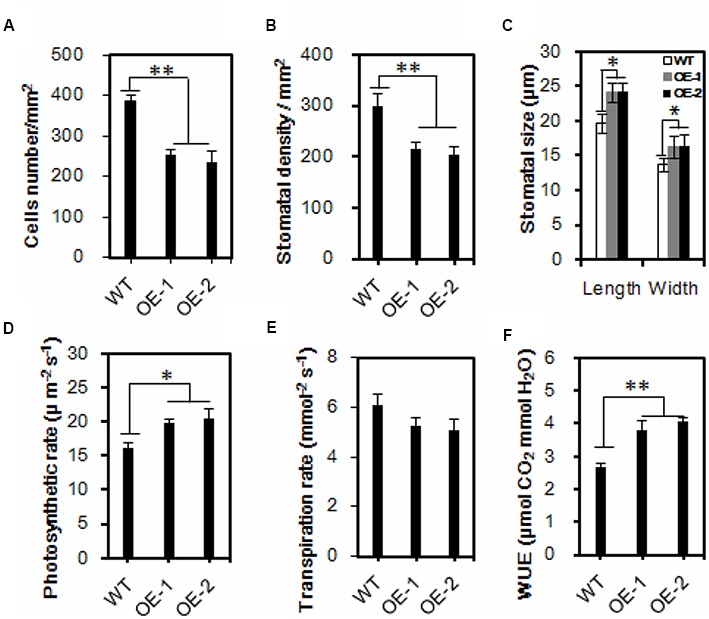
Reduced leaf stomatal density, enlarged stomatal size and increased water use efficiency in the transgenic Chinese kale. (A,B) Comparisons of cells and stomatal density. Three leaves were sampled for each plant, and five plants were sampled for both the control and the transgenic lines. Values are mean ± SD (∗∗p < 0.01). (C) Comparisons of stomatal dimension in transgenic and wild-type plants. Values are mean ± SD (n = 50, ∗∗p < 0.01). (D–F) Comparisons of photosynthesis rate (D), transpiration rate (E), and WUE (F) in wild-type and transgenic Chinese kale plants. Values are mean ± SD (∗P < 0.05, ∗∗P < 0.01). Asterisks indicate Student’s t-test significant differences.
Reduced stomatal density is known to affect water and CO2 exchange. We thus measured photosynthesis and transpiration rates of both the wild-type and the AtEDT1/HDG11-overexpressing Chinese kale plants at the seedling stage. Interestingly, the photosynthesis rate of transgenic line OE-1 and OE-2 was increased 22.5 and 26.8%, respectively, compared to the wild-type control (Figure 4D). Nevertheless, the transpiration rate in transgenic lines is lower than in wild-type plants (Figure 4E). Consequently, the water use efficiency (WUE) was significantly higher than in the wild-type control (Figure 4F).
Transgenic Chinese Kale Was Hypersensitive to ABA
During the seedling stage, the AtEDT1/HDG11-overexpressing lines showed higher sensitivity to ABA (Figure 5A). A test was performed to investigate the response of transgenic and wild-type plants to ABA. Without ABA, the germination rate of transgenic plants did not show an apparent difference from that in wild-type plants (data not shown). However, the transgenic plants demonstrated a dramatically delayed and reduced level of germination in the medium containing ABA, which indicated hypersensitivity of the transgenic plants to ABA (Figures 5B–D). After treatment with 3 μM ABA, only 50.4% of lines OE-1 and 44.7% of lines OE-2 germinated compared to 69.6% of the wild-type. Moreover, under the 1.0 μM ABA treatment, 70% of the wild-type germinated at day 4 compared to only 28.2 and 32.5% of lines OE-1 and OE-2, respectively.
FIGURE 5.
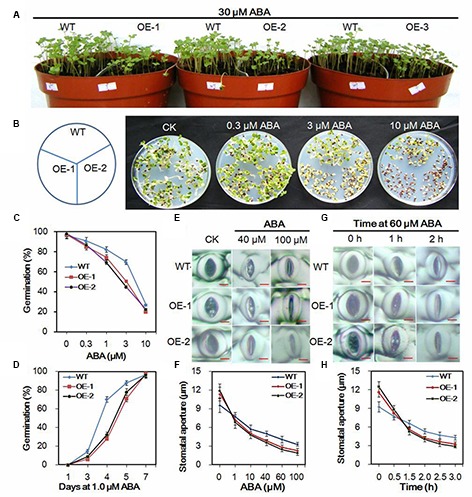
Responses of transgenic and wild-type Chinese kale plants to ABA. (A) Transgenic and wild-type seedlings treated with 30 μM ABA. (B) Seeds of Chinese kale sowed on MS agar plates with designated concentrations of ABA for 5 days. (C) Germination rate of Chinese kale with different concentrations of ABA for 5 days. (D) Germination in the presence of 1.0 μM ABA. (E) ABA-induced stomatal closing in transgenic and wild-type plants. Bar = 5 μm. (F) Stomatal apertures as a function of ABA concentration. Values are mean ± SD (n = 30–50 stomata per data point). (G) ABA (60 μM)-induced stomatal closing. Bar = 5 μm. (H) Stomatal aperture in 60 μM ABA at indicated time point. Values are mean ± SD (n = 30–50 stomata per data point).
Stomatal dynamic variation is a crucial ABA-regulated process. To investigate the ABA induced stomatal closure, experiments were performed at the concentration of 1–100 μM ABA to assay the stomatal closing. Under normal conditions, the stomatal apertures in transgenic plants were larger than in the wild-type plants (Figure 5E). However, after treatment with indicated ABA for 2 h, the guard cells were dramatically changed, and the stomatal apertures were smaller in transgenic lines than those in the wild-type plants (Figures 5E,F). Accordingly, when treated with 60 μM ABA for more than 1 h, the stomatal apertures in transgenic were smaller than in the wild-type plants (Figures 5G,H). Results further indicated that the stomatal closure in the transgenic plants was faster than in the wild-type under the ABA treatment conditions (Figures 5E–H).
Expression Analysis of Phenotype- and the Drought Stress-Related Genes
To uncover phenotype-related molecular mechanisms, the expression levels of 12 phenotype-related genes were investigated. YUC3, YUC5, YUC6, YUC7, and YUC8 belong to the YUC gene family, which encodes the flavin monooxygenases proteins, which play essential roles in auxin biosynthesis and plant development (Cheng et al., 2006), were found to be significantly upregulated in the AtEDT1/HDG11-overexpressing plants (Figure 6A). Moreover, the PIN1, PIN2, PIN3, PIN4, and PIN7 encoding auxin efflux facilitator proteins, which control auxin distribution and regulate cell division and cell expansion (Blilou et al., 2005), were increased. In addition, EXPA5, the cell-wall-loosening protein gene which controls root elongation (Xu et al., 2014) was also significantly upregulated in the transgenic plants (Figure 6A). ERECTA, which encodes a signaling molecule, and is a member of the family of the cell surface receptor kinase, has been demonstrated to enhance vegetative and reproductive organ development and stomata behavior (Torii et al., 1996; Yokoyama et al., 1998; Douglas et al., 2002; Shpak et al., 2004; Masle et al., 2005; Woodward et al., 2005; Bemis et al., 2013). Thus, up-regulation of auxin biosynthesis and response genes may influence the development in transgenic Chinese kale (Figure 6A).
FIGURE 6.
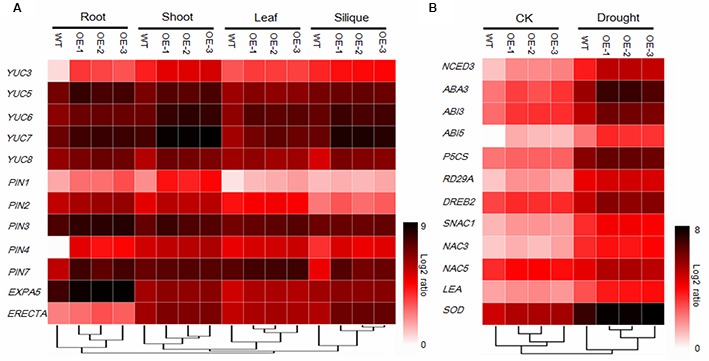
Gene expression analysis of 12 development-related genes (A) and 12 stress-related genes under normal (CK) and drought stress conditions (B) using 384-well block high-throughput qRT-PCR. Relative transcript levels after normalization to reference gene tubulin8. Values are the mean of three independent experiments and expressed as log2 ratios relative to expression in WT, results shown as a heat map. Gene expression profiles were organized by hierarchical clustering.
To better understand the mechanisms of drought and osmotic tolerance in the AtEDT1/HDG11-overexpressing plants, the expression levels of 12 stress-related genes were also investigated. As shown in Figure 6B, the expression levels of NCED3 and LOS5/ABA3, encoding the key enzymes in the ABA synthesis pathway, were up-regulated in the transgenic plants under both normal and drought stress conditions. RD29A, a stress responsive marker gene, was up-regulated. P5CS, encoding a key enzyme in the proline biosynthesis, SOD, encoding the key enzyme in Cu/Zn-SOD synthesis, and LEA, encoding the late embryogenesis abundant (LEA) protein, were strongly induced in the transgenic plants under drought stress. DREB2, encoding the dehydration responsive element binding protein, which induces a set of abiotic stress-related genes and imparts stress endurance to plants, was also up-regulated in transgenic plants under both normal and drought conditions compared to the corresponding wild-type. Moreover, ABA-Insensitive3 (ABI3) and ABA-Insensitive5 (ABI5), which encode proteins for ABA signaling response, were up-regulated in the transgenic plants. In addition, SNAC1, NAC3, and NAC5, which belong to the NAC family of transcription factors and have been well studied in response to drought and/or salt stress, were up-regulated under both the normal and stress conditions.
AtEDT1/HDG11 Can Target the Promoter of ABI3, ABI5, EXPA5, and YUC6
HD-binding cis-elements (also called L1 box cis-elements; Figure 7A) can directly bind to the HD-ZIP IV class proteins (Shan et al., 2014; Xu et al., 2014; Cai et al., 2015). In the studied phenotype, AtEDT1/HDG11 expression was modulated. The promoters of ABI3, ABI5, EXPA5, and YUC6 genes were isolated and analyzed using the PLACE and Plant-CARE databases. Consistent with real-time quantitative PCR results, each of the AtEDT1/HDG11-overexpressing lines up-regulated genes containing HD-binding cis-elements in their promoters (Figure 7B). This result revealed that AtEDT1/HDG11 can directly or indirectly regulate these genes as a transcription activator.
FIGURE 7.
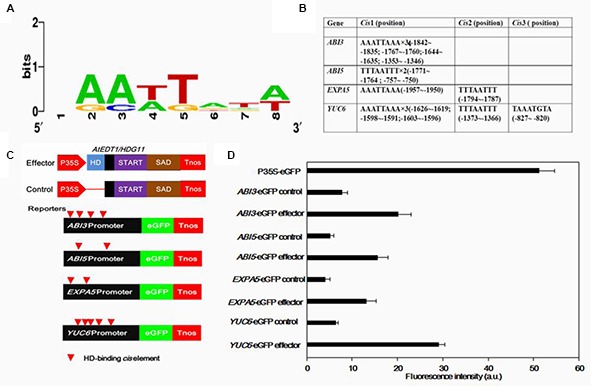
Transient assays for the transactivation of ABI3, ABI5, EXPA5, and YUC6 promoters by AtEDT1/HDG11 in onion epidermal cells. (A) Conservation logo of the HD-binding cis-element (L1-box). (B) Analysis of HD-binding cis-elements in promoters of ABI3, ABI5, EXPA5 and YUC6. (C) Effector containing 35S-AtEDT1/HDG11 or a modified 35S-AtEDT1/HDG11 without the HD domain deleted server as a negative control. Enhanced green fluorescent protein (eGFP) reporter constructs containing the ABI3, ABI5, EXPA5, and YUC6 promoter, respectively. The effector and reporter were transiently transformed into the onion. The 35S-eGFP construct served as a positive control. After incubation for 24 h, GFP fluorescence was observed by confocal microscope. The experiment was repeated at least three times, and a typical result is presented. (D) Quantification fluorescence intensity. Error bars indicate SD. a.u., arbitrary units.
To further demonstrate that AtEDT1/HDG11 acts as a transcription activator, the promoters of ABI3, ABI5, EXPA5, and YUC6 were fused to an enhanced green fluorescent protein (eGFP) reporter (Figure 7C). These reporter constructs were co-transfected into onion cells with either the 35S-AtEDT1/HDG11 construct as effector, or a modified 35S-AtEDT1/HDG11 with the HD domain removed, as a negative effector control. Results indicated that the promoter activity (i.e., increased GFP intensity) of ABI3, ABI5, EXPA5, and YUC6 was significantly improved in the presence of the 35S-AtEDT1/HDG11 effector, but not in the negative control (Figure 7D). In a positive control, the GFP signal was constitutively observed in onion cells which were transfected with the CaMV35S promoter-driven construct P35S-eGFP. The result indicated that the transcription factor AtEDT1/HDG11 was able to target four genes and activated their expression in vivo.
Discussion
Improved Drought, Salinity, and Osmotic Tolerance of the AtEDT1/HDG11-Overexpressing Chinese Kale Was Caused by Multiple Determinants
Chinese kale is an originally Chinese vegetable belonging to the Brassicaceae family, and is widely cultivated in Southeast Asia; however, it is present in relatively small quantities in Europe and America (Sagwansupyakorn, 1994). It is relatively vulnerable to abiotic stresses, such as drought and high salinity, and is classified as an environmentally sensitive crop along with other leafy vegetables. In this study, we evaluated the AtEDT1/HDG11-overexpressing lines of Chinese kale. Our results indicated that drought and osmotic tolerance were significantly improved in transgenic Chinese kale plants. Furthermore, we investigated the factors contributed to the improved drought, salinity and osmotic tolerance in overexpressed lines.
Similar to the AtHDG11 overexpression in Arabidopsis and pepper, the drought tolerance of AtHDG11-overexpressing Chinese kale is contributed to by multiple determinants. The AtEDT1/HDG11-overexpressing Chinese kale plants have a dramatically improved root system, enhanced root hair, lateral root numbers and primary root length (Figure 1; Supplementary Figure 3). To avoid drought-induced stress, plants improve the depth and thickness of root systems allowing them to extract water and nutrients from deep soil layers and thus to minimize the adverse abiotic stress on plant growth (de Dorlodot et al., 2007; Uga et al., 2013). The larger root system in transgenic plants should allow the enhanced root water uptake from the soil to meet the transpirational requirements of the plant (Werner et al., 2010). In addition, the water loss from transgenic plants was significantly decreased (Figure 2E). The majority of plant transpiration occurs through stomata and the transpiration rate can be regulated by stomatal density and/or movement (Yu et al., 2008; Kim et al., 2010). Under water deficit conditions, leaf stomatal density and guard cell size will change in response to water status (Xu and Zhou, 2008). Therefore, the lower transpiration in transgenic Chinese kale was partially attributable to a reduction in stomatal density (Masle et al., 2005; Yu et al., 2008; Yoo et al., 2010).
Likewise, stomatal aperture size is another important determinant for transpiration (Huang et al., 2009). Stomatal closure is one of the crucial ABA-regulated processes activated by dehydration conditions (Leung and Giraudat, 1998; Gonzalez-Guzman et al., 2012). Under water deficit conditions, increased cellular ABA is thought to provoke a reduction in turgor pressure of the guard cells, leading to stomatal closure and subsequent restricted transpiration as a mechanism to adapt to water deficiency (Zhu, 2002). Compared to the corresponding wild-type plants, the transgenic lines guard cells were more sensitive to ABA and drought stress, which led to quick stomatal closure. The smaller stomatal aperture may contribute to reduced water loss from the plant cells, thus enhancing osmotic stress tolerances in the transgenic plants (Huang et al., 2009). The observation that HDG11 up-regulation leads to the ABA hypersensitivity phenotype has not been reported previously until this study; such observation can help us better understand the mechanism of reduced water loss in the transformed lines compared to the untransformed lines.
Drought and osmotic stress can cause oxidative and osmotic damage in plants (Zhu, 2002; Carvalho, 2008). The transgenic plants were better protected from osmotic and oxidative damage by increasing proline and SOD (Figures 2H,I and 3E,F). ROS was overproduced under various environmental stressors such as drought and osmotic. The significantly reduced H2O2 levels detected in the transgenic Chinese kale, under both drought and osmotic conditions (Figures 2J and 3G), indicate that they are more efficient in oxidative scavenging, which contributed to reduced oxidative damage in the transgenic plants.
Molecular Mechanisms Underlying the Drought Tolerance and Auxin Overproduction Phenotypes of AtEDT1/HDG11-Overexpressing Lines
The HD-ZIP transcription factor, AtHDG11, was first found to be involved in trichome branching and other aspects of development (Nakamura et al., 2006; Khosla et al., 2014). Homeodomain transcription factor can directly target HD-binding cis-elements (also called L1 box cis-elements), which has been widely documented (Shan et al., 2014). Transcriptomes were compared between the wild-type and edt1D Arabidopsis roots, and the results revealed that several gene families of cell-wall-loosening proteins and jasmonate biosynthesis and signaling pathways were upregulated in the edt1D root (Xu et al., 2014; Cai et al., 2015). Most of these genes contain HD-binding cis-elements in their promoters predominantly with the TTTAATTT sequence, which can be bound by HDG11 in vitro and in vivo. Similar to Arabidopsis edt1D we also detected EXPANSIN A5 (EXPA5) up-regulated in AtHDG11 overexpression lines. This may contribute, in part, to improved primary root elongation in transformed Chines kale plants. Similarly, in the cotton HD-ZIP, the transcription factor GhHOX3 controls cotton fiber elongation directly by regulating wall loosening protein genes GhRDL1 and GhEXPA1 (Shan et al., 2014). This indicates homeodomain transcription factor has a conserved function in regulation, regulating downstream genes via directly targeting the L1-box cis-element.
The AtEDT1/HDG11-overexpressing lines improvement of drought and osmotic stress was partly attributable to the regulation of a set of stress-related genes. In this study, we found AtEDT1/HDG11 significantly up-regulated the stress response of related genes and transcription factors containing at least one L1-box cis-element (data not shown). We found that ABA synthesis pathways (NCED3 and LOS5/ABA3), the Proline synthesis gene P5CS and stress tolerance-related genes RD29A, LEA, and Cu/Zn SOD were significantly upregulated in transgenic plants under both normal and drought stress conditions (Figure 6B), similarly to previous studies on Arabidopsis and rice (Yu et al., 2008, 2013). In addition, the expression levels of six stress related transcription factors ABI3, ABI5, DREB2, SNAC1, NAC3, and NAC5 (Liu et al., 1998; Finkelstein and Lynch, 2000; Lopez-Molina et al., 2002; Hu et al., 2006), were also found to be significantly up-regulated in AtEDT1/HDG11-overexpressing plants (Figure 6B). It is known that ABI3, a B3-domain transcription factor and ABI5, a basic leucine zipper transcription factor, have been assigned roles in ABA signaling largely based on their involvement in late seed development, particularly in the ABA-dependent induction of LEA genes in the final stages of seed development (Finkelstein and Lynch, 2000; Battaglia et al., 2008). Studies in transgenic plants indicate overexpression of the transcriptional regulators ABI3 or ABI5 that confer hypersensitivity to ABA (Himmelbach et al., 2003; Zou et al., 2008). Moreover, since ABI3 and 14–3–3 proteins are able to form complexes with the bZIP protein ABI5, the resulting dimers interact to regulate the expression of ABA-controlled genes via binding to ABA responsive elements (ABREs) located in their promoters (Finkelstein et al., 2002). It is possible that up-regulation of ABI3 and ABI5 in transgenic Chinese kale augments the ability to respond to stress signals and regulates ABA-induced gene expression (Yu et al., 2008). Interestingly, digital gene expression profile (DGE) analysis of ABI3 and ABI5 genes in the ZH11 wild type and AtEDT1/HDG11-overexpressing rice did not detect significant results, potentially because these are different species.
We found that the auxin biosynthesis YUC gene family and auxin transport PIN gene family were upregulated in AtEDT1/HDG11-overexpressing Chinese kale lines, though not reported in Arabidopsis and rice (Yu et al., 2008, 2013; Xu et al., 2014; Cai et al., 2015). The YUC gene family encodes flavin monooxygenase enzymes which catalyze a rate-limiting step in auxin biosynthesis. It is known that auxin has profound effects on plant growth and development (Zhao, 2010; Kim et al., 2013). Overexpression of the YUC family gene in Arabidopsis and other species always leads to similar auxin overproduction phenotypes, which is consistent with observations in the present study (Figure 1; Supplementary Figure 2). Similar to the auxin biosynthesis gene, we found transport gene PINs were also upregulated in AtEDT1/HDG11-overexpressing lines. It widely known that PINs plays an important role in controlling auxin polar transport and regulating cell division and cell expansion in the primary root (Blilou et al., 2005; Wiśniewska et al., 2006). This may be attributed, to an extent, to the formation of a larger root system in transformed lines. In addition to regulating development, auxin biosynthesis, and transport genes were also involved in stress tolerance. Many auxin biosynthesis, transporter and response genes are involved in plant responses to biotic and abiotic stresses, including drought, high salinity, and pathogen infection (Woo et al., 2007; Ye et al., 2009; Blomster et al., 2011; Cheol Park et al., 2013). Overexpression of AtYUC6 and AtYUC7 in Arabidopsis result in auxin-overproduction phenotypes and up-regulation of stress-response genes, significantly elevating tolerance to drought stress (Lee et al., 2012; Ke et al., 2015). Similarly, the ectopic expression of AtYUC6 in potato also involved auxin overproduction phenotypes and enhanced drought tolerance (Kim et al., 2013). On the contrary, the T-DNA insertional rice mutants in a CONSTITUTIVELY WILTED1 (COW1) gene, which encodes a new member of the YUC protein family, exhibit water-deficient phenotypes of rolled leaves and reduced leaf widths that could result from lower root to shoot ratios which ultimately lead to insufficient water uptake (Woo et al., 2007). Recently, the Arabidopsis plants overexpressing YUC6 displayed enhanced IAA-related phenotypes and exhibited improved drought stress tolerance, low rates of water loss and controlled ROS accumulation under drought and oxidative stresses (Cha et al., 2015). This result demonstrated a double function of YUC6, which acts as a flavin monooxygenase in auxin biosynthesis and as a FAD- and NADPH-dependent thiol-reductase in the stress response. These studies demonstrate that the stress-related phenotype observed in plants that overexpress YUC6 is not based on IAA overproduction but on its activity as a thiol-reductase. In this study, the regulation of YUC6 and its family members by HDG11 suggests that up-regulation of YUCs in the HDG11 Brassica overexpression lines may not only control auxin biosynthesis but also enhance ROS scavenging under drought and osmotic stress conditions (Figures 2J and 3G).
Taken together, our study indicated that overexpressed AtEDT1/HDG11 in the Chinese kale enhances drought, salinity, and osmotic tolerance and improves biomass. We have analyzed expression patterns of several important auxins, ABA, and stress-related genes. Future transcriptome analysis of the wild-type and the transgenic Chinese kale should help to uncover other key genes involved in abiotic tolerance and plant development.
Author Contributions
Conceived and designed the experiments: ZZ, JL, and CC. Performed the experiments: ZZ, XX, and BS. Analyzed the data: HC, ZZ, and LZ. Wrote the paper: ZZ, JL, and CC. Revised the manuscript: CC and BC.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are kindly thankful to Prof. Xiang Chengbin (University of Science and Technology of China) for providing the pCB2004-HDG11 plasmid vector.
Funding. This work was supported by the key project of Guangdong Science and Technology Section (2010A020102001, 2013B051000069 and 2014B020202005), the key project of Guangzhou Science and Technology Section and Agricultural Section (2011Y2-0001511BppZXbb3140003), the National Natural Science Foundation of China (31401874) and the Principal Foundation of South China Agricultural University (K13012) Guangzhou Collaborative Innovation of Industry-Study-Research projects (201508030021).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01285
References
- Abràmoff M. D., Magalhães P. J., Ram S. J. (2004). Image processing with ImageJ. Biophotonics Int. 11 36–42. [Google Scholar]
- Agalou A., Purwantomo S., Overnas E., Johannesson H., Zhu X., Estiati A., et al. (2008). A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol. Biol. 66 87–103. 10.1007/s11103-007-9255-7 [DOI] [PubMed] [Google Scholar]
- Ariel F. D., Manavella P. A., Dezar C. A., Chan R. L. (2007). The true story of the HD-Zip family. Trends Plant Sci. 12 419–426. 10.1016/j.tplants.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Ashraf M. (2010). Inducing drought tolerance in plants: recent advances. Biotech. Adv. 28 169–183. 10.1016/j.biotechadv.2009.11.005 [DOI] [PubMed] [Google Scholar]
- Bates L. S., Waldren R. P., Teare I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39 205–207. 10.1007/BF00018060 [DOI] [Google Scholar]
- Battaglia M., Olvera-Carrillo Y., Garciarrubio A., Campos F., Covarrubias A. A. (2008). The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 148 6–24. 10.1104/pp.108.120725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis S. M., Lee J. S., Shpak E. D., Torii K. U. (2013). Regulation of floral patterning and organ identity by Arabidopsis ERECTA-family receptor kinase genes. J. Exp. Bot. 64 5323–5333. 10.1093/jxb/ert270 [DOI] [PubMed] [Google Scholar]
- Berger D., Altmann T. (2000). A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 14 1119–1131. 10.1101/gad.14.9.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., et al. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433 39–44. 10.1038/nature03184 [DOI] [PubMed] [Google Scholar]
- Blomster T., Salojärvi J., Sipari N., Brosché M., Ahlfors R., Keinänen M., et al. (2011). Apoplastic reactive oxygen species transiently decrease auxin signaling and cause stress-induced morphogenic response in Arabidopsis. Plant Physiol. 157 1866–1883. 10.1104/pp.111.181883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X. T., Xu P., Wang Y., Xiang C. B. (2015). Activated expression of AtEDT1/HDG11 promotes lateral root formation in Arabidopsis mutant edt1 by upregulating jasmonate biosynthesis. J. Integr. Plant Biol. 57 1017–1030. 10.1111/jipb.12347 [DOI] [PubMed] [Google Scholar]
- Carvalho M. D. (2008). Drought stress and reactive oxygen species. Plant Signal. Behav. 3 156–165. 10.4161/psb.3.3.5536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J. Y., Kim W. Y., Kang S. B., Kim J. I., Baek D., Jung I. J., et al. (2015). A novel thiol-reductase activity of Arabidopsis YUC6 confers drought tolerance independently of auxin biosynthesis. Nat. Commun. 6:8041 10.1038/ncomms9041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Liu S., Wu J., Fang L., Sun S., Liu B., et al. (2011). BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 11:1 10.1186/1471-2229-11-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Gene Dev. 20 1790–1799. 10.1101/gad.1415106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheol Park H., Cha J. Y., Yun D. J. (2013). Roles of YUCCAs in auxin biosynthesis and drought stress responses in plants. Plant signal. Behav. 8 337–349. 10.4161/psb.24495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Dorlodot S., Forster B., Pagès L., Price A., Tuberosa R., Draye X. (2007). Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci. 12 474–481. 10.1016/j.tplants.2007.08.012 [DOI] [PubMed] [Google Scholar]
- Douglas S. J., Chuck G., Dengler R. E., Pelecanda L., Riggs C. D. (2002). KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell, 14 547–558. 10.1105/tpc.010391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. R., Gampala S. S., Rock C. D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14 S15–S45. 10.1105/tpc.010441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. R., Lynch T. J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12 599–609. 10.1105/tpc.12.4.599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M., Pizzio G. A., Antoni R., Vera-Sirera F., Merilo E., Bassel G. W., et al. (2012). Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24 2483–2496. 10.1105/tpc.112.098574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A., Yang Y., Grill E. (2003). Relay and control of abscisic acid signaling. Curr. Opin. Plant Biol. 6 470–479. 10.1016/S1369-5266(03)00090-6 [DOI] [PubMed] [Google Scholar]
- Hu H., Dai M., Yao J., Xiao B., Li X., Zhang Q., et al. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. U.S.A. 103 12987–12992. 10.1073/pnas.0604882103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. Y., Chao D. Y., Gao J. P., Zhu M. Z., Shi M., Lin H. X. (2009). A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 23 1805–1817. 10.1101/gad.1812409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J. S., Kim Y. S., Baek K. H., Jung H., Ha S. H., Do Choi Y., et al. (2010). Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 153 185–197. 10.1104/pp.110.154773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J. S., Kim Y. S., Redillas M. C., Jang G., Jung H., Bang S. W., et al. (2013). OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 11 101–114. 10.1111/pbi.12011 [DOI] [PubMed] [Google Scholar]
- Ke Q., Wang Z., Ji C. Y., Jeong J. C., Lee H. S., Li H., et al. (2015). Transgenic poplar expressing Arabidopsis YUCCA6 exhibits auxin-overproduction phenotypes and increased tolerance to abiotic stress. Plant Physiol. Bio. 94 19–27. 10.1016/j.plaphy.2015.05.003 [DOI] [PubMed] [Google Scholar]
- Khosla A., Boehler A. P., Bradley A. M., Neumann T. R., Schrick K. (2014). HD-Zip proteins GL2 and HDG11 have redundant functions in Arabidopsis trichomes, and GL2 activates a positive feedback loop via MYB23. Plant Cell 26 2184–2200. 10.1105/tpc.113.120360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I. J., Baek D., Park H. C., Chun H. J., Oh D. H., Lee M. K., et al. (2013). Overexpression of Arabidopsis YUCCA6 in potato results in high-auxin developmental phenotypes and enhanced resistance to water deficit. Mol. plant 6 337–349. 10.1093/mp/sss100 [DOI] [PubMed] [Google Scholar]
- Kim T. H., Böhmer M., Hu H., Nishimura N., Schroeder J. I. (2010). Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61 561–591. 10.1146/annurev-arplant-042809-112226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M., Murty M. V., Aragones D. V. (2000). Characteristics of root growth and water uptake from soil in upland rice and maize under water stress. Soil. Sci. Plant Nutr. 46 721–732. 10.1080/00380768.2000.10409137 [DOI] [Google Scholar]
- Lee M., Jung J. H., Han D. Y., Seo P. J., Park W. J., Park C. M. (2012). Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis. Planta 235 923–938. 10.1007/s00425-011-1552-3 [DOI] [PubMed] [Google Scholar]
- Leung J., Giraudat J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Biol. 49 199–222. 10.1146/annurev.arplant.49.1.199 [DOI] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., et al. (1998). Two transcription factors, DREB1 and DREB 2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought-and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10 1391–1406. 10.1105/tpc.10.8.1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., McLachlin D. T., Chait B. T., Chua N. H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32 317–328. 10.1046/j.1365-313X.2002.01430.x [DOI] [PubMed] [Google Scholar]
- Loukehaich R., Wang T., Ouyang B., Ziaf K., Li H., Zhang J., et al. (2012). SpUSP, an annexin-interacting universal stress protein, enhances drought tolerance in tomato. J. Exp. Bot. 63 5593–5606. 10.1093/jxb/ers220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masle J., Gilmore S. R., Farquhar G. D. (2005). The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature 436 866–870. 10.1038/nature03835 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Katsumata H., Abe M., Yabe N., Komeda Y., Yamamoto K. T., et al. (2006). Characterization of the class IV homeodomain-leucine zipper gene family in Arabidopsis. Plant Physiol. 141 1363–1375. 10.1104/pp.106.077388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A., Engström P., Söderman E. (2004). The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol. Biol. 55 663–677. 10.1007/s11103-004-1581-4 [DOI] [PubMed] [Google Scholar]
- Ouyang S. Q., Liu Y. F., Liu P., Lei G., He S. J., Ma B., et al. (2010). Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 62 316–329. 10.1111/j.1365-313X.2010.04146.x [DOI] [PubMed] [Google Scholar]
- Pan X., Welti R., Wang X. (2010). Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 5 986–992. 10.1038/nprot.2010.37 [DOI] [PubMed] [Google Scholar]
- Price A. H., Tomos A. D. (1997). Genetic dissection of root growth in rice (Oryza sativa L.). II: mapping quantitative trait loci using molecular markers. Theor. Appl. Genet. 95 143–152. 10.1007/s001220050542 [DOI] [Google Scholar]
- Ré D. A., Capella M., Bonaventure G., Chan R. L. (2014). Arabidopsis AtHB7 and AtHB12 evolved divergently to fine tune processes associated with growth and responses to water stress. BMC Plant Biol. 14:1 10.1186/1471-2229-14-150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagwansupyakorn C. (1994). Brassica oleracea L. cv. group. Chinese kale. Plant Resour. South East Asia 8 115–117. [Google Scholar]
- Shan C. M., Shangguan X. X., Zhao B., Zhang X. F., Chao L. M., Yang C. Q., et al. (2014). Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat. Commun. 5 5519 10.1038/ncomms6519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58 221–227. 10.1093/jxb/erl164 [DOI] [PubMed] [Google Scholar]
- Shpak E. D., Berthiaume C. T., Hill E. J., Torii K. U. (2004). Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131 1491–1501. 10.1242/dev.01028 [DOI] [PubMed] [Google Scholar]
- Shpak E. D., McAbee J. M., Pillitteri L. J., Torii K. U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309 290–293. 10.1126/science.1109710 [DOI] [PubMed] [Google Scholar]
- Söderman E., Mattsson J., Engström P. (1996). The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. Plant J. 10 375–381. 10.1046/j.1365-313X.1996.10020375.x [DOI] [PubMed] [Google Scholar]
- Torii K. U., Mitsukawa N., Oosumi T., Matsuura Y., Yokoyama R., Whittier R. F., et al. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8 735–746. 10.1105/tpc.8.4.735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga Y., Sugimoto K., Ogawa S., Rane J., Ishitani M., Hara N., et al. (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45 1097–1102. 10.1038/ng.2725 [DOI] [PubMed] [Google Scholar]
- Von Groll U., Berger D., Altmann T. (2002). The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 14 1527–1539. 10.1105/tpc.001016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Nehnevajova E., Köllmer I., Novák O., Strnad M., Krämer U., et al. (2010). Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 22 3905–3920. 10.1105/tpc.109.072694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewska J., Xu J., Seifertová D., Brewer P. B., Rùžièka K., Blilou I., et al. (2006). Polar PIN localization directs auxin flow in plants. Science 312 883–883. 10.1126/science.1123542 [DOI] [PubMed] [Google Scholar]
- Woo Y. M., Park H. J., Su’udi M., Yang J. I., Park J. J., Back K., et al. (2007). Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol. Biol. 65 125–136. 10.1007/s11103-007-9203-6 [DOI] [PubMed] [Google Scholar]
- Woodward C., Bemis S. M., Hill E. J., Sawa S., Koshiba T., Torii K. U. (2005). Interaction of auxin and ERECTA in elaborating Arabidopsis inflorescence architecture revealed by the activation tagging of a new member of the YUCCA family putative flavin monooxygenases. Plant Physiol. 139 192–203. 10.1104/pp.105.063495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Frugis G., Colgan D., Chua N. H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14 3024–3036. 10.1101/gad.852200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Cai X. T., Wang Y., Xing L., Chen Q., Xiang C. B. (2014). HDG11 upregulates cell-wall-loosening protein genes to promote root elongation in Arabidopsis. J. Exp. Bot. 65 4285–4295. 10.1093/jxb/eru202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Zhou G. (2008). Responses of leaf stomatal density to water status and its relationship with photosynthesis in a grass. J. Exp. Bot. 59 3317–3325. 10.1093/jxb/ern185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Kang B. G., Osburn L. D., Li Y., Max C. Z. M. (2009). Identification of the flavin-dependent monooxygenase-encoding YUCCA gene family in Populus trichocarpa and their expression in vegetative tissues and in response to hormone and environmental stresses. Plant Cell Tiss. Org. 97 271–283. 10.1007/s11240-009-9526-x [DOI] [Google Scholar]
- Yokoyama R., Takahashi T., Kato A., Torii K. U., Komeda Y. (1998). The Arabidopsis ERECTA gene is expressed in the shoot apical meristem and organ primordia. Plant J. 15 301–310. 10.1046/j.1365-313X.1998.00203.x [DOI] [PubMed] [Google Scholar]
- Yoo C. Y., Pence H. E., Jin J. B., Miura K., Gosney M. J., Hasegawa P. M., et al. (2010). The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell 22 4128–4141. 10.1105/tpc.110.078691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Chen X., Hong Y. Y., Wang Y., Xu P., Ke S. D., et al. (2008). Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20 1134–1151. 10.1105/tpc.108.058263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Chen X., Wang Z., Wang S., Wang Y., Zhu Q., et al. (2013). Arabidopsis enhanced drought tolerance1/HOMEODOMAIN GLABROUS11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol. 162 1378–1391. 10.1104/pp.113.217596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L. H., Wu S. J., Peng Y. S., Liu R. N., Chen X., Zhao P., et al. (2016). Arabidopsis EDT1/HDG11 improves drought and salt tolerance in cotton and poplar and increases cotton yield in the field. Plant Biotechnol. J. 14 72–84. 10.1111/pbi.12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Haider I., Kohlen W., Jiang L., Bouwmeester H., Meijer A. H., et al. (2012). Function of the HD-Zip I gene Oshox22 in ABA-mediated drought and salt tolerances in rice. Plant Mol. Biol. 80 571–585. 10.1007/s11103-012-9967-1 [DOI] [PubMed] [Google Scholar]
- Zhao Y. (2010). Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61 49–64. 10.1146/annurev-arplant-042809-112308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53 247–273. 10.1146/annurev.arplant.53.091401.14332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Xu X., Cao B., Chen C., Chen Q., Xiang C., et al. (2015). Pyramiding of AtEDT1/HDG11 and Cry2Aa2 into pepper (Capsicum annuum L.) enhances drought tolerance and insect resistance without yield decrease. Plant Cell Tiss. Org. 120 919–932. 10.1007/s11240-014-0600-7 [DOI] [Google Scholar]
- Zou M., Guan Y., Ren H., Zhang F., Chen F. (2008). A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol. Biol. 66 675–683. 10.1007/s11103-008-9298-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


