Abstract
Broad phenotypic variations were obtained previously in derivatives from the asymmetric somatic hybridization of cauliflower “Korso” (Brassica oleracea var. botrytis, 2n = 18, CC genome) and black mustard “G1/1” (Brassica nigra, 2n = 16, BB genome). However, the mechanisms underlying these variations were unknown. In this study, 28 putative introgression lines (ILs) were pre-selected according to a series of morphological (leaf shape and color, plant height and branching, curd features, and flower traits) and physiological (black rot/club root resistance) characters. Multi-color fluorescence in situ hybridization revealed that these plants contained 18 chromosomes derived from “Korso.” Molecular marker (65 simple sequence repeats and 77 amplified fragment length polymorphisms) analysis identified the presence of “G1/1” DNA segments (average 7.5%). Additionally, DNA profiling revealed many genetic and epigenetic differences among the ILs, including sequence alterations, deletions, and variation in patterns of cytosine methylation. The frequency of fragments lost (5.1%) was higher than presence of novel bands (1.4%), and the presence of fragments specific to Brassica carinata (BBCC 2n = 34) were common (average 15.5%). Methylation-sensitive amplified polymorphism analysis indicated that methylation changes were common and that hypermethylation (12.4%) was more frequent than hypomethylation (4.8%). Our results suggested that asymmetric somatic hybridization and alien DNA introgression induced genetic and epigenetic alterations. Thus, these ILs represent an important, novel germplasm resource for cauliflower improvement that can be mined for diverse traits of interest to breeders and researchers.
Keywords: introgression lines, genetic diversity, Brassica oleracea, Brassica nigra, somatic hybridization, epigenetic variation, cauliflower
Introduction
Cauliflower (Brassica oleracea var. botrytis, 2n = 18, CC genome) is a major vegetable crop valued worldwide for its nutrition and flavor. As with other modern crop species, intense selection for preferred traits, and a tendency for inbreeding have resulted in low genetic diversity among cauliflower breeding resources. To address this problem, related species such as black mustard (Brassica nigra, 2n = 16, BB genome), with a large reservoir of genes conferring desirable characteristics, have been proposed as a valuable source of genetic diversity for Brassica crop improvement. In principle, this diversity can be transferred into crops via sexual hybridization and subsequent backcrossing. However, in practice, genetic manipulation using distantly related plants has been severely restricted by difficulties in creating the initial sexual hybrid and by sterility issues in the early backcross generations (Glimelius, 1999; Jauhar, 2006). Asymmetric somatic hybridization is a potential alternative for gene transfer from relatives to cultivated crops, especially when wide crosses are not applicable (Gerdemann et al., 1994; Xia, 2009). The Brassicaceae is a model plant family commonly used for somatic hybridization (Glimelius, 1999; Navrátilová, 2004), resulting in interspecies or even intertribal hybrids. Moreover, agronomically important traits, such as disease resistance and specific fatty acid compositions, have been successfully integrated into the crops (Gerdemann et al., 1994; Hansen and Earle, 1994, 1995, 1997; Wang et al., 2003; Tu et al., 2008; Scholze et al., 2010).
Brassica is also an excellent model for the study of allotetraploid speciation. Brassica is closely related to the classic plant model Arabidopsis and a wealth of germplasm exists from species in this genus. Three diploid species are widely cultivated: Brassica rapa (AA, x = 10), B. nigra (BB, x = 8), and B. oleracea (CC, x = 9). Recent interspecific hybridization among these species has resulted in polyploidization and the production of three allotetraploid species: Brassica napus (AACC, x = 19), Brassica juncea (AABB, x = 18), and Brassica carinata (BBCC, x = 17) (Nagaharu, 1935).
The formation of allotetraploids is an influential mode of speciation in flowering plants, often accompanied by rapid, extensive genomic and epigenetic changes that globally alter gene expression, termed “genomic shock.” These include fragment gain and loss through chromosome rearrangement or the activation of transposable elements (Kashkush et al., 2003; Kraitshtein et al., 2010), extensive alteration of DNA cytosine methylation (Zhao et al., 2011; Gautam et al., 2016), histone modification (Ha et al., 2011), and changes to small RNA (Ha et al., 2009). However, genomic-shock-induced changes during sexual polyploid synthesis should be distinct from the changes that occur from genomic shock during somatic hybridization. Somatic hybrids combine both the nuclear and cytoplasmic genomes within a single cell. Therefore, the introgression of chromatin segments via asymmetric somatic hybridization likely occurs via non-homologous end-joining of fragmented genome pieces, rather than by the homologous recombination that occurs through sexual reproduction (Liu et al., 2015). Moreover, the epigenetic states of somatic cells and gametal cells tend to differ, given that gametal cells are more conserved to ensure genetic fidelity (Bird, 1997, 2002; Liu et al., 2015). Thus, the variations induced by “somatic genomic shock” likely have unique genetic and epigenetic characteristics compared with “sexual genomic shock.” However, little data exist on the exact nature of these differences, or if the types of genomic shock indeed differ in their effects.
A number of hybrid progenies have been regenerated from asymmetric somatic hybrids between cauliflower “Korso” and black mustard “G1/1.” The hetero-cytoplasmic nature of these hybrids was confirmed through the finding that most progenies have chloroplast genomic components from “G1/1” and mitochondrial DNA from “Korso,” while mitochondrial genome recombination occurred in a few hybrids (Wang G. X. et al., 2011). Among the progenies, dozens of lines containing 18 chromosomes showed some obvious characters of “G1/1” origin, such as waxless leaves with lobes and ears, as well as green petioles; however, they were also densely covered with trichomes and exhibited purple stems and leaf veins, characteristic of “Korso.” This combination of characters indicated that they were putative introgression lines (ILs). Although hybrid synthesis primarily focused on the transfer of disease-resistance genes from “G1/1” into cultivated cauliflower (Wang G. X. et al., 2011) because the former is resistant to several Brassica pathogens (Westman et al., 1999), the putative ILs contained considerable genetic diversity for many traits beyond disease resistance. Therefore, these lines should possess many characters that differ from near-isogenic lines and feature discrete portions of “G1/1” chromatin in a “Korso” genetic background. Additionally, asymmetric somatic introgression should induce further diverse genomic variations involving change in DNA sequences and epigenetic modifications. To verify this, we used 12 putative ILs shown to be phenotypically stable for several generations to characterize genetic and epigenetic alterations from “somatic genomic shock.”
Materials and methods
Plant materials
Asymmetric somatic hybridization between cauliflower “Korso” and black mustard “G1/1” resulted in the establishment of 117 individual hybrids in the field; of these, 13 fertile plants with preferred traits were selected for continued selfing and backcrossing (Wang G. X. et al., 2011). Hundreds of derivatives were obtained following year-by-year selection (since 2006) that combined phenotypic observation, cytological and molecular analysis, as well as pathogen-resistance assays (Figure 1). In this study, 28 putative ILs were chosen for phenotypic diversity and chromosomal constitution analysis, based on possession of cauliflower-like morphology and phenotypic stability over three generations. Next, 12 putative ILs (IL1-12) were used for genetic an epigenetic analysis. Chinese cabbage “Asko” (B. rapa, AA, 2n = 20) and tetraploid B. carinata (BBCC, 2n = 34) were chosen as reference materials for molecular and chromosomal analysis. The selfed seeds from ILs, parents, and B. carinata were sown in a greenhouse and are available upon request.
Figure 1.
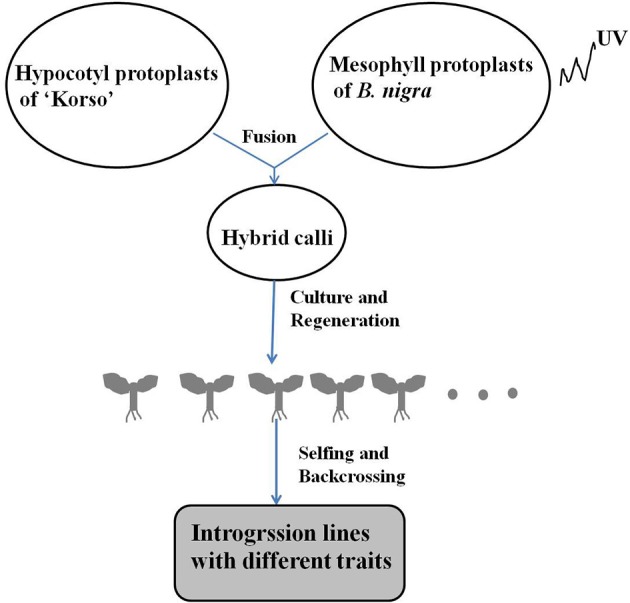
Generation of introgression lines via somatic hybridization.
Multi-color FISH analysis
Multi-color FISH was conducted on meiosis metaphase I chromosomes of parent lines and putative ILs to confirm their chromosomal composition. B. nigra and CentBr (centromeric-specific tandem repeats of Brassica; accession numbers: CW978699 and CW978837, respectively; Wang G. et al., 2011) DNA were selected as probes and labeled with biotin (dig)-14-dUTP (Roche, Indianapolis, IN) using nick translation with an average length of 500 bp. Slides were prepared following a previously published protocol (Zhong et al., 1996) with minor modifications. To decompose the cell walls of pollen mother cells, anthers approximately 1–3 mm in length were digested at 37°C for 3 h in an enzyme mixture containing 6% cellulose R-10 and 6.5% pectinase (Sigma, solution in 40% glycerol). Previously described methods (Wang G. et al., 2011) were followed for FISH analysis. Hybridized probes were visualized using an FITC-conjugated avidin antibody or rhodamine-conjugated anti-digoxin antibody (Roche, Indianapolis, IN). Chromosomes were counterstained using 0.1 mg/mL DAPI (Vector Laboratories, Burlingame, CA). Images of the signals and chromosomes were captured using a CCD camera (QIMAGING, RETIGA-SRV, FAST1394) attached to a Nikon Eclipse 80i epifluorescence microscope (Tokyo, Japan). Image contrast and brightness were adjusted in Adobe Photoshop (8.0).
SSR, AFLP, and MSAP fingerprinting
Simple sequences repeat (SSR), amplified fragment length polymorphism (AFLP), and methylation-sensitive amplification polymorphism (MSAP) fingerprinting were performed following previously described methods (Shaked et al., 2001; Liu et al., 2015). Sixty-five SSR, 77 AFLP, and 16 MSAP primer combinations were used; for SSR and AFLP, markers were collected from the A genome of B. rapa due to the limited number of cytological and genetic studies on the B genome of B. nigra (Tables S1–S3). For AFLP and MSAP, two independent technical replicates were performed for each of the three biological replicates. Only clear and reproducible bands larger than 100 bp were scored.
Results
Phenotypic diversity of introgression lines
The 28 ILs were extremely diverse in multiple morphological and physiological traits (Table 1), including distinct variations in leaf or flower morphology, fertility, and resistance to pathogens (i.e., black rot or clubroot). Here we describe diagnostic differences in flower heads as an example. Four phenotypic groups were apparent based on flower curd characters (Figure 2). In the first group (type I), flower heads were white, compact, and hemispheric, with a wheel-like radial arrangement; flower buds were small, granular, and close. The second group (type II) possessed flat, loose flower heads with no obvious wheel-like arrangement; flower buds were light yellow, tiny, and soft, while the pedicel was green and slightly longer than Type I pedicels. The third group (type III) possessed relatively flat flower heads and pale yellow-green color; flower buds were small, loose, soft, and distributed in clusters, but with no obvious ball flower formation. The pedicel was green and long. The flower heads of the fourth group (type IV) were relatively flat, yellow green, with small flower buds that were fine and soft.
Table 1.
Phenotypic traits associated with introgression lines (IL1-12) and their parents.
| Materials | Leaf traits | Curd traits | Other special traits | Curd morphology |
|---|---|---|---|---|
| “Korso” | Ovate; shallowly serrated margins; lacking auricles; waxy, glossy, and hairless surface | White and firm curd; short pedicel | Susceptible to both black rot Xcc15 and clubroot race 4; potassium content: 156 mg/100 g; phosphorus content: 36 mg/100 g |  |
| “G1/1” | Almost triangular; possesses auricles; green; and wax-free surface | No curd; direct flowering | Resistant to both Xcc15 and clubroot race 4 |  |
| IL1 | Grayish green; possesses auricles | Yellow and firm curd; short pedicel |  |
|
| IL2 | Pale grayish green; shallowly serrated margins; possesses auricles | White and firm curd; purple florets, short pedicel | Resistant to Xcc15 |  |
| IL3 | Green; serrated margins; no auricles | Pale and greenish curd; long pedicel | Early inflorescent; phosphorus content: 73 mg/100 g |  |
| IL4 | Light grayish green; serrated margins; no auricles | White and firm curd; short pedicel |  |
|
| IL5 | light green; wavy margin; possesses auricles | Pale green and soft curd; long pedicel |  |
|
| IL6 | Pale grayish green; wavy margin; wing-like | White; firm, medium curd; white florets | Resistant to clubroot race 4 |  |
| IL7 | Pale grayish green; elongated; no auricles | No obvious curd; flower buds in clusters with leaf folder |  |
|
| IL8 | Light grayish green; serrated margins; no auricles | Pale yellow and scattered curd; long pedicel |  |
|
| IL9 | Green; serrated margins; possesses auricles | Yellowish and loose curd; long pedicel | Potassium content: 236 mg/100 g |  |
| IL10 | Pale grayish green; serrated margins; possesses auricles | Creamy white and loose curd; purple flower buds; long pedicel |  |
|
| IL11 | Similar to parent “Korso” | Creamy white and soft tomentose curd; short pedicel | Resistant to Xcc15 |  |
| IL12 | Pale grayish green; serrated margins; possesses auricles | Yellowish green and soft curd; purple flower buds; long pedicel | Early inflorescent |  |
Figure 2.
Four types (I–IV) of representative flower heads in introgression lines. C: “Korso,” B: “G1/1.”
Chromosomal constitution of the putative introgression lines
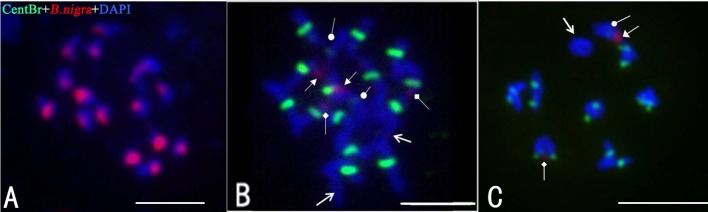
Figure 3 shows the results of multi-color FISH on chromosomes of “G1/1,” “Korso,” and 28 putative ILs. The two parent lines exhibited different hybridization patterns with both probes. As expected, when using “G1/1” genomic DNA, all 16 “G1/1” chromosome pairs exhibited red signals (Figure 3A). In “Korso,” red signals (2 strong and 2 weak; Figure 3B) were detected on only two pairs of satellite chromosomes that likely correspond to the hybridization signals of 45SrDNA (Fukui et al., 1998).
Figure 3.
Fluorescence in situ hybridization (FISH) analysis of the introgression lines using CentBr and “G1/1” genomic DNA as probes. (A) Meiosis metaphase I of “G1/1.” (B) Mitosis metaphase I of “Korso.” (C) Meiosis prometaphase I of IL2. Triangle- and diamond-headed pointers indicate strong and weak red signals, respectively; circle-headed pointers indicate weak green signals; regular arrows indicate chromosomes with no signals. Bar = 5 μm.
No signals were detected on “G1/1” chromosomes using the CentBr probe. In contrast, seven pairs of strong signals and one pair of weak signals were detected on “Korso” chromosomes (Figure 3B); the latter was hybridized to chromosomes that also exhibited strong red signals. Only 1 “Korso” chromosome pair exhibited no signals (Figure 3B).
Most cells in the putative ILs contained 18 chromosomes and exhibited hybridization patterns identical to “Korso.” The results of genomic in situ hybridization (GISH) found no “G1/1” chromosomal segments present in these putative ILs, likely because GISH cannot detect introgressions from Brassica chromosomes due to their small size and compact structure.
Genetic sequence variation analyzed using SSR and AFLP markers
We analyzed the sequence introgression and variation in 12 ILs (IL1–12) using 65 SSR and 77 AFLP markers that were polymorphic between the parents. The results of SSR and AFLP profiling revealed 1799 loci from both parents, including 682 (37.9%) those were polymorphic. The IL profiles were similar to the “Korso” profile, but loss of 5–13 “Korso”-specific bands were common. Additionally, all putative ILs were confirmed to possess anywhere from 3 to 33 “G1/1”-derived fragments. However, we noted that in 3 ILs, SSR markers failed to detect “G1/1”-derived bands but AFLP profiling revealed 20–25 “G1/1”-specific loci, indicating that the latter method is more effective when the introgression size is small. Next, all 12 ILs contained B. carinata-specific loci (35–49), and we were able to amplify 1–7 new bands from them.
These results clearly showed that all 12 lines were ILs and that genomic-sequence variations were common among them. Notably, IL genomes were not simply similar to the “Korso” genome but with some “G1/1”-derived fragments added. For example, we noted that some ILs lacked bands present in “Korso,” or possessed fragments lacking in “Korso.” Moreover, several B. carinata-specific bands were present in all ILs, hinting at the occurrence of common changes in the early stages of both somatic hybridization and naturally occurring hybridization.
DNA profiling of the ILs revealed 1493 “Korso,” 1423 “G1/1,” and 1559 B. carinata fragments; 1117 of these were shared by both parents (Figure 4). The number of fragments present in the profiles of the 12 representative ILs ranged from 3 to 33, with an overall frequency of 7.5%, whereas the number of fragments lost ranged from 7 to 32, with an overall frequency of 5.1% (Table 2). Note that the percentages of “G1/1”-fragment presence and “Korso”-fragment loss were respectively based on the 306 “G1/1” fragments that did not co-migrate with “Korso” and the 376 “Korso” fragments that did not co-migrate with “G1/1.” In addition, the 12 ILs possessed 1–7 new fragments (1.4%) that were not present in the “Korso” profile, and 4–49 (15.5%) B. carinata-specific fragments from the 226 that did not co-migrate with “Korso” and “G1/1.”
Figure 4.

Amplified fragment length polymorphism (AFLP) fingerprinting of the parents and ILs, with primer pair E45–M57. C: “Korso,” B: “G1/1,” BC: B. carinata, 1–11: introgression lines. Arrows indicate “Korso”-specific bands, diamond-headed pointers indicate “G1/1”-specific bands, and circle-headed pointers indicate B. carinata-specific bands.
Table 2.
DNA profiles and phenotypic summary of partial introgression lines (IL1-12).
| Introgression lines (ILs) | No. of B. nigra-specific bands and markers | Deletions in ILs | New bands in ILs | B. carinata-specific bands and markers | Phenotype |
|---|---|---|---|---|---|
| IL1 | 25: (23 AFLP), BrID10227, nia-m037a | 12: (8 AFLP), BrID10205, nia-m037a, At3g55005a, cnu-m472a | 7: (6 AFLP), BrID10223 | 40: (32 AFLP), BrID10205, BrID101161, BrID10223, At3g55005a, cnu-m472a, cnu-m052a | Leaf color and leaf type like the cauliflower; small, white, and firm flower head |
| IL2 | 26: (25 AFLP), BrID10227 | 13: (8 AFLP), BrID10205, BrID10381, BrID10173, nia-m037a | 6 AFLP | 35: (32 AFLP), BrID10985, BrID101161, BrID10381 | Leaf color and leaf type like the cauliflower; small, white, and firm flower head |
| IL3 | 31: (29 AFLP), BrFLC2(R2), BrID10227 | 16: (9 AFLP), BrID10277, BrID10205, BrFLC2(R2), nia-m037a, cnu-m472a, nia-m063 | 6 AFLP | 49: (41 AFLP), BrID10277, BrID10205, BrFLC2(R2), BrID101161, cnu-m472a, cnu-m052a | Leaf color and leaf type like the cauliflower; small, white, and firm flower head |
| IL4 | 20: (18 AFLP), BrFLC2(R2), BrID101081 | 23: (8 AFLP), BrID10277, BrID10205, BrFLC2(R2), BrID10881, BrID101157, BrID101183, BrID90311, BrID10493, BrID10223, BrID10173, nia-m037a, nia-m063a | 6 AFLP | 40: (32 AFLP), BrID10277, BrFLC2(R2), BrID90430, BrID101161, cnu-m052a | Leaf color and leaf type like the cauliflower; small, white, and firm flower head |
| IL5 | 22: (21 AFLP), At4g36960 | 26: (9 AFLP), BrID10277, BrID10205, BrID10881, BrID101157, BrID101161, BrID10971, BrID10381, BrID90311, BrID10493, BrID10173, nia-m037a, At3g55005a, nia-m063a, At3g51260 | 6 AFLP | 40: (33 AFLP), BrID90430, BrID10381, BrID10493, At3g55005a, nia-m063a, cnu-m052a | Leaf color and leaf type like the cauliflower; loose, light yellow flower head; some light purple areas |
| IL6 | 27: (25 AFLP), BrID10227, At4g36960 | 11: (7 AFLP), BrID10205, BrID10493, cnu-m472a | 6 AFLP | 35: (31 AFLP), BrID10493, cnu-m472a, cnu-m052a | Leaf color and leaf type like the cauliflower; small, white, and firm flower head |
| IL7 | 32: (31 AFLP), At4g36960 | 30: (9 AFLP), BrID10277, BrID10205, BrID10881, BrID10381, BrID101183, BrID10971, BrID90311, BrID10103, BrID10493, BrID10101, BrID10275, BrID101199, nia-m037a, At3g55005a, cnu-m472a, At3g51260 | 6 AFLP | 48: (40 AFLP), BrID10277, BrID90430, BrID101161, BrID10381, At3g55005a, cnu-m472a, cnu-m052a | Leaf color and leaf type like the cauliflower; white and firm flower head |
| IL8 | 27: (25 AFLP), BrID10985, BrFLC2(R2) | 32: (13 AFLP), BrID10205, BrFLC2(R2), BrID101157, BrID101183, BrID90311, BrID10493, BrID10101, BrID10971, BrID10275, BrID101199, BrID10173, At1g58220, nia-m037a, At3g55005a, At4g36960, At3g51260 | 6 AFLP | 49: (41 AFLP), BrID10277, BrID10985, BrID10205, BrFLC2(R2), BrID90430, At3g55005a, cnu-m052a | Waxy leaf and loose flower head |
| IL9 | 33: (31 AFLP), BrFLC2(R2), BrID10227 | 17: (11 AFLP), BrID10277, BrID10205, BrFLC2(R2), BrID101157, nia-m037a, cnu-m472a | 6 AFLP | 45: (38 AFLP), BrID10277, BrID10205, BrFLC2(R2), BrID101161, cnu-m472a, cnu-m052a | Loose flower head |
| IL10 | 25 AFLP | 17: (8 AFLP), BrID10881, BrID101157, BrID101183, BrID10971, BrID10493, nia-m037a, At3g55005a, At3g51260 | 6 AFLP | 34: (31 AFLP), BrID101161, At3g55005a, At3g51260 | Waxy leaf and loose flower head |
| IL11 | 3: BrID10227, nia-m037a, At3g55005a | 7: BrID10277, BrID10205, BrID101157, BrID10381, nia-m037a, At3g55005a, Ni4-B10 | 1: Ni4-B10 | 4: BrID10277, BrID10205, BrID10381 | Ligule present; small, tight flower head |
| IL12 | 5: BrID10277, BrID10985, nia-m037a, At3g55005a, At2g38130 | 27: BrID10205, BrFLC2(R2), BrID10881, BrID101157, BrID101183, BrID90311, BrID10971, BrID10103, BrID10493, BrID10101, BrID10275, BrID101199, BrID10173, At1g58220, nia-m037a, At3g55005a, cnu-m472a, nia-m063a, Ni4-B10, At3g51260 | 2: BrID10223, Ni4-B10 | 5: BrID10277, BrID10223, BrID90430, At2g38130, nia-m063a | Gray-green leaf color; firm, small flower head |
| Average | 23.0 | 19.3 | 5.3 | 35.3 | |
| Frequency | 7.5% | 5.1% | 1.4% | 15.5% |
Epigenetic changes in methylation patterns analyzed with MSAP
The MSAP analysis used isoschizomers MspI and HpaII, which differ in their sensitivity to cytosine methylation (Shaked et al., 2001; Liu et al., 2015). Our comparison of the profiles generated from EcoRI-MspI and EcoRI-HpaII digestion revealed three kinds of band classifications (Figure 5). First, A type was characterized by fragments from both the EcoRI-MspI and EcoRI-HpaII digestions, representing non-methylated sites (or methylation within a single strand). Second, B type was characterized by half-methylation sites, present in EcoRI-HpaII digestion but not in EcoRI-MspI digestion. Third, C-type bands stemmed from complete methylation sites present in EcoRI-MspI digestion but not in EcoRI-HpaII digestion. Using 16 MSAP primer combinations amplifying clear and stable bands, we detected 401 fragments in parents, with 290 present in “Korso” but not in “G1/1.”
Figure 5.
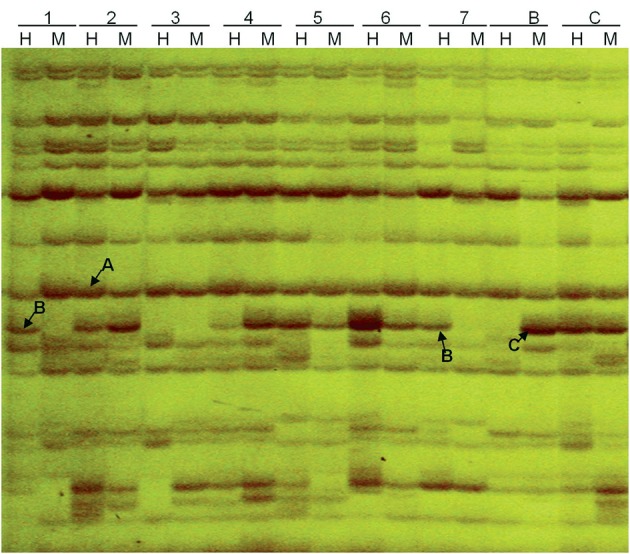
Methylation-sensitive amplified polymorphism (MSAP) profiling based on primer combinations ETTC + HMTCC. 1–7: introgression lines, B: “G1/1,” C: “Korso,” H: DNA cleaved with EcoRI + HpaII, M: DNA cleaved with EcoRI + MspI. A, B, and C indicate three separate band types.
Table 3 summarizes variation in methylation patterns and their heredity in parents and the ILs. The average frequency of hypermethylated loci in different ILs was 12.4%, while the average frequency of hypomethylated loci was 4.8%.
Table 3.
Variation in cytosine methylation across the partial introgression lines (IL1-12).
| Introgression lines (ILs) | Hypermethylation in ILs | Frequency (%) | Hypomethylation in ILs | Frequency (%) |
|---|---|---|---|---|
| IL1 | 42 | 14.5 | 7 | 2.4 |
| IL2 | 36 | 12.4 | 9 | 3.1 |
| IL3 | 35 | 12.1 | 8 | 2.8 |
| IL4 | 39 | 13.5 | 22 | 7.6 |
| IL5 | 28 | 9.7 | 18 | 6.2 |
| IL6 | 32 | 11.0 | 17 | 5.9 |
| IL7 | 38 | 13.1 | 12 | 4.1 |
| IL8 | 28 | 9.7 | 10 | 3.5 |
| IL9 | 34 | 11.7 | 8 | 2.8 |
| IL10 | 42 | 14.5 | 22 | 7.6 |
| IL11 | 40 | 13.8 | 20 | 6.9 |
| IL12 | 36 | 12.4 | 13 | 4.5 |
| Average | 35.8 | 12.4 | 13.8 | 4.8 |
Discussion
Somatic hybridization and introgression can potentially increase genetic diversity
The low genetic diversity in modern Brassica varieties is concerning because it reduces potential genetic gains in breeding programs. To combat this problem, ILs containing fragments from related species can be used to generate improved cultivars. This method has proven successful in many crops, including wheat (Liu et al., 2007), rice (Rangel et al., 2008), potato (Chavez et al., 1988), eggplant (Mennella et al., 2010), B. napus (Primard-Brisset et al., 2005; Leflon et al., 2007), barley (Johnston et al., 2009), tomato (Menda et al., 2014), and rye grass (Roderick et al., 2003).
In this study, we generated a set of ILs using somatic hybridization between cauliflower and black mustard. The original objective was to select disease- and drought-resistant ILs for use in a Brassica breeding program. In fact, these ILs showed considerable genetic diversity in many morphological and physiological traits beyond resistance. Some of these traits are useful for increasing cauliflower adaptation to stressors. For instance, IL2 and IL6 respectively exhibited strong resistance to black rot and clubroot, major Brassica diseases with worldwide distribution. Other ILs exhibited commercially desirable traits such as early maturation and high nutritional quality, especially high phosphorus and potassium content (IL3 and IL9), as well as less desirable traits such as loose flower heads, Early inflorescent, and long pedicel. This diversity indicates that all of the ILs could be a source of new alleles for developing Brassica cultivars. Thus, somatic hybridization provides an effective and efficient means of achieving introgression into a crop species from its wild relatives and should continue to be important in producing novel germplasm for breeding programs.
Combining cytological and molecular methods effectively identified brassica introgression lines
Although GISH has been successfully used to investigate genomic relationships in several agriculturally important genera, including Brassica (e.g., Lysak et al., 2005), and has detected introgressions in crops such as wheat (e.g., Liu et al., 2015), GISH was unable to detect black mustard chromosomal segments in our ILs. We believe this outcome was likely due to the small and compact Brassica chromosomes or else very small introgression sizes. We dealt with this issue by first using multi-color FISH to identify chromosome composition and then combining SSR and AFLP markers to detect “G1/1” sequence introgression. Primer sources and amplification results from the genomes of B. rapa, B. nigra, B. oleracea, and B. carinata are shown in Table S1. As those data indicate, the primers are usable in multiple Brassica species and can detect abundant polymorphism. Moreover, compared with the SSR primers, AFLP markers were more effective when working with small introgressions, such as those in our ILs (Table 2). In sum, our study demonstrated that Brassica ILs can be clearly identified with a combination of cytological and molecular methods.
Genetic changes from somatic hybridization in the brassica introgression lines
The extensive phenotypic variation of the ILs was caused by genetic and epigenetic changes in the ILs relative to their parents. However, black mustard introgression was actually more limited (7.5%; Table 2) than other ILs, such as the somatic hybrids between wheat and tall wheatgrass (Liu et al., 2015). We believe the low introgression was probably due to suboptimal UV treatment. Thus, treatment factors such as irradiation time and intensity will require further testing and validation to achieve higher introgression rates. At the same time, the amount of introgression observed may be enough to achieve considerable genomic diversity. For example, in stable rice introgression lines derived from intergeneric hybridization (followed by successive selfing) between rice (Oryza sativa L.) and a wild relative (Zizania latifolia Griseb.), very low introgression (0.1%) by foreign DNA was enough to trigger up to 30% of the genomic changes (Wang et al., 2005). The mechanism underlying these changes is unclear, but some researchers have suggested that they happen very early, through a cryptic pathway that differs from conventional or unorthodox meiotic recombination of homeoalleles between rice and Zizania (Zhang et al., 2013).
Similar to previous findings (Wang et al., 2005), we also observed sequence loss and new bands (Table 2) following somatic hybridization, backcrossing, and self-pollination (Figure 1). The frequency of fragments lost (5.1%) was higher than presence of novel bands (1.4%). It is not entirely clear why the percentage of novel bands was so much lower, but one reason may be that some of them were B. carinata-specific, and B. carinata-specific fragments occurred at a relatively high frequency (15.5%). Thus, future investigations may need to confirm the exact nature of the lost and novel bands, as well as exclude any B. carinata-specific loci.
Our results were consistent with previous reports in artificial hybrid B. carinata (BBCC, x = 17), produced via crossing and polyploidization between B. nigra (BB, x = 8) and B. oleracea (CC, x = 9). Around 47% of the hybrid B. carinata (BBCC) genome possessed isoenzyme sites specific to natural allotetraploid B. carinata (Jourdan and Salazar, 1993). Given this similarity, we propose that the early generations of artificial synthesis and natural polyploid evolution of both genomes may have experienced the same change events. Supporting this idea, previous analyses of recombination in other genera, such as the homoploid hybrid sunflower (Helianthus anomalus), showed that newly synthesized hybrids converged on the linkage pattern of wild hybrids within five generations (Rieseberg et al., 1996).
Based on our results and those of previous studies, we propose that genetic changes in Brassica ILs occur via the following mechanisms. First, alien genetic elements are incorporated into cauliflower early in hybridization, through the activation of transposable elements or DNA methylation, as suggested in rice introgression lines (Wang et al., 2005). Next, chromosomal rearrangements occur through intergenomic translocations or transpositions (Udall et al., 2005) and homoeologous pairing (Leflon et al., 2006; Nicolas et al., 2007). During Brassica evolution, ancestral group rearrangements contributed to the high homology found especially in the three linkage groups of the B genome (B5, B6, and B4) and the A genome (A5, A6, and A4) (Panjabi et al., 2008). Finally, consecutive selection results in rapid sequence elimination, as reported in synthetic hybrids and hybrids less related to the original parents (Osborn et al., 2003; Pires et al., 2004; Gaeta et al., 2007; Zhang et al., 2013).
Changes to DNA methylation in the brassica introgression lines
Cytosine methylation plays an important role in epigenetic gene regulation at both the transcriptional and the post-transcriptional levels (Paszkowski and Whitham, 2001). Our ILs exhibited fairly high proportions of change to methylation patterns (17.2%; Table 3), compared with several other plant hybrid/allopolyploid systems analyzed to date using MSAP. For instance, in resynthesized allotetraploid Arabidopsis suecica, 8.3% of the fragments experienced methylation changes (Madlung et al., 2002), whereas in newly synthesized allohexaploid wheat, ~13% of the loci saw alterations to cytosine methylation (Shaked et al., 2001). In contrast, a 23.6% change to cytosine methylation was observed in asymmetric somatic wheat introgressions (Liu et al., 2015). Taken together, these data allow us to conclude that somatic hybridization induced a broad spectrum of cytosine-methylation changes that perturbed gene expression to a larger extent than allopolyploidization.
Consistent with previous findings (Zhang et al., 2013), changes to hypermethylation (9.7–13.5%; Table 3) occurred more frequently than changes to hypomethylation (2.4–7.6%). Zhang et al. (2013) proposed that this pattern may be caused by the activation and subsequent silencing of some transposable elements after hybridization, but this hypothesis requires more data for confirmation. Additionally, the methylation patterns in ILs may be related to changes in enzymatic machinery, as well as the expression of siRNAs and long noncoding RNAs (Goll and Bestor, 2005; Wierzbicki et al., 2008; Zhang et al., 2013).
In conclusion, somatic hybridization mimics many of the genetic alterations induced by polyploidization or sexual wide hybridization, but to a stronger extent and with considerably less time. Therefore, somatic hybridization is both effective and efficient in achieving introgression of a crop species and its wild relatives. Moreover, this approach provides a potential means to explore the genetic and epigenetic events induced by “somatic genomic shock” (Liu et al., 2015).
Author contributions
GW conceived the research work and wrote the paper. FL provided plant materials and intellectual advice on the project. JL performed molecular and FISH experiments. JZ and SH performed the field management and character survey. XZ performed the methylation-sensitive amplified polymorphism analysis. NG participated in the technical guidance of relative experiment. MZ and YZ performed materials classification and preservation. YW provided technical guidance for pathogen-resistance assays.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Natural Science Foundation of China (No. 31000538), the Natural Science Foundation of Beijing, China (No. 6142012), the Project of Technology Innovation Ability from Beijing Academy of Agriculture and Forestry Sciences (KJCX20140111), and the Youth Science Research Foundation of Beijing Academy of Agriculture and Forestry Sciences (No. QNJJ201601). National Key Research and Development Project (2016YFD0100204-14).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01258
References
- Bird A. P. (1997). Does DNA methylation control transposition of selfish elements in the germline. Trends Genet. 13, 469–470. 10.1016/S0168-9525(97)01310-3 [DOI] [PubMed] [Google Scholar]
- Bird A. P. (2002). DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21. 10.1101/gad.947102 [DOI] [PubMed] [Google Scholar]
- Chavez R., Brown C. R., Iwanaga M. (1988). Application of interspecific sesquiploidy to introgression of PLRV resistance from non-tuber-bearing Solanum etuberosum to cultivated potato germplasm. Theor. Appl. Genet. 76, 497–500. 10.1007/bf00260898 [DOI] [PubMed] [Google Scholar]
- Fukui K., Nakayama S., Ohmido N., Yoshiaki H., Yamabe M. (1998). Quantitative karyotyping of three diploid Brassica species by imaging methods and localization of 45S rDNA loci on the identified chromosomes. Theor. Appl. Genet. 96, 325–330. 10.1007/s001220050744 [DOI] [PubMed] [Google Scholar]
- Gaeta R. T., Pires J. C., Iniguez-Luy F., Osborn L. E. (2007). Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19, 3403–3417. 10.1105/tpc.107.054346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam M., Dang Y., Ge X., Shao Y., Li Z. (2016). Genetic and epigenetic changes in oilseed rape (Brassica napus L.) extracted from intergeneric allopolyploid and additions with Orychophragmus. Front. Plant Sci. 7:438. 10.3389/fpls.2016.00438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdemann M. K., Sacristán M. D., Braatz C., Schieder O. (1994). Utilization of asymmetric somatic hybridization for the transfer of disease resistance from Brassica nigra to Brassica napus. Plant Breed. 113, 106–113. 10.1111/j.1439-0523.1994.tb00712.x [DOI] [Google Scholar]
- Glimelius K. (1999). Somatic hybridization, in Biology of Brassica Coenospecies, ed Gomez-Campo C. (Amsterdam: Elsevier; ), 107–148. [Google Scholar]
- Goll M. G., Bestor T. H. (2005). Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514. 10.1146/annurev.biochem.74.010904.153721 [DOI] [PubMed] [Google Scholar]
- Ha M., Lu J., Tian L., Ramachandran V., Kasschau K. D., Chapman E. J., et al. (2009). Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc. Natl. Acad. Sci. U.S.A. 106, 17835–17840. 10.1073/pnas.0907003106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M., Ng D. W., Li W. H., Chen Z. J. (2011). Coordinated histone modifications are associated with gene expression variation within and between species. Genome Res. 21, 590–598. 10.1101/gr.116467.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen L. N., Earle E. D. (1994). Novel flowering and fatty acid characters in rapid cycling Brassica napus L. resynthesized by protoplast fusion. Plant Cell Rep. 14, 151–156. 10.1007/bf00233780 [DOI] [PubMed] [Google Scholar]
- Hansen L. N., Earle E. D. (1995). Transfer of resistance to Xanthomonas campestris pv. campestris into Brassica oleracea L. by protoplast fusion. Theor. Appl. Genet. 91, 1293–1300. 10.1007/BF00220944 [DOI] [PubMed] [Google Scholar]
- Hansen L. N., Earle E. D. (1997). Somatic hybrids between Brassica oleracea l. and Sinapis alba L. with resistance to Alternaria brassicae (Berk) Sacc. Theor. Appl. Genet. 94, 1078–1085. 10.1007/s001220050518 [DOI] [Google Scholar]
- Jauhar P. P. (2006). Modern biotechnology as an integral supplement to conventional plant breeding: the prospects and challenges. Crop Sci. 46, 1841–1859. 10.2135/cropsci2005.07-0223 [DOI] [Google Scholar]
- Johnston P. A., Timmerman-Vaughan G. M., Farnden K. J., Pickering R. (2009). Marker development and characterisation of Hordeum bulbosum introgression lines: a resource for barley improvement. Theor. Appl. Genet. 118, 1429–1437. 10.1007/s00122-009-0992-7 [DOI] [PubMed] [Google Scholar]
- Jourdan P., Salazar E. (1993). Brassica carinata resynthesized by protoplast fusion. Theor. Appl. Genet. 86, 567–572. 10.1007/BF00838710 [DOI] [PubMed] [Google Scholar]
- Kashkush K., Feldman M., Levy A. A. (2003). Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33, 102–106. 10.1038/ng1063 [DOI] [PubMed] [Google Scholar]
- Kraitshtein Z., Yaakov B., Khasdan V., Kashkush K. (2010). Genetic and epigenetic dynamics of a retrotransposon after allopolyploidization of wheat. Genetics 186, 801–812. 10.1534/genetics.110.120790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leflon M., Brun H., Eber F., Delourme R., Lucas M. O., Vallée P., et al. (2007). Detection, introgression and localization of genes conferring specific resistance to Leptosphaeria maculans from Brassica rapa into B. napus. Theor. Appl. Genet. 115, 897–906. 10.1007/s00122-007-0616-z [DOI] [PubMed] [Google Scholar]
- Leflon M., Eber F., Letanneur J. C., Chelysheva L., Coriton O., Huteau V., et al. (2006). Pairing and recombination at meiosis of Brassica rapa (AA) and Brassica napus (AACC) hybrids. Theor. Appl. Genet. 113, 1467–1480. 10.1007/s00122-006-0393-0 [DOI] [PubMed] [Google Scholar]
- Liu S., Li F., Kong L., Sun Y., Qin L., Chen S., et al. (2015). Genetic and epigenetic changes in somatic hybrid introgression lines between wheat and tall wheatgrass. Genetics 199, 1035–1045. 10.1534/genetics.114.174094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. W., Zhao S. Y., Chen F. G., Xia G. M. (2007). Generation of novel high quality HMW-GS genes in two introgression lines of Triticum aestivum/Agropyron elongatum. BMC Evol. Biol. 7:76. 10.1186/1471-2148-7-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak M. A., Koch M. A., Pecinka A., Schubert I. (2005). Chromosome triplication found across the tribe Brassiceae. Genome Res. 15, 516–525. 10.1101/gr.3531105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung A., Masuelli R., Watson B., Reynolds S., Davison J., Comai L. (2002). Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol. 129, 733–746. 10.1104/pp.003095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menda N., Strickler S. R., Edwards J. D., Bombarely A., Dunham D. M., Martin G. B., et al. (2014). Analysis of wild-species introgressions in tomato inbreds uncovers ancestral origins. BMC Plant Biol. 28:287. 10.1186/s12870-014-0287-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella G., Rotino G. L., Fibiani M., D'Alessandro A., Francese G., Toppino L., et al. (2010). Characterization of health-related compounds in eggplant (Solanum melongena L.) lines derived from introgression of allied species. J. Agric. Food Chem. 58, 7597–7603. 10.1021/jf101004z [DOI] [PubMed] [Google Scholar]
- Nagaharu U. (1935). Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 7, 389–452. [Google Scholar]
- Navrátilová B. (2004). Protoplast cultures and protoplast fusion focused on Brassicaceae-a review. Hortic. Sci. 31, 140–157. Available online at: http://81.0.228.28/publicFiles/51556.pdf; http://www.agriculturejournals.cz/publicFiles/51556.pdf [Google Scholar]
- Nicolas S. D., Le Mignon G., Eber F., Coriton O., Momod H., Clouet V., et al. (2007). Homeologous recombination plays a major role in chromosome rearrangements that occur during meiosis of Brassica napus haploids. Genetics 175, 487–503. 10.1534/genetics.106.062968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn T. C., Butrulle D. V., Sharpe A. G., Pickering K. J., Parkin I. A., Parkin I. A. P., et al. (2003). Detection and effects of a homeologous reciprocal transposition in Brassica napus. Genetics 165, 1569–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panjabi P., Jagannath A., Bisht N. C., Padmaja K. L., Sharma S., Gupta V., et al. (2008). Comparative mapping of Brassica juncea and Arabidopsis thaliana using Intron Polymorphism (IP) markers: homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genomics 9:113. 10.1186/1471-2164-9-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski J., Whitham S. A. (2001). Gene silencing and DNA methylation processes. Curr. Opin. Plant Biol. 4, 123–129. 10.1016/S1369-5266(00)00147-3 [DOI] [PubMed] [Google Scholar]
- Pires J. C., Zhao J. W., Schranz M. E., Leon E. J., Quijada P. A., Lukens L. N., et al. (2004). Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae). Biol. J. Linn. Soc. 82, 675–688. 10.1111/j.1095-8312.2004.00350.x [DOI] [Google Scholar]
- Primard-Brisset C., Poupard J. P., Horvais R., Eber F., Pelletier G., Renard M., et al. (2005). A new recombined double low restorer line for the Ogu-INRA cms in rapeseed (Brassica napus L.). Theor. Appl. Genet. 111, 736–746. 10.1007/s00122-005-2059-8 [DOI] [PubMed] [Google Scholar]
- Rangel P. N., Brondani R. P., Rangel P. H., Brondani C. (2008). Agronomic and molecular characterization of introgression lines from the interspecific cross Oryza sativa (BG90-2) x Oryza glumaepatula (RS-16). Genet. Mol. Res. 7, 184–195. 10.4238/vol7-1gmr406 [DOI] [PubMed] [Google Scholar]
- Rieseberg L. H., Sinervo B., Linder C. R., Ungerer M. C., Arias D. M. (1996). Role of gene interactions in hybrid speciation: evidence from ancient and experimental hybrids. Science 272, 741–745. 10.1126/science.272.5262.741 [DOI] [PubMed] [Google Scholar]
- Roderick H. W., Morgan W. G., Harper J. A., Thomas H. M. (2003). Introgression of crownrust (Puccinia coronata) resistance from meadow fescue (Festuca pratensis) into Italian rye grass (Lolium multiflorum) and physical mapping of the locus. Heredity 91, 396–400. 10.1038/sj.hdy.6800344 [DOI] [PubMed] [Google Scholar]
- Scholze P., Kramer R., Ryschka U., Klocke E., Schumann G. (2010). Somatic hybrids of vegetable brassicas as source for new resistances to fungal and virus diseases. Euphytica 176, 1–14. 10.1007/s10681-010-0205-0 [DOI] [Google Scholar]
- Shaked H., Kashkush K., Ozkan H., Feldman M., Levy A. A. (2001). Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13, 1749–1759. 10.1105/tpc.13.8.1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y. Q., Sun J., Liu Y., Ge X. H., Zhao Z. G., Yao X. C., et al. (2008). Production and characterization of intertribal somatic hybrids of Raphanus sativus and Brassica rapa with dye and medicinal plant Isatis indigotica. Plant Cell Rep. 27, 873–883. 10.1007/s00299-008-0513-1 [DOI] [PubMed] [Google Scholar]
- Udall J. A., Quijada P. A., Osborn T. C. (2005). Detection of chromosomal rearrangements derived from homologous recombination in four mapping populations of Brassica napus L. Genetics 169, 967–979. 10.1534/genetics.104.033209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., He Q., Liu F., Cheng Z., Talbert P. B., Jin W. (2011). Characterization of CENH3 proteins and centromere-associated DNA sequences in diploid and allotetraploid Brassica species. Chromosoma 120, 353–365. 10.1007/s00412-011-0315-z [DOI] [PubMed] [Google Scholar]
- Wang G. X., Tang Y., Yan H., Sheng X. G., Hao W. W., Zhang L., et al. (2011). Production and characterization of interspecific somatic hybrids between Brassica oleracea var. botrytis and B. nigra and their progenies for the selection of advanced pre-breeding materials. Plant Cell Rep. 30, 1811–1821. 10.1007/s00299-011-1088-9 [DOI] [PubMed] [Google Scholar]
- Wang Y. M., Dong Z. Y., Zhang Z. J., Lin X. Y., Shen Y., Zhou D., et al. (2005). Extensive de novo genomic variation in rice induced by introgression from wild rice (Zizania latifolia Griseb.). Genetics 170, 1945–1956. 10.1534/genetics.105.040964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. P., Sonntag K., Rudloff E. (2003). Development of rapeseed with high erucic acid content by asymmetric somatic hybridization between Brassica napus and Crambe abyssinica. Theor. Appl. Genet. 106, 1147–1155. 10.1007/s00122-002-1176-x [DOI] [PubMed] [Google Scholar]
- Westman A. L., Kresovich S., Dickson M. H. (1999). Regional variation in Brassica nigra and other weedy crucifers for disease reaction to Alternaria brassicicola and Xanthomonas campestris pv. campestris. Euphytica 106, 253–259. 10.1023/A:1003544025146 [DOI] [Google Scholar]
- Wierzbicki A. T., Haag J. R., Pikaard C. S. (2008). Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135, 635–648. 10.1016/j.cell.2008.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G. M. (2009). Progress of chromosome engineering mediated by asymmetric somatic hybridization. J. Genet. Genomics 36, 547–556. 10.1016/S1673-8527(08)60146-0 [DOI] [PubMed] [Google Scholar]
- Zhang X., Ge X., Shao Y., Sun G., Li Z. (2013). Genomic change, retrotransposon mobilization and extensive cytosine methylation alteration in Brassica napus introgressions from two intertribal hybridizations. PLoS ONE 8:e56346. 10.1371/journal.pone.0056346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N., Zhu B., Li M. J., Wang L., Xu L. Y., Zhang H., et al. (2011). Extensive and heritable epigenetic remodeling and genetic stability accompany allohexaploidization of wheat. Genetics 188, 499–509. 10.1534/genetics.111.127688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X. B., Hans de Jong J., Zabel P. (1996). Preparation of tomato meiotic pachytene and mitotic metaphase chromosomes suitable for fluorescence in situ hybridization (FISH). Chromosome Res. 4, 24–28. 10.1007/BF02254940 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.