Abstract
Serpina family A member 4 (SERPINA4), also known as kallistatin, exerts important effects in inhibiting tumor growth and angiogenesis in many malignancies. However, the precise role of SERPINA4 in CRC has not been fully elucidated. The present study aimed to investigate the expression of SERPINA4 and its clinical significance in CRC. Quantitative real-time polymerase chain reaction (qRT-PCR) and western blot analyses showed that the mRNA and protein expression of SERPINA4 in colorectal cancer (CRC) specimens was significantly decreased than that in adjacent normal mucosa. Immunohistochemistry (IHC) was conducted to characterize the expression pattern of SERPINA4 by using a tissue microarray (TMA) containing 327 archived paraffin-embedded CRC specimens. Statistical analyses revealed that decreased SERPINA4 expression was significantly associated with invasion depth, nodal involvement, distant metastasis, American Joint Committee on Cancer (AJCC) stage, and tumor differentiation. SERPINA4 was also an independent prognostic indicator of disease-free survival and overall survival in patients with CRC. Furthermore, the impact of altered SERPINA4 expression on CRC cells was analyzed with a series of in vitro and in vivo assays. The results demonstrated that SERPINA4 significantly inhibits malignant tumor progression and serves as a novel prognostic indicator and a potential therapeutic target for CRC.
Keywords: Colorectal cancer, SERPINA4, progression, prognosis, therapy
Introduction
Colorectal cancer (CRC) is the third most common cancer and the third leading cause of cancer death worldwide [1,2]. At present, the gold standard for early detection of CRC is colonoscopy and surgical resection remains the main treatments for patients with CRC [3]. Many advancements have been made in CRC screening, detection, and targeted therapy in recent years [4-7]; However, the long-term survival rate is unsatisfactory [3], which may be attributed to late diagnosis and tumor recurrence. Therefore, in order to improve the survival rate of CRC, it is vital to continue to explore specific diagnostic biomarkers for early detection and targeted therapy.
Serpina family A member 4 (SERPINA4), also known as KAL, KST, or kallistatin, was first identified as a tissue kallikrein-binding protein in the 1900s [8,9]. SERPINA4 is a novel anti-angiogenesis agent, which exerts multiple effects on inflammation, angiogenesis, and tumor growth [10-13]. SERPINA4 suppressed angiogenesis and inflammation via inhibition of TNF-α-induced NF-κB activation and by blocking VEGF signaling pathway, inducing apoptosis of cultured human endothelial cells [14-16]. SERPINA4 also inhibited xenograft tumor growth in mice by antagonizing VEGF-related cell proliferation, migration, and invasion of endothelial cells [11,17]. Moreover, previous studies showed that SERPINA4 not only suppresses tumor growth by its anti-angiogenesis activity [18,19], but also directly inhibits cancer cell proliferation, migration and invasion by adjusting cancer cell signaling in many malignancies such as lung, breast, and hepatocellular carcinoma [20-23]. However, the precise role of SERPINA4 in CRC progression and prognosis has not been fully elucidated.
In the present study, we investigated the expression and clinical significance of SERPINA4 in CRC. First, we evaluated SERPINA4 expression at both the transcriptional and translational levels in CRC tumor tissues and paired adjacent normal mucosa. We then studied the relationship between SERPINA4 expression and survival of patients with CRC using a tissue microarray (TMA) analysis. We last examined the function of SERPINA4 on CRC cell proliferation, migration and invasion, and cell cycle progression by overexpressing SERPINA4.
Materials and methods
Patients and specimens
Fresh primary cancer specimens and matched normal mucosa were collected from 40 CRC patients (25 males and 15 females) who underwent surgery without prior chemotherapy or radiotherapy at the Department of General Surgery of Shanghai General Hospital. The tissues were collected after surgical resection, transferred immediately into RNA keeper Tissue Stabilizer (Vazyme Biotech Co, Ltd, Jiangsu, China), stored at 4°C overnight, and then transferred to -80°C prior to RNA and protein extraction.
Three hundred twenty seven paired samples were obtained from patients with primary CRC (169 males and 158 females; median age, 67.52 y, range 24-92 y) at the time of tumor resection by Shanghai General Hospital between January 2003 and December 2005. Specimens were paraffin embedded for TMA construction. Patients did not receive chemotherapy or radiotherapy before surgery. The tumor staging was determined in accordance with the American Joint Committee on Cancer (AJCC). The disease-free survival (DFS) and overall survival (OS) durations were defined as the period from initial surgery to clinically proven metastasis or recurrence and death, respectively. The diagnoses were confirmed by at least two pathologists. All patients provided written informed consent before enrollment in the study. This study was approved by the Institutional Review Boards of Shanghai Jiao Tong University and the Shanghai General Hospital.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cultured cells or frozen tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). After assessing RNA intensity and purity, 2 μg of total RNA were reverse transcribed into cDNA using PrimeScript/tm RT Master Mix (Takara, Shiga, Japan) according to the manufacturer’s instructions. Then, 1 μg of cDNA was used as a template for qRT-PCR with SYBR Premix Ex Taq (Takara, Shiga, Japan) according to the manufacturer′s instructions. The amplification protocol used was as follows: an initial denaturation step for 2 min at 95°C followed by 40 cycles of denaturation for 10 s at 95°C, annealing for 30 s at 62°C, and elongation for 30 s at 72°C, and a final extension step at 72°C for 30 s. The following primers were used for the qRT-PCR: GAPDH sense, 5’-GGAGC GAGAT CCCTC CAAAA T-3’ and antisense 5’-GGCTG TTGTC ATACT TCTCA TGG-3’; SERPINA4 sense, 5’-CGAGC TGTCT GAGTC CGATG-3’ and antisense, 5’-GCCCA CAGTG TCGTA GAAGT T-3’. GAPDH was used as the internal control. Each reaction was performed in triplicate. The relative SERPINA4 mRNA expression was calculated by using the 2-ΔΔCt comparative method.
Western blot analysis
Total protein was isolated from CRC cells and tissue samples using RIPA Lysis Buffer (Beyotime Biotechnology, Jiangsu, China). The protein concentration was measured using the BCA protein assay kit (Beyotime Biotechnology). Equal amounts of protein (30 μg) were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were blocked in 5% fat-free milk solution containing 0.1% Tween-20 for 1 h at room temperature, followed by incubation with the appropriate primary antibodies: SERPINA4 (1:2000 dilution, Abcam, Cambridge, UK) and GAPDH (1:1000 dilution, Abcam) at 4°C overnight. The membranes were then incubated with the secondary detection antibody (1:5000 dilution, Abgent, San Diego, CA, USA) against rabbit IgG-HRP for 2 h at room temperature. The blots were visualized with Immobilon Western Chemiluminescent HRP Substrate (Millipore, Billerica, MA, USA) according to the manufacturer’s instructions.
TMA construction and immunohistochemistry (IHC)
TMA slides were constructed as previously described [4]. After dewaxing and rehydration, paraffin-embedded sections were immersed in boiled citrate buffer (0.01 M, pH 6.0) for antigen retrieval. Then immunohistochemistry staining was performed using the SERPINA4 primary antibody (1:300 dilution, Abcam) at 4°C overnight, followed by incubation with the anti-mouse or anti-rabbit EnVision two-step Visualization System (Gene Tech, Shanghai, China) for 30 min at room temperature. Finally, tissue sections were counterstained with Mayer’s hematoxylin and covered with coverslips. Two researchers who were blinded to patient prognosis evaluated the slides independently. The staining intensity for SERPINA4 was scored as 0 (no staining), 1 (weak staining), 2 (moderated staining), and 3 (strong staining). The staining area was scored as 0 (0%), 1 (1-25%), 2 (26-50%), 3 (51-75%), 4 (76-100%) on the basis of the percentage of positively stained cells. The final staining scores, which is the sum of the intensity and extension scores, were divided into three groups as followed: 0-2, negative expression; 3-4, weak expression; and 5-7, positive expression.
Cell culture and reagent
Human CRC cell lines, HCT116, RKO, SW620, SW480, Caco2, LoVo, HT29, and HCT8, were obtained from the Type Culture Collection of the Chinese Academy of Science (Shanghai, China). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA) and 1% penicillin-streptomycin (Gibco) in a humidified atmosphere containing 5% CO2 at 37°C.
Transfection of SERPINA4 in CRC cell lines
To restore the expression of SERPINA4 in CRC cell lines, human full length SERPINA4 cDNA was inserted into the GV260 vector (Jikai Gene Chemical Co, Ltd, Shanghai, China). Caco2 cells and RKO cells (5 × 105/well) were seeded in 6-well plates and cultured to 70-80% confluence. Cells were then transfected with GV260-SERPINA4 or empty vector controls using Lipofectamine 2000 (Invitrogen) following with the manufacturer’s procedure. SERPINA4 expression was confirmed using quantitative PCR and western blot analysis.
Cell counting Kit-8 (CCK-8) assays
The effect of SERPINA4 overexpression on the proliferation of CRC cells (Caco2 and RKO) was evaluated by measuring the absorbance at 450 nm following the manufacturer’s instructions. Briefly, cells were plated in triplicate in 96-well cell culture plates at a density of 2 × 103 cells/well. At the appropriate time (12, 24, 48, and 72 h), the cells were incubated with 10 μL CCK-8 solution for 2 h at 37°C. The absorbance was measured on a Gen5 microplate reader (BioTek, Winooski, VT, USA). The experiment was performed in triplicate.
Cell colony formation assays
To evaluate colony-formation assays, 800 log-phase cells were seeded in six-well plates. The cells were cultured at 37°C under a humidified atmosphere containing 5% CO2. After 14 days, the cells were fixed in methyl alcohol for 15 min and stained with Giemsa solution for 20 min. Colonies were then counted and photographed. The experiment was performed in triplicate.
Nude mice xenograft models
CRC xenografts were established in 4-week-old male BALB/C nude mice purchased from the Institute of Zoology, Chinese Academy of Science Shanghai. First, the mice were randomly divided into 2 groups (n = 5). Then, RKO-SERPINA4 or RKO-Control cells (5 × 106) were suspended in 100 μL of PBS and injected subcutaneously into their abdomen. All mice were euthanized 3 weeks after injection and tumor weight was measured. Paraffin sections of the xenograft tissues were prepared for hematoxylin and eosin (H&E) staining and IHC with SERPINA4 antibody (1:300 dilution, Abcam) and Ki67 antibody (1:500 dilution, Abcam). All of the animal procedures were conducted following the Shanghai General Hospital Animal Care guidelines. All efforts were made to minimize animal suffering.
Cell migration and invasion assays
The migration and invasion ability of CRC cells was evaluated by using Transwell chamber (Millipore). In brief, 1 × 105 cells in serum-free medium were seeded in the upper compartment of the Transwell chamber. The transwell membrane was either coated with Matrigel (for invasion; BD, CA, USA) or without (for migration). Then, the bottom chamber was filled with 600 μL of basal medium containing 10% FBS. After incubation at 37°C in a humidified atmosphere containing 5% CO2 for 24 h, the migrated or invasion cells on the bottom chamber were fixed with methanol, stained with crystal violet. The stained cells were counted under a microscope in four randomly selected files at a magnification of 200 ×. The experiment was performed in triplicate.
Flow cytometry analysis of the cell cycle
The cell cycle was analyzed using the Cell Cycle Kit (BD Biosciences, San Jose, CA, USA). Cells were trypsinized, washed in phosphate buffered saline (PBS) for 5 min, and collected by centrifugation at 1200 revolutions per minute (rpm) for 5 min. Then cells were fixed with 5 mL of prechilled 70% ethanol for 2 h at 4°C. The cells were then centrifuged, washed, and resuspended in a solution containing RNase and propidium iodide (PI). After 20 min of incubation, the cells were analyzed by flow cytometry (BD Accuri C6, BD Biosciences). The experiment was performed in triplicate.
Statistical analysis
All statistical analyses were carried out by using the SPSS 19.0 statistical software package (SPSS Inc, Chicago, IL, USA). The differences between two groups were compared with the 2-tailed Student’s t-test, χ2 test, or Fisher’s exact test, as appropriate. Survival rates were calculated using the Kaplan-Meier method and the log-rank test was used to compare the survival curves. The Cox proportional hazards model was used to calculate the univariate and multivariate hazard ratios for the variables. A P value of less than 0.05 was considered statistically significant.
Results
SERPINA4 expression is significantly downregulated in CRC
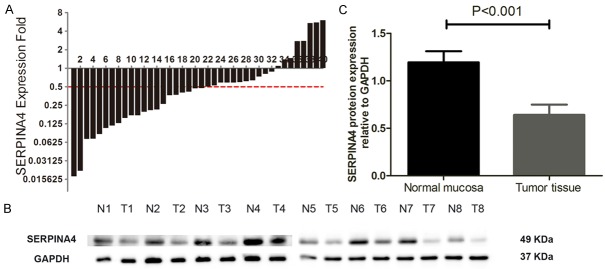
We analyzed 40 paired CRC specimens to evaluated SERPINA4 mRNA and protein expression. As shown in Figure 1A, SERPINA4 was downregulated in 80% (32/40) of CRC tissues compared with the matched adjacent normal tissues (P < 0.001), and the relative level of SERPINA4 mRNA in 22/40 (55%) tumor tissues was below 0.5-fold compared with that of the paired adjacent normal mucosa. The average SERPINA4 expression (ΔCt value) in the colon tumor group was 11.03 ± 1.11 whereas in the normal tissue group, it was 9.73 ± 1.32 (P < 0.001), indicating that the SERPINA4 mRNA level was downregulated in cancer tissues than in paired normal mucosa. Subsequently, western blot analysis showed that SERPINA4 protein expression was significantly lower in tumor tissues compared with that in the paired adjacent normal mucosa (0.64 ± 0.04 vs. 1.20 ± 0.30, respectively; P < 0.001) (Figure 1B, 1C). These results confirmed that SERPINA4 expression decreased both at the transcriptional and translational levels.
Figure 1.
Expression of SERPINA4 in human colorectal cancer tissues. A. SERPINA4 mRNA expression in 40 tumor tissues and paired adjacent normal mucosa detected by qRT-PCR. The relative SERPINA4 mRNA level was normalized to GAPDH expression. A logarithmic scale of 2-ΔΔCt was used to represent the fold-change. B. Western blot analysis of SERPINA4 protein expression in eight paired CRC tissues, with GAPDH being the loading control. C. SERPINA4 protein is lower in tumor tissues than in paired adjacent normal mucosa. ‘N’ represents normal tissue; ‘T’ represents tumor tissue.
Association of SERPINA4 staining with clinicopathological parameters of CRC
To analyze the clinicopathological parameters of SERPINA4 expression, IHC was used to detect SERPINA4 protein expression in a TMA containing specimens from 327 cases of primary CRC paired with normal mucosa. As shown in Figure 2, SERPINA4 was prominently localized in the membrane and cytoplasm of the colorectal epithelium. Of the 327 normal mucosa specimens in the TMA, 49/327 (15.0%) were negative for SERPINA4 expression, 115/327 (35.2%) specimens presented a weak staining, and 163/327 (49.8%) specimens exhibited a strong staining. In contrast, in the CRC mucosa, 165/327 (50.5%) specimens were negative for SERPINA4, 107/327 (32.7%) presented a weak staining, and 55/327 (16.8%) presented a strong staining (Table 1). These results revealed that the distribution of SERPINA4 expression was significantly different between colorectal normal mucosa and tumor mucosa. SERPINA4 was significantly downregulated in the CRC tissues than in the matched normal mucosa (P < 0.001).
Figure 2.
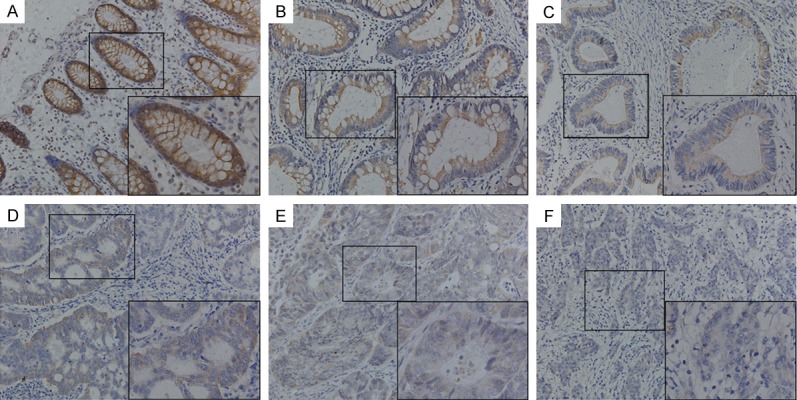
Immunohistochemical staining for SERPINA4 expression in tissue microarray. (A) Positive SERPINA4 expression in normal colonic epithelium, (B) adjacent tumor tissue, and (C) well-differentiated tumor tissue. (D, E) Weak SERPINA4 staining in moderately differentiated colorectal cancer tissues. (F) Negative SERPINA4 staining in poorly differentiated colorectal cancer tissue. Original magnification, 200 × (400 × for insets).
Table 1.
SERPINA4 immunohistochemical staining
| Tissue sample | n | SERPINA4 expression | P value | ||
|---|---|---|---|---|---|
|
| |||||
| Negative (%) | Weak (%) | Positive (%) | |||
| Normal mucosa | 327 | 49 (15.0) | 115 (35.2) | 163 (49.8) | < 0.001* |
| CRC tissue | 327 | 165 (50.5) | 107 (32.7) | 55 (16.8) | |
Indicates statistical difference, P value is based on the chi-square test.
The associations between SERPINA4 expression and clinicopathological parameters for the 327 subjects are summarized in Table 2. SERPINA4 expression showed a positive result with the invasion depth (T stage) (P = 0.001), nodal involvement (N stage) (P = 0.015), distant metastasis (M stage) (P = 0.008), American Joint Committee on Cancer (AJCC) stage (P < 0.001), and tumor differentiation (P = 0.022), whereas no relationship was observed between SERPINA4 expression and the age, gender, or tumor location (Table 2).
Table 2.
Association between clinicopathological parameters and SERPINA4 expression
| parameters | n | SERPINA4 expression | P value | ||
|---|---|---|---|---|---|
|
| |||||
| Negative (n = 165) (%) | Weak (n = 107) (%) | Positive (n = 55) (%) | |||
| Age, years (%) | 0.456 | ||||
| < 65 | 123 | 60 (36.4) | 45 (42.1) | 18 (32.7) | |
| ≥ 65 | 204 | 105 (63.6) | 62 (57.9) | 37 (67.3) | |
| Gender (%) | 0.259 | ||||
| Male | 169 | 80 (48.5) | 57 (51.8) | 32 (61.5) | |
| Female | 158 | 85 (51.5) | 53 (48.2) | 20 (38.5) | |
| Location (%) | 0.102 | ||||
| Right | 77 | 41 (24.8) | 26 (24.3) | 10 (18.2) | |
| Transverse | 21 | 9 (5.5) | 11 (10.3) | 1 (1.8) | |
| Left | 23 | 14 (8.5) | 6 (5.6) | 2 (3.6) | |
| Sigmoid colon | 85 | 39 (23.6) | 24 (22.4) | 23 (41.8) | |
| Rectum | 121 | 62 (37.6) | 40 (37.4) | 19 (34.5) | |
| T stage (%) | 0.001* | ||||
| T1 | 20 | 2 (1.2) | 11 (10.3) | 7 (12.7) | |
| T2 | 69 | 35 (21.2) | 22 (20.6) | 12 (21.8) | |
| T3 | 137 | 63 (38.2) | 52 (48.6) | 22 (40.0) | |
| T4 | 101 | 65 (39.4) | 22 (20.6) | 14 (25.5) | |
| N stage (%) | |||||
| N0 | 253 | 116 (70.3) | 92 (86.0) | 46 (83.6) | 0.015* |
| N1 | 62 | 43 (26.1) | 11 (10.3) | 7 (12.7) | |
| N2 | 12 | 6 (3.6) | 4 (3.7) | 2 (3.6) | |
| M stage (%) | 0.008* | ||||
| M0 | 308 | 149 (90.3) | 104 (97.2) | 55 (100.0) | |
| M1 | 19 | 16 (9.7) | 3 (2.8) | 0 (0.0) | |
| AJCC stage (%) | < 0.001* | ||||
| I | 83 | 24 (14.5) | 37 (34.6) | 22 (40.0) | |
| II | 159 | 76 (46.1) | 58 (54.2) | 25 (45.5) | |
| III | 67 | 50 (30.3) | 9 (8.4) | 8 (14.5) | |
| IV | 18 | 15 (9.1) | 3 (2.8) | 0 (0.0) | |
| Differentiation (%) | 0.022* | ||||
| High | 88 | 40 (24.2) | 31 (29.0) | 17 (30.9) | |
| Moderate | 192 | 91 (55.2) | 69 (64.5) | 32 (58.2) | |
| Low | 47 | 34 (20.6) | 7 (6.5) | 6 (10.9) | |
P < 0.05 indicates a significant association among the variables.
Survival analysis and prognostic significance of SERPINA4 expression
To assess the possible association between SERPINA4 expression in CRC tumors and patient survival, Kaplan-Meier curves with a log-rank test for DFS and OS were built in 327 patients who accepted radical colectomy (Figure 3). Patients with SERPINA4-negative tumors had a poorer DFS (log-rank test, P < 0.001) and OS (log-rank test, P < 0.001) than that in patients with SERPINA4-positive tumors. Kaplan-Meier curves also showed that the rate of recurrence was significantly elevated in patients negative for SERPINA4 expression. In these recurrence cases, patients with a negative SERPINA4 expression had a higher recurrence rate than in patients with a weak or positive expression [negative, 85/165 (51.5%); weak, 19/107 (17.8%); positive, 3/55 (5.5%); P < 0.001].
Figure 3.
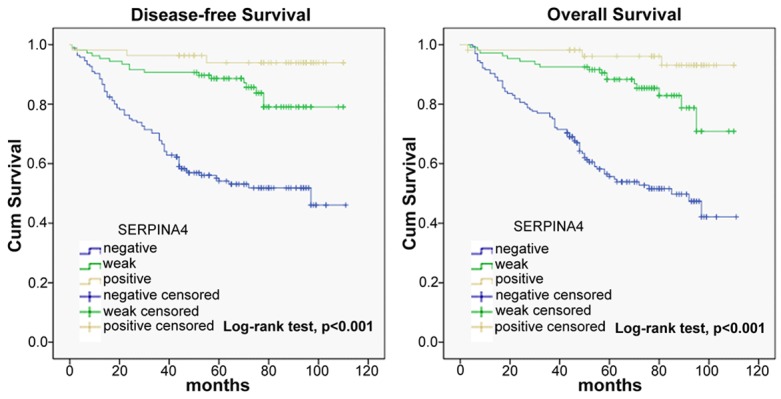
Kaplan-Meier analysis of disease-free survival (DFS) and overall survival (OS) in 327 CRC patients. DFS and OS of patients according to SERPINA4 expression levels determined by immunohistochemical staining. The survival rate of patients with SERPINA4-positive tumors was significantly higher than that of patients with SERPINA4-negative tumors (P < 0.001).
In univariate analysis, patients with negative tumor SERPINA4 expression had a significantly lower DFS and OS rate than that in patients with positive tumor SERPINA4 expression (DFS, HR = 11.727, 95% CI: 3.691-37.257, P < 0.001; OS, HR = 12.268, 95% CI: 3.858-39.007, P < 0.001) (Table 3). In addition, both DFS and OS were significantly associated with pT stage, pN stage, pM stage, and AJCC stage. In multivariate analysis with clinicopathological parameters, the expression of SERPINA4 was an independent prognostic marker to predict tumor recurrence (Table 4).
Table 3.
Univariate analysis for disease-free survival (DFS) and overall survival (OS)
| Variable | DFS | OS | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Age, years | ||||||
| < 65 | ----- | ----- | ||||
| ≥ 65 | 1.333 | 0.869-2.045 | 0.188 | 1.354 | 0.883-2.077 | 0.165 |
| Gender | ||||||
| Male | ----- | ----- | ||||
| Female | 0.826 | 0.552-1.235 | 0.352 | 0.829 | 0.555-1.240 | 0.362 |
| Location | ||||||
| Right | 1.022 | 0.598-1.748 | 0.936 | 1.054 | 0.617-1.803 | 0.846 |
| Transverse | 1.192 | 0.528-2.691 | 0.672 | 1.201 | 0.532-2.711 | 0.659 |
| Left | 1.301 | 0.576-2.938 | 0.527 | 1.402 | 0.620-3.170 | 0.417 |
| Sigmoid colon | 1.053 | 0.632-1.754 | 0.844 | 1.082 | 0.649-1.802 | 0.764 |
| Rectum | ----- | ----- | ||||
| T stage | ||||||
| T1 | 0.086 | 0.120-0.625 | 0.015* | 0.093 | 0.013-0.675 | |
| T2 | 0.363 | 0.200-0.660 | 0.001* | 0.370 | 0.204-0.672 | 0.001* |
| T3 | 0.460 | 0.296-0.715 | 0.001* | 0.485 | 0.312-0.754 | 0.001* |
| T4 | ----- | ----- | ||||
| N stage | ||||||
| N0 | 0.176 | 0.090-0.346 | < 0.001* | 0.190 | 0.097-0.372 | < 0.001* |
| N1 | 0.346 | 0.166-0.721 | 0.005* | 0.355 | 0.171-0.738 | 0.006* |
| N2 | ----- | ----- | ||||
| M stage | ||||||
| M0 | ----- | ----- | ||||
| M1 | 3.891 | 2.119-7.145 | < 0.001* | 3.536 | 1.928-6.485 | < 0.001* |
| AJCC stage | ||||||
| I | 0.126 | 0.058-0.278 | < 0.001* | 0.142 | 0.065-0.311 | < 0.001* |
| II | 0.226 | 0.119-0.432 | < 0.001* | 0.259 | 0.136-0.493 | < 0.001* |
| III | 0.406 | 0.207-0.798 | 0.009* | 0.441 | 0.225-0.866 | 0.017* |
| IV | ----- | ----- | ||||
| Differentiation | ||||||
| High | 0.889 | 0.469-1.687 | 0.719 | 0.930 | 0.490-1.766 | 0.825 |
| Moderate | 0.922 | 0.522-1.631 | 0.781 | 0.966 | 0.546-1.709 | 0.906 |
| Low | ----- | ----- | ||||
| SERPINA4 | ||||||
| Negative | 11.727 | 3.691-37.257 | < 0.001* | 12.268 | 3.858-39.007 | < 0.001* |
| Weak | 3.117 | 0.913-10.650 | 0.050* | 3.219 | 0.941-11.010 | 0.042* |
| Positive | ----- | ----- | ||||
HR hazard ratio, CI confidence interval.
P < 0.05 indicates that the 95% CI of the HR did not include 1.
Table 4.
Multivariate analysis for DFS and OS
| Variable | DFS | OS | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| HR | 95% CI | P value | HR | 95% CI | P value | |
| T stage | 1.326 | 0.897-1.960 | 0.157 | 1.297 | 0.874-1.926 | 0.197 |
| N stage | 1.527 | 0.985-2.368 | 0.059 | 1.453 | 0.936-2.255 | 0.096 |
| M stage | 3.090 | 1.215-7.861 | 0.018* | 2.590 | 0.999-6.715 | 0.050* |
| AJCC stage | 0.859 | 0.499-1.479 | 0.583 | 0.882 | 0.506-1.535 | 0.656 |
| SERPINA4 | 0.339 | 0.221-0.519 | < 0.001* | 0.329 | 0.215-0.503 | < 0.001* |
HR hazard ratio, CI confidence interval.
P < 0.05 indicates that the 95% CI of the HR did not include 1.
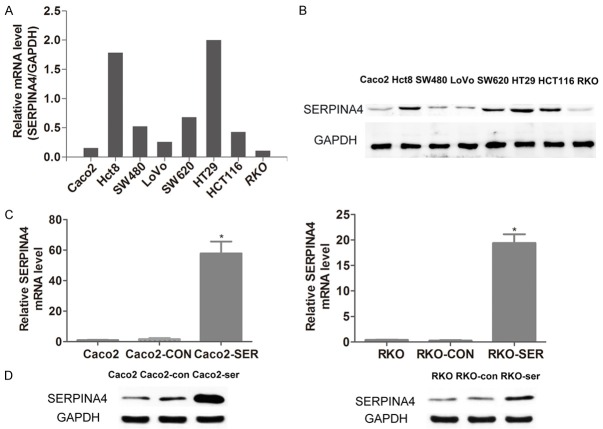
SERPINA4 overexpression inhibits CRC cell proliferation
To explore the function of SERPINA4 in CRC, we elevated the expression of SERPINA4. Quantification by qRT-PCR and western blot demonstrated that SERPINA4 expression level was lower in Caco2 and RKO cells than in other CRC cell lines (Figure 4A, 4B). Subsequently, a SERPINA4 expression plasmid was transfected into Caco2 and RKO cells. SERPINA4 overexpression was confirmed in transfected cells by qRT-PCR and western blot analysis (Figure 4C, 4D). Next, we performed CCK-8 and plate colony formation assays to evaluate the effect of SERPINA4 in CRC cell growth. As shown in Figure 5A, a significant inhibition of cell growth was detected in the SERPINA4 overexpression group compared with that in the control group at 12, 24, 48, and 72 h after transfection (P < 0.05). In addition, the colony formation assays showed that SERPINA4 overexpression inhibited cancer cell clonogenicity when compared with the control group (Figure 5B, 5C; P < 0.01). These results indicated that SERPINA4 overexpression significantly inhibited CRC cell proliferation.
Figure 4.
SERPINA4 expression in cell lines. (A) SERPINA4 mRNA levels and (B) protein levels in CRC cell lines. Caco2 and RKO cells showed lower SERPINA4 expression than that of the other tested cell lines. (C) qRT-PCR analysis and (D) western blot analysis of SERPINA4 expression in Caco2 and RKO cell lines stably overexpressing SERPINA4 (*P < 0.05).
Figure 5.
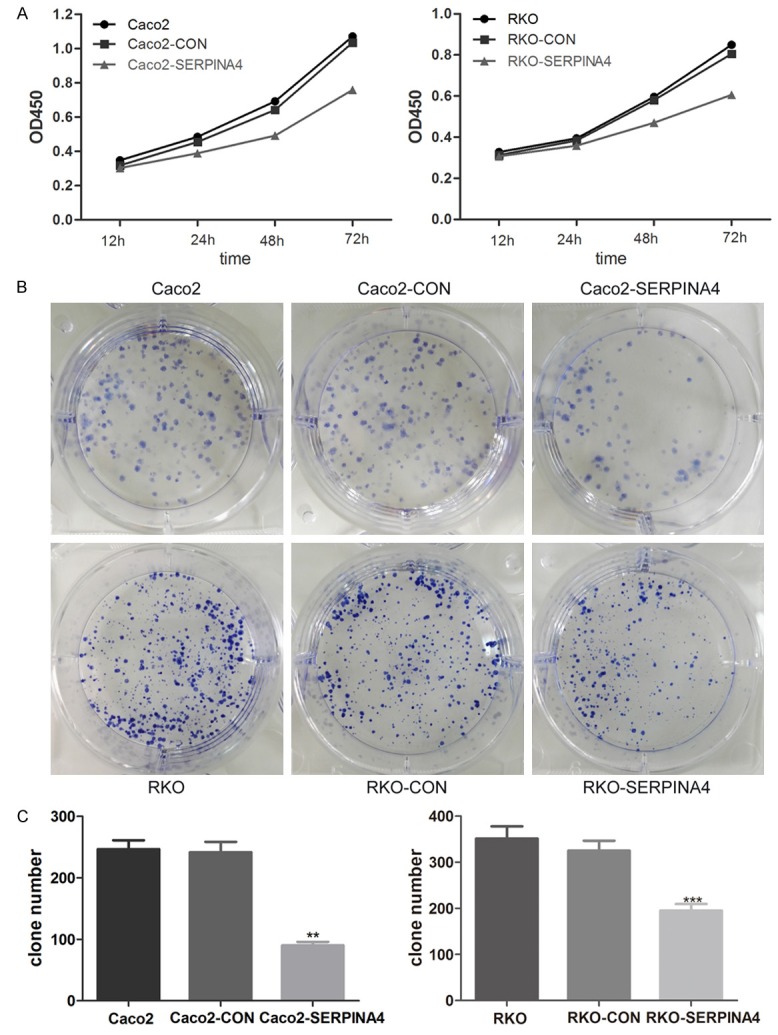
The effects of SERPINA4 overexpression in CRC cell proliferation. A. The effect of SERPINA4 overexpression on cell growth was evaluated by Cell Counting kit-8 assays. B. A plate colony formation assay was performed to assess the impact of SERPINA4 overexpression on cell growth. C. The number of CRC cell clones was quantified. (**P < 0.01, ***P < 0.001).
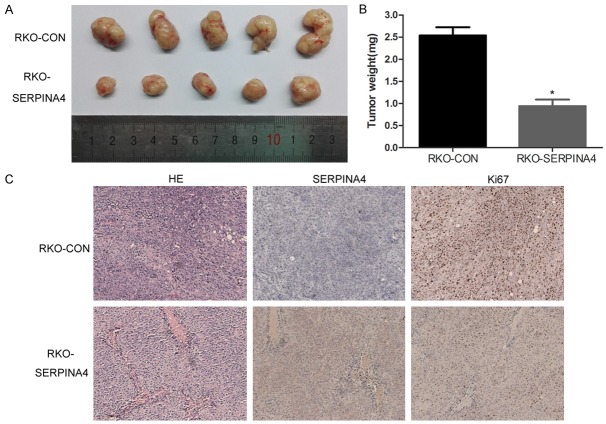
SERPINA4 overexpression inhibits xenograft tumor growth
Based on the in vitro findings described above, RKO-SERPINA4 cells and RKO-Control cells were subcutaneously injected into the abdomen of nude mice to investigate the role of SERPINA4 in tumorigenesis. As shown in Figure 6A, the growth index of the RKO-SERPINA4 tumors was significantly lower than that of the RKO-Control tumors. Specifically, the average tumor weight was 0.94 ± 0.33 mg vs. 2.54 ± 0.41 mg, respectively (Figure 6B). In addition, we utilized IHC to detect SERPINA4 and Ki67 in the xenograft tissues. The expression of Ki67 was significantly weaker in the RKO-SERPINA4 xenografts than in the RKO-Control xenografts (Figure 6C). These data showed that SERPINA4 overexpression suppressed CRC tumor growth in vivo.
Figure 6.
SERPINA4 overexpression suppresses tumorigenicity in vivo. RKO-SERPINA4 cells and RKO-Control cells were injected subcutaneously into nude mice. A. After tumor development, tumors were removed from mice after euthanasia. The RKO-SERPINA4 tumors were smaller than the RKO-Control tumors. B. The tumors from different groups were weighted. C. H&E- and IHC-stained paraffin-embedded xenograft sections. Ki67 expression was weaker in the RKO-SERPINA4 group than in the RKO-Control group (*P < 0.05).
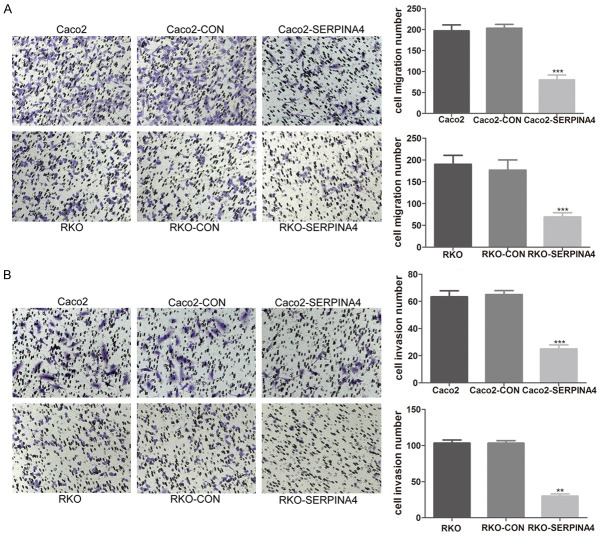
SERPINA4 overexpression inhibits CRC cell migration and invasion
Cell migration and invasion are necessary for tumor development and metastasis. We used transwell chambers either uncoated or precoated with Matrigel to assess the effect of SERPINA4 expression on cell migration and invasion, respectively. SERPINA4 overexpression inhibited the migration and invasion of CRC cells compared with that of the control cells (Figure 7A, 7B; P < 0.001). These findings indicated that SERPINA4 decreases the migrant and invasive behavior of these cells.
Figure 7.
SERPINA4 overexpression significantly inhibits CRC cell migration and invasion. A. The effect of SERPINA4 overexpression on migration using transwell chambers without Matrigel and the number of migrating CRC cells was quantified. B. The effect of SERPINA4 overexpression on invasion using transwell chambers with Matrigel and the number of migrating CRC cells was quantified (**P < 0.01, ***P < 0.001).
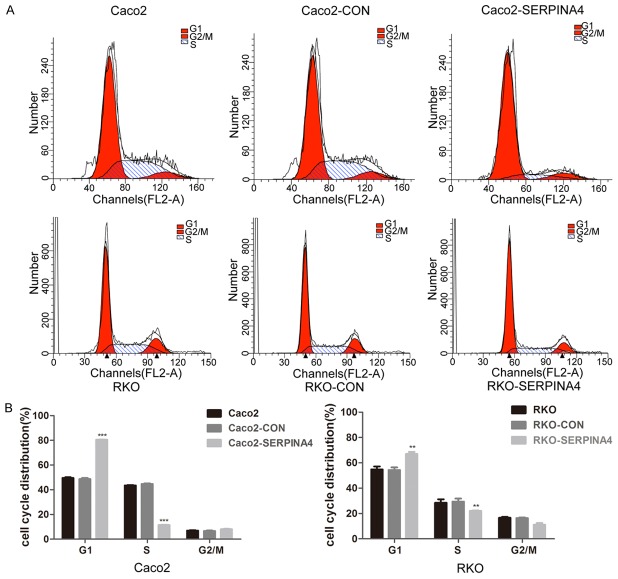
SERPINA4 overexpression changes the CRC cell cycle
Then we explored the role of SERPINA4 in the CRC cell cycle by using flow cytometry analysis and found that SERPINA4 overexpression cells were arrested in the G1 phase and the number of S phase cells had a reciprocal decrease in compared with that in the control group (Figure 8A, 8B). These findings indicated that SERPINA4 overexpression induced an accumulation of CRC cells in G1 phase and a reduction in the number of cells in S phase.
Figure 8.
SERPINA4 overexpression induces marked cell cycle arrest. A. Flow cytometry analysis showing the cell phase distribution of CRC cells. SERPINA4 overexpressing cells showed G1 phase arrest and a reduced number of cells in S phase compared to cells in the control groups. B. The percentages of CRC cells in each phase were quantified (***P < 0.001).
Discussion
Serine proteinase inhibitors (serpins) are single chain proteins containing a conserved domain structure of 370-390 residues and involved in a variety of physiologic events such as coagulation, fibrinolysis, complements activation, and phagocytosis by interacting with their target proteinase [24]. Many serpins such as PEDF and maspin have anti-angiogenic activities [25,26]. As a member of the serpin family, SERPINA4 also shows antiangiogenic properties, and has been identified as an endogenous angiogenic inhibitor that suppresses tumor growth [12,13,18]. However, the bio-functional and prognostic value of SERPINA4 in CRC has not been fully expounded.
In the present study, we showed for the first time that SERPINA4 was downregulated in cancerous colorectal tissues compared with normal epithelium at the transcriptional as well as translational level. Downexpression of SERPINA4 was associated with cancer progression and metastasis. Individuals with SERPINA4-negatively stained tumors experienced shorter OS and DFS, indicating SERPINA4 as a novel prognostic biomarker of CRC outcome. Finally, by univariate and multivariate Cox model analyses, we confirmed SERPINA4 to be an independent prognostic factor for DFS and OS in CRC.
To further understand the role of SERPINA4 in tumor progression, GV260 vector was used to restore the expression of SERPINA4 in CRC cell lines. In accordance with discoveries of Tse et al. in hepatocellular carcinoma cells [23], SERPINA4 overexpressed CRC cell lines exhibited impaired proliferative potency using CCK-8 and colony formation assays. Moreover, SERPINA4 overexpression also led to a significant reduction in xenograft tumor formation, indicating that SERPINA4 significantly suppresses CRC cell proliferation both in vitro and in vivo. Recently, Spinetti et al. [27] showed SERPINA4 has a potent effect on cell invasion in circulating proangiogenic cells. Coincidentally, our study revealed SERPINA4 overexpression in CRC cells suppressed migration and invasion, which indicates the potential involvement of SERPINA4 in tumor metastasis. In addition, SERPINA4 was recently identified as a unique inhibitor of expression of cyclin D1 [17,28], and cyclin D1 is a major regulator of the progression of cells into the proliferative stage of the cell cycle [29]. Thus, we analyzed the effect of SERPINA4 on the CRC cell cycle, and our results demonstrated SERPINA4 suppressed cancer cell cycle progression through the regulation to G1-S-phase transition. Judging by the observations aforementioned, targeting on SERPINA4 is viable for anti-cancer therapy.
In conclusion, our data suggest for the first time, that decreased expression of SERPINA4 is significantly correlated with aggressive phenotype and poor clinical outcome in CRC. SERPINA4 is a novel independent prognostic indicator for patients with CRC. SERPINA4 suppresses cancer progression and serves as a potential therapeutic target for CRC. Further investigation into related signaling pathways and precise mechanisms underlying the anti-tumor activity of SERPINA4 are required in the future study.
Acknowledgements
This project was supported by the funds: National Natural Science Foundation of China (81472238, 81220108021), National High Technology Research and Development Program (SS2014AA020803), Project of Shanghai Science and Technology Commission (14411950502).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 3.DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, Alteri R, Robbins AS, Jemal A. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64:252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 4.Yan DW, Li DW, Yang YX, Xia J, Wang XL, Zhou CZ, Fan JW, Wen YG, Sun HC, Wang Q, Qiu GQ, Tang HM, Peng ZH. Ubiquitin D is correlated with colon cancer progression and predicts recurrence for stage II-III disease after curative surgery. Br J Cancer. 2010;103:961–969. doi: 10.1038/sj.bjc.6605870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang M, Cui F, Lu S, Lu H, Xue Y, Wang J, Chen J, Zhao S, Ma S, Zhang Y, Yu Y, Peng Z, Tang H. Developmental pluripotency-associated 4: a novel predictor for prognosis and a potential therapeutic target for colon cancer. J Exp Clin Cancer Res. 2015;34:60. doi: 10.1186/s13046-015-0176-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Wei Y, Feng Q, Ren L, He G, Chang W, Zhu D, Yi T, Lin Q, Tang W, Xu J, Qin X. Ribosomal protein S15A promotes malignant transformation and predicts poor outcome in colorectal cancer through misregulation of p53 signaling pathway. Int J Oncol. 2016;48:1628–1638. doi: 10.3892/ijo.2016.3366. [DOI] [PubMed] [Google Scholar]
- 7.Yuan G, Zhang B, Yang S, Jin L, Datta A, Bae S, Chen X, Datta PK. Novel role of STRAP in progression and metastasis of colorectal cancer through Wnt/beta-catenin signaling. Oncotarget. 2016;7:16023–16037. doi: 10.18632/oncotarget.7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao J, Chai KX, Chen LM, Xiong W, Chao S, Woodley-Miller C, Wang LX, Lu HS, Chao L. Tissue kallikrein-binding protein is a serpin. I. Purification, characterization, and distribution in normotensive and spontaneously hypertensive rats. J Biol Chem. 1990;265:16394–16401. [PubMed] [Google Scholar]
- 9.Chao J, Tillman DM, Wang MY, Margolius HS, Chao L. Identification of a new tissue-kallikrein-binding protein. Biochem J. 1986;239:325–331. doi: 10.1042/bj2390325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang CR, Chen SY, Wu CL, Liu MF, Jin YT, Chao L, Chao J. Prophylactic adenovirus-mediated human kallistatin gene therapy suppresses rat arthritis by inhibiting angiogenesis and inflammation. Arthritis Rheum. 2005;52:1319–1324. doi: 10.1002/art.20991. [DOI] [PubMed] [Google Scholar]
- 11.Miao RQ, Chen V, Chao L, Chao J. Structural elements of kallistatin required for inhibition of angiogenesis. Am J Physiol Cell Physiol. 2003;284:C1604–1613. doi: 10.1152/ajpcell.00524.2002. [DOI] [PubMed] [Google Scholar]
- 12.Miao RQ, Agata J, Chao L, Chao J. Kallistatin is a new inhibitor of angiogenesis and tumor growth. Blood. 2002;100:3245–3252. doi: 10.1182/blood-2002-01-0185. [DOI] [PubMed] [Google Scholar]
- 13.Zhu B, Lu L, Cai W, Yang X, Li C, Yang Z, Zhan W, Ma JX, Gao G. Kallikrein-binding protein inhibits growth of gastric carcinoma by reducing vascular endothelial growth factor production and angiogenesis. Mol Cancer Ther. 2007;6:3297–3306. doi: 10.1158/1535-7163.MCT-06-0798. [DOI] [PubMed] [Google Scholar]
- 14.Huang KF, Yang HY, Xing YM, Lin JS, Diao Y. Recombinant human kallistatin inhibits angiogenesis by blocking VEGF signaling pathway. J Cell Biochem. 2014;115:575–584. doi: 10.1002/jcb.24693. [DOI] [PubMed] [Google Scholar]
- 15.Huang KF, Huang XP, Xiao GQ, Yang HY, Lin JS, Diao Y. Kallistatin, a novel anti-angiogenesis agent, inhibits angiogenesis via inhibition of the NF-kappaB signaling pathway. Biomed Pharmacother. 2014;68:455–461. doi: 10.1016/j.biopha.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 16.Yin H, Gao L, Shen B, Chao L, Chao J. Kallistatin inhibits vascular inflammation by antagonizing tumor necrosis factor-alpha-induced nuclear factor kappaB activation. Hypertension. 2010;56:260–267. doi: 10.1161/HYPERTENSIONAHA.110.152330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Yang Z, Li P, Bledsoe G, Chao L, Chao J. Kallistatin antagonizes Wnt/beta-catenin signaling and cancer cell motility via binding to low-density lipoprotein receptor-related protein 6. Mol Cell Biochem. 2013;379:295–301. doi: 10.1007/s11010-013-1654-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu L, Yang Z, Zhu B, Fang S, Yang X, Cai W, Li C, Ma JX, Gao G. Kallikrein-binding protein suppresses growth of hepatocellular carcinoma by anti-angiogenic activity. Cancer Lett. 2007;257:97–106. doi: 10.1016/j.canlet.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 19.Diao Y, Ma J, Xiao WD, Luo J, Li XY, Chu KW, Fung P, Habib N, Farzaneh F, Xu RA. Inhibition of angiogenesis and HCT-116 xenograft tumor growth in mice by kallistatin. World J Gastroenterol. 2007;13:4615–4619. doi: 10.3748/wjg.v13.i34.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang NQ, Zou J, Diao Y. Plasmid-mediated expression of kallistatin and its biological activity in lung cancer related cells. Yao Xue Xue Bao. 2013;48:359–365. [PubMed] [Google Scholar]
- 21.Shiau AL, Teo ML, Chen SY, Wang CR, Hsieh JL, Chang MY, Chang CJ, Chao J, Chao L, Wu CL, Lee CH. Inhibition of experimental lung metastasis by systemic lentiviral delivery of kallistatin. BMC Cancer. 2010;10:245. doi: 10.1186/1471-2407-10-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li P, Guo Y, Bledsoe G, Yang Z, Chao L, Chao J. Kallistatin induces breast cancer cell apoptosis and autophagy by modulating Wnt signaling and microRNA synthesis. Exp Cell Res. 2016;340:305–314. doi: 10.1016/j.yexcr.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tse LY, Sun X, Jiang H, Dong X, Fung PW, Farzaneh F, Xu R. Adeno-associated virus-mediated expression of kallistatin suppresses local and remote hepatocellular carcinomas. J Gene Med. 2008;10:508–517. doi: 10.1002/jgm.1180. [DOI] [PubMed] [Google Scholar]
- 24.Potempa J, Korzus E, Travis J. The serpin superfamily of proteinase inhibitors: structure, function, and regulation. J Biol Chem. 1994;269:15957–15960. [PubMed] [Google Scholar]
- 25.Dawson DW, Volpert OV, Gillis P, Crawford SE, Xu H, Benedict W, Bouck NP. Pigment epithelium-derived factor: a potent inhibitor of angiogenesis. Science. 1999;285:245–248. doi: 10.1126/science.285.5425.245. [DOI] [PubMed] [Google Scholar]
- 26.Zhang M, Volpert O, Shi YH, Bouck N. Maspin is an angiogenesis inhibitor. Nat Med. 2000;6:196–199. doi: 10.1038/72303. [DOI] [PubMed] [Google Scholar]
- 27.Spinetti G, Fortunato O, Cordella D, Portararo P, Krankel N, Katare R, Sala-Newby GB, Richer C, Vincent MP, Alhenc-Gelas F, Tonolo G, Cherchi S, Emanueli C, Madeddu P. Tissue kallikrein is essential for invasive capacity of circulating proangiogenic cells. Circ Res. 2011;108:284–293. doi: 10.1161/CIRCRESAHA.110.236786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang B, Abreu JG, Zhou K, Chen Y, Hu Y, Zhou T, He X, Ma JX. Blocking the Wnt pathway, a unifying mechanism for an angiogenic inhibitor in the serine proteinase inhibitor family. Proc Natl Acad Sci U S A. 2010;107:6900–6905. doi: 10.1073/pnas.0906764107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]