Abstract
Enhancer of zeste homolog 2 (EZH2) has been emerged as novel anticancer target. Various EZH2 small-molecule inhibitors have been developed in recent years. A major class of EZH2 inhibitors are S-adenosyl-L-methionine (SAM)-competitive inhibitors, such as EPZ005687, EI1, GSK126, UNC1999 and GSK343. Autophagy, a physiological process of self-digestion, is involved in the turnover of proteins or intracellular organelles. It can serve as cytoprotective or cytotoxic function in cancer. Our previous study has found that UNC1999 and GSK343 are potent autophagy inducers. In this study, the underlying molecular mechanisms were further investigated. Our results showed that UNC1999 and GSK343 transcriptionally upregulated autophagy of human colorectal cancer (CRC) cells through inducing LC3B gene expression. Besides, UNC1999/GSK343-induced autophagy was partially dependent on ATG7 but independent to EZH2 inhibition. Microarray and PCR array analyses identified that UNC1999 and GSK343 also induced endoplasmic reticulum (ER) stress and unfolded protein response (UPR). UNC1999/GSK343-induced ER stress/UPR contributed to the survival of cancer cells, which was opposite to UNC1999/GSK343-induced autophagy that promoted cell death.
Keywords: Autophagy, colorectal cancer, ER stress, EZH2, unfolded protein response
Introduction
Autophagy is a process for the degradation of proteins and organelles through the lysosomal pathway. It acts as a temporary survival mechanism in response to nutrient starvation through self-digestion to provide an alternative energy source [1]. Autophagy involves three stages including initiation (phagophore formation), elongation (autophagosome growth and closure) and maturation (autophagosome-lysosome fusion) [2,3]. Various signaling pathways have been implicated in the regulation of autophagy. Autophagy is inhibited by mammalian target of rapamycin (mTOR)-dependent signaling and interruption of mTOR signaling by rapamycin is known to stimulate autophagy [4]. The biochemical mechanism by which mTOR inhibits autophagy is involved a protein complex associated with the kinase ATG1 (ULK1) [5]. The PI3K pathway is important for autophagy [6]. Class I and class III PI3K differently regulate autophagy. The class I PI3K is a negative regulator of autophagy. Activation of class I PI3K and AKT leads to the activation of mTOR and inhibits autophagy. By contrast, the class III PI3K promotes autophagy through interaction with Beclin 1 [7,8]. The energy-sensing enzyme AMP-activated protein kinase (AMPK) plays a major role in the regulation of cellular lipid and protein metabolism [9]. Activation of AMPK is reported to induce autophagy through phosphorylation and activation of the tuberous sclerosis protein 1 (TSC1)/TSC2 complex that negatively regulates mTOR complex [10]. In addition, the autophagy-initiating kinase ULK1 is recently reported to be phosphorylated and activated by AMPK [11].
Overexpression of enhancer of zeste homolog 2 (EZH2), a histone H3 lysine 27 (H3K27)-specific methyltransferase, has been found in tumors to inhibit the expression of tumor suppressor genes [12-14]. Inhibition of EZH2 to reactivate tumor suppressive genes is regarded as an attractive anticancer strategy [15,16]. Several potent inhibitors of EZH2 have been developed in recent years [17]. S-adenosyl-L-methionine (SAM) is a universal methyl donor for catalytic reactions of histone methyltransferases. A major class of EZH2 inhibitors belong to SAM-competitive inhibitors, such as EPZ005687, EI1, GSK126, GSK343 and UNC1999 [18-22]. Our recent study has demonstrated for the first time that EZH2 inhibitors, GSK343 and UNC1999, induce autophagy in an EZH2-dependent manner, leading to cell death of cancer cells [23]. However, the underlying molecular mechanisms are still unclear. In this study, we demonstrated that EZH2 inhibitors transcriptionally induced autophagy in human colorectal cancer (CRC) cells through the upregulation of LC3B gene expression. In parallel, EZH2 inhibitor activated PERK/eIF2α arm of the UPR pathways that promoted cell survival. Therefore, inhibitor of ER stress enhanced EZH2 inhibitor-induced cytotoxicity.
Materials and methods
Materials
RPMI-1640 medium, L-glutamine, sodium pyruvate, and Antibiotic: Antimycotic Solution (penicillin G, streptomycin and amphotericin B) were purchased from Life Technologies (Gaithersburg, MD, USA). Fetal bovine serum (FBS) was purchased from GIBCO (Grand Island, NY, USA). EZH2, LC3B, ATG5, ULK1, GRP78, TRB3, GAPDH, Tubulin, and β-Actin antibodies were purchased from GeneTex (Hsinchu, Taiwan). ATG7 and eIF2α antibodies was purchased from Santa Cruz (Island, CA, USA). Phospho-eIF2α (p-eIF2α) antibody was purchased from Cell Signaling Technology (Beverly, MA, USA). ATF4 antibody was purchased from ProteinTech Group (Chicago, IL, USA). Horseradish peroxidase-labeled goat anti-rabbit and anti-mouse secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, PA, USA). pCMV-EZH2 plasmid was purchased from Addgene (Cambridge, MA, USA). PolyJet™ In Vitro DNA Tranfection Reagent was purchased from SignaGen Laboratories (Ijamsville, MD, USA). siGENOME human EZH2 and ATG5 SMARTpool siRNAs, siGENOME Non-Targeting human siRNA Pool, ON-TARGETplus human LC3B SMARTpool siRNA, ON-TARGETplus human Non-Targeting siRNA Pool, and DharmaFECT 4 siRNA Transfection Reagent were purchased from Dharmacon (Lafayette, CO, USA). UNC1999 and 3-methyladenine (3-MA) were purchased from Cayman Chemical (Ann Arbor, MI, USA). GSK343 was purchased from BioVision (Mountain View, CA, USA). Bafilomycin A1 was purchased from LC Laboratories (Woburn, MA, USA). GSK2606414 was purchased from ApexBio Technology (Houston, TX, USA). Dimethyl sulfoxide (DMSO) and 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) were purchased from Sigma Chemical (St. Louis, MO, USA). Protease and phosphatase inhibitor cocktails were purchased from Roche (Indianapolis, IN, USA). Other chemicals or reagents not specified were purchased from OneStar Biotechnology (New Taipei City, Taiwan).
Cell culture
Human colorectal cancer cells (HCT116, LoVo, HCT-15, and DLD-1) were kindly provided by Prof. Ya-Wen Cheng (Taipei Medical University, Taipei, Taiwan). ATG7-wildtype (ATG7-WT), ATG7-knockout (ATG7-KO), ULK1-wildtype (ULK1-WT) and ULK1-dominant-negative mutant (ULK1-DN) DLD-1 cells were purchased from Horizon Discovery (Cambridge, UK). These cells were cultured in RPMI-1640 medium supplemented with 10% FBS, 1 mM sodium pyruvate, 1% L-glutamine, 1% Antibiotic:Antimycotic Solution, and incubated at 37°C in a humidified incubator containing 5% CO2.
Cell viability assay
Cell viability was measured with an MTT assay. Cells were plated in 96-well plates and treated with drugs. After 72 h of incubation, 0.5 mg/mL of MTT was added to each well for an additional 4 h. The blue MTT formazan precipitate was then dissolved in 200 μL of DMSO. The absorbance at 550 nm was measured on a multiwell plate reader.
Western blot analysis
Cells were lysed in an ice-cold buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM MgCl2, 2 mM EDTA, 1% NP-40, 10% glycerol, 1 mM DTT, 1 × protease inhibitor cocktail and 1 × phosphatase inhibitor cocktail at 4°C for 30 min. Cell lysates were separated on a sodium dodecylsulfate (SDS)-polyacrylamide gel, and then transferred electrophoretically onto the Hybond-C Extra nitrocellulose membrane (GE Healthcare, Piscataway, NJ, USA). The membrane was pre-hybridized in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.05% Tween-20 (TBST buffer), and 5% skim milk for 1 h, and then transferred to a solution containing 1% bovine serum albumin (BSA)/TBST and a primary antibody and incubated overnight at 4°C. After washing with the TBST buffer, the membrane was submerged in 1% BSA/TBST containing a horseradish peroxidase-conjugated secondary antibody for 1 h. The membrane was washed with TBST buffer, and then developed with an enhanced chemiluminescence (ECL) system (Perkin-Elmer, Boston, MA, USA) and exposed to x-ray film (Roche, Indianapolis, IN, USA).
Fluorescence microscopic analysis of autophagic vacuoles
Formation of autophagic vacuoles was monitored using a Cyto-ID Autophagy Detection Kit (Enzo Life Sciences, Farmingdale, NY, USA) following the manufacturer’s protocol. Briefly, cells were washed twice in phosphate-buffered saline (PBS) containing 5% FBS and then stained with Cyto-ID Detection Reagent and Hoechst 33342. After 30 min of incubation at 37°C, cells were washed and examined by fluorescence microscopy.
Transient transfection
For EZH2 overexpression, human EZH2-overexpressing (pCMV-EZH2) and its control (pCMV) plasmids were transiently transfected into cells with PolyJet™ In Vitro DNA Tranfection Reagent according to the manufacturer’s instructions. For siRNA knockdown analysis, human EZH2, ATG5, LC3B and control siRNAs were transiently transfected into cells with DharmaFECT 4 siRNA Transfection Reagent according to the manufacturer’s instructions. Twenty-four hours after transfection, the transfection mixture was replaced with fresh complete medium and cells were used for further experiments.
Microarray analysis and gene set enrichment analysis (GSEA)
Total RNA was extracted from HCT116 cells that were treated with 5 μM UNC1999 and 10 μM GSK343 for 4 h by the GENEzol TriRNA Pure Kit (Geneaid Biotech; New Taipei City, Taiwan). Microarray analysis using Agilent SurePrint G3 Human GE 8 × 60 K Microarray (Agilent Technologies) was performed by Welgene Biotech Company (Taipei, Taiwan). The raw data were deposited in NCBI GEO database (GSE83633). Gene set enrichment analysis (GSEA) was performed using GSEA v2.2.2 (http://www.broadinstitute.org/gsea/) provided by the Broad Institute of MIT and Harvard (Cambridge, MA, USA) [24,25]. Enrichment analysis of the 50 cancer hallmarks from the Molecular Signatures Database (MSigDB) v5.1 was performed with default parameter setting [25,26].
Real-time quantitative PCR (qPCR)
Total RNA (1 μg) was reverse-transcribed for 30 min at 42°C with the iScript cDNA Synthesis Kit according to the supplier’s standard protocol (Bio-Rad Laboratories; Richmond, CA, USA). qPCR was performed using the following conditions: 10 min at 95°C and 45 cycles of 10 sec at 95°C and 30 sec at 60°C. The 2 × SYBR Green PCR Master Mix (Roche) and 200 nM of forward and reverse primers were used (human LC3B: forward 5’-AACGGGCTGTGTGAGAAAAC-3’ and reverse 5’-AGTGAGGACTTTGGGTGTGG-3’; human β-Actin: forward 5’-GTTGCTATCCAGGCTGTGCT-3’ and reverse 5’-AGGGCATACCCCTCGTAGAT-3’). Each assay was performed on a LightCycler Nano Real-Time PCR System (Roche) in triplicate, and the fold-changes in expression were derived using the comparative CT method calculated by LightCycler Nano Software v1.1 (Roche).
PCR array
Human Unfolded Protein Response Plus RT2 Profiler PCR Array (Qiagen; Valencia, CA, USA) containing primers for 84 key genes of the UPR were used to examine the effects of EZH2 inhibitors the according to the manufacturer’s protocol. Total RNA (0.5 μg) was reverse transcribed by a RT2 First Strand Kit (Qiagen), prior to amplification using the RT2 Profiler PCR Array. PCR amplifications were performed on a StepOne Plus Real-Time PCR System thermocycler (Life Technologies). The results were analyzed using a supplied online software. The comparative Ct method was used for relative transcript quantification against the average ΔCt derived from internal controls (β-Actin, β-2-microglobulin, GAPDH, HPRT1, and RPLP0).
Statistical analysis
Means and standard deviations of samples were calculated from the numerical data (at least 3 replica) generated in this study. Data were analyzed using Student’s t-test, and p values of < 0.05 were considered significant (*).
Results
Characterization of autophagy induced by the EZH2 inhibitors in human colorectal cancer cells
Our previous study shows that SAM-competitive EZH2 inhibitors, GSK343 and UNC1999 (Figure 1A), induce autophagy in cancer cells [23]. The involved molecular mechanisms were further investigated in this study. First, we confirmed that GSK343 and UNC1999 induced autophagy in human colorectal cancer (CRC) cells. LoVo, HCT-15, and DLD-1 cells were treated with various doses of UNC1999 for 24 h, and autophagy was evaluated by the accumulation of LC3-II. As shown in Figure 1B, UNC1999 induced LC3-II accumulation in these cells. Because impaired autophagosome-lysosome fusion can result in LC3-II accumulation, autophagic flux was analyzed by treating bafilomycin A1, a vacuolar-type H+-ATPase inhibitor that blocks autophagosome-lysosome fusion [27]. As shown in Figure 1C, UNC1999 induced more accumulation of LC3-II in the presence of bafilomycin A1, suggesting that the increase of LC3-II by UNC1999 was not due to the blockade of autophagic degradation. Furthermore, the formation of autophagic vacuoles was monitored using Cyto-ID Autophagic Detection Kit. UNC1999 increased the Cyto-ID fluorescence in DLD-1 cells (Figure 1D). Similarly, GSK343 also induced LC3-II accumulation in HCT116 and DLD-1 cells (Figure 1E). Therefore, EZH2 inhibitors can induce autophagy in human CRC cells. To investigate whether EZH2 inhibition was responsible for the effect of UNC1999 on inducing autophagy, LoVo, HCT-15 and DLD-1 cells were transfected with EZH2-overexpressing plasmids, and then treated with UNC1999 for 24 h. As shown in Figure 1F, UNC1999-induced LC3-II accumulation was not rescued by EZH2 overexpression. In addition, knockdown of EZH2 by siRNA was not sufficient to induce LC3-II accumulation in DLD-1 cells (Figure 1G). Therefore, UNC1999 induces autophagy of human CRC cells in an EZH2-independent manner.
Figure 1.
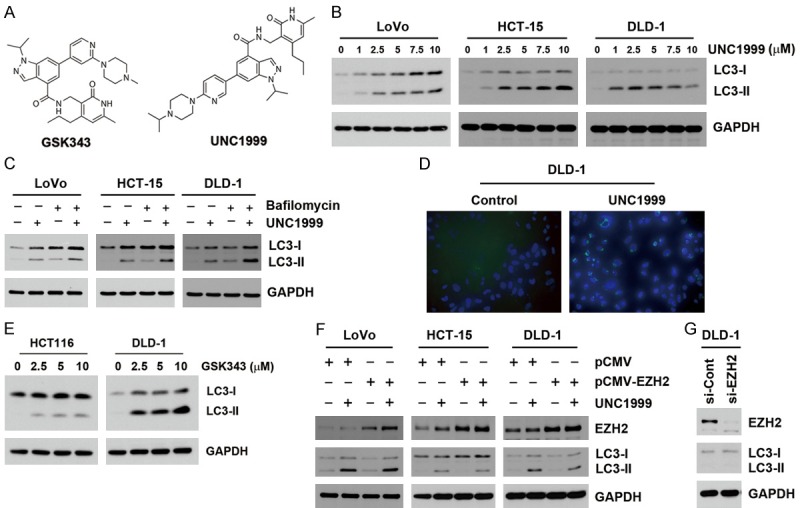
UNC1999 induced autophagy in human CRC cells in an EZH2-independent manner. A. The chemical structures of GSK343 and UNC1999. B. LoVo, HCT-15 and DLD-1 cells were treated with indicated doses of UNC1999 for 24 h. The protein expressions were analyzed by Western blots. C. LoVo, HCT-15 and DLD-1 cells were treated with 2.5 μM UNC1999 for 24 h in the absence or presence of 20 nM bafilomycin A1 (post-treatment for 4 h). The protein expressions were analyzed by Western blots. D. DLD-1 cells were treated with 2.5 μM UNC1999 for 24 h, and then stained with Cyto-ID Autophagy Detection Kit. The Cyto-ID fluorescence was observed by fluorescent microscopy. E. HCT116 and DLD-1 cells were treated with indicated doses of GSK343 for 24 h. The protein expressions were analyzed by Western blots. F. LoVo, HCT-15 and DLD-1 cells were transiently transfected with an EZH2-overexpressing (pCMV-EZH2) or a control (pCMV) plasmid for 48 h, and then treated with 2.5 μM UNC1999 for 24 h. The protein expressions were analyzed by Western blots. G. DLD-1 cells were transiently transfected with EZH2 siRNA for 72 h. The protein expressions were analyzed by Western blots.
EZH2 inhibitor-induced autophagy is partially dependent on ATG7
To investigate how EZH2 inhibitors induced autophagy in human CRC cells, a class III PI3K inhibitor, 3-methyladenine (3-MA), was used. Surprisingly, 3-MA enhanced UNC1999- and GSK343-induced LC3-II accumulation (Figure 2A and 2B). To confirm the effect of 3-MA, an siRNA against ATG5 was transfected into DLD-1 cells. Consistently, inhibition of ATG5 expression potentiated UNC1999-induced LC3-II accumulation (Figure 2C). Furthermore, ULK1-dominant-negative mutant (ULK1-DN) and ATG7-knockout (ATG7-KO) DLD-1 cells were used. As shown in Figure 2D and 2E, UNC1999 could still induce LC3-II accumulation in ULK1-DN and ATG7-KO DLD-1 cells. However, we noticed that UNC1999-induced LC3-II accumulation was partially inhibited in ATG7-KO cells (Figure 2E). To confirm this results, cells were treated with UNC1999 with or without bafilomycin A. As shown in Figure 2F, the level of LC3-II was attenuated in ATG7-KO cells compared to ATG7-WT cells. Therefore, these results suggest that EZH2 inhibitors induce autophagy in human CRC cells, which was partially dependent on ATG7.
Figure 2.
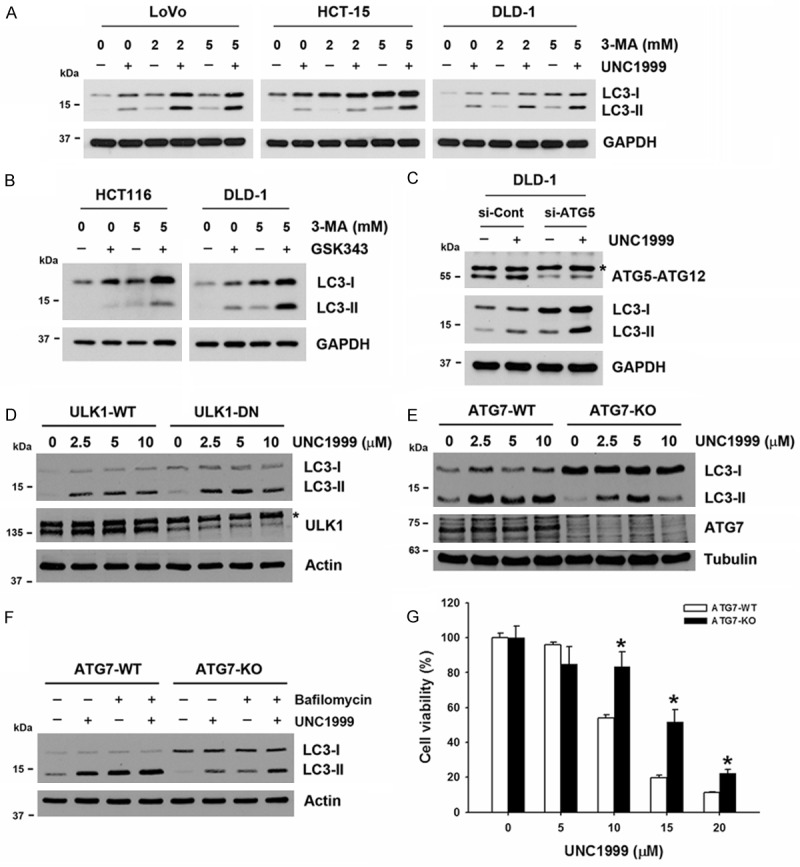
UNC1999 induced partially ATG7-dependent autophagy. A. LoVo, HCT-15 and DLD-1 cells were pretreated with 2 and 5 mM 3-MA for 1 h, and then exposed to 2.5 μM UNC1999 for 24 h. The protein expressions were analyzed by Western blots. B. HCT116 and DLD-1 cells were pretreated with 5 mM 3-MA for 1 h, and then exposed to 5 μM GSK343 for 24 h. The protein expressions were analyzed by Western blots. C. DLD-1 cells were transfected with ATG5 siRNA for 48 h, and then exposed to 2.5 μM UNC1999 for 24 h. The protein expressions were analyzed by Western blots. *nonspecific band. D and E. ATG7-KO and ULK1-DN DLD-1, as well as their control wildtype (WT) cells were treated with indicated doses of UNC1999 for 24 h. The protein expressions were analyzed by Western blots. F. ATG7-WT and ATG7-KO cells were treated with 5 μM UNC1999 for 24 h in the absence or presence of 20 nM bafilomycin A1 (post-treatment for 4 h). The protein expressions were analyzed by Western blots. G. ATG7-WT and ATG7-KO cells were treated with indicated doses of UNC1999 for 72 h. The cell viability was analyzed by an MTT assay. *P < 0.05 indicates significant differences between ATG7-WT and ATG7-KO cells.
EZH2 inhibitor-induced autophagy is associated with the upregulation of LC3B gene transcription
In addition to the cytosolic events that can regulate autophagy, accumulating evidences suggest the existence of transcriptional control of autophagy [28]. To investigate whether transcription and translation were involved in UNC1999-induced autophagy, a transcription inhibitor, antinomycin D, and a protein synthesis inhibitor, cycloheximide, were used. As shown in Figure 3A, UNC1999- and GSK343-induced LC3-II accumulation was inhibited by these two drugs, indicating that induction of gene expression and de novo protein synthesis are required for UNC1999- and GSK343-induced autophagy. Because treatment of 3-MA and ATG5 siRNA, as well as ATG7- and ULK1-deficiency did not inhibit EZH2 inhibitor-induced autophagy, we proposed that EZH2 inhibitors may directly induce the transcription of LC3B to trigger autophagy. Indeed, UNC1999 and GSK343 transiently increased the mRNA level of LC3B gene in HCT116 cells (Figure 3B). To investigate the role of autophagy in the anticancer activity of EZH2 inhibitors, endogenous LC3B expression was knocked down by transfecting siRNA (Figure 3C), and then cell viability was examined by MTT assay. As shown in Figure 3D, knockdown of LC3B gene rescued cell growth inhibition by UNC1999. Therefore, UNC1999 induced autophagy through transcriptional upregulation of LC3B gene. In addition, the partial ATG7-dependency of UNC1999-induced autophagy (Figure 2E and 2F) may be due to that ATG7 is responsible for the LC3 lipidation [29].
Figure 3.
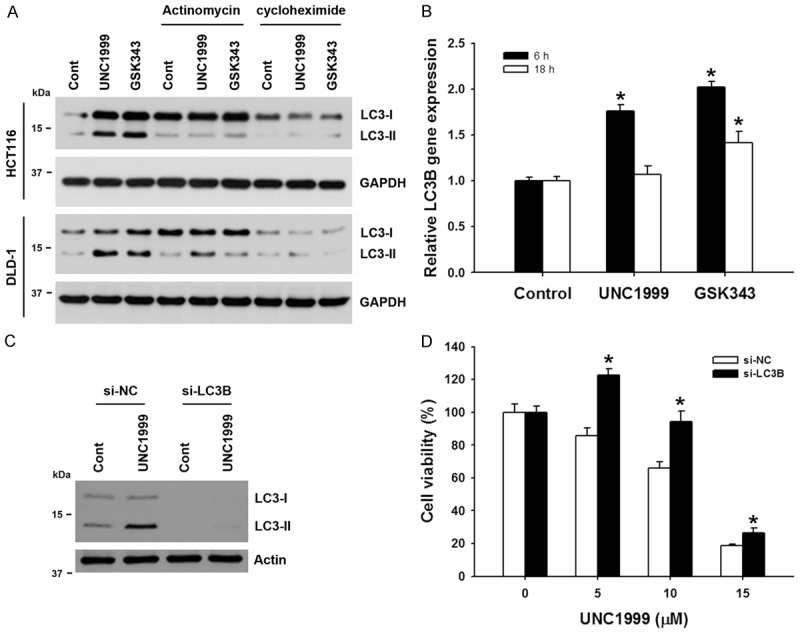
EZH2 inhibitors transcriptionally induced autophagy. A. HCT116 and DLD-1 cells were treated with 2 μg/mL antinomycin or 5 μg/mL cycloheximide for 1 h, and then exposed to 2.5 μM UNC1999 or 5 μM GSK343 for additional 24 h. The protein expressions were analyzed by Western blots. B. HCT116 cells were treated with 5 μM UNC1999 or 10 μM GSK343 for 6 or 18 h. The LC3B mRNA expression was analyzed by qPCR. *P<0.05 indicates significant differences between drug-treated and control cells. C. HCT116 cells were transiently transfected with EZH2 siRNA for 48 h, and then treated with 5 μM UNC1999 for 24 h. The protein expressions were analyzed by Western blots. C. HCT116 cells were transiently transfected with EZH2 siRNA for 24 h, and then treated with indicated doses of UNC1999 for 72 h. The cell viability was analyzed by an MTT assay. *P<0.05 indicates significant differences between si-NC- and si-LC3B-transfected cells.
Microarray analysis reveals that EZH2 inhibitors upregulate genes associated with an unfolded protein response
To further identify the biological pathways affected by UNC1999 and GSK343, microarray analyses were performed and analyzed by the Gene Set Enrichment Analysis (GSEA) software using hallmark gene sets [24-26]. The results indicated 4 out of 50 gene sets are upregulated in response to UNC1999 and GSK343 (Table 1). The top-ranking pathways with statistical significance (p value < 0.05%) included unfolded protein responses (UPR) and cholesterol biosynthesis (Figure 4A and Table 1). Activation of UPR drew our attention because UPR is known to connect ER stress to autophagy [30]. ER stress/UPR consists of three major arms (PERK/eIF2α, IRE1/XBP1, and ATF6). Each UPR pathway induces different target genes. ATF6 binds to ER-stress response elements (ERSEs) and induces transcription of several genes, including GRP78, CHOP and XBP1. XBP1 binds to UPR elements (UPREs) and activates many genes that are crucial for secretory function. One of these gene products, p58IPK, inhibits PERK activity. PERK-mediated phosphorylation of eIF2α suppresses global translation except the ATF4 mRNA. ATF4 can upregulate the third set of UPR target genes, one of which is CHOP that can induce apoptosis in cells with irrecoverable levels of ER stress [31]. The alterations of ER stress/UPR-related genes by UNC1999 and GSK343 were visualized by the Ingenuity Pathway Analysis (IPA). The results indicated that UNC1999 predominantly upregulated genes associated with PERK/eIF2α pathways (Figure 4B). However, GSK343 induced the expression of genes related to the three arms of UPR (Figure 5). Therefore, EZH2 inhibitors may induce autophagy through the activation of UPR pathways.
Table 1.
The Gene Set Enrichment Analysis (GSEA) for pathways enriched in both UNC1999- and GSK343-treated HCT116 cells
| Pathways | Number of Genes in Pathway | Number of Pathway Genes Differentially Expressed (% of Total) | p value |
|---|---|---|---|
| HALLMARK_UNFOLDED_PROTEIN_RESPONSE | 81 | 18 (22%) | 0.000 |
| HALLMARK_CHOLESTEROL_HOMEOSTASIS | 61 | 13 (21%) | 0.000 |
| HALLMARK_MTORC1_SIGNALING | 148 | 27 (18%) | 0.500 |
| HALLMARK_PI3K_AKT_MTOR_SIGNALING | 72 | 7 (10%) | 0.125 |
Figure 4.

Microarray analysis revealed the induction of ER stress/UPR by EZH2 inhibitors. A. HCT116 cells were treated with 5 μM UNC1999 or and then microarray analysis was performed. UNC1999/GSK343-regulated genes were analyzed by GSEA. The enrichment plots of “HALLMARK_UNFOLDED_PROTEIN_RESPONSE” and “HALLMARK_CHOLESTEROL_HOMEOSTASIS” were shown. B. The alterations of genes associated with UPR in UNC1999-treated HCT116 cells were analyzed and visualized by IPA. The red and green colors indicate the upregulation and downregulation by UNC1999, respectively.
Figure 5.
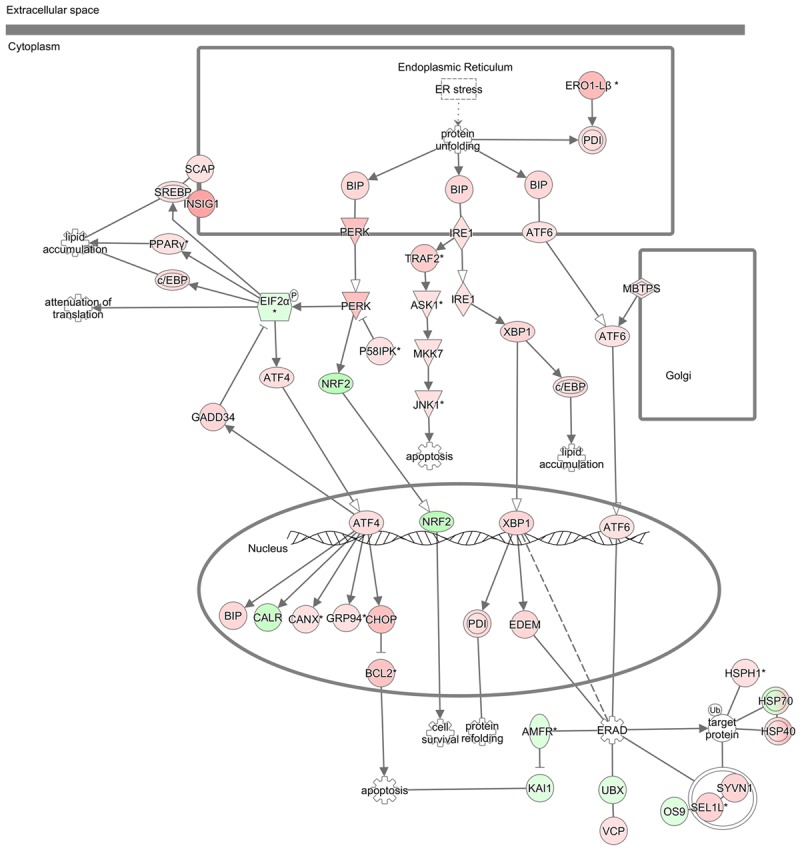
Alteration of ER stress/UPR-related genes by GSK343. The alterations of genes associated with UPR in GSK343-treated HCT116 cells were analyzed and visualized by IPA. The red and green colors indicate the upregulation and downregulation by GSK343, respectively.
EZH2 inhibitors activate the PERK/eIF2α arm of the unfolded protein response pathways
To confirm the result of microarray analysis, the Human Unfolded Protein Response Plus PCR Array analysis was performed. As shown in Figure 6A, the common genes induced by UNC1999 and GSK343 were ATF4 and DDIT3 (CHOP/GADD153) genes at 6 h, and TRB3 gene at 6 and 18 h. Induction of ATF4 and CHOP represents the downstream of PERK/eIF2α arm of UPR [32]. TRB3, a mammalian homolog of Drosophila tribbles, functions as a negative modulator of AKT/mTOR [33]. TRB3 is induced by ATF4/CHOP pathway and is involved in CHOP-dependent cell death during ER stress [34]. It has been reported that ATF4 and CHOP directly bind and regulate LC3B gene promoter to promote autophagy [35]. Our result indicated that the transient induction of ATF4 and DDIT3/CHOP gene expression was correlated with the upregulation of LC3B genes (Figure 3B). Therefore, we proposed that EZH2 inhibitors can activate autophagy through PERK/eIF2α signaling axis. Indeed, UNC1999 induced the phosphorylation of eIF2α, as well as the expression of ATF4 (Figure 6B). In addition, the expression of GRP78, a central regulator for ER stress [36], was also induced by UNC1999 (Figure 6B). Therefore, UNC1999 indeed induces ER stress and UPR in human CRC cells. To investigate the role of ER stress in UNC1999-induced autophagy, a PERK inhibitor, GSK2606414, was used. As shown in Figure 6C, phosphorylation of eIF2α by UNC1999 was attenuated by GSK2606414. However, GSK2606414 did not alter UNC1999-induced LC3-II accumulation. Therefore, UNC1999 induced autophagy in an ER stress-independent manner.
Figure 6.
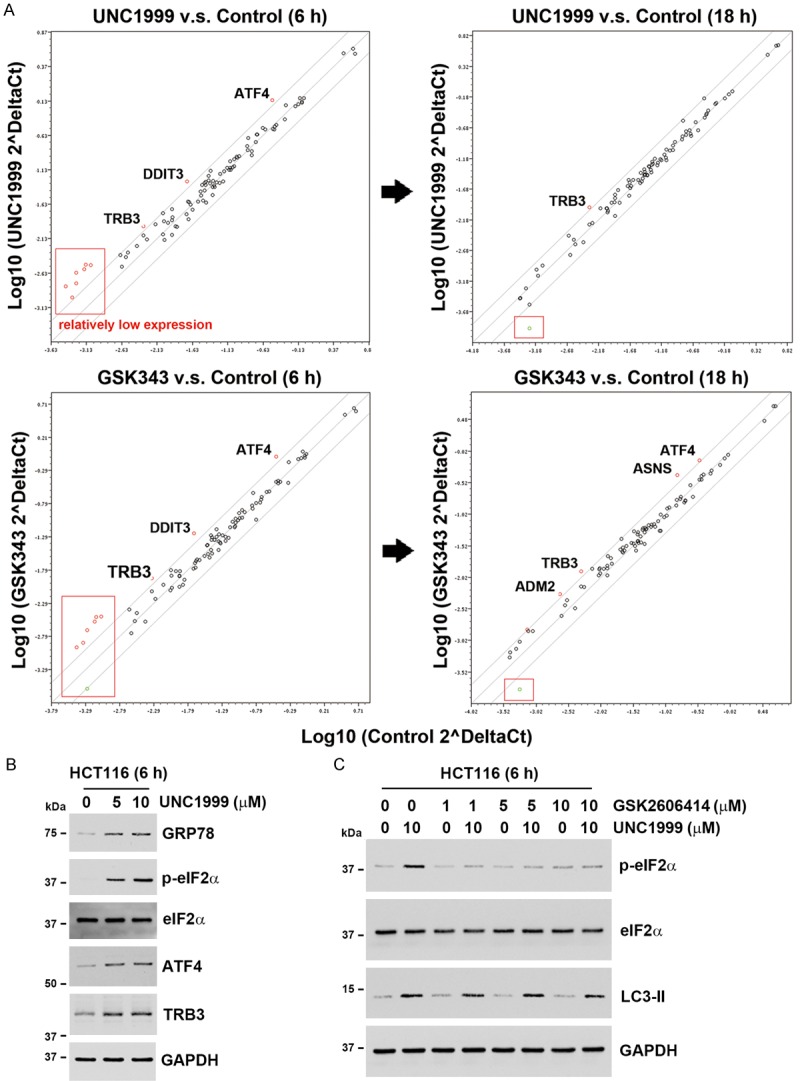
PCR array analysis identified the activation of the PERK/eIF2α arm by EZH2 inhibitors. A. HCT116 cells were treated with 5 μM UNC1999 or 10 μM GSK343 for 6 and 18 h, and the ER stress/UPR-associated genes were analyzed by the Human Unfolded Protein Response Plus PCR Array. The scatter plot compared normalized expression of every gene on the array between control and test samples. The central line indicates unchanged gene expression. The fold regulation cut-off (boundary) was set to “2-fold”. The red circles indicated that genes were up-regulated and the green circles indicated that genes were down-regulated. The genes (red square) with relatively higher average threshold cycle (>30) were not considered significant. The names of significant genes were shown in diagrams. B. HCT116 cells were treated with indicated doses of UNC1999 for 6 h. The protein expressions were analyzed by Western blots. C. HCT116 cells were treated with indicated doses of UNC1999 with or without GSK2606414 for 6 h. The protein expressions were analyzed by Western blots.
Inhibition of ER stress enhances UNC1999-induced cell death
To investigate the role of ER stress/UPR in the anticancer activity of EZH2 inhibitors in CRC cells, HCT116 cells were treated with GSK2606414, and then exposed to UNC1999 for 48 h. The cell viability was examined by a MTT assay. As shown in Figure 7A, synergistic inhibition of cell viability by GSK2606414 and UNC1999 was found. The inhibition of ER stress by GSK2606414 was ascertained by reduction of the eIF2α phosphorylation (Figure 7B). To confirm the enhancement of UNC1999-inducedcell death by GSK2606414, cell apoptosis was examined by the cleavage of poly (ADP-ribose) polymerase (PARP). Indeed, UNC1999-induced PARP cleavage was augmented in the presence of GSK2606414. In addition, UNC1999-induced autophagy (LC3-II accumulation) was not altered by GSK2606414 (Figure 7B). Therefore, ER stress/UPR plays a cytoprotective role in the anticancer activity of UNC1999.
Figure 7.
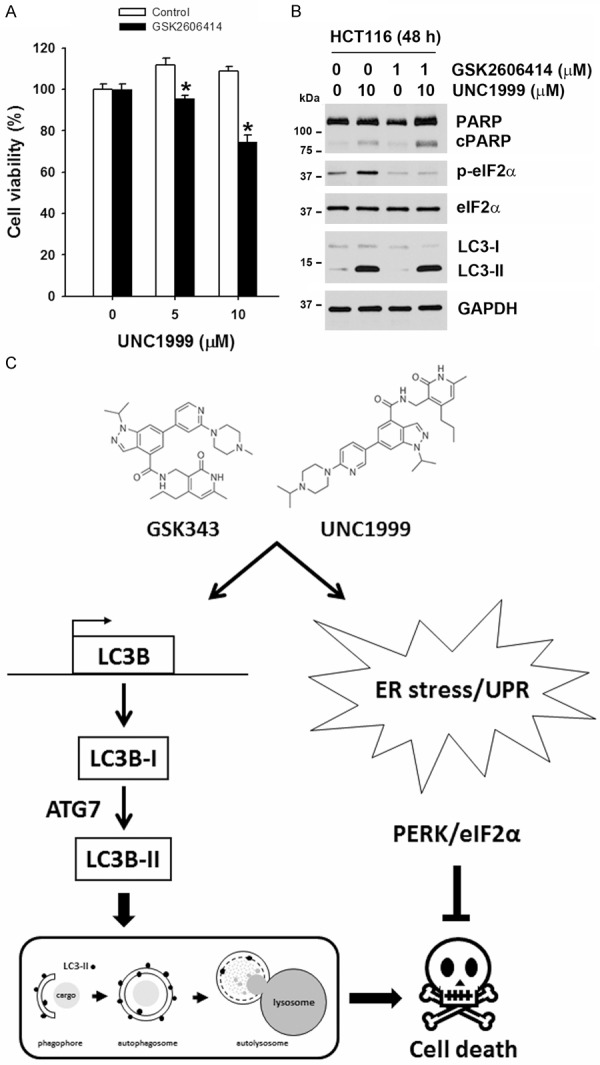
Inhibition of ER stress enhanced UNC1999-induced cell death. (A and B) HCT116 cells were treated with indicated doses of UNC1999 with or without 10 μM GSK2606414 for 48 h. The cell viability was analyzed by an MTT assay (A). The protein expressions were analyzed by Western blots (B). *P < 0.05 indicates significant differences between GSK2606414-treated and control cells. (C) A working model for the action mechanism of EZH2 inhibitors.
Discussion
Recently, non-canonical autophagy has been characterized [37-41]. Unlike canonical autophagy, the formation of the double-membraned autophagosome does not require the hierarchical intervention of all of the ATG proteins [39]. For example, non-canonical autophagy can occur in the Beclin 1/VPS34-, ATG5/ATG7- and ULK1/ULK2-independent manners [37,40,41]. Even so, non-canonical autophagy pathways and structures have the same function as canonical autophagy in sequestering some of the cytoplasm and compartmentalizing pathogens. In addition, material sequestered by non-canonical autophagy is ultimately degraded in the lysosomal compartment [42]. However, no specific markers are able to distinguish between the non-canonical and canonical autophagic pathways. In our study, EZH2 inhibitor induced 3-MA-insenstitive, as well as ATG5- and ULK1-indpendent autophagy despite the partial dependency of ATG7, suggesting that EZH2 inhibitors may also induce non-canonical autophagy. Further investigation is required to clarify the underlying molecular pathways.
Despite the fact that the execution of autophagy includes a unique set of cytoplasmic events, nuclear events, in particular transcriptional programs, have emerged as an important regulator of this process [28,43]. Particularly, recent studies have suggested the existence of epigenetic mechanism for autophagy. For example, the methyltransferase G9a, that catalyzes the dimethylation of histone H3 lysine 9 (H3K9-me2), directly represses genes known to participate in the autophagic process [44]. In addition, induction of autophagy is coupled with the reduction of histone H4 lysine 16 acetylation (H4K16-ac) through downregulation of the histone acetyltransferase hMOF (also called KAT8 or MYST1). H4K16 deacetylation is associated predominantly with the downregulation of autophagy-related genes, and antagonizing H4K16-ac downregulation upon autophagy induction results in the promotion of cell death [45]. Most recently, EZH2 has been shown to inhibit autophagy through epigenetically repressing the negative regulators of mTOR [46]. However, this study is controversial to our results showing that knockdown of EZH2 by siRNA was not sufficient to induce autophagy and overexpression of EZH2 cannot block EZH2 inhibitor-induced autophagy. In addition, our previous study demonstrated that 3-deazaneplanocin A (DZNep), an indirect EZH2 inhibitor through depleting EZH2 [16], fails to induce autophagy [23]. It is likely that cell type or context specificity for EZH2-dependent regulation of autophagy may exist.
In addition to a basic role in the turnover of proteins and organelles, autophagy is observed under pathological conditions including myopathy, neuronal degeneration, infectious disease, and cancer [47,48]. Previous studies have reported that either blockage of autophagy or induction of autophagy could lead to tumor growth. The impact of autophagy appears to vary with intrinsic properties of the tumor [49]. Accumulating evidences indicate that autophagy facilitates the resistance of cancer cells to chemotherapy and radiation [50]. On the other hand, total destruction of the cells by autophagy is served as type II programmed cell death [51]. Autophagic cell death has been reported to be activated in cancer cells in response to various anticancer therapies. Therefore, investigating the role of autophagy in cancer therapy and then modulating the activity of autophagy will improve the efficacy of cancer therapy, which relies on more understanding the mechanism of autophagy. In this notion, our previous and current studies showed that UNC1999 and GSK343 induced autophagic cell death [23]. Therefore, enhancement of autophagy by small molecules, such as rapamycin, might enhance the anticancer activity of UNC1999 and GSK343.
ER stress/UPR could result in both adaptive and apoptotic outputs and is associated with a wide range of diseases including cancers [52]. Generally, the ability of cells to respond to ER stress is critical for cell survival. However, chronic or unresolved ER stress can lead to apoptosis [53]. Our study showed that induction of ER stress/UPR belonged to an immediate response to EZH2 inhibitors, which play a protective role from cancer cell death. Besides, microarray and hallmark gene set analyses found that, in addition to UPR, EZH2 inhibitors may also activate mTOR, PI3K/AKT survival signaling pathways despite of no statistical significance.
Taken together, we proposed a working model (Figure 7C) to describe the action mechanism of EZH2 inhibitors (UNC1999 and GSK343). EZH2 inhibitors exhibit anti-CRC activity through inducing autophagic cell death. EZH2 inhibitors induce autophagy via transcriptionally upregulated the expression of LC3 gene. In contrast, EZH2 inhibitors also trigger ER stress to activate PERK/eIF2α pathway, which plays a protective role in cancer cells. Thus, combination therapy of ER stress and EZH2 inhibitors may have synergistic anti-CRC activity.
Acknowledgements
This work was supported by the Ministry of Science and Technology (MOST103-2311-B-038-003 and MOST104-2320-B-038-005); Taipei Medical University-Shuang Ho Hospital (103TMU-SHH-02 and 104TMU-SHH-03); Research team of prevention and therapy of colorectal cancer in Taipei Medical University (TMU-T104-01); and Comprehensive Cancer Center of Taipei Medical University fund by the Health and welfare surcharge of tobacco products (MOHW105-TDU-B-212-134001).
Disclosure of conflict of interest
None.
References
- 1.Meijer AJ, Dubbelhuis PF. Amino acid signalling and the integration of metabolism. Biochem Biophys Res Commun. 2004;313:397–403. doi: 10.1016/j.bbrc.2003.07.012. [DOI] [PubMed] [Google Scholar]
- 2.Levine B. Cell biology: autophagy and cancer. Nature. 2007;446:745–747. doi: 10.1038/446745a. [DOI] [PubMed] [Google Scholar]
- 3.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 7.Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3’-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 9.Hardie DG, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- 10.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 11.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, Sabel MS, Livant D, Weiss SJ, Rubin MA, Chinnaiyan AM. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raaphorst FM, Meijer CJ, Fieret E, Blokzijl T, Mommers E, Buerger H, Packeisen J, Sewalt RA, Otte AP, van Diest PJ. Poorly differentiated breast carcinoma is associated with increased expression of the human polycomb group EZH2 gene. Neoplasia. 2003;5:481–488. doi: 10.1016/s1476-5586(03)80032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 15.Croonquist PA, Van Ness B. The polycomb group protein enhancer of zeste homolog 2 (EZH 2) is an oncogene that influences myeloma cell growth and the mutant ras phenotype. Oncogene. 2005;24:6269–6280. doi: 10.1038/sj.onc.1208771. [DOI] [PubMed] [Google Scholar]
- 16.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RK, Tan PB, Liu ET, Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan JZ, Yan Y, Wang XX, Jiang Y, Xu HE. EZH2: biology, disease, and structure-based drug discovery. Acta Pharmacol Sin. 2014;35:161–174. doi: 10.1038/aps.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, Sacks JD, Raimondi A, Majer CR, Song J, Scott MP, Jin L, Smith JJ, Olhava EJ, Chesworth R, Moyer MP, Richon VM, Copeland RA, Keilhack H, Pollock RM, Kuntz KW. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 19.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A 3rd, Diaz E, LaFrance LV, Mellinger M, Duquenne C, Tian X, Kruger RG, McHugh CF, Brandt M, Miller WH, Dhanak D, Verma SK, Tummino PJ, Creasy CL. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 20.Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, Zeng J, Li M, Fan H, Lin Y, Gu J, Ardayfio O, Zhang JH, Yan X, Fang J, Mi Y, Zhang M, Zhou T, Feng G, Chen Z, Li G, Yang T, Zhao K, Liu X, Yu Z, Lu CX, Atadja P, Li E. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci U S A. 2012;109:21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verma SK, Tian X, LaFrance LV, Duquenne C, Suarez DP, Newlander KA, Romeril SP, Burgess JL, Grant SW, Brackley JA, Graves AP, Scherzer DA, Shu A, Thompson C, Ott HM, Aller GSV, Machutta CA, Diaz E, Jiang Y, Johnson NW, Knight SD, Kruger RG, McCabe MT, Dhanak D, Tummino PJ, Creasy CL, Miller WH. Identification of Potent, Selective, Cell-Active Inhibitors of the Histone Lysine Methyltransferase EZH2. ACS Med Chem Lett. 2012;3:1091–1096. doi: 10.1021/ml3003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konze KD, Ma A, Li F, Barsyte-Lovejoy D, Parton T, Macnevin CJ, Liu F, Gao C, Huang XP, Kuznetsova E, Rougie M, Jiang A, Pattenden SG, Norris JL, James LI, Roth BL, Brown PJ, Frye SV, Arrowsmith CH, Hahn KM, Wang GG, Vedadi M, Jin J. An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1. ACS Chem Biol. 2013;8:1324–1334. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu TP, Lo HL, Wei LS, Hsiao HH, Yang PM. S-Adenosyl-L-methionine-competitive inhibitors of the histone methyltransferase EZH2 induce autophagy and enhance drug sensitivity in cancer cells. Anticancer Drugs. 2015;26:139–47. doi: 10.1097/CAD.0000000000000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, Airoldi EM, Ait-Si-Ali S, Akematsu T, Akporiaye ET, Al-Rubeai M, Albaiceta GM, Albanese C, Albani D, Albert ML, Aldudo J, Algül H, Alirezaei M, Alloza I, Almasan A, Almonte-Beceril M, Alnemri ES, Alonso C, Altan-Bonnet N, Altieri DC, Alvarez S, Alvarez-Erviti L, Alves S, Amadoro G, Amano A, Amantini C, Ambrosio S, Amelio I, Amer AO, Amessou M, Amon A, An Z, Anania FA, Andersen SU, Andley UP, Andreadi CK, Andrieu-Abadie N, Anel A, Ann DK, Anoopkumar-Dukie S, Antonioli M, Aoki H, Apostolova N, Aquila S, Aquilano K, Araki K, Arama E, Aranda A, Araya J, Arcaro A, Arias E, Arimoto H, Ariosa AR, Armstrong JL, Arnould T, Arsov I, Asanuma K, Askanas V, Asselin E, Atarashi R, Atherton SS, Atkin JD, Attardi LD, Auberger P, Auburger G, Aurelian L, Autelli R, Avagliano L, Avantaggiati ML, Avrahami L, Awale S, Azad N, Bachetti T, Backer JM, Bae DH, Bae JS, Bae ON, Bae SH, Baehrecke EH, Baek SH, Baghdiguian S, Bagniewska-Zadworna A, Bai H, Bai J, Bai XY, Bailly Y, Balaji KN, Balduini W, Ballabio A, Balzan R, Banerjee R, Bánhegyi G, Bao H, Barbeau B, Barrachina MD, Barreiro E, Bartel B, Bartolomé A, Bassham DC, Bassi MT, Bast RC Jr, Basu A, Batista MT, Batoko H, Battino M, Bauckman K, Baumgarner BL, Bayer KU, Beale R, Beaulieu JF, Beck GR Jr, Becker C, Beckham JD, Bédard PA, Bednarski PJ, Begley TJ, Behl C, Behrends C, Behrens GM, Behrns KE, Bejarano E, Belaid A, Belleudi F, Bénard G, Berchem G, Bergamaschi D, Bergami M, Berkhout B, Berliocchi L, Bernard A, Bernard M, Bernassola F, Bertolotti A, Bess AS, Besteiro S, Bettuzzi S, Bhalla S, Bhattacharyya S, Bhutia SK, Biagosch C, Bianchi MW, Biard-Piechaczyk M, Billes V, Bincoletto C, Bingol B, Bird SW, Bitoun M, Bjedov I, Blackstone C, Blanc L, Blanco GA, Blomhoff HK, Boada-Romero E, Böckler S, Boes M, Boesze-Battaglia K, Boise LH, Bolino A, Boman A, Bonaldo P, Bordi M, Bosch J, Botana LM, Botti J, Bou G, Bouché M, Bouchecareilh M, Boucher MJ, Boulton ME, Bouret SG, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady N, Braga VM, Brancolini C, Braus GH, Bravo-San Pedro JM, Brennan LA, Bresnick EH, Brest P, Bridges D, Bringer MA, Brini M, Brito GC, Brodin B, Brookes PS, Brown EJ, Brown K, Broxmeyer HE, Bruhat A, Brum PC, Brumell JH, Brunetti-Pierri N, Bryson-Richardson RJ, Buch S, Buchan AM, Budak H, Bulavin DV, Bultman SJ, Bultynck G, Bumbasirevic V, Burelle Y, Burke RE, Burmeister M, Bütikofer P, Caberlotto L, Cadwell K, Cahova M, Cai D, Cai J, Cai Q, Calatayud S, Camougrand N, Campanella M, Campbell GR, Campbell M, Campello S, Candau R, Caniggia I, Cantoni L, Cao L, Caplan AB, Caraglia M, Cardinali C, Cardoso SM, Carew JS, Carleton LA, Carlin CR, Carloni S, Carlsson SR, Carmona-Gutierrez D, Carneiro LA, Carnevali O, Carra S, Carrier A, Carroll B, Casas C, Casas J, Cassinelli G, Castets P, Castro-Obregon S, Cavallini G, Ceccherini I, Cecconi F, Cederbaum AI, Ceña V, Cenci S, Cerella C, Cervia D, Cetrullo S, Chaachouay H, Chae HJ, Chagin AS, Chai CY, Chakrabarti G, Chamilos G, Chan EY, Chan MT, Chandra D, Chandra P, Chang CP, Chang RC, Chang TY, Chatham JC, Chatterjee S, Chauhan S, Che Y, Cheetham ME, Cheluvappa R, Chen CJ, Chen G, Chen GC, Chen G, Chen H, Chen JW, Chen JK, Chen M, Chen M, Chen P, Chen Q, Chen Q, Chen SD, Chen S, Chen SS, Chen W, Chen WJ, Chen WQ, Chen W, Chen X, Chen YH, Chen YG, Chen Y, Chen Y, Chen Y, Chen YJ, Chen YQ, Chen Y, Chen Z, Chen Z, Cheng A, Cheng CH, Cheng H, Cheong H, Cherry S, Chesney J, Cheung CH, Chevet E, Chi HC, Chi SG, Chiacchiera F, Chiang HL, Chiarelli R, Chiariello M, Chieppa M, Chin LS, Chiong M, Chiu GN, Cho DH, Cho SG, Cho WC, Cho YY, Cho YS, Choi AM, Choi EJ, Choi EK, Choi J, Choi ME, Choi SI, Chou TF, Chouaib S, Choubey D, Choubey V, Chow KC, Chowdhury K, Chu CT, Chuang TH, Chun T, Chung H, Chung T, Chung YL, Chwae YJ, Cianfanelli V, Ciarcia R, Ciechomska IA, Ciriolo MR, Cirone M, Claerhout S, Clague MJ, Clària J, Clarke PG, Clarke R, Clementi E, Cleyrat C, Cnop M, Coccia EM, Cocco T, Codogno P, Coers J, Cohen EE, Colecchia D, Coletto L, Coll NS, Colucci-Guyon E, Comincini S, Condello M, Cook KL, Coombs GH, Cooper CD, Cooper JM, Coppens I, Corasaniti MT, Corazzari M, Corbalan R, Corcelle-Termeau E, Cordero MD, Corral-Ramos C, Corti O, Cossarizza A, Costelli P, Costes S, Cotman SL, Coto-Montes A, Cottet S, Couve E, Covey LR, Cowart LA, Cox JS, Coxon FP, Coyne CB, Cragg MS, Craven RJ, Crepaldi T, Crespo JL, Criollo A, Crippa V, Cruz MT, Cuervo AM, Cuezva JM, Cui T, Cutillas PR, Czaja MJ, Czyzyk-Krzeska MF, Dagda RK, Dahmen U, Dai C, Dai W, Dai Y, Dalby KN, Dalla Valle L, Dalmasso G, D'Amelio M, Damme M, Darfeuille-Michaud A, Dargemont C, Darley-Usmar VM, Dasarathy S, Dasgupta B, Dash S, Dass CR, Davey HM, Davids LM, Dávila D, Davis RJ, Dawson TM, Dawson VL, Daza P, de Belleroche J, de Figueiredo P, de Figueiredo RC, de la Fuente J, De Martino L, De Matteis A, De Meyer GR, De Milito A, De Santi M, de Souza W, De Tata V, De Zio D, Debnath J, Dechant R, Decuypere JP, Deegan S, Dehay B, Del Bello B, Del Re DP, Delage-Mourroux R, Delbridge LM, Deldicque L, Delorme-Axford E, Deng Y, Dengjel J, Denizot M, Dent P, Der CJ, Deretic V, Derrien B, Deutsch E, Devarenne TP, Devenish RJ, Di Bartolomeo S, Di Daniele N, Di Domenico F, Di Nardo A, Di Paola S, Di Pietro A, Di Renzo L, DiAntonio A, Díaz-Araya G, Díaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dickey CA, Dickson RC, Diederich M, Digard P, Dikic I, Dinesh-Kumar SP, Ding C, Ding WX, Ding Z, Dini L, Distler JH, Diwan A, Djavaheri-Mergny M, Dmytruk K, Dobson RC, Doetsch V, Dokladny K, Dokudovskaya S, Donadelli M, Dong XC, Dong X, Dong Z, Donohue TM Jr, Doran KS, D’Orazi G, Dorn GW 2nd, Dosenko V, Dridi S, Drucker L, Du J, Du LL, Du L, du Toit A, Dua P, Duan L, Duann P, Dubey VK, Duchen MR, Duchosal MA, Duez H, Dugail I, Dumit VI, Duncan MC, Dunlop EA, Dunn WA Jr, Dupont N, Dupuis L, Durán RV, Durcan TM, Duvezin-Caubet S, Duvvuri U, Eapen V, Ebrahimi-Fakhari D, Echard A, Eckhart L, Edelstein CL, Edinger AL, Eichinger L, Eisenberg T, Eisenberg-Lerner A, Eissa NT, El-Deiry WS, El-Khoury V, Elazar Z, Eldar-Finkelman H, Elliott CJ, Emanuele E, Emmenegger U, Engedal N, Engelbrecht AM, Engelender S, Enserink JM, Erdmann R, Erenpreisa J, Eri R, Eriksen JL, Erman A, Escalante R, Eskelinen EL, Espert L, Esteban-Martínez L, Evans TJ, Fabri M, Fabrias G, Fabrizi C, Facchiano A, Færgeman NJ, Faggioni A, Fairlie WD, Fan C, Fan D, Fan J, Fang S, Fanto M, Fanzani A, Farkas T, Faure M, Favier FB, Fearnhead H, Federici M, Fei E, Felizardo TC, Feng H, Feng Y, Feng Y, Ferguson TA, Fernández ÁF, Fernandez-Barrena MG, Fernandez-Checa JC, Fernández-López A, Fernandez-Zapico ME, Feron O, Ferraro E, Ferreira-Halder CV, Fesus L, Feuer R, Fiesel FC, Filippi-Chiela EC, Filomeni G, Fimia GM, Fingert JH, Finkbeiner S, Finkel T, Fiorito F, Fisher PB, Flajolet M, Flamigni F, Florey O, Florio S, Floto RA, Folini M, Follo C, Fon EA, Fornai F, Fortunato F, Fraldi A, Franco R, Francois A, François A, Frankel LB, Fraser ID, Frey N, Freyssenet DG, Frezza C, Friedman SL, Frigo DE, Fu D, Fuentes JM, Fueyo J, Fujitani Y, Fujiwara Y, Fujiya M, Fukuda M, Fulda S, Fusco C, Gabryel B, Gaestel M, Gailly P, Gajewska M, Galadari S, Galili G, Galindo I, Galindo MF, Galliciotti G, Galluzzi L, Galluzzi L, Galy V, Gammoh N, Gandy S, Ganesan AK, Ganesan S, Ganley IG, Gannagé M, Gao FB, Gao F, Gao JX, García Nannig L, García Véscovi E, Garcia-Macía M, Garcia-Ruiz C, Garg AD, Garg PK, Gargini R, Gassen NC, Gatica D, Gatti E, Gavard J, Gavathiotis E, Ge L, Ge P, Ge S, Gean PW, Gelmetti V, Genazzani AA, Geng J, Genschik P, Gerner L, Gestwicki JE, Gewirtz DA, Ghavami S, Ghigo E, Ghosh D, Giammarioli AM, Giampieri F, Giampietri C, Giatromanolaki A, Gibbings DJ, Gibellini L, Gibson SB, Ginet V, Giordano A, Giorgini F, Giovannetti E, Girardin SE, Gispert S, Giuliano S, Gladson CL, Glavic A, Gleave M, Godefroy N, Gogal RM Jr, Gokulan K, Goldman GH, Goletti D, Goligorsky MS, Gomes AV, Gomes LC, Gomez H, Gomez-Manzano C, Gómez-Sánchez R, Gonçalves DA, Goncu E, Gong Q, Gongora C, Gonzalez CB, Gonzalez-Alegre P, Gonzalez-Cabo P, González-Polo RA, Goping IS, Gorbea C, Gorbunov NV, Goring DR, Gorman AM, Gorski SM, Goruppi S, Goto-Yamada S, Gotor C, Gottlieb RA, Gozes I, Gozuacik D, Graba Y, Graef M, Granato GE, Grant GD, Grant S, Gravina GL, Green DR, Greenhough A, Greenwood MT, Grimaldi B, Gros F, Grose C, Groulx JF, Gruber F, Grumati P, Grune T, Guan JL, Guan KL, Guerra B, Guillen C, Gulshan K, Gunst J, Guo C, Guo L, Guo M, Guo W, Guo XG, Gust AA, Gustafsson ÅB, Gutierrez E, Gutierrez MG, Gwak HS, Haas A, Haber JE, Hadano S, Hagedorn M, Hahn DR, Halayko AJ, Hamacher-Brady A, Hamada K, Hamai A, Hamann A, Hamasaki M, Hamer I, Hamid Q, Hammond EM, Han F, Han W, Handa JT, Hanover JA, Hansen M, Harada M, Harhaji-Trajkovic L, Harper JW, Harrath AH, Harris AL, Harris J, Hasler U, Hasselblatt P, Hasui K, Hawley RG, Hawley TS, He C, He CY, He F, He G, He RR, He XH, He YW, He YY, Heath JK, Hébert MJ, Heinzen RA, Helgason GV, Hensel M, Henske EP, Her C, Herman PK, Hernández A, Hernandez C, Hernández-Tiedra S, Hetz C, Hiesinger PR, Higaki K, Hilfiker S, Hill BG, Hill JA, Hill WD, Hino K, Hofius D, Hofman P, Höglinger GU, Höhfeld J, Holz MK, Hong Y, Hood DA, Hoozemans JJ, Hoppe T, Hsu C, Hsu CY, Hsu LC, Hu D, Hu G, Hu HM, Hu H, Hu MC, Hu YC, Hu ZW, Hua F, Hua Y, Huang C, Huang HL, Huang KH, Huang KY, Huang S, Huang S, Huang WP, Huang YR, Huang Y, Huang Y, Huber TB, Huebbe P, Huh WK, Hulmi JJ, Hur GM, Hurley JH, Husak Z, Hussain SN, Hussain S, Hwang JJ, Hwang S, Hwang TI, Ichihara A, Imai Y, Imbriano C, Inomata M, Into T, Iovane V, Iovanna JL, Iozzo RV, Ip NY, Irazoqui JE, Iribarren P, Isaka Y, Isakovic AJ, Ischiropoulos H, Isenberg JS, Ishaq M, Ishida H, Ishii I, Ishmael JE, Isidoro C, Isobe K, Isono E, Issazadeh-Navikas S, Itahana K, Itakura E, Ivanov AI, Iyer AK, Izquierdo JM, Izumi Y, Izzo V, Jäättelä M, Jaber N, Jackson DJ, Jackson WT, Jacob TG, Jacques TS, Jagannath C, Jain A, Jana NR, Jang BK, Jani A, Janji B, Jannig PR, Jansson PJ, Jean S, Jendrach M, Jeon JH, Jessen N, Jeung EB, Jia K, Jia L, Jiang H, Jiang H, Jiang L, Jiang T, Jiang X, Jiang X, Jiang X, Jiang Y, Jiang Y, Jiménez A, Jin C, Jin H, Jin L, Jin M, Jin S, Jinwal UK, Jo EK, Johansen T, Johnson DE, Johnson GV, Johnson JD, Jonasch E, Jones C, Joosten LA, Jordan J, Joseph AM, Joseph B, Joubert AM, Ju D, Ju J, Juan HF, Juenemann K, Juhász G, Jung HS, Jung JU, Jung YK, Jungbluth H, Justice MJ, Jutten B, Kaakoush NO, Kaarniranta K, Kaasik A, Kabuta T, Kaeffer B, Kågedal K, Kahana A, Kajimura S, Kakhlon O, Kalia M, Kalvakolanu DV, Kamada Y, Kambas K, Kaminskyy VO, Kampinga HH, Kandouz M, Kang C, Kang R, Kang TC, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kantorow M, Kaparakis-Liaskos M, Kapuy O, Karantza V, Karim MR, Karmakar P, Kaser A, Kaushik S, Kawula T, Kaynar AM, Ke PY, Ke ZJ, Kehrl JH, Keller KE, Kemper JK, Kenworthy AK, Kepp O, Kern A, Kesari S, Kessel D, Ketteler R, Kettelhut Ido C, Khambu B, Khan MM, Khandelwal VK, Khare S, Kiang JG, Kiger AA, Kihara A, Kim AL, Kim CH, Kim DR, Kim DH, Kim EK, Kim HY, Kim HR, Kim JS, Kim JH, Kim JC, Kim JH, Kim KW, Kim MD, Kim MM, Kim PK, Kim SW, Kim SY, Kim YS, Kim Y, Kimchi A, Kimmelman AC, Kimura T, King JS, Kirkegaard K, Kirkin V, Kirshenbaum LA, Kishi S, Kitajima Y, Kitamoto K, Kitaoka Y, Kitazato K, Kley RA, Klimecki WT, Klinkenberg M, Klucken J, Knævelsrud H, Knecht E, Knuppertz L, Ko JL, Kobayashi S, Koch JC, Koechlin-Ramonatxo C, Koenig U, Koh YH, Köhler K, Kohlwein SD, Koike M, Komatsu M, Kominami E, Kong D, Kong HJ, Konstantakou EG, Kopp BT, Korcsmaros T, Korhonen L, Korolchuk VI, Koshkina NV, Kou Y, Koukourakis MI, Koumenis C, Kovács AL, Kovács T, Kovacs WJ, Koya D, Kraft C, Krainc D, Kramer H, Kravic-Stevovic T, Krek W, Kretz-Remy C, Krick R, Krishnamurthy M, Kriston-Vizi J, Kroemer G, Kruer MC, Kruger R, Ktistakis NT, Kuchitsu K, Kuhn C, Kumar AP, Kumar A, Kumar A, Kumar D, Kumar D, Kumar R, Kumar S, Kundu M, Kung HJ, Kuno A, Kuo SH, Kuret J, Kurz T, Kwok T, Kwon TK, Kwon YT, Kyrmizi I, La Spada AR, Lafont F, Lahm T, Lakkaraju A, Lam T, Lamark T, Lancel S, Landowski TH, Lane DJ, Lane JD, Lanzi C, Lapaquette P, Lapierre LR, Laporte J, Laukkarinen J, Laurie GW, Lavandero S, Lavie L, LaVoie MJ, Law BY, Law HK, Law KB, Layfield R, Lazo PA, Le Cam L, Le Roch KG, Le Stunff H, Leardkamolkarn V, Lecuit M, Lee BH, Lee CH, Lee EF, Lee GM, Lee HJ, Lee H, Lee JK, Lee J, Lee JH, Lee JH, Lee M, Lee MS, Lee PJ, Lee SW, Lee SJ, Lee SJ, Lee SY, Lee SH, Lee SS, Lee SJ, Lee S, Lee YR, Lee YJ, Lee YH, Leeuwenburgh C, Lefort S, Legouis R, Lei J, Lei QY, Leib DA, Leibowitz G, Lekli I, Lemaire SD, Lemasters JJ, Lemberg MK, Lemoine A, Leng S, Lenz G, Lenzi P, Lerman LO, Lettieri Barbato D, Leu JI, Leung HY, Levine B, Lewis PA, Lezoualc’h F, Li C, Li F, Li FJ, Li J, Li K, Li L, Li M, Li M, Li Q, Li R, Li S, Li W, Li W, Li X, Li Y, Lian J, Liang C, Liang Q, Liao Y, Liberal J, Liberski PP, Lie P, Lieberman AP, Lim HJ, Lim KL, Lim K, Lima RT, Lin CS, Lin CF, Lin F, Lin F, Lin FC, Lin K, Lin KH, Lin PH, Lin T, Lin WW, Lin YS, Lin Y, Linden R, Lindholm D, Lindqvist LM, Lingor P, Linkermann A, Liotta LA, Lipinski MM, Lira VA, Lisanti MP, Liton PB, Liu B, Liu C, Liu CF, Liu F, Liu HJ, Liu J, Liu JJ, Liu JL, Liu K, Liu L, Liu L, Liu Q, Liu RY, Liu S, Liu S, Liu W, Liu XD, Liu X, Liu XH, Liu X, Liu X, Liu X, Liu Y, Liu Y, Liu Z, Liu Z, Liuzzi JP, Lizard G, Ljujic M, Lodhi IJ, Logue SE, Lokeshwar BL, Long YC, Lonial S, Loos B, López-Otín C, López-Vicario C, Lorente M, Lorenzi PL, Lõrincz P, Los M, Lotze MT, Lovat PE, Lu B, Lu B, Lu J, Lu Q, Lu SM, Lu S, Lu Y, Luciano F, Luckhart S, Lucocq JM, Ludovico P, Lugea A, Lukacs NW, Lum JJ, Lund AH, Luo H, Luo J, Luo S, Luparello C, Lyons T, Ma J, Ma Y, Ma Y, Ma Z, Machado J, Machado-Santelli GM, Macian F, MacIntosh GC, MacKeigan JP, Macleod KF, MacMicking JD, MacMillan-Crow LA, Madeo F, Madesh M, Madrigal-Matute J, Maeda A, Maeda T, Maegawa G, Maellaro E, Maes H, Magariños M, Maiese K, Maiti TK, Maiuri L, Maiuri MC, Maki CG, Malli R, Malorni W, Maloyan A, Mami-Chouaib F, Man N, Mancias JD, Mandelkow EM, Mandell MA, Manfredi AA, Manié SN, Manzoni C, Mao K, Mao Z, Mao ZW, Marambaud P, Marconi AM, Marelja Z, Marfe G, Margeta M, Margittai E, Mari M, Mariani FV, Marin C, Marinelli S, Mariño G, Markovic I, Marquez R, Martelli AM, Martens S, Martin KR, Martin SJ, Martin S, Martin-Acebes MA, Martín-Sanz P, Martinand-Mari C, Martinet W, Martinez J, Martinez-Lopez N, Martinez-Outschoorn U, Martínez-Velázquez M, Martinez-Vicente M, Martins WK, Mashima H, Mastrianni JA, Matarese G, Matarrese P, Mateo R, Matoba S, Matsumoto N, Matsushita T, Matsuura A, Matsuzawa T, Mattson MP, Matus S, Maugeri N, Mauvezin C, Mayer A, Maysinger D, Mazzolini GD, McBrayer MK, McCall K, McCormick C, McInerney GM, McIver SC, McKenna S, McMahon JJ, McNeish IA, Mechta-Grigoriou F, Medema JP, Medina DL, Megyeri K, Mehrpour M, Mehta JL, Mei Y, Meier UC, Meijer AJ, Meléndez A, Melino G, Melino S, de Melo EJ, Mena MA, Meneghini MD, Menendez JA, Menezes R, Meng L, Meng LH, Meng S, Menghini R, Menko AS, Menna-Barreto RF, Menon MB, Meraz-Ríos MA, Merla G, Merlini L, Merlot AM, Meryk A, Meschini S, Meyer JN, Mi MT, Miao CY, Micale L, Michaeli S, Michiels C, Migliaccio AR, Mihailidou AS, Mijaljica D, Mikoshiba K, Milan E, Miller-Fleming L, Mills GB, Mills IG, Minakaki G, Minassian BA, Ming XF, Minibayeva F, Minina EA, Mintern JD, Minucci S, Miranda-Vizuete A, Mitchell CH, Miyamoto S, Miyazawa K, Mizushima N, Mnich K, Mograbi B, Mohseni S, Moita LF, Molinari M, Molinari M, Møller AB, Mollereau B, Mollinedo F, Mongillo M, Monick MM, Montagnaro S, Montell C, Moore DJ, Moore MN, Mora-Rodriguez R, Moreira PI, Morel E, Morelli MB, Moreno S, Morgan MJ, Moris A, Moriyasu Y, Morrison JL, Morrison LA, Morselli E, Moscat J, Moseley PL, Mostowy S, Motori E, Mottet D, Mottram JC, Moussa CE, Mpakou VE, Mukhtar H, Mulcahy Levy JM, Muller S, Muñoz-Moreno R, Muñoz-Pinedo C, Münz C, Murphy ME, Murray JT, Murthy A, Mysorekar IU, Nabi IR, Nabissi M, Nader GA, Nagahara Y, Nagai Y, Nagata K, Nagelkerke A, Nagy P, Naidu SR, Nair S, Nakano H, Nakatogawa H, Nanjundan M, Napolitano G, Naqvi NI, Nardacci R, Narendra DP, Narita M, Nascimbeni AC, Natarajan R, Navegantes LC, Nawrocki ST, Nazarko TY, Nazarko VY, Neill T, Neri LM, Netea MG, Netea-Maier RT, Neves BM, Ney PA, Nezis IP, Nguyen HT, Nguyen HP, Nicot AS, Nilsen H, Nilsson P, Nishimura M, Nishino I, Niso-Santano M, Niu H, Nixon RA, Njar VC, Noda T, Noegel AA, Nolte EM, Norberg E, Norga KK, Noureini SK, Notomi S, Notterpek L, Nowikovsky K, Nukina N, Nürnberger T, O’Donnell VB, O’Donovan T, O’Dwyer PJ, Oehme I, Oeste CL, Ogawa M, Ogretmen B, Ogura Y, Oh YJ, Ohmuraya M, Ohshima T, Ojha R, Okamoto K, Okazaki T, Oliver FJ, Ollinger K, Olsson S, Orban DP, Ordonez P, Orhon I, Orosz L, O'Rourke EJ, Orozco H, Ortega AL, Ortona E, Osellame LD, Oshima J, Oshima S, Osiewacz HD, Otomo T, Otsu K, Ou JH, Outeiro TF, Ouyang DY, Ouyang H, Overholtzer M, Ozbun MA, Ozdinler PH, Ozpolat B, Pacelli C, Paganetti P, Page G, Pages G, Pagnini U, Pajak B, Pak SC, Pakos-Zebrucka K, Pakpour N, Palková Z, Palladino F, Pallauf K, Pallet N, Palmieri M, Paludan SR, Palumbo C, Palumbo S, Pampliega O, Pan H, Pan W, Panaretakis T, Pandey A, Pantazopoulou A, Papackova Z, Papademetrio DL, Papassideri I, Papini A, Parajuli N, Pardo J, Parekh VV, Parenti G, Park JI, Park J, Park OK, Parker R, Parlato R, Parys JB, Parzych KR, Pasquet JM, Pasquier B, Pasumarthi KB, Patschan D, Patterson C, Pattingre S, Pattison S, Pause A, Pavenstädt H, Pavone F, Pedrozo Z, Peña FJ, Peñalva MA, Pende M, Peng J, Penna F, Penninger JM, Pensalfini A, Pepe S, Pereira GJ, Pereira PC, Pérez-de la Cruz V, Pérez-Pérez ME, Pérez-Rodríguez D, Pérez-Sala D, Perier C, Perl A, Perlmutter DH, Perrotta I, Pervaiz S, Pesonen M, Pessin JE, Peters GJ, Petersen M, Petrache I, Petrof BJ, Petrovski G, Phang JM, Piacentini M, Pierdominici M, Pierre P, Pierrefite-Carle V, Pietrocola F, Pimentel-Muiños FX, Pinar M, Pineda B, Pinkas-Kramarski R, Pinti M, Pinton P, Piperdi B, Piret JM, Platanias LC, Platta HW, Plowey ED, Pöggeler S, Poirot M, Polčic P, Poletti A, Poon AH, Popelka H, Popova B, Poprawa I, Poulose SM, Poulton J, Powers SK, Powers T, Pozuelo-Rubio M, Prak K, Prange R, Prescott M, Priault M, Prince S, Proia RL, Proikas-Cezanne T, Prokisch H, Promponas VJ, Przyklenk K, Puertollano R, Pugazhenthi S, Puglielli L, Pujol A, Puyal J, Pyeon D, Qi X, Qian WB, Qin ZH, Qiu Y, Qu Z, Quadrilatero J, Quinn F, Raben N, Rabinowich H, Radogna F, Ragusa MJ, Rahmani M, Raina K, Ramanadham S, Ramesh R, Rami A, Randall-Demllo S, Randow F, Rao H, Rao VA, Rasmussen BB, Rasse TM, Ratovitski EA, Rautou PE, Ray SK, Razani B, Reed BH, Reggiori F, Rehm M, Reichert AS, Rein T, Reiner DJ, Reits E, Ren J, Ren X, Renna M, Reusch JE, Revuelta JL, Reyes L, Rezaie AR, Richards RI, Richardson DR, Richetta C, Riehle MA, Rihn BH, Rikihisa Y, Riley BE, Rimbach G, Rippo MR, Ritis K, Rizzi F, Rizzo E, Roach PJ, Robbins J, Roberge M, Roca G, Roccheri MC, Rocha S, Rodrigues CM, Rodríguez CI, de Cordoba SR, Rodriguez-Muela N, Roelofs J, Rogov VV, Rohn TT, Rohrer B, Romanelli D, Romani L, Romano PS, Roncero MI, Rosa JL, Rosello A, Rosen KV, Rosenstiel P, Rost-Roszkowska M, Roth KA, Roué G, Rouis M, Rouschop KM, Ruan DT, Ruano D, Rubinsztein DC, Rucker EB 3rd, Rudich A, Rudolf E, Rudolf R, Ruegg MA, Ruiz-Roldan C, Ruparelia AA, Rusmini P, Russ DW, Russo GL, Russo G, Russo R, Rusten TE, Ryabovol V, Ryan KM, Ryter SW, Sabatini DM, Sacher M, Sachse C, Sack MN, Sadoshima J, Saftig P, Sagi-Eisenberg R, Sahni S, Saikumar P, Saito T, Saitoh T, Sakakura K, Sakoh-Nakatogawa M, Sakuraba Y, Salazar-Roa M, Salomoni P, Saluja AK, Salvaterra PM, Salvioli R, Samali A, Sanchez AM, Sánchez-Alcázar JA, Sanchez-Prieto R, Sandri M, Sanjuan MA, Santaguida S, Santambrogio L, Santoni G, Dos Santos CN, Saran S, Sardiello M, Sargent G, Sarkar P, Sarkar S, Sarrias MR, Sarwal MM, Sasakawa C, Sasaki M, Sass M, Sato K, Sato M, Satriano J, Savaraj N, Saveljeva S, Schaefer L, Schaible UE, Scharl M, Schatzl HM, Schekman R, Scheper W, Schiavi A, Schipper HM, Schmeisser H, Schmidt J, Schmitz I, Schneider BE, Schneider EM, Schneider JL, Schon EA, Schönenberger MJ, Schönthal AH, Schorderet DF, Schröder B, Schuck S, Schulze RJ, Schwarten M, Schwarz TL, Sciarretta S, Scotto K, Scovassi AI, Screaton RA, Screen M, Seca H, Sedej S, Segatori L, Segev N, Seglen PO, Seguí-Simarro JM, Segura-Aguilar J, Seki E, Sell C, Seiliez I, Semenkovich CF, Semenza GL, Sen U, Serra AL, Serrano-Puebla A, Sesaki H, Setoguchi T, Settembre C, Shacka JJ, Shajahan-Haq AN, Shapiro IM, Sharma S, She H, Shen CK, Shen CC, Shen HM, Shen S, Shen W, Sheng R, Sheng X, Sheng ZH, Shepherd TG, Shi J, Shi Q, Shi Q, Shi Y, Shibutani S, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shin DW, Shinohara ML, Shintani M, Shintani T, Shioi T, Shirabe K, Shiri-Sverdlov R, Shirihai O, Shore GC, Shu CW, Shukla D, Sibirny AA, Sica V, Sigurdson CJ, Sigurdsson EM, Sijwali PS, Sikorska B, Silveira WA, Silvente-Poirot S, Silverman GA, Simak J, Simmet T, Simon AK, Simon HU, Simone C, Simons M, Simonsen A, Singh R, Singh SV, Singh SK, Sinha D, Sinha S, Sinicrope FA, Sirko A, Sirohi K, Sishi BJ, Sittler A, Siu PM, Sivridis E, Skwarska A, Slack R, Slaninová I, Slavov N, Smaili SS, Smalley KS, Smith DR, Soenen SJ, Soleimanpour SA, Solhaug A, Somasundaram K, Son JH, Sonawane A, Song C, Song F, Song HK, Song JX, Song W, Soo KY, Sood AK, Soong TW, Soontornniyomkij V, Sorice M, Sotgia F, Soto-Pantoja DR, Sotthibundhu A, Sousa MJ, Spaink HP, Span PN, Spang A, Sparks JD, Speck PG, Spector SA, Spies CD, Springer W, Clair DS, Stacchiotti A, Staels B, Stang MT, Starczynowski DT, Starokadomskyy P, Steegborn C, Steele JW, Stefanis L, Steffan J, Stellrecht CM, Stenmark H, Stepkowski TM, Stern ST, Stevens C, Stockwell BR, Stoka V, Storchova Z, Stork B, Stratoulias V, Stravopodis DJ, Strnad P, Strohecker AM, Ström AL, Stromhaug P, Stulik J, Su YX, Su Z, Subauste CS, Subramaniam S, Sue CM, Suh SW, Sui X, Sukseree S, Sulzer D, Sun FL, Sun J, Sun J, Sun SY, Sun Y, Sun Y, Sun Y, Sundaramoorthy V, Sung J, Suzuki H, Suzuki K, Suzuki N, Suzuki T, Suzuki YJ, Swanson MS, Swanton C, Swärd K, Swarup G, Sweeney ST, Sylvester PW, Szatmari Z, Szegezdi E, Szlosarek PW, Taegtmeyer H, Tafani M, Taillebourg E, Tait SW, Takacs-Vellai K, Takahashi Y, Takáts S, Takemura G, Takigawa N, Talbot NJ, Tamagno E, Tamburini J, Tan CP, Tan L, Tan ML, Tan M, Tan YJ, Tanaka K, Tanaka M, Tang D, Tang D, Tang G, Tanida I, Tanji K, Tannous BA, Tapia JA, Tasset-Cuevas I, Tatar M, Tavassoly I, Tavernarakis N, Taylor A, Taylor GS, Taylor GA, Taylor JP, Taylor MJ, Tchetina EV, Tee AR, Teixeira-Clerc F, Telang S, Tencomnao T, Teng BB, Teng RJ, Terro F, Tettamanti G, Theiss AL, Theron AE, Thomas KJ, Thomé MP, Thomes PG, Thorburn A, Thorner J, Thum T, Thumm M, Thurston TL, Tian L, Till A, Ting JP, Titorenko VI, Toker L, Toldo S, Tooze SA, Topisirovic I, Torgersen ML, Torosantucci L, Torriglia A, Torrisi MR, Tournier C, Towns R, Trajkovic V, Travassos LH, Triola G, Tripathi DN, Trisciuoglio D, Troncoso R, Trougakos IP, Truttmann AC, Tsai KJ, Tschan MP, Tseng YH, Tsukuba T, Tsung A, Tsvetkov AS, Tu S, Tuan HY, Tucci M, Tumbarello DA, Turk B, Turk V, Turner RF, Tveita AA, Tyagi SC, Ubukata M, Uchiyama Y, Udelnow A, Ueno T, Umekawa M, Umemiya-Shirafuji R, Underwood BR, Ungermann C, Ureshino RP, Ushioda R, Uversky VN, Uzcátegui NL, Vaccari T, Vaccaro MI, Váchová L, Vakifahmetoglu-Norberg H, Valdor R, Valente EM, Vallette F, Valverde AM, Van den Berghe G, Van Den Bosch L, van den Brink GR, van der Goot FG, van der Klei IJ, van der Laan LJ, van Doorn WG, van Egmond M, van Golen KL, Van Kaer L, van Lookeren Campagne M, Vandenabeele P, Vandenberghe W, Vanhorebeek I, Varela-Nieto I, Vasconcelos MH, Vasko R, Vavvas DG, Vega-Naredo I, Velasco G, Velentzas AD, Velentzas PD, Vellai T, Vellenga E, Vendelbo MH, Venkatachalam K, Ventura N, Ventura S, Veras PS, Verdier M, Vertessy BG, Viale A, Vidal M, Vieira HL, Vierstra RD, Vigneswaran N, Vij N, Vila M, Villar M, Villar VH, Villarroya J, Vindis C, Viola G, Viscomi MT, Vitale G, Vogl DT, Voitsekhovskaja OV, von Haefen C, von Schwarzenberg K, Voth DE, Vouret-Craviari V, Vuori K, Vyas JM, Waeber C, Walker CL, Walker MJ, Walter J, Wan L, Wan X, Wang B, Wang C, Wang CY, Wang C, Wang C, Wang C, Wang D, Wang F, Wang F, Wang G, Wang HJ, Wang H, Wang HG, Wang H, Wang HD, Wang J, Wang J, Wang M, Wang MQ, Wang PY, Wang P, Wang RC, Wang S, Wang TF, Wang X, Wang XJ, Wang XW, Wang X, Wang X, Wang Y, Wang Y, Wang Y, Wang YJ, Wang Y, Wang Y, Wang YT, Wang Y, Wang ZN, Wappner P, Ward C, Ward DM, Warnes G, Watada H, Watanabe Y, Watase K, Weaver TE, Weekes CD, Wei J, Weide T, Weihl CC, Weindl G, Weis SN, Wen L, Wen X, Wen Y, Westermann B, Weyand CM, White AR, White E, Whitton JL, Whitworth AJ, Wiels J, Wild F, Wildenberg ME, Wileman T, Wilkinson DS, Wilkinson S, Willbold D, Williams C, Williams K, Williamson PR, Winklhofer KF, Witkin SS, Wohlgemuth SE, Wollert T, Wolvetang EJ, Wong E, Wong GW, Wong RW, Wong VK, Woodcock EA, Wright KL, Wu C, Wu D, Wu GS, Wu J, Wu J, Wu M, Wu M, Wu S, Wu WK, Wu Y, Wu Z, Xavier CP, Xavier RJ, Xia GX, Xia T, Xia W, Xia Y, Xiao H, Xiao J, Xiao S, Xiao W, Xie CM, Xie Z, Xie Z, Xilouri M, Xiong Y, Xu C, Xu C, Xu F, Xu H, Xu H, Xu J, Xu J, Xu J, Xu L, Xu X, Xu Y, Xu Y, Xu ZX, Xu Z, Xue Y, Yamada T, Yamamoto A, Yamanaka K, Yamashina S, Yamashiro S, Yan B, Yan B, Yan X, Yan Z, Yanagi Y, Yang DS, Yang JM, Yang L, Yang M, Yang PM, Yang P, Yang Q, Yang W, Yang WY, Yang X, Yang Y, Yang Y, Yang Z, Yang Z, Yao MC, Yao PJ, Yao X, Yao Z, Yao Z, Yasui LS, Ye M, Yedvobnick B, Yeganeh B, Yeh ES, Yeyati PL, Yi F, Yi L, Yin XM, Yip CK, Yoo YM, Yoo YH, Yoon SY, Yoshida K, Yoshimori T, Young KH, Yu H, Yu JJ, Yu JT, Yu J, Yu L, Yu WH, Yu XF, Yu Z, Yuan J, Yuan ZM, Yue BY, Yue J, Yue Z, Zacks DN, Zacksenhaus E, Zaffaroni N, Zaglia T, Zakeri Z, Zecchini V, Zeng J, Zeng M, Zeng Q, Zervos AS, Zhang DD, Zhang F, Zhang G, Zhang GC, Zhang H, Zhang H, Zhang H, Zhang H, Zhang J, Zhang J, Zhang J, Zhang J, Zhang JP, Zhang L, Zhang L, Zhang L, Zhang L, Zhang MY, Zhang X, Zhang XD, Zhang Y, Zhang Y, Zhang Y, Zhang Y, Zhang Y, Zhao M, Zhao WL, Zhao X, Zhao YG, Zhao Y, Zhao Y, Zhao YX, Zhao Z, Zhao ZJ, Zheng D, Zheng XL, Zheng X, Zhivotovsky B, Zhong Q, Zhou GZ, Zhou G, Zhou H, Zhou SF, Zhou XJ, Zhu H, Zhu H, Zhu WG, Zhu W, Zhu XF, Zhu Y, Zhuang SM, Zhuang X, Ziparo E, Zois CE, Zoladek T, Zong WX, Zorzano A, Zughaier SM. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fullgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol. 2014;15:65–74. doi: 10.1038/nrm3716. [DOI] [PubMed] [Google Scholar]
- 29.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoyer-Hansen M, Jaattela M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14:1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- 31.Todd DJ, Lee AH, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol. 2008;8:663–674. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- 32.Wu J, Kaufman RJ. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- 33.Du K, Herzig S, Kulkarni RN, Montminy M. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 2003;300:1574–1577. doi: 10.1126/science.1079817. [DOI] [PubMed] [Google Scholar]
- 34.Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 2005;24:1243–1255. doi: 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, Lambin P, van der Kogel AJ, Koritzinsky M, Wouters BG. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest. 2010;120:127–141. doi: 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Ni M, Lee B, Barron E, Hinton DR, Lee AS. The unfolded protein response regulator GRP78/BiP is required for endoplasmic reticulum integrity and stress-induced autophagy in mammalian cells. Cell Death Differ. 2008;15:1460–1471. doi: 10.1038/cdd.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scarlatti F, Maffei R, Beau I, Codogno P, Ghidoni R. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 2008;15:1318–1329. doi: 10.1038/cdd.2008.51. [DOI] [PubMed] [Google Scholar]
- 38.Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, Cook LJ, Rubinsztein DC. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170:1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Codogno P, Mehrpour M, Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol. 2012;13:7–12. doi: 10.1038/nrm3249. [DOI] [PubMed] [Google Scholar]
- 40.Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci U S A. 2011;108:11121–11126. doi: 10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 42.Dupont N, Codogno P. Non-canonical Autophagy: Facts and Prospects. Current Pathobiology Reports. 2013;1:263–271. [Google Scholar]
- 43.Fullgrabe J, Klionsky DJ, Joseph B. Histone post-translational modifications regulate autophagy flux and outcome. Autophagy. 2013;9:1621–3. doi: 10.4161/auto.25803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Artal-Martinez de Narvajas A, Gomez TS, Zhang JS, Mann AO, Taoda Y, Gorman JA, Herreros-Villanueva M, Gress TM, Ellenrieder V, Bujanda L, Kim DH, Kozikowski AP, Koenig A, Billadeau DD. Epigenetic regulation of autophagy by the methyltransferase g9a. Mol Cell Biol. 2013;33:3983–3993. doi: 10.1128/MCB.00813-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fullgrabe J, Lynch-Day MA, Heldring N, Li W, Struijk RB, Ma Q, Hermanson O, Rosenfeld MG, Klionsky DJ, Joseph B. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500:468–471. doi: 10.1038/nature12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei FZ, Cao Z, Wang X, Wang H, Cai MY, Li T, Hattori N, Wang D, Du Y, Song B, Cao LL, Shen C, Wang L, Wang H, Yang Y, Xie D, Wang F, Ushijima T, Zhao Y, Zhu WG. Epigenetic regulation of autophagy by the methyltransferase EZH2 through an MTOR-dependent pathway. Autophagy. 2015;11:2309–2322. doi: 10.1080/15548627.2015.1117734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larsen KE, Sulzer D. Autophagy in neurons: a review. Histol Histopathol. 2002;17:897–908. doi: 10.14670/HH-17.897. [DOI] [PubMed] [Google Scholar]
- 48.Nishino I. Autophagic vacuolar myopathies. Curr Neurol Neurosci Rep. 2003;3:64–69. doi: 10.1007/s11910-003-0040-y. [DOI] [PubMed] [Google Scholar]
- 49.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen S, Rehman SK, Zhang W, Wen A, Yao L, Zhang J. Autophagy is a therapeutic target in anticancer drug resistance. Biochim Biophys Acta. 2010;1806:220–229. doi: 10.1016/j.bbcan.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 51.Clarke PG. Developmental cell death: morphological diversity and multiple mechanisms. Anat Embryol (Berl) 1990;181:195–213. doi: 10.1007/BF00174615. [DOI] [PubMed] [Google Scholar]
- 52.Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest. 2005;115:2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13:184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]


