Abstract
Inflammatory Breast Cancer (IBC) is the most lethal form of breast cancer with a 35% 5-year survival rate. The accurate and early diagnosis of IBC and the development of targeted therapy against this deadly disease remain a great medical challenge. Plasma membrane proteins (PMPs) such as E-cadherin and EGFR, play an important role in the progression of IBC. Because the critical role of PMPs in the oncogenic processes they are the perfect candidates as molecular markers and targets for cancer therapies. In the present study, Stable Isotope Labeling with Amino Acids in Cell Culture (SILAC) followed by mass spectrometry analysis was used to compare the relative expression levels of membrane proteins (MP) between non-cancerous mammary epithelial and IBC cells, MCF-10A and SUM-149, respectively. Six of the identified PMPs were validated by immunoblotting using the membrane fractions of non-IBC and IBC cell lines, compared with MCF-10A cells. Immunohistochemical analysis using IBC, invasive ductal carcinoma or normal mammary tissue samples was carried out to complete the validation method in nine of the PMPs. We identified and quantified 278 MPs, 76% of which classified as PMPs with 1.3-fold or higher change. We identified for the first time the overexpression of the novel plasminogen receptor, PLGRKT in IBC and of the carrier protein, SCAMP3. Furthermore, we describe the positive relationship between L1CAM expression and metastasis in IBC patients and the role of SCAMP3 as a tumor-related protein. Overall, the membrane proteomic signature of IBC reflects a global change in cellular organization and suggests additional strategies for cancer progression. Together, this study provides insight into the specialized IBC plasma membrane proteome with the potential to identify a number of novel therapeutic targets for IBC.
Keywords: SILAC, IBC, proteomics, membrane, markers, IDC, SCAMP3, L1CAM
Introduction
Inflammatory breast cancer (IBC) is characterized by its rapid and aggressive behavior, where patients have a 43% increased risk of death compared to women with stage-matched non-IBC advanced breast cancer [1]. The hallmark of IBC is the formation of tumor emboli which invade the vascular and lymphatic systems, and are responsible for the inflammatory phenotype, and the high rate of metastasis [2]. Cells comprising tumor emboli maintain aggregation through the overexpression of the transmembrane glycoprotein E-cadherin, forming an overactive complex with alpha/beta-catenin [3]. Paradoxically, in other types of breast cancer, loss of E-cadherin is associated with an epithelial to mesenchymal transition linked with aggressive tumor invasion and metastasis. Besides the overexpression of E-cadherin, other plasma membrane proteins (PMP) such as EGFR and HER2 are overexpressed in 60% of IBC tumors, both in association with rapid tumor growth rate, invasion and metastasis via the activation of PI3K/AKT and ERK oncogenic pathways [4-6].
Plasma membrane proteins are critical for cell structure, to carry out functions such as membrane-cytoskeleton interactions, extracellular matrix interactions with adjacent cells, sensors of external signals and their downstream intracellular transmission, and as transporters of molecules. Due to their function, PMPs play an important role in oncogenic processes are targets of approximately 70% of cancer therapies in use or under study [7]. A proteomic analysis for membrane protein (MP) identification is a powerful tool used to identify novel biomarkers in breast cancer. Of such methods, SILAC is a simple and accurate approach for identification and quantitation of complex protein mixtures. Many proteomics studies using SILAC have examined the membrane proteome in various cancers, such as breast cancer [7,8] and lymphoma [9]. Recently, Ziegler et al. examined the PM proteome of several non-IBC cell lines with different molecular subtypes [7]. Results from this study reflected overexpression of tyrosine kinases, cellular adhesion molecules and structural proteins.
The accurate and early diagnosis of IBC and the development of targeted therapy against this deadly disease remain a great medical challenge. The identification of membrane proteins from the cell surface and from organelles can shed light on the formation, progression and metastasis processes of IBC. Thus, defining the membrane proteomic profile of IBC has potential for identifying novel molecular markers that will help in the advancement of early diagnosis and subsequent development of therapeutic targets. The present study is the first to identify and quantify the membrane proteome of IBC. This novel study allows characterization and comparison of the PMP profile of the well-studied model of IBC, SUM-149, and non-cancerous mammary epithelial MCF-10A cells. Our data describes the complex image of PMPs present on IBC cells, reflecting the multiple strategies IBC uses to promote highly lymphovascular invasion, and rapid metastatic activity.
Materials and methods
Cell culture and reagents
The patient derived IBC cell line SUM-149 and the SUM-102 cell line were obtained from Dr. Steven Ethier, Medical University of South Carolina Charleston, SC, USA. KPL-4 and MDA-IBC-3 cells were kindly provided by Dr. Kurebayashi (Kawasaki Medical School, Japan) and Dr. Wendy Woodward, University of Texas MD Anderson Cancer Center (Houston, TX), respectively. Cells were grown as described previously [10]. MCF-7 and MCF-10A were obtained from American Type Culture Collection (ATCC) and were cultured in DMEM, 10% FBS or DMEM/F-12 containing 10% Horse Serum, respectively. SILAC™ Protein Identification and Quantitation D-MEM/F-12-Flex Media Kit was purchased from Life Technologies. All kits and developing substrates for immunohistochemistry (IHC) analysis were obtained from Vector Laboratories. Antibodies to C1QBP, Flotilin-1, Metadherin and ITGβ5 were purchased from Cell Signaling Technologies. L1CAM, MCAM and MST1R antibodies were obtained from Abcam. Antibodies to PLGRKT and SCAMP3 were acquired from Sigma.
Cell labeling
MCF-10A and SUM-149 cells were cultured in 60mm dishes and maintained in their appropriate culture media. To initiate the incorporation of light or heavy labels, 1×105 MCF-10A cells were harvested and suspended in 3 mL of advanced DMEM/F-12-Flex media supplemented with 10% dialyzed FBS, 20 ng/mL EGF, and 0.1 mg/mL heavy lysine ([U13C6] L-lysine and heavy [U13C6] L-arginine. Following the same procedure, SUM-149 cells were suspended in modified DMEM/F-12-Flex supplemented with 10% dFBS and 0.1 mg/mL light L-lysine and light L-arginine. Every three days the media was replaced with the corresponding labeling medium and cells were allowed to expand for at least six doubling times to achieve 99% incorporation of labeled amino acid into the proteins. After six doublings, 106 cells of each cell line were harvested to determine the efficiency of incorporation. 2×106 of each cell line were mixed at 1:1 ratio and lysed on ice for 30 min following the procedure described in [8]. Membrane pellet was dissolved in 20 µL of 4X NuPAGE LDS Sample Buffer containing DTT and heated at 70°C for 10 mins, and analyzed by 1D SDS-NuPAGE and stained with Coomassie Brilliant Blue R-250. For proteomic analysis, each of the cell lines was analyzed in three biologic replicates.
Tryptic digestion and peptide fractionation
The entire gel lane for each sample was collected and divided in 10 gel sections. Each gel section was subjected to in-gel tryptic digestion by overnight incubation with trypsin in 50 mM NH4HCO3 at 37°C. Digested peptides were then extracted with 60% acetonitrile (ACN) and 0.1% trifluoroacetic acid (TFA), dried on a speedvac and resuspended in 0.5% TFA. All samples were purified using C18 ZipTips (Millipore) according to manufacturer’s recommendations and re-suspended in 2% ACN with 0.1% formic acid prior to LC-MS/MS analysis.
LC-MS/MS analysis
Sample fractions were dissolved in 25 µL of 2% ACN in 0.1% TFA prior to injection on LC-MS/MS. A 3.0 µL aliquot was directly injected onto a custom packed 2 cm×100 µm C18 Magic 5 µm particle trap column. Peptides were then eluted and electro sprayed from a custom packed emitter (75 µm×25 cm C18 Magic 3 µm particle) with a linear gradient from 95% solvent A (0.1% FA in water) to 35% solvent B (0.1% FA in ACN) in 35 min at a flow rate of 300 nL/min on a Waters Nano Acquity UPLC system. Data dependent acquisitions were performed on a Q Exactive mass spectrometer (Thermo Scientific) according to an experiment where full MS scans from 300-1750 m/z were acquired at a resolution of 70,000 followed by 12 MS/MS scans acquired under HCD fragmentation at a resolution of 17,500 with an isolation width of 1.2 Da.
Data analysis
Raw data files were peak processed with Proteome Discoverer (Thermo, v.1.3) prior to searching with Mascot Server (Matrix Sciences Inc., version 2.4) against the SwissProt Human database (v.050113). Search parameters utilized were fully tryptic with 2 missed cleavages, parent mass tolerances of 10 ppm and fragment mass tolerances of 0.05 Da. A fixed modification of carbamidomethyl cysteine and variable modifications of 13C6 on lysine and arginine, acetyl (protein N-term), pyro glutamic for N-term glutamine, oxidation of methionine was considered. Search results were loaded into the Scaffold Viewer (Proteome Software, Inc.) for assessment of protein identification probabilities. SILAC peptides ratios were calculated using the ProteoIQ software (Nusep, Inc., v.2.6). Protein identifications were accepted if they could be established at >90.0% probability and contained at least 2 identified peptides.
Tissue samples
Breast tissues were kindly provided by Dr. Robert J. Schneider, (NYU, School of Medicine, NY, NY). The Institutional Review Board at NYU approved the informed consent forms for tissue collection. Breast cancer tissues consisted of 17 IBC and 24 invasive ductal carcinoma (IDC) tumors and 10 normal breast tissues.
Immunohistochemistry
Paraffin-embedded tissues were deparaffinized, rehydrated and subjected to antigen retrieval using a citrate based solution. Sections were incubated in 5% hydrogen peroxide for 30 min before staining using the Universal Vectastain-ABC horseradish peroxidase kit and incubated with the indicated antibodies. Slides were developed with the DAB substrate kit and counterstained with haematoxylin. We classified the intensity of staining using weak, moderate or strong staining intensities. We identified the percent of stained cells using a quantitative score defined as: “+” less than 10% cells positive staining, “++”, 10-50% cells positive staining and “+++” more than 50% cells positive staining. Location of protein expression was classified as nuclear (N), cytoplasmic (C), nuclear and cytoplasmic (NC), membranous and cytoplasmic (MC) or membranous (M).
Immunoblotting
Breast cancer cells were lysed and equal total protein was resolved via SDS-PAGE and immunoblotted as described in [11] using the indicated antibodies.
Statistical analysis
Student’s t-test statistical analyses for immunoblotting studies were done using GraphPad Prism® v.6.0 (San Diego, CA). To analyze the IHC raw data and assess its distribution, univariate statistics, frequencies and percentages were employed. Evaluation for normality assumptions was done prior to the application of any bivariate statistical tests using the Shapiro-Wilk estimate. Differences among group proportions were assessed using Chi-square distribution statistics or Fisher’s exact test. Differences among group means within the patient data were evaluated using an independent samples t-test approach with Levene’s statistic. All analyses were considered significant at P≤0.05. A bivariate association model of seven independent correlation matrices was evaluated using the Pearson product-moment analysis. Each matrix confronted two pairs of possible combinations between: staining, lymphatic invasion, metastasis and invasion. To account for the distribution of all the variables in the dataset, a normality diagnostic test was performed using the Shapiro-Francia estimator. The significance level (α) was set to ≤ 0.05, except for the normality diagnostic test (P>0.05). IBM Statistical Package for Social Sciences (IBM-SPSS, Chicago, IL) v.23.0 for Windows was used.
Results
Quantitative analysis of differential membrane proteome expression in IBC SUM-149 vs. MCF-10A cells
To quantitatively analyze membrane proteome alterations in IBC cells, we performed a SILAC-based proteomic analysis. In triplicate experiments we identified and quantified a total of 2,102, 1,869, and 2,002 proteins (false discovery rate ≤1% and at least two identified peptides), respectively, excluding possible sample contaminants (i.e., trypsin, keratins and cytokeratins). Although it is well known that SUM-149 cells express cytokeratins 8, 18 and 19, we excluded them in order to decrease false results from contamination by sample handling [12]. By carrying out an analysis of variance, we excluded from the analysis those proteins that were not replicated in the three independent experiments. After performing this assessment, we successfully identified and quantified a total of 634 proteins with 278 (44%) MPs. A change of 1.3-fold (ratio) was used as a cut-off value for significance, which is convention in SILAC proteomic approaches [13]. Among the 278 MPs, 212 (76%) increased at least 1.3-fold and three were downregulated (Table 1). To gain a deeper understanding of the contribution of PMPs to IBC, we grouped 55 proteins in this category showing 1.3-fold or higher upregulation in differential expression between MCF-10A and SUM-149 cells (Table 2). Proteins in this category include cell adhesion proteins (MCAM, L1CAM, ITGB5, MTDH), receptors, (MST1R, C1QBP), including the novel membrane plasminogen receptor, PLGRKT. Also, we identified intracellular signaling proteins such as Ras related proteins and transport proteins (SCAMP3, FLOT1, VAMP, ABCC3) among others. Although some listed proteins can exist in multiple cell locations, there is good evidence of their PM association.
Table 1.
Membrane proteins displaying a 1.3 or higher fold change in differential expression between normal and IBC cells
| Gene | Protein name | UniProtKB accession number | SILAC ratio |
|---|---|---|---|
| Down-regulated Proteins | |||
| MAN1B1 | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase | Q9UKM7 | -1.7 |
| SLC25A31 | ADP/ATP translocase 4 | Q9H0C2 | -4.5 |
| TBL2 | Transducin beta-like protein 2 | Q9Y4P3 | -1.3 |
| Up-regulated Proteins | |||
| ABCC3 | Canalicular multispecific organic anion transporter 2 | O15438 | 1.4 |
| ABCD3 | ATP-binding cassette, sub-family D (ALD), member 3 | P28288 | 2.0 |
| ACBD5 | Acyl-coa binding domain containing protein 5 | Q5T8D3 | 2.0 |
| AGK | Acylglycerol kinase | Q53H12 | 1.6 |
| AGPS | Alkylglycerone phosphate synthase | O00116 | 2.0 |
| AIFM1 | Apoptosis-inducing factor 1, mitochondrial | O95831 | 1.7 |
| ALDH18A1 | Delta-1-pyrroline-5-carboxylate synthase | P54886 | 1.7 |
| ALDH3A2 | Fatty aldehyde dehydrogenase | P51648 | 1.6 |
| APMAP | Adipocyte plasma membrane associated protein | Q9HDC9 | 1.6 |
| ARL8A | ADP-ribosylation factor-like 8A | Q96BM9 | 1.3 |
| ATAD1 | ATPase family AAA domain-containing protein 1 | Q8NBU5 | 2.2 |
| ATAD3A/ATAD3B | Atpase family, AAA domain containing 3A | Q5T9A4 | 1.9 |
| ATL3 | Atlastin-3 | Q6DD88 | 1.6 |
| ATP13A1 | Manganese-transporting ATPase 13A1 | Q9HD20 | 1.3 |
| ATP2A2 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | P16615 | 1.4 |
| ATP2A3 | ATPase, Ca++ transporting, ubiquitous | Q93084 | 1.3 |
| ATP5A1 | ATP synthase subunit alpha, mitochondrial | P25705 | 1.8 |
| ATP5B | ATP synthase subunit beta, mitochondrial | P06576 | 1.8 |
| ATP5C1 | ATP synthase subunit gamma, mitochondrial | P36542 | 1.7 |
| ATP5D | ATP synthase subunit delta, mitochondrial | P30049 | 1.8 |
| ATP5E | ATP synthase subunit epsilon, mitochondrial | P56381 | 1.9 |
| ATP5EP2 | ATP synthase subunit epsilon-like protein, mitochondrial | Q5VTU8 | 1.9 |
| ATP5F1 | ATP synthase F(0) complex subunit B1, mitochondrial | P24539 | 1.7 |
| ATP5H | ATP synthase subunit d, mitochondrial | O75947 | 1.7 |
| ATP5J | ATP synthase-coupling factor 6, mitochondrial | P18859 | 1.7 |
| ATP5L | ATP synthase subunit g, mitochondrial | O75964 | 1.8 |
| ATP5O | ATP synthase subunit O, mitochondrial | P48047 | 1.8 |
| ATP6V0D1 | V-type proton ATPase subunit d 1 | P61421 | 1.4 |
| ATP6V1A | V-type proton ATPase catalytic subunit A | P38606 | 2.1 |
| ATP6V1G1 | V-type proton ATPase subunit G 1 | O75348 | 1.7 |
| B3GAT3 | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase3 | O94766 | 2.3 |
| C14orf2 | 6.8 kDa mitochondrial proteolipid | P56378 | 2.0 |
| C19orf70 | MICOS complex subunit MIC13 | Q5XKP0 | 2.7 |
| C1QBP | Complement component 1, Q subcomponent binding protein | Q07021 | 2.9 |
| CANX | Calnexin | P27824 | 2.0 |
| CAP1 | Adenylyl cyclase-associated protein 1 | Q01518 | 1.5 |
| CCDC47 | Coiled-coil domain containing 47 | Q96A33 | 2.2 |
| CCSMST1 | Protein CCSMST1 | Q4G0I0 | 2.0 |
| CERS2 | Ceramide synthase 2 | Q96G23 | 1.3 |
| CISD2 | CDGSH iron sulfur domain 2 | Q8N5K1 | 1.6 |
| CKMT1A | Creatine kinase U-type, mitochondrial | P12532 | 1.5 |
| CLCC1 | Chloride channel CLIC-like 1 | Q96S66 | 2.3 |
| COQ5 | 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondria | Q5HYK3 | 2.0 |
| COX20 | Cytochrome c oxidase protein 20 homolog | Q5RI15 | 2.7 |
| COX5A | cytochrome c oxidase subunit 5A | P20674 | 1.5 |
| COX5B | Cytochrome c oxidase subunit 5B, mitochondrial | P10606 | 1.3 |
| COX6C | Cytochrome c oxidase subunit 6C | P09669 | 1.3 |
| CPOX | Oxygen-dependent coproporphyrinogen-III oxidase, mitochondrial | P36551 | 1.5 |
| CPT2 | Carnitine palmitoyltransferase 2 | P23786 | 1.7 |
| CTAGE9 (includes others) | CTAGE family, member 9 | A4FU28 | 1.3 |
| CYB5B | Cytochrome b5 type B | O43169 | 1.6 |
| CYC1 | Cytochrome c1, heme protein, mitochondrial | P08574 | 1.5 |
| CYP51A1 | Lanosterol 14-alpha demethylase | Q16850 | 2.0 |
| DAD1 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit DAD1 | P61803 | 1.9 |
| DDOST | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit | P39656 | 2.0 |
| DERL1 | Derlin-1 | Q9BUN8 | 1.9 |
| DHRS1 | Dehydrogenase/reductase (SDR family) member 1 | Q96LJ7 | 2.0 |
| DNAJC19 | Mitochondrial import inner membrane translocase subunit TIM14 | Q96DA6 | 2.4 |
| EBP | 3-beta-hydroxysteroid-Delta (8), Delta (7)-isomerase | Q15125 | 2.1 |
| EMD | Emerin | P50402 | 1.8 |
| ENO1 | Enolase 1, (alpha) | P06733 | 2.1 |
| ERLIN2 | Erlin-2 | O94905 | 1.4 |
| ERO1L | ERO1-like protein alpha | Q96HE7 | 2.7 |
| ESYT2 | Extended synaptotagmin-2 | A0FGR8 | 1.3 |
| FKBP2 | Peptidyl-prolyl cis-trans isomerase FKBP2 | P26885 | 2.6 |
| FKBP8 | Peptidyl-prolyl cis-trans isomerase FKBP8 | Q14318 | 2.9 |
| FLOT1 | Flotillin-1 | O75955 | 1.7 |
| GALNT3 | Polypeptide N-acetylgalactosaminyltransferase 3 | Q14435 | 2.8 |
| GALNT7 | N-acetylgalactosaminyltransferase 7 | Q86SF2 | 1.7 |
| GBAS | Protein NipSnap homolog 2 | O75323 | 2.5 |
| GOLGA7 | Golgin subfamily A member 7 | Q7Z5G4 | 1.7 |
| GOSR1 | Golgi SNAP receptor complex member 1 | O95249 | 2.9 |
| GOT2 | Aspartate aminotransferase, mitochondrial | P00505 | 1.8 |
| HK1 | Hexokinase-1 | P19367 | 1.6 |
| HK2 | Hexokinase-2 | P52789 | 2.0 |
| HLA-A | HLA class I histocompatibility antigen, A-29 | P30512 | 1.9 |
| HLA-A | HLA class I histocompatibility antigen, A-31 | P16189 | 2.0 |
| HLA-A | HLA class I histocompatibility antigen, A-33 | P16190 | 2.1 |
| HLA-A | HLA class I histocompatibility antigen, A-69 | P10316 | 1.7 |
| HLA-A | HLA class I histocompatibility antigen, A-68 | P01891 | 1.5 |
| HLA-A | HLA class I histocompatibility antigen, A-2 | P01892 | 1.7 |
| HLA-E | HLA class I histocompatibility antigen, alpha chain E | P13747 | 2.1 |
| HSD17B12 | Very-long-chain 3-oxoacyl-CoA reductase | Q53GQ0 | 3.0 |
| HSP90AA1 | Heat shock protein HSP 90-alpha | P07900 | 2.2 |
| HSPA2 | Heat shock-related 70 kDa protein 2 | P54652 | 2.1 |
| IFITM3 | Interferon induced transmembrane protein 3 | Q01628 | 3.6 |
| IMMT | MICOS complex subunit MIC60 | Q16891 | 2.4 |
| ITGB5 | Integrin, beta 5 | P18084 | 2.3 |
| ITPR3 | Inositol 1,4,5-trisphosphate receptor type 3 | Q14573 | 1.3 |
| KTN1 | Kinectin | Q86UP2 | 1.5 |
| L1CAM | Neural cell adhesion molecule L1 | P32004 | 1.7 |
| LETM1 | Leucine zipper-EF-hand containing transmembrane protein 1 | O95202 | 1.6 |
| LMAN1 | Protein ERGIC-53 | P49257 | 2.0 |
| LRPPRC | Leucine-rich PPR motif-containing protein, mitochondrial | P42704 | 2.6 |
| LRRC59 | Leucine rich repeat containing 59 | Q96AG4 | 2.8 |
| M6PR | Cation-dependent mannose-6-phosphate receptor | P20645 | 2.3 |
| MCAM | Cell surface glycoprotein MUC18 | P43121 | 2.2 |
| MCU | mitochondrial calcium uniporter | Q8NE86 | 1.5 |
| MFF | Mitochondrial fission factor | Q9GZY8 | 1.8 |
| MGST1 | Microsomal glutathione S-transferase 1 | P10620 | 1.9 |
| MIA3 | Melanoma inhibitory activity protein 3 | Q5JRA6 | 1.4 |
| MLEC | Malectin | Q14165 | 2.0 |
| MMGT1 | Membrane magnesium transporter 1 | Q8N4V1 | 2.7 |
| MOGS | Mannosyl-oligosaccharide glucosidase | Q13724 | 2.5 |
| MSN | Moesin | P26038 | 1.8 |
| MST1R | Macrophage-stimulating protein receptor | Q04912 | 1.6 |
| MTDH | Protein LYRIC | Q86UE4 | 1.7 |
| MTX1 | Metaxin-1 | Q13505 | 2.7 |
| MTX2 | Metaxin-2 | O75431 | 2.1 |
| MXRA7 | Matrix-remodelling associated 7 | P84157 | 2.3 |
| MYO1C | Unconventional myosin-Ic | O00159 | 1.5 |
| NAPA | Alpha-soluble NSF attachment protein | P54920 | 1.6 |
| NDUFA3 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 3 | O95167 | 1.8 |
| NDUFA5 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 | Q16718 | 2.0 |
| NDUFA8 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 | P51970 | 1.9 |
| NDUFB1 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 1 | O75438 | 1.7 |
| NDUFB10 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 | O96000 | 2.0 |
| NDUFB4 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4 | O95168 | 1.4 |
| NDUFB5 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 5, mitochondrial | O43674 | 2.3 |
| NDUFB8 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 | O95169 | 1.4 |
| NDUFC2 | NADH dehydrogenase [ubiquinone] 1 subunit C2 | O95298 | 1.5 |
| NDUFS2 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitocondrial | O75306 | 1.5 |
| NDUFS3 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitocondrial | O75489 | 1.8 |
| NIPSNAP1 | Protein NipSnap homolog 1 | Q9BPW8 | 2.3 |
| NOMO1 | NODAL modulator 1 | Q15155 | 1.6 |
| NOMO2 | Nodal modulator 2 | Q5JPE7 | 1.6 |
| NOMO3 | Nodal modulator 3 | P69849 | 1.6 |
| PDIA6 | Protein disulfide-isomerase A6 | Q15084 | 3.1 |
| PGAM5 | Serine/threonine-protein phosphatase PGAM5, mitochondrial | Q96HS1 | 1.5 |
| PGRMC1 | Progesterone receptor membrane component 1 | O00264 | 2.3 |
| PHB | Prohibitin | P35232 | 2.2 |
| PHB2 | Prohibitin-2 | Q99623 | 2.3 |
| PKP2 | Plakophilin-2 | Q99959 | 1.5 |
| PLGRKT | Plasminogen receptor (KT) | Q9HBL7 | 3.3 |
| PLOD1 | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 | Q02809 | 1.7 |
| PLOD2 | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 | O00469 | 1.4 |
| PLOD3 | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 | O60568 | 1.4 |
| PNPT1 | Polyribonucleotide nucleotidyltransferase 1, mitochondrial | Q8TCS8 | 2.0 |
| PREB | Prolactin regulatory element binding protein | Q9HCU5 | 2.2 |
| PTPN1 | Tyrosine-protein phosphatase non-receptor type 1 | P18031 | 2.3 |
| RAB14 | Ras-related protein Rab-14 | P61106 | 1.7 |
| RAB18 | Ras-related protein Rab-18 | Q9NP72 | 1.7 |
| RAB1C | Putative Ras-related protein Rab-1C | Q92928 | 1.5 |
| RAB2B | Ras-related protein Rab-2B | Q8WUD1 | 1.4 |
| RAB3D | Ras-related protein Rab-3D | O95716 | 1.7 |
| RAB6A | Ras-related protein Rab-6A | P20340 | 2.3 |
| RAB8A | Ras-related protein Rab-8A | P61006 | 1.7 |
| RAB8B | Ras-related protein Rab-8B | Q92930 | 1.7 |
| RDH11 | Retinol dehydrogenase 11 | Q8TC12 | 2.3 |
| RPN1 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 1 | P04843 | 1.9 |
| RPN2 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2 | P04844 | 2.1 |
| SCAMP3 | Secretory carrier-associated membrane protein 3 | O14828 | 1.6 |
| SCD | Acyl-CoA desaturase | O00767 | 1.8 |
| SDHA | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | P31040 | 3.0 |
| SDHB | Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial | P21912 | 2.8 |
| SEC22B | Vesicle-trafficking protein SEC22b | O75396 | 2.9 |
| SEC61G | Protein transport protein Sec61 subunit gamma | P60059 | 2.1 |
| SFXN4 | Sideroflexin 4 | Q6P4A7 | 2.5 |
| SHMT2 | serine hydroxymethyltransferase 2, mitochondrial | P34897 | 1.5 |
| SIGMAR1 | Sigma non-opioid intracellular receptor 1 | Q99720 | 2.8 |
| SLC16A3 | Monocarboxylate transporter 4 | O15427 | 1.9 |
| SLC25A11 | Mitochondrial 2-oxoglutarate/malate carrier protein | Q02978 | 2.4 |
| SLC25A19 | Mitochondrial thiamine pyrophosphate carrier | Q9HC21 | 2.9 |
| SLC25A22 | Mitochondrial glutamate carrier 1 | Q9H936 | 2.0 |
| SLC25A24 | Calcium-binding mitochondrial carrier protein SCaMC-1 | Q6NUK1 | 2.0 |
| SLC25A3 | Phosphate carrier protein, mitochondrial | Q00325 | 2.1 |
| SLC35E1 | solute carrier family 35, member E1 | Q96K37 | 1.4 |
| SLC38A10 | Putative sodium-coupled neutral amino acid transporter 10 | Q9HBR0 | 3.3 |
| SPCS2 | Signal peptidase complex subunit 2 | Q15005 | 2.7 |
| SPCS3 | Signal peptidase complex subunit 3 | P61009 | 2.7 |
| SPTLC1 | Serine palmitoyltransferase | O15269 | 1.7 |
| SRPR | Signal recognition particle receptor subunit alpha | P08240 | 1.5 |
| SSR4 | Translocon-associated protein subunit delta | P51571 | 2.5 |
| SURF4 | Surfeit locus protein 4 | O15260 | 1.9 |
| SYNJ2BP | Synaptojanin 2 binding protein | P57105 | 1.7 |
| SYPL1 | Synaptophysin-like 1 | Q16563 | 2.0 |
| TAP1 | Antigen peptide transporter 1 | Q03518 | 2.8 |
| TAPBP | Tapasin | O15533 | 3.2 |
| TIMM23 | Mitochondrial import inner membrane translocase subunit Tim23 | O14925 | 2.2 |
| TIMM23B | Putative mitochondrial import inner membrane translocase subunit Tim23B | Q5SRD1 | 2.2 |
| TIMM44 | Mitochondrial import inner membrane translocase subunit TIM44 | O43615 | 2.2 |
| TM9SF1 | Transmembrane 9 superfamily member 1 | O15321 | 1.6 |
| TMED2 | Transmembrane emp24 domain-containing protein 2 | Q15363 | 1.6 |
| TMED4 | Transmembrane emp24 domain-containing protein 4 | Q7Z7H5 | 1.9 |
| TMEM109 | Transmembrane protein 109 | Q9BVC6 | 2.0 |
| TMEM205 | Transmembrane protein 205 | Q6UW68 | 3.5 |
| TMEM258 | Transmembrane protein 258 | P61165 | 2.4 |
| TMEM33 | Transmembrane protein 33 | P57088 | 2.2 |
| TMEM43 | transmembrane protein 43 | Q9BTV4 | 1.5 |
| TMUB1 | Transmembrane and ubiquitin-like domain containing protein 1 | Q9BVT8 | 2.1 |
| TMX1 | Thioredoxin-related transmembrane protein 1 | Q9H3N1 | 2.4 |
| TMX3 | Protein disulfide-isomerase TMX3 | Q96JJ7 | 1.8 |
| TOMM70A | Mitochondrial import receptor subunit TOM70 | O94826 | 1.9 |
| TPBG | Trophoblast glycoprotein | Q13641 | 1.4 |
| TRAM1 | Translocating chain-associated membrane protein 1 | Q15629 | 2.0 |
| TRAP1 | Heat shock protein 75 kDa, mitochondrial | Q12931 | 2.2 |
| UQCRB | Cytochrome b-c1 complex subunit 7 | P14927 | 1.6 |
| UQCRC1 | Cytochrome b-c1 complex subunit 1, mitochondrial | P31930 | 1.4 |
| UQCRC2 | Cytochrome b-c1 complex subunit 2, mitochondrial | P22695 | 1.4 |
| UQCRFS1 | Cytochrome b-c1 complex subunit Rieske, mitochondria | P47985 | 1.5 |
| USMG5 | Up-regulated during skeletal muscle growth protein 5 | Q96IX5 | 1.9 |
| VAMP2 | Vesicle-associated membrane protein 2 | P63027 | 1.7 |
| VAMP3 | Vesicle-associated membrane protein 3 | Q15836 | 1.7 |
| VAPB | Vesicle-associated membrane protein-associated protein B/C | O95292 | 2.1 |
| VDAC2 | Voltage-dependent anion channel 2 | P45880 | 1.7 |
| VIMP | Selenoprotein S | Q9BQE4 | 2.0 |
| VMA21 | Vacuolar ATPase assembly integral membrane protein VMA21 | Q3ZAQ7 | 3.4 |
| YIPF5 | Protein YIPF5 | Q969M3 | 1.4 |
| YIPF6 | Protein YIPF6 | Q96EC8 | 2.2 |
| YME1L1 | ATP-dependent zinc metalloprotease YME1L1 | Q96TA2 | 2.2 |
| ZDHHC5 | Palmitoyltransferase ZDHHC5 | Q9C0B5 | 1.3 |
| ZMPSTE24 | CAAX prenyl protease 1 homolog | O75844 | 2.2 |
Table 2.
Plasma membrane proteins displaying 1.3-fold or higher fold-change in differential expression between normal and IBC cells
| Gene | Protein name | UniProtKB accession number | SILAC Ratio |
|---|---|---|---|
| IFITM3 | Interferon induced transmembrane protein 3 | Q01628 | 3.6 |
| TMEM205 | Transmembrane protein 205 | Q6UW68 | 3.5 |
| PLGRKT | Plasminogen receptor (KT) | Q9HBL7 | 3.3 |
| SLC38A10 | Solute Carrier Family 38, Member 10 | Q9HBR0 | 3.3 |
| PDIA6 | Protein disulfide-isomerase A6 | Q15084 | 3.1 |
| C1QBP | Complement component 1Q subcomponent binding protein | Q07021 | 2.9 |
| SIGMAR1 | Sigma Non-Opioid Intracellular Receptor 1 | Q99720 | 2.8 |
| GBAS | Protein NipSnap homolog 2 | O75323 | 2.5 |
| TMEM258 | Transmembrane protein 258 | P61165 | 2.4 |
| ITGB5 | Integrin, beta 5 | P18084 | 2.3 |
| ATAD1 | ATPase family AAA domain-containing protein 1 | Q8NBU5 | 2.2 |
| MCAM | Cell surface glycoprotein MUC18 | P43121 | 2.2 |
| PHB | Prohibitin | P35232 | 2.2 |
| TMEM33 | Transmembrane protein 33 | P57088 | 2.2 |
| ATP6V1A | V-type proton atpase catalytic subunit A | P38606 | 2.1 |
| ENO1 | Enolase 1, (alpha) | P06733 | 2.1 |
| HLA-A | HLA class I histocompatibility antigen, A-33 alpha chain | P16190 | 2.1 |
| HLA-A | HLA class I histocompatibility antigen, A-31 alpha chain | P16189 | 2.1 |
| HLA-E | HLA class I histocompatibility antigen, alpha chain E | P13747 | 2.1 |
| SYPL1 | Synaptophysin-like 1 | Q16563 | 2.0 |
| HLA-A | HLA class I histocompatibility antigen, A-29 alpha chain | P30512 | 1.9 |
| SLC16A3 | Monocarboxylate transporter 4 | O15427 | 1.9 |
| ATP5A1 | ATP synthase subunit alpha, mitochondrial | P25705 | 1.8 |
| ATP5B | ATP synthase subunit beta, mitochondrial | P06576 | 1.8 |
| ATP5O | ATP synthase subunit O, mitochondrial | P48047 | 1.8 |
| MSN | Moesin | P26038 | 1.8 |
| ATP6V1G1 | V-type proton atpase subunit G 1 | O75348 | 1.7 |
| FLOT1 | Flotillin 1 | O75955 | 1.7 |
| HLA-A | HLA class I histocompatibility antigen, A-69 alpha chain | P10316 | 1.7 |
| HLA-A | HLA class I histocompatibility antigen, A-2 alpha chain | P01892 | 1.7 |
| L1CAM | Neural cell adhesion molecule L1 | P32004 | 1.7 |
| MTDH | Protein LYRIC | Q86UE4 | 1.7 |
| RAB18 | Ras-related protein Rab-18 | Q9NP72 | 1.7 |
| RAB3D | Ras-related protein Rab-3D | O95716 | 1.7 |
| RAB8A | Ras-related protein Rab-8A | P61006 | 1.7 |
| RAB8B | Ras-related protein Rab-8B | Q92930 | 1.7 |
| VAMP2 | Vesicle-associated membrane protein 2 | P63027 | 1.7 |
| VAMP3 | Vesicle-associated membrane protein 3 | Q15836 | 1.7 |
| APMAP | Adipocyte plasma membrane associated protein | Q9HDC9 | 1.6 |
| MST1R | Macrophage-stimulating protein receptor | Q04912 | 1.6 |
| NOMO1 | Nodal modulator 1 | Q15155 | 1.6 |
| NOMO3 | Nodal modulator 3 | P69849 | 1.6 |
| SCAMP3 | Secretory carrier-associated membrane protein 3 | O14828 | 1.6 |
| CAP1 | Adenylyl cyclase-associated protein 1 | Q01518 | 1.5 |
| HLA-A | HLA class I histocompatibility antigen, A-68 alpha chain | P01891 | 1.5 |
| KTN1 | Kinectin | Q86UP2 | 1.5 |
| PKP2 | Plakophilin-2 | Q99959 | 1.5 |
| ABCC3 | Canalicular multispecific organic anion transporter 2 | O15438 | 1.4 |
| ATP6V0D1 | V-type proton atpase subunit d 1 | P61421 | 1.4 |
| RAB2B | Ras-related protein Rab-2B | Q8WUD1 | 1.4 |
| SLC35E1 | Solute carrier family 35, member E1 | Q96K37 | 1.4 |
| TPBG | Trophoblast glycoprotein | Q13641 | 1.4 |
| ESYT2 | Extended synaptotagmin-2 | A0FGR8 | 1.3 |
| ITPR3 | Inositol 1,4,5-trisphosphate receptor type 3 | Q14573 | 1.3 |
| ZDHHC5 | Palmitoyltransferase ZDHHC5 | Q9C0B5 | 1.3 |
Proteins in Bold were selected for validation via IHC or immunoblotting analysis.
Although, PMPs such as E-cadherin and EGFR are overexpressed in SUM-149 cells, they are not listed in Table 2. Our raw data (data not shown) did not reveal a difference of 1.3 fold or greater of E-cadherin expression. Meanwhile, the overexpression of EGFR in SUM-149 cells was evidenced in one of our experiments in a ratio of 2.2 [14]. It’s important to highlight that E-cadherin and EGFR have been found overexpressed in IBC when compared with non-IBC breast cancer cells. In our model, we are comparing SUM-149 cells with non-cancerous mammary epithelial cells (MCF-10A) and both cell lines overexpress EGFR.
Validation of SILAC results by immunoblotting
To assure that we isolated only membrane proteins, the cytoplasmic fraction from MCF-10A and SUM-149 cells were investigated by SILAC. We assessed the expression of β-actin and E-cadherin as cytoplasmic and membrane protein controls, respectively. We observed expression of β-actin but not of E-cadherin in the cytoplasm, indicating a successful fractionation (Figure 1A). Since the key objective of this study was to identify potential biomarkers for IBC, we selected six biologically important PMPs for validation by immunoblotting (FLOT1, ITGB5, L1CAM, MCAM, MTDH, and PLGRKT). These candidates were chosen because there is no evidence from previous reports demonstrating differential expression or a role in IBC. To confirm the difference in expression between SUM-149 vs. MCF-10A cells, and to assess the expression of these proteins in several breast cancer cell lines, we used IBC (KPL-4, MDA-IBC-3 and SUM-149) and non-IBC (MCF-7 and SUM-102) cells with different molecular characteristics (Table 3), compared to MCF-10A cells. Cytoskeleton proteins, such as actins and tubulins were highly represented in our data (data not shown). β-actin has been associated with the plasma membrane for cell organization in the process of cancer proliferation and metastasis [15]. In Figure 1A, β-actin expression was detected in the cytoplasmic cell fraction and was not differentially expressed in SUM-149 cells compared to the MCF-10A PM fraction. For this reason, we chose β-actin as our loading control. SUM-149 cell protein expression was significantly higher for FLOT1 (2.1: P≤0.05), ITGB5 (2.5: P≤0.05), L1CAM (1.9: P≤0.001), MCAM (5.9: P≤0.05), MTDH (1.3: P≤0.01) and PLGRKT (1.6: P≤0.05), in agreement with SILAC results. Furthermore, the IBC KPL-4 cell line also overexpressed FLOT1 (2.1: P≤0.05), ITGB5 (2.7: P≤0.05), and MTDH (1.5: P≤0.05). Non-IBC MCF-7 cells overexpressed FLOT1 (P≤0.05) and L1CAM (P≤0.05). Meanwhile, SUM-102 showed downregulation of ITGB5 (P≤0.05) protein expression (Figure 1B, 1C).
Figure 1.
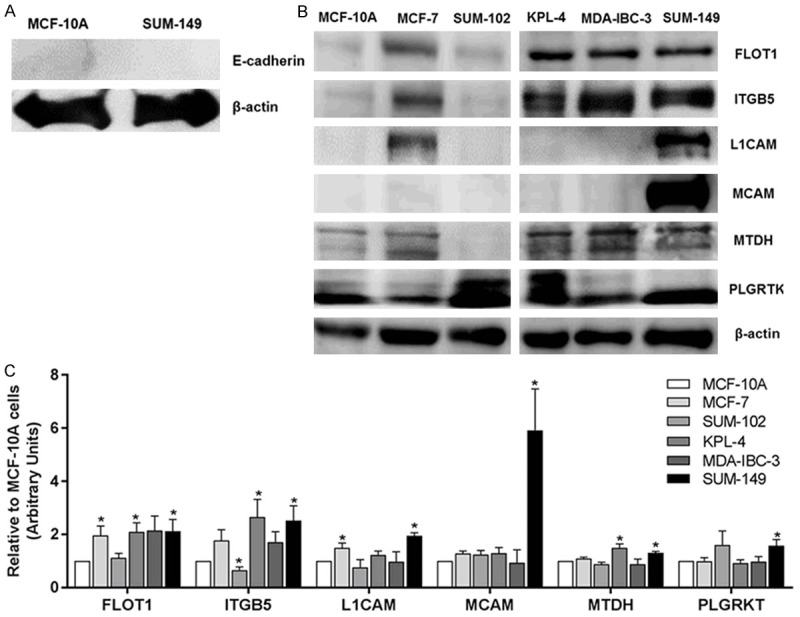
Immunoblotting validation of candidate PMPs identified by SILAC. A. Cytoplasmic fraction from MCF-10A and SUM-149 cells. β-actin and E-cadherin were used as controls for cytoplasmic and membrane proteins, respectively. B. Expression of six representatives upregulated PMPs identified by SILAC on IBC and non-IBC cell lines. C. Densitometric analysis using Image J software. β-actin was used as a loading control. Data is expressed relative to MCF-10A cells. Bars represent mean ± SEM of quadruplicates. *P≤0.05.
Table 3.
Molecular subtypes of cell lines used for SILAC validation
| Molecular subtype | Cell line |
|---|---|
| ER+, PR+, HER2- | MCF-7 |
| ER-, PR-, HER2+, | KPL-4, MDA-IBC-3 |
| ER-, PR-, HER2-, EGFR+ | MCF-10A, SUM-102, SUM-149 |
ER: Estrogen Receptor. PR: Progesterone Receptor. HER2: Human Epidermal Growth Factor Receptor 2. EGFR: Epidermal Growth Factor Receptor.
Protein expression and cellular distribution
The clinical and demographic details of the 17 IBC and 24 IDC patients are shown in Table 4. As expected, IBC patients presented clinically at a younger age than non-IBC patients (P≤0.01). IBC, like non-IBCs, is a heterogeneous disease and can occur as any of the six molecular breast cancer subtypes. However, IBC as a highly lethal cancer is most commonly triple negative or ER-, PR- and HER2+. Clinical data comparing IBC to IDC patients revealed no significant difference in HER2 or ER status between the two groups, but showed a higher expression of PR in IDCs than IBC patients (P≤0.05).
Table 4.
Clinical and pathological characterization of IBC versus IDC patients
| Characteristic | IBC patients (n=17) | IDC patients (n=24) | P-valuea |
|---|---|---|---|
| Age (years) | |||
| Range | 34-62 | 39-94 | 0.0083b |
| Mean ± SD | 47.73 ± 9.7 | 60.38 ± 15.4 | |
| ER | |||
| Positive | 9 (52.9%) | 17 (70.8%) | >0.05 |
| Negative | 8 (47.1%) | 7 (29.2%) | |
| PR | |||
| Positive | 4 (23.5%) | 15 (62.5%) | 0.025c |
| Negative | 13 (76.5%) | 9 (37.5%) | |
| Her2 | |||
| Positive | 7 (41.2%) | 8 (33.3%) | >0.05 |
| Negative | 9 (52.9%) | 15 (62.5%) | |
| Unknown | 1 (5.9%) | 1 (4.2%) | |
| Death | |||
| Dead | 8 (47.1%) | 10 (41.2%) | >0.05 |
| Alive | 9 (52.9%) | 14 (58.3%) | |
Significant P value (P≤ 0.05).
Student’s t-test.
Fisher’s exact test.
As shown in Table 5 and Figure 2A, 2B, antibodies recognizing C1QBP, L1CAM, MCAM, MST1R, MTDH, PLGRKT and SCAMP3 were used to confirm the results of previous analyses and to assess protein distributions. Eight of seventeen IBC tissues, stained strongly and all normal breast tissues (NBT) stained weakly (10/10) for C1QBP (P<0.0001). The presence of C1QBP was also detected in lymphatic vessels in IBC tissue samples. In addition, >50% of cells stained positive with the distribution of C1QBP cytoplasmic in both type of tissues. When we compare IDC to NBT, 12/17 IDC cases stained moderate or strong in C1QBP, showing a statistically significant difference in stain intensity (P≤0.01) between the two groups, while the percent of stained cells and location does not defer. There was no statistical difference in intensity, percent of stained cells or location between IBC and IDC samples for C1QBP, making C1QBP a protein that stains stronger in tumor tissue.
Table 5.
Expression and distribution of validated plasma membrane proteins in tumor and normal breast tissues
| Type of tissue | Protein | Intensitya | Cells stainedb | Locationc |
|---|---|---|---|---|
| IBC | C1QBP | 1/17: Weak | 14/17: +++ | 14/17: C |
| 5/17: Moderate | ||||
| 8/17: Strong | ||||
| 3/17: NTT | ||||
| L1CAM | 8/17: Weak | 8/17: + | 9/17: C | |
| 1/17: Moderate | 1/17: ++ | |||
| 6/17: NS | ||||
| 2/17: NTT | ||||
| MCAM | 4/17: Weak | 1/17: + | 2/17: C | |
| 10/17: NS | 3/17: +++ | 2/17: MC | ||
| 3/17: NTT | ||||
| MST1R | 6/17: Weak | 2/17: + | 3/17: C | |
| 8/17: Moderate | 2/17: ++ | 10/17: NC | ||
| 1/17: Strong | 11/17: +++ | 1/17: MC | ||
| 2/17: NTT | 1/17: M | |||
| MTDH | 6/15: Weak | 1/15: + | 14/15: C | |
| 4/15: Moderate | 13/15: +++ | |||
| 4/15: Strong | ||||
| 1/15: NTT | ||||
| PLGRKT | 4/17: Weak | 1/17: + | 8/17: C | |
| 4/17: Moderate | 3/17: ++ | 4/17: NC | ||
| 4/17: Strong | 8/17: +++ | |||
| 1/17: NS | ||||
| 4/17: NTT | ||||
| SCAMP3 | 12/17: Weak | 9/17: + | 9/17: C | |
| 2/17: NS | 1/17: ++ | 1/17: NC | ||
| 3/17: NTT | 2/17: +++ | 2/17: M | ||
| Control | C1QBP | 10/10: Weak | 10/10: +++ | 10/10: C |
| L1CAM | 10/10: NS | _ | _ | |
| MCAM | 10/10: Weak | 4/10: + | 9/10: C | |
| 1/10: ++ | 1/10: MC | |||
| 5/10: +++ | ||||
| MST1R | 8/10: Weak | 9/10: +++ | 9/10: NC | |
| 1/10: Moderate | ||||
| 1/10: NS | ||||
| MTDH | 10/10: Moderate | 10/10: +++ | 10/10: NC | |
| PLGRKT | 10/10: Moderate | 10/10: +++ | 10/10: NC | |
| SCAMP3 | 10/10: NS | _ | _ | |
| DC | C1QBP | 5/17: Weak | 17/17: +++ | 17/17: C |
| 6/17: Moderate | ||||
| 6/17: Strong | ||||
| L1CAM | 8/18: Weak | 4/18: + | 8/18: C | |
| 2/18: Moderate | 5/18: ++ | 1/18: MC | ||
| 6/18: NS | 1/18: +++ | 1/18: M | ||
| 2/18: NTT | ||||
| MCAM | 17/17: NS | _ | _ | |
| MST1R | 12/17: Weak | 1/17: + | 12/17: C | |
| 3/17: Moderate | 2/17: ++ | 2/17: NC | ||
| 1/17: Strong | 13/17: +++ | 2/17: MC | ||
| 1/17: NS | ||||
| MTDH | 7/17: Weak | 16/17: +++ | 16/17: C | |
| 9/17: Moderate | ||||
| 1/17: NTT | ||||
| PLGRKT | 12/17: Weak | 2/17: + | 9/17: C | |
| 1/17: Moderate | 2/17: ++ | 4/17: NC | ||
| 4/17: NS | 9/17: +++ | |||
| SCAMP3 | 10/19: NS | 5/17: + | 8/17: C | |
| 8/19: Weak | 1/17: ++ | |||
| 1/19: NTT | 2/17: +++ |
NS = no staining, NTT = no tumor tissue. “+” = Less than 10% cells positive staining, “++” = 10-50% cells positive staining, “+++” = More than 50% cells positive staining. C, NC, MC, M means cytoplasmic, nuclear and cytoplasmic, membranous and cytoplasmic, and membranous, respectively.
Figure 2.
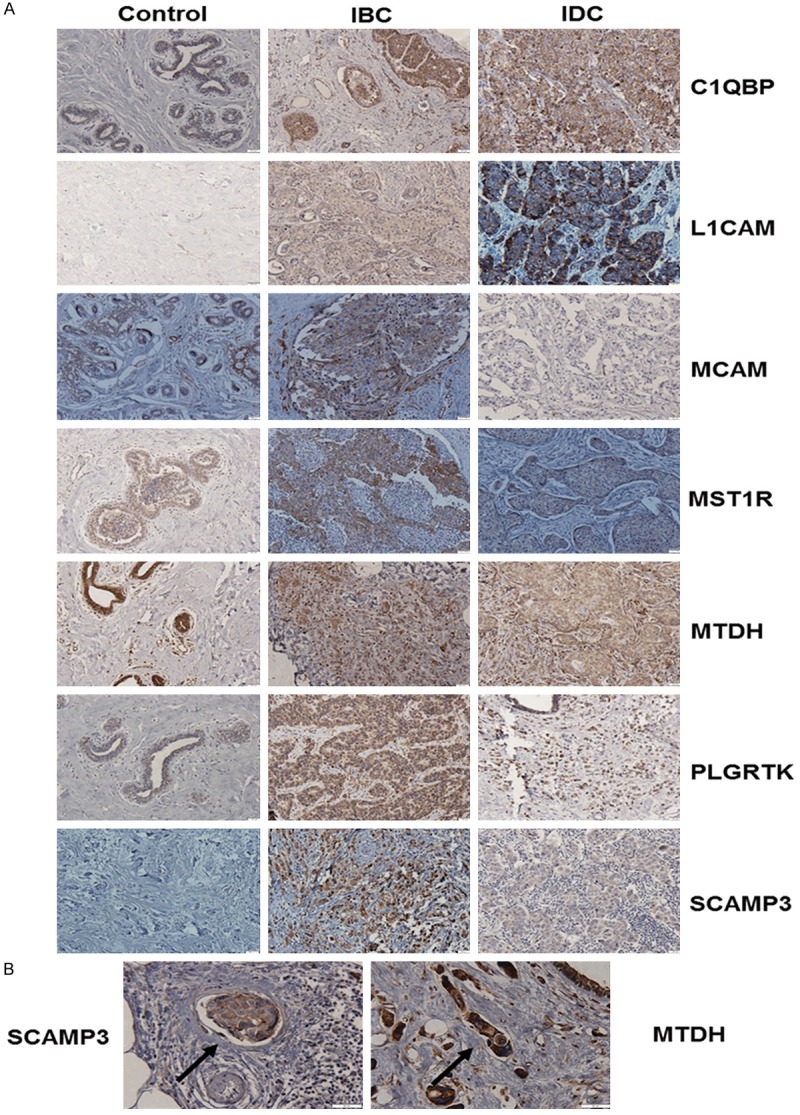
Protein expression and cellular distribution of selected proteins. A. Immunohistochemical analysis using antibodies against C1QBP, L1CAM, MCAM, MST1R, MTDH, PLGRTK and SCAMP3 in normal breast tissues (n=10), IBC (n=17) and IDC (n=24). B. MTDH and SCAMP3 expression in tumor emboli cells. Black arrows point to emboli. Micrographs were captured using an Olympus inverted microscope. Scale bar = 20 µm.
IHC analysis shows that cytoplasmic L1CAM was expressed in 60% and 44% of IBC and IDC tumor tissues, respectively. Interestingly, 11% of IDC samples show expression of L1CAM in the membrane while no membranous expression was observed in IBCs. However, this cell adhesion molecule was not expressed in control samples vs IBC and IDC (P≤0.01). Our correlation analyses evidence that a positive relationship exists between L1CAM staining intensity and metastasis (P≤0.04; P≤0.02) in IBC patients (Table 6). IHC results demonstrate that IBC MCAM-stained tissues display cytoplasmic or membranous/cytoplasmic expression. However, weak cytoplasmic expression of this protein was also detected in 100% of controls (P≤0.001). Meanwhile, no expression of MCAM was detected in IDC tissues. Interestingly, a negative correlation between MCAM and lymphatic invasion was observed in women with IBC (P≤0.04) (Table 6), suggesting that MCAM might be acting as a tumor suppressor. Overall, these data suggest that L1CAM is a tumor-associated protein, while MCAM is negatively associated with lymphovascular invasion in IBC patients.
Table 6.
Correlation analyses
| Protein | Variables | Pearson’s r | P-value |
|---|---|---|---|
| L1CAM | Staining vs metastasis | .556 | 0.039 |
| Intensity vs metastasis | .611 | 0.020 | |
| MCAM | Staining vs LI | -.550 | 0.042 |
LI = Lymphatic Invasion.
Nine cases of IBC stained moderate or strong for MST1R compared to 8/10 control cases that stained weakly (P≤0.05) in more than 50% of cells (P≤0.05). Significant differences were observed in location in IBCs vs. IDCs (P≤0.01). Although membranous staining of MST1R was detected in IDCs, its main distribution was observed in the cytoplasm contrasting thereby with controls where all samples stained NC (P≤0.0001).
Forty percent of IBC and IDC cases stained weakly for MTDH, while 100% of controls stained moderately (P≤0.01). IHC results showed a cytoplasmic distribution of MTDH in IBCs and IDCs but nuclear and cytoplasmic in control tissues (P≤0.0001). Staining intensity was significantly different between IBCs and IDCs (P≤0.05). Strong expression of MTDH was also observed in lymphatic vessels and in tumor emboli in IBC tissues (Figure 2B).
Twelve IDC tissues displayed weak staining of PLGRTK in comparison with IBCs and controls where 8/17 displayed moderate and strong staining (P≤0.01) and 10/10 moderate intensity (P≤0.0001), respectively. Moreover, a significant difference in staining intensity was observed between IBC vs. controls (P≤0.01). IBC and IDC tissues demonstrated cytoplasmic staining in comparison with nuclear and cytoplasmic distribution in controls (P≤0.001). Weak staining of SCAMP3 was identified in 86% of the IBC cases. SCAMP3 was identified in the membrane and cytoplasm of tumor emboli cells and in lymphatic vessels (Figure 2B), while cytoplasmic expression was found in >40% of IDCs (P≤0.05). Interestingly, as shown in Figure 2A, no expression of this protein was detected in controls (vs. IBC, P≤0.0001, vs. IDC, P≤0.05). Therefore, our results suggest an important role for SCAMP3 in IBC invasion.
Interaction analyses
Functional networks analysis of upregulated PMPs was performed using Ingenuity Pathway Analysis. The top network functions identified as upregulated proteins in IBC cells were involved in Cell Morphology, Cellular Assembly and Organization, Immunological Diseases (26 proteins), Hereditary Disorder, Cellular Assembly, Organization, Function, Maintenance (13 proteins) and Gene Expression, RNA Damage and Repair, RNA Post-Transcriptional Modification (8 proteins) (Figures 3A, 4, 5 and 6). These findings indicate that the SUM-149 cell PM proteome was mostly associated with cell morphology, organization and maintenance. Thus, the interaction potential of selected proteins was further analyzed. Interaction analysis identified direct and indirect relationships of eight verified proteins (C1QBP, FLOT1, ITGB5, L1CAM, MCAM, MST1R, MTDH, and SCAMP3) with central molecules that have an important role in breast cancer (EGFR, AKT and ERK). However, no direct or indirect interactions were found between PLGRKT and selected proteins or incorporated molecules into network (see Figure 6 for PLGRKT interacting proteins). This network shows the direct binding interaction between MST1R and EGFR and their capacity for AKT activation. Furthermore, C1QBP and SCAMP3 cause activation of AKT and EGFR, respectively (Figure 3B). Since, EGFR, AKT and ERK pathways are key for the IBC development and progression, the interaction of the validated proteins with these pathways suggest their potential role in IBC pathogenesis.
Figure 3.
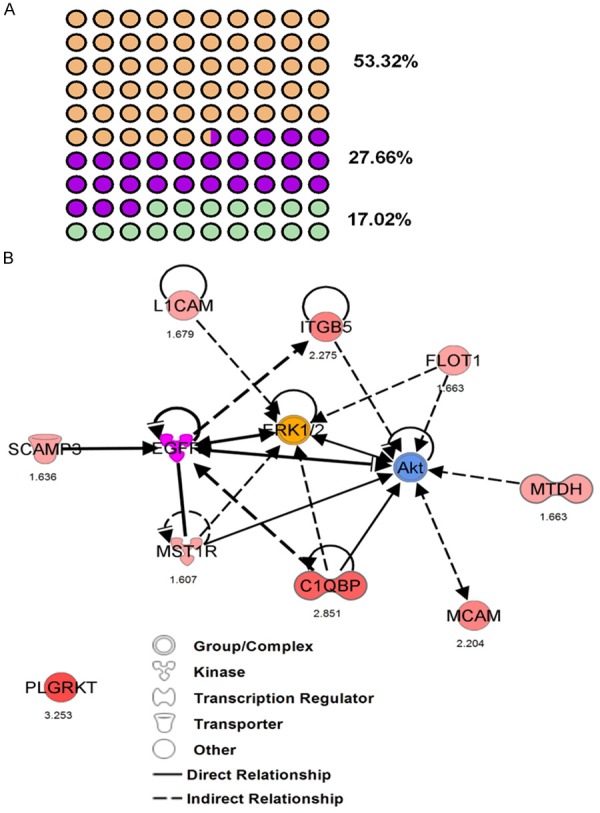
Functional network analysis of differentially upregulated PMPs. A. Top network functions identified as upregulated proteins in IBC cells. Network 1: Cell Morphology, Cellular Assembly and Organization, Immunological Disease (orange dots). Network 2: Hereditary Disorder, Cellular Assembly, Organization, Function, Maintenance (purple dots). Network 3: Gene Expression, RNA Damage and Repair, RNA Post-Transcriptional Modification (green dots). B. The image was created using the Ingenuity Pathways Analysis (IPA) platform (Ingenuity Systems; ©2000-2015 QIAGEN) by overlaying the PMPs detected by SILAC (red) onto a molecular network from the Ingenuity knowledgebase. Red indicates high SILAC ratios, and purple, yellow and blue indicates proteins that were not identified by SILAC but form part of this network. For each identified protein, the number corresponds to the protein quantification (log2 ratio). Legend indicates the function of each protein and the interactions between them.
Figure 4.
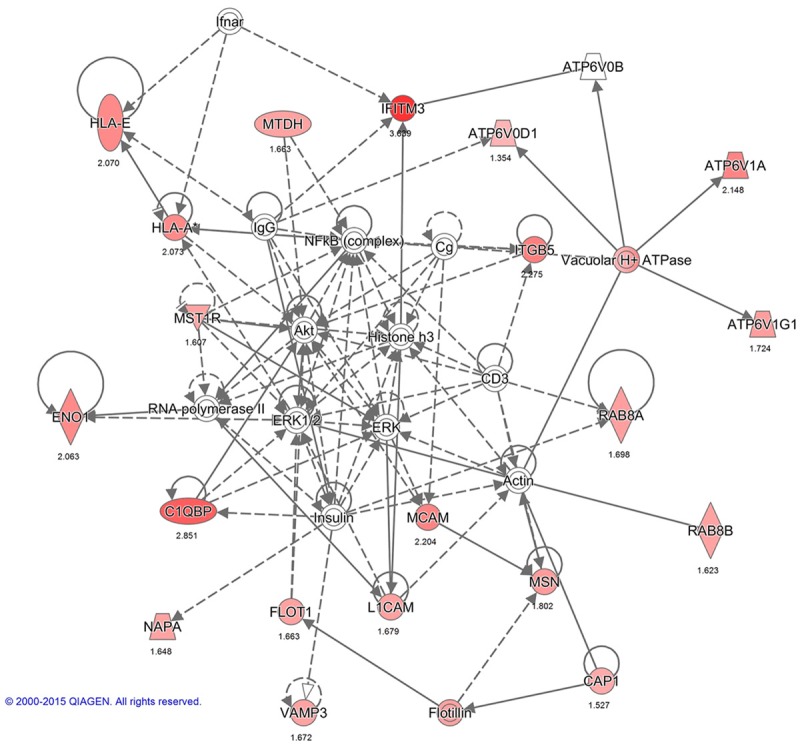
Interactions between PMPs in network 1: Cell Morphology, Cellular Assembly and Organization, Immunological Disease. The image was created using the Ingenuity Pathways Analysis (IPA) platform (Ingenuity Systems; ©2000-2015 QIAGEN) by overlaying the membrane proteins detected by SILAC onto a molecular network from the Ingenuity knowledgebase. Red indicates high SILAC ratios, and gray indicates proteins that were not identified by SILAC but form part of this network. For each identified protein, the number corresponds to the protein quantification (log2 ratio).
Figure 5.
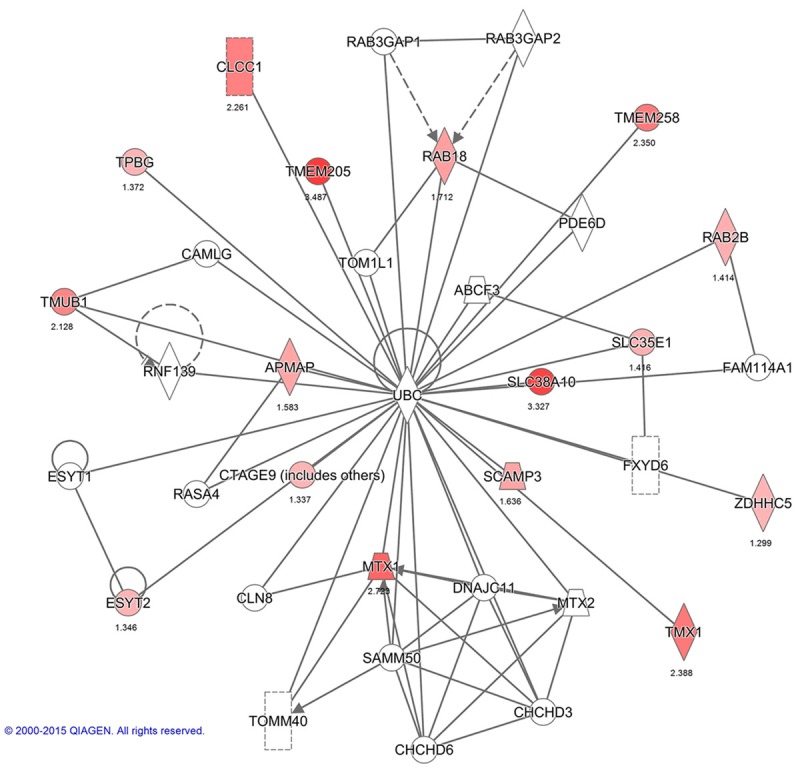
Interactions between PMPs in network 2: Hereditary Disorder, Cellular Assembly, Organization, Function, and Maintenance. The image was created using the IPA platform (Ingenuity Systems; ©2000-2015 QIAGEN) by overlaying the membrane proteins detected by SILAC onto a molecular network from the Ingenuity knowledgebase. Red indicates high SILAC ratios, and gray indicates proteins that were not identified by SILAC but form part of this network. For each identified protein, the number corresponds to the protein quantification (log2 ratio).
Figure 6.
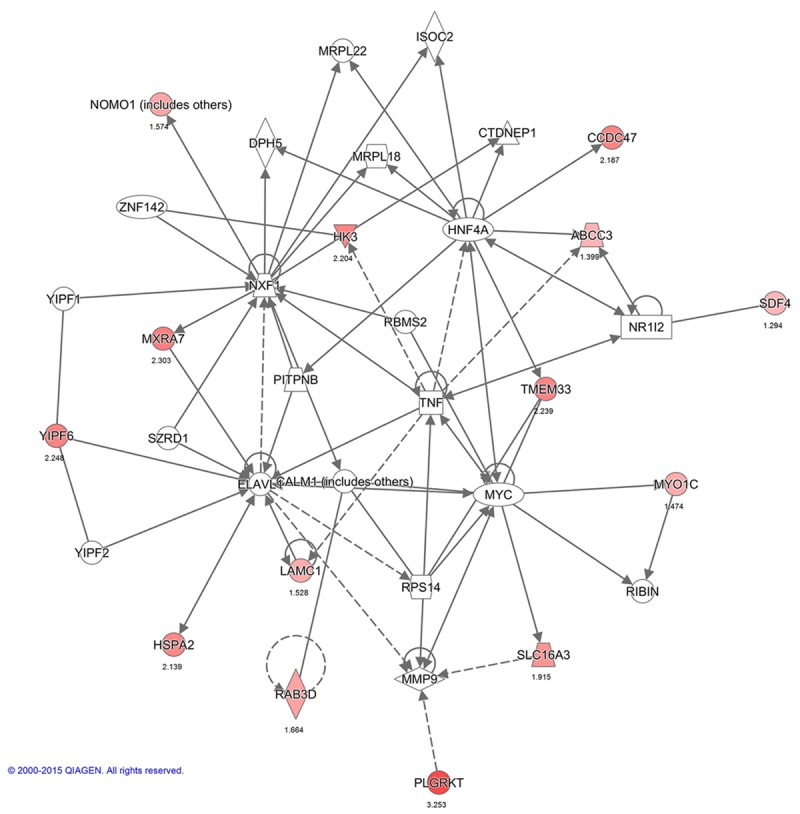
Interactions between PMPs in network 3: Gene Expression, RNA Damage and Repair, RNA Post-Transcriptional Modification. The image was created using the IPA platform (Ingenuity Systems; ©2000-2015 QIAGEN) by overlaying the membrane proteins detected by SILAC onto a molecular network from the Ingenuity knowledgebase. Red indicates high SILAC ratios, and gray indicates proteins that were not identified by SILAC but form part of this network. For each identified protein, the number corresponds to the protein quantification (log2 ratio).
Discussion
Identification of PM-associated proteins is an important first step in the development of cancer-targeted therapies. In this report we quantify, identify and define for the first time the IBC membrane proteome. By comparing IBC cells with non-cancerous breast cancer cells using SILAC, we were able to identify strategies that IBC cells and tumors might use to proliferate, invade and progress to metastasis. Finally, we could establish similarities and differences between non-IBCs and IBCs comparing multiple breast cancer cell lines and tumors with different molecular profiles.
SUM-149 and MCF-10A cells are the most used and well known models to examine the cell and molecular biology of IBC and non-cancerous mammary epithelia, respectively. However, some limitations exist in in vitro models, such as nutrient requirements or culture conditions. For this reason, using patient tumor tissues is the most accurate scenario to study protein expression and function. Since, tumors are influenced by a range of biological factors, which are necessary for tumor development and progression, using only the membrane fraction of cells to assess protein expression and quantification could be a limiting factor. It is well known that the selected proteins are PMPs or plasma membrane interacting proteins; however, IHC results also show expression of these proteins in other cell locations (i.e., cytoplasm or nucleus). This difference in location can be explained by the effect of several cellular stimuli in the tumor microenvironment. It is important to underline that we classified the protein expression distribution taking in consideration the location of the protein in the greatest number of stained cells. Here we discuss the distribution and the function of each protein in the different cell compartments.
This is the first proteomic study where PLGRKT expression is described in IBC and non-IBC cells and tissues. PLGRKT is a novel integral membrane plasminogen receptor with an exposed C-terminal lysine to the cell surface which promotes plasminogen activation by the urokinase receptor and tissue plasminogen activator (uPA) [16]. PLGRKT is involved in regulation of inflammatory response and regulates monocyte/macrophage migration and matrix metalloproteinase activation [17]. Recent studies in IBC have evidenced that an increase in macrophage infiltration and an interaction with human monocytes promote tumor-progression and invasion of IBC [18-20]. The colocalization of the protease Cathepsin B, uPA and uPAR with caveolin-1 in the caveolae has been associated with metastasis to lymph nodes in IBC [21,22]. Moreover, caveolin-1 overexpression mediates IBC cell invasion via AKT and RhoC GTPase [23]. Similarly, to caveolins, lipid rafts associated flotillins are involved in the transport of key molecules in breast cancer. Since, IBC interaction with macrophages/monocytes and overexpression of caveolin-1, uPA and uPAR have been associated with invasion and metastasis and our data reveal the overexpression of flotilin-1 and PLGRKT is feasible to hypothesize that these proteins play an important the role in IBC progression.
MCAM and L1CAM, have been associated with cancer progression via the activation of PI3K/AKT and ERK signaling cascades [24]. Up-regulation of MCAM promotes motility, invasion, and tumorigenesis and is associated with a poor prognosis in breast cancer [25,26]. Immunoblotting data showed MCAM overexpression in SUM-149 IBC cells similar to a published study [27]. Our IHC results show MCAM expression was detected in only 29% of IBC tissues. Although MCAM has been associated with oncogenesis, other studies show that its overexpression suppresses tumor growth establishing a controversial dual role [28,29]. Herein, 100% of NBTs express MCAM while IDCs did not show expression suggesting a tumor suppressor role. Interestingly, we demonstrated a negative relationship between MCAM and lymphovascular invasion in IBC patients. Since, this is the first study evaluating the expression of MCAM in IBC, further investigation using a larger subset of samples are necessary to elucidate the role of this protein in IBC. On the other hand, we validated published results that establish L1CAM overexpression in IBC [30]. This overexpression has been associated with IBC cell survival and invasion [31,32]. Importantly, in this study we also evidenced that L1CAM expression correlates with metastasis establishment in women with IBC. Cleavage of the L1CAM ectodomain proximal to the PM is mediated by metalloproteinases yielding a C-terminal stub that is a γ-secretase substrate. This γ-secretase processed fragment results in the release of a soluble L1CAM intracellular domain into the cytoplasm, which has been implicated in breast cancer cell adhesion and migration [33,34]. This phenomenon could explain our IHC results, which show expression of L1CAM at the cytoplasm in IBC instead of PM localization.
The function of secretory carrier membrane protein (SCAMP3) has not been characterized in detail yet; however evidence demonstrates that it acts as a regulator of EGFR trafficking within endosomal membranes enhancing the recycling of the receptor and decreasing its degradation [35]. This is the first study where the protein expression of SCAMP3 has been assessed in breast cancer. Our findings demonstrate that SCAMP3 is expressed in almost 90% of IBC tissues, lymphatic vessels and tumor emboli cells. Although, further studies are necessary, SCAMP3 promises to be a molecular marker for the diagnosis or treatment of IBC.
Membrane, nuclear and/or cytoplasmic Metadherin (MTDH) is overexpressed in about 45% of the primary tumors and is significantly correlated with clinical stage, tumor size, metastasis and poor survival through the activation of multiple oncogenic pathways such as PI3K/AKT, Wnt/β-catenin and MAPK [36,37]. In IBC, high ratios of HER2 transcripts were associated with increased proteomic levels of MTDH in SUM-190 cells [38]. Here, we show overexpression of MTDH in SUM-149 and KPL-4 IBC cells. Although, studies demonstrate that MTDH is expressed in low levels or is absent in most of normal human breast tissues [39], IHC data showed moderate nuclear and cytoplasmic expression in NBTs and strong cytoplasmic expression in IBCs, suggesting a redistribution of MTDH from nucleus to cytoplasm. Moreover, as well as SCAMP3, MTDH might be associated with lymphovascular invasion and metastasis in IBC.
Our SILAC data revealed the overexpression of the receptor tyrosine kinase (RTK), macrophage-stimulating protein receptor. MST1R is a RTK of the c-Met family that activates several signaling cascades including RAS-ERK and PI3K-AKT. MST1R is overexpressed in approximately 50% of breast cancers and is associated with proliferation, metastasis and poor prognosis but barely detectable in normal breast epithelia [40,41]. In accordance with previous findings, our results show weak expression in NBTs and overexpression in IBC cells and tissues. The MST1R precursor protein is synthesized as a single chain and remains in the cytoplasm where is cleaved to produce a functional heterodimer with other RTKs [42,43]. MST1R/MET crosstalk with β-catenin pathway facilitating its nuclear translocation leading in the transcription of oncogenic mRNAs [43]. Although we show membranous localization of MST1R, the most IBC tissues stained in the nucleus. Recent data suggest that MST1R/EGFR translocate to the nucleus, acting as a transcriptional regulator of c-JUN to promote survival of cancer cells in hypoxic conditions [44,45]. In IBC, overexpression of eIF4G1 increases the translation of VEGF, which accounts for resistance to hypoxia required for IBC cell survival [46,47]. We could hypothesize that the translocation of MST1R to the nucleus in IBC might play a role in IBC tumor survival under hypoxic conditions.
Complement 1q binding protein (C1QBP), is mainly distributed in mitochondria but it can also be detected in the cytosol and cell surface by the activation of ERK [48]. Recently, elevated expression of cytoplasmic C1QBP was correlated with poor survival, lymphovascular invasion and metastasis to lymph nodes in breast and endometrial cancer patients [49,50]. Our SILAC results show overexpression of C1QBP in SUM-149 IBC cells. Furthermore, IHC analysis demonstrated significantly elevated C1QBP protein levels in tumors and in accordance with published studies its expression in IBC lymphatic vessels might be directly associated with lymphovascular invasion process.
The present study is the first to identify the altered protein expression of membrane-proteins in IBC. The established proteomic differences between controls, non-IBCs and IBCs evidence the heterogeneity of breast cancer and suggest the use of diverse strategies for tumor formation and development. Furthermore, our data validate the central role of EGFR, AKT and ERK pathways in the oncogenic process of IBC and reveal the importance of continuing studies to assess the function of identified proteins in the localized cell compartments. Finally, we have presented potential biomarkers of IBC that will not only benefit accurate and early diagnosis of this intractable disease but also could be targets for further development of therapies.
Acknowledgements
We thank Dr. Scott Schaffer from the Proteomics and Mass Spectrometry Facility in UMASS for the MS analysis. We would like to acknowledge the UCC-SOM RCMI Common Instrumentation Area. This work was supported by National Cancer Institute #CA174307 (ISA), National Institute of General Medical Sciences #GM111171 (Martínez-Montemayor), National Center for Research Resources #RR003035, National Institute on Minority Health and Health Disparities #MD007583 (MMM, GMM), Na-tional Center for Research Resources #RR016470, GM103475 (UPR-pilot Martínez-Montemayor), National Institute on Minority Health and Health Disparities #MD008149 (MMM), National Institute on Minority Health and Health Disparities #MD007587 (MMM), National Institute on Minority Health and Health Disparities #MD007600 (JPL), National Institute of General Medical Sciences #GM110513 (LAC), Title-V-PPOHA #P031M105050 and Title-V-Cooperative #P031S130068 U.S. Department of Education (LAC). New York State Stem Cell Program #CO28126, The Breast Cancer Research Foundation, National Cancer Institute CA178509, IBCRF (RJS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the U.S. Department of Education.
Disclosure of conflict of interest
None.
References
- 1.Dawood S, Ueno NT, Valero V, Woodward WA, Buchholz TA, Hortobagyi GN, Gonzalez-Angulo AM, Cristofanilli M. Differences in survival among women with stage III inflammatory and noninflammatory locally advanced breast cancer appear early: a large population-based study. Cancer. 2011;117:1819–1826. doi: 10.1002/cncr.25682. [DOI] [PubMed] [Google Scholar]
- 2.Walker R. Rosen’s Breast Pathology. J C Path. 1997;50:1036–1036. [Google Scholar]
- 3.Tomlinson JS, Alpaugh ML, Barsky SH. An Intact Overexpressed E-cadherin/α, β-Catenin Axis Characterizes the Lymphovascular Emboli of Inflammatory Breast Carcinoma. Cancer Res. 2001;61:5231–5241. [PubMed] [Google Scholar]
- 4.Cabioglu N, Gong Y, Islam R, Broglio KR, Sneige N, Sahin A, Gonzalez-Angulo AM, Morandi P, Bucana C, Hortobagyi GN, Cristofanilli M. Expression of growth factor and chemokine receptors: new insights in the biology of inflammatory breast cancer. Ann Oncol. 2007;18:1021–1029. doi: 10.1093/annonc/mdm060. [DOI] [PubMed] [Google Scholar]
- 5.Dawood S, Broglio K, Gong Y, Yang WT, Cristofanilli M, Kau SW, Meric-Bernstam F, Buchholz TA, Hortobagyi GN, Gonzalez-Angulo AM Inflammatory Breast Cancer Research Group. Prognostic significance of HER-2 status in women with inflammatory breast cancer. Cancer. 2008;112:1905–1911. doi: 10.1002/cncr.23350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kleer CG, van Golen KL, Merajver SD. Molecular biology of breast cancer metastasis. Inflammatory breast cancer: clinical syndrome and molecular determinants. Breast Cancer Res. 2000;2:423–429. doi: 10.1186/bcr89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ziegler YS, Moresco JJ, Tu PG, Yates JR 3rd, Nardulli AM. Plasma Membrane Proteomics of Human Breast Cancer Cell Lines Identifies Potential Targets for Breast Cancer Diagnosis and Treatment. PLoS One. 2014;9:e102341. doi: 10.1371/journal.pone.0102341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang X, Zhao J, Hajivandi M, Wu R, Tao J, Amshey JW, Pope RM. Quantification of membrane and membrane-bound proteins in normal and malignant breast cancer cells isolated from the same patient with primary breast carcinoma. J Proteome Res. 2006;5:2632–2641. doi: 10.1021/pr060125o. [DOI] [PubMed] [Google Scholar]
- 9.Deeb SJ, Cox J, Schmidt-Supprian M, Mann M. N-linked glycosylation enrichment for in-depth cell surface proteomics of diffuse large B-cell lymphoma subtypes. Mol Cell Proteomics. 2014;13:240–251. doi: 10.1074/mcp.M113.033977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suárez-Arroyo IJ, Rios-Fuller TJ, Feliz-Mosquea YR, Lacourt-Ventura M, Leal-Alviarez DJ, Maldonado-Martinez G, Cubano LA, Martínez-Montemayor MM. Ganoderma lucidum Combined with the EGFR Tyrosine Kinase Inhibitor, Erlotinib Synergize to Reduce Inflammatory Breast Cancer Progression. J Cancer. 2016;7:500–511. doi: 10.7150/jca.13599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Montemayor MM, Acevedo RR, Otero-Franqui E, Cubano LA, Dharmawardhane SF. Ganoderma lucidum (Reishi) inhibits cancer cell growth and expression of key molecules in inflammatory breast cancer. Nutr Cancer. 2011;63:1085–1094. doi: 10.1080/01635581.2011.601845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forozan F, Veldman R, Ammerman CA, Parsa NZ, Kallioniemi A, Kallioniemi OP, Ethier SP. Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br J Cancer. 1999;81:1328–1334. doi: 10.1038/sj.bjc.6695007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim MH, Jung SY, Ahn J, Hwang SG, Woo HJ, An S, Nam SY, Lim DS, Song JY. Quantitative proteomic analysis of single or fractionated radiation-induced proteins in human breast cancer MDA-MB-231 cells. Cell Biosci. 2015;5:2. doi: 10.1186/2045-3701-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ethier SP, Kokeny KE, Ridings JW, Dilts CA. erbB Family Receptor Expression and Growth Regulation in a Newly Isolated Human Breast Cancer Cell Line. Cancer Res. 1996;56:899–907. [PubMed] [Google Scholar]
- 15.Yamaguchi H, Shiraishi M, Fukami K, Tanabe A, Ikeda-Matsuo Y, Naito Y, Sasaki Y. MARCKS regulates lamellipodia formation induced by IGF-I via association with PIP2 and beta-actin at membrane microdomains. J Cell Physiol. 2009;220:748–755. doi: 10.1002/jcp.21822. [DOI] [PubMed] [Google Scholar]
- 16.Andronicos NM, Chen EI, Baik N, Bai H, Parmer CM, Kiosses WB, Kamps MP, Yates JR, Parmer RJ, Miles LA. Proteomics-based discovery of a novel, structurally unique, and developmentally regulated plasminogen receptor, Plg-R(KT), a major regulator of cell surface plasminogen activation. Blood. 2010;115:1319–1330. doi: 10.1182/blood-2008-11-188938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lighvani S, Baik N, Diggs JE, Khaldoyanidi S, Parmer RJ, Miles LA. Regulation of macrophage migration by a novel plasminogen receptor Plg-RKT. Blood. 2011;118:5622–5630. doi: 10.1182/blood-2011-03-344242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohamed MM, El-Ghonaimy EA, Nouh MA, Schneider RJ, Sloane BF, El-Shinawi M. Cytokines secreted by macrophages isolated from tumor microenvironment of inflammatory breast cancer patients possess chemotactic properties. Int J Biochem Cell Biol. 2014;46:138–147. doi: 10.1016/j.biocel.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jhaveri K, Teplinsky E, Silvera D, Valeta-Magara A, Arju R, Giashuddin S, Sarfraz Y, Alexander M, Darvishian F, Levine PH, Hashmi S, Zolfaghari L, Hoffman HJ, Singh B, Goldberg JD, Hochman T, Formenti S, Esteva FJ, Moran MS, Schneider RJ. Hyperactivated mTOR and JAK2/STAT3 Pathways: Molecular Drivers and Potential Therapeutic Targets of Inflammatory and Invasive Ductal Breast Cancers After Neoadjuvant Chemotherapy. Clin Breast Cancer. 2015;16:113–122. doi: 10.1016/j.clbc.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohamed MM, Cavallo-Medved D, Sloane BF. Human monocytes augment invasiveness and proteolytic activity of inflammatory breast cancer. Biol Chem. 2008;389:1117–1121. doi: 10.1515/BC.2008.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Victor BC, Anbalagan A, Mohamed MM, Sloane BF, Cavallo-Medved D. Inhibition of cathepsin B activity attenuates extracellular matrix degradation and inflammatory breast cancer invasion. Breast Cancer Res. 2011;13:R115. doi: 10.1186/bcr3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nouh MA, Mohamed MM, El-Shinawi M, Shaalan MA, Cavallo-Medved D, Khaled HM, Sloane BF. Cathepsin B: a potential prognostic marker for inflammatory breast cancer. J Transl Med. 2011;9:1. doi: 10.1186/1479-5876-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joglekar M, Elbazanti WO, Weitzman MD, Lehman HL, van Golen KL. Caveolin-1 mediates inflammatory breast cancer cell invasion via the Akt1 pathway and RhoC GTPase. J Cell Biochem. 2015;116:923–933. doi: 10.1002/jcb.25025. [DOI] [PubMed] [Google Scholar]
- 24.Lei X, Guan CW, Song Y, Wang H. The multifaceted role of CD146/MCAM in the promotion of melanoma progression. Cancer Cell Int. 2015;15:3. doi: 10.1186/s12935-014-0147-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng Gf, Cai Sx, Wu GJ. Up-regulation of METCAM/MUC18 promotes motility, invasion, and tumorigenesis of human breast cancer cells. BMC Cancer. 2011;11:113–113. doi: 10.1186/1471-2407-11-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zabouo G, Imbert AM, Jacquemier J, Finetti P, Moreau T, Esterni B, Birnbaum D, Bertucci F, Chabannon C. CD146 expression is associated with a poor prognosis in human breast tumors and with enhanced motility in breast cancer cell lines. Breast Cancer Res. 2009;11:R1–R1. doi: 10.1186/bcr2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mostert B, Kraan J, Bolt-de Vries J, van der Spoel P, Sieuwerts AM, Schutte M, Timmermans AM, Foekens R, Martens JW, Gratama JW, Foekens JA, Sleijfer S. Detection of circulating tumor cells in breast cancer may improve through enrichment with anti-CD146. Breast Cancer Res Treat. 2011;127:33–41. doi: 10.1007/s10549-010-0879-y. [DOI] [PubMed] [Google Scholar]
- 28.Shih LM, Hsu MY, Palazzo JP, Herlyn M. The cell-cell adhesion receptor Mel-CAM acts as a tumor suppressor in breast carcinoma. Am J Pathol. 1997;151:745–751. [PMC free article] [PubMed] [Google Scholar]
- 29.Ouhtit A, Gaur RL, Abd Elmageed ZY, Fernando A, Thouta R, Trappey AK, Abdraboh ME, El-Sayyad HI, Rao P, Raj MG. Towards understanding the mode of action of the multifaceted cell adhesion receptor CD146. Biochim Biophys Acta. 2009;1795:130–136. doi: 10.1016/j.bbcan.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 30.Willmarth NE, Ethier SP. Autocrine and juxtacrine effects of amphiregulin on the proliferative, invasive, and migratory properties of normal and neoplastic human mammary epithelial cells. J Biol Chem. 2006;281:37728–37737. doi: 10.1074/jbc.M606532200. [DOI] [PubMed] [Google Scholar]
- 31.Buchheit CL, Angarola BL, Steiner A, Weigel KJ, Schafer ZT. Anoikis evasion in inflammatory breast cancer cells is mediated by Bim-EL sequestration. Cell Death Differ. 2015;22:1275–1286. doi: 10.1038/cdd.2014.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehman HL, Van Laere SJ, van Golen CM, Vermeulen PB, Dirix LY, van Golen KL. Regulation of inflammatory breast cancer cell invasion through Akt1/PKBalpha phosphorylation of RhoC GTPase. Mol Cancer Res. 2012;10:1306–1318. doi: 10.1158/1541-7786.MCR-12-0173. [DOI] [PubMed] [Google Scholar]
- 33.Kiefel H, Bondong S, Hazin J, Ridinger J, Schirmer U, Riedle S, Altevogt P. L1CAM: A major driver for tumor cell invasion and motility. Cell Adh Migr. 2012;6:374–384. doi: 10.4161/cam.20832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Galileo DS. Soluble L1CAM promotes breast cancer cell adhesion and migration in vitro, but not invasion. Cancer Cell Int. 2010;10:34–34. doi: 10.1186/1475-2867-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aoh QL, Castle AM, Hubbard CH, Katsumata O, Castle JD. SCAMP3 Negatively Regulates Epidermal Growth Factor Receptor Degradation and Promotes Receptor Recycling. Mol Biol Cell. 2009;20:1816–1832. doi: 10.1091/mbc.E08-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu G, Chong RA, Yang Q, Wei Y, Blanco MA, Li F, Reiss M, Au JL, Haffty BG, Kang Y. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell. 2009;15:9–20. doi: 10.1016/j.ccr.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Zhang N, Song LB, Liao WT, Jiang LL, Gong LY, Wu J, Yuan J, Zhang HZ, Zeng MS, Li M. Astrocyte elevated gene-1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin Cancer Res. 2008;14:3319–3326. doi: 10.1158/1078-0432.CCR-07-4054. [DOI] [PubMed] [Google Scholar]
- 38.Zhang EY, Cristofanilli M, Robertson F, Reuben JM, Mu Z, Beavis RC, Im H, Snyder M, Hofree M, Ideker T, Omenn GS, Fanayan S, Jeong SK, Paik YK, Zhang AF, Wu SL, Hancock WS. Genome wide proteomics of ERBB2 and EGFR and other oncogenic pathways in inflammatory breast cancer. J Proteome Res. 2013;12:2805–2817. doi: 10.1021/pr4001527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown DM, Ruoslahti E. Metadherin, a cell surface protein in breast tumors that mediates lung metastasis. Cancer Cell. 2004;5:365–374. doi: 10.1016/s1535-6108(04)00079-0. [DOI] [PubMed] [Google Scholar]
- 40.Maggiora P, Marchio S, Stella MC, Giai M, Belfiore A, De Bortoli M, Di Renzo MF, Costantino A, Sismondi P, Comoglio PM. Overexpression of the RON gene in human breast carcinoma. Oncogene. 1998;16:2927–2933. doi: 10.1038/sj.onc.1201812. [DOI] [PubMed] [Google Scholar]
- 41.Lee WY, Chen HH, Chow NH, Su WC, Lin PW, Guo HR. Prognostic significance of co-expression of RON and MET receptors in node-negative breast cancer patients. Clin Cancer Res. 2005;11:2222–2228. doi: 10.1158/1078-0432.CCR-04-1761. [DOI] [PubMed] [Google Scholar]
- 42.Ronsin C, Muscatelli F, Mattei MG, Breathnach R. A novel putative receptor protein tyrosine kinase of the met family. Oncogene. 1993;8:1195–1202. [PubMed] [Google Scholar]
- 43.Wang MH, Zhang R, Zhou YQ, Yao HP. Pathogenesis of RON receptor tyrosine kinase in cancer cells: activation mechanism, functional crosstalk, and signaling addiction. J Biomed Res. 2013;27:345–356. doi: 10.7555/JBR.27.20130038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang HY, Liu HS, Lai MD, Tsai YS, Tzai TS, Cheng HL, Chow NH. Hypoxia promotes nuclear translocation and transcriptional function in the oncogenic tyrosine kinase RON. Cancer Res. 2014;74:4549–4562. doi: 10.1158/0008-5472.CAN-13-3730. [DOI] [PubMed] [Google Scholar]
- 45.Liu HS, Hsu PY, Lai MD, Chang HY, Ho CL, Cheng HL, Chen HT, Lin YJ, Wu TJ, Tzai TS, Chow NH. An unusual function of RON receptor tyrosine kinase as a transcriptional regulator in cooperation with EGFR in human cancer cells. Carcinogenesis. 2010;31:1456–1464. doi: 10.1093/carcin/bgq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silvera D, Schneider RJ. Inflammatory breast cancer cells are constitutively adapted to hypoxia. Cell Cycle. 2009;8:3091–3096. doi: 10.4161/cc.8.19.9637. [DOI] [PubMed] [Google Scholar]
- 47.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 48.Majumdar M, Meenakshi J, Goswami SK, Datta K. Hyaluronan binding protein 1 (HABP1)/C1QBP/p32 is an endogenous substrate for MAP kinase and is translocated to the nucleus upon mitogenic stimulation. Biochem Biophys Res Commun. 2002;291:829–837. doi: 10.1006/bbrc.2002.6491. [DOI] [PubMed] [Google Scholar]
- 49.Niu M, Sun S, Zhang G, Zhao Y, Pang D, Chen Y. Elevated expression of HABP1 is correlated with metastasis and poor survival in breast cancer patients. Am J Cancer Res. 2015;5:1190–1198. [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao J, Liu T, Yu G, Wang J. Overexpression of HABP1 correlated with clinicopathological characteristics and unfavorable prognosis in endometrial cancer. Tumour Biol. 2015;36:1299–1306. doi: 10.1007/s13277-014-2761-8. [DOI] [PubMed] [Google Scholar]


