Abstract
Background
Changes in gut microbiota composition and activity have been associated with different metabolic disorders, including obesity, diabetes, and cardiometabolic disorders. Recent evidence suggests that different organs are directly under the influence of bacterial metabolites that may directly or indirectly regulate physiological and pathological processes.
Scope of review
We reviewed seminal as well as recent papers showing that gut microbes influence energy, glucose and lipid homeostasis by controlling different metabolic routes such as endocrine, enteric and central nervous system. These dialogues are discussed in the context of obesity and diabetes but also for brain pathologies and neurodegenerative disorders.
Major conclusions
The recent advances in gut microbiota investigation as well as the discovery of specific metabolites interacting with host cells has led to the identification of novel inter-organ communication during metabolic disturbances. This suggests that gut microbes may be viewed as “novel” future therapeutic partners.
This article is part of a special issue on microbiota.
Keywords: Gut microbiota, Metabolites, Immune system, Enteric nervous system, Central nervous system, Enteroendocrine cells
1. Introduction
Our human evolutionary history is a long process that continues to progress. This complex biological evolution also explains the current way we live. We should keep in mind that we are likely the “end product” of a billion years-long process of permanent interaction with our environment. Many evolutionary theories have been proposed and discussed (e.g., Darwinism, creationism), but we have to acknowledge that all current scientific data are largely based on the fact that our environment has played a major role in the way we have evolved. Among the environmental factors, unequivocal evidence shows that microbes have colonized plants, soils and animals. Because microbes colonize many areas of vertebrates (i.e., both outside and inside the body), they have evolved with vertebrates, and they all have established a complex host-microbial relationship, thereby shaping their own genotype and, more importantly, their phenotype. As humans, we are providing them “board and lodging”, whereas, in turn, they are conferring on us numerous biological functions that we are unable to perform through our own metabolism. This symbiotic relationship may influence not only our health but also the risk of developing disease when the communication between these “organs” and our organs is disordered.
In this review, we will focus our attention on specific mechanisms by which gut microbes regulate physiological processes in the context of energy and glucose homeostasis. We will discuss how specific “chemical dialogues” may take place between the gut microbiota and target host cells. We will also highlight that this is a bidirectional communication system with a putative impact on host health.
2. Gut microbiota communication and gut peptides: impact on host metabolism
Gut microbiota composition and activity is under the influence of different factors. Among them, specific host-dependent factors, such as genetic background, sex, age, and the immune system, play a key role that is difficult to change on a voluntary basis. Conversely, specific behaviors may directly influence the gut microbiota, such as the use of antibiotics, anti-acid, anti-diabetic, or specific surgical procedures (gastric bypass). Finally, over the last 20 years, numerous data have undoubtedly shown that diet and nutrients strongly contribute to shape the gut microbiota composition (e.g., fatty acids, non-digestible carbohydrates, prebiotics, polyphenols) [1], [2], [3], [4], [5], [6], [7], [8], [9].
2.1. Microbial metabolites as triggering factors: focus on short-chain fatty acids
Depending on the substrates (amino acids, lipids, carbohydrates) present in the gut lumen, gut bacteria can generate specific metabolites. For example, organic acids such as short-chain fatty acids (SCFAs), or branched-SCFAs and specific bile acid derivatives and vitamins are continuously produced.
The microbial fermentation of carbohydrates in the gut typically produces acetate, propionate, butyrate, and lactate, which are specific SCFAs. The relative proportion of SCFAs and, eventually, their relative abundance and ratio may result in a specific host response. It is important to note that this metabolic collaboration is dependent on the presence of a particular genus of bacteria because all substrates (nutrients) are not equally transformed into SCFAs upon carbohydrate fermentation. In addition, not all the SCFAs have the same metabolic impact. For example, butyrate is known to be a primary energy source for colonocytes [10], [11]. Acetate is in theory used as a cholesterol or fatty acid precursor, whereas propionate is gluconeogenic in the liver and the gut, but it may also neutralize lipogenesis from acetate or glucose in the liver [12], [13].
In addition to this direct role in the de novo production of nutrients, it also has been demonstrated that these SCFAs can bind to specific receptors, such as G-protein coupled receptors FFAR2 and FFAR3, (also called GPR43 and 41). These two receptors are structurally related to each other and activated by SCFAs. More than a decade ago, Brown et al. identified the endogenous ligand of these receptors [14]. They are encoded by genes that are located close to each other in the genome. Moreover, these receptors exhibit some overlapping expression but also partially share signaling pathways (Gαi/o and/or Gq). Thus, for example, the stimulation of GPR43 by SCFAs reduces cAMP production and activates ERK (extracellular signal-regulated kinase) cascade via Gαi/o dependent mechanism, or increases intracellular Ca2+ levels and promotes activation of MAPK pathway (mitogen-activated protein kinase) via interactions with the Gq family. Since this finding, numerous reports have shown that these receptors are expressed in a wide variety of tissues and cells types (immune cells, endocrine cells and adipocytes) [15], [16]. For example, GPR43 mRNA is expressed in white adipose tissues as well as in cellular models (e.g., 3T3-L1 differentiated in adipocyte and mature adipocytes). In addition, several studies have shown that GPR43 is highly expressed in the adipose tissue during high-fat diet (HFD)-induced obesity compared with control normal chow diet-fed mice [17], [18], [19]. SCFAs are also able to suppress cAMP-induced lipolysis (isoproterenol) in a concentration dependent manner [17]. By using GPR43 knockout mice, Ge and colleagues found that this is an effect dependent on GPR43 [20]. SCFAs also have been shown to be involved in the management of inflammation, by mechanisms comprising the control of neutrophil chemotaxis but also by acting on the proliferation of T regulatory cells (Treg) [21], [22]. For example, different reports show that GPR43 contribute to the recruitment of immune cells and their activity may impact on the regulation of inflammatory processes in intestinal inflammation [23].
More recently, De Vadder and colleagues have shown that gut microbes improve various features of energy metabolism (e.g., insulin sensitivity) via mechanisms depending of a SCFAs-induced intestinal gluconeogenesis in the intestine. Specifically, they found that propionate acts on GPR43 in the periportal afferent neural system to induce intestinal gluconeogenesis via a gut-brain neural circuit and, eventually, has beneficial effects on host physiology [13].
Because, the bacterial fermentation of dietary fibers in the intestine is the major source of the SCFAs, these discoveries have led to the unequivocal demonstration of molecular mechanisms by which gut microbes dialog with organs and contribute to control host metabolism through the regulation of several intracellular cascades. Hence, SCFAs are considered to be key messengers through which bacteria are able to talk to organs and thereby modulate host metabolism.
2.2. Gut microbes may control food intake
Over the last 15 years, different researchers have contributed to decoding the mechanisms explaining how the ingestion of non-digestible carbohydrates (e.g., inulin-type fructans, arabinoxylans, chitin glucan, resistant starches) improves metabolic disorders through a gut microbiota-dependent pathway [24], [25], [26]. In 2004, it was reported that changing the gut microbiota in rats using three different prebiotics (inulin-type fructans) that varied according to their chemical structure reduced food intake, body weight, and fat mass [24]. This discovery raised key questions: how can we explain that changing the gut microbes by using prebiotics affects the control of a brain-controlled factors such as food intake? Are there any other putative factors involved? These questions will be addressed in a different part of this review.
2.3. Gut microbes and gut peptides
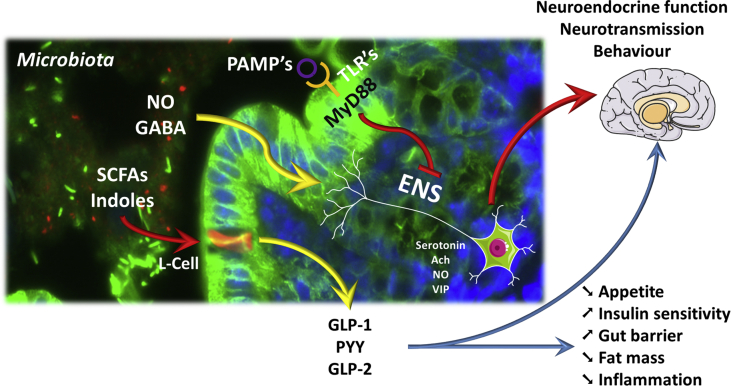
Searching for a mechanism of action, we reasoned that because the vast majority of microbes residing in the gut are located in the ileum and in the colon, the beneficial effects of prebiotics might be related to this area of the gastro-intestinal tract. Interestingly, this portion of the intestine is precisely where most of the enteroendocrine L-cells are located. Because L-cells produce anorexigenic peptides such as glucagon-like peptide-1 (GLP-1) and Peptide YY (PYY), we decided to quantify the concentration of these peptides in the portal vein of rodents in which the microbial composition was changed by using prebiotics. We discovered that the levels of GLP-1 and PYY were increased in portal vein blood [24], [27]. This effect was associated with an increase in the GLP-1 and PYY precursors (preproglucagon and pre-proPYYmRNA expression, respectively) in the ileum and in the colon. We next discovered that affecting the gut microbiota composition and activity strongly decreased the orexigenic hormone ghrelin in the blood of rats treated with prebiotics [24] (Figure 1).
Figure 1.
Overview of the different interactions existing between microbial metabolites, endocrine and nervous routes. Gut microbes interact with host cells using different mechanisms. SCFAs (short chain fatty acids) are metabolites produced by the microbial fermentation of different nutrients; these SCFAs are recognized by specific G-protein coupled receptors expressed at the surface of enteroendocrine cells such as L-cells, producing GLP-1, GLP-2, and PYY. Indoles are also bacterial metabolites of tryptophan degradation involved in the control of GLP-1 release and appetite control. The secretion of such hormones control appetite, gut barrier, and glucose homeostasis (e.g., insulin sensitivity) via direct interactions with organs but also through nervous routes. Similar to what is observed in the brain, different neurotransmitters or molecules (produced by intestinal microbes), such as nitric oxide (NO) as well as γ-aminobutyric acid (GABA), act through the enteric nervous system (ENS). Secondary messengers, including NO, serotonin, acetylcholine (Ach) or vasoactive intestinal polypeptide (VIP) release, are involved in the gut to peripheral organ and brain communication, leading to the control of different behaviors (e.g., food intake, anxiety, stress). Pathogen-associated molecular patterns (PAMPs) are recognized by pathogen recognition receptors such as Toll-Like receptors (TLR's) that are for most of them signaling through the central adaptor molecule myeloid differentiation primary response gene 88 (MyD88). The intestinal abundance of PAMPs and the activation of different TLR's at the intestinal epithelial surface or at the level of the ENS regulate numerous metabolic functions such as for instance leptin sensitivity, gut hormones signaling to the brain, hence controlling whole-body energy homeostasis.
These observations were the first linking gut microbiota activity, and hence a phenomenon occurring in the lower part of the gut, with signals integrated into the brain to control food intake. Most of these findings have been confirmed with different non-digestible carbohydrates (i.e., resistant starches and arabinoxylans) and will not be discussed in the present review [28], [29], [30], [31], [32], [33], [34].
Interestingly, prebiotic (inulin-type fructans) fermentation increased the abundance of SCFAs (i.e., propionate and butyrate) [35], [36]. GPR41 and 43 are expressed on enteroendocrine L-cells producing GLP-1 and PYY [37]. As discussed earlier in this review, SCFAs are ligands for these receptors. Therefore, it is easy to consider that SCFA activation of both GPR's promotes the secretion of GLP-1 and peptide YY, as shown by several reports [37], [38], [39].
To further investigate whether the beneficial effects observed between changes in gut microbes and metabolism were explained by a mechanism involving GLP-1 production, Cani et al. have used two different approaches. They first used genetic and pharmacological manipulations and found that mice lacking the GLP-1 receptor (GLP-1R) were not sensitive to the impact of prebiotics [40]. In other words, in the absence of GLP-1R, mice remained obese, resistant to insulin, and did not reduce their food intake. The same observation was made when using a pharmacological blockade of the GLP-1R, 4 weeks chronic infusion of the GLP-1R antagonist exendin 9-39. This treatment completely abolished the beneficial effects of prebiotics on food intake, hepatic insulin sensitivity, and glucose tolerance [40].
Therefore, these studies clearly show that the metabolic effects observed following the microbiota-increased endogenous production of GLP-1 require functional GLP-1 receptors.
Besides, the increased expression of the precursor of GLP-1 and PYY, we also found that the gut microbiota controls the differentiation of stem cells into enteroendocrine cells and thereby increases the number of L-cells secreting GLP-1, GLP-2 and PYY in both the jejunum and in the colon [41], [42], [43]. However, the exact mechanism explaining how the gut bacteria shape the fate of stem cells into enteroendocrine cells remains unknown (Figure 1).
Because of the beneficial systemic effects of gut peptides on energy and glucose homeostasis, deciphering the mechanisms involved in the regulation of the secretion of these peptides and L-cells may provide beneficial effects to obese and type 2 diabetic patients.
2.4. Human evidence linking gut bacteria and appetite
Numerous studies have shown that alterations to gut microbiota composition and activity may change food intake as well as gut peptide production in humans. However, the impact of GLP-1 is less conclusive. Briefly, in 1996, Ropert et al. first reported that the ingestion of lactulose (a compound fermented by microbes) or the intracolonic administration of SCFAs in healthy volunteers was linked to the endogenous production of gut peptides [44]. In 2003, Piche and colleagues reported a significant increase in plasma GLP-1 following a meal one week after a daily ingestion of oligofructose (20 g per day) [45]. Moreover, Archer et al. and Whelan et al. observed that gut microbiota modulation using non-digestible carbohydrates reduced food intake and daily energy intake. Nevertheless, none of these studies investigated gut peptides [46], [47]. In 2006, it was shown that microbial fermentation of oligofructose (16 g/day for 2 weeks) significantly increased satiety, reduced hunger, and reduced prospective food desire to ingest food [48]. These results were confirmed and enriched by the demonstration that the increased satiety and reduced hunger were associated with changes in plasma GLP-1 and PYY levels [49]. This has also been established in obese patients who also exhibited a decrease in circulating ghrelin [50]. Since these studies were performed, several other studies have reported that the ingestion and modulation of microbiota activity using different non-digestible carbohydrates affect appetite sensations [51], [52], [53], [54], [55], [56], [57], [58].
Altogether, these studies strongly suggest the existence of a gut-brain axis involving enteroendocrine relay. However, recent evidence in rodents has shown that SCFAs such as acetate also directly reach the brain to control food intake.
Gut microbes also are able to transform specific amino acids into specific products that can change the secretion of GLP-1. Recent data have shown that the by-product of tryptophan, namely indole, triggers GLP-1 secretion by a mechanism dependent on the membrane potential of an action and more specifically by the modulation of intracellular Ca2+ induced after the inhibition of voltage-gated K+ channels (31). It is worth noting that this study also found that a longer period of stimulation of the cells by indole might inhibit mitochondrial metabolism and thereby lowers intracellular ATP concentration. This effect induces ATP-sensitive K+ (K-ATP) channels to open and a hyperpolarizing of the plasma membrane, eventually slowing down GLP-1 release (31).
2.5. Gut microbes and leptin production
Leptin is an adipokine that is proportionally secreted in adipose tissue mass and informs the brain of the whole-body nutritional status of the subject [59], [60]. Leptin is involved in the regulation of energy homeostasis but also has been connected to the regulation of glucose homeostasis and numerous gastrointestinal functions, such as GLP-1 secretion [61].
Leptin-resistance is a well-known hallmark of obesity. In 2011, Everard et al. were the first to show that gut microbiota control leptin action. More precisely, they chronically fed mice with a HFD in order to induce leptin resistance. The resistance to leptin action in obese mice was assessed by the lack of effect of exogenous leptin administration on food intake, body weight gain, and lipogenesis marker (i.e., acetyl-CoA carboxylase) expression after leptin administration. Although chronically treated with a HFD, the mice that ate a HFD supplemented with prebiotics exhibited a physiological response to leptin. Prebiotic treated mice exhibited lower food intake and body weight and reduced lipogenesis in the adipose tissue as compared to mice receiving the HFD only. This observation suggests that changing the gut microbiota composition using prebiotics improved leptin sensitivity in diet-induced obesity and type 2 diabetic mice [42]. By using germ-free mice, Schéle et al. confirmed that gut microbiota contributed to regulating leptin sensitivity [62]. They found that upon colonizing the gut of the axenic mice with gut microbiota (i.e., conventionalization), the conventionalized mice exhibited a lower expression of proglucagon. The same group also found that the presence of microbiota induces a lower expression of brain-derived neurotrophic factor (BDNF), which is also considered to be involved in the regulation of energy homeostasis by hypothalamic and brainstem circuits [63]
Although the mechanisms are not fully elucidated, several hypotheses have been proposed. First, data have shown that changing the gut microbiota by using prebiotics reduces low-grade inflammation [40], [42], [64], [65], but also that reducing inflammation improves leptin sensitivity [66]. Therefore, reducing both peripheral and central inflammation may help to dampen leptin resistance during obesity. Because leptin and GLP-1 are intertwined, we may not exclude that under prebiotic feeding, the improved leptin sensitivity also could contribute to the higher secretion of GLP-1 and, thereby improved glucose and energy homeostasis. As discussed earlier in this review, SCFAs regulate the production of several enteroendocrine peptides, but SCFAs also may regulate the production of leptin. Prebiotics have been shown to strongly increase the production of SCFAs in both the cecum and in the blood. Studies have shown that the secretion [36], [67] of leptin by adipocytes is tightly regulated by SCFAs and GPR41/43 activation [68], [69]. Therefore, we propose that targeted modulations of the gut microbiota by use of prebiotics or specific bacteria that produce SCFAs could be viewed as a novel therapeutic target to reset leptin sensitivity during obesity.
Finally, this example further supported that gut microbiota-derived products act as key messengers between bacteria and different peripheral organs, including adipose tissue and the brain. Nevertheless, the role of specific bacterial genera or species in these processes remains to be elucidated.
In a very informative study, Frost and colleagues found that direct acetate and acetate derived from the colon through microbial fermentation of prebiotics (using C13 labeled carbohydrates) induces an anorectic signal involving glutamate–glutamine transcellular cycle and GABA production in the hypothalamic arcuate nucleus (ARC) [70] (Figure 1). Thus, two questions arise: Are the bacteria able to communicate with the brain? If so, excepting the SCFAs, what are the other mechanisms involved?
2.6. Gut microbes and communication with the nervous system
Bi-directional gut-brain interactions are well-described mechanisms involved in the maintenance of homeostasis, and this dialog implies involvement of gut microbiota (for review [71]). Efferent signals from the brain may modulate peripheral gut functions, such as the release of gut factors (hormones, neurotransmitters, immune factors) and/or gastrointestinal contractions, which, in turn, modify the composition of gut microflora. On the other hand, gut microbes may interact with the enteric nervous system (ENS), afferent nerves (vagal sensory neurons, spinal sensory neurons and intrinsic primary afferent neurons (IPANs)), and the central nervous system (CNS) to control the production and/or release of neurotransmitters by direct or indirect actions on neurons. In fact, Toll-Like Receptors (TLR) 3, 4 and 7 are expressed in the myenteric and submucosal plexi of murine intestine and human ileum and also in the dorsal root ganglia [72]. This last result suggests that bacterial LPS may directly target enteric neurons to control “local” functions (i.e., gastrointestinal motility and/or secretion) and to inform the brain of gut variations (e.g., nutrient sensing, presence of pathogens). Reinforcing the existence of a direct communication between microbiota and ENS is the fact that TLR2, which is a recognized peptidoglycan from gram-positive bacteria, is also expressed in ENS neurons [73]. TLR2 knockout mice had alterations in ENS architecture and neurochemical profile, suffered from intestinal dysmotility, and had abnormal mucosal secretions [73].
Another possibility is that gut microbes are sensed by the ENS is the presence of IPAN. IPANs are ideally placed to respond to luminal bacteria as suggested by Forsythe and Kunze [74], because their neurites are exposed to the intestinal lumen to detect gut microbiota. This close inter-relation between IPANs and gut microbiota is reinforced by the work of McVey Neufeld et al. [75], which demonstrates that absence of microflora is associated with a decrease in gut IPAN excitability in the mouse.
Another cellular partner of microbiota in controlling ENS activity in the gut is the glial cell, because this cell type expresses TLRs receptors [72], [73]. Different types of enteric glial cells are present in the intestinal wall. One of them, the mucosal enteric glial cell, is implicated in neuroprotection, maintenance of the intestinal epithelial barrier, and the regulation of immune responses in the mucosa. By using immunohistochemistry studies, Kabouridis et al. have recently demonstrated that homeostasis of glial cells is under the influence of gut microbiota [76], [77]. Here, the authors discovered that microbiota are required for both the initial establishment (i.e., migration) and for the postnatal development of enteric glial cells in the intestine.
Gut microbes are also able to control the function of glial cells in the central nervous system, particularly the maturation and function of microglia. To demonstrate this, Erny et al. [78] used germ-free or antibiotic-treated mice. Germ-free mice present alterations in microglia function including changes in their genetic profile and morphology, disturbance of the cellular network, and diminution of microglial response to viral infection. Moreover, antibiotic treatment in normal mice induces immature and malformed microglia, which could be reversed by microbiota or SCFAs that result from bacterial fermentation.
Currently, alterations of gut microbiota are associated with the development of numerous pathologies that directly or indirectly target the nervous system.
In a recent study, Breton et al. discovered that specific bacterial proteins produced by a specific Enterobacteriaceae, such as E. coli, may physiologically explain and eventually directly link bacteria present in the microbiota to the control of appetite [79], [80]. They found that the intestinal infusion of proteins from E. coli increased the secretion of GLP-1 or PYY. In addition, they demonstrate that acute intraperitoneal administration of E. coli proteins decreased food intake and activated c-Fos in the arcuate nucleus (ARC). Along the same line, the same group has shown that the bacterial protein ClpB, a bacterial protein mimetic of α-MSH, stimulated the firing rate of ARC POMC neurons [80]. Altogether, these data suggest that changes of gut microbiota composition and possibly bacteria expressing the same kind proteins may influence energy homeostasis by targeting the brain, via direct (i.e., POMC neurons and α-MSH) or indirect routes (i.e., GLP-1, PYY).
2.7. Gut microbes as a source of “local” neurotransmitter
It is now well established that the gut microbes interact with the brain to modulate behavior and global physiology. Despite the knowledge that this communication involves various systems (autonomic nervous system, enteric nervous system, immune system and neuroendocrine system), the molecular actors and mechanisms implicated here have only begun to be explored. One expected molecular partner between the microbiota and the brain could be the neurotransmitters of bacterial origin.
The role of gut microbes as a potential source of neurotransmitters begins in the 1980s and 1990s with the association of two major discoveries. First, enteric bacteria are able to synthesize and release nitric oxide (NO) from reduction of nitrite [81], [82]. Second, NO is considered a major neurotransmitter in the brain [83]. Another possibility for bacteria to produce NO is bacterial nitric oxide synthase [84]. The role of NO from microbial origin is not clearly established, but some data support the hypothesis that bacterial NO is implicated in the development of antibiotic resistance [85]. Whether bacterial NO is implicated in the control of other gut physiological or pathological functions remains to be determined.
In 2014, Williams et al. [86] demonstrated that Clostridium sporogenes are able to decarboxylate tryptophan to tryptamine. The authors showed that this bacterium expresses tryptophan decarboxylases to generate tryptamine, which is known to induce the release of serotonin from enterochromaffin cells to stimulate gastro-intestinal motility by acting on ENS neurons [87]. The work of Williams et al. [86] raises the hypothesis that gut microbiota can transform tryptophan from the diet to various metabolites, including tryptamine, to modify whole body homeostasis. The study could explain, to cite the authors, “that the reduction of plasma tryptophan would decrease the production of serotonin in the brain and could represent one mechanism by which microbiota influence behaviour”. This research presents the possibility of new therapeutic strategies to target brain pathologies, an idea that is reinforced by the fact that tryptophan decarboxylases are present in at least 10% of the human samples tested from the NIH Human Microbiome Project [86].
Another neurotransmitter produced by microbiota is γ-aminobutyric acid (GABA). GABA is produced by Lactobacillus and Bifidobacterium with large interspecies variations [88]. The physiological impact of bacterial GABA could be multiplied in the gut, because intestinal GABA is implicated in the control of intestinal motility, gastric emptying, gastric acid secretion, and visceral pain [89].
The number of studies that have demonstrated the capacity of gut microbes to synthesize neurotransmitter continues to grow. Additionally, some bacteria can produce monoamines such as noradrenaline, dopamine, and serotonin [90]. This last actor is one of the most studied neurotransmitters in the disturbance observed in pathologies that affect the nervous system.
2.8. Gut microbes and brain pathologies
Based on the data described above and in the literature, modifications of the cross talk between gut microbes and the brain could generate pathologies that modify social comportment (i.e., aggressiveness, depression, autism), general behavior (i.e., stress, food intake), visceral pain or neuropathologies (e.g., Alzheimer's or Parkinson's disease). In addition to peripheral disturbances generated by dysbiosis, mouse embryos from germ-free mothers have a significant increase in the permeability of the blood–brain barrier [91]. This blood–brain barrier permeability is still conserved in adult germ-free mice but could be restored in response to a colonization of germ-free mice by a conventional murine microbiota. Similar results were observed with mono-colonization of SCFA-producing microbes. This last result suggests that gut microbiota participates in central homeostasis to maintain blood–brain barrier permeability.
The list of diseases linked to the “gut microbiota-to-brain axis” is long, and the molecular players involved can vary. For example, a close correlation exists between autism spectrum disorders (ASD) and gut microbiota. In a very complete review, Li and Zhou [92] explain that gut microbiota composition is largely disturbed in ASD children who have a significant increase in fecal Sutterella spp compared to children without ASD. As SCFAs could cross the blood–brain barrier to modulate neuronal activity, this bacterial factor may participate in the establishment of ASD. Supporting this, oral gavage or intracerebroventricular injection of the SCFA propionic acid in rodents induced behavioral deficits that were consistent to that observed in human subjects with ASD [95] [92], [96]. In fact, the levels of SCFAs in the feces could be correlated negatively [93] or positively [94] with ASD.
The hypothalamic–pituitary–adrenal (HPA) axis is also under the influence of gut microbes. The absence of gut microbes in mice is associated with a significant increase in plasma ACTH and corticosterone levels in response to restrain stress [97]. This phenotype is reversed by reconstitution with Bifidobacterium infantis. In the same way, the over-activated HPA axis of germ-free mice is partly corrected by reconstitution with specific pathogen-free feces at an early stage but not by any reconstitution exerted at a later stage [97]. Of course, the effect of probiotics and/or prebiotics on stress/anxiety/depression could imply afferent messages. Nevertheless, Bercik et al. [98] have demonstrated that gut microbes may influence brain chemistry and behavior independently of the autonomic nervous system. The authors suggest that gut dysbiosis could contribute to the establishment of psychiatric disorders. Again, modulation of gut microbiota is associated with alterations of peripheral and central serotonin transmission. Germ-free mice have a significant decrease in tryptophan hydroxylase, an enzyme that permits serotonin synthesis, which could explain the lower level of plasma serotonin compared to control mice [99]. The absence of gut microbiota also affects serotonin levels in the brain [100]. As serotonin is implicated in the control of numerous physiological functions, alterations of serotonin transmission provoked by intestinal dysbiosis could influence CNS and ENS development, intestinal immune system, visceral sensitivity, and behavior (stress, depression, cognition) [100].
Recently, researchers have focused their attention on the role of gut microbes on neurodegenerative pathologies such as Alzheimer's disease and Parkinson's disease. Although a direct link between microbiota and these diseases has not been clearly established. Immunosenescence is largely associated with the development of age-related health problems, such as dementia or Alzheimer's disease [101]. During aging, the population of gut microbiota changes with an increase in Bacteroidetes versus Firmicutes proportion and a reduction of Bifidobacteria numbers. Perez Martinez et al. [101] suggested that direct manipulation of gut microbes could improve the response of the immune system to counterbalance the deleterious effect of immunosenescence. Concerning Parkinson's disease, a close correlation is observed between gut microbes (and more particular Prevotellaceae) and the motor phenotype of patients [102], [103]. Here, the authors suggest a link between microbiota and α-synuclein. In fact, the ENS and parasympathetic nerves are amongst the structures earliest and most frequently affected by α-synuclein pathology. Thus, ENS neurons and afferent nerves could again serve as the molecular pathway between gut microbes and the development of brain pathologies.
To conclude, modulation of gut microbes in various brain pathologies, including neurodevelopmental (autism), neurodegenerative (Alzheimer, Parkinson) and metabolic (type 2 diabetes) disorders, is considered to be a promising therapeutic target [104]. Understanding the molecular links between gut microbes and the development of brain pathologies constitutes the next scientific step to unravel the complex dialog observed in the gut to brain axis.
3. Gut microbes and interorgan communication: focus on LPS
Obesity and associated metabolic disorders are characterized by low-grade inflammation. In 2004, it was discovered that a high-fat diet (HFD) changes the gut microbiota (for review [105]). However, we are living with a tremendous amount of bacteria in the gut (up to 100 trillion) and few, if any, are really crossing the gut barrier. In 2007, Cani et al. demonstrated that lipopolysaccharide (LPS), a constituent of gram-negative bacteria, was more present in the blood of HFD fed mice and genetically obese and diabetic mice [1], [65], [106]. This phenomenon was called “metabolic endotoxemia”. By using different models (i.e., toll-like receptors knockout mice (CD14/TLR-4)), they demonstrated that LPS is a triggering factor involved in the onset of insulin resistance, inflammation, and possibly alteration of food intake and fat mass development [1], [106].
Thus, the gut is likely the largest barrier between the outside world and the inner part of the body. We and others have shown that bacteria contribute to maintaining a perfect gut lining. For example, besides GLP-1, L-cells produce glucagon-like peptide 2 (GLP-2) that reinforces gut barrier function. Data have shown that changing the microbiota composition using prebiotics reduces gut permeability, plasma LPS and bacterial translocation in obese animals [65], and possibly in obese humans [2], [107]. This observation further supports the assumption that constituents of bacteria may regulate metabolic functions such as food intake and glucose homeostasis, as described previously in this review. Interestingly, TLRs that recognize bacterial products are also expressed on colonic epithelia and co-localize with enteroendocrine cells [108]. More importantly, these receptors (e.g., TLR-4) have been shown to be expressed on vagal afferent neurons suggesting a possible direct sensing of bacterial products by visceral afferent nerves and supporting an additional mechanism of the gut to brain axis [109]. Interestingly, de La Serre and colleagues have elegantly demonstrated that chronic LPS exposure, mimicking metabolic endotoxemia, increased food intake in rats. They found that metabolic endotoxemia markedly affected vagal afferent neuron function, with reduced vagal afferent leptin signaling [109].
This important finding offers an additional mechanism explaining how gut bacteria and metabolites or products may contribute to the development of hyperphagia, obesity and metabolic disorders (Figure 1).
4. Pathogen recognition receptors and energy metabolism: brief focus on MYD88
The symbiosis that exists between hosts and microbes occurs because of a finely regulated system that can induce the destruction of a pathogen or conversely the tolerance of commensals [110], [111]. Different molecules coming from pathogens called “pathogen-associated molecular patterns” (PAMPs), such as LPS, glycoproteins, peptidoglycans, lipoteichoic acid, double-stranded RNA, and bacterial DNA, may be recognized by pathogen recognition receptors (PRRs). Among them, as described earlier in this review, we discuss the TLRs. Myeloid differentiation primary response gene 88 (MyD88) is a central adaptor molecule for the majority of these TLRs (with the exception of TLR3 and certain TLR4 signals) and for interleukin-1 receptor (IL-1R) and interleukin-18 receptor (IL-18R). Therefore, we may consider MyD88 to be a protein that plays a role at the interface of the interaction between the host and different microorganisms. Strikingly, MyD88 is also present in virtually all organs and cell types, including in organs distant from the gastrointestinal tract, thereby suggesting that during evolution, physiology and immunity have co-evolved to shape the interaction between microorganisms and host cells. Numerous studies have shown that whole-body deletion of specific TLRs or MyD88 alters the gut microbiota and energy homeostasis, thereby highlighting a key role for the immune system in this context [112], [113], [114], [115], [116]. For example, mice lacking MyD88 have an altered microbial composition in the gut [112], [114]; however, in these models, it is difficult to delineate whether the gut microbiota contributed to the phenotype observed. In other words, the change in the host immune response may occur from birth (i.e., when the gut microbiota starts to colonize the gut) or via adaptive processes such as antimicrobial peptide production. Data have shown that the type 1 diabetes prone non-obese mice (NOD) mice are protected from the onset of diabetes if MyD88 is deleted. The same protection has also been shown to be transferred to germ-free recipient mice [114]. In a different context, Hosoi et al. have shown that deleting MyD88 significantly increases the development of type 2 diabetes in mice exposed to HFD [115]. Another study has shown that MyD88 knockout mice are more sensitive to HFD-induced obesity and hepatic steatosis [117]. Altogether, these data suggest that there may be a cell or organ specific role of Myd88 signaling, and eventually a different phenotype according to the PAMPs and/or the organ reached.
According to this assumption, Everard et al. recently examined the hypothesis that intestinal epithelial cells act as sensors because of their direct interactions with nutrients and gut microbes [118]. For this purpose, the researchers generated a mouse model lacking MyD88 only in intestinal epithelial cells that was tamoxifen inducible. This powerful model allowed for the generation of mice with a normal immune system and gut microbiota. Then the researchers deleted MyD88 in adult rodents or after the induction of obesity. During normal diet feeding, deletion of MyD88 in the intestinal epithelial cells did not modify energy metabolism or glucose metabolism and inflammation. However, in the intestinal epithelium, more regulatory T (Treg) cells were found [118]. Strikingly, during HFD feeding mice lacking MyD88 in the intestinal epithelium became partially resistant to HFD-induced obesity, adiposity, and insulin resistance. The effect on fat mass and body weight was likely due to higher energy expenditure measured using metabolic chambers, whereas energy intake and excretion (i.e., energy content in the feces and feces volume) were not affected [118]. Interestingly, in this model, the researchers found that the gut microbiota was changed in the mice exhibiting the intestinal epithelial cells MYD88 deletion and that transferring the microbiota also partially transferred the protection to the recipient wild-type germ-free mice. Thus, this led the researchers to concluded that intestinal epithelial MyD88 is a sensor that changes host metabolism according to the diet [118].
This finding strongly suggests that fat feeding requires, at least in part, a signal coming from the host intestinal cells to induce obesity and metabolic disorders. The authors have demonstrated that that this effect was restricted to the intestinal epithelial cells, because mice lacking MYD88 in myeloid cells (i.e., monocytes, matures macrophages and granulocytes) were sensitive to HFD-induced obesity and metabolic disorders just like wild-type mice [118].
In accordance with their hypothesis that the role of MYD88 may be different according to the targeted organ, the scientists recently discovered that the absence of MYD88 in hepatocytes strongly affected glucose and lipid metabolism without affecting energy homeostasis. Mice lacking MYD88 in hepatocytes are prone to develop hepatic insulin resistance, inflammation and diabetes, whereas they exhibited the same adiposity and body weight gain that wild-type mice fed with a HFD. This research was able to decouple adiposity and obesity from glucose and lipid metabolism. The mechanisms are related to a strong modulation of the transcriptional activity of several genes involved in bile acid metabolism and different bioactive lipids. Moreover, hepatocyte deletion of MYD88 changed not only gut microbes but also plasma and liver metabolome, thereby inducing a phenotype that resembled those observed in diet-induced obese and diabetic mice. Importantly, similar to the data found in hepatocyte MYD88 knock out mice, Duparc et al. found that in human obese subjects who developed nonalcoholic steatohepatitis (NASH), different factors involved in the production of bioactive lipids synthesis (i.e., different cytochromes P450) decreased. Together, these data also highlight that the crosstalk between microorganisms and host cells such as hepatocytes can tightly control glucose, lipid metabolism and inflammation through the action of different bioactive lipid compounds [119].
As described in this review, gut microbes may control leptin sensitivity and therefore energy homeostasis via a possible gut to brain axis. However, in addition to the role of specific PAMPs and the activation of specific TLRs, several previous studies have also proposed that specific fatty acids (i.e., dietary fatty acids but also components of LPS) may bind to TLR-4 and activate a MYD88-dependent signaling and inflammation, but this remains controversial [120], [121], [122], [123]. In a very elegant study, Kleinridders and colleagues have demonstrated that MyD88 from the central nervous system (CNS) may elicit signals that directly controls leptin sensitivity and thereby food intake. They found that mice lacking MYD88 in the CNS are protected against HFD-induced insulin resistance [116].
5. Conclusion
Evidence suggests that signals coming from the gastrointestinal tract and/or involving different metabolites of nutrients, neurotransmitters, and PRRs may act as metabolic sensors involved in the interaction between nutrients, gut bacteria, and the host by acting on different organs and eventually the brain to control energy homeostasis.
Acknowledgments
PDC is a research associate at FRS-FNRS (Fonds de la Recherche Scientifique), Belgium. PDC was the recipient of grants from FNRS (convention J.0084.15, convention 3.4579.11), PDR (Projet de Recherche, convention: T.0138.14) and ARC (Action de Recherche Concertée – Communauté française de Belgique convention: 12/17-047). This work was supported by the Fonds de la Recherche Scientifique – FNRS for the FRFS-WELBIO under grant: WELBIO-CR-2012S-02R. This work was supported in part by the Funds Baillet Latour (Grant for Medical Research 2015). PDC was a recipient of an ERC Starting Grant in 2013 (European Research Council, Starting grant 336452-ENIGMO). CK was the recipient of grants from the Societe Francaise de Nutrition (SFN), the Fondation Recherche Médicale (FRM) (Grant ING20150532586), and the Societe Francophone du Diabète (Allocations Exceptionnelles 2016).
Contributor Information
Patrice D. Cani, Email: patrice.cani@uclouvain.be.
Claude Knauf, Email: claude.knauf@inserm.fr.
Conflict of interest
None declared.
References
- 1.Cani P.D., Amar J., Iglesias M.A., Poggi M., Knauf C., Bastelica D. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 2.Dewulf E.M., Cani P.D., Claus S.P., Fuentes S., Puylaert P.G., Neyrinck A.M. Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut. 2013;62:1112–1121. doi: 10.1136/gutjnl-2012-303304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeevi D., Korem T., Zmora N., Israeli D., Rothschild D., Weinberger A. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163:1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Gibson G.R., Roberfroid M.B. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. Journal of Nutrition. 1995;125:1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 5.Bindels L.B., Delzenne N.M., Cani P.D., Walter J. Towards a more comprehensive concept for prebiotics. Nature Reviews. Gastroenterology & Hepatology. 2015;12:303–310. doi: 10.1038/nrgastro.2015.47. [DOI] [PubMed] [Google Scholar]
- 6.Delzenne N.M., Neyrinck A.M., Backhed F., Cani P.D. Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nature reviews. Endocrinology. 2011;7:639–646. doi: 10.1038/nrendo.2011.126. [DOI] [PubMed] [Google Scholar]
- 7.Anhe F.F., Roy D., Pilon G., Dudonne S., Matamoros S., Varin T.V. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut. 2015;64:872–883. doi: 10.1136/gutjnl-2014-307142. [DOI] [PubMed] [Google Scholar]
- 8.Wu G.D., Chen J., Hoffmann C., Bittinger K., Chen Y.Y., Keilbaugh S.A. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.David L.A., Maurice C.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe BE Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Lahham S.H., Peppelenbosch M.P., Roelofsen H., Vonk R.J., Venema K. Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochimica et Biophysica Acta. 2010;1801:1175–1183. doi: 10.1016/j.bbalip.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Cummings J.H., Pomare E.W., Branch W.J., Naylor C.P., Macfarlane G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delzenne N.M., Williams C.M. Prebiotics and lipid metabolism. Current Opinion in Lipidology. 2002;13:61–67. doi: 10.1097/00041433-200202000-00009. [DOI] [PubMed] [Google Scholar]
- 13.De Vadder F., Kovatcheva-Datchary P., Goncalves D., Vinera J., Zitoun C., Duchampt A. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156:84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 14.Brown A.J., Goldsworthy S.M., Barnes A.A., Eilert M.M., Tcheang L., Daniels D. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. The Journal of Biological Chemistry. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- 15.Le Poul E., Loison C., Struyf S., Springael J.Y., Lannoy V., Decobecq M.E. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. The Journal of Biological Chemistry. 2003;278:25481–25489. doi: 10.1074/jbc.M301403200. [DOI] [PubMed] [Google Scholar]
- 16.Kimura I., Inoue D., Hirano K., Tsujimoto G. The SCFA receptor GPR43 and energy metabolism. Frontiers in Endocrinology. 2014;5:85. doi: 10.3389/fendo.2014.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong Y.H., Nishimura Y., Hishikawa D., Tsuzuki H., Miyahara H., Gotoh C. Acetate and propionate short chain fatty acids stimulate adipogenesis via GPCR43. Endocrinology. 2005;146:5092–5099. doi: 10.1210/en.2005-0545. [DOI] [PubMed] [Google Scholar]
- 18.Dewulf E.M., Cani P.D., Neyrinck A.M., Possemiers S., Holle A.V., Muccioli G.G. Inulin-type fructans with prebiotic properties counteract GPR43 overexpression and PPARgamma-related adipogenesis in the white adipose tissue of high-fat diet-fed mice. The Journal of Nutritional Biochemistry. 2011;22:712–722. doi: 10.1016/j.jnutbio.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Dewulf E.M., Ge Q., Bindels L.B., Sohet F.M., Cani P.D., Brichard S.M. Evaluation of the relationship between GPR43 and adiposity in human. Nutrition & Metabolism. 2013;10:11. doi: 10.1186/1743-7075-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge H., Li X., Weiszmann J., Wang P., Baribault H., Chen J.L. Activation of G protein-coupled receptor 43 in adipocytes leads to inhibition of lipolysis and suppression of plasma free fatty acids. Endocrinology. 2008;149:4519–4526. doi: 10.1210/en.2008-0059. [DOI] [PubMed] [Google Scholar]
- 21.Furusawa Y., Obata Y., Fukuda S., Endo T.A., Nakato G., Takahashi D. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 22.Maslowski K.M., Vieira A.T., Ng A., Kranich J., Sierro F., Yu D. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macia L., Thorburn A.N., Binge L.C., Marino E., Rogers K.E., Maslowski K.M. Microbial influences on epithelial integrity and immune function as a basis for inflammatory diseases. Immunological Reviews. 2012;245:164–176. doi: 10.1111/j.1600-065X.2011.01080.x. [DOI] [PubMed] [Google Scholar]
- 24.Cani P.D., Dewever C., Delzenne N.M. Inulin-type fructans modulate gastrointestinal peptides involved in appetite regulation (glucagon-like peptide-1 and ghrelin) in rats. The British Journal of Nutrition. 2004;92:521–526. doi: 10.1079/bjn20041225. [DOI] [PubMed] [Google Scholar]
- 25.Delzenne N.M., Neyrinck A.M., Cani P.D. Gut microbiota and metabolic disorders: how prebiotic can work? The British Journal of Nutrition. 2013;109(Suppl 2):S81–S85. doi: 10.1017/S0007114512004047. [DOI] [PubMed] [Google Scholar]
- 26.Birt D.F., Boylston T., Hendrich S., Jane J.L., Hollis J., Li L. Resistant starch: promise for improving human health. Advances in Nutrition. 2013;4:587–601. doi: 10.3945/an.113.004325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delzenne N.M., Cani P.D., Daubioul C., Neyrinck A.M. Impact of inulin and oligofructose on gastrointestinal peptides. The British Journal of Nutrition. 2005;93(Suppl 1):S157–S161. doi: 10.1079/bjn20041342. [DOI] [PubMed] [Google Scholar]
- 28.Aziz A.A., Kenney L.S., Goulet B., Abdel-Aal E. Dietary starch type affects body weight and glycemic control in freely fed but not energy-restricted obese rats. Journal of Nutrition. 2009;139:1881–1889. doi: 10.3945/jn.109.110650. [DOI] [PubMed] [Google Scholar]
- 29.Charrier J.A., Martin R.J., McCutcheon K.L., Raggio A.M., Goldsmith F., Goita M. High fat diet partially attenuates fermentation responses in rats fed resistant starch from high-amylose maize. Obesity (Silver Spring) 2013 doi: 10.1002/oby.20362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keenan M.J., Zhou J., McCutcheon K.L., Raggio A.M., Bateman H.G., Todd E. Effects of resistant starch, a non-digestible fermentable fiber, on reducing body fat. Obesity (Silver Spring) 2006;14:1523–1534. doi: 10.1038/oby.2006.176. [DOI] [PubMed] [Google Scholar]
- 31.Shen L., Keenan M.J., Martin R.J., Tulley R.T., Raggio A.M., McCutcheon K.L. Dietary resistant starch increases hypothalamic POMC expression in rats. Obesity (Silver Spring) 2009;17:40–45. doi: 10.1038/oby.2008.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J., Hegsted M., McCutcheon K.L., Keenan M.J., Xi X., Raggio A.M. Peptide YY and proglucagon mRNA expression patterns and regulation in the gut. Obesity (Silver Spring) 2006;14:683–689. doi: 10.1038/oby.2006.77. [DOI] [PubMed] [Google Scholar]
- 33.Zhou J., Martin R.J., Tulley R.T., Raggio A.M., McCutcheon K.L., Shen L. Dietary resistant starch upregulates total GLP-1 and PYY in a sustained day-long manner through fermentation in rodents. American Journal of Physiology – Endocrinology and Metabolism. 2008;295:E1160–E1166. doi: 10.1152/ajpendo.90637.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neyrinck A.M., Van Hee V.F., Piront N., De Backer F., Toussaint O., Cani P.D. Wheat-derived arabinoxylan oligosaccharides with prebiotic effect increase satietogenic gut peptides and reduce metabolic endotoxemia in diet-induced obese mice. Nutrition & Diabetes. 2012;2:e28. doi: 10.1038/nutd.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberfroid M., Gibson G.R., Hoyles L., McCartney A.L., Rastall R., Rowland I. Prebiotic effects: metabolic and health benefits. The British Journal of Nutrition. 2010;104:S1–S63. doi: 10.1017/S0007114510003363. [DOI] [PubMed] [Google Scholar]
- 36.Everard A., Lazarevic V., Gaia N., Johansson M., Stahlman M., Backhed F. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. The ISME Journal. 2014;8:2116–2130. doi: 10.1038/ismej.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nohr M.K., Pedersen M.H., Gille A., Egerod K.L., Engelstoft M.S., Husted A.S. GPR41/FFAR3 and GPR43/FFAR2 as cosensors for short-chain fatty acids in enteroendocrine cells vs FFAR3 in enteric neurons and FFAR2 in enteric leukocytes. Endocrinology. 2013;154:3552–3564. doi: 10.1210/en.2013-1142. [DOI] [PubMed] [Google Scholar]
- 38.Samuel B.S., Shaito A., Motoike T., Rey F.E., Backhed F., Manchester J.K. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tolhurst G., Heffron H., Lam Y.S., Parker H.E., Habib A.M., Diakogiannaki E. Short-chain fatty acids stimulate glucagon-like Peptide-1 secretion via the g-protein-coupled receptor FFAR2. Diabetes. 2012;61:364–371. doi: 10.2337/db11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cani P.D., Knauf C., Iglesias M.A., Drucker D.J., Delzenne N.M., Burcelin R. Improvement of glucose tolerance and hepatic insulin sensitivity by oligofructose requires a functional glucagon-like peptide 1 receptor. Diabetes. 2006;55:1484–1490. doi: 10.2337/db05-1360. [DOI] [PubMed] [Google Scholar]
- 41.Cani P.D., Hoste S., Guiot Y., Delzenne N.M. Dietary non-digestible carbohydrates promote L-cell differentiation in the proximal colon of rats. The British Journal of Nutrition. 2007;98:32–37. doi: 10.1017/S0007114507691648. [DOI] [PubMed] [Google Scholar]
- 42.Everard A., Lazarevic V., Derrien M., Girard M., Muccioli G.G., Neyrinck A.M. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes. 2011;60:2775–2786. doi: 10.2337/db11-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wichmann A., Allahyar A., Greiner T.U., Plovier H., Lunden G.O., Larsson T. Microbial modulation of energy availability in the colon regulates intestinal transit. Cell Host & Microbe. 2013;14:582–590. doi: 10.1016/j.chom.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 44.Ropert A., Cherbut C., Roze C., Le Quellec A., Holst J.J., Fu-Cheng X. Colonic fermentation and proximal gastric tone in humans. Gastroenterology. 1996;111:289–296. doi: 10.1053/gast.1996.v111.pm8690193. [DOI] [PubMed] [Google Scholar]
- 45.Piche T., des Varannes S.B., Sacher-Huvelin S., Holst J.J., Cuber J.C., Galmiche J.P. Colonic fermentation influences lower esophageal sphincter function in gastroesophageal reflux disease. Gastroenterology. 2003;124:894–902. doi: 10.1053/gast.2003.50159. [DOI] [PubMed] [Google Scholar]
- 46.Archer B.J., Johnson S.K., Devereux H.M., Baxter A.L. Effect of fat replacement by inulin or lupin-kernel fibre on sausage patty acceptability, post-meal perceptions of satiety and food intake in men. The British Journal of Nutrition. 2004;91:591–599. doi: 10.1079/BJN20031088. [DOI] [PubMed] [Google Scholar]
- 47.Whelan K., Efthymiou L., Judd P.A., Preedy V.R., Taylor M.A. Appetite during consumption of enteral formula as a sole source of nutrition: the effect of supplementing pea-fibre and fructo-oligosaccharides. The British Journal of Nutrition. 2006;96:350–356. doi: 10.1079/bjn20061791. [DOI] [PubMed] [Google Scholar]
- 48.Cani P.D., Joly E., Horsmans Y., Delzenne N.M. Oligofructose promotes satiety in healthy human: a pilot study. The European Journal of Clinical Nutrition. 2006;60:567–572. doi: 10.1038/sj.ejcn.1602350. [DOI] [PubMed] [Google Scholar]
- 49.Cani P.D., Lecourt E., Dewulf E.M., Sohet F.M., Pachikian B.D., Naslain D. Gut microbiota fermentation of prebiotics increases satietogenic and incretin gut peptide production with consequences for appetite sensation and glucose response after a meal. The American Journal of Clinical Nutrition. 2009;90:1236–1243. doi: 10.3945/ajcn.2009.28095. [DOI] [PubMed] [Google Scholar]
- 50.Parnell J.A., Reimer R.A. Weight loss during oligofructose supplementation is associated with decreased ghrelin and increased peptide YY in overweight and obese adults. The American Journal of Clinical Nutrition. 2009;89:1751–1759. doi: 10.3945/ajcn.2009.27465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peters H.P., Boers H.M., Haddeman E., Melnikov S.M., Qvyjt F. No effect of added beta-glucan or of fructooligosaccharide on appetite or energy intake. The American Journal of Clinical Nutrition. 2009;89:58–63. doi: 10.3945/ajcn.2008.26701. [DOI] [PubMed] [Google Scholar]
- 52.Tarini J., Wolever T.M. The fermentable fibre inulin increases postprandial serum short-chain fatty acids and reduces free-fatty acids and ghrelin in healthy subjects. Applied Physiology, Nutrition, and Metabolism. 2010;35:9–16. doi: 10.1139/H09-119. [DOI] [PubMed] [Google Scholar]
- 53.Klosterbuer A.S., Thomas W., Slavin J.L. Resistant starch and pullulan reduce postprandial glucose, insulin, and GLP-1, but have no effect on satiety in healthy humans. Journal of Agricultural and Food Chemistry. 2012;60:11928–11934. doi: 10.1021/jf303083r. [DOI] [PubMed] [Google Scholar]
- 54.Frost G., Brynes A., Leeds A. Effect of large bowel fermentation on insulin, glucose, free fatty acids, and glucagon-like peptide 1 (7-36) amide in patients with coronary heart disease. Nutrition. 1999;15:183–188. doi: 10.1016/s0899-9007(98)00177-4. [DOI] [PubMed] [Google Scholar]
- 55.Bird A.R., Conlon M.A., Christophersen C.T., Topping D.L. Resistant starch, large bowel fermentation and a broader perspective of prebiotics and probiotics. Beneficial Microbes. 2010;1:423–431. doi: 10.3920/BM2010.0041. [DOI] [PubMed] [Google Scholar]
- 56.Robertson M.D. Dietary-resistant starch and glucose metabolism. Current Opinion in Clinical Nutrition and Metabolic Care. 2012;15:362–367. doi: 10.1097/MCO.0b013e3283536931. [DOI] [PubMed] [Google Scholar]
- 57.Nilsson A., Johansson E., Ekstrom L., Bjorck I. Effects of a brown beans evening meal on metabolic risk markers and appetite regulating hormones at a subsequent standardized breakfast: a randomized cross-over study. PLoS One. 2013;8:e59985. doi: 10.1371/journal.pone.0059985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Daud N.M., Ismail N.A., Thomas E.L., Fitzpatrick J.A., Bell J.D., Swann J.R. The impact of oligofructose on stimulation of gut hormones, appetite regulation and adiposity. Obesity (Silver Spring) 2014;22:1430–1438. doi: 10.1002/oby.20754. [DOI] [PubMed] [Google Scholar]
- 59.Spiegelman B.M., Flier J.S. Obesity and the regulation of energy balance. Cell. 2001;104:531–543. doi: 10.1016/s0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- 60.Considine R.V., Sinha M.K., Heiman M.L., Kriauciunas A., Stephens T.W., Nyce M.R. Serum immunoreactive-leptin concentrations in normal-weight and obese humans. The New England Journal of Medicine. 1996;334:292–295. doi: 10.1056/NEJM199602013340503. [DOI] [PubMed] [Google Scholar]
- 61.Anini Y., Brubaker P.L. Role of leptin in the regulation of glucagon-like peptide-1 secretion. Diabetes. 2003;52:252–259. doi: 10.2337/diabetes.52.2.252. [DOI] [PubMed] [Google Scholar]
- 62.Schele E., Grahnemo L., Anesten F., Hallen A., Backhed F., Jansson J.O. The gut microbiota reduces leptin sensitivity and the expression of the obesity-suppressing neuropeptides proglucagon (Gcg) and brain-derived neurotrophic factor (Bdnf) in the central nervous system. Endocrinology. 2013;154:3643–3651. doi: 10.1210/en.2012-2151. [DOI] [PubMed] [Google Scholar]
- 63.Unger T.J., Calderon G.A., Bradley L.C., Sena-Esteves M., Rios M. Selective deletion of Bdnf in the ventromedial and dorsomedial hypothalamus of adult mice results in hyperphagic behavior and obesity. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2007;27:14265–14274. doi: 10.1523/JNEUROSCI.3308-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cani P.D., Neyrinck A.M., Fava F., Knauf C., Burcelin R.G., Tuohy K.M. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50:2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- 65.Cani P.D., Possemiers S., Van de W.T., Guiot Y., Everard A., Rottier O. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58:1091–1103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Milanski M., Arruda A.P., Coope A., Ignacio-Souza L.M., Nunez C.E., Roman E.A. Inhibition of hypothalamic inflammation reverses diet-induced insulin resistance in the liver. Diabetes. 2012;61:1455–1462. doi: 10.2337/db11-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bindels L.B., Porporato P., Dewulf E.M., Verrax J., Neyrinck A.M., Martin J.C. Gut microbiota-derived propionate reduces cancer cell proliferation in the liver. British Journal of Cancer. 2012;107:1337–1344. doi: 10.1038/bjc.2012.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zaibi M.S., Stocker C.J., O'Dowd J., Davies A., Bellahcene M., Cawthorne M.A. Roles of GPR41 and GPR43 in leptin secretory responses of murine adipocytes to short chain fatty acids. FEBS Letter. 2010;584:2381–2386. doi: 10.1016/j.febslet.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 69.Xiong Y., Miyamoto N., Shibata K., Valasek M.A., Motoike T., Kedzierski R.M. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1045–1050. doi: 10.1073/pnas.2637002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frost G., Sleeth M.L., Sahuri-Arisoylu M., Lizarbe B., Cerdan S., Brody L. The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nature Communications. 2014;5:3611. doi: 10.1038/ncomms4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rhee S.H., Pothoulakis C., Mayer E.A. Principles and clinical implications of the brain-gut-enteric microbiota axis. Nature Reviews. Gastroenterology & Hepatology. 2009;6:306–314. doi: 10.1038/nrgastro.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barajon I., Serrao G., Arnaboldi F., Opizzi E., Ripamonti G., Balsari A. Toll-like receptors 3, 4, and 7 are expressed in the enteric nervous system and dorsal root ganglia. Journal of Histochemistry & Cytochemistry. 2009;57:1013–1023. doi: 10.1369/jhc.2009.953539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brun P., Giron M.C., Qesari M., Porzionato A., Caputi V., Zoppellaro C. Toll-like receptor 2 regulates intestinal inflammation by controlling integrity of the enteric nervous system. Gastroenterology. 2013;145:1323–1333. doi: 10.1053/j.gastro.2013.08.047. [DOI] [PubMed] [Google Scholar]
- 74.Forsythe P., Kunze W.A. Voices from within: gut microbes and the CNS. Cellular and Molecular Life Sciences: CMLS. 2013;70:55–69. doi: 10.1007/s00018-012-1028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McVey Neufeld K.A., Mao Y.K., Bienenstock J., Foster J.A., Kunze W.A. The microbiome is essential for normal gut intrinsic primary afferent neuron excitability in the mouse. Neurogastroenterology and Motility: The Official Journal of the European Gastrointestinal Motility Society. 2013;25:183–e188. doi: 10.1111/nmo.12049. [DOI] [PubMed] [Google Scholar]
- 76.Kabouridis P.S., Lasrado R., McCallum S., Chng S.H., Snippert H.J., Clevers H. Microbiota controls the homeostasis of glial cells in the gut lamina propria. Neuron. 2015;85:289–295. doi: 10.1016/j.neuron.2014.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rao M., Gershon M.D. Bugs, guts, and glia: how microbiota influence enteric gliogenesis and migration. Neuron. 2015;85:229–230. doi: 10.1016/j.neuron.2014.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Erny D., Hrabe de Angelis A.L., Jaitin D., Wieghofer P., Staszewski O., David E. Host microbiota constantly control maturation and function of microglia in the CNS. Nature Neuroscience. 2015;18:965–977. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Breton J., Tennoune N., Lucas N., Francois M., Legrand R., Jacquemot J. Gut commensal E. coli proteins activate host satiety pathways following nutrient-induced bacterial growth. Cell Metabolism. 2016;23:324–334. doi: 10.1016/j.cmet.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 80.Tennoune N., Chan P., Breton J., Legrand R., Chabane Y.N., Akkermann K. Bacterial ClpB heat-shock protein, an antigen-mimetic of the anorexigenic peptide alpha-MSH, at the origin of eating disorders. Translational Psychiatry. 2014;4:e458. doi: 10.1038/tp.2014.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ji X.B., Hollocher T.C. Reduction of nitrite to nitric oxide by enteric bacteria. Biochemical and Biophysical Research Communications. 1988;157:106–108. doi: 10.1016/s0006-291x(88)80018-4. [DOI] [PubMed] [Google Scholar]
- 82.Salzman A.L. Nitric oxide in the gut. New Horizons. 1995;3:352–364. [PubMed] [Google Scholar]
- 83.Garthwaite J., Charles S.L., Chess-Williams R. Endothelium-derived relaxing factor release on activation of NMDA receptors suggests role as intercellular messenger in the brain. Nature. 1988;336:385–388. doi: 10.1038/336385a0. [DOI] [PubMed] [Google Scholar]
- 84.Gusarov I., Starodubtseva M., Wang Z.Q., McQuade L., Lippard S.J., Stuehr D.J. Bacterial nitric-oxide synthases operate without a dedicated redox partner. The Journal of Biological Chemistry. 2008;283:13140–13147. doi: 10.1074/jbc.M710178200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gusarov I., Shatalin K., Starodubtseva M., Nudler E. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science. 2009;325:1380–1384. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Williams B.B., Van Benschoten A.H., Cimermancic P., Donia M.S., Zimmermann M., Taketani M. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host & Microbe. 2014;16:495–503. doi: 10.1016/j.chom.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Takaki M., Mawe G.M., Barasch J.M., Gershon M.D., Gershon M.D. Physiological responses of guinea-pig myenteric neurons secondary to the release of endogenous serotonin by tryptamine. Neuroscience. 1985;16:223–240. doi: 10.1016/0306-4522(85)90059-4. [DOI] [PubMed] [Google Scholar]
- 88.Barrett E., Ross R.P., O'Toole P.W., Fitzgerald G.F., Stanton C. Gamma-Aminobutyric acid production by culturable bacteria from the human intestine. Journal of Applied Microbiology. 2012;113:411–417. doi: 10.1111/j.1365-2672.2012.05344.x. [DOI] [PubMed] [Google Scholar]
- 89.Hyland N.P., Cryan J.F. A gut feeling about GABA: focus on GABA(B) receptors. Frontiers in Pharmacology. 2010;1:124. doi: 10.3389/fphar.2010.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Clarke G., Stilling R.M., Kennedy P.J., Stanton C., Cryan J.F., Dinan T.G. Minireview: gut microbiota: the neglected endocrine organ. Molecular Endocrinology. 2014;28:1221–1238. doi: 10.1210/me.2014-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Braniste V., Al-Asmakh M., Kowal C., Anuar F., Abbaspour A., Toth M. The gut microbiota influences blood-brain barrier permeability in mice. Science Translational Medicine. 2014;6:263ra158. doi: 10.1126/scitranslmed.3009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li Q., Zhou J.M. The microbiota-gut-brain axis and its potential therapeutic role in autism spectrum disorder. Neuroscience. 2016;324:131–139. doi: 10.1016/j.neuroscience.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 93.Adams J.B., Johansen L.J., Powell L.D., Quig D., Rubin R.A. Gastrointestinal flora and gastrointestinal status in children with autism–comparisons to typical children and correlation with autism severity. BMC Gastroenterology. 2011;11:22. doi: 10.1186/1471-230X-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang L., Christophersen C.T., Sorich M.J., Gerber J.P., Angley M.T., Conlon M.A. Elevated fecal short chain fatty acid and ammonia concentrations in children with autism spectrum disorder. Digestive Diseases and Sciences. 2012;57:2096–2102. doi: 10.1007/s10620-012-2167-7. [DOI] [PubMed] [Google Scholar]
- 95.El-Ansary A.K., Ben Bacha A., Kotb M. Etiology of autistic features: the persisting neurotoxic effects of propionic acid. Journal of Neuroinflammation. 2012;9:74. doi: 10.1186/1742-2094-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Macfabe D.F. Short-chain fatty acid fermentation products of the gut microbiome: implications in autism spectrum disorders. Microbial Ecology in Health and Disease. 2012;23 doi: 10.3402/mehd.v23i0.19260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sudo N., Chida Y., Aiba Y., Sonoda J., Oyama N., Yu X.N. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. The Journal of Physiology. 2004;558:263–275. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bercik P., Denou E., Collins J., Jackson W., Lu J., Jury J. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 2011;141 doi: 10.1053/j.gastro.2011.04.052. 599–609, 609 e591–593. [DOI] [PubMed] [Google Scholar]
- 99.Yano J.M., Yu K., Donaldson G.P., Shastri G.G., Ann P., Ma L. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.O'Mahony S.M., Clarke G., Borre Y.E., Dinan T.G., Cryan J.F. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behavioural Brain Research. 2015;277:32–48. doi: 10.1016/j.bbr.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 101.Perez Martinez G., Bauerl C., Collado M.C. Understanding gut microbiota in elderly's health will enable intervention through probiotics. Beneficial Microbes. 2014;5:235–246. doi: 10.3920/BM2013.0079. [DOI] [PubMed] [Google Scholar]
- 102.Scheperjans F., Aho V., Pereira P.A., Koskinen K., Paulin L., Pekkonen E. Gut microbiota are related to Parkinson's disease and clinical phenotype. Movement Disorders. 2015;30:350–358. doi: 10.1002/mds.26069. [DOI] [PubMed] [Google Scholar]
- 103.Wood H. Parkinson disease. Gut reactions–can changes in the intestinal microbiome provide new insights into Parkinson disease? Nature Review Neurology. 2015;11:66. doi: 10.1038/nrneurol.2014.256. [DOI] [PubMed] [Google Scholar]
- 104.Ghaisas S., Maher J., Kanthasamy A. Gut microbiome in health and disease: linking the microbiome-gut-brain axis and environmental factors in the pathogenesis of systemic and neurodegenerative diseases. Pharmacology & Therapeutics. 2016;158:52–62. doi: 10.1016/j.pharmthera.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cani P.D., Everard A. Talking microbes: when gut bacteria interact with diet and host organs. Molecular Nutrition & Food Research. 2016;60:58–66. doi: 10.1002/mnfr.201500406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cani P.D., Bibiloni R., Knauf C., Waget A., Neyrinck A.M., Delzenne N.M. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 107.Malaguarnera M., Vacante M., Antic T., Giordano M., Chisari G., Acquaviva R. Bifidobacterium longum with fructo-oligosaccharides in patients with non alcoholic steatohepatitis. Digestive Diseases and Sciences. 2011 doi: 10.1007/s10620-011-1887-4. [DOI] [PubMed] [Google Scholar]
- 108.Bogunovic M., Dave S.H., Tilstra J.S., Chang D.T., Harpaz N., Xiong H. Enteroendocrine cells express functional Toll-like receptors. American Journal of Physiology. Gastrointestinal and Liver Physiology. 2007;292:G1770–G1783. doi: 10.1152/ajpgi.00249.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.de La Serre C.B., de Lartigue G., Raybould H.E. Chronic exposure to low dose bacterial lipopolysaccharide inhibits leptin signaling in vagal afferent neurons. Physiology & Behavior. 2015;139:188–194. doi: 10.1016/j.physbeh.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Adolph T.E., Tomczak M.F., Niederreiter L., Ko H.J., Bock J., Martinez-Naves E. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–276. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rosenstiel P. Stories of love and hate: innate immunity and host-microbe crosstalk in the intestine. Current Opinion in Gastroenterology. 2013;29:125–132. doi: 10.1097/MOG.0b013e32835da2c7. [DOI] [PubMed] [Google Scholar]
- 112.Vijay-Kumar M., Aitken J.D., Carvalho F.A., Cullender T.C., Mwangi S., Srinivasan S. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim K.A., Gu W., Lee I.A., Joh E.H., Kim D.H. High fat diet-induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PLoS One. 2012;7:e47713. doi: 10.1371/journal.pone.0047713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wen L., Ley R.E., Volchkov P.Y., Stranges P.B., Avanesyan L., Stonebraker A.C. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hosoi T., Yokoyama S., Matsuo S., Akira S., Ozawa K. Myeloid differentiation factor 88 (MyD88)-deficiency increases risk of diabetes in mice. PLoS One. 2010;5:e12537. doi: 10.1371/journal.pone.0012537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kleinridders A., Schenten D., Konner A.C., Belgardt B.F., Mauer J., Okamura T. MyD88 signaling in the CNS is required for development of fatty acid-induced leptin resistance and diet-induced obesity. Cell Metabolism. 2009;10:249–259. doi: 10.1016/j.cmet.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yokoyama S., Hosoi T., Ozawa K. Stearoyl-CoA Desaturase 1 (SCD1) is a key factor mediating diabetes in MyD88-deficient mice. Gene. 2012;497:340–343. doi: 10.1016/j.gene.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 118.Everard A., Geurts L., Caesar R., Van Hul M., Matamoros S., Duparc T. Intestinal epithelial MyD88 is a sensor switching host metabolism towards obesity according to nutritional status. Nature Communications. 2014;5:5648. doi: 10.1038/ncomms6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Duparc T., Plovier H., Marrachelli V.G., Van Hul M., Essaghir A., Stahlman M. Hepatocyte MyD88 affects bile acids, gut microbiota and metabolome contributing to regulate glucose and lipid metabolism. Gut. 2016 doi: 10.1136/gutjnl-2015-310904. . pii: gutjnl-2015-310904.[in press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Galbo T., Perry R.J., Jurczak M.J., Camporez J.P., Alves T.C., Kahn M. Saturated and unsaturated fat induce hepatic insulin resistance independently of TLR-4 signaling and ceramide synthesis in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:12780–12785. doi: 10.1073/pnas.1311176110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shi H., Kokoeva M.V., Inouye K., Tzameli I., Yin H., Flier J.S. TLR4 links innate immunity and fatty acid-induced insulin resistance. Journal of Clinical Investigation. 2006;116:3015–3025. doi: 10.1172/JCI28898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Erridge C., Samani N.J. Saturated fatty acids do not directly stimulate Toll-like receptor signaling. Arteriosclerosis, Thrombosis, and Vascular Biology. 2009;29:1944–1949. doi: 10.1161/ATVBAHA.109.194050. [DOI] [PubMed] [Google Scholar]
- 123.Pal D., Dasgupta S., Kundu R., Maitra S., Das G., Mukhopadhyay S. Fetuin-A acts as an endogenous ligand of TLR4 to promote lipid-induced insulin resistance. Nature Medicine. 2012;18:1279–1285. doi: 10.1038/nm.2851. [DOI] [PubMed] [Google Scholar]