Abstract
Epigenetic modifications are essential regulators of biological processes. Decreased DNA methylation of OAS2 (2′-5′-Oligoadenylate Synthetase 2), encoding an antiviral protein, has been seen in psoriasis. To provide further insight into the epigenetic regulation of OAS2, we performed pyrosequencing to detect OAS2 DNA methylation status at 11 promoter and first exon located CpG sites in psoriasis (n = 12) and two common subtypes of squamous cell carcinoma (SCC) of the head and neck: tongue (n = 12) and tonsillar (n = 11). Compared to corresponding controls, a general hypomethylation was seen in psoriasis. In tongue and tonsillar SCC, hypomethylation was found at only two CpG sites, the same two sites that were least demethylated in psoriasis. Despite differences in the specific residues targeted for methylation/demethylation, OAS2 expression was upregulated in all conditions and correlations between methylation and expression were seen in psoriasis and tongue SCC. Distinctive methylation status at four successively located CpG sites within a genomic area of 63 bp reveals a delicately integrated epigenetic program and indicates that detailed analysis of individual CpGs provides additional information into the mechanisms of epigenetic regulation in specific disease states. Methylation analyses as clinical biomarkers need to be tailored according to disease-specific sites.
DNA methylation of the fifth position of cytosine is an important regulatory mechanism of genome function. Methylation is subject to dynamic changes and essential for the regulation of gene expression, cellular differentiation and is commonly altered in human disease1,2,3. Genome-wide array- and sequencing-based techniques are increasingly applied to investigate DNA methylation, providing a broader view of global methylation patterns, giving a better understanding of the functional elements controlling gene expression and identifying numerous disease-associated differentially methylated CpG sites4,5,6. The exact mode of epigenetic modifications for particular genes and their role in disease, however, is yet to be completely understood.
Overexpression of OAS2 (2′-5′-oligoadenylate synthetase 2) has been reported in patients with inflammatory, autoimmune and malignant diseases, whereas its role in these conditions remains poorly understood7,8. The OAS2 protein is a well-known innate immune activated antiviral enzyme catalyzing synthesis of 2′-5′-oligoadenylate for RNase L activation and inhibition of viral propagation9. More recent studies show that it also participates in other biological processes. In pancreatic β cells, OAS2 could be induced and activated by in vitro transcribed cellular RNAs, leading to cell proliferation inhibition and apoptosis10. In acute monocytic leukemia cells THP-1, NOD2 (Nucleotide-binding and oligomerization domain-2, an immune receptor to intracellular bacterial lipopolysaccharides) was found to interact with OAS2 enhancing RNase-L function, indicating a connection between OAS2 and other innate immune signaling pathways11. Extracellular OAS2 has also been reported as a negative regulator of T-cell function in oral cancer, promoting tumour progression by modulating anti-tumour immune response8. Interestingly, in psoriasis, a chronic inflammatory skin disease, overexpression of OAS2 was found to be associated with differential DNA methylation12,13,14.
Squamous cell carcinoma of the head and neck (SCCHN), the sixth most common malignant tumour worldwide, constitutes an anatomically heterogeneous group of neoplasms arising within the head and neck area15. Several risk factors have been well characterized, such as tobacco smoking and alcohol consumption, and oncogenic viruses have also been suggested as a cause for the development of a subset of SCCHN16,17. Infection with high-risk human papillomavirus (HPV, double-stranded DNA viruses infecting epithelial cells), is most commonly found in tonsillar SCC (66.4%) and least in tongue (25.7%) and pharyngeal (15.3%) SCC18. Recent investigations on the incidence of HPV infection in SCCHN in northern Sweden identified HPV positivity in 91% of tonsillar SCC19, whereas no evidence of HPV infection was observed in SCC of the mobile tongue20. Although overexpression of OAS2 has been reported in SCCHN8, the status of OAS2 in these distinct subtypes of SCCHN is not well known.
Pyrosequencing is a sequencing-by-synthesis method that quantitatively measures DNA methylation based on the detection of pyrophosphate released upon nucleotide incorporation21. As a cost-effective and efficient method to quantify DNA methylation it is widely used to validate high-throughput methylation array data, providing a clearer picture of methylation status for defined DNA regions4. Therefore in this study, we performed pyrosequencing to detect DNA methylation of OAS2 in psoriasis, SCC of the mobile tongue and SCC of the tonsil. Unexpectedly, we found that distinct epigenetic features were notable at 4 successively located CpG sites within a genomic area of 63 bp in these different pathological conditions. Exploring mechanisms of epigenetic changes in OAS2 will be useful for illustrating the role of OAS2 in human diseases. In a broader context, our data provide novel insight into the sophisticated epigenetic machinery and reinforce that methylation analyses as clinical biomarkers will need to be tailored according to disease-specific sites.
Materials and Methods
Patients and samples
This is a retrospective study of 58 patients plus control samples. Twelve patients were diagnosed with moderate-severe psoriasis, 12 with SCC of the mobile tongue and 34 with SCC of the tonsil. For all patients with SCC of the mobile tongue and 11 patients with SCC of the tonsil collected for this study, biopsies were taken from tumour and adjacent tumour-free tissue prior to treatment (clinical data are shown in Supplementary Table S1). DNA samples from 23 tonsillar samples have been used in another study19. The status of HPV infection and p16 expression (a proposed surrogate marker for high risk HPV infection22) in these tonsillar tumor samples had been determined (17/23 were HPV-positive and 21/23 showed p16 expression)19. The quick score system was applied to evaluate levels of p16 expression. The quick score produces values ranging from 0 to 18 by multiplying the percentage of p16 positive cells (scored as 0–6) with intensity (scored as 1–3)23. Quick scores ranging from 0 to 12 were seen in these samples. For the psoriasis group, 12 patients diagnosed with moderate-severe psoriasis and matched healthy individuals were the same as included in a previous study24. Clinical data on patients with ready-to-use DNA samples are shown in Supplementary Table S2. This study was approved by the Regional Ethics Review Board, Umeå, Sweden (Dnr 08-108 M and Dnr 08-003 M) and performed in accordance with the Declaration of Helsinki. Written informed consent was obtained from all subjects.
DNA/RNA isolation
Biopsies were fresh-frozen in liquid nitrogen and stored at −80 °C until DNA extraction. AllPrep DNA/RNA/miRNA Universal Kit (Qiagen, Hilden, Germany) was used to simultaneously isolate DNA and RNA from tumour (T) and tumour-free (TF) samples from patients with tongue and tonsillar SCC. Briefly, the fresh frozen biopsies (less than 20 mg) were homogenized in 600 μl Buffer RLT PLT Plus using the Precellys Tissue homogenizer (Bertin Technologies, Artigus Pres Boreaux, France). Tissue lysates were processed according to the Qiagen protocol and eluted twice in a total of 60 ul RNase-free water for RNA isolation and twice in a total of 150 Buffer EB for DNA isolation. The final yields range from 4.86 to 77.04 μg for RNA and 3.45 to 71.69 μg for DNA. Quantity and purity of DNA/RNA was measured using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). DNA quality was confirmed by gel electrophoresis and RNA quality by Agilent RNA 6000 Nano kit (Agilent 2100 Bioanalyzer, Agilent Technologies, Santa Clara, CA, USA). Another 23 DNA samples of tonsillar SCC had been extracted from paraffin embedded diagnostic biopsies (percentage of tumour cells range from 25 to 95%) using the QIAamp DNA FFPE Tissue Kit or QIAamp Mini Kit (Qiagen, Valencia, CA, USA)19. Following the manufacturer’s instructions, eight sections with a thickness of 10 μm were cut from each paraffin-embedded tumour block for DNA preparation. Purified DNA was eluted in 50 μl of Buffer ATE (supplied with the QIAamp DNA FFPE Tissue Kit) or Buffer AE (supplied with the QIAamp Mini Kit). The final DNA yields range from 4 to 50.60 μg. DNA from psoriatic epidermis and healthy controls was isolated using the PureLink® Genomic DNA Kits (Life Technologies, Carlsbad, CA, USA)24.
Bisulfite treatment and pyrosequencing
Based on our previously published methylation 450 K array data on psoriasis (accession number: GSE63315)24, 13 probes for OAS2 were found. Among these CpG sites, three were differentially methylated in psoriasis compared to matched controls (|delta-beta| > 0.1 and adjusted P-value < 0.01) (Supplementary Table S3). Of these, two were located 391 and 213 bp upstream of the transcription start site (TSS) respectively and one in the first exon. In order to validate the array data and evaluate its representativity of the surrounding area, four commercially available PyroMark CpG assays from Qiagen were used to determine methylation levels in two areas in which these three CpG sites were located (Supplementary Fig. S1). Information of the four PyroMark CpG assays detecting a total of 11 CpG sites is summarized in Supplementary Table S4. A schematic diagram of CpG sites spanning −500 bp upstream of TSS to the first exon (+317 bp) is shown in Fig. 1a.
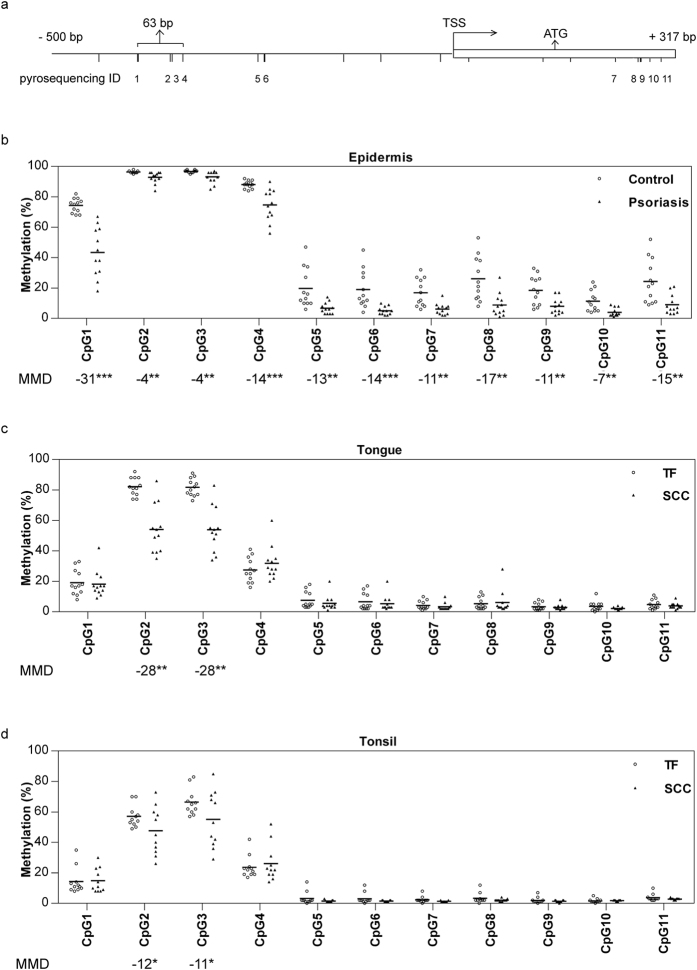
Figure 1. Hypomethylation of OAS2 in psoriasis and SCCHN.
(a) Schematic diagram of CpG sites spans −500 bp upstream of transcription start site (TSS) to the first exon (+317 bp). CpG sites are indicated by vertical ticks. The CpG sites covered by PyroMark CpG assays are numbered in order from 1 to 11 (pyrosequencing ID). Array identified hypomethylated CpG sites in psoriasis are highlighted by thick vertical ticks. (b–d) Methylation status of 11 CpG sites in psoriasis, tongue SCC and tonsillar SCC determined by pyrosequencing analysis. Individual methylation data are shown as dot plots and the horizontal lines indicate means. Mean methylation difference (MMD) between disease and controls is shown under each CpG sites. Mann-Whitney U test was performed for comparing psoriasis with controls and Wilcoxon signed ranks test for comparing tumour with adjacent tumour-free samples (***P < 0.001, **P < 0.01, *P < 0.5).
Five hundred ng of genomic DNA was used for bisulfite conversion using EpiTect Fast DNA Bisulfite kit (Qiagen). Bisulfite converted DNA samples were cleaned up hereafter and finally eluted in 20 μl Buffer EB. According to the PyoMark PCR protocol, 1 μl bisulfite converted DNA were amplified in 25 μl PCR reactions and 10 μl PCR product was used for pyrosequencing. Pyrosequencing was performed using the PyroMark Q24 advanced machine and following the instructions for PyroMark Q24 Advanced CpG Reagents from Qiagen. Degree of methylation was expressed as methylated cytosines divided by total cytosines (the sum of methylated and unmethylated cytosines), ranging from 0 to 100%, corresponding to array beta value 0 to 1.
Real time RT-PCR
Real-time RT-PCR was performed to detect OAS2 mRNA levels in tumour and adjacent tumour-free controls. Complementary DNA (cDNA) was synthesized from 500 ng of total RNA with oligo (dT) primer, according to the instructions for the RevertAid H minus first strand cDNA synthesis kit (Fermentas, Thermo Scientific, Wilmington, DE, USA). Real time RT-PCR was performed using an IQ5 multicolor real-time PCR detection system with IQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA). Primers for OAS2 were ordered from Bio-rad (Bio-Rad, assay ID qHsaCED0037726). NormFinder software was used to analyze a set of reference gene candidates based on our own whole-genome expression data, and a total of four genes were confirmed to be good reference genes in our study, including LAD1 (Forward: CCTCCCACCCGTCACACT, Reverse: CTGCTGTAGGTTCGCTGTGT), RPS12 (Forward: TGCTGCTGGAGGTGTAATGG, Reverse: GCACACAAAGATGGGCTTGG), GAPDH and USB). Primers for GAPDH and USB were ordered from Primerdesign Ltd (Southhampton, United Kingdom) and the sequences of primers were not provided. For real time RT-PCR data normalization, the geometric mean of these reference genes were calculated. The cycling conditions were set as follows: enzyme activation at 95 °C for 3 min, 40 cycles of denaturation at 95 °C for 15 sec and annealing/extension at 60 °C for 60 sec. For OAS2 mRNA levels in psoriasis, we used our previous published gene expression array data of the same material25.
DNA Methylation data from Epigenome Roadmap
The Roadmap Epigenomics Project has produced 127 reference epigenomes spanning diverse cell and tissue types, providing the largest collection so far of human epigenomes representative of all major lineages in the human body26. A total of 37 reference methylomes by whole genome bisulphite sequencing (WGBS) were available, representing multiple brain, heart, muscle, gastrointestinal tract, adipose, skin and reproductive samples, as well as immune lineages, ES (embryonic stem) cells and iPS (induced pluripotent stem) cells, and differentiated lineages derived from ES cells26. OAS2 DNA methylation data in the promoter region (1000 bp upstream of TSS and the first exon, a sequence of 1317 bp) was extracted using UCSC Table Browser27. A total of 25 CpG dinucleotides were found in the defined promoter region using the February 2009 (CrCh37/hg19) build of the human genome.
Cell culture and stimuli
Human adult epidermal keratinocytes (HEKa, Life technologies, Carlsbad, USA) were maintained in medium 154 with human keratinocyte growth supplement (HKGS) (Life technologies) and cultured at 37 °C with 5% CO2. The day before stimulation, cells between passage 3 to 6 were plated in 6-well plates at 2 × 105 cells/well in 2 ml of complete growth medium. At the time of stimulation, cells were treated with 1 μg/ml double-stranded RNA (dsRNA) analog poly(I:C) (Polyinosinic acid: Polycytidylic acid) or 1 μg/ml double-stranded DNA (dsDNA) analog poly(dA:dT) (poly(deoxyadenylic-deoxythymidylic) acid sodium salt) (Invivogen, San Diego, CA, USA). Experiments were performed in duplicate and repeated in two independent experiments. Cells were collected 24 hours after stimulation and RNA/DNA co-isolated using ZR-Duet™ DNA/RNA MiniPrep from Zymo Research, according to the manufacturer’s manual (Irvine, CA, USA).
Statistics
To identify differentially methylated CpG sites, DNA methylation levels in diseased tissue were compared with controls, either skin from healthy volunteers for the psoriasis samples or self-paired tumour-free samples for cancer patients. Non-parametric two-tailed tests were used and the significance level was set at 5%. Wilcoxon signed-rank test for two related samples was performed to compare differences between tumour and adjacent tumour-free samples. Mann-Whitney U test for two independent samples was performed to compare differences between psoriatic epidermis and epidermis from healthy individuals. Significantly differentially methylated sites (P < 0.05) with more than 10% decrease in absolute methylation were defined as hypomethylated. Spearman correlation coefficient (rho) was calculated to evaluate the strength of correlation. All statistical tests were conducted in IBM SPSS Statistics 21.
Results
Pyrosequencing confirmed hypomethylation of OAS2 in psoriasis
Methylation status of 11 OAS2 CpG sites in psoriasis and control skin samples were successfully quantified using pyrosequencing. As both 450K array24 and pyrosequencing data were available for CpG1, 2, 6 and 9, we first evaluated the correlation between data determined by pyrosequencing and methylation array. Spearman correlation analysis indicated good correlation between the two platforms, with the strongest correlation seen for CpG1 (rho = 0.963, P = 0.000) (Supplementary Table S5). Individual methylation levels at the 11 CpG sites are presented in Fig. 1b showing a high degree of inter-individual and inter-locus variation. In general, high methylation levels were seen at CpG1 to 4 and low levels at CpG5 to 11. When comparing psoriasis to control skin, an overall demethylation was observed and a total of 8 CpG sites were identified as hypomethylated (mean methylation difference >10%, P < 0.01). The most hypomethylated site being CpG1, with a mean methylation difference of 31% between psoriatic epidermis and controls (P = 0.000).
Only two OAS2 CpG sites were hypomethylated in SCCHN
Methylation status of the same 11 CpG sites was investigated in SCCHN. In Fig. 1c (mobile tongue SCC) and 1D (tonsillar SCC) we can see that, similar to psoriatic epidermis, methylation levels at these 11 CpG sites exhibited inter-individual and inter-locus variations. The highest methylation levels in both tumour and tumour-free tissue were seen at CpG2 and 3, whereas methylation levels at CpG5 to 11 were less than 10%. When comparing tongue tumour with tumour-free tissue pairwise, significant methylation differences were seen only at CpG2 (P = 0.002) and CpG3 (P = 0.002). Similarly, significant decreases in DNA methylation were seen in tonsillar SCC compared to tumor-free tissues at CpG2 (P = 0.023) and CpG3 (P = 0.023). Notably, methylation levels at these two CpG sites were the least changed in psoriasis versus control skin. Furthermore, the degree of hypomethylation in mobile tongue SCC was higher than that in tonsillar SCC.
HPV infection and DNA methylation of OAS2
DNA methylation levels of OAS2 in another 23 tonsillar cancer samples were also studied. HPV status and p16 expression (based on quickscore ranging from 0 to 18) in these samples have been investigated previously19. In this sample group, 17 tumour samples were HPV-positive, and all of these HPV-positive tumours were p16-positive (quickscore ≥ 6). Among the six HPV-negative tumours, both negative and positive staining for p16 were seen (quickscore from 0 to 12). When comparing HPV-positive with HPV-negative tumours, no difference in OAS2 DNA methylation was found based on HPV status (Supplementary Fig. S2). Similarly, no correlation between p16 staining and DNA methylation of OAS2 was identified.
Correlation between DNA methylation and gene expression
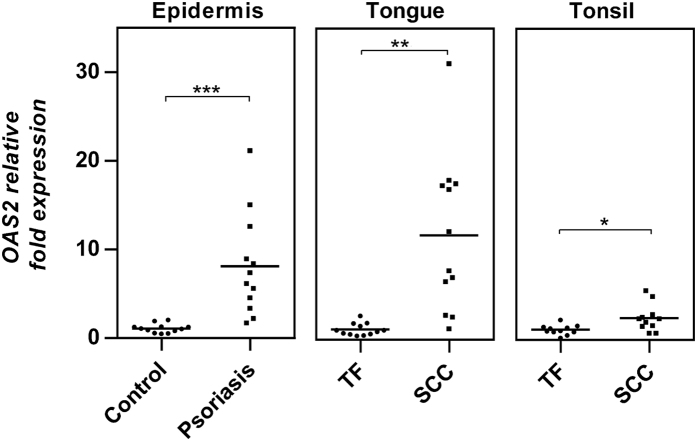
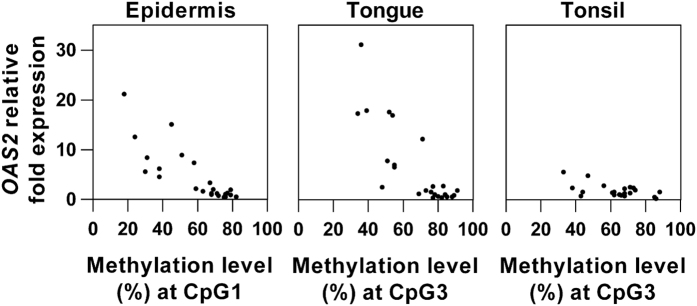
In order to evaluate whether OAS2 DNA methylation could be involved in regulating gene expression, correlation between DNA methylation and gene expression was investigated using our previous gene expression profiling data for psoriasis (accession number GSE53431). OAS2 expression in SCCHN was studied using real time RT-PCR. Expression data are from the same subjects as the methylation data. In Fig. 2, we can see that similar to overexpression of OAS2 in psoriasis (P = 0.000, mean fold change = 8.1), OAS2 was significantly overexpressed in tongue SCC compared to tumour-free tissues (P = 0.003, mean fold change = 11.8). Overexpression of OAS2 was also seen in tonsillar SCC (P = 0.013, mean fold change = 2.4) but levels were much lower than in psoriasis and tongue SCC. Methylation levels at each CpG site were then correlated to gene expression data (Supplementary Table S6). In psoriatic epidermis, OAS2 mRNA was significantly correlated with methylation levels of all 11 CpG sites, with the strongest correlation seen between CpG1 methylation and gene expression (Spearman correlation coefficient rho = −0.845, P = 0.000) (Fig. 3a). In tongue SCC, methylation levels at CpG2 and CpG3 were significantly correlated with mRNA levels (Spearman correlation coefficient rho = −0.738 (CpG2) and −0.769 (CpG3), P = 0.000) (Fig. 3b), whereas in tonsillar SCC OAS2 hypomethylation at CpG2 and 3 was not correlated with gene overexpression (Fig. 3c).
Figure 2. OAS2 was significantly upregulated in psoriasis and SCCHN.
Levels of OAS2 mRNA in psoriasis were determined previously using Illumina HumanHT-12 v4 Expression BeadChip. Significant up-regulation of OAS2 was seen in psoriasis compared to matched controls (***P < 0.001). Levels of OAS2 mRNA in SCCHN and adjacent tumour-free tissues were determined using real time RT-PCR, and found to be significantly upregulated in tongue SCC (**P < 0.01) and tonsillar SCC (*P < 0.05). LAD1, RPS12, GAPDH and USB were used as internal controls for real time RT-PCR analysis.
Figure 3. Correlation between OAS2 DNA methylation and gene expression.
Spearman correlation test was performed to study the correlation between DNA methylation and gene expression. In psoriasis, DNA methylation at each CpG site was negatively correlated with gene expression. The strongest correlation was seen between CpG1 methylation and gene expression (rho = −0.845, P = 0.000). In tongue SCC, DNA methylation at CpG2 and 3 was negatively correlated with gene expression. The higher correlation between CpG3 methylation and gene expression is shown (rho = −0.769, P = 0.000). No correlation between OAS2 DNA methylation and gene expression was found in tonsillar SCC (rho = −0.307, P = 0.164).
Methylation levels at CpG 2 and 3 demonstrate least variation across different tissues
As DNA methylation could be tissue-specific26,28, we sought to investigate whether the difference in methylation between psoriasis and SCCHN arises merely from tissue-dependent methylation modifications. OAS2 methylation levels in the proximal promoter region were obtained from 37 reference methylomes. Methylation status across different tissues/cells for each CpG site is shown in Supplementary Fig. S3 and level of variability for each site measured by calculating the coefficient of variation (CV, Supplementary Table S7). A high degree of tissue- and/or developmental-specific variations in DNA methylation was seen for the majority of CpG sites (Supplementary Fig. S3A). Focusing on methylation status of 18 adult tissues/cells (Supplementary Fig. S3B), high methylation variations were also seen, indicating tissue-specific methylation status for most CpG sites. We then inspected methylation status for the two CpG sites (CpG2 and 3) that were hypomethylated in SCC but not in psoriasis. Interestingly, methylation levels at these two CpG sites were more consistent across different tissue/cells compared to nearby CpG sites. Our pyrosequencing data also showed that methylation levels at these two CpG sites were similar between healthy epidermis and tumour-free head and neck tissues, whereas at the two most nearby CpG sites (CpG1 and CpG4), tissue-specific methylation levels were seen.
Innate immune response was not accompanied by altered methylation of OAS2
As OAS2 is involved in the innate immune response to viral infection we performed cell line experiments to study gene expression and DNA methylation in response to innate immune triggers. The methylation status for OAS2 in primary HEKa cells corresponded closely to normal epidermal cells and OAS2 mRNA was increased upon poly(dA:dT) or poly(I:C) stimulation, whereas DNA methylation levels were not affected (Supplementary Fig. S4).
Discussion
We previously profiled DNA methylation in psoriatic epidermis using the Illumina 450K BeadChip platform and found hypomethylation of OAS2 in psoriasis compared to healthy controls. Though covering key features of the human genome, only 2% of the human genome CpG sites were targeted and hybridization probes spanning 50 bases in the 450K platform limits the identification of heterogeneous methylation and informative single CpG sites4,29. Therefore, further characterization of methylation changes at single-base resolution and with broader CpG coverage is required to elucidate details of epigenetic modifications. In this study, by using the quantitative pyrosequencing method, we have clarified OAS2 methylation status in two genomic regions where the three array identified hypomethylated CpG dinucleotides are located. Our data demonstrated that methylation changes did not occur in a coordinated manner, and notably, distinct methylation levels were seen at 4 successively located CpG sites within a genomic area of 63 bp. One common DNA methylation analysis task is to identify Differentially Methylated Regions (DMRs) between two groups as it is widely accepted that DNA methylation alterations at multiple adjacent CpG sites is of biological relevance30. On the other hand, several studies have identified single CpG dinucleotides important for gene expression and disease development, and demethylation pressure at specific CpGs has been suggested31,32. Therefore, the importance of epigenetic modifications in diverse biological processes should be carefully evaluated based on methylation results with single-base resolution.
Similar to the strong inverse correlation between OAS2 DNA methylation and gene expression in psoriasis reported by Roberson et al.14, we also found out that gene expressions in psoriatic epidermis and tongue SCC were highly correlated with DNA methylation. Interestingly, we demonstrated that distinct CpG sites were targeted for methylation modification in different diseases. The relationship between genetic variation, DNA methylation and gene regulation is complex and can be of different nature depending on tissue and genomic region33. As different types of tissues are investigated in this study, it is not unexpected that methylation changes at different CpG sites are required for gene regulation in different tissues. Interestingly, when focusing on the 63 bp region where CpG1 to 4 are located, differentially methylated CpG sites in psoriasis and SCCHN were mutually exclusive. CpG1 and 4 were hypomethylated in psoriatic lesions compared with controls, whereas CpG2 and 3 were hypomethylated in SCCHN when compared with adjacent tumour-free tissue. By comparing the methylation status at different CpG sites across different normal tissues/cells using the Roadmap reference data, high degree of tissue-dependent variation in OAS2 DNA methylation was clearly seen. Noteworthy, methylation levels at CpG 2 and 3 are least variegated across normal tissue/cells, but turn out to be the only two CpG sites that are demethylated in tongue and tonsillar SCC. Therefore, methylation levels at these two CpG sites seems to be specifically modified only in some diseases.
As reverse correlations between DNA methylation and gene expression were seen in psoriatic epidermis and tongue SCC, and transcription factors could be important drivers of methylation changes5,34, it seems likely that under distinct pathological conditions, different transcriptional factors targeting different genomic elements are recruited for activating OAS2 expression. The general hypomethylation of OAS2 seen in psoriasis might functionally be related to OAS2 mRNA up-regulation, whereas in SCC of the mobile tongue, local decreased methylation at only two CpG sites (CpG2 and 3) seems sufficient to affect regulation of OAS2 expression. Though it is far from understood how a specific CpG dinucleotide is marked for modification, distinct epigenetic modes underlying common overexpression of OAS2 in benign hyper-proliferative and malignant conditions indicate that understanding of epigenetics could offer useful information for understanding the pathogenesis of different diseases.
When treating primary HEKa cells with dsRNA or dsDNA analogues, we found potent OAS2 mRNA induction whereas DNA methylation levels remained unchanged (Fig. S4). Thus, it seems that epigenetic mechanisms are not required for transcriptional activation of OAS2 in keratinocytes in short-term antiviral response. By comparing HPV-positive with HPV-negative tonsillar SCC, no difference in methylation level was found. Though a direct connection between HPV infection and OAS2 expression has not yet been established, we speculate that in psoriasis and SCCHN, the classical antivirus pathway was not responsible for overexpression of OAS2. Besides the well-known antiviral function of the OAS family, OAS proteins are implicated in other cellular events such as gene induction, regulation of apoptosis, cell growth and differentiation7,35. Very recently, it was shown that extracellular OAS2 could lead to immune dysfunction in oral SCC8, suggesting a cancer-promoting role for OAS2. As OAS2 expression levels differ between the subtypes of SCCHN, it remains to be elucidated whether this is a common function of OAS2 in SCCHN.
In summary, we used a high resolution quantitative method for analysis of OAS2 DNA methylation in psoriasis and SCCHN. Four successive CpG sites located in a 63 bp promoter area of OAS2 were differentially methylated in a site- and disease- specific manner. Our results provide further insight into how epigenetic programming integrates delicately with genomic elements. Unraveling the epigenetic mechanisms involved in regulation of OAS2 could aid our understanding of the role of OAS2 in different cellular contexts. More generally, DNA methylation analysis has promise for diagnosis, prognosis and prediction of therapeutic response in cancer and other diseases36,37. Our data indicate disease- and/or tissue-specific epigenetic regulation of individual genes, such that accurate analysis of methylation requires detailed mapping of individual CpG sites to identify the changes that characterize different disease states. In turn, identifying specific sites and how they are targeted for methylation-mediated regulation may improve our understanding of the underlying biology of normal and diseased tissues. From a clinical viewpoint, analysis of methylation at single CpG sites represents a simple assay and can provide predictive biomarkers31, but these assays may need to be tailored to different CpG sites in different disease states.
Additional Information
How to cite this article: Gu, X. et al. Epigenetic regulation of OAS2 shows disease-specific DNA methylation profiles at individual CpG sites. Sci. Rep. 6, 32579; doi: 10.1038/srep32579 (2016).
Supplementary Material
Acknowledgments
This study was supported by grants from Hudfonden, Lion’s Cancer Research Foundation, Umeå University, The Swedish Cancer Society contract number 15 0637, Umeå University and project MEYS-NPSI-LO1413 from the Ministry of Education, Youth and Sports and GACR P206/12/G151 (PC) in the Czech Republic.
Footnotes
Author Contributions X.G. designed and performed experiments, analyzed data and wrote the manuscript. L.B. performed experiments. P.J.C. wrote the manuscript. R.F. designed experiments. E.N., C.L., K.O., L.N.S. and O.G. provided medical materials. K.N. supervisor the project and wrote the manuscript. All authors commented on the manuscript.
References
- Ziller M. J. et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 500, 477–481 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Z. D. & Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220 (2013). [DOI] [PubMed] [Google Scholar]
- Jones P. A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492 (2012). [DOI] [PubMed] [Google Scholar]
- Plongthongkum N., Diep D. H. & Zhang K. Advances in the profiling of DNA modifications: cytosine methylation and beyond. Nat. Rev. Genet. 15, 647–661 (2014). [DOI] [PubMed] [Google Scholar]
- Schubeler D. Function and information content of DNA methylation. Nature 517, 321–326 (2015). [DOI] [PubMed] [Google Scholar]
- Bock C. Analysing and interpreting DNA methylation data. Nat. Rev. Genet. 13, 705–719 (2012). [DOI] [PubMed] [Google Scholar]
- Choi U. Y., Kang J. S., Hwang Y. S. & Kim Y. J. Oligoadenylate synthase-like (OASL) proteins: dual functions and associations with diseases. Exp. Mol. Med. 47, e144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar A. A. et al. Extracellular 2′5′-oligoadenylate synthetase 2 mediates T-cell receptor CD3-zeta chain down-regulation via caspase-3 activation in oral cancer. Immunology 147, 251–264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozzi A. et al. OASes and STING: adaptive evolution in concert. Genome Biol. Evol. 7, 1016–1032 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan M., Zheng D., Field L. L. & Bonnevie-Nielsen V. Induction and activation of antiviral enzyme 2′,5′-oligoadenylate synthetase by in vitro transcribed insulin mRNA and other cellular RNAs. Mol. Biol. Rep. 39, 7813–7822 (2012). [DOI] [PubMed] [Google Scholar]
- Dugan J. W. et al. Nucleotide oligomerization domain-2 interacts with 2′-5′-oligoadenylate synthetase type 2 and enhances RNase-L function in THP-1 cells. Mol. immunol. 47, 560–566 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera G. K., Di Meglio P. & Nestle F. O. Psoriasis. Annu. rev. pathol. 7, 385–422 (2012). [DOI] [PubMed] [Google Scholar]
- Keermann M. et al. Transcriptional landscape of psoriasis identifies the involvement of IL36 and IL36RN. BMC Genomics 16, 322 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson E. D. et al. A subset of methylated CpG sites differentiate psoriatic from normal skin. J. Invest. Dermatol. 132, 583–592 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamangar F., Dores G. M. & Anderson W. F. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J. Clin. Oncol. 24, 2137–2150 (2006). [DOI] [PubMed] [Google Scholar]
- Pezzuto F. et al. Update on Head and Neck Cancer: Current Knowledge on Epidemiology, Risk Factors, Molecular Features and Novel Therapies. Oncology 89, 125–136 (2015). [DOI] [PubMed] [Google Scholar]
- Gupta K. & Metgud R. Evidences suggesting involvement of viruses in oral squamous cell carcinoma. Pathol. Res. Int. 2013, 642496 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abogunrin S., Di Tanna G. L., Keeping S., Carroll S. & Iheanacho I. Prevalence of human papillomavirus in head and neck cancers in European populations: a meta-analysis. BMC Cancer 14, 968 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loizou C. et al. Incidence of tonsillar cancer in northern Sweden: impact of human papilloma virus. Oncol. Lett. (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgaramella N. et al. Expression of p16 in squamous cell carcinoma of the mobile tongue is independent of HPV infection despite presence of the HPV-receptor syndecan-1. Br. J. Cancer 113, 321–326 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella S., Shen L., Baggerly K. A., Issa J. P. & Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. BioTechniques 35, 146–150 (2003). [DOI] [PubMed] [Google Scholar]
- Hoffmann M. et al. HPV DNA, E6*I-mRNA expression and p16INK4A immunohistochemistry in head and neck cancer - how valid is p16INK4A as surrogate marker? Cancer Lett. 323, 88–96 (2012). [DOI] [PubMed] [Google Scholar]
- Detre S., Saclani Jotti G. & Dowsett M. A “quickscore” method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J. Clin. Pathol. 48, 876–878 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Nylander E., Coates P. J., Fahraeus R. & Nylander K. Correlation between Reversal of DNA Methylation and Clinical Symptoms in Psoriatic Epidermis Following Narrow-Band UVB Phototherapy. J. Invest. Dermatol. 135, 2077–2083 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Nylander E., Coates P. J. & Nylander K. Oxidation reduction is a key process for successful treatment of psoriasis by narrow-band UVB phototherapy. Acta Derm.-Venereol. 95, 140–146 (2015). [DOI] [PubMed] [Google Scholar]
- Roadmap Epigenomics C. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D. et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokk K. et al. DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome biol. 15, r54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M. et al. High density DNA methylation array with single CpG site resolution. Genomics 98, 288–295 (2011). [DOI] [PubMed] [Google Scholar]
- Peters T. J. et al. De novo identification of differentially methylated regions in the human genome. Epigenet. Chromatin 8, 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus R. et al. Quantitative DNA methylation analysis identifies a single CpG dinucleotide important for ZAP-70 expression and predictive of prognosis in chronic lymphocytic leukemia. J. Clin. Oncol. 30, 2483–2491 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusgen N. et al. Inter-locus as well as intra-locus heterogeneity in LINE-1 promoter methylation in common human cancers suggests selective demethylation pressure at specific CpGs. Clin. epigenet. 7, 17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Arcelus M. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Life 2, e00523 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal C. & Miotto B. Emerging concept in DNA methylation: role of transcription factors in shaping DNA methylation patterns. J. Cell. Physiol. 230, 743–751 (2015). [DOI] [PubMed] [Google Scholar]
- Hovanessian A. G. & Justesen J. The human 2′-5′oligoadenylate synthetase family: unique interferon-inducible enzymes catalyzing 2′-5′ instead of 3′-5′ phosphodiester bond formation. Biochimie 89, 779–788 (2007). [DOI] [PubMed] [Google Scholar]
- Heyn H. & Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat. Rev. Genet. 13, 679–692 (2012). [DOI] [PubMed] [Google Scholar]
- Mikeska T. & Craig J. M. DNA methylation biomarkers: cancer and beyond. Genes (Basel) 5, 821–864 (2014). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.