Abstract
Inactivation of Noggin, a secreted antagonist of Bone Morphogenetic Proteins (BMPs), in mice leads, among others, to severe malformations of the appendicular skeleton and defective skeletal muscle fibers. To determine the molecular basis of the phenotype, we carried out a histomorphological and molecular analysis of developing muscles Noggin−/− mice. We show that in 18.5 dpc embryos there is a marked reduction in muscle fiber size and a failure of nuclei migration towards the cell membrane. Molecularly, the absence of Noggin results in an increased BMP signaling in muscle tissue as shown by the increase in SMAD1/5/8 phosphorylation, concomitant with the induction of BMP target genes such as Id1, 2, 3 as well as Msx1. Finally, upon removal of Noggin, the number of mesenchymal Pax7+ muscle precursor cells is reduced and they are more prone to differentiate into adipocytes in vitro. Thus, our results highlight the importance of Noggin/BMP balance for myogenic commitment of early fetal progenitor cells.
Noggin is a glycosylated, secreted 64 kDa protein homodimer and one of the many extracellular antagonists of the Bone Morphogenetic Protein (BMP) signaling pathway. Similar to Chordin, Chordin-like, Follistatin, Fsrp and Cerberus, Noggin encodes a protein that directly binds to the BMP (BMP2, BMP4, BMP5, BMP7) or Growth Differentiation Factor (GDF5, GDF6) ligands with various affinities1,2. By preventing interaction of the ligands with their receptor, Noggin interferes with the downstream activation of the signaling cascade.
Loss of Noggin in mice resulted in an embryonic lethal phenotype that is characterized by different developmental defects such as exencephaly, axial outgrowth defects with a loss of the caudal vertebrae, failure of neural tube closure, excessive cartilage formation and fused joints to mention a few3,4.
During embryonic development, the bilaterally organized somites differentiate into a ventral-medial part, called the sclerotome, and a dorsal-lateral part, called the dermomyotome. The sclerotome gives rise to the cartilage and the bone of the vertebral column, while the dermomyotome develops into muscle, endothelia, cartilage, connective tissue and dermis.
At 9.5 dpc, delamination and migration of Pax3 positive (Pax3+) cells originating from the dermomyotome enables the differentiation of muscle progenitor cells in the myotome and in the limb. At 10.5–12.5 dpc, the first wave of myogenesis (embryonic myogenesis) takes place. Embryonic myoblasts fuse with each other and differentiate into large primary myofibers5. As most of the myoblasts remain in a committed and undifferentiated state, the number of myofibers produced in this first wave is limited. These primary fibers serve to form the basic muscle pattern6. Another cell type, which is Pax7+, remains undifferentiated. These cells are present from early stages onwards and give rise to fetal myoblasts7. Their proliferation is triggered by mitogens secreted by the primary fibers and they will differentiate into many smaller secondary myofibers during the secondary wave of myogenesis (14.5–16.5 dpc), or fetal myogenesis8.
Yet not all Pax7+ cells proliferate and differentiate but some remain in an undifferentiated state and become activated in the postnatal life following triggers like trauma or physical exercise. These cells are called satellite cells when they can be morphologically identified as mononucleated cells residing between the myofiber plasma membrane and the basal lamina (from 17.5 dpc onwards). They are considered to form the stem cell niche responsible for the growth and restoration of the muscle9.
The dermomyotomal and sclerotomal somitic populations are subject to the intricate crosstalk of several signaling cascades including WNT, Sonic hedgehog (SHH), and Bone Morphogenetic proteins (BMPs), ensuring a regulated differentiation of these lineages. WNT signaling from the overlying epidermis and the roof plate of the neural tube induces the expression of dermomyotome specific genes, while SHH signaling from the notochord and the floor plate of the neural tube induces sclerotomal gene expression10. In addition, BMP expression in the epidermis, the roof plate of the neural tube and the lateral plate mesoderm prevent the differentiation of myogenic lineage11. Differently, Noggin, present in the roof and floor plate of the neural tube, blocks this BMP action and therefore allows for the myogenic precursors to differentiate12,13. This balance between multiple signaling pathways results, among others, in the restricted expression of myogenic regulatory factors (MRF) and Pax genes in myogenic cell populations14.
Besides its role during the patterning of the somite, BMP signaling also affects the differentiation of myofibers. The effect of BMP signaling was shown to depend on the developmental stage and the progression along the myogenic program. Whereas the differentiation of embryonic myoblasts was shown to be insensitive to BMP signals, the fetal myoblasts and the Pax7+ precursors require a decrease of the BMP signaling in order to allow further myogenic differentiation8.
We have reported before that the Noggin null (Noggin−/−) mice presented a muscle defect during the final stages of in utero development4. From early stages onwards the size of the muscles was reduced. At 18.5 dpc, the Noggin−/− embryos displayed a dramatic muscle phenotype characterized by disorganized myofibers that failed to align. In addition, the number of multinucleated myofibers was reduced, more mononucleated myoblasts were found and the nuclei failed to migrate towards the plasma membrane in the Noggin−/− muscles. The defects appear late during the development since we could not detect them at 16.5 dpc, indicating that the phenotype was induced between 16.5 dpc and 18.5 dpc (fetal myogenesis). While we made that observation, the molecular basis of that phenotype was not investigated. Here we focused on the molecular mechanisms underlying the phenotype. Our results suggest that the absence of Noggin, and consequently BMP hypersignaling, leads to the decrease of the number of muscle progenitor cells and their phenotypic shift towards adipocytes in vitro.
Results
Noggin −/− muscle fibers display a reduced fiber thickness
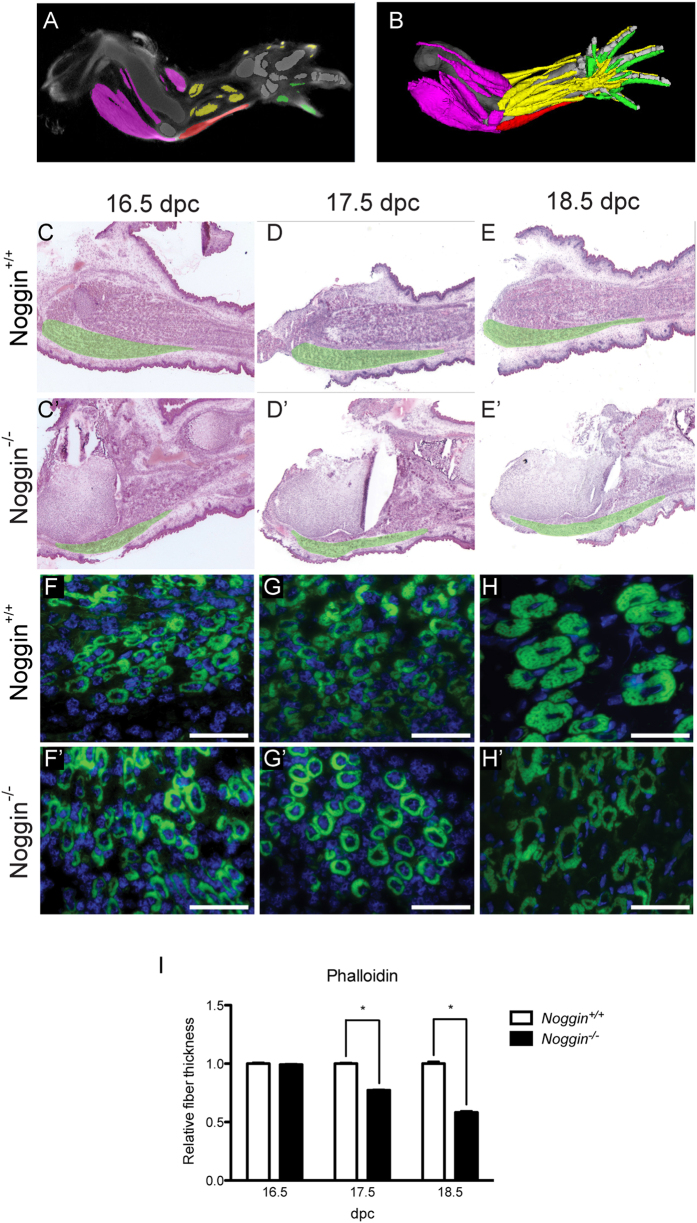
The limb muscles of Noggin−/− mice are strongly affected, with some of them missing and others severely malformed. Thus, to carry out a meaningful phenotypic analysis that would focus on the primary molecular differences rather than consequences of severe muscle malformation, we needed to select anatomically the same muscle in both genotypes. We performed H&E staining and compared the forelimb anatomy using the Jatlasviewer (www.emouseatlas.org/emap/analysis_tools_resources/software/jatlasviewer.html). We identified musculus flexor carpi ulnaris (indicated in red in Fig. 1A,B) as the muscle clearly identifiable and the least malformed in both wild type and Noggin null genotypes (Fig. 1C,E). For the analysis, we focused on three different developmental stages: one without an apparent defect (16.5 dpc), a dramatic defect (18.5 dpc) and the stage in between (17.5 dpc) (Fig. 1C–E’).
Figure 1. Analysis of the muscle fiber thickness.
(A,B) The limb at 15–16 dpc using Jatlasviewer. The musculus flexor carpi ulnaris is colored in red. (C–E’) H&E staining on sagittal sections of the limbs at the indicated stages and genotype. The musculus flexor carpi ulnaris is digitally indicated in green. (F–H’) Actin immunofluorescence on cross-sections of muscles at the indicated stages. (I) Quantification using ImageJ of the thickness of the fiber. Values plotted as mean ± sem; over 100 fibers of at least 3 different mice embryos were analyzed per condition; *p < 0.05
Since the histological appearance of the muscle suggested defective fiber thickness, we performed an F-Actin staining, using Phalloidin, and measured thickness of muscle fibers by measuring the muscle fiber diameter using ImageJ software. No difference in the overall morphology or in fiber thickness could be detected at 16.5 dpc in both genotypes. At 17.5 dpc Noggin−/− myofibers were slightly but significantly thinner and this trend persisted until 18.5 dpc where the difference was apparent (Fig. 1F–H’). Quantitative analysis of this observation confirmed the histological phenotype (Fig. 1I).
The loss of Noggin increases BMP signaling in embryonic muscles
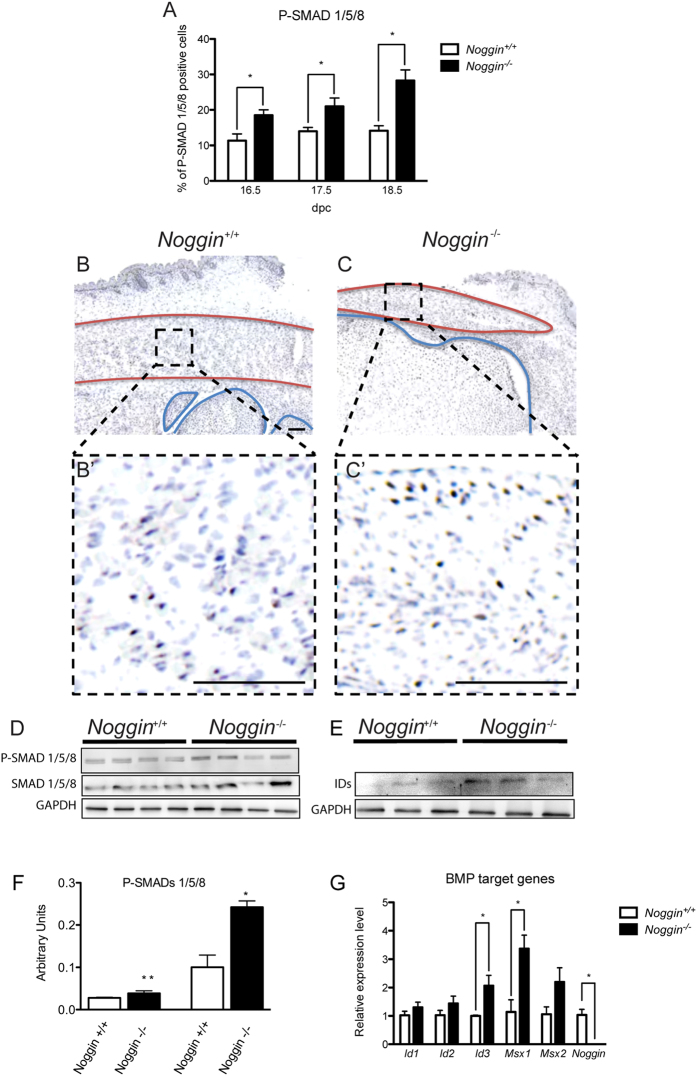
Canonical BMP signaling phosphorylates SMAD1/5/8 and this phosphorylation status is used to detect active BMP signaling. Since Noggin is a BMP antagonist, its absence should result in augmented BMP signaling. Therefore we investigated the activation of the SMAD1/5/8 proteins. The immunohistochemical analysis for the phosphorylated SMAD1/5/8 (P-SMAD1/5/8) displayed an increase in the number of positive cells in the muscle already from 16.5 dpc onwards as quantified in Fig. 2A and shown in Fig. 2B–C’. To confirm that observation we carried out Western Blot (WB) analysis for both P-SMAD1/5/8 and IDs proteins levels (Fig. 2D,E respectively) in the same muscle. The results of the P-SMAD1/5/8 WB has been quantified (Fig. 2F).
Figure 2. Analysis of the BMP signaling pathway.
(A) Quantification of P-SMAD1/5/8 immunohistochemistry on sagittal sections of musculus flexor carpi ulnaris of Noggin+/+ and Noggin−/− mice at the three different stages investigated. Values plotted as mean ± SEM; n = 5; *p < 0.05. Representative immunohistochemistry for P-SMAD1/5/8 at 16.5 dpc in Noggin+/+ (B) and Noggin−/− muscle (C). Enlargement of muscle area delineated in black in B’ and C’. Muscle is delineated in red, cartilage and bone in blue. Scale bar: 100 μm. (D) WB analysis of the levels of P-SMAD1/5/8 compared to total SMAD1/5/8 and (E) IDs, normalized for GAPDH. (F) Quantification of the WB, data are representative of 3 independent experiments and values are expressed as mean ± sd; *p < 0.05, **p < 0.01. (G) qPCR analysis of BMP target genes at E16.5 dpc. Values plotted as mean ± SEM; n = 5; *p < 0.05.
To verify the presence of active BMP signaling in the Noggin−/− musculus flexor carpi ulnaris, we investigated the expression of BMP immediate target genes. Using qRT-PCR at 16.5 dpc, we detected apparent upregulation of Msx2 and significant upregulation of Id3 and Msx1 (Fig. 2G).
Thus, at 16.5 dpc we detected the onset of molecular changes preceding the histological defects detectable 24 hours later.
The loss of Noggin does not affect the rate of proliferation
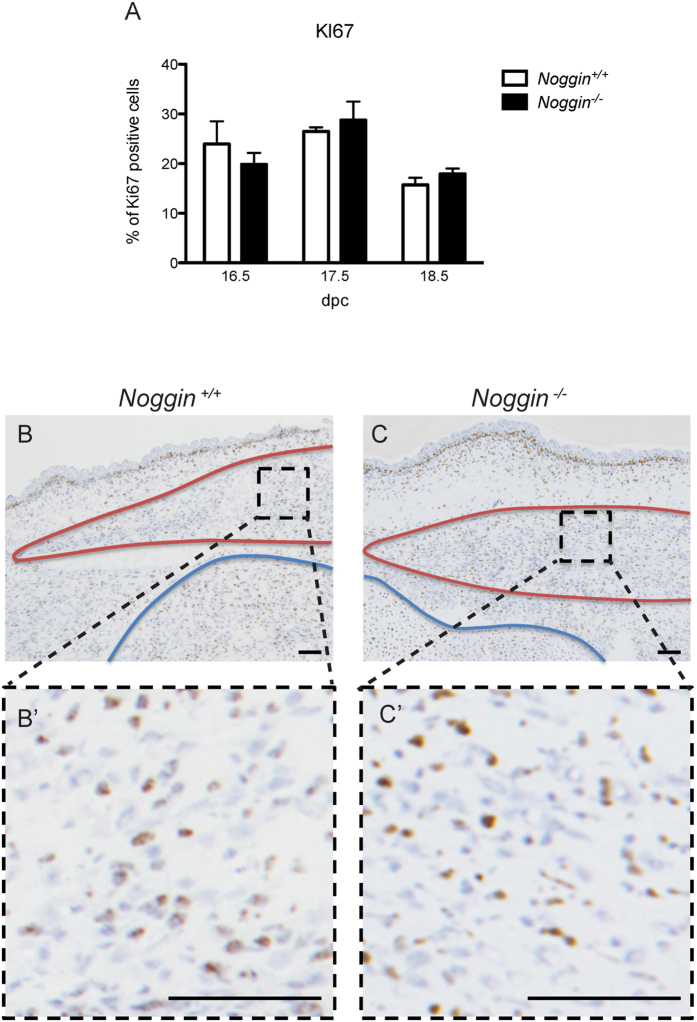
One of the functions of BMP signaling is to promote the exit from the cell cycle15. Thus one might expect that in the absence of Noggin, the cellular proliferation could be impaired leading to a decrease in the size of the muscle16. To address this issue, we analyzed the mitotic activity of the limb muscle cells using a KI67 antibody. KI67 is a nuclear protein present mainly in the G2, M, and at the end of the S phase and its presence is associated with active cell proliferation17. Despite the differences in muscle size and fiber thickness, no significant difference in cell proliferation could be detected in Noggin−/− muscles between the 16.5 dpc (no observable muscle phenotype) and 18.5 dpc (a strong phenotype) (Fig. 3).
Figure 3. Analysis of the mitotic activity.
(A) Quantification of KI67 immunohistochemistry on sagittal sections of musculus flexor carpi ulnaris of Noggin+/+ and Noggin−/− mice at the three different stages investigated. Values plotted as mean ± SEM; n = 5; *p < 0.05. Representative immunohistochemistry for Ki67 at 16.5 dpc in Noggin+/+ (B) and Noggin−/− (C) muscle. Muscle is delineated in red, cartilage and bone in blue. (B’–C’) Enlargement of muscle area delineated in black in B and C. Scale bar: 100 μm.
Noggin −/− muscles have less Pax7+ progenitor cells
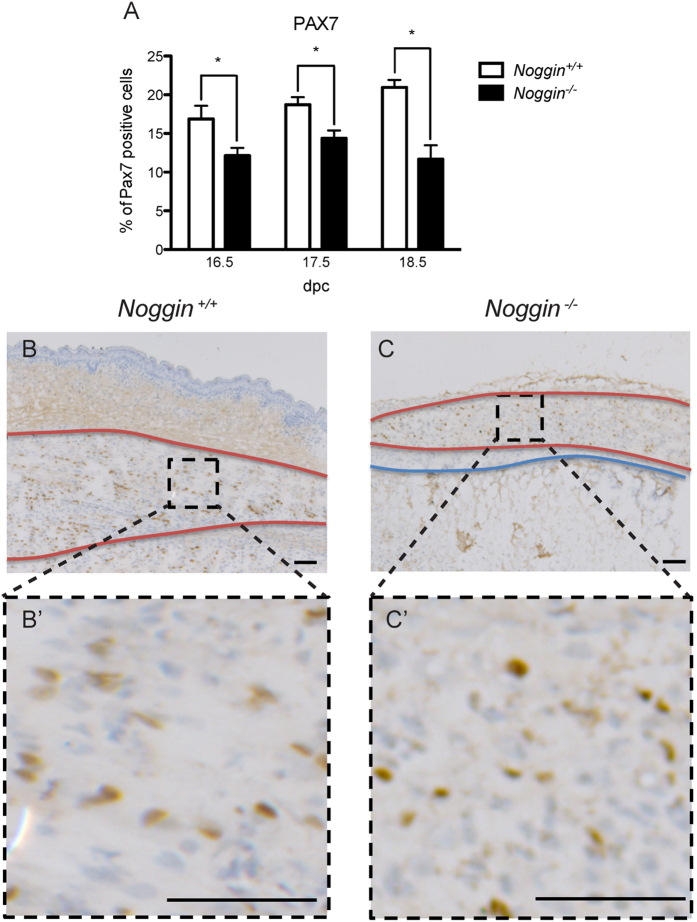
The cell proliferation was not affected in the muscle of Noggin null mice and the onset of muscle phenotype was relatively late during embryogenesis. Thus we hypothesized that perhaps not the muscle induction but rather muscle growth could be affected by excessive BMP signaling. During the muscle growth, Pax7+ cells provide the reservoir of stem cells that selectively enter myogenic differentiation and contribute to the growth of the muscle fiber during late embryonic development and in postnatal muscle repair processes18,19. Since BMP has a known negative effect on myogenesis (reviewed in ref. 20), we hypothesized that perhaps the number of Pax7+ cells in the late embryonic muscle could be reduced in the absence of Noggin. To investigate that, we carried out immunohistochemical analysis of the developing limb muscle for the expression of Pax7. Following the staining, we counted the number of positive cells using ImageJ software. Indeed, the number of Pax7+ cells was significantly reduced in Noggin−/− muscles throughout the stages investigated (Fig. 4).
Figure 4. Analysis of the satellite cells.
(A) Quantification of Pax7 immunohistochemistry on sagittal sections of musculus flexor carpi ulnaris of Noggin+/+ and Noggin−/− mice at the three different stages investigated. Values plotted as mean ± SEM; n≤5; *p < 0.05. Representative immunohistochemistry for Pax7 at 18 dpc in Noggin+/+ (B) and Noggin−/− (C) limbs. Muscle is delineated in red, cartilage and bone in blue. (B’-C’) Enlargement of muscle area delineated in black. Scale bar: 100 μm.
Impairment of myogenic potential in Noggin null progenitors
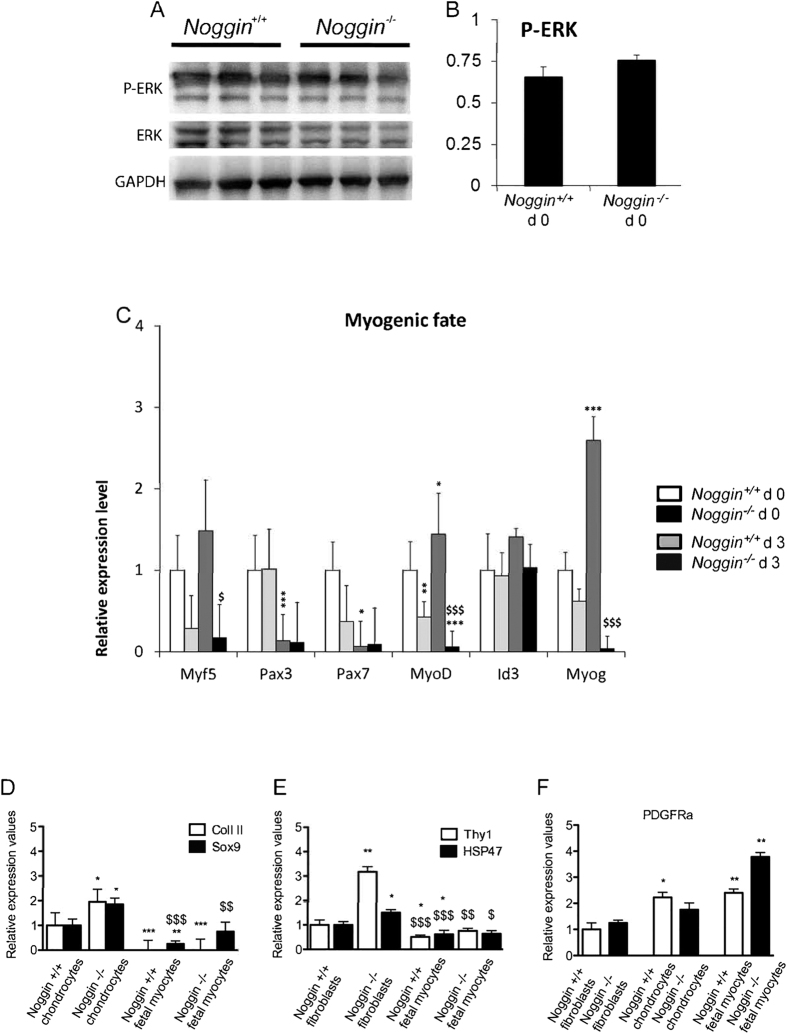
The cell proliferation was not affected in Noggin−/− mice, but the number of Pax7+ progenitor cells was reduced. To exclude a defective proliferation, we analyzed the levels of P-ERK21,22. To accomplish that, fetal myoblast progenitors from Noggin−/− and wild type embryos were isolated from 16.5 dpc limbs and cultured under myogenic conditions. No significant differences were detected in the levels of P-ERK, suggesting no defects in the proliferation rate were present in the muscle progenitors of Noggin−/− mice (Fig. 5A,B).
Figure 5. Myogenic differentiation of Noggin−/− early fetal progenitors.
(A) WB analysis and (B) protein quantification for the expression levels of P-ERK normalized on the total protein content and GAPDH for Noggin−/− compared to Noggin+/+ 16.5 dpc muscle early fetal progenitors. (C) Expression levels of genes (Myf5, Pax3, Pax7, Id3, MyoD and Myogenin) involved in myogenesis at 0 and 3 days (d0, d3) of differentiation for Noggin+/+ and Noggin−/− cells. (D) Expression levels of chondrocytes (Coll II, Sox9), fibroblasts (Thy1, Hsp47) or mesenchymal stem cell (Pdgfra) in Noggin+/+ and Noggin−/− early fetal progenitor cells. Results are expressed as ΔΔct normalized to specific housekeeping genes (Gapdh, Hprt and Tbp). Data are representative of 3 independent experiments and values are expressed as mean ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001 Noggin+/+, $p < 0.05, $$p < 0.01, $$$p < 0.001 Noggin−/−.
Next, to address the differentiation potential of progenitor cells, Noggin−/− cells were analyzed by qRT-PCR at 0 and 3 days of differentiation in vitro (Fig. 5C). At day 0, no significant differences in the expression levels of myogenic markers between Noggin+/+ and Noggin−/− cells were detected, with the exception of MyoD mRNA. After 3 days in culture, the expression of the myogenic progenitor markers, Pax3 and Pax7, decreased significantly irrespective of the genetic background. The expression levels of myogenic markers like MyoD, Myf5 and Myogenin increased in the wild type while decreased significantly in the Noggin−/− mouse cells (Fig. 5C). Thus as expected23,24, under the in vitro differentiation conditions used, the Pax3+ and Pax7+ cells did not maintain growth. At the same time, in the absence of Noggin, myogenic differentiation was repressed.
To ensure that the analyzed cell population was free of other cells that might affect the outcome of the experiment, we tested for the presence of cell types commonly contaminating such cultures, such as fibroblasts or chondrocytes. No large differences in the expression levels of Collagen II (Coll II) and SOX925 (Fig. 5D), Thy126 and HSP4727 (Fig. 5E), PDGFRa28 (Fig. 5F) were detected in the mRNA extracted from Noggin+/+ and Noggin−/− cells, thus excluding any contamination of chondrocytes, fibroblasts or mesenchymal/endothelial stem cells respectively.
Loss of myogenic differentiation in Noggin −/− progenitor cells
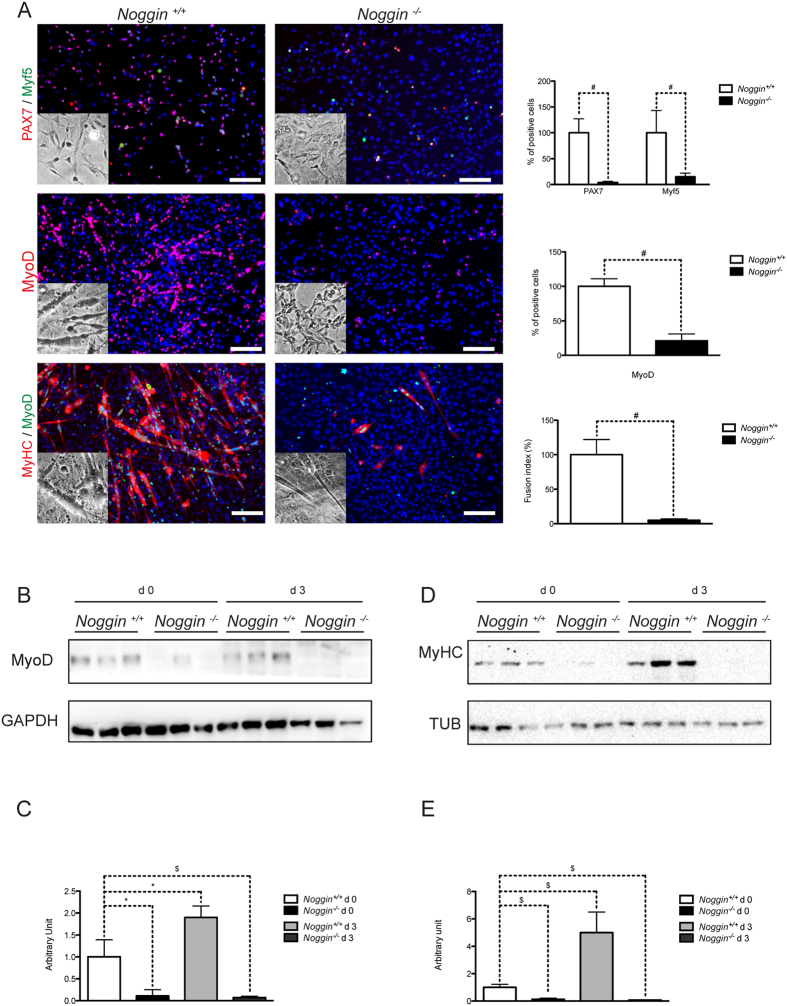
Next, we analyzed the cellular distribution of Pax7 and Myf5. Dual immunofluorescence revealed an apparent decrease in number of nuclei positive for Pax7 and Myf5 in Noggin−/− fetal myoblasts as compared to Noggin+/+. The percentage of Pax7+ and Myf5+ nuclei in Noggin−/− cultures was reduced to respectively 4% ± 2.26 and 15.8% ± 7.8 of Noggin+/+ values (Fig. 6A). After three days under the differentiation condition the number of MyoD+ cells was significantly reduced in Noggin−/− fetal myoblasts as compared to controls (8% ± 4 of Noggin+/+ cells; Fig. 6A). The myoblasts during the later stages of differentiation fuse to form myotubes (Suppl Fig. 1A). Thus we determined the fusion index calculated as the percentage of nuclei inside of MyHC+ and α-Sarcomeric Actinin+ myotubes to the number of total nuclei of Noggin−/− cultures and showed that it was also markedly decreased (Fig. 6A and Suppl. Fig. 1B).
Figure 6. Quantification of myogenic differentiation of Noggin−/− early fetal progenitors.
(A) Upper panels: IF analysis for Pax7+ (red), Myf5+ (green) nuclei of early fetal myoblasts in growing medium (day 0 of differentiation) from both Noggin+/+ and Noggin−/− embryos. Nuclei were stained with HOECHST (blue). The quantification of Pax7+ Myf5+ cells is reported in the upper right panel. Middle panels: IF analysis for MyoD+ (red) in Noggin+/+ and Noggin−/− cells at day 0 of differentiation; histogram with the percentage of MyoD+ nuclei in Noggin−/− cells with respect to Noggin+/+ cells are reported in the middle right panel. Lower panels: IF analysis for MyHC+ (red) myotubes and MyoD+ (green) nuclei in cells maintained during three days in differentiation medium. Fusion index (FI) analysis of differentiated Noggin−/− cells compared to Noggin+/+ cells is shown in the right panel. Data are representative of 3 independent experiments and values are expressed as mean ± sd, relative to Noggin+/+; #p < 0.001 vs Noggin+/+. An inset in each panel shows DIC images of cells. (B) WB analysis at 0 and 3 days of differentiation for MyoD; (C) protein quantification is reported in the lower panel. (D) WB analysis at 0 and 3 days of differentiation for MyHC; (E) protein quantification is reported in the lower panel. GAPDH and tubulin (TUB) were used as loading controls in B and C respectively. Data are representative of 3 independent experiments and values are expressed as mean ± sd; *p < 0.05, $p < 0.01.
To semi-quantify these observations we carried out a protein expression analysis by WB. While MyoD and MyHC protein levels were detectable in Noggin+/+ cells, they were almost undetectable in Noggin−/− cells at early (d0) and late (d3) stage of differentiation (Fig. 6B–E).
In addition, the levels of embryonic MyHC (eMyHC) strongly expressed in Noggin+/+ cells late stage, were barely detectable in Noggin−/− cells at both time points of differentiation (Suppl Fig. 1C).
Aberrant adipogenic potential in Noggin −/− progenitors
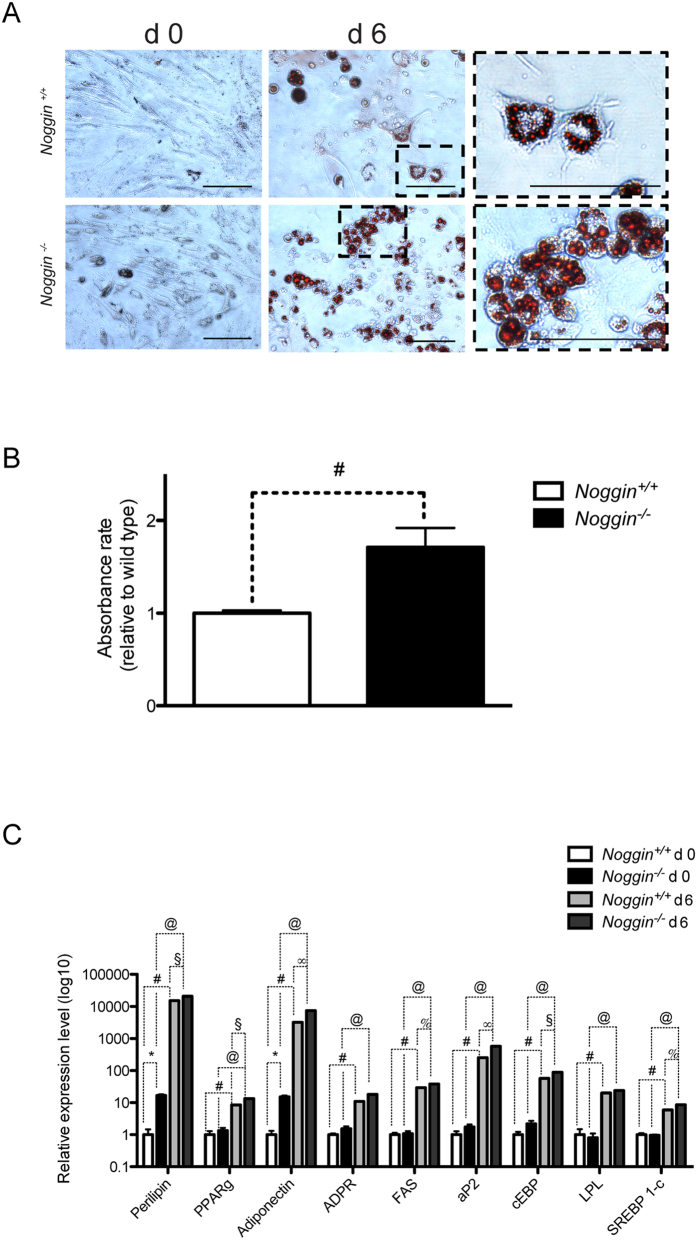
It has been reported that BMP signaling pathway is also involved in adipocyte commitment and differentiation in vitro and in vivo29,30. To investigate this aspect of cellular differentiation in Noggin−/− cells, we tested their adipocyte commitment in vitro by growing them in adipogenic medium. Noggin+/+ progenitor cells were able to shift towards an adipocyte fate, even if they were still able to generate myotubes (Suppl Fig. 1D). Noggin−/− cells, however, only differentiated towards adipocytes, even more efficiently compared to Noggin+/+ cells (1.7 ar ± 0.2 vs. 1 ar ± 0.03) as shown by quantification of Oil-Red-O staining (Fig. 7A,B).
Figure 7. Adipogenic induction of Noggin−/− early fetal progenitors.
(A) Oil-Red-O staining of Noggin+/+ (upper panels) and Noggin−/− (lower panels) early fetal progenitors cultivated for 6 days under adipogenic condition; higher magnifications of the inset are reported in right panels. (B) Lipid quantification of Noggin+/+ and Noggin−/− cells. (C) qRT-PCR analysis of adipogenic markers at d0 and d6 of differentiation. Data of at least 5 independent experiments are expressed as ΔΔct values normalized to specific housekeeping genes (Gapdh, Hprt and Tbp); values reported as mean ± SEM, normalized to Noggin+/+ values. *p < 0.01, #p < 0.001; @ p < 0.001; ∞p < 0.05, §p < 0.01, % p < 0.001.
The molecular signature of these cells further supported their adipogenic phenotype. Noggin−/− cells showed a significant increase in the expression of the majority of the adipocyte genes after 6 days of adipogenic induction as shown by qRT-PCR data (Fig. 7C and Table 1- please note the logarithmic scale on the Figure). Notably, at day 0 Perilipin (166% ± 1.5) and Adiponectin (151% ± 1.6) were already present at increased levels as compared to the Noggin−/− cells (Fig. 7C).
Table 1. ΔΔct values normalized to specific housekeeping genes of qRT-PCR analysis reported in Fig. 7C.
| Perilipin | PPARg | Adipo-nectin | ADPR | FAS | aP2 | cEBP | LPL | SREBP1 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Noggin+/+ | d0 | 1.0 ± 0.5 | 1.0 ± 0.3 | 1.0 ± 0.3 | 1.0 ± 0.1 | 1.0 ± 0.1 | 1.0 ± 0.3 | 1.0 ± 0.2 | 1.0 ± 0.5 | 1.0 ± 0.1 |
| Noggin−/− | d0 | 16.6 ± 1.5** | 1.3 ± 0.3 | 15.2 ± 1.6** | 1.5 ± 0.3 | 1.1 ± 0.2 | 1.7 ± 0.3 | 2.2 ± 0.5 | 0.8 ± 0.2 | 0.9 ± 0.1 |
| Noggin+/+ | d6 | 15.2 × 103 ± 0.4*** | 8.5 ± 0.3*** | 3.2 × 103 ± 0.4*** | 10.9 × 103 ± 0.3*** | 29.4 ± 0.2*** | 253.8 ± 0.4*** | 57.4 ± 0.2*** | 20.1 ± 0.2*** | 5.9 ± 0.0*** |
| Noggin−/− | d6 | 20.7 × 103 ± 0.3*** $$ °°° | 13.5 ± 0.2*** $ °°° | 7.4 × 103 ± 0.3*** $ °° | 17.9 ± 0.4*** °°° | 38.1 ± 0.4*** $$$°°° | 569.2 ± 0.1*** $ °°° | 88.5 ± 0.5*** $$ °°° | 23.9 ± 0.3*** °°° | 8.5 ± 0.4*** $$$ °°° |
Discussion
We have previously reported on a muscle defect in Noggin−/− mice4. Here we investigated the molecular basis of that phenotype. Interestingly, the defect has a late onset, indicating that the initial muscle induction and formation occurs normally. At the later stages however, when the muscle acquires more mass, it becomes sensitive to the absence of Noggin. Our analysis indicates that the loss of Noggin in the mouse embryonic muscle resulted in an increased BMP signaling together with a decrease in the relative number of Pax7+ cells. Intriguingly, the differentiation defect persisted in in vitro setting suggesting that it was cell autonomous. Specifically the cells lost the ability to enter the myogenic pathway while maintained the adipogenic one.
Our analysis shows that the loss of Noggin during mouse embryonic development results in an increased BMP signaling in developing muscle. Molecularly it has been shown that the inhibitory effect of BMP signaling on the myogenic program is a result of the sequestering of the E-proteins by the immediate BMP target genes (Id1, 2, 3)31,32. Thereby the ID proteins block the MRF-mediated activation of the myogenic program, which is dependent on the binding of the MRFs to these E-proteins. Furthermore, Msx1 was shown to block cellular differentiation by preventing cell cycle exit and to antagonize the myogenic activity of Pax3 in migrating limb muscle precursors33,34. Since the BMP target genes are upregulated in the Noggin−/− mice, the sequestering of the E-proteins, the inhibition of cell cycle exit and/or the loss of differentiation of the migrating limb precursors are all potential mechanisms underlying the Noggin−/− muscle phenotype.
The BMP signaling was shown to regulate the patterning of the somite and the myogenic program3, thus a muscle defect in BMP antagonist mutants could have been anticipated. However, the extent of the muscle defect in the Noggin−/− mice and the late onset was far greater than reported for other BMP antagonist mutants35. Reshef et al. did suggest that the epaxial musculature in the Noggin−/− mice was largely absent in the posterior part of the embryo, but no further research was conducted on the muscles of the Noggin−/− mice36. The work presented by Lassar’s group did in fact lay a foundation for our work. Reshef et al. have shown that the precise regulation of BMP signaling by Noggin and other antagonists, controls the entry of Pax3+ cells into myogenic differentiation program defined by the expression levels of MyoD and Myf5. This regulatory loop is therefore essential for the myogenesis within the somites in vitro and in ovo. We have extended the work of Reshef et al. by showing that the same regulatory loop used also at later stages of development in mouse. Specifically, we show that, in Noggin−/− embryos, not only the muscle fibers exhibit a morphological defect but the maintenance of the fetal myogenic progenitor pool is impaired as well. In fact, we show for the first time that Noggin−/− myogenic progenitors are unable to properly differentiate into the myogenic lineage, but display a more efficient commitment to adipogenic lineage.
In vitro, BMP signaling was shown to balance the proliferation and differentiation of muscle satellite cells. Upon differentiation, BMP signaling is downregulated resulting in the activation of the myogenic program and formation of myofibers37,38. In contrast, when BMP was applied ectopically in the chicken wing, the number of muscle fibers and Pax7+ cells increased14,16. However, when BMP was applied during earlier stages of chicken development, similar chondrogenesis and myogenesis defects were seen as in the Noggin−/− mice39. These results strongly suggest that the timing and the localization of the BMP signal and the differentiation status of the target cells determines the molecular response explaining the apparent contradiction in the results published by us and among these research groups.
It was suggested that cells in the somitic environment could have different intrinsic properties as they respond differently to the same signals40. For example, embryonic myoblasts were shown to be insensitive to BMP signaling allowing them to stop proliferating and differentiate into the primary myofibers. For fetal myoblasts and satellite cells, however, it was shown that the level of BMP signaling has to be reduced, by either reducing the expression of BMP ligands, or antagonizing the signaling, in order to let differentiation and fusion into secondary fibers occur16. This potentially explains the Noggin−/− muscle phenotype, as the increased BMP signaling in the muscle would have no effect on the differentiation of the primary fibers, but it could inhibit the generation of the secondary fibers. Therefore fetal myoblasts, which normally give rise to the secondary fibers, maintain the committed but undifferentiated state. Indeed, we observe mononucleated myoblasts in the Noggin−/− muscle, concomitant with a reduction of multinucleated myofibers. The limited number of primary fibers that is formed in the absence of Noggin results in a muscle that fails to maintain its organization.
Another explanation of the observed phenotype could be based on a possibility of lineage switch. In fact, the myogenic potential of C2C12 cells can be fully blocked by the addition of BMP ligands, which then promote osteochondrogenic differentiation of this cell line41,42 and reviewed in ref. 43. Thus we hypothesize that a number of muscle progenitor cells are drawn into the chondrogenic lineage in the Noggin−/− limb44,45. This would deplete the pool of muscle precursors in a way that initially enough cells are left to maintain the muscle organization, but at later stages, when the muscles grow rapidly, there are not enough cells to supply the muscle. In this view, the loss of Noggin induces SMAD-dependent increase in BMP signaling that results in an inhibition of fetal myoblast differentiation and a reduction in the number of satellite cells, which causes a severe late-onset muscle defect. At the same time, the prechondrogenic pool increases, leading to excessive chondrogenesis.
To test this hypothesis we investigated the molecular and cellular properties of mononuclear myogenic progenitors isolated from Noggin+/+ and Noggin−/− littermates. Interestingly, the transcript levels of early myogenic markers (Pax3 and Pax7) had the same dynamics in Noggin+/+ and Noggin−/− progenitors. However, protein levels of Myf5 and MyoD were significantly reduced in Noggin−/− progenitors after 3 days of myogenic induction. Indeed, it has been previously reported that no significant differences were detected for Pax3, Pax7 and Myf5 expression in Noggin−/− embryonic muscles13. The regulation of Pax7 protein levels is complex and not yet completely defined. In principle we cannot exclude that in our conditions, as recently reported46 and reviewed in ref. 23, a post-translational modification could induce the ubiquitination and consequently a cleavage of Pax7.
In this view a complete inhibition of the MyoD and possibly Myogenin proteins is probably the main cause of the lack in differentiation of early fetal myoblasts in our cultures as it was previously reported47.
One of the key findings of this work is that MyoD and consequently embryonic and adult MyHC protein levels are strongly decreased in Noggin−/− progenitors. Recently, the pattern of expression of MyHC isoforms has been fully characterized and defined as a transition form from one cohort of MyHC isoforms (embryonal, neonatal and type I MyHC) to another (fast type II isoforms), occurring during muscle cell differentiation48. Thus, eMyHC dramatically decreased in Noggin−/− cells at both time points considered, could not support further expression of the adult MyHC isoform.
Intriguingly, the response of the Noggin−/− muscle progenitors to adipocytogenic conditions dramatically differs from the wild type cells. The absence of Noggin and thus overexposure to BMP signaling during embryonic development induces the muscle progenitor cells to be significantly more responsive to adipogenic activation than the corresponding wild type cells. Indeed, Adiponectin and Perilipin genes normally expressed in adult white adipose tissue49, are also highly transcribed in these aberrant progenitor cells.
These results are in line with previous reports showing the participation of BMP4 in adipocyte lineage commitment and the rescue induced with Noggin exposure in vivo50. Moreover, it has been recently demonstrated that murine myoblasts (C2C12) display lipid accumulation upon BMP6 stimulation51.
Taken together our results stress the importance of balance between Noggin and BMPs for myogenic commitment of early progenitor cells during limb development. The absence of Noggin could induce a concomitant BMP over-signaling, likely responsible for the impaired myogenic commitment of Noggin−/− progenitors. Indeed, BMP2, in cooperation with SMAD1/4 and C/EBPα, can induce the expression of PPARγ2, key transcription factor for adipocyte differentiation52. Our data also indicate the power of cell memory, where the level of BMP exposure is remembered during later differentiation stages, even in vitro, and even when the actual signal is not present anymore.
Further studies are necessary to define the role of other BMP inhibitors53, e.g. Chordin or Follistatin, Xnr3 or Cerberus, which could eventually compensate the absence of Noggin and be involved in the aberrant differentiation ability observed in Noggin−/− muscle progenitors54. It could be also interesting to investigate if the alteration of Noggin/BMP balance is involved in the adipogenic conversion of skeletal muscle precursors observed in chronic muscle degenerations.
Materials and Methods
Embryo processing
All the mouse experiments were carried out in accordance with European guidelines on animal research and approved by the ethics committee at the University of Leuven. Noggin heterozygous mice were maintained in CD1 background as described before4. Mice were sacrificed and embryos were collected at 16.5, 17.5 and 18.5 days post coitus (dpc). Forelimbs were isolated using forceps and subsequently embedded in Tissue-Tek (Laborimpex), snap frozen in liquid nitrogen and stored at −80 °C. Hind limbs were processed for beta-galactosidase (beta-gal) genotyping. Frozen sections (5 μm) for immunological and histological staining (Haematoxylin and eosin; standard protocol) were made using a cryostat (Prosan).
Beta-galactosidase genotyping
Targeted Noggin inactivation was achieved by replacing Noggin exon with LacZ expression cassette3 permitting the genotyping by beta-gal staining. The well-described phenotypic differences were used to discriminate between heterozygous and homozygous animals (4,55). Hind limbs were fixed in 4% PFA in PBS for 10 minutes and washed twice with LacZ1 solution (PBS, 2mM MgCl2, 5 mM EGTA) for 10 minutes at RT, followed by another washing step with LacZ2 (PBS, 0.1 M Na3PO4, 2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% NP-40; pH 7.2–7.4) for 5 minutes at RT. Staining was performed by applying staining solution (5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 0.5 mg/ml, X-gal in LacZ2 buffer for 2–3 hours at 37 °C or ON at RT. After washing steps with PBS and clearing with 2% KOH, limbs were treated with an increasing gradient of glycerol/KOH before storage.
Immunohistochemistry
Frozen sections were fixed with methanol for 10 minutes at −20 °C. After quenching in 3% H202/MQ and washing in TBST, antigen retrieval was performed with sodium citrate buffer (pH6), followed by additional washing with TBST. Sections were blocked with Normal Goat Serum (Millipore) (1/5 in TBST) and O/N incubated with the primary antibody (P-SMAD1/5/8, Cell Signaling, 9511S; 1/100) at 4 °C. After washing, the biotinylated secondary antibody was applied for 30 minutes at RT. The signal was amplified with the ABC reagent (Vectastain, dilution according to manufacturer’s protocol) for 30 minutes at RT. Phalloidin staining was performed using manufacturers protocol (Sigma). After washing, the color was developed using DAB (DAKO, dilution according to manufacturer’s protocol) and sections were mounted using Pertex.
The P-SMAD1/5/8 protocol was used with following modifications. Frozen sections were fixed with ethanol and methanol for 5 minutes and quenched in 0.5% H202/MeOH. Washing steps were performed with PBS, a KI67 antibody (DAKO TEC3; 1/100) or Pax7 antibody (Hybridoma bank, Iowa, USA; 1/400) was used and blocking was done with 10%NGS/PBS. ImageJ software was used to measure muscle fiber thickness and the quantification of the percentage of P-SMAD1/5/8, KI67 or Pax7+ cells. The significance of the difference in expression was analyzed using 2way ANOVA test with Bonferroni comparison and significance was achieved with p-values less than 0.05 (Student’s T-test).
Gene expression analysis
For muscle tissue, RNA was isolated by scraping the muscles of at least 10 frozen sections 16.5 dpc embryonic limbs using the High Pure RNA Isolation Kit (Roche Applied Science). At least three RNA isolations were performed per condition and subsequently reverse transcribed using Primescript RT Reagent (Takara; manufacturers protocol). qRT-PCR was performed in duplicate with gene specific primers (Table 2) with SYBR Premix Ex Taq II (Takara; manufacturers protocol) using Rotor-gene 6000 detection system (Corbett Research, Westburg). Gene expression was normalized to the housekeeping gene Hprt1 and presented as a ratio to control embryos. The significance of the difference in expression was analyzed using the Student’s T-test.
Table 2. list of primers.
| Gene | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| Adiponectin56 | GGAGAGAAAGGAGATGCAGGT | CTTTCCTGCCAGGGGTTC |
| ADRP57 | CTTGTGTCCTCCGCTTATGTCAGT | CTGCTCCTTTGGTCTTATCCACCA |
| aP258 | GGAAGCTTGTCTCCAGTGAA | GCGGTGATTTCATCGAATTC |
| C/EBPα58 | TGCGCAAGAGCCGAGATAAA | CCTTGACCAAGGAGCTCTCA |
| Coll II | CCAGGATGCCCGAAAATTAG | TTCTCCCTTGTCACCACGAT |
| FAS59 | AGTGTCCACCAACAAGCG | GATGCCGTCAGGTTTCAG |
| GAPDH60 | TGGTGAAGGTCGGTGTGAAC | GCTCCTGGAAGATGGTGATGG |
| HPRT60 | TGGATACAGGCCAGACTTTGTT | CAGATTCAACTTGCGCTCATC |
| Hprt1 | TGCTGACCTGCTGGATTACA | TATGTCCCCCGTTGACTGAT |
| Hsp 4761 | GCAGCAGCAAGCAACACTACAACT | AGAACATGGCGTTCACAAGCAGTG |
| Id1 | GAGTCTGAAGTCGGGACCAC | AACACATGCCGCCTCGG |
| Id2 | CCTGCATCACCAGAGACCTG | GGGAATTCAGATGCCTGCAA |
| Id362 | TGCTACGAGGCGGTGTGCTG | TGTCGTCCAAGAGGCTAAGAGGCT |
| LPL63 | CTGCTGGCGTAGCAGGAAGT | GCTGGAAAGTGCCTCCATTG |
| Msx1 | GCCAGAAGATGCTCTGGTGA | TCAGCCGTCTGGCTGGG |
| Msx2 | GCCTCGGTCAAGTCGGAAAA | GGCTCATATGTCTGGGCGG |
| Myf564 | ACAGAGCTGCTCTGAGCCCACC | TACATCAGGACAGTAGATGCTGTC |
| MyoD65 | GAGCAAAGTGAATGAGGCCTT | CACTGTAGTAGGCGGTGTCGT |
| Myog65 | ATGGAGCTGTATGAGACATCCCC | CGACACAGACTTCCTCTTACAC |
| Noggin | GGGGCGAAGTAGCCATAAAG | GGGGCGAAGTAGCCATAAAG |
| Pax364 | ACTACCCAGACATTTACACCAGG | AATGAGATGGTTGAAAGCCATCAG |
| Pax765 | GACGACGAGGAAGGAGACAA | ACATCTGAGCCCTCATCCAG |
| PDGFRa65 | CCTGTAACTGACACGCTCCG | CCATTTCCAAACCGCACA |
| Perilipin58 | TTGGGGATGGCCAAAGAGAC | CTCACAAGGCTTGGTTTGGC |
| PPARγ66 | GCCCTTTGGTGACTTTATGGA | GCAGCAGGTTGTCTTGGATG |
| Sox967 | CAAGAACAAGCCACACGTCA | TGTAATCGGGGTGGTCTTTC |
| SREBP-1c59 | CTGGAGACATCGCAAACAAGC | ATGGTAGACAACAGCCGCATC |
| TBP60 | CAAACCCAGAATTGTTCTCCTT | ATGTGGTCTTCCTGAATCCCT |
| Thy 168 | GCCGCCATGAGAATAACA | GCTAGGGTAAGGACCTTGAT |
For the cells, Noggin+/+ and Noggin−/− early fetal myoblasts, chondrocytes and fibroblasts were used for RNA extraction with RNA Mini Kit, removing genomic DNA traces by Turbo DNase. 1ug RNA was reverse-transcribed with SuperScript III kit and qPCR was performed in 384 well plates (10ul final volume; thermal profile, 95 °C 15″−60 °C 45″ (40x); ViiA 7 qPCR plate reader), using Platinum Sybr Green Mix, 1ul 1:4 diluted cDNA and 250 nM primers (all kits, reagents and plate reader by Life Technologies, Carlsbad, CA, USA). Proliferating MEF cells as well as fibroblasts from adult mice or 3T3-L1 preadipocytes (ATCC, Manassas, VA, USA) were used as positive controls.
Fetal myoblasts isolation and myogenic differentiation
Early fetal myoblasts were isolated from forelimbs of E16.5 Noggin−/− and control embryos (after careful removal of bones and skin) as previously described40. Briefly, tissues were dissociated one hour at 37 °C with 0.06% Collagenase (from Clostridium Histolyticum, Sigma-Aldrich St. Louis, Mo, USA, for all reagents excepted when notified) and 0.04% Pancreatin in PBS (Gibco-Invitrogen, Carlsbad, CA, USA). After centrifugation, cells were seeded in collagen-coated dishes in presence of Dulbecco’s modified Eagle’s medium (DMEM)-high glucose and 20% fetal bovine serum (FBS; Gibco-Life Technologies, Carlsbad, CA, USA), supplemented with 1% Chicken Embryo Extract (Sera Laboratories International, UK), 100 mg/ml Sodium Pyruvate, 100 IU/ml penicillin and streptomycin, 2 mM L-glutamine. The plates were incubated at 37 °C, 5% CO2, 5% O2 and the medium refreshed every two days. When cells reached confluence, the differentiation was induced by shifting medium to DMEM high glucose supplemented with 2% horse serum (Gibco-Invitrogen, Carlsbad, CA, USA) and 100 mg/ml Sodium Pyruvate (differentiation medium; DM).
Adipogenic differentiation
Early fetal myoblasts were dispended on collagen coated dishes and expanded till confluence, in presence of STEMPRO Adipogenesis Differentiation Medium (Gibco-Lyfe Technologies, Carlsbad, CA, USA) to enhance adipogenesis, as described from the manufacturer. 3T3-L1 preadipocytes (ATCC) used as control for adipocyte differetiation were grown in DMEM supplemented with 10% (v/v) fetal calf serum (Gibco-Lyfe Technologies, Carlsbad, CA, USA), 25 mM HEPES (pH 7.0), 100 U/ml penicillin and streptomycin, and 25 μg/ml Fungizone Antimycotic (Gibco-Lyfe Technologies, Carlsbad, CA, USA). To induce adipocyte differentiation, confluent cells were cultured in DMEM with 10% (v/v) FBS (Gibco-Lyfe Technologies, Carlsbad, CA, USA), 25 mM HEPES (pH 7.0), 100 U/ml penicillin and streptomycin, and 25 μg/ml fungizone, plus 0.5 mM isobutylmethyl-xanthine, 1 μM dexamethasone, and 1 μg/ml insulin (MDI). MDI was replaced 48 h after, with insulin (1 μg/ml), and the medium changed every two days.
Immunofluorescence analysis
Cells at day 0 and 3 of differentiation were fixed with 4% PFA (Polysciences Europe GmbH, Germany) and permeabilized with 0.2% PBS-Triton, 1% Bovine Serum Albumin (BSA). A blocking solution containing donkey serum was applied and primary antibodies were incubated overnight (O/N) in PBS supplemented with 2.5% w/v BSA. Primary antibodies: 1:50 anti-rabbit Myf5 (c-20, Santa-Cruz Byotechnology, Heidelberg, Germany), 1:3 mouse anti-PAX7 (Developmental Studies Hybridoma Bank, DSHB, Iowa, USA), 1:50 mouse anti-MyoD (Dako, Agilent Technologies, Denmark), 1:30 rabbit anti-MyoD (Santa Cruz Byotechnology, Heidelberg, Germany), 1:10 mouse anti-MyHC (clone MF20, Developmental Studies Hybridoma Bank, DSHB, Iowa, USA), anti-mouse α-Sarcomeric Actinin (Abcam, Cambridge, UK). Secondary Alexa Fluor donkey antibodies (Life Technologies, Carlsbad, CA, USA) were diluted at 4 μg/ml in PBS supplemented with 2.5% w/v BSA and the nuclei counterstained with 10 ug/ml Hoechst. Fluorescence images were acquired at 10x magnification via Eclipse Ti inverted microscope (Nikon). Fusion index (number of myotube nuclei vs total number of nuclei) such as the percentage of myotube positive for a specific marker were calculated for a minimum of four random fields from Noggin+/+ and Noggin−/− embryo cells of different embryos (n = 7, three different litters).
Western blot (WB) analysis
WB analysis were performed on cell lysates in RIPA buffer supplemented with 10 mM Sodium Fluoride, 0.5 mM Sodium Orthovanadate, 1:100 Protease Inhibitor Cocktail and 1 mM Phenylmethanesulfonyl Fluoride. Equal amounts of protein (30 μg) were heat-denatured in sample-loading buffer (50 mM Tris-HCl, pH 6.8, 100 mM DTT, 2% SDS, 0.1% bromophenol blue, 10% glycerol), resolved by SDS-PAGE and transferred to nitrocellulose membranes (GE Healthcare Bio-Sciences, Pittsburg, USA). The filters were blocked with Tris-buffered saline (TBS) containing 0.05% Tween and 5% non-fat dry milk and then incubated overnight with the different primary antibodies: rabbit 1:1000 anti-P-SMAD1/5/8 (Cell Signaling); rabbit 1:100 anti-SMAD1/5/8 (ThermoScientific); rabbit 1:100 anti-IDs (SantaCruz); rabbit 1:100 anti-P-ERK (SantaCruz); mouse 1:200 anti-ERK (SantCruz); mouse 1:100 anti-MyoD (Dako, Agilent Technologies, Denmark); mouse 1:10 anti-MyHC and mouse 1:5 anti-eMyHC (Developmental Studies Hybridoma Bank, DSHB, Iowa, USA), mouse 1:1000 anti-Tubulin beta, clone KMX-1 (Millipore, Chemicon, Billerica, MA, USA), anti-rabbit GAPDH. All secondary horseradish peroxidase (HRP)-conjugated antibodies (Santa Cruz Biotechnology, CA, USA) were diluted 1:5000 in TBS-Tween and 2.5% non-fat dry milk. After incubation with SuperSignal Dura Chemiluminescence substrate (Thermo Scientific, Waltham, MA USA), WB analysis was performed with GelDoc chemiluminescence detection system (BioRad, Temse, Belgium). Quantification by relative densitometry was obtained normalizing versus background and GAPDH or Tubulin, using the QuantityOne software (BioRad).
Oil-Red-O staining
Lipid accumulation in early fetal myoblasts was assessed by Oil-Red-O staining. Briefly, cells were fixed for 15 min in 4% PFA, washed three times in PBS, and stained for 30 min with the lipophilic dye Oil-Red-O in 65% isopropanol, followed by washing steps and drying. Bright field Oil-Red-O images were captured at 10x and 20x magnification using an Eclipse Ti inverted microscope (Nikon). Lipids extracted with a petrol ether/isopropanol mixture (3:2) were quantified for their absorbance at 490 nm with Victor spectrophotometer (PerkinElmer, Massachussets, USA).
Statistical analysis
All results were expressed as mean ± SD, with the exception of gene expression (mean ± SEM). Representative WB show independent samples. When two groups were compared, a Student’s t-test was used. When three or more groups were compared a two-way ANOVA was used. The statistical significance of the differences between percentage values was assessed using a Kruskal-Wallis one-way analysis of variance by ranks test. All statistical tests were performed via Prism software (GraphPad).
Additional Information
How to cite this article: Costamagna, D. et al. Noggin inactivation affects the number and differentiation potential of muscle progenitor cells in vivo. Sci. Rep. 6, 31949; doi: 10.1038/srep31949 (2016).
Supplementary Material
Acknowledgments
This research was supported by a research grant from FWO G.0214.07 to PT and HM and IWT grant to HM. MS has been supported with the contribution of “Opening The Future” Campaign (EJJ-OPTFUT-02010), CARIPLO 2015_0634, FWO (#G060612N, #G0A8813N, #G088715N), GOA (EJJ-C2161-GOA/11/012), and OT#09-053 (EJJ-C0420-OT/09/053). DC is supported by University of Turin, Italy. In addition, PT and MS are recipient of the Interuniversity Attraction Poles grant, IUAP-VII/07 (EJJ-C4851-17/07-P). We thank Inge Van Hoven for excellent mouse husbandry, Christina Vochten and Vicky Raets for professional administrative assistance. We would also like Rondoufonds voor Duchenne Onderzoek for kind donation.
Footnotes
Author Contributions H.M. and D.C. carried out all the experiments. P.T. and H.M. conceived the work, H.M., D.C., M.S. and P.T. wrote and edited the manuscript. Authors declare no competing financial interests.
References
- Gazzerro E. & Canalis E. Bone morphogenetic proteins and their antagonists. Reviews in endocrine & metabolic disorders 7, 51–65, doi: 10.1007/s11154-006-9000-6 (2006). [DOI] [PubMed] [Google Scholar]
- Merino R. et al. Expression and function of Gdf-5 during digit skeletogenesis in the embryonic chick leg bud. Developmental biology 206, 33–45, doi: 10.1006/dbio.1998.9129 (1999). [DOI] [PubMed] [Google Scholar]
- Brunet L. J., McMahon J. A., McMahon A. P. & Harland R. M. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science 280, 1455–1457 (1998). [DOI] [PubMed] [Google Scholar]
- Tylzanowski P., Mebis L. & Luyten F. P. The Noggin null mouse phenotype is strain dependent and haploinsufficiency leads to skeletal defects. Developmental Dynamics 235, 1599–1607, doi: Doi 10.1002/Dvdy.20782 (2006). [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy L. et al. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev 19, 1426–1431, doi: 10.1101/gad.345505 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schienda J. et al. Somitic origin of limb muscle satellite and side population cells. Proceedings of the National Academy of Sciences of the United States of America 103, 945–950, doi: 10.1073/pnas.0510164103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F. et al. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol 172, 91–102, doi: 10.1083/jcb.200508044 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biressi S. et al. Myf5 expression during fetal myogenesis defines the developmental progenitors of adult satellite cells. Dev Biol 379, 195–207, doi: 10.1016/j.ydbio.2013.04.021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P. et al. Pax7 is required for the specification of myogenic satellite cells. Cell 102, 777–786 (2000). [DOI] [PubMed] [Google Scholar]
- Marcelle C., Stark M. R. & Bronner-Fraser M. Coordinate actions of BMPs, Wnts, Shh and noggin mediate patterning of the dorsal somite. Development 124, 3955–3963 (1997). [DOI] [PubMed] [Google Scholar]
- Lee K. J., Dietrich P. & Jessell T. M. Genetic ablation reveals that the roof plate is essential for dorsal interneuron specification. Nature 403, 734–740 (2000). [DOI] [PubMed] [Google Scholar]
- Yusuf F. & Brand-Saberi B. The eventful somite: patterning, fate determination and cell division in the somite. Anatomy and embryology 211 Suppl 1, 21–30, doi: 10.1007/s00429-006-0119-8 (2006). [DOI] [PubMed] [Google Scholar]
- McMahon J. A. et al. Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev. 12, 1438–1452 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amthor H., Christ B. & Patel K. A molecular mechanism enabling continuous embryonic muscle growth - a balance between proliferation and differentiation. Development 126, 1041–1053 (1999). [DOI] [PubMed] [Google Scholar]
- Ericson J., Norlin S., Jessell T. M. & Edlund T. Integrated FGF and BMP signaling controls the progression of progenitor cell differentiation and the emergence of pattern in the embryonic anterior pituitary. Development 125, 1005–1015 (1998). [DOI] [PubMed] [Google Scholar]
- Wang H. et al. Bmp signaling at the tips of skeletal muscles regulates the number of fetal muscle progenitors and satellite cells during development. Developmental cell 18, 643–654, doi: 10.1016/j.devcel.2010.02.008 (2010). [DOI] [PubMed] [Google Scholar]
- Silvestrini R., Costa A., Veneroni S., Del Bino G. & Persici P. Comparative analysis of different approaches to investigate cell kinetics. Cell and tissue kinetics 21, 123–131 (1988). [DOI] [PubMed] [Google Scholar]
- Rudnicki M. A., Le Grand F., McKinnell I. & Kuang S. The molecular regulation of muscle stem cell function. Cold Spring Harbor symposia on quantitative biology 73, 323–331, doi: 10.1101/sqb.2008.73.064 (2008). [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy L. et al. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes & development 19, 1426–1431, doi: 10.1101/gad.345505 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori R. et al. BMP signaling controls muscle mass. Nature genetics 45, 1309–1318, doi: 10.1038/ng.2772 (2013). [DOI] [PubMed] [Google Scholar]
- Rommel C. et al. Differentiation stage-specific inhibition of the Raf-MEK-ERK pathway by Akt. Science 286, 1738–1741 (1999). [DOI] [PubMed] [Google Scholar]
- Miyake T. et al. Cardiotrophin-1 maintains the undifferentiated state in skeletal myoblasts. The Journal of biological chemistry 284, 19679–19693, doi: 10.1074/jbc.M109.017319 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olguin H. C. & Pisconti A. Marking the tempo for myogenesis: Pax7 and the regulation of muscle stem cell fate decisions. Journal of cellular and molecular medicine 16, 1013–1025, doi: 10.1111/j.1582-4934.2011.01348.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olguin H. C., Yang Z., Tapscott S. J. & Olwin B. B. Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination. The Journal of cell biology 177, 769–779, doi: 10.1083/jcb.200608122 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Eberspaecher H., Lefebvre V. & De Crombrugghe B. Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Developmental dynamics: an official publication of the American Association of Anatomists 209, 377–386, doi: (1997). [DOI] [PubMed] [Google Scholar]
- Piersma A. H. et al. Characterization of fibroblastic stromal cells from murine bone marrow. Experimental hematology 13, 237–243 (1985). [PubMed] [Google Scholar]
- Kuroda K. & Tajima S. HSP47 is a useful marker for skin fibroblasts in formalin-fixed, paraffin-embedded tissue specimens. J Cutan Pathol 31, 241–246, doi: DOI 10.1111/j.0303-6987.2003.00166.x (2004). [DOI] [PubMed] [Google Scholar]
- Tsutsumi N. et al. Essential role of PDGFRalpha-p70S6K signaling in mesenchymal cells during therapeutic and tumor angiogenesis in vivo: role of PDGFRalpha during angiogenesis. Circulation research 94, 1186–1194, doi: 10.1161/01.RES.0000126925.66005.39 (2004). [DOI] [PubMed] [Google Scholar]
- Huang H. et al. BMP signaling pathway is required for commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proceedings of the National Academy of Sciences of the United States of America 106, 12670–12675, doi: 10.1073/pnas.0906266106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q. Q. & Lane M. D. Adipogenesis: from stem cell to adipocyte. Annual review of biochemistry 81, 715–736, doi: 10.1146/annurev-biochem-052110-115718 (2012). [DOI] [PubMed] [Google Scholar]
- Clever J. L., Sakai Y., Wang R. A. & Schneider D. B. Inefficient skeletal muscle repair in inhibitor of differentiation knockout mice suggests a crucial role for BMP signaling during adult muscle regeneration. American journal of physiology. Cell physiology 298, C1087–C1099, doi: 10.1152/ajpcell.00388.2009 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta J. & Kessler J. A. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development 131, 4131–4142, doi: 10.1242/dev.01273 (2004). [DOI] [PubMed] [Google Scholar]
- Hu G., Lee H., Price S. M., Shen M. M. & Abate-Shen C. Msx homeobox genes inhibit differentiation through upregulation of cyclin D1. Development 128, 2373–2384 (2001). [DOI] [PubMed] [Google Scholar]
- Hughes S. M. Muscle development: reversal of the differentiated state. Current biology: CB 11, R237–R239 (2001). [DOI] [PubMed] [Google Scholar]
- Matzuk M. M. et al. Multiple defects and perinatal death in mice deficient in follistatin. Nature 374, 360–363, doi: 10.1038/374360a0 (1995). [DOI] [PubMed] [Google Scholar]
- Reshef R., Maroto M. & Lassar A. B. Regulation of dorsal somitic cell fates: BMPs and Noggin control the timing and pattern of myogenic regulator expression. Genes & development 12, 290–303 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y. et al. BMP signalling permits population expansion by preventing premature myogenic differentiation in muscle satellite cells. Cell death and differentiation 18, 222–234, doi: 10.1038/cdd.2010.95 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichs M. et al. BMP signaling balances proliferation and differentiation of muscle satellite cell descendants. BMC cell biology 12, 26, doi: 10.1186/1471-2121-12-26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprez D. M., Coltey M., Amthor H., Brickell P. M. & Tickle C. Bone morphogenetic protein-2 (BMP-2) inhibits muscle development and promotes cartilage formation in chick limb bud cultures. Developmental biology 174, 448–452, doi: 10.1006/dbio.1996.0087 (1996). [DOI] [PubMed] [Google Scholar]
- Biressi S., Molinaro M. & Cossu G. Cellular heterogeneity during vertebrate skeletal muscle development. Developmental biology 308, 281–293, doi: 10.1016/j.ydbio.2007.06.006 (2007). [DOI] [PubMed] [Google Scholar]
- Tylzanowski P., Verschueren K., Huylebroeck D. & Luyten F. Role of BMP receptors, Smads and Smad interacting protein Sip1 in the onset of osteogenesis in C2C12 cell line. Journal of Bone and Mineral Research 15, S239–S239 (2000). [Google Scholar]
- Li G. et al. Differential Effect of BMP4 on NIH/3T3 and C2C12 Cells: Implications for Endochondral Bone Formation. J.Bone Miner.Res. 20, 1611–1623 (2005). [DOI] [PubMed] [Google Scholar]
- Brazil D. P., Church R. H., Surae S., Godson C. & Martin F. BMP signalling: agony and antagony in the family. Trends in cell biology, doi: 10.1016/j.tcb.2014.12.004 (2015). [DOI] [PubMed] [Google Scholar]
- Katagiri T. et al. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. The Journal of cell biology 127, 1755–1766 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray S. S., Murray E. J., Glackin C. A. & Urist M. R. Bone morphogenetic protein inhibits differentiation and affects expression of helix-loop-helix regulatory molecules in myoblastic cells. Journal of cellular biochemistry 53, 51–60, doi: 10.1002/jcb.240530107 (1993). [DOI] [PubMed] [Google Scholar]
- Olguin H. C. Regulation of Pax7 protein levels by caspase-3 and proteasome activity in differentiating myoblasts. Biological research 44, 323–327, doi: /S0716-97602011000400002 (2011). [PubMed] [Google Scholar]
- Sassoon D. et al. Expression of two myogenic regulatory factors myogenin and MyoD1 during mouse embryogenesis. Nature 341, 303–307, doi: 10.1038/341303a0 (1989). [DOI] [PubMed] [Google Scholar]
- Brown D. M., Parr T. & Brameld J. M. Myosin heavy chain mRNA isoforms are expressed in two distinct cohorts during C2C12 myogenesis. Journal of muscle research and cell motility 32, 383–390, doi: 10.1007/s10974-011-9267-4 (2012). [DOI] [PubMed] [Google Scholar]
- Lee B. & Shao J. Adiponectin and lipid metabolism in skeletal muscle. Acta Pharmaceutica Sinica B 2, 335–340, doi: 10.1016/j.apsb.2012.06.008 (2012). [DOI] [Google Scholar]
- Bowers R. R., Kim J. W., Otto T. C. & Lane M. D. Stable stem cell commitment to the adipocyte lineage by inhibition of DNA methylation: role of the BMP-4 gene. Proceedings of the National Academy of Sciences of the United States of America 103, 13022–13027, doi: 10.1073/pnas.0605789103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A. et al. Brown fat determination and development from muscle precursor cells by novel action of bone morphogenetic protein 6. PloS one 9, e92608, doi: 10.1371/journal.pone.0092608 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W. et al. Schnurri-2 controls BMP-dependent adipogenesis via interaction with Smad proteins. Developmental cell 10, 461–471, doi: 10.1016/j.devcel.2006.02.016 (2006). [DOI] [PubMed] [Google Scholar]
- Dal-Pra S., Furthauer M., Van-Celst J., Thisse B. & Thisse C. Noggin1 and Follistatin-like2 function redundantly to Chordin to antagonize BMP activity. Dev Biol 298, 514–526, doi: 10.1016/j.ydbio.2006.07.002 (2006). [DOI] [PubMed] [Google Scholar]
- Shi S. et al. BMP antagonists enhance myogenic differentiation and ameliorate the dystrophic phenotype in a DMD mouse model. Neurobiology of disease 41, 353–360, doi: 10.1016/j.nbd.2010.10.003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijgerde M., Karp S., McMahon J. & McMahon A. P. Noggin antagonism of BMP4 signaling controls development of the axial skeleton in the mouse. Dev Biol 286, 149–157, doi: 10.1016/j.ydbio.2005.07.016 (2005). [DOI] [PubMed] [Google Scholar]
- McInnes K. J. et al. Deletion of the androgen receptor in adipose tissue in male mice elevates retinol binding protein 4 and reveals independent effects on visceral fat mass and on glucose homeostasis. Diabetes 61, 1072–1081, doi: 10.2337/db11-1136 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadinger S. E., Bucher N. L., Schreiber B. M. & Farmer S. R. PPARgamma2 regulates lipogenesis and lipid accumulation in steatotic hepatocytes. Am J Physiol Endocrinol Metab 288, E1195–E1205, doi: 10.1152/ajpendo.00513.2004 (2005). [DOI] [PubMed] [Google Scholar]
- Tung E. W., Boudreau A., Wade M. G. & Atlas E. Induction of adipocyte differentiation by polybrominated diphenyl ethers (PBDEs) in 3T3-L1 cells. PLoS One 9, e94583, doi: 10.1371/journal.pone.0094583 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D. F., Zhang X. G., Yang H. L. & Sun C. Differential expression of lipid metabolism genes in the liver and adipose tissue of mice treated with evodiamine. Genet Mol Res 12, 1501–1510, doi: 10.4238/2013.January.4.13 (2013). [DOI] [PubMed] [Google Scholar]
- Penna F. et al. Autophagic degradation contributes to muscle wasting in cancer cachexia. Am J Pathol 182, 1367–1378, doi: 10.1016/j.ajpath.2012.12.023 (2013). [DOI] [PubMed] [Google Scholar]
- Wong M. M. et al. Sirolimus stimulates vascular stem/progenitor cell migration and differentiation into smooth muscle cells via epidermal growth factor receptor/extracellular signal-regulated kinase/beta-catenin signaling pathway. Arterioscler Thromb Vasc Biol 33, 2397–2406, doi: 10.1161/ATVBAHA.113.301595 (2013). [DOI] [PubMed] [Google Scholar]
- Perry H. M. et al. Helix-loop-helix factor inhibitor of differentiation 3 regulates interleukin-5 expression and B-1a B cell proliferation. Arterioscler Thromb Vasc Biol 33, 2771–2779, doi: 10.1161/ATVBAHA.113.302571 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales A. M. & Orlando R. A. Role of adipocyte-derived lipoprotein lipase in adipocyte hypertrophy. Nutr Metab (Lond) 4, 22, doi: 10.1186/1743-7075-4-22 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A. P. & Wagers A. J. Pax3 induces differentiation of juvenile skeletal muscle stem cells without transcriptional upregulation of canonical myogenic regulatory factors. J Cell Sci 123, 2632–2639, doi: 10.1242/jcs.061606 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W. A. et al. NF-kappaB-mediated Pax7 dysregulation in the muscle microenvironment promotes cancer cachexia. J Clin Invest 123, 4821–4835, doi: 10.1172/JCI68523 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbagha N. G. et al. Alternative splicing in Acad8 resulting a mitochondrial defect and progressive hepatic steatosis in mice. Pediatr Res 70, 31–36, doi: 10.1038/pr.2011.25610.1203/PDR.0b013e31821b89ee (2011). [DOI] [PubMed] [Google Scholar]
- Kim E. J. et al. Ihh and Runx2/Runx3 signaling interact to coordinate early chondrogenesis: a mouse model. PloS one 8, e55296, doi: 10.1371/journal.pone.0055296 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z. et al. Epigenetic regulation of Thy-1 gene expression by histone modification is involved in lipopolysaccharide-induced lung fibroblast proliferation. J Cell Mol Med 17, 160–167, doi: 10.1111/j.1582-4934.2012.01659.x (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.