Abstract
Background
Non-alcoholic fatty liver (NAFLD) is a common, multi-factorial, and poorly understood liver disease whose incidence is globally rising. NAFLD is generally asymptomatic and associated with other manifestations of the metabolic syndrome. Yet, up to 25% of NAFLD patients develop a progressive inflammatory liver disease termed non-alcoholic steatohepatitis (NASH) that may progress towards cirrhosis, hepatocellular carcinoma, and the need for liver transplantation.
In recent years, several lines of evidence suggest that the gut microbiome represents a significant environmental factor contributing to NAFLD development and its progression into NASH. Suggested microbiome-associated mechanisms contributing to NAFLD and NASH include dysbiosis-induced deregulation of the gut endothelial barrier function, which facilitates systemic bacterial translocation, and intestinal and hepatic inflammation. Furthermore, increased microbiome-modulated metabolites such as lipopolysaccharides, short chain fatty acids (SCFAs), bile acids, and ethanol, may affect liver pathology through multiple direct and indirect mechanisms.
Scope of review
Herein, we discuss the associations, mechanisms, and clinical implications of the microbiome's contribution to NAFLD and NASH. Understanding these contributions to the development of fatty liver pathogenesis and its clinical course may serve as a basis for development of therapeutic microbiome-targeting approaches for treatment and prevention of NAFLD and NASH.
Major conclusions
Intestinal host–microbiome interactions play diverse roles in the pathogenesis and progression of NAFLD and NASH. Elucidation of the mechanisms driving these microbial effects on the pathogenesis of NAFLD and NASH may enable to identify new diagnostic and therapeutic targets of these common metabolic liver diseases.
This article is part of a special issue on microbiota.
Keywords: NAFLD, NASH, Microbiome, Liver
1. Introduction
Non-alcoholic fatty liver (NAFLD) is defined by pathologic accumulation of fat in the liver and is regarded the most common liver disease worldwide, with an estimated prevalence of around 25–30%. The prevalence of NAFLD is greatly increased in patients suffering from other pre-existing manifestations of the metabolic syndrome, such as obesity, type 2 diabetes, hyperlipidemia, and hypertension [1]. While NAFLD is generally asymptomatic, NAFLD patients feature an increased risk for development of other manifestations of the metabolic syndrome and accompanying complications such as cardiovascular diseases [2]. With that said, NAFLD can also occur in lean patients with normal BMI without abdominal obesity, and its prevalence is rapidly rising in countries such as India [3]. NAFLD and its associated manifestations were linked to elevated insulin resistance [3] and increased oxidized LDL to HDL ratio [4].
In up to 25% of NAFLD patients, the disease may evolve into a progressive form of liver disease named non-alcoholic steatohepatitis (NASH). NASH is defined as an inflammatory response to hepatic fat accumulation, resulting in chronic liver damage, scarring, and fibrosis. Continuous liver fibrosis may progress to cirrhosis, in which hepatocyte loss may lead to functional impairment [2]. Patients suffering of cirrhosis are predisposed to life risking complications including portal hypertension and increased risk for hepatocellular carcinoma [2]. Only limited options of pharmacotherapy are available for the treatment of NAFLD and NASH (vitamin K, metformin), and the advised treatment is a change in life style including weight reduction, enhanced exercise, and control of metabolic risk factors with glucose and lipid lowering agents [5], [6].
While the pathogenesis of NAFLD is unknown, it is believed to involve abnormal lipid metabolism associated with obesity and the metabolic syndrome. Risk factors contributing to NAFLD development and progression include dietary fat consumption, genetic predisposition, excess visceral adiposity, insulin resistance, elevated serum free fatty acids, and excessive pro-inflammatory mediators. Additional liver intrinsic factors include modified hepatic glucose metabolism, insulin resistance, and altered lipid metabolism. Together, these factors lead to hepatic steatosis and, in some cases, chronic hepatic inflammation, lipotoxicity, and hepatocyte damage, which may progress into chronic hepatitis and cirrhosis [2].
Multiple animal models have been developed for the study of NAFLD and NASH. All models contain features common to some, but not all, NAFLD and NASH human manifestations. Dietary models include high-fat diet (HFD), methionine-choline deficient diet (MCDD), and high-fructose diet, all leading to NAFLD development and progression in rodents. HFD leads to the development of many metabolic syndrome-associated manifestations including ectopic accumulation of fat in the liver, leading to steatosis and associated insulin resistance, but in the absence of liver inflammation. Choline-deficient diet leads to decreased levels of VLDL and hepatic beta oxidation, resulting in accumulation of liver fatty acids and cholesterol, an intense inflammatory reaction, but little or no insulin resistance [7], [8]. High-fructose diet induces steatosis, along with other metabolic abnormalities associated with this diet, such as weight gain, insulin resistance, and hyperlipidemia [9]. Genetic NAFLD models include mutations associated with NAFLD predisposition, such as the ones noted in the PNPLA3 gene [10]. It is worth noting that there are phenotypic differences between PNPLA3 associated mouse models (deletion, induced expression/insertion, and transgenic) highlighting a potential caveat of using genetic models to delineate mechanisms involved in human disease [11]. In homozygous carriers of PNPLA3, the prevalence of NAFLD is twofold higher as compared to non-carriers [12], [13]. Another NAFLD risk gene is phosphatidylethanolamine N-methyltransferase (PEMT), which is involved in phosphatidylcholine synthesis. PEMT deficient mice fed with MCDD feature severe hepatic steatosis [14] that is partially recovered by choline supplementation [15]. Interestingly, loss of function PMET mutations were also found in some NAFLD subjects [16].
2. The gut microbiome
Following the decoding of the human genome sequence [17], the visionary call for a second human genome project by Relman and Falkow [18] called for characterization of the genetic component of the microorganisms that colonize eukaryotes. A better understanding of microbial pathogenesis was a prelude to the realization of the importance of the microbiome to human physiology. Jeffrey Gordon and his group pioneered the understanding of factors affecting the structure of bacterial communities [19] and how microbial compositional structure may affect the risk of disease in mammals [20], [21]. The microbiota communities, of which the gut microbiota is the most extensively studied, were found to play a crucial role in many aspects of development, metabolism and physiology [22], [23], [24], [25]. Gut microbial composition is not homogeneous among individuals, and is characterized by a substantial inter-individual heterogeneity [26]. This represents a conundrum when searching for an association between disease and a deviation of bacterial community composition from the “normal” state. In a seminal study by the group of Jeffrey Gordon [27], a “core microbiome” was characterized on the basis of a particular gene and inferred metabolic pathway content, identified through metagenomics and parallel sequencing approaches [27]. Analyses encompassing characterization of microbiome composition predominantly on the basis of 16s rRNA sequence identity has indicated that the gut microbiome comprises over 1000 species of bacteria, with the most common Phyla including Firmicutes, Bacteroidetes, Actinobacteria and Proteobacteria [28]. Microbiota populations are compositionally dynamic, and its changes in microbial population structure can occur under multiple environmental, immune, and nutritional circumstances. Dysbiosis, in turn, can have profound effects on the host and has been associated with a number of human pathological conditions [27], [29], [30], [31], [32].
Germ free (GF) and antibiotic-treated mice models have become indispensable tools in determining the role and contribution of the microbiota to health and disease, including metabolic diseases such as NAFLD. GF mice are bred under sterile conditions, hence eliminating colonization by microorganisms. The procedure of generating these GF animals can be a challenge, technically and in terms of required infrastructure. In general, all steps in the process are designed to eliminate microorganism exposure. A donor pregnant female mouse is disinfected in a sterile hood and undergoes a C-section. The uterine sack is surgically removed, placed in liquid disinfectant, and transferred to a sterile GF isolator. In the isolator, the uterine sack is opened, and pups are removed and introduced to GF foster mothers. Following the initial procedure, the animals are raised within the sterile environment and are given sterile food and water. A more broadly applied alternative is microbiome depletion in mice through oral administration of wide spectrum antibiotics. In these models, introduction of specific communities or individual bacteria identified and isolated in healthy or in disease states into GF or antibiotic-treated mice is performed in order to study the microbiota contribution and function in normal host physiology and in disease progression [33], [34], [35].
Using metagenomics and GF platforms, significant associations have been made between compositional and functional alterations in the microbiome (termed dysbiosis) and the propensity to a variety of multi-factorial diseases, including obesity and its associated metabolic abnormalities such as NAFLD [21] in animal models and in humans. Indeed, in obesity and its associated metabolic complications, evidence of the microbiome as a contributing factor has been repeatedly featured [20], [36]. In these studies, HFD GF mice were found to gain less weight than conventional mice. Colonization of GF mice with microbiota from conventional mice resulted in replenished weight gain [20], [36]. Genetically obese mice and HFD mice had a shift in their gut microbial composition to one that is able to harvest dietary energy at a higher capacity [21]. This obesity phenotype was transmissible upon fecal transplantation into GF mice, resulting in significantly enhanced weight gain and total body fat as compared to GF mice receiving fecal transplantation of lean mice [21]. Proposed mechanisms for these effects include increased microbiome ability and efficiency for carbohydrate metabolism and production of short chain fatty acid [20], [21], [36], reduction in conjugated bile acids [35], and augmented systemic and adipose inflammation [37]. It is worth noting that similar changes were identified in the gut microbiota of obese humans [38], although some of the results were conflicting among different studies [39], [40].
3. Associations between dysbiosis and NAFLD
Portal blood flow constitutes an important link between the intestine and liver, with the majority of the liver blood supply derived from the intestine. The intestinal blood supply exposes the liver to a multitude of intestinal metabolites and food products [41]. In recent years, evidence suggested an involvement of the microbiota in NAFLD development [42]. An indication of this involvement may have come as early as 1982 when Drenick et al. [43] studied hepatic steatosis development in patients undergoing gastric bypass surgery that coincided with bacterial overgrowth. In this early study, a regression in hepatic steatosis was noted when patients were treated with the antibiotic metronidazole, suggesting a potential role of the gut microbial community in fatty liver development. Subsequently, small intestinal bacterial overgrowth has been shown to be more prevalent in patients with NASH than in healthy controls [44]. An accumulating number of studies in animal models and humans have followed to broaden our understanding of the microbiota role in the development and pathogenesis of NAFLD [42]. In the interest of clarity towards the taxonomic organization of bacteria described in this review, Table 1 illustrates the major categories of relevant microbiota. This table is not intended to provide a comprehensive taxonomic organization, but rather an indicative one relevant to the studies described herein.
Table 1.
Taxonomic representation of major bacterial categories of microbiota, from phylum to species. Bacteria referred to in the text are indicated in bold.
| Phylum | Class | Order | Family | Genus | Species |
|---|---|---|---|---|---|
| Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | Bacteroides vulgatus |
| Bacteroides fragilis | |||||
| Bacteroides acidifaciens | |||||
| Barnesiellaceae | Barnesiella | Barnesiella intestinihominis | |||
| Porphyromonadaceae | Paludibacter | Paludibacter propionicigenes | |||
| Parabacteroides | Parabacteroides distasonis | ||||
| Parabacteroides eggerthii | |||||
| Prevotellaceae | Prevotella | Prevotella copri | |||
| Prevotella brevis | |||||
| Cytophagia | Cytophagales | Cytophagaceae | Emticicia | Emticicia oligotrophica | |
| Flavobacteria | Flavobacteriales | Flavobacteriaceae | Capnocytophaga | Capnocytophaga canimorsus | |
| Flavobacterium | Flavobacterium denitrificans | ||||
| Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus | Bacillus cereus |
| Staphylococcaceae | Staphylococcus | Staphylococcus aureus | |||
| Lactobacillales | Aerococcaceae | Aerococcus | Aerococcus viridans | ||
| Enterococcaceae | Enterococcus | Enterococcus avium | |||
| Lactobacillaceae | Lactobacillus | Lactobacillus acidophilus | |||
| Lactobacillus reuteri | |||||
| Pediococcus | Pediococcus acidilactici | ||||
| Clostridia | Clostridiales | Christensenellaceae | Christensenella | Christensenella minuta | |
| Oscillospiraceae | Oscillibacter | Oscillibacter valericigenes | |||
| Clostridiaceae | Clostridium | Clostridium difficile | |||
| Anaerosporobacter | Anaerosporobacter mobolis | ||||
| Lachnospiraceae | Blautia | Blautia coccoides | |||
| Lachnobacterium | Lachnobacterium bovis | ||||
| Lachnospira | Lachnospira multipara | ||||
| Ruminococcaceae | Faecalibacterium | Faecalibacterium prausnitzii | |||
| Oscillospira | Oscillospira valericigenes | ||||
| Ruminococcus | Ruminococcus albus | ||||
| Syntrophomonas | Syndrophomonas palmitatica | ||||
| Veillonellaceae | Allisonella | Allisonella histaminiformans | |||
| Anaerovibrio | Anaerovibrio lipolytica | ||||
| Megamonas | Megamonas hypermegale | ||||
| Erysipelotrichia | Erysipelotrichales | Erysipelotrichaceae | Bulleidia | Bulleidia extructa | |
| Coprobacillus | Coprobacillus cateniformis | ||||
| Actinomyces | Actinomyces hyovaginalis | ||||
| Actinobacteria | Actinobacteria | Actinomycetales | Actinomycetaceae | Actinobaculum | Actinobaculum massiliense |
| Micrococcaceae | Acaricomes | Acaricomes phytoseiuli | |||
| Bifidobacteriales | Bifidobacteriaceae | Bifidobacterium | Bifidobacterium bifidum | ||
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Rhizobiaceae | Rhizobium | Rhizobium agrobacterium |
| Rhodobacterales | Rhodobacteraceae | Amaricoccus | Amaricoccus macauensis | ||
| Rickettsiales | Rickettsiaceae | Rickettsia | Rickettsia conorii | ||
| Betaproteobacteria | Burkholderiales | Alcaligenaceae | Brackiella | Brackiella oedipodis | |
| Neisseriales | Comamonadaceae | Comamonas | Comamonas composti | ||
| Neisseriaceae | Aquaspirillum | Aquaspirillum bengal | |||
| Gammaproteobacteria | Aeromonadales | Succinivibrionaceae | Ruminobacter | Ruminobacter amylophilus | |
| Enterobacteriales | Enterobacteriaceae | Escherichia | Escherichia coli | ||
| Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | Desulfovibrio | Desulfovibrio aminophilus | |
| Epsilonproteobacteria | Campylobacterales | Campylobacteraceae | Campylobacter | Campylobacter coli | |
| Helicobacteraceae | Helicobacter | Helicobacter pylori |
3.1. Animal studies
Multiple studies in animal models suggested that the gut microbiome might play a role in the pathogenesis of NAFLD [45], [46], [47], [48]. Of note, one of these studies demonstrated that compared to conventionalized mice, GF mice administered HFD are resistant to hepatic steatosis and dyslipidemia, while displaying improved glucose tolerance with enhanced insulin sensitivity [45]. A direct involvement of the microbiome in NAFLD development was indicated by the observation that NAFLD is transmissible to GF mice upon fecal microbiome transplantation [46]. In this study, C57BL/6J mice fed with HFD featured a variable metabolic response to this diet. The first group (designated “responders”) developed characteristics of metabolic syndrome and NAFLD. However, some mice (designated “non-responders”) did not display this metabolic phenotype upon exposure to HFD and did not develop hyperglycemia, systemic inflammation or liver steatosis. Colonization of intestinal microbiota from the responder and non-responder groups into GF mice resulted in a propensity for NAFLD coupled with a detectable increase in expression of genes involved in lipogenesis only in the responder group [46]. Furthermore, microbiota population characterization by 16s rRNA gene analysis demonstrated a unique bacterial population profile in the responder as compared to the non-responder group. In particular, two bacterial species, Barnesiella intestinihominis and lachnospiraceae, were identified to be overrepresented in the GF mice colonized with responder microbiome, suggesting an associated role in the developed phenotype. Conversely, the bacterium Bacteroides vulgatus was found to be overrepresented in the GF mice colonized by non-responders suggesting a potential protective effect. Overall, this strategy suggested a direct involvement of the microbiota in NAFLD development.
3.2. Human studies
Dietary habits are a strong determinant of gut microbial composition [49], [50] and are linked to the metabolic syndrome and its associated diseases. While no single bacterium has been mechanistically associated with the development of steatosis, human studies have suggested that a dysbiotic environment exists in NAFLD patients [51]. Most studies have focused on identifying differences in the bacterial community composition between healthy individuals and NAFLD patients. Michail et al. [52] described microbial alterations in obese pediatric patients with NAFLD, as compared with obese children without NAFLD and lean healthy children. Taxonomic characterization of bacteria in feces of these subjects was carried out through 16s rRNA gene analysis using a microarray analysis approach. Children with NAFLD featured a higher representation of Gammaproteobacteria and Epsilonproteobacteria than healthy lean and obese children [52]. At the genus level, children with NAFLD had a greater presence of Prevotella as compared to healthy controls (Table 2). Metagenomic analysis by shotgun sequencing suggested that, compared to healthy subjects, children with NAFLD had a greater number of pathways involved in energy metabolism and lipid synthesis, possibly pointing towards a microbiome with more efficient energy metabolism capabilities in NAFLD patients. This was further validated through a metaproteomics analysis that identified proteins involved in energy metabolism (e.g. NAD-dependent aldehyde dehydrogenase) that were highly expressed in NAFLD patients [52]. In another study, Spencer et al. [53] investigated the microbiota contribution to NAFLD development under low-choline diet conditions. Patients were exposed to 10 days of normal diet (baseline) followed by 42 days on a choline-depleted diet, to initiate fatty liver, and then returned to a normal diet for 10 more days. Patients were analyzed for gut microbiota compositional changes by multiple time-point stool collections, accompanied by assessment of liver fat accumulation and characterization of host PEMT mutations associated with NAFLD. Changes in microbiota composition during choline-depleted diet were evident mostly at the class level, while a higher abundance of baseline Gammaproteobacteria correlated with lower risk of developing fatty liver (Table 2). In contrary, higher abundance of baseline Erysipelotrichia correlated with a higher risk of developing fatty liver [53]. Moreover, host PEMT genotype data as well as abundance of Gammaproteobacteria and Erysipelotrichia were combined into a linear model that accurately predicted the degree of NAFLD development. Other studies similarly pursued a fecal bacterial community characterization strategy using 16s rRNA gene sequencing and showed an increase in the level of some Firmicutes phyla belonging to Lactobacillus (Table 2) among others and a reduction in the level of other Firmicutes phyla belonging for example to Oscillibacter [54], [55].
Table 2.
Human studies investigating microbiota involvement in the development of NAFLD and NASH.
| Study | Technique | Groups | Samples | Main findings | |
|---|---|---|---|---|---|
| NAFLD | Michail et al., 2015 [52] | 16s rRNA Microarray microbial community profiling | 13 obese children with NAFLD 11 obese children without NAFLD 26 healthy children |
Stool | Obese children with NAFLD: ↑ Gammaproteobacteria ↑ Epsilonproteobacteria ↑ Prevotella |
| Spencer et al., 2011 [53] | 16s rRNA V1–V2 region sequence analysis | 15 individuals: 10 days normal diet (baseline), then 42 days choline-depleted diet. Back to 10 days normal diet |
Multiple stool samples from multiple time points | Baseline samples: ↑ Gammaproteobacter at baseline correlates to lower risk of developing fatty liver on low-choline diet. ↑ Erysipelotrichia at baseline correlates to higher risk of developing fatty liver on low-choline diet. |
|
| Raman et al., 2013 [55] | 16s rRNA V1–V2 region sequence analysis | 30 obese NAFLD patients 30 healthy controls |
stool | Obese NAFLD versus healthy controls: ↑ Lactobacillus ↓ Firmicutes ↓ Oscillibacteria |
|
| NASH | Zhu et al., 2013 [56] | 16s rRNA V4–V5 region sequence analysis | 22 NASH children 25 obese children 16 healthy controls |
stool | Obese and NASH versus Healthy controls: ↑ Bacteroidetes ↑ Prevotella NASH versus obese and healthy controls ↑ Proteobacter ↑ Enterobacteriaceae ↑ Escherichia |
| Wong et al., 2013 [136] | 16s rRNA V1–V2 region sequence analysis | 16 NASH patients 22 Healthy controls |
stool | NASH versus healthy controls: ↓ Firmicutes No Change – Bacteroidetes ↑ Parabacteroides ↑ Allisonella ↓ Faecalibacterium ↓ Anaerosporobacter |
|
| Boursier et al., 2016 [59] | 16s rRNA V4 region sequence analysis | 22 NAFLD patients 35 NASH patients |
stool | NASH versus NAFLD: ↑ Bacteroidetes |
A number of studies focused on microbiota alterations in NASH development. A pediatric study by Zhu et al. [56] recruiting obese and NASH patients found that the relative abundance of Bacteroidetes, predominantly Prevotella, increases in obese and NASH as compared to lean children (Table 2). Furthermore, this study revealed an elevated presence of alcohol-producing bacteria in NASH as compared to obese patients without NASH, suggesting a possible role for the elevated concentration of alcohol produced by the bacteria as discussed below [56]. Specifically, an increased abundance of the phylum Proteobacteria was observed in NASH patients compared to obese individuals. Within Proteobacteria, the family Enterobacteriaceae and genus Escherichia were found to be at a higher relative abundance in NASH patients compared to obese individuals (Table 2). Mouzaki et al. [57] performed a quantitative real time-PCR approach to estimate total bacterial concentrations in stool from healthy individuals and NASH patients. Their analysis found a lower percentage of Bacteroidetes to be present in NASH patients as compared to healthy controls (Table 2). The differences were independent of diet or body mass index (BMI). Interestingly, these observations were contradictory to those reported by Zhu et al. [56], in which a higher abundance of Bacteroidetes was found in NASH patients. A study by Wong et al. [58] explored the potential dysbiosis in NASH patients as compared to healthy controls and found a decrease in Firmicutes abundance in patients as compared to controls, with no changes noted in Bacteroidetes levels between the two groups. At the level of bacterial genera, the study showed an increase in Parabacteroides and Allisonella and a decrease in Faecalibacterium and Anaerosporobacter in NASH patients as compared to controls (Table 2). A study by Boursier et al. [59] indicated that the involvement of gut microbiota in disease severity progression from NAFLD to NASH may contribute to liver fibrosis and cirrhosis [59]. In this study, a 16s rRNA gene characterization approach was coupled with metagenomics analysis in predicting the metagenomics composition and functionality [60]. An association was found between increased Bacteroides concentrations and NASH development (Table 2), and an increase in Ruminococcus concentrations and risk of liver fibrosis [59].
4. Mechanisms for gut microbiota effect on NAFLD
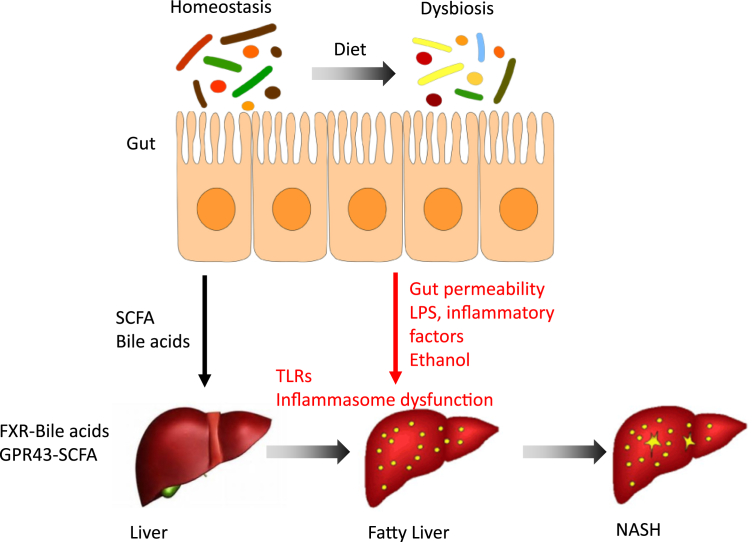
Several mechanisms have been suggested for the microbiome role in NAFLD and its complications. These include microbiome-induced regulation of gut barrier and inflammatory responses and metabolites produced or modified by the microbiota such as SCFAs, bile acids, and ethanol (Figure 1).
Figure 1.
Suggested mechanisms for the effect of gut microbiome in NAFLD development and progression to NASH. Bacterial metabolites such as SCFA and bile acids may be potently involved in normal liver function and reduced liver lipogenesis and inflammation. Aberrations in commensal microbiome composition, diversity, and function may lead to increased gut permeability, production of LPS and other inflammatory factor, recued diversity of bile acids, and production of ethanol. All these metabolites and factors in combination with lipids derived from the diet can cause liver steatosis, inflammation and damage, which may lead to hepatic fibrosis, scarring, and NASH development.
4.1. Intestinal barrier dysfunction
The gastrointestinal tract mucosal epithelia form a mechanistic barrier that prevents the trillions of commensal microorganisms from entering the sterile host milieu, where they may activate a systemic immune response. This is achieved through the intestine unique mechanical structure as well as its complex mucosal immunological components [61]. Mechanical structural components include tight junctions that inter-connect adjacent epithelial cells and are involved in regulation of intestinal permeability [62]. The intestinal mucosal immune system consists of a complex network of innate and adaptive cell populations [62], [63]. The cross talk between microbiota components and the immune system is important in tolerance establishment on the surface of the intestinal mucosa and also in maintaining the gut epithelial barrier function [64]. Taken together, a delicate balance is established that maintains intestinal functionality (for example nutrient and water absorptive capability) while preventing a non-specific immunological response against invasive commensal microbes. Dysregulation of the gastrointestinal immune epithelial network can disrupt tight junction functionality and lead to a “leaky gut” facilitating bacterial translocation [62].
A number of liver diseases including NAFLD [65], [66] and other metabolic syndrome manifestations [67], [68], [69] were suggested to be associated with increased gut permeability [70]. The association between increased gut permeability and human NAFLD was first shown by Miele et al. [65]. Immunohistochemistry assays indicated that increased permeability was linked to dysregulation of epithelial tight junction formation. Furthermore, small intestinal bacterial overgrowth was associated with NAFLD in this study, suggesting a microbial contribution to NAFLD pathology through increased bacterial translocation [65]. Another study utilized HFD mice with dextran sulfate sodium (DSS)-induced colitis as means of impairing gut barrier integrity during generation of hepatic steatosis [71]. As compared to hepatosteatosis in HFD mice, HFD DSS-administered mice aggravated hepatosteatosis and fibrosis accompanied with hepatic inflammation, suggesting a role of barrier dysfunction in disease severity and NASH development. Portal endotoxin levels were elevated in HFD mice but more significantly elevated in HFD DSS-administered mice, suggesting a possible enhanced bacterial translocation to contribute to NAFLD severity [71]. Recently, Luther et al. [72] performed a meta-analysis comprehensively assessing the association between intestinal permeability and risk of developing NAFLD and progressing into NASH. Indeed, patients with NAFLD and NASH were more likely to display enhanced intestinal permeability (Figure 1). To mechanistically understand the association between gut permeability and NAFLD development, clinical data were correlated with observations made in mice fed with methionine-choline deficient diet (MCDD) to induce NAFLD. Interestingly, in the MCDD model, liver damage was found to precede enhancement in gut permeability. The authors suggest that the observed hepatic injury may contribute to permeability defects through mechanisms that remain elusive [72].
4.2. Inflammation
Pathogens, such as bacteria and viruses, are recognized through specialized recognition receptors that include toll-like receptors (TLRs), inflammasome forming and non-forming NOD-like receptors and C type lectin receptors [73]. TLR signaling is activated upon pathogen and tissue damage recognition inducing a signaling cascade leading to production of inflammatory cytokines [74]. Additionally, pathogen and damage-associated molecules may induce the formation of a cytoplasmic multi-protein complex termed the inflammasome, which may consist of nod-like receptors (NLRs) and ASC (PYCARD), promoting proximity cleavage of Caspase-1 and catalytic activation of IL-1β and IL-18, as well as a specialized cell death termed pyroptosis. Other non-canonical inflammasomes induced by LPS cause Caspase-11 cleavage leading to IL-1α processing, pyroptosis, and further activation of the canonical inflammasomes. Inflammasome signaling has been suggested to either contribute or ameliorate fatty liver. NAFLD development demonstrated in MCDD mice led to hepatic induction of the inflammasome activation, leading to IL-1β secretion, induction of liver inflammation, and fibrosis [75], [76]. Inflammasome dysfunction or deficiency results in aggravated hepatic inflammatory response, liver damage, fibrosis and cell death [47], [76]. A role of the NLRP3 inflammasome has been suggested in NAFLD development and progression to liver fibrosis and NASH as demonstrated in NLRP3 deficient mice fed with MCDD featuring enhanced propensity for NAFLD and liver damage as compared to WT controls [76]. A recent study by Yang et al. [77] demonstrated that oral administration of sulforaphane to long term HFD mice alleviates liver steatosis by inhibition of NLRP3 inflammasome signaling. The possible association between inflammasome activation and NAFLD development and progression may be explained by hepatic influx of saturated fatty acids and LPS that are abundantly found in MCDD and HFD mice that may induce NLRP3 inflammasome activation [75]. However, this activation was mainly demonstrated in primary hepatocytes and hepatic cells lines with the in vivo relevance remaining unclear.
In other studies, our group has demonstrated a link between intestinal inflammation driven by gastro-intestinal alterations in NLRP6 inflammasome signaling and a risk for enhanced NAFLD progression [47]. Strikingly, these hepatic effects were mediated by NLRP6 modulation of the gut microbiota. Inflammasome-deficient mice displayed changes in their gut microbiota composition and aggravated hepatic steatosis driven by massive influx of TLR4 and TLR9 agonists into the portal circulation, leading to increased hepatic TNFα secretion and resultant hepatic damage and inflammation. The importance of the crosstalk between gut microbes and host in NAFLD was highlighted in inflammasome-deficient mice co-housed with MCDD or obese mice, leading to microbial transfer from inflammasome-deficient mice to co-housed recipient mice, the later developing an exacerbated NAFLD phenotype. Taken together, alterations of host and gut microbiome interactions through defective inflammasome sensing, disrupted inflammatory response, and dysbiosis play a pivotal role in hepatic steatosis and its progression to NASH (Figure 1).
4.3. Metabolites
Metabolites produced, degraded, or modulated by gut commensals are considered pivotal components of the communication networks between the host and its microbiota [78], [79]. Consequently, bacterial metabolites may contribute to disease development and progression as demonstrated in several disease models including colitis [80] and metabolic syndrome related disorders [81].
4.3.1. Lipopolysaccharides
One of the most studied microbial-modulated components that affect host pathology through pattern recognition systems is lipopolysaccharide (LPS). LPS, also known as endotoxin, is a component of gram-negative bacterial cell wall. LPS consists of three main parts: Lipid A, a core oligosaccharide and O side chain [82], which mainly trigger pro-inflammatory responses leading to the activation of an immune response. Bacterial LPS is a ligand for the LPS-binding protein, which, in turn, interacts with CD14 located on hepatic cells including Kupffer cells in the lining sinusoids. Kupffer cells are mononuclear, phagocytic cells involved in responding to pathogens and contribute to development of inflammation in liver injury and in NAFLD progression [83]. CD14 is associated with TLR4 on the cell surface and this interaction results in a cascade of inflammatory events, leading to secretion of pro-inflammatory cytokines and generation of oxidative stress [84], [85]. Modestly but chronically, elevated amounts of LPS, ranging up to around 10 times higher than healthy controls, were detected in metabolic syndrome mice models and termed as “metabolic endotoxemia” [86]. Specifically, mice models of NAFLD showed elevated levels of portal LPS with increased levels of TLR4 and CD14 expression in hepatic Kupffer cells [87]. HFD mice and genetically obese leptin deficient mice develop metabolic endotoxemia [88], indicated by an increase in LPS that was associated with insulin resistance and increased fat deposition. Metabolic endotoxemia was ameliorated by administering antibiotics to the mice, resulting in major changes in microbial community structure coinciding with reduction in body weight gain and inflammation, indicating the possible involvement of gut microbiota in metabolic endotoxemia [88].
The association between endotoxemia and NAFLD in humans was characterized by determining the serum levels of endotoxin and other inflammatory markers, correlating them to clinical manifestations. A recent prospective study showed an association between elevated levels of endotoxin markers and risk of NAFLD development [58]. In a study by Harte et al. [89], serum endotoxin was elevated in patients with NAFLD (Figure 1), which correlated with an increase in the inflammatory markers soluble CD14 and TNFRII. Mehta et al. [90] induced endotoxemia in healthy adults by intravenous administration of LPS and found that it induced insulin resistance, a phenotype closely associated with NAFLD [2]. However, other reports did not reveal an association between endotoxemia and NAFLD/NASH development and suggesting that endotoxemia may not be the sole driver of disease progression in all patients [56].
4.3.2. Short-chain fatty acids
Western diets are typically high in carbohydrates, and up to an estimated 20–60 g of carbohydrates reach the colon on a daily basis, where they can undergo fermentation by the gut microbial populations. Short chain fatty acids (SCFA), including acetate, propionate, and butyrate, are produced by bacterial fermentation of polysaccharides [91]. SCFA involvement in NAFLD development and pathogenesis may derive from their potential contribution to the maintenance of body weight, intestinal homeostasis, and improved metabolism of glucose and lipids [92], [93], [94]. Turnbaugh et al. [21] showed that the cecal content of obese mice is enriched in SCFA. Similar results were reported in obese compared to lean individuals, where elevated SCFA concentration, in particular propionate, is correlated with a higher BMI [39]. Thus, it has been proposed that SCFAs regulate the development of NAFLD, although the role of SCFA in NAFLD/NASH development and pathogenesis remains unclear.
Butyrate and propionate bind the G-protein coupled receptors GPR41 (FFAR3), GPR43 (FFAR2), and GPR109A that are mainly expressed in colonic epithelium, adipose tissue, liver, and pancreatic beta cells. Mice lacking GPR43 and fed with HFD gained more weight and showed increased adiposity, fatty liver, and insulin resistance, whereas GPR43 overexpression in adipose tissues exhibited no change in weight gain in response to HFD and no evident signs of liver steatosis [95]. Notably, GF conditions or administration of antibiotics abolished all the metabolic syndrome-related phenotypes of GPR43 null mice, including dyslipidemia and fatty liver, indicating that the gut microbiota are required for GPR43 function probably due to bacterial synthesis of SCFAs, which are GPR43 agonists [95]. GPR43–SCFA interactions play a central role in suppression of inflammatory responses in models of colitis, arthritis and asthma [96]. GF mice showed exacerbated inflammatory processes in these disease models that were ameliorated by SCFAs [96]. Since inflammation drives NAFLD progression into NASH, GPR43 signaling may be involved in regulating liver inflammation and NAFLD progression. Additionally, SCFA–GPR43 interaction in the gut may help in maintaining normal intestinal permeability while suppressing mucosal inflammation. Hence, it may limit hepatic damage caused by microbial products and dysbiosis (Figure 1). However, a direct link between GPR43 role as an inhibitor of inflammatory responses and NAFLD development and its related pathologies has not been established to date.
4.3.3. Bile acids
Bile acids derived from hepatic cholesterol catabolism and de novo synthesis are conjugated and transported into the gallbladder. Postprandial contraction of the gallbladder drives bile acids into the intestinal lumen [97], [98], [99]. In addition to their role in facilitating dietary fat digestion, bile acids are now recognized as important regulators of lipid metabolism, energy and glucose homeostasis [100].
Gut microbial enzymes can transform the primary bile acids into conjugated bile acids, facilitating dietary fats digestion and absorption through formation of micelles. Several groups have shown that GF mice or antibiotic treated mice had low concentrations of conjugated bile acids, pointing to a central role of gut microbiota in regulating bile acid composition, conjugation, and diversity [101], [102], [103], [104]. Bile acids bind the nuclear receptor farnesoid X receptor (FXR, also known as NR1H4), which is a transcription factor that controls their endogenous synthesis and release, as well as other metabolic functions by directed changes in transcriptional gene expression [105], [106]. In the intestine, FXR binds bile acids, resulting in the activation of its target gene fibroblast grown factor 15 (FGF15). In turn, FGF15 inhibits the expression of hepatic cholesterol 7-α-hydroxylase (Cyp7a1), a rate-limiting enzyme in bile acid biosynthesis [103]. Obese and insulin resistant mice show decreased gut microbiota diversity, accompanied by a reduction in bile acids composition and abundance, increased FXR and FGF15 expression in the ileum and decreased hepatic Cyp7a1 [35].
In the liver, bile acids directly bind to FXR leading to suppression of bile acids synthesis, composition and pool size. Moreover, FXR-bile acid interaction in the liver contributes to liver regeneration [107], reduces accumulation of fat in the liver, and improves glucose and cholesterol metabolism [108], [109]. Accordingly, activation of hepatic FXR by bile acids agonists was found to be beneficial for reducing liver steatosis and rescuing liver damage. In one recent study, the FLINT trial [110], the FXR agonist, obeticholic acid (OCA) showed clear beneficial effects in alleviating NAFLD activity including accumulation of lipids, liver inflammation, and injury. Some improvement in fibrosis was detected in patients receiving OCA but more statistical power is needed to determine its effect on more severe conditions such as NASH and advanced fibrosis. Thus, activation of hepatic FXR represents an interesting therapeutic candidate that is currently being tested for treatment of NASH in clinical trials [110], [111].
The complex effect of bile acids on metabolic homeostasis is demonstrated using FXR deficient mice (also named as Nr1h4−/− mice). The initial characterization of FXR null mice fed with high cholesterol diet showed elevated hepatic lipids and steatosis with increased cholesterol, triglycerides and fatty acids [112], [113]. Thus suggesting a positive role of FXR activation in maintaining lipid homeostasis and protecting from hepatic steatosis. In concert with this suggestion, treatment of HFD mice with FXR agonists led to a significant reduction in liver steatosis and plasma triglycerides and cholesterol [114], [115]. This reduction in the systemic lipid profile can be explained in part by FXR induction of hepatic genes involved in lipoprotein clearance. These genes include the HDL receptor Scrab1, VLDL receptor, and ApoCII, a cofactor of lipoprotein lipase. FXR also decreases hepatic SREBP-1c, a transcription factor required for fatty acids and triglyceride synthesis [109]. The primary, but not sole, site for FXR activation is the ileum. Other tissues such as liver and kidneys, and possibly adipose tissues, are also activated [116]. To dissect the tissue-specific function of FXR-bile acid in the gut, intestinal-specific FXR deficient mice [117], [118],were fed with HFD. These intestinal FXR deficient HFD mice, but not liver-specific-FXR deficient mice [112], had lower hepatic triglycerides and reduced liver steatosis as compared to controls [118]. In a complementary approach, two recent studies used HFD mice receiving an orally synthetic FXR agonist. This agonist is poorly absorbed into the circulation, resulting in intestinally restricted FXR activation [117], [119]. As a result, the mice had significant lower liver steatosis and reduced expression of genes involved in hepatic lipogenesis and lower ceramides levels along with other improvements in metabolic homeostasis [117], [119]. Together, all these studies suggest that inhibition of FXR activation in the gut can be used as a therapeutic approach to alleviate liver steatosis and hepatic lipogenesis.
Several groups utilizing GF and antibiotic-treated mice showed that gut microbiota not only regulate bile acid composition and diversity but also modulate FXR and FXR-related genes, including hepatic Cyp7a1 and intestinal FGF15, thus controlling bile acid synthesis [101], [102], [103], [104]. The association between FXR-bile acids, the gut microbiome, and metabolic homeostasis was further demonstrated by Ryan et al. [120], who showed that the beneficial effects of bariatric surgery on metabolism were associated with changes in gut microbiota and diminished in FXR-deficient mice. Together, these studies point towards a dominant role of the gut microbiota in regulating bile acids diversity via FXR signaling, which, in turn, regulates obesity and its related metabolic manifestations including NAFLD (Figure 1). A recent study by Mouzaki et al. [121] shows that NASH patients had a reduction in the secondary bile acids pool and in fecal levels of Bacteroidetes and Clostridium leptum. However, a direct role of the gut microbiome in controlling FXR-bile acids signaling in NAFLD and NASH development has not been elucidated. Recently, our group has shown that the conjugated bile acid taurine alters the microbiome composition, leading to activation of NLRP6 inflammasome, secretion of anti-microbial peptides, and protection from colitis [80]. Therefore, it will be interesting to test the effect of taurine on the gut microbiota in NAFLD development and NASH progression.
4.4. Ethanol
One of the mechanisms suggested for the association between NAFLD and dysbiosis could include microbial production of ethanol as a possible liver toxin. Zhu L. et al. [56] examined gut microbial composition and ethanol levels in the blood of NASH, obese, and healthy children. Only a few differences were evident in the gut microbiome composition of NASH as compared with obese patients and included differences across phyla, families, and genera in Proteobacteria, Enterobacteriaceae, and Escherichia, respectively. Some of these microbiome changes included alcohol-producing bacteria, and, accordingly, a significant increase in ethanol levels were found in NAFLD subjects as compared to both obese and healthy children. Furthermore, increased levels of ethanol were detected in correlation with NASH [122]. These results suggest that production of ethanol by the gut microbiota may serve as a hepatotoxin, contributing to development of NAFLD and its progression to NASH (Figure 1).
5. Microbiome-based treatments for NAFLD
The etiology of NAFLD has not been clearly elucidated; therefore, treatment options remain limited and disappointing. Treatment approaches for NAFLD patients most commonly involve intensive lifestyle modifications including recommendations to enhance physical exercise and perform dietary modifications [123], [124]. Additional treatments include anti-inflammatory drugs, anti-oxidants, lipid-lowering agents, and insulin sensitizers, as well as supplements such as vitamins [125]. Recently, attention has been drawn towards developing treatments targeting the microbiome in NAFLD patients. Microbiome manipulation is most commonly carried out by antibiotics, prebiotics, probiotics, and fecal microbiota transplantation (FMT).
5.1. Antibiotics
Only a few trials in rodents and humans with NAFLD have been performed utilizing antibiotics, since their long-term use may result in predisposition to side-affects stemming from elimination of functionally important commensal bacteria and emergence of resistant strains [126]. Oral antibiotics can dramatically affect the gut microbial configuration [126], [127] and may modify microbial drivers of NAFLD. Administration of the antibiotic polymyxin B and neomycin to high fructose diet-fed mice led to a reduction in hepatic lipid accumulation [128], [129]. A recent study shows that HFD mice treated with a combination of three antibiotics (bacitracin, neomycin, and streptomycin) for four months exhibited a significant reduction in liver triglycerides and lipid accumulation as well as serum ceramides production [118]. However, contradictory evidence exists on the efficacy of antimicrobial treatment for fatty liver disease in humans. For example, in one study, six months of alternating Norfloxacin and neomycin treatment decreased small intestinal bacterial overgrowth and improved liver function of patients with liver cirrhosis [130]. Conversely, another study measuring inflammatory parameters of NAFLD patients found no effect of Norfloxacin treatment on these immune biomarkers [131].
5.2. Probiotics
As defined by the World Health Organization, probiotics are “live microorganisms which when administered in adequate amounts confer a health benefit to the host”. While the efficacy of probiotics was only demonstrated in a few diseases, popularity of its consumption is mounting as an empirical means of disease prevention and enhanced well-being [132]. While more than one type of probiotic was studied for NAFLD treatment, most of these treatments included combinations of Bifidiobacteria and Lactobacilli [133].
A decade ago, Loguercio et al. [134] and Li et al. [135] described that probiotic treatment improved some parameters of liver damage. Loguercio et al. [134] showed that NAFLD patients treated with a mixed-species probiotic treatment (Bifidiobacterium and Lactobacillus) had improved serum alanine aminotransferase activity (ALT) and reduced markers of oxidative stress and of the inflammatory cytokine TNFα [134]. Li et al. [135] used genetically obese (ob/ob) mice fed on HFD and treated with either anti-TNF antibodies or the probiotic VSL #3. Both treatments showed comparable improvement in hepatic histology, decreased serum ALT activity, and improved hepatic insulin resistance [135]. VSL #3 is a probiotic combination of eight bacterial species (Bifidobacterium breve, Bifidobacterium longum, Bifidobacterium infantis, Lactobacillus acidophilus, Lactobacillus plantarum, Lactobacillus paracasei, Lactobacillus bulgaricus, and Streptococcus thermophiles) that has become popular in studies and clinical trials for NAFLD and NASH patients. A study by Wong et al. [136] separated NASH patients into two groups, one receiving a probiotic and prebiotic formula for a period of 6 months and the other receiving supportive care for the same time period. Probiotic/prebiotic treatment consisted of a combination of five bacterial species (L. plantarum, Lactobacillus delbrueckii ssp bulgaricus, L. acidophilus, Lactobacillus rhamnosus, and Bifidobacterium bifidum) and fructose-oligosaccharides (prebiotics). Patient clinical parameters that were analyzed following treatment included intrahepatic triglyceride (IHTG) content, alanine transaminase (ALT), aspartate aminotransferase (AST), fasting glucose, and liver stiffness. In addition, stool samples were analyzed to determine effects on microbiota composition. Following the 6-month probiotics/supportive care, IHTG was reduced in the probiotic group. Furthermore, probiotic use reduced AST levels but no significant changes were seen in the other biochemical and metabolic parameters. Microbiota analysis from stool samples revealed that most of the NASH patients receiving probiotics showed an expansion in Bacteroidetes and a decrease in Firmicutes abundance that correlated with reduced intrahepatic triglyceride content. In addition, two meta-analyses were performed to summarize the various clinical trials assessing probiotics for NAFLD and NASH patients [137], [138]. Both demonstrated a statistically significant improvement in metabolic and inflammatory parameters in probiotic-treated patients as compared to placebo-treated controls, suggesting that probiotics may be used as potential treatment in NAFLD/NASH [137], [138]. However, further prospective trials are needed in order to further corroborate these findings and search for a mechanism of activity.
6. Conclusions and perspectives
Increasing NAFLD and NASH prevalence has become a major burden on global health. As we viewed here, evidence on the gut microbiota association and involvement in NAFLD development is accumulating. Further mechanistic studies assessing microbial contribution to disease pathogenesis in animal models and in human patients may provide invaluable information in understanding the roots of NAFLD and NASH, and uncovering new treatment strategies against this common disease. In future studies, a combination of multi-omics approaches in NAFLD mice models and NAFLD patients [139] should be applied to identify bacterial community and host changes on the level of species abundance (16S ribosomal RNA gene sequencing), gene abundance (shotgun metagenomic sequencing), transcript abundance (bacterial and host RNA sequencing), and metabolite abundance (metabolomics profiling). Such analyses may help in the design of new interventions based on supplementation or inhibition of disease-associated metabolites tailored to the individual. In addition, fecal microbiome transplantation (FMT) is a ‘microbiome replacing’ approach that was recently found to be highly effective in drug resistant Clostridium difficile infection and potentially may be similarly efficacious in NAFLD and NASH. Taken together, deciphering and modulating the dysbiotic gut microbiota in NAFLD may allow for comprehensive mechanistic elucidation of the molecular basis of gut microbiota–host interactions that governs NAFLD progression, allowing for a rational design of microbiome-targeting therapeutics for this common and cureless disorder.
Acknowledgments
We thank the members of the Elinav laboratory for discussions and apologize to authors whose work not included in the review due to space constraints. E.E. is supported by: Y. and R. Ungar; the Abisch Frenkel Foundation for the Promotion of Life Sciences; the Gurwin Family Fund for Scientific Research; the Leona M. and Harry B. Helmsley Charitable Trust; the Crown Endowment Fund for Immunological Research; the estate of J. Gitlitz; the estate of L. Hershkovich; the Benoziyo Endowment Fund for the Advancement of Science; the Adelis Foundation; J. L. and V. Schwartz; A. and G. Markovitz; A. and C. Adelson; the French National Center for Scientific Research (CNRS); D. L. Schwarz; The V. R. Schwartz Research Fellow Chair; L. Steinberg; J. N. Halpern; A. Edelheit, and by grants funded by the European Research Council; a Marie Curie Integration grant; the German–Israeli Foundation for Scientific Research and Development; the Israel Science Foundation; the Minerva Foundation; the Rising Tide Foundation; the Helmholtz Foundation; and the European Foundation for the Study of Diabetes. E.E. is the incumbent of the Rina Gudinski Career Development Chair.
Conflict of interest
None declared.
References
- 1.Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of non-alcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence and outcomes. Hepatology. Dec 28 2015 doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 2.Rinella M.E. Nonalcoholic fatty liver disease: a systematic review. Journal of the American Medical Association. Jun 9 2015;313(22):2263–2273. doi: 10.1001/jama.2015.5370. [DOI] [PubMed] [Google Scholar]
- 3.Das K., Das K., Mukherjee P.S., Ghosh A., Ghosh S., Mridha A.R. Nonobese population in a developing country has a high prevalence of nonalcoholic fatty liver and significant liver disease. Hepatology. May 2010;51(5):1593–1602. doi: 10.1002/hep.23567. [DOI] [PubMed] [Google Scholar]
- 4.Ampuero J., Ranchal I., Gallego-Duran R., Pareja M.J., Del Campo J.A., Pastor-Ramirez H. Oxidized LDL antibodies/HDL-c ratio is linked to advanced disease in NAFLD lean patients. Journal of Gastroenterology and Hepatology. Mar 6 2016 doi: 10.1111/jgh.13335. [DOI] [PubMed] [Google Scholar]
- 5.Corey K.E., Rinella M.E. Medical and surgical treatment options for nonalcoholic steatohepatitis. Digestive Diseases and Sciences. May 2016;61(5):1387–1397. doi: 10.1007/s10620-016-4083-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam B., Younossi Z.M. Treatment options for nonalcoholic fatty liver disease. Therapeutic Advances in Gastroenterology. Mar 2010;3(2):121–137. doi: 10.1177/1756283X09359964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smallwood T., Allayee H., Bennett B.J. Choline metabolites: gene by diet interactions. Current Opinion in Lipidology. Feb 2016;27(1):33–39. doi: 10.1097/MOL.0000000000000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al Rajabi A., Castro G.S., da Silva R.P., Nelson R.C., Thiesen A., Vannucchi H. Choline supplementation protects against liver damage by normalizing cholesterol metabolism in Pemt/Ldlr knockout mice fed a high-fat diet. Journal of Nutrition. Mar 2014;144(3):252–257. doi: 10.3945/jn.113.185389. [DOI] [PubMed] [Google Scholar]
- 9.Collison K.S., Saleh S.M., Bakheet R.H., Al-Rabiah R.K., Inglis A.L., Makhoul N.J. Diabetes of the liver: the link between nonalcoholic fatty liver disease and HFCS-55. Obesity (Silver Spring) Nov 2009;17(11):2003–2013. doi: 10.1038/oby.2009.58. [DOI] [PubMed] [Google Scholar]
- 10.Veena J., Muragundla A., Sidgiddi S., Subramaniam S. Non-alcoholic fatty liver disease: need for a balanced nutritional source. British Journal of Nutrition. Dec 14 2014;112(11):1858–1872. doi: 10.1017/S0007114514002591. [DOI] [PubMed] [Google Scholar]
- 11.Smagris E., BasuRay S., Li J., Huang Y., Lai K.M., Gromada J. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology. Jan 2015;61(1):108–118. doi: 10.1002/hep.27242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romeo S., Kozlitina J., Xing C., Pertsemlidis A., Cox D., Pennacchio L.A. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nature Genetics. Dec 2008;40(12):1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sookoian S., Pirola C.J. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. Jun 2011;53(6):1883–1894. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 14.Walkey C.J., Yu L., Agellon L.B., Vance D.E. Biochemical and evolutionary significance of phospholipid methylation. Journal of Biological Chemistry. Oct 16 1998;273(42):27043–27046. doi: 10.1074/jbc.273.42.27043. [DOI] [PubMed] [Google Scholar]
- 15.Waite K.A., Cabilio N.R., Vance D.E. Choline deficiency-induced liver damage is reversible in Pemt(-/-) mice. Journal of Nutrition. Jan 2002;132(1):68–71. doi: 10.1093/jn/132.1.68. [DOI] [PubMed] [Google Scholar]
- 16.Song J., da Costa K.A., Fischer L.M., Kohlmeier M., Kwock L., Wang S. Polymorphism of the PEMT gene and susceptibility to nonalcoholic fatty liver disease (NAFLD) FASEB Journal. Aug 2005;19(10):1266–1271. doi: 10.1096/fj.04-3580com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J. Initial sequencing and analysis of the human genome. Nature. Feb 15 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 18.Relman D.A., Falkow S. The meaning and impact of the human genome sequence for microbiology. Trends in Microbiology. May 2001;9(5):206–208. doi: 10.1016/s0966-842x(01)02041-8. [DOI] [PubMed] [Google Scholar]
- 19.Rawls J.F., Mahowald M.A., Ley R.E., Gordon J.I. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. Oct 20 2006;127(2):423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Backhed F., Manchester J.K., Semenkovich C.F., Gordon J.I. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proceedings of the National Academy of Sciences of the United States of America. Jan 16 2007;104(3):979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. Dec 21 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 22.Flint H.J., Scott K.P., Duncan S.H., Louis P., Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. Jul–Aug 2012;3(4):289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamada N., Chen G.Y., Inohara N., Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nature Immunology. Jul 2013;14(7):685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kau A.L., Ahern P.P., Griffin N.W., Goodman A.L., Gordon J.I. Human nutrition, the gut microbiome and the immune system. Nature. Jun 16 2011;474(7351):327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto M., Yamaguchi R., Munakata K., Takashima K., Nishiyama M., Hioki K. A microarray analysis of gnotobiotic mice indicating that microbial exposure during the neonatal period plays an essential role in immune system development. BMC Genomics. 2012;13:335. doi: 10.1186/1471-2164-13-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. Jun 14 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E. A core gut microbiome in obese and lean twins. Nature. Jan 22 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D.R. Enterotypes of the human gut microbiome. Nature. May 12 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frank D.N., St Amand A.L., Feldman R.A., Boedeker E.C., Harpaz N., Pace N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proceedings of the National Academy of Sciences of the United States of America. Aug 21 2007;104(34):13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larsen N., Vogensen F.K., van den Berg F.W., Nielsen D.S., Andreasen A.S., Pedersen B.K. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PloS One. 2010;5(2):e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang F., Kaplan J.L., Gold B.D., Bhasin M.K., Ward N.L., Kellermayer R. Detecting microbial dysbiosis associated with pediatric Crohn disease despite the high variability of the gut microbiota. Cell Reports. Feb 2 2016;14(4):945–955. doi: 10.1016/j.celrep.2015.12.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z., Klipfell E., Bennett B.J., Koeth R., Levison B.S., Dugar B. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. Apr 7 2011;472(7341):57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown K., Godovannyi A., Ma C., Zhang Y., Ahmadi-Vand Z., Dai C. Prolonged antibiotic treatment induces a diabetogenic intestinal microbiome that accelerates diabetes in NOD mice. ISME Journal. Feb 2016;10(2):321–332. doi: 10.1038/ismej.2015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reikvam D.H., Erofeev A., Sandvik A., Grcic V., Jahnsen F.L., Gaustad P. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PloS One. 2011;6(3):e17996. doi: 10.1371/journal.pone.0017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ridaura V.K., Faith J.J., Rey F.E., Cheng J., Duncan A.E., Kau A.L. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. Sep 6 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Backhed F., Ding H., Wang T., Hooper L.V., Koh G.Y., Nagy A. The gut microbiota as an environmental factor that regulates fat storage. Proceedings of the National Academy of Sciences of the United States of America. Nov 2 2004;101(44):15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caesar R., Tremaroli V., Kovatcheva-Datchary P., Cani P.D., Backhed F. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metabolism. Oct 6 2015;22(4):658–668. doi: 10.1016/j.cmet.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Microbial ecology: human gut microbes associated with obesity. Nature. Dec 21 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 39.Schwiertz A., Taras D., Schafer K., Beijer S., Bos N.A., Donus C. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) Jan 2010;18(1):190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H., DiBaise J.K., Zuccolo A., Kudrna D., Braidotti M., Yu Y. Human gut microbiota in obesity and after gastric bypass. Proceedings of the National Academy of Sciences of the United States of America. Feb 17 2009;106(7):2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abdel-Misih S.R., Bloomston M. Liver anatomy. Surgical Clinics of North America. Aug 2010;90(4):643–653. doi: 10.1016/j.suc.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abu-Shanab A., Quigley E.M. The role of the gut microbiota in nonalcoholic fatty liver disease. Nature Reviews Gastroenterology & Hepatology. Dec 2010;7(12):691–701. doi: 10.1038/nrgastro.2010.172. [DOI] [PubMed] [Google Scholar]
- 43.Drenick E.J., Fisler J., Johnson D. Hepatic steatosis after intestinal bypass – prevention and reversal by metronidazole, irrespective of protein-calorie malnutrition. Gastroenterology. Mar 1982;82(3):535–548. [PubMed] [Google Scholar]
- 44.Wigg A.J., Roberts-Thomson I.C., Dymock R.B., McCarthy P.J., Grose R.H., Cummins A.G. The role of small intestinal bacterial overgrowth, intestinal permeability, endotoxaemia, and tumour necrosis factor alpha in the pathogenesis of non-alcoholic steatohepatitis. Gut. Feb 2001;48(2):206–211. doi: 10.1136/gut.48.2.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rabot S., Membrez M., Bruneau A., Gerard P., Harach T., Moser M. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB Journal. Dec 2010;24(12):4948–4959. doi: 10.1096/fj.10-164921. [DOI] [PubMed] [Google Scholar]
- 46.Le Roy T., Llopis M., Lepage P., Bruneau A., Rabot S., Bevilacqua C. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. Dec 2013;62(12):1787–1794. doi: 10.1136/gutjnl-2012-303816. [DOI] [PubMed] [Google Scholar]
- 47.Henao-Mejia J., Elinav E., Jin C., Hao L., Mehal W.Z., Strowig T. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. Feb 9 2012;482(7384):179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng H., Liu J., Jackson M.I., Zhao F.Q., Yan L., Combs G.F., Jr. Fatty liver accompanies an increase in lactobacillus species in the hind gut of C57BL/6 mice fed a high-fat diet. Journal of Nutrition. May 2013;143(5):627–631. doi: 10.3945/jn.112.172460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu G.D., Chen J., Hoffmann C., Bittinger K., Chen Y.Y., Keilbaugh S.A. Linking long-term dietary patterns with gut microbial enterotypes. Science. Oct 7 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeevi D., Korem T., Zmora N., Israeli D., Rothschild D., Weinberger A. Personalized nutrition by prediction of glycemic responses. Cell. Nov 19 2015;163(5):1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 51.Schnabl B., Brenner D.A. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. May 2014;146(6):1513–1524. doi: 10.1053/j.gastro.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michail S., Lin M., Frey M.R., Fanter R., Paliy O., Hilbush B. Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease. FEMS Microbiology Ecology. Feb 2015;91(2):1–9. doi: 10.1093/femsec/fiu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spencer M.D., Hamp T.J., Reid R.W., Fischer L.M., Zeisel S.H., Fodor A.A. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology. Mar 2011;140(3):976–986. doi: 10.1053/j.gastro.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang W., Wu N., Wang X., Chi Y., Zhang Y., Qiu X. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Scientific Reports. 2015;5:8096. doi: 10.1038/srep08096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raman M., Ahmed I., Gillevet P.M., Probert C.S., Ratcliffe N.M., Smith S. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clinical Gastroenterology and Hepatology. Jul 2013;11(7):868–875. doi: 10.1016/j.cgh.2013.02.015. e861–863. [DOI] [PubMed] [Google Scholar]
- 56.Zhu L., Baker S.S., Gill C., Liu W., Alkhouri R., Baker R.D. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. Feb 2013;57(2):601–609. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]
- 57.Mouzaki M., Comelli E.M., Arendt B.M., Bonengel J., Fung S.K., Fischer S.E. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. Jul 2013;58(1):120–127. doi: 10.1002/hep.26319. [DOI] [PubMed] [Google Scholar]
- 58.Wong V.W., Wong G.L., Chan H.Y., Yeung D.K., Chan R.S., Chim A.M. Bacterial endotoxin and non-alcoholic fatty liver disease in the general population: a prospective cohort study. Alimentary Pharmacology & Therapeutics. Sep 2015;42(6):731–740. doi: 10.1111/apt.13327. [DOI] [PubMed] [Google Scholar]
- 59.Boursier J., Mueller O., Barret M., Machado M., Fizanne L., Araujo-Perez F. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. Mar 2016;63(3):764–775. doi: 10.1002/hep.28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langille M.G., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology. Sep 2013;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klatt N.R., Funderburg N.T., Brenchley J.M. Microbial translocation, immune activation, and HIV disease. Trends in Microbiology. Jan 2013;21(1):6–13. doi: 10.1016/j.tim.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peterson L.W., Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nature Reviews. Immunology. Mar 2014;14(3):141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 63.Brenchley J.M., Douek D.C. HIV infection and the gastrointestinal immune system. Mucosal Immunology. Jan 2008;1(1):23–30. doi: 10.1038/mi.2007.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Honda K., Littman D.R. The microbiome in infectious disease and inflammation. Annual Review of Immunology. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miele L., Valenza V., La Torre G., Montalto M., Cammarota G., Ricci R. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology. Jun 2009;49(6):1877–1887. doi: 10.1002/hep.22848. [DOI] [PubMed] [Google Scholar]
- 66.Giorgio V., Miele L., Principessa L., Ferretti F., Villa M.P., Negro V. Intestinal permeability is increased in children with non-alcoholic fatty liver disease, and correlates with liver disease severity. Digestive and Liver Disease. Jun 2014;46(6):556–560. doi: 10.1016/j.dld.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 67.Everard A., Cani P.D. Diabetes, obesity and gut microbiota. Best Practice & Research. Clinical Gastroenterology. Feb 2013;27(1):73–83. doi: 10.1016/j.bpg.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 68.Tremaroli V., Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. Sep 13 2012;489(7415):242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 69.Brun P., Castagliuolo I., Di Leo V., Buda A., Pinzani M., Palu G. Increased intestinal permeability in obese mice: new evidence in the pathogenesis of nonalcoholic steatohepatitis. American Journal of Physiology. Gastrointestinal and Liver Physiology. Feb 2007;292(2):G518–G525. doi: 10.1152/ajpgi.00024.2006. [DOI] [PubMed] [Google Scholar]
- 70.Llorente C., Schnabl B. The gut microbiota and liver disease. Cellular and Molecular Gastroenterology and Hepatology. May 1 2015;1(3):275–284. doi: 10.1016/j.jcmgh.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gabele E., Dostert K., Hofmann C., Wiest R., Scholmerich J., Hellerbrand C. DSS induced colitis increases portal LPS levels and enhances hepatic inflammation and fibrogenesis in experimental NASH. Journal of Hepatology. Dec 2011;55(6):1391–1399. doi: 10.1016/j.jhep.2011.02.035. [DOI] [PubMed] [Google Scholar]
- 72.Luther J., Garber J.J., Khalili H., Dave M., Bale S.S., Jindal R. Hepatic injury in nonalcoholic steatohepatitis contributes to altered intestinal permeability. Cellular and Molecular Gastroenterology and Hepatology. Mar 2015;1(2):222–232. doi: 10.1016/j.jcmgh.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miura K., Ohnishi H. Role of gut microbiota and Toll-like receptors in nonalcoholic fatty liver disease. World Journal of Gastroenterology. Jun 21 2014;20(23):7381–7391. doi: 10.3748/wjg.v20.i23.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakamoto N., Kanai T. Role of toll-like receptors in immune activation and tolerance in the liver. Frontiers in Immunology. 2014;5:221. doi: 10.3389/fimmu.2014.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Csak T., Ganz M., Pespisa J., Kodys K., Dolganiuc A., Szabo G. Fatty acid and endotoxin activate inflammasomes in mouse hepatocytes that release danger signals to stimulate immune cells. Hepatology. Jul 2011;54(1):133–144. doi: 10.1002/hep.24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dixon L.J., Berk M., Thapaliya S., Papouchado B.G., Feldstein A.E. Caspase-1-mediated regulation of fibrogenesis in diet-induced steatohepatitis. Laboratory Investigation. May 2012;92(5):713–723. doi: 10.1038/labinvest.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang G., Lee H.E., Lee J.Y. A pharmacological inhibitor of NLRP3 inflammasome prevents non-alcoholic fatty liver disease in a mouse model induced by high fat diet. Scientific Reports. 2016;6:24399. doi: 10.1038/srep24399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shapiro H., Thaiss C.A., Levy M., Elinav E. The cross talk between microbiota and the immune system: metabolites take center stage. Current Opinion in Immunology. Oct 2014;30:54–62. doi: 10.1016/j.coi.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 79.Rooks M.G., Garrett W.S. Gut microbiota, metabolites and host immunity. Nature Reviews. Immunology. May 27 2016;16(6):341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Levy M., Thaiss C.A., Zeevi D., Dohnalova L., Zilberman-Schapira G., Mahdi J.A. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell. Dec 3 2015;163(6):1428–1443. doi: 10.1016/j.cell.2015.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu W., Gregory J.C., Org E., Buffa J.A., Gupta N., Wang Z. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell. Mar 24 2016;165(1):111–124. doi: 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raetz C.R., Whitfield C. Lipopolysaccharide endotoxins. Annual Review of Biochemistry. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dixon L.J., Barnes M., Tang H., Pritchard M.T., Nagy L.E. Kupffer cells in the liver. Comprehensive Physiology. Apr 2013;3(2):785–797. doi: 10.1002/cphy.c120026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Asehnoune K., Strassheim D., Mitra S., Kim J.Y., Abraham E. Involvement of reactive oxygen species in Toll-like receptor 4-dependent activation of NF-kappa B. Journal of Immunology. Feb 15 2004;172(4):2522–2529. doi: 10.4049/jimmunol.172.4.2522. [DOI] [PubMed] [Google Scholar]
- 85.Lu Y.C., Yeh W.C., Ohashi P.S. LPS/TLR4 signal transduction pathway. Cytokine. May 2008;42(2):145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 86.Cani P.D., Amar J., Iglesias M.A., Poggi M., Knauf C., Bastelica D. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. Jul 2007;56(7):1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 87.Rivera C.A., Adegboyega P., van Rooijen N., Tagalicud A., Allman M., Wallace M. Toll-like receptor-4 signaling and Kupffer cells play pivotal roles in the pathogenesis of non-alcoholic steatohepatitis. Journal of Hepatology. Oct 2007;47(4):571–579. doi: 10.1016/j.jhep.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cani P.D., Bibiloni R., Knauf C., Waget A., Neyrinck A.M., Delzenne N.M. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. Jun 2008;57(6):1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 89.Harte A.L., da Silva N.F., Creely S.J., McGee K.C., Billyard T., Youssef-Elabd E.M. Elevated endotoxin levels in non-alcoholic fatty liver disease. Journal of Inflammation (London) 2010;7:15. doi: 10.1186/1476-9255-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mehta N.N., McGillicuddy F.C., Anderson P.D., Hinkle C.C., Shah R., Pruscino L. Experimental endotoxemia induces adipose inflammation and insulin resistance in humans. Diabetes. Jan 2010;59(1):172–181. doi: 10.2337/db09-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bach Knudsen K.E. Microbial degradation of whole-grain complex carbohydrates and impact on short-chain fatty acids and health. Advances in Nutrition. Mar 2015;6(2):206–213. doi: 10.3945/an.114.007450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.De Vadder F., Kovatcheva-Datchary P., Goncalves D., Vinera J., Zitoun C., Duchampt A. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. Jan 16 2014;156(1–2):84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 93.den Besten G., van Eunen K., Groen A.K., Venema K., Reijngoud D.J., Bakker B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. Journal of Lipid Research. Sep 2013;54(9):2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Puertollano E., Kolida S., Yaqoob P. Biological significance of short-chain fatty acid metabolism by the intestinal microbiome. Current Opinion in Clinical Nutrition & Metabolic Care. Mar 2014;17(2):139–144. doi: 10.1097/MCO.0000000000000025. [DOI] [PubMed] [Google Scholar]
- 95.Kimura I., Ozawa K., Inoue D., Imamura T., Kimura K., Maeda T. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nature Communications. 2013;4:1829. doi: 10.1038/ncomms2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Maslowski K.M., Vieira A.T., Ng A., Kranich J., Sierro F., Yu D. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. Oct 29 2009;461(7268):1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ridlon J.M., Kang D.J., Hylemon P.B. Bile salt biotransformations by human intestinal bacteria. Journal of Lipid Research. Feb 2006;47(2):241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 98.Thomas C., Pellicciari R., Pruzanski M., Auwerx J., Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nature Reviews Drug Discovery. Aug 2008;7(8):678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 99.Chiang J.Y. Bile acids: regulation of synthesis. Journal of Lipid Research. Oct 2009;50(10):1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thomas C., Gioiello A., Noriega L., Strehle A., Oury J., Rizzo G. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metabolism. Sep 2009;10(3):167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Claus S.P., Tsang T.M., Wang Y., Cloarec O., Skordi E., Martin F.P. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Molecular Systems Biology. 2008;4:219. doi: 10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kuribayashi H., Miyata M., Yamakawa H., Yoshinari K., Yamazoe Y. Enterobacteria-mediated deconjugation of taurocholic acid enhances ileal farnesoid X receptor signaling. European Journal of Pharmacology. Dec 15 2012;697(1–3):132–138. doi: 10.1016/j.ejphar.2012.09.048. [DOI] [PubMed] [Google Scholar]
- 103.Sayin S.I., Wahlstrom A., Felin J., Jantti S., Marschall H.U., Bamberg K. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metabolism. Feb 5 2013;17(2):225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 104.Swann J.R., Want E.J., Geier F.M., Spagou K., Wilson I.D., Sidaway J.E. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proceedings of the National Academy of Sciences of the United States of America. Mar 15 2011;108(Suppl 1):4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee F.Y., Lee H., Hubbert M.L., Edwards P.A., Zhang Y. FXR, a multipurpose nuclear receptor. Trends in Biochemical Sciences. Oct 2006;31(10):572–580. doi: 10.1016/j.tibs.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 106.Yuan L., Bambha K. Bile acid receptors and nonalcoholic fatty liver disease. World Journal of Hepatology. Dec 8 2015;7(28):2811–2818. doi: 10.4254/wjh.v7.i28.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Huang W., Ma K., Zhang J., Qatanani M., Cuvillier J., Liu J. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. Apr 14 2006;312(5771):233–236. doi: 10.1126/science.1121435. [DOI] [PubMed] [Google Scholar]
- 108.Cipriani S., Mencarelli A., Palladino G., Fiorucci S. FXR activation reverses insulin resistance and lipid abnormalities and protects against liver steatosis in Zucker (fa/fa) obese rats. Journal of Lipid Research. Apr 2010;51(4):771–784. doi: 10.1194/jlr.M001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.de Aguiar Vallim T.Q., Tarling E.J., Edwards P.A. Pleiotropic roles of bile acids in metabolism. Cell Metabolism. May 7 2013;17(5):657–669. doi: 10.1016/j.cmet.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Neuschwander-Tetri B.A., Loomba R., Sanyal A.J., Lavine J.E., Van Natta M.L., Abdelmalek M.F. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet. Mar 14 2015;385(9972):956–965. doi: 10.1016/S0140-6736(14)61933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cariou B. The farnesoid X receptor (FXR) as a new target in non-alcoholic steatohepatitis. Diabetes & Metabolism. Dec 2008;34(6 Pt 2):685–691. doi: 10.1016/S1262-3636(08)74605-6. [DOI] [PubMed] [Google Scholar]
- 112.Prawitt J., Abdelkarim M., Stroeve J.H., Popescu I., Duez H., Velagapudi V.R. Farnesoid X receptor deficiency improves glucose homeostasis in mouse models of obesity. Diabetes. Jul 2011;60(7):1861–1871. doi: 10.2337/db11-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sinal C.J., Tohkin M., Miyata M., Ward J.M., Lambert G., Gonzalez F.J. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. Sep 15 2000;102(6):731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 114.Zhang Y., Lee F.Y., Barrera G., Lee H., Vales C., Gonzalez F.J. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proceedings of the National Academy of Sciences of the United States of America. Jan 24 2006;103(4):1006–1011. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Watanabe M., Horai Y., Houten S.M., Morimoto K., Sugizaki T., Arita E. Lowering bile acid pool size with a synthetic farnesoid X receptor (FXR) agonist induces obesity and diabetes through reduced energy expenditure. Journal of Biological Chemistry. Jul 29 2011;286(30):26913–26920. doi: 10.1074/jbc.M111.248203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Houten S.M., Volle D.H., Cummins C.L., Mangelsdorf D.J., Auwerx J. In vivo imaging of farnesoid X receptor activity reveals the ileum as the primary bile acid signaling tissue. Molecular Endocrinology. Jun 2007;21(6):1312–1323. doi: 10.1210/me.2007-0113. [DOI] [PubMed] [Google Scholar]
- 117.Jiang C., Xie C., Lv Y., Li J., Krausz K.W., Shi J. Intestine-selective farnesoid X receptor inhibition improves obesity-related metabolic dysfunction. Nature Communications. 2015;6:10166. doi: 10.1038/ncomms10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jiang C., Xie C., Li F., Zhang L., Nichols R.G., Krausz K.W. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. Journal of Clinical Investigation. Jan 2015;125(1):386–402. doi: 10.1172/JCI76738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fang S., Suh J.M., Reilly S.M., Yu E., Osborn O., Lackey D. Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nature Medicine. Feb 2015;21(2):159–165. doi: 10.1038/nm.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ryan K.K., Tremaroli V., Clemmensen C., Kovatcheva-Datchary P., Myronovych A., Karns R. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature. May 8 2014;509(7499):183–188. doi: 10.1038/nature13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mouzaki M., Wang A.Y., Bandsma R., Comelli E.M., Arendt B.M., Zhang L. Bile acids and dysbiosis in non-alcoholic fatty liver disease. PloS One. 2016;11(5):e0151829. doi: 10.1371/journal.pone.0151829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Volynets V., Kuper M.A., Strahl S., Maier I.B., Spruss A., Wagnerberger S. Nutrition, intestinal permeability, and blood ethanol levels are altered in patients with nonalcoholic fatty liver disease (NAFLD) Digestive Diseases and Sciences. Jul 2012;57(7):1932–1941. doi: 10.1007/s10620-012-2112-9. [DOI] [PubMed] [Google Scholar]
- 123.Yang M., Gong S., Ye S.Q., Lyman B., Geng L., Chen P. Non-alcoholic fatty liver disease in children: focus on nutritional interventions. Nutrients. Nov 2014;6(11):4691–4705. doi: 10.3390/nu6114691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chalasani N., Younossi Z., Lavine J.E., Diehl A.M., Brunt E.M., Cusi K. The diagnosis and management of non-alcoholic fatty liver disease: practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology. Jun 2012;55(6):2005–2023. doi: 10.1002/hep.25762. [DOI] [PubMed] [Google Scholar]
- 125.Eslamparast T., Eghtesad S., Poustchi H., Hekmatdoost A. Recent advances in dietary supplementation, in treating non-alcoholic fatty liver disease. World Journal of Hepatology. Feb 27 2015;7(2):204–212. doi: 10.4254/wjh.v7.i2.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dethlefsen L., Huse S., Sogin M.L., Relman D.A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biology. Nov 18 2008;6(11):e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dethlefsen L., Relman D.A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proceedings of the National Academy of Sciences of the United States of America. Mar 15 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bergheim I., Weber S., Vos M., Kramer S., Volynets V., Kaserouni S. Antibiotics protect against fructose-induced hepatic lipid accumulation in mice: role of endotoxin. Journal of Hepatology. Jun 2008;48(6):983–992. doi: 10.1016/j.jhep.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 129.Vos M.B., Lavine J.E. Dietary fructose in nonalcoholic fatty liver disease. Hepatology. Jun 2013;57(6):2525–2531. doi: 10.1002/hep.26299. [DOI] [PubMed] [Google Scholar]
- 130.Madrid A.M., Hurtado C., Venegas M., Cumsille F., Defilippi C. Long-term treatment with cisapride and antibiotics in liver cirrhosis: effect on small intestinal motility, bacterial overgrowth, and liver function. American Journal of Gastroenterology. Apr 2001;96(4):1251–1255. doi: 10.1111/j.1572-0241.2001.03636.x. [DOI] [PubMed] [Google Scholar]
- 131.Soza A., Riquelme A., GonzÁlez R., Alvarez M., PÉrez-ayuso R.M., Glasinovic J.C. Increased orocecal transit time in patients with nonalcoholic fatty liver disease. Digestive Diseases and Sciences. 2005;50(6):1136–1140. doi: 10.1007/s10620-005-2720-8. [DOI] [PubMed] [Google Scholar]
- 132.Gareau M.G., Sherman P.M., Walker W.A. Probiotics and the gut microbiota in intestinal health and disease. Nature Reviews Gastroenterology & Hepatology. Sep 2010;7(9):503–514. doi: 10.1038/nrgastro.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Paolella G., Mandato C., Pierri L., Poeta M., Di Stasi M., Vajro P. Gut-liver axis and probiotics: their role in non-alcoholic fatty liver disease. World Journal of Gastroenterology. Nov 14 2014;20(42):15518–15531. doi: 10.3748/wjg.v20.i42.15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Loguercio C., De Simone T., Federico A., Terracciano F., Tuccillo C., Di Chicco M. Gut-liver axis: a new point of attack to treat chronic liver damage? American Journal of Gastroenterology. Aug 2002;97(8):2144–2146. doi: 10.1111/j.1572-0241.2002.05942.x. [DOI] [PubMed] [Google Scholar]
- 135.Li Z., Yang S., Lin H., Huang J., Watkins P.A., Moser A.B. Probiotics and antibodies to TNF inhibit inflammatory activity and improve nonalcoholic fatty liver disease. Hepatology. Feb 2003;37(2):343–350. doi: 10.1053/jhep.2003.50048. [DOI] [PubMed] [Google Scholar]
- 136.Wong V.W., Tse C.H., Lam T.T., Wong G.L., Chim A.M., Chu W.C. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis – a longitudinal study. PloS One. 2013;8(4):e62885. doi: 10.1371/journal.pone.0062885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gao X., Zhu Y., Wen Y., Liu G., Wan C. Efficacy of probiotics in nonalcoholic fatty liver disease in adult and children: a meta-analysis of randomized controlled trials. Hepatology Research. Feb 11 2016 doi: 10.1111/hepr.12671. [DOI] [PubMed] [Google Scholar]
- 138.Ma Y.Y., Li L., Yu C.H., Shen Z., Chen L.H., Li Y.M. Effects of probiotics on nonalcoholic fatty liver disease: a meta-analysis. World Journal of Gastroenterology. Oct 28 2013;19(40):6911–6918. doi: 10.3748/wjg.v19.i40.6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Del Chierico F., Nobili V., Vernocchi P., Russo A., De Stefanis C., Gnani D. Gut microbiota profiling of pediatric NAFLD and obese patients unveiled by an integrated meta-omics based approach. Hepatology. Mar 29 2016 doi: 10.1002/hep.28572. [DOI] [PubMed] [Google Scholar]