Abstract
Background
Diabetes, obesity, and the metabolic syndrome are multifactorial diseases dependent on a complex interaction of host genetics, diet, and other environmental factors. Increasing evidence places gut microbiota as important modulators of the crosstalk between diet and development of obesity and metabolic dysfunction. In addition, host genetics can have important impact on the composition and function of gut microbiota. Indeed, depending on the genetic background of the host, diet and other environmental factors may produce different changes in gut microbiota, have different impacts on host metabolism, and create different interactions between the microbiome and the host.
Scope of review
In this review, we highlight how appropriate animal models can help dissect the complex interaction of host genetics with the gut microbiome and how diet can lead to different degrees of weight gain, levels of insulin resistance, and metabolic outcomes, such as diabetes, in different individuals. We also discuss the challenges of identifying specific disease-associated microbiota and the limitations of simple metrics, such as phylogenetic diversity or the ratio of Firmicutes to Bacteroidetes.
Major conclusions
Understanding these complex interactions will help in the development of novel treatments for microbiome-related metabolic diseases. This article is part of a special issue on microbiota.
Keywords: Obesity, Metabolic syndrome, Microbiome, Microbiota, Microbial diversity, Host genetics, Environment
The potential importance of gut microbiota in human disease was recognized more than 1000 years ago in China where fecal transplants, referred to as “yellow soup”, were used to treat gastrointestinal disorders. Over the past two decades, the scientific community has begun to rediscover the fact that not all gut bacteria are harmful and that transplantation of intestinal or fecal bacteria can be clinically useful in treatment of recurrent Clostridium difficile infection and possibly other diseases [1]. A key question is how important are gut microbiota in the control of metabolism and in the pathogenesis of obesity and diabetes.
The human body is a complex ecosystem hosting trillions of microbiota amounting to about 1.5 kg in mass. These colonize all surfaces of the body but especially the gastrointestinal tract. Using genetic techniques such as 16S sequencing, more than 1000 bacteria have been identified in the intestine, with ∼200 defining the core gut microbiome, i.e., constituting at least 0.5% of the microbial sequences detected [2], [3]. These bacteria exhibit important functions in the defense against foreign pathogens and the breakdown of otherwise indigestible dietary polysaccharides to produce short chain fatty acids, such as acetate, butyrate, and propionate, which can serve as important metabolites, a direct energy source for intestinal epithelial cells, modifiers of insulin resistance and modulators of insulin secretion [4], [5], [6]. Moreover, bacteria produce a wide range of other metabolites, as well as modifying human produced metabolites, such as bile acids, that can be taken up into the bloodstream where they have the potential to modulate host metabolism and other functions, including even behavioral and neural functions [7], [8], [9], [10], [11]. The microbiome can also influence the immune system [12], [13] and the integrity of the intestinal epithelium allowing bacterial products, including endotoxins, to enter the blood stream, leading to insulin resistance and other immune mediated disorders [4], [14].
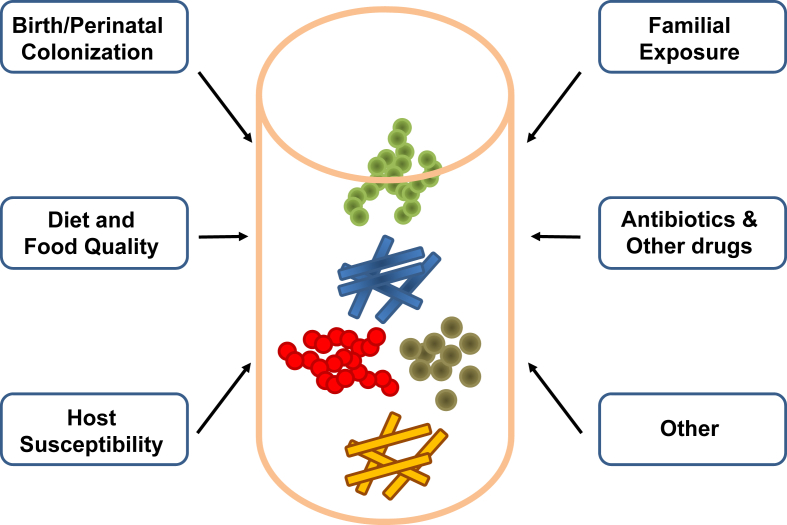
The core gut microbiome in humans is established within the first three years of life and is subject to multiple influences (Figure 1) [15], [16]. The initial colonization occurs during birth with vaginal microbiota from the mother, or, in the case of children born via cesarean section, with skin microbiota [17]. Not surprisingly, breast versus formula feeding also results in differences in the gut microbiota [18], [19]. The gut microbiota acquired during early life are further modified by environmental factors, such as diet, antibiotic treatment, and the microbiome of close family members. This early transfer of the microbiome allows it to potentially contribute to what is viewed as the inheritability of a disease [20]. These can be distinguished by twin studies, since, in contrast to classical genetic traits, transmission of gut microbiota is similar between mono- and dizygotic twins [21]. However, in most genetic studies, such as those used for genome wide association (GWAS) analysis, how transfer of microbiota between family members affects interpretation of data on the genetics of diabetes and obesity remains to be determined. Likewise, to what extent these initial differences in colonization result in altered risk for developing obesity or other diseases later in life remains controversial since the microbiome continues to remodel. However, as discussed below, this programming of the microbiome can lead to persistent effects even generations later.
Figure 1.
Factors contributing to the development of the microbiome. The development and composition of the gut microbiome is highly dependent on a multitude of environmental and host factors, especially those present in early life. Although core components of the gut microbiome tend to remain stable in adults, they continue to rapidly respond to alterations in the envrionment such as diet, medication and other factors.
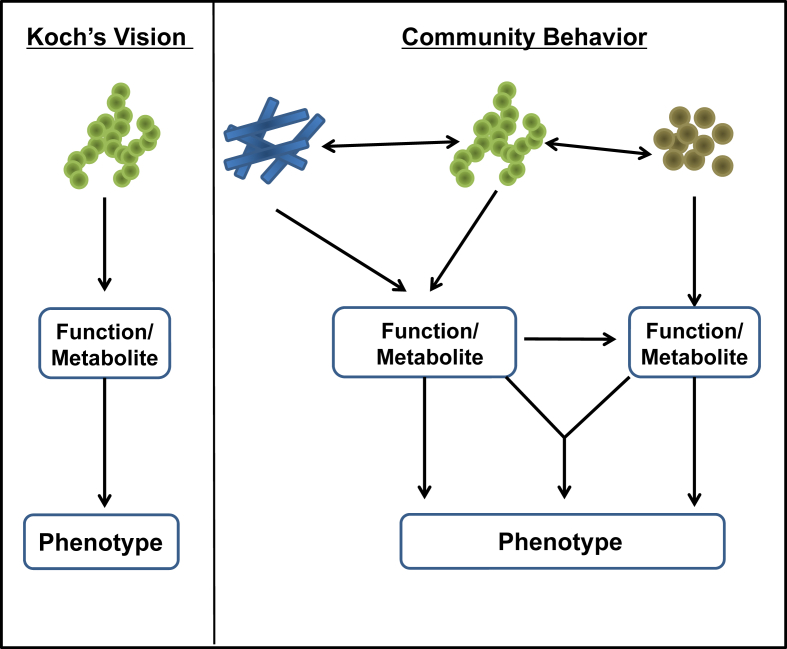
As the number of studies measuring microbiota composition in states of health and disease steadily rises, we are beginning to understand both the complexity of this system and its potential to interact with many physiological and pathological processes. Despite the ability of fecal transfer to mimic some disease characteristics, establishing causality, remains a challenge when evaluating the impact of individual gut microbiota on the regulation of host metabolism and metabolic disease. In an ideal case, a potential causative gut microbe involved in diabetes/obesity should be able to fulfill Koch's postulates, i.e., one can demonstrate that the specific microorganism is present in diabetic and/or obese individuals and that after isolation from an affected individual, it produces the same disease when reintroduced into a normal recipient (Figure 2, left panel). For metabolic diseases, such as diabetes, obesity, and metabolic syndrome, however, it appears that the effect of the microbiome is mediated through an interaction with larger microbial communities that together create a functional network. Moreover, many gene functions contributed by the gut microbiome are likely not present in just one microbe but are found in several bacterial strains. Conversely, the machinery involved in metabolic pathways may be split among different species of microbes, requiring the co-occurrence of multiple species to metabolize dietary components into specific metabolites (Figure 2 right panel). Furthermore, this community may interact with critical host factors and other environmental factors ultimately modifying disease development and progression. Determining the relative impact of these components, their hierarchy and interactions is a challenging task.
Figure 2.
Co-dependency of gut microbiota to modulate host metabolism. To identify individual bacterial taxa causally linked to specific host phenotype, one might anticipate a model which fulfills Koch's postulates, much like traditional views from genetics with one gene yielding one mRNA and then one protein with a defined and unique function. However, like modern cell biology the association of microbial taxa with host phenotypes is likely complex and impacted by the interaction of the microbiota themselves supporting their growth. Moreover, individual functions and metabolic pathways are either shared or split among different microbiota, and in most cases several microbiota and their functions may be required to impact specific host phenotypes.
1. Genetics and environment in the pathogenesis of obesity and diabetes
Obesity and its associated morbidities, such as type 2 diabetes, cardiovascular disease, and metabolic syndrome, are increasing in numbers worldwide. Despite the fact that knockout or overexpression of many single genes can lead to obesity and/or diabetes in mice [22], [23], [24], in humans less than 5–7% of individuals appear to have disease due to single gene alteration [25], [26]. Indeed, genome-wide association studies in humans have identified a large number of genetic polymorphisms associated with increased risk for obesity and diabetes, with each gene having very small effects [27]. In fact, even if one considers all common gene variants combined, they explain only a small part of the development of obesity and type 2 diabetes. These findings point to the strong role of environmental factors driving the growth in disease prevalence. Much epidemiological evidence suggests that two important environmental drivers are a more sedentary lifestyle and a changing diet. Together, increased consumption of refined sugars, saturated fats, and, most importantly, total calories strongly correlate with the increased prevalence of obesity, diabetes, and other components of the metabolic syndrome.
Within the population, however, it is clear that there is significant individual variation in response to these environmental challenges. Perhaps the best example is the study by Bouchard et al. in which identical twins were kept relatively sedentary and challenged by overfeeding 1000 kilocalories (4.2 MJ) per day, six out of seven days for 100 days [28]. While weight gain between individuals in a given twin pair correlated very closely, there was more than a two-fold difference in weight gain between the twins that gained the most weight and those that gained the least weight, indicating that some factor(s) accounts for major differences in caloric efficiency. Likewise, many other studies have shown that once weight gain occurs, while there is a strong correlation between excessive body fat as measured by BMI and the development of insulin resistance, there are many individuals who are very insulin resistant despite being only minimally overweight and a number of very obese individuals who remain metabolically healthy [29]. This indicates that in addition to factors that regulate food intake, insulin sensitivity, and beta cell function, among individuals, there are important differences in how caloric excess relates to weight gain and in the relationship between excess body fat and the development of insulin resistance.
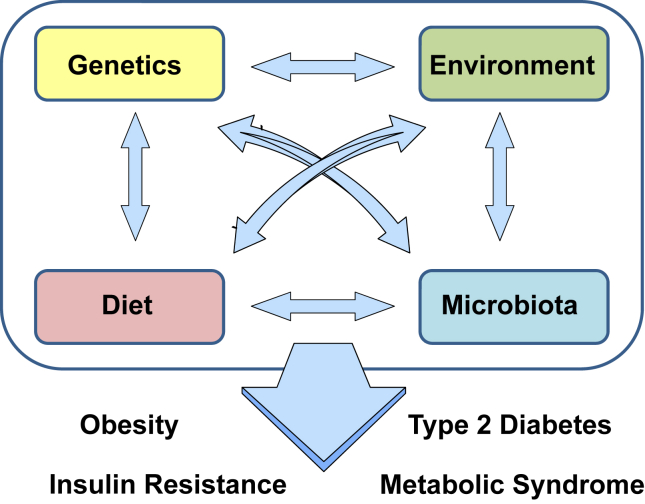
At least three factors that contribute to these differences are the individual differences in basal energy expenditure, probably related to the ability to induce brown/beige energy burning fat [30], [31], [32], [33], intrinsic genetic differences in insulin resistance, and how body fat is distributed [34]. Thus, central obesity correlates best with insulin resistance while accumulation of subcutaneous fat may have a protective effect. Indeed, we and others have demonstrated that transplantation of subcutaneous adipose tissue into the abdominal cavity can exert beneficial metabolic effects, although increasing total fat mass, suggesting differences in adipose tissue depots beyond their anatomical location [35], [36]. Subcutaneous and intraabdominal fat display distinct gene expression signatures with developmental genes linked to important metabolic functions [37]. These findings are further substantiated by studies in mice demonstrating that maintaining subcutaneous adipocyte function during weight gain results in a metabolically healthier obese phenotype [38], [39]. Hence, whereas the development of type 2 diabetes, insulin resistance, and obesity are linked, the individual risk determining the speed and severity of progression is dependent upon a complex mixture of genes and environmental interactions. Host genetics, diet, and other environmental factors interact with each other in the regulation of weight gain and insulin resistance but also impact the composition of the gut microbiota (Figure 3). While recent advances in integrating metabolomics, genomic, and taxonomic data suggest solutions to the challenge of establishing causal relationships between microbiota and host phenotypes [40], [41], [42], a major question remains as to which aspects of this complex pathophysiology are directly regulated or modified by the gut microbiome, which reflect co-regulation by the microbiome and host factors, and which are intrinsic to the host or are driven by factors independent of the microbiome.
Figure 3.
Obesity and its associated co-morbidities are the result of a complex interaction of host genetics, environment and gut micrbiota. The development of obesity, insulin resistance, type 2 diabetes and the metabolic syndrome in general are the consequence of a complex multidirectional interaction between host genetics, environment, diet and the gut microbiota.
2. Developing murine models to study gene/environment/microbiome interactions
To begin to define the role of the microbiome in gene/environment interaction in obesity, diabetes, and metabolic syndrome, we set out to create a mouse model system that captures some of the complexity observed in human populations. C57Bl/6J mice, or B6J for short, are the most commonly used mouse strain to study metabolism. B6J mice develop rapid and reproducible features of the metabolic syndrome when exposed to a high fat diet (HFD). They also develop adipose tissue inflammation, hepatosteatosis, insulin resistance, and hyperglycemia, all of which can be greatly enhanced by high fat diet feeding. However, compared to other strains, C57Bl/6J mice do not develop beta cell failure and uncontrolled type 2 diabetes. Rather, they develop a massive expansion of beta cells, which at least partially compensates for the peripheral insulin resistance [43].
In contrast, 129/Sv mice, a strain traditionally used to generate knockout animals due to the availability of well-established ES cell lines, are resistant to diet induced obesity and the development of the metabolic syndrome [44], [45], [46], [47], [48], [49]. 129 mice also remain insulin sensitive on HFD and are protected from the development of type 2 diabetes even when they are made genetically insulin resistant by inactivation of one allele of both the insulin receptor and IRS-1. This is in contrast to B6J mice in which 90% of the double heterozygous IR/IRS1 KOs develop overt diabetes by 6 months of age [50].
Mapping of quantitative trait loci (QTL) from F2 intercrosses between C57Bl/6J and 129Sv mice revealed several loci linked to the development of diabetes [44]. One QTL on chromosome 14 showed the highest association with the development of hyperinsulinemia and insulin resistance. One of the genes in this locus, PKCδ, was identified as being differentially expressed between B6J and 129 mice, with higher levels in the B6J mice on chow diet and a more robust increase in level following HFD feeding. Subsequent studies identified PKCδ, and other members of the novel subclass of PKCs, as an important regulator of hepatic insulin sensitivity through its direct action on insulin signaling [45], [51]. Conversely, several other factors tended to protect 129 mice from obesity and insulin resistance, including a lower tendency to develop adipose tissue inflammation than B6J mice with aging or HFD and an increased presence of brown/beige adipose tissue, leading to higher metabolic rates, which protects 129 mice against the development of obesity [44], [45], [46], [47], [48], [49]. Thus, some of the metabolic differences between B6J and 129Sv mice are the result of genetically driven risk and protective variations.
2.1. Where does the gut microbiome fit in the pathogenesis of obesity and insulin resistance?
The first studies to focus on the role of the microbiome in obesity and diabetes in humans and rodents revealed significant differences between lean and obese subjects in the two most abundant phyla of intestinal microbiota, most notably an increase in the ratio of Firmicutes to Bacteroidetes [51], [52], [53]. This difference, however, is not observed in all studies [54], [55], [56], and, in fact, the obesity-resistant 129Sv mice have a higher ratio of Firmicutes to Bacteroidetes than do B6J mice ([57] and see below). Nonetheless, Turnbaugh and colleagues demonstrated that transfer of gut microbiota from obese humans or obese mice to germfree C57Bl/6 mice resulted in greater weight gain in the recipient mice compared to mice receiving microbiota from lean individuals [58], suggesting a causal relationship between these alterations in the gut microbiota and the development of obesity.
No universal microbe has been identified leading to the difference in weight gain, but, in various studies, associations of weight gain with different individual bacterial taxa, including Akkermansia muciniphila, Faecalibacterium prausnitzii [14], [59], [60], [61] and Lactobacillus reuteri and Roseburia intestinalis [57] have been observed. Some of the complexity in determining cause and effect is illustrated by a recent study by Goodrich and colleagues [62b], which demonstrated a strong association of BMI in humans with microbiota of the Christensenellaceae family. Transplantation of microbiota from donors into germfree mice produced reduced weight gain upon the presence of Christensenellaceae, which could be mimicked by administration of one member of this family Christensenella minuta. Despite this apparent link, the observed effects do not appear to be due to the Christensenellaceae themselves but to the altered composition of the microbial ecosystem, which occurred in presence or absence of these bacteria. Indeed, other studies have shown that identification of individual bacterial families or species associated with a specific disease or host function, such as obesity, is strongly impacted by the ethnicity, socioeconomic status, and geographical region of the subjects, making replication of some of the associations observed in specific studies difficult [62], [63], [64], [65], [66].
Recent research has suggested that rather than specific organisms, it is a loss in microbial diversity, which is related to the development of obesity and other metabolic diseases [67]. A loss of microbial diversity has also been identified as a risk factor for inflammatory bowel disease and other non-metabolic disorders [68], [69]. As will be apparent in the discussion below, whether this low diversity is cause or consequence of metabolic disease is difficult to address as animals with similar diversity scores can have very different propensity to obesity and diabetes [57]. Reduced dietary diversity, which is common in most countries with raising obesity rates, can result in a similar loss of microbial diversity [70], again indicating the strong interactions among the four driving forces in the pathogenesis of metabolic disorders (Figure 3).
3. Developing more complete models of the complex interaction between genetics and the microbiome in mice
One of the first attempts to investigate the relationship of intestinal microbial community structure to host genetics took advantage of the “collaborative cross”, a study involving a diversity outbred population of mice developed from 8 inbred mouse strains (C57BL/6J, A/J, NOD/ShiLtJ, NZO/HILtJ, WSB/EiJ, CAST/EiJ, PWK/PhJ, and 129S1/SvImJ) [71]. 16S RNA profiling of enteric microbial communities revealed strain-driven differences in enteric microbial communities, which were retained with dietary intervention. Diet-strain interactions were seen for a core group of microbial taxa, and some of these could be linked to cardiometabolic phenotypes. Using QTL mapping in different outbred strains of mice, several loci were identified that were associated with immune pathways that impacted on intestinal microbiota [72], [73].
To more precisely define the interaction between host genetics, environment (including both housing conditions and diet), and the microbiome, we developed a new model comparing the C57Bl/6J (B6J for short) from Jackson Laboratories to two substrains of 129Sv mice, one the 129S1/SvlmJ (or 129J) also from Jackson Laboratories and the other the 129S6/SvEvTac (129T) mice from Taconic Farms [57]. These two 129 mouse lines are nearly genetically identical; however, these substrains have now been reared apart for over 30 years and are bred and housed in different commercial facilities, which use different diets and other nutritional supplements. Interestingly, while 129J mice are both obesity- and diabetes-resistant and insulin sensitive when challenged with a HFD, 129T mice become as obese as B6J mice when fed a HFD but retain insulin sensitivity and normal glucoses similar to those in the obesity-resistant 129J mice (Figure 4A, bottom panel). However, when both substrains of 129 mice were bred in a common animal facility for three generations to “normalize” their environments, the 129T mice lost their tendency to become obese and became resistant to HFD-induced similar to 129J mice, highlighting the significance of gene/environment/microbiome interactions in the phenotypes of these two substrains of 129 mice. By using these three strains/substrains of mice before and after environmental normalization (i.e., as they come from the vendor vs. after breeding in a common facility for 3 generations) and challenging them with HFD, we were able create a novel “Interaction Model” with 12 independent, but interrelated, properties (3 genetic backgrounds × 2 sites of breeding × 2 diets). This allowed us to dissect the interaction between host genetics, diet, and the gut microbiome on key metabolic traits, such as obesity, diabetes, and insulin resistance.
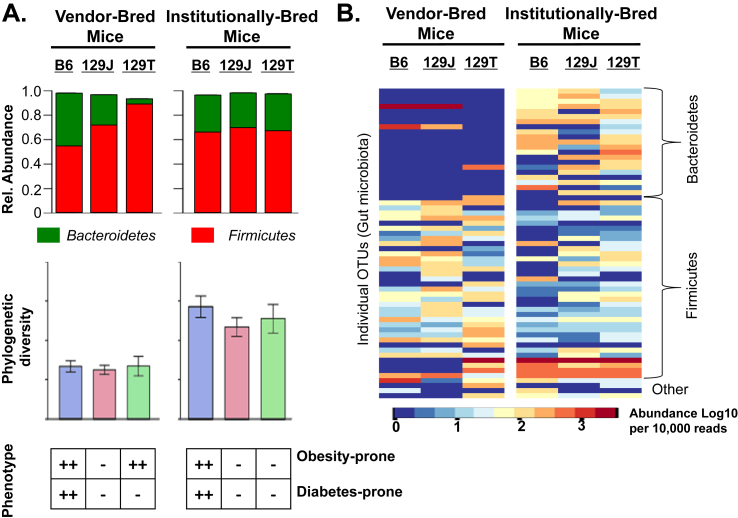
Figure 4.
Host genetics and environment shape personalized microbiota. The impact of gut microbiota on host physiology depends on a complex interaction of host genetics, environment, diet. A) Scores such as phylogenetic diversity or ratio of Firmicutes to Bacteroidetes (F/B) may appear as robust metrics predicting metabolic fitness in a genetically homogenous population. However, in different genetic backgrounds similar diversity scores or F/B ratios can reflect very different microbial communities, with distinct impact on host metabolism. B) Assessment of individual taxa or operational taxonomic units (OTUs) can provide more insight into the true nature and diversity of gut microbiota.
This Interaction Model provides insight into a number of important aspects regarding the gut microbiome in the pathogenesis of metabolic syndrome. First, while many early studies focused on the ratio of Firmicutes to Bacteroidetes (F/B) as a marker of an obesogenic microbiome with a high F/B ratio being more likely to support obesity, this is clearly not true when one considers these mice. Thus, when these three strains of mice arrive from the vendor, the F/B ratio is lowest in the B6J mice from Jackson (about 60/40) and is highest in the 129T mice from Taconic (about 90/10), although both strains are equally prone to development of obesity on HFD, whereas the obesity-resistant 129J mice are intermediate between the other strains (about 70/30) (Figure 4A, top). Following breeding in a common environment, all three strains have similar F/B ratios (about 70/30), despite the fact that one (the B6J) remains obesity-prone, while the two 129Sv substrains are obesity-resistant. Likewise, although low diversity of gut microbes is said to be a risk factor for metabolic and other diseases, both the obesity- and diabetes-prone mice and the obesity- and diabetes-resistant mice had similar low phylogenetic diversity when bred at the commercial vendor site and similar higher phylogenetic diversity when bred in our academic facility (Figure 4A, middle panel). Indeed, phylogenetic diversity increased over time in the vendor-bred mice as they were housed in our academic facility whether they became obese or not, although it did increase more slowly or to a lesser extent in mice on HFD [57]. Hence, neither changes in the Firmicutes/Bacteroidetes ratio nor diversity score were reliable predictors of metabolic outcome in the host, but much more reflected differences in diet and/or environment, irrespective of their impact on host metabolism.
Importantly, when complete 16S RNA sequencing was performed, despite very similar diversity scores, the actual composition of the microbiota, i.e., the specific taxa or operational taxonomic units (OTUs) detected, differed greatly among strains (Figure 4B). This was true whether the mice were bred at the commercial facilities or bred for several generations in a single mouse facility. What was also observed is that the “environmentally normalized” mice maintained some microbial markers of their previous environmental history, i.e., their provenance, even after three generations of breeding in a new site and on a new diet. Thus, in this study, community-based metrics, such as diversity and F/B ratio, seem to better reflect environmental alterations and their interaction with host genetics than predicting metabolic outcome. If we are to find cause and effect relationships between the gut microbiome and metabolic disease phenotypes, it is important to consider the specific collection of taxa that are present and their respective contributions to the bacterial effects rather than just their phylogenetic diversity or balance between the two dominant phyla. Going forward, this will require more detailed metagenomics analyses, more complete bacterial sequencing, or ways to assess the function of the microbiome, not just its composition, such as PICRUSt analysis [74].
The Interaction Model also provides a view into the dynamic nature of the gut microbiome, which is extremely important to consider in both laboratory-based animal studies and human studies (Figure 5). Thus, time course studies demonstrate that the microbiome can be very dynamic, even in adult animals, depending on diet, genetic background of the host, and other environmental conditions. For example, frequent sampling of mice imported from commercial vendors to a typical research laboratory (e.g., our laboratory) reveal that within days of arrival there is a rapid remodeling of the microbiome, which proceeds with different kinetics for different organisms and different diets (Figure 5). Furthermore, the direction and kinetics of change of individual microbial taxa depends itself on an interaction with the host. These data demonstrate how timing of sample collection in relation to the occurrence of a given phenotype needs to be considered, in addition to host genetics, when performing correlative studies. Thus, studying the abundance and dynamics of individual taxa or microbial communities as they might relate to being causal factors in specific host phenotypes, i.e., fulfilling Koch's postulates, is complex. For example, when studying the response to diet, it is not clear whether the most relevant microbial pattern is the pattern before the dietary challenge, at some specific time during the dietary challenge, or at the end of the dietary challenge. What is clear, however, is that the microbiome at these three time points is likely to be quite different. Likewise, the dynamic nature of the microbiome makes it difficult to define interactions among members of the microbial community that might be related to specific metabolic phenotypes.
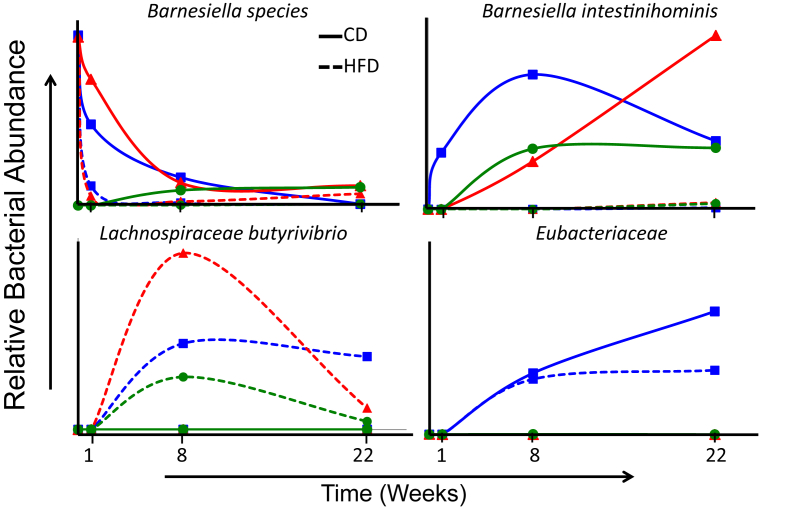
Figure 5.
Dynamic nature of diet, host and microbiota interactions. Gut microbiota rapidly reconfigure with highly individual kinetics among taxa upon environmental changes such as different housing conditions or changes in diet. Some bacterial taxa such as the Barnesiella species illustrated in the upper left panel are present in the microbiota of one specific vendor versus another [compare B6J (blue line) and 129J (red line) from Jackson to 129T (green line) from Taconic Farms]. In this example, this microbial species in the two Jackson-bred strains is rapidly lost in the new environment, but the rate of loss depends on the diet with faster kinetics upon high fat diet feeding versus chow diet (dashed versus solid lines). Other bacterial taxa such as Barnesiella intestinihominis or Lachnospiraceae Butyrivibrio are absent from all the vendor bred mice, but rapidly colonize the gut upon introduction into a common new environment. In the case of B. intestinihominis (upper right panel) the microbe only colonizes in the chow diet fed group, whereas with Lachnospiraceae Butyrivibrio (lower left panel) colonization occurs only in the HFD fed group. Some taxa, such as a specific OTU of the Eubacteriaceae (lower right panel) colonize the gut upon introduction into a new environment independent of type of the diet, but specific to a single genetic background, in this case B6J. Thus, the association of certain groups of microbiota with specific host phenotypes is the result of a complex interaction of host genetics and various environmental factors at a specific time.
The complexity, dynamic nature, and redundant functions of gut microbiota highlight the need to better understand the impact of both individual bacteria and bacterial communities on the function of the microbiome and the impact of the whole bacterial ecosystem. Investigating the entire microbiome, i.e., the sum of all microbial genes, instead of the phylogenetic assignment, and defining functional gene groups bioinformatically or experimentally is emerging as a promising way to bypass the high degree of heterogeneity at the taxa level between individual subjects. This strategy allows for defining a network of genes provided by potentially unrelated microbiota that might be necessary for a specific function, such as bile acid metabolism or short-chain fatty acid metabolism. This function can then be evaluated for its impact on host metabolism by changing not only the bacterial landscape but also by providing or removing key metabolic intermediates that might mimic the effects of the microbiota on metabolism.
4. Fecal transplantation – is it the gold standard for metabolic disease?
Fecal transplantation is generally regarded as the gold standard for dissecting functional interactions of microbial communities with the host. Indeed, this is the most direct attempt to assign a cause effect relationship in gut microbiota related physiological effects. Transplantation of feces from the three vendor derived strains used in the Interaction Model into germfree B6J mice resulted in increased blood glucose, increased liver weight (reflecting fat in the liver), and decreased glucose tolerance when the donor feces came from B6J and 129T mice, consistent with a causal effect. However, there was no statistically significant difference in body weight, and the changes in glucose were modest. It is also important to keep in mind that B6J mice represent only one of the potential host backgrounds, and that in this background, host genetics is a very important driver of obesity and insulin resistance that may dominate over microbial effects. To fully explore the role of the microbiome and the importance of host–microbiome interactions, one would have to put each of the different donor microbiota into each of the different potential host backgrounds starting in a germ-free condition – a truly daunting task.
One also needs to be cautious in interpretation of metabolic effects in germ-free mice, since germ-free mice are known to have abnormalities in intestinal epithelial development and in energy balance [75]. From the study comparing the three strains of mice reared in two different environments and fed either chow and HFD, it appears that genetics are a dominant driver of obesity and insulin resistance in mice which have a high genetic predisposition for these phenotypes, such as C57Bl/6J (B6J) mice, and that alterations in the gut microbiota have limited impact. In contrast, in permissive genetic backgrounds, such as 129S6 (129T) mice, the gut microbiome may have a stronger impact on the development of obesity. Thus, future germ-free studies should consider using mice of this and other permissive genetic backgrounds to explore the impact of the microbiome. Similar considerations need to be taken into account in analysis of human data as the microbiome may have greater or lesser effects depending of genetic and ethnic background.
5. Conclusion
Obesity and the metabolic syndrome are multifactorial diseases caused by a complex interaction of host genetics and a multitude of environmental factors. Among the many factors determining metabolic homeostasis and health, diet and total calories consumed are undoubtedly the most significant drivers of obesity and its metabolic complications. However, it has become increasingly apparent that amount and nature of calories taken up by the intestine are strongly impacted by the microbiota colonizing it. These observations have sparked a tremendous number of studies detailing the connections between food, gut microbiota, and metabolic disease that have deepened our understanding of how microbial communities are formed and reconfigure upon environmental changes. These studies in animals or humans have also shown that inter-individual variations are not random, but depend on a close relationship between microbiota, host genetics, and individual habits and behavior. Our Interaction Model demonstrates that, at least in permissive genetic backgrounds, gut microbiota play a significant role in modulating the development of obesity, insulin resistance, and other phenotypes and that groups of specific bacterial taxa may be involved. However, the field is still young, and our understanding is still increasing, almost exponentially. Clearly we need to move beyond just describing effects of the microbiome on host metabolism and determine the molecular mechanisms that might underlie these interactions. This knowledge will eventually allow us to put together these trillions of pieces of the puzzle and assemble a functional understanding of host–microbiome interactions so that we can utilize to manipulate this system to support a metabolically healthier life.
Contributor Information
Siegfried Ussar, Email: Siegfried.ussar@helmholtz-muenchen.de.
C. Ronald Kahn, Email: C.Ronald.Kahn@joslin.harvard.edu.
Conflict of interest
The authors declare that there is no conflict of interest.
References
- 1.Kelly C.R., Kahn S., Kashyap P., Laine L., Rubin D., Atreja A. Update on fecal microbiota transplantation 2015: indications, methodologies, mechanisms, and outlook. Gastroenterology. 2015;149:223–237. doi: 10.1053/j.gastro.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tremaroli V., Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 4.Kelly C.J., Zheng L., Campbell E.L., Saeedi B., Scholz C.C., Bayless A.J. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. 2015;17:662–671. doi: 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perry R.J., Peng L., Barry N.A., Cline G.W., Zhang D., Cardone R.L. Acetate mediates a microbiome–brain–beta-cell axis to promote metabolic syndrome. Nature. 2016;534:213–217. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan J., McKenzie C., Potamitis M., Thorburn A.N., Mackay C.R., Macia L. The role of short-chain fatty acids in health and disease. Advances in Immunology. 2014;121:91–119. doi: 10.1016/B978-0-12-800100-4.00003-9. [DOI] [PubMed] [Google Scholar]
- 7.Buffington S.A., Di Prisco G.V., Auchtung T.A., Ajami N.J., Petrosino J.F., Costa-Mattioli M. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell. 2016;165:1762–1775. doi: 10.1016/j.cell.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gacias M., Gaspari S., Santos P.M., Tamburini S., Andrade M., Zhang F. Microbiota-driven transcriptional changes in prefrontal cortex override genetic differences in social behavior. Elife. 2016;5 doi: 10.7554/eLife.13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryan K.K., Tremaroli V., Clemmensen C., Kovatcheva-Datchary P., Myronovych A., Karns R. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature. 2014;509:183–188. doi: 10.1038/nature13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sayin S.I., Wahlstrom A., Felin J., Jantti S., Marschall H.U., Bamberg K. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metabolism. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Wahlstrom A., Sayin S.I., Marschall H.U., Backhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metabolism. 12 Jul 2016;24(1):41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Arpaia N., Campbell C., Fan X., Dikiy S., van der Veeken J., deRoos P. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung H., Pamp S.J., Hill J.A., Surana N.K., Edelman S.M., Troy E.B. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everard A., Belzer C., Geurts L., Ouwerkerk J.P., Druart C., Bindels L.B. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koenig J.E., Spor A., Scalfone N., Fricker A.D., Stombaugh J., Knight R. Succession of microbial consortia in the developing infant gut microbiome. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wopereis H., Oozeer R., Knipping K., Belzer C., Knol J. The first thousand days – intestinal microbiology of early life: establishing a symbiosis. Pediatric Allergy and Immunology: Official Publication of the European Society of Pediatric Allergy and Immunology. 2014;25:428–438. doi: 10.1111/pai.12232. [DOI] [PubMed] [Google Scholar]
- 17.Dominguez-Bello M.G., De Jesus-Laboy K.M., Shen N., Cox L.M., Amir A., Gonzalez A. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nature Medicine. 2016;22(3):250–253. doi: 10.1038/nm.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S.A., Lim J.Y., Kim B.S., Cho S.J., Kim N.Y., Kim O.B. Comparison of the gut microbiota profile in breast-fed and formula-fed Korean infants using pyrosequencing. Nutrition Research and Practice. 2015;9:242–248. doi: 10.4162/nrp.2015.9.3.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z., Roy N.C., Guo Y., Jia H., Ryan L., Samuelsson L. Human breast milk and infant formulas differentially modify the intestinal microbiota in human infants and host physiology in rats. The Journal of Nutrition. 2016;146:191–199. doi: 10.3945/jn.115.223552. [DOI] [PubMed] [Google Scholar]
- 20.Faith J.J., Colombel J.F., Gordon J.I. Identifying strains that contribute to complex diseases through the study of microbial inheritance. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:633–640. doi: 10.1073/pnas.1418781112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim M.Y., You H.J., Yoon H.S., Kwon B., Lee J.Y., Lee S. The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut. 2016 doi: 10.1136/gutjnl-2015-311326. [DOI] [PubMed] [Google Scholar]
- 22.Biddinger S.B., Kahn C.R. From mice to men: insights into the insulin resistance syndromes. Annual Review of Physiology. 2006;68:123–158. doi: 10.1146/annurev.physiol.68.040104.124723. [DOI] [PubMed] [Google Scholar]
- 23.Bunner A.E., Chandrasekera P.C., Barnard N.D. Knockout mouse models of insulin signaling: relevance past and future. World Journal of Diabetes. 2014;5:146–159. doi: 10.4239/wjd.v5.i2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tschop M., Heiman M.L. Rodent obesity models: an overview. Experimental and Clinical Endocrinology & Diabetes. 2001;109:307–319. doi: 10.1055/s-2001-17297. [DOI] [PubMed] [Google Scholar]
- 25.Farooqi S., O'Rahilly S. Genetics of obesity in humans. Endocrine Reviews. 2006;27:710–718. doi: 10.1210/er.2006-0040. [DOI] [PubMed] [Google Scholar]
- 26.Semple R.K., Savage D.B., Cochran E.K., Gorden P., O'Rahilly S. Genetic syndromes of severe insulin resistance. Endocrine Reviews. 2011;32:498–514. doi: 10.1210/er.2010-0020. [DOI] [PubMed] [Google Scholar]
- 27.Karaderi T., Drong A.W., Lindgren C.M. Insights into the genetic susceptibility to type 2 diabetes from genome-wide association studies of obesity-related traits. Current Diabetes Reports. 2015;15:83. doi: 10.1007/s11892-015-0648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouchard C., Tremblay A., Despres J.P., Nadeau A., Lupien P.J., Theriault G. The response to long-term overfeeding in identical twins. New England Journal of Medicine. 1990;322:1477–1482. doi: 10.1056/NEJM199005243222101. [DOI] [PubMed] [Google Scholar]
- 29.Stefan N., Haring H.U., Hu F.B., Schulze M.B. Metabolically healthy obesity: epidemiology, mechanisms, and clinical implications. The Lancet Diabetes & Endocrinology. 2013;1:152–162. doi: 10.1016/S2213-8587(13)70062-7. [DOI] [PubMed] [Google Scholar]
- 30.Cypess A.M., Lehman S., Williams G., Tal I., Rodman D., Goldfine A.B. Identification and importance of brown adipose tissue in adult humans. New England Journal of Medicine. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Min S.Y., Kady J., Nam M., Rojas-Rodriguez R., Berkenwald A., Kim J.H. Human ‘brite/beige’ adipocytes develop from capillary networks, and their implantation improves metabolic homeostasis in mice. Nature Medicine. 2016;22:312–318. doi: 10.1038/nm.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito M., Okamatsu-Ogura Y., Matsushita M., Watanabe K., Yoneshiro T., Nio-Kobayashi J. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Marken Lichtenbelt W.D., Vanhommerig J.W., Smulders N.M., Drossaerts J.M., Kemerink G.J., Bouvy N.D. Cold-activated brown adipose tissue in healthy men. New England Journal of Medicine. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 34.Shungin D., Winkler T.W., Croteau-Chonka D.C., Ferreira T., Locke A.E., Magi R. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanford K.I., Middelbeek R.J., Townsend K.L., Lee M.Y., Takahashi H., So K. A novel role for subcutaneous adipose tissue in exercise-induced improvements in glucose homeostasis. Diabetes. 2015;64:2002–2014. doi: 10.2337/db14-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran T.T., Yamamoto Y., Gesta S., Kahn C.R. Beneficial effects of subcutaneous fat transplantation on metabolism. Cell Metabolism. 2008;7:410–420. doi: 10.1016/j.cmet.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gesta S., Bluher M., Yamamoto Y., Norris A.W., Berndt J., Kralisch S. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6676–6681. doi: 10.1073/pnas.0601752103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J.Y., vandeWall E., Laplante M., Azzara A., Trujillo M.E., Hofmann S.M. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. Journal of Clinical Investigation. 2007;117:2621–2637. doi: 10.1172/JCI31021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kusminski C.M., Holland W.L., Sun K., Park J., Spurgin S.B., Lin Y. MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nature Medicine. 2012;18:1539–1549. doi: 10.1038/nm.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eng A., Borenstein E. An algorithm for designing minimal microbial communities with desired metabolic capacities. Bioinformatics. 2016;32:2008–2016. doi: 10.1093/bioinformatics/btw107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Granger B.R., Chang Y.C., Wang Y., DeLisi C., Segre D., Hu Z. Visualization of metabolic interaction networks in microbial communities using VisANT 5.0. PLoS Computational Biology. 2016;12:e1004875. doi: 10.1371/journal.pcbi.1004875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noecker C., Eng A., Srinivasan S., Theriot C.M., Young V.B., Jansson J.K. Metabolic model-based integration of microbiome taxonomic and metabolomic profiles elucidates mechanistic links between ecological and metabolic variation. mSystems. 2016;1 doi: 10.1128/mSystems.00013-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kluth O., Matzke D., Schulze G., Schwenk R.W., Joost H.G., Schurmann A. Differential transcriptome analysis of diabetes-resistant and -sensitive mouse islets reveals significant overlap with human diabetes susceptibility genes. Diabetes. 2014;63:4230–4238. doi: 10.2337/db14-0425. [DOI] [PubMed] [Google Scholar]
- 44.Almind K., Kahn C.R. Genetic determinants of energy expenditure and insulin resistance in diet-induced obesity in mice. Diabetes. 2004;53:3274–3285. doi: 10.2337/diabetes.53.12.3274. [DOI] [PubMed] [Google Scholar]
- 45.Bezy O., Tran T.T., Pihlajamaki J., Suzuki R., Emanuelli B., Winnay J. PKCdelta regulates hepatic insulin sensitivity and hepatosteatosis in mice and humans. The Journal of Clinical Investigation. 2011;121:2504–2517. doi: 10.1172/JCI46045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fink B.D., Herlein J.A., Almind K., Cinti S., Kahn C.R., Sivitz W.I. Mitochondrial proton leak in obesity-resistant and obesity-prone mice. American Journal of Physiology Regulatory Integrative and Comparative Physiology. 2007;293:R1773–R1780. doi: 10.1152/ajpregu.00478.2007. [DOI] [PubMed] [Google Scholar]
- 47.Kokkotou E., Jeon J.Y., Wang X., Marino F.E., Carlson M., Trombly D.J. Mice with MCH ablation resist diet-induced obesity through strain-specific mechanisms. American Journal of Physiology Regulatory Integrative and Comparative Physiology. 2005;289:R117–R124. doi: 10.1152/ajpregu.00861.2004. [DOI] [PubMed] [Google Scholar]
- 48.Lin C., Theodorides M.L., McDaniel A.H., Tordoff M.G., Zhang Q., Li X. QTL analysis of dietary obesity in C57BL/6byj X 129P3/J F2 mice: diet- and sex-dependent effects. PLoS One. 2013;8:e68776. doi: 10.1371/journal.pone.0068776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mori M.A., Liu M., Bezy O., Almind K., Shapiro H., Kasif S. A systems biology approach identifies inflammatory abnormalities between mouse strains prior to development of metabolic disease. Diabetes. 2010;59:2960–2971. doi: 10.2337/db10-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kulkarni R.N., Almind K., Goren H.J., Winnay J.N., Ueki K., Okada T. Impact of genetic background on development of hyperinsulinemia and diabetes in insulin receptor/insulin receptor substrate-1 double heterozygous mice. Diabetes. 2003;52:1528–1534. doi: 10.2337/diabetes.52.6.1528. [DOI] [PubMed] [Google Scholar]
- 51.(a) Ley R.E., Backhed F., Turnbaugh P., Lozupone C.A., Knight R.D., Gordon J.I. Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Samuel V.T., Liu Z.X., Wang A., Beddow S.A., Geisler J.G., Kahn M. Inhibition of protein kinase C epsilon prevents hepatic insulin resistance in nonalcoholic fatty liver disease. The Journal of Clinical Investigation. 2007;117:739–745. doi: 10.1172/JCI30400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 53.Million M., Maraninchi M., Henry M., Armougom F., Richet H., Carrieri P. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. International Journal of Obesity. 2012;36:817–825. doi: 10.1038/ijo.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D.R. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Collado M.C., Isolauri E., Laitinen K., Salminen S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. American Journal of Clinical Nutrition. 2008;88:894–899. doi: 10.1093/ajcn/88.4.894. [DOI] [PubMed] [Google Scholar]
- 56.Schwiertz A., Taras D., Schafer K., Beijer S., Bos N.A., Donus C. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 57.Ussar S., Griffin N.W., Bezy O., Fujisaka S., Vienberg S., Softic S. Interactions between gut microbiota, host genetics and diet modulate the predisposition to obesity and metabolic syndrome. Cell Metabolism. 2015;22:516–530. doi: 10.1016/j.cmet.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turnbaugh P.J., Backhed F., Fulton L., Gordon J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dao M.C., Everard A., Aron-Wisnewsky J., Sokolovska N., Prifti E., Verger E.O. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2016;65:426–436. doi: 10.1136/gutjnl-2014-308778. [DOI] [PubMed] [Google Scholar]
- 60.Hippe B., Remely M., Aumueller E., Pointner A., Magnet U., Haslberger A.G. Faecalibacterium prausnitzii phylotypes in type two diabetic, obese, and lean control subjects. Benef Microbes. 2016:1–8. doi: 10.3920/BM2015.0075. [DOI] [PubMed] [Google Scholar]
- 61.Schneeberger M., Everard A., Gomez-Valades A.G., Matamoros S., Ramirez S., Delzenne N.M. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Scientific Reports. 2015;5:16643. doi: 10.1038/srep16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.(a) Chong C.W., Ahmad A.F., Lim Y.A., Teh C.S., Yap I.K., Lee S.C. Effect of ethnicity and socioeconomic variation to the gut microbiota composition among pre-adolescent in Malaysia. Scientific Reports. 2015;5:13338. doi: 10.1038/srep13338. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Goodrich J.K., Waters J.L., Poole A.C., Sutter J.L., Koren O., Blekhman R. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kemppainen K.M., Ardissone A.N., Davis-Richardson A.G., Fagen J.R., Gano K.A., Leon-Novelo L.G. Early childhood gut microbiomes show strong geographic differences among subjects at high risk for type 1 diabetes. Diabetes Care. 2015;38:329–332. doi: 10.2337/dc14-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lozupone C.A., Stombaugh J., Gonzalez A., Ackermann G., Wendel D., Vazquez-Baeza Y. Meta-analyses of studies of the human microbiota. Genome Research. 2013;23:1704–1714. doi: 10.1101/gr.151803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller G.E., Engen P.A., Gillevet P.M., Shaikh M., Sikaroodi M., Forsyth C.B. Lower neighborhood socioeconomic status associated with reduced diversity of the colonic microbiota in healthy adults. PLoS One. 2016;11:e0148952. doi: 10.1371/journal.pone.0148952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhernakova A., Kurilshikov A., Bonder M.J., Tigchelaar E.F., Schirmer M., Vatanen T. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lozupone C.A., Stombaugh J.I., Gordon J.I., Jansson J.K., Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prideaux L., Kang S., Wagner J., Buckley M., Mahar J.E., De Cruz P. Impact of ethnicity, geography, and disease on the microbiota in health and inflammatory bowel disease. Inflammatory Bowel Diseases. 2013;19:2906–2918. doi: 10.1097/01.MIB.0000435759.05577.12. [DOI] [PubMed] [Google Scholar]
- 69.Willing B.P., Dicksved J., Halfvarson J., Andersson A.F., Lucio M., Zheng Z. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139 doi: 10.1053/j.gastro.2010.08.049. 1844–1854 e1841. [DOI] [PubMed] [Google Scholar]
- 70.Heiman M.L., Greenway F.L. A healthy gastrointestinal microbiome is dependent on dietary diversity. Molecular Metabolism. 2016;5:317–320. doi: 10.1016/j.molmet.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O'Connor A., Quizon P.M., Albright J.E., Lin F.T., Bennett B.J. Responsiveness of cardiometabolic-related microbiota to diet is influenced by host genetics. Mammalian Genome: Official Journal of the International Mammalian Genome Society. 2014;25:583–599. doi: 10.1007/s00335-014-9540-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Benson A.K., Kelly S.A., Legge R., Ma F., Low S.J., Kim J. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McKnite A.M., Perez-Munoz M.E., Lu L., Williams E.G., Brewer S., Andreux P.A. Murine gut microbiota is defined by host genetics and modulates variation of metabolic traits. PLoS One. 2012;7:e39191. doi: 10.1371/journal.pone.0039191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langille M.G., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Backhed F., Manchester J.K., Semenkovich C.F., Gordon J.I. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]