Abstract
The genome of Chromobacterium subtsugae strain PRAA4-1, a betaproteobacterium producing insecticidal compounds, was sequenced and compared with the genome of C. violaceum ATCC 12472. The genome of C. subtsugae displayed a reduction in genes devoted to capsular and extracellular polysaccharide, possessed no genes encoding nitrate reductases, and exhibited many more phage-related sequences than were observed for C. violaceum. The genomes of both species possess a number of gene clusters predicted to encode biosynthetic complexes for secondary metabolites; these clusters suggest they produce overlapping, but distinct assortments of metabolites.
Keywords: Chromobacterium subtsugae, Genome sequence, Non-ribosomal peptide synthases, Polyketide synthases
1. Direct link to deposited data
2. Introduction
Until recently, purple-pigmented bacteria of the genus Chromobacterium were represented by a single species, Chromobacterium violaceum Bergonzini 1881. Chromobacterium violaceum is best known for production of the purple pigment violacein, which has exhibited diverse antimicrobial and antitumor activities. Considerable research has been conducted on the biosynthesis of violacein and the quorum sensing system which regulates it, and the genome sequence of C. violaceum has been reported by the Brazilian National Genome Project Consortium [1]. In 2007, Martin et al. [2] described Chromobacterium subtsugae, which in addition to violacein, also produced insecticidal factors that were active against a variety of insect pests. More recently, an extract of C. subtsugae was approved by the EPA for use as an organic insecticide that is now commercially available as Grandevo® (Marrone BioInnovations Incorporated, Davis, CA, USA). Since the initial description of C. subtsugae, six additional species of Chromobacterium have been described: C. aquaticum, C. haemolyticum, C. piscinae, C. pseudoviolaceum, C. vaccinii, and C. amazonense [3], [4], [5], [6], [7]. The biology of these new species, and the metabolites they produce, remains largely unexplored.
In an effort to begin examining the metabolic and biosynthetic diversity of the genus Chromobacterium, we have sequenced and assembled the genome of C. subtsugae and compared it with that of the type strain of C. violaceum (ATCC 12472). Key differences distinguishing the species are discussed.
3. Materials and methods
Version 2.9 of the Roche/454 gsAssembler software was used to assemble 792,321 C. subtsugae 454 reads into a draft genome comprising 76 contigs of at least 200 bp in length. The overall 4,805,598 bp assembly had an N50 of 222,962 bp, with minimum and maximum contig lengths of 201 and 462,591 bp, respectively. The concentration of G + C in the assembled genome was 64. 8%.
The C. subtsugae assembly and the published C. violaceum genome (GenBank identifier AE 016825.1) were both submitted to the RAST server for parallel annotation [8]. Annotated protein sets for each genome were compared, and subsets unique to each species were investigated in detail. In addition, secondary metabolite biosynthesis gene clusters were predicted using version 2.0 of the antiSMASH utility [9]. Whole-genome alignments were performed using Mauve [10].
4. Results and discussion
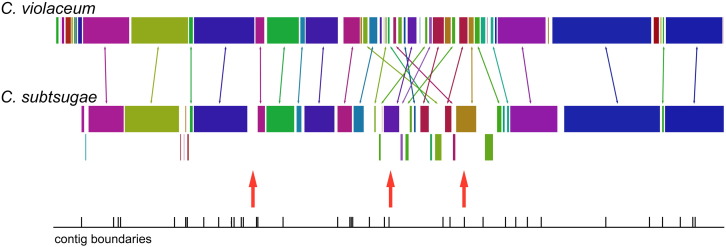
The assembled genomic sequences for C. subtsugae were deposited in GenBank with the accession numbers JYKA01000001 through JYKA01000076. The C. subtsugae annotation revealed 4492 coding sequences, of which, 2268 could be assigned a specific function. Approximately 70% of C. subtsugae genes assigned by the RAST server had greater than 70% sequence identity with their apparent C. violaceum orthologs. Comparing the annotations for both species revealed substantial differences in certain metabolic and biosynthetic activities. C. subtsugae had a considerable reduction in genes devoted to capsular and extracellular polysaccharide production, as well as gram-negative cell wall components. It was also apparent that C. subtsugae had many fewer genes devoted to Nitrogen metabolism, particularly in reduction of nitrate and nitrite. C. subtsugae also displays many more phage-related gene sequences than C. violaceum, containing three major clusters of phage-related sequences not shared with C. violaceum. Whole-genome comparisons indicated substantial chromosomal rearrangements between the taxa (Fig. 1), which may be the result of activities mediated by phage and/or phage-like gene transfer agent particles [11], [12].
Fig. 1.
Alignment of the C. subtsugae PRAA4–1 genome with that of C. violaceum ATCC 12472. Red arrows indicate clusters of phage sequences unique to PRAA4-1.
4.1. General metabolism
C. subtsugae appears broadly similar to C. violaceum in terms of general metabolic pathways, including enzymes involved in glycolysis and gluconeogenesis, the citric acid cycle, pentose phosphate pathway, and Entner-Doudoroff pathway. Gene complements for amino acid biosynthetic pathways appear nearly identical, as are those for purine and pyrimidine metabolism.
One substantial metabolic difference between the two species is in their capacity to utilize nitrate for anaerobic respiration. Unlike C. violaceum, C. subtsugae apparently lacks both the assimilatory nitrate reductase that permits the bacteria to produce usable ammonia from environmental nitrate, and also lacks the operon encoding the membrane associated respiratory nitrate reductase that allows nitrate to serve as the terminal electron acceptor under anaerobic conditions. C. subtsugae appears able to utilize fumarate as the terminal electron acceptor under anaerobic conditions, a trait shared with C. violaceum.
4.2. Capsular polysaccharide and biofilm production
The genome sequence of C. subtsugae suggests significant differences from C. violaceum in both biofilm production and biosynthesis of capsular lipopolysaccharide decorations. C. subtsugae appears to lack several suites of C. violaceum genes that are involved in biofilm production, such as the pgaABCD operon that is required for the synthesis and transport of the biofilm adhesin poly-B-1, 6-N-acetyl-D-glucosamine [13], and also lacks a fap operon that allows the synthesis of amyloid fibrils, which have been shown in certain Pseudomonads to increase aggregation and biofilm formation [14]. C. violaceum also appears to possess an operon (locus tags CV_0304–CV_0309 of accession no. AE016825.1) encoding a large protein adhesin and the Type I secretion system required to transport it out of the cell, similar to that found in Pseudomonas fluorescens [15], which is seemingly absent in C. subtsugae. An apparent cluster of lipopolysaccharide biosynthesis genes, while present in both species, is highly modified in C. subtsugae; genes in the C. violaceum chromosome spanning the region CV_4008–CV_4024 are highly substituted in C. subtsugae, and while still appearing to involve lipopolysaccharide synthesis, suggest substantial changes to the carbohydrate content. In particular, two genes encoding UDP-N-acetyl glucosamine 2-epimerase, needed for synthesis of the capsule precursor UDP-N-acetyl-mannosaminouronic acid, are absent from this cluster in C. subtsugae, and do not appear elsewhere in the genome.
4.3. Membrane transport and secretion systems
Both species have highly similar ABC transporters for amino acids and peptides and share highly similar complements of Type II secretory elements. Both also possess a similar Type IV secretory system for fimbrial proteins.
Type III secretory systems allow delivery of proteins across the inner and outer membranes of the bacteria, and some can deliver effector proteins directly into the cytoplasm of a host cell. The genome sequence of C. violaceum revealed the presence of two such Type III secretion systems that resembled those of Salmonella enterica pathogenicity islands 1 and 2 (Spi-1 and Spi-2) [16]. Designated Cpi-1 and Cpi-2 in S. enterica, the presence of these operons may help explain how C. violaceum periodically infects mammals. In C. violaceum, Cpi-1 is divided into Cpi-1A and Cpi-1B, with Cpi-1A located approximately 200 kb upstream of Cpi-1B. Cpi-2 is situated between Cpi-1A and Cpi-1B, immediately upstream of Cpi-1B. While C. subtsugae has a largely intact and similarly separated Cpi-1A and Cpi-1B, genes contained in Cpi-2, including its Type III secretory system, are not evident.
Other differences in secretory apparatus relate to proteins involved with biofilm features mentioned previously; neither the Type I secretion system for a putative large biofilm adhesin protein nor the Type VIII system associated with fap amyloid fibrils were observed in C. subtsugae.
4.4. Secondary metabolites
Both C. violaceum and C. subtsugae have the ability to produce a number of interesting secondary metabolites, such as violacein. Both species also appear to have the ability to synthesize an enterobactin-like siderophore and associated transport system, allowing uptake of ferric iron from their environmental surroundings.
Both C. violaceum and C. subtsugae possess a large number of apparent operons encoding non-ribosomal peptide synthases and polyketide synthases that may be involved in the synthesis of additional siderophores, antibiotics, and toxins. antiSMASH v2.0 [9] predicted 22 gene clusters in C. subtsugae that are likely involved in the synthesis of secondary metabolites, and predicted 17 such clusters in C. violaceum. Many of these, for instance gene clusters involved in bacteriocin, indole, and terpene biosynthesis, are shared with C. violaceum. However, comparing the two species, a number of predicted biosynthetic gene clusters appear highly modified or unique to C. subtsugae, and may contribute to its insecticidal properties.
In a particularly intriguing case, a cluster of 15 predicted genes (locus tags MY55_RS12975 through MY55_13045 of JYKA01000009.1), and their organization, are highly similar to a cluster of 15 genes found in the insect pathogen Photorhabdus temperata subsp. khanii strain NC19. The G + C content of the 42 kb region encoding this cluster was found to be 52.2%, which contrasts with the 64.8% ratio for the entire C. subtsugae genome, indicating that this gene cluster was acquired recently. The G + C content of the P. temperata subsp. khanii NC19 was calculated to be 43.3%. Similar predicted genes appear to be associated with other insect associated bacteria such as Xenorhabdus spp., Paenibacillus larvae and Paenibacillus alvei. These genes appear to encode a non-ribosomal peptide synthase - type 1 polyketide synthase, predicted by antiSMASH v2.0 to produce a core sequence of ser-asn-malonate-asp/asn-malonate-pro-ser-leu-lys. At the periphery of this large cluster are genes for Pfa synthases that produce long-chain omega-3 polyunsaturated fatty acids similar to those encountered in certain marine bacteria and in the heterocyst glycolipids of nitrogen-fixing cyanobacteria [17]. Whether the lipid products of these genes are involved in modification of the peptide is not clear.
Genomic comparison of C. subtsugae with more closely related species in the Chromobacterium genus, or disruption of key genes present in likely biosynthetic clusters, should reveal those genetic elements involved in the production of insecticidal compounds.
Conflict of interest statement
Drs. Blackburn and Gundersen-Rindal are co-inventors on a patent assigned to The United States of America (US7244607), covering the use of Chromobacterium subtsugae for insect control. The patent is licensed by Marrone Bio Innovations.
Acknowledgments
This research was funded by the Agricultural Research Service of the USDA.
References
- 1.Brazilian National Genome Project Consortium The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc. Natl. Acad. Sci. 2003;100:11660–11665. doi: 10.1073/pnas.1832124100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin P.A., Gundersen-Rindal D.E., Blackburn M.B., Buyer J. Chromobacterium subtsugae sp. nov., a betaproteobacterium toxic to Colorado potato beetle and other insect pests. Int. J. Syst. Evol. Microbiol. 2007;57:993–999. doi: 10.1099/ijs.0.64611-0. [DOI] [PubMed] [Google Scholar]
- 3.Young C.C., Arun A.B., Lai W.A., Chen W.M., Chao J.H., Shen F.T., Rekha P.D., Kämpfer P. Chromobacterium aquaticum sp. nov., isolated from spring water samples. Int. J. Syst. Evol. Microbiol. 2008;58:877–880. doi: 10.1099/ijs.0.65573-0. [DOI] [PubMed] [Google Scholar]
- 4.Han X.Y., Han F.S., Segal J. Chromobacterium haemolyticum sp. nov., a strongly haemolytic species. Int. J. Syst. Evol. Microbiol. 2008;58:1398–1403. doi: 10.1099/ijs.0.64681-0. [DOI] [PubMed] [Google Scholar]
- 5.Kämpfer P., Busse H.J., Scholz H.C. Chromobacterium piscinae sp. nov. and Chromobacterium pseudoviolaceum sp. nov., from environmental samples. Int. J. Syst. Evol. Microbiol. 2009;59:2486–2490. doi: 10.1099/ijs.0.008888-0. [DOI] [PubMed] [Google Scholar]
- 6.Soby S.D., Gadagkar S.R., Contreras C., Caruso F.L. Chromobacterium vaccinii sp. nov., isolated from native and cultivated cranberry (Vaccinium macrocarpon Ait.) bogs and irrigation ponds. Int. J. Syst. Evol. Microbiol. 2013;63:1840–1846. doi: 10.1099/ijs.0.045161-0. [DOI] [PubMed] [Google Scholar]
- 7.Menezes C.B.A., Tonin M.F., Corrêa D.B.A., Parma M., de Melo I.S., Zucchi T.D., Destéfano S.A.L., Fantinatti-Garboggini F. Chromobacterium amazonense sp. nov. isolated from water samples from the Rio Negro, Amazon, Brazil. Antonie Van Leeuwenhoek. 2015;107:1057–1063. doi: 10.1007/s10482-015-0397-3. [DOI] [PubMed] [Google Scholar]
- 8.Overbeek R., Olson R., Pusch G.D., Olsen G.J., Davis J.J., Disz T., Edwards R.A., Gerdes S., Parrello B., Shukla M., Vonstein V., Wattam A.R., Xia F., Stevens R. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blin K., Medema M.H., Kazempour D., Fischbach M.A., Breitling R., Takano E., Weber T. antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt449. (p.gkt449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darling A.E., Mau B., Perna, N.T . progressiveMauve: multiple genome alignment with gene gain, loss, and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brüssow H., Canchaya C., Hardt W.D. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lang A.S., Zhaxybayeva O., Beatty J.T. Gene transfer agents: phage-like elements of genetic exchange. Nat. Rev. Microbiol. 2012;10:472–482. doi: 10.1038/nrmicro2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X., Preston J.F., Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 2004;186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dueholm M.S., Søndergaard M.T., Nilsson M., Christiansen G., Stensballe A., Overgaard M.T., Givskov M., Tolker-Nielsen T., Otzen D.E., Nielsen P.H. Expression of Fap amyloids in Pseudomonas aeruginosa, P. fluorescens, and P. putida results in aggregation and increased biofilm formation. MicrobiologyOpen. 2013;2:365–382. doi: 10.1002/mbo3.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinsa S.M., Espinosa-Urgel M., Ramos J.L., O'Toole G.A. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 2003;49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- 16.Betts H.J., Chaudhuri R.R., Pallen M.J. An analysis of type-III secretion gene clusters in Chromobacterium violaceum. Trends Microbiol. 2004;12:476–482. doi: 10.1016/j.tim.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 17.Shulse C.N., Allen E.E. Widespread occurrence of secondary lipid biosynthesis potential in microbial lineages. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020146. [DOI] [PMC free article] [PubMed] [Google Scholar]