Phospholipids are required to bind to two distinct sites on the inward rectifier potassium channel for maximal efficacy. Lee et al. show that a membrane-associating tryptophan residue in the second site can mimic the effect of phospholipid binding and cause a conformational change to reveal the primary binding site.
Abstract
Inward rectifier potassium (Kir) channel activity is controlled by plasma membrane lipids. Phosphatidylinositol-4,5-bisphosphate (PIP2) binding to a primary site is required for opening of classic inward rectifier Kir2.1 and Kir2.2 channels, but interaction of bulk anionic phospholipid (PL−) with a distinct second site is required for high PIP2 sensitivity. Here we show that introduction of a lipid-partitioning tryptophan at the second site (K62W) generates high PIP2 sensitivity, even in the absence of PL−. Furthermore, high-resolution x-ray crystal structures of Kir2.2[K62W], with or without added PIP2 (2.8- and 2.0-Å resolution, respectively), reveal tight tethering of the C-terminal domain (CTD) to the transmembrane domain (TMD) in each condition. Our results suggest a refined model for phospholipid gating in which PL− binding at the second site pulls the CTD toward the membrane, inducing the formation of the high-affinity primary PIP2 site and explaining the positive allostery between PL− binding and PIP2 sensitivity.
INTRODUCTION
Inwardly rectifying potassium (Kir) channels play a critical role in stabilizing membrane potentials and hence controlling numerous physiological phenomena in excitable and inexcitable cells (Nichols and Lopatin, 1997; Hibino et al., 2010). Phosphatidylinositol-4,5-bisphosphate (PIP2) in the inner membrane leaflet is required for the activation of all Kir channel subtypes (Hilgemann and Ball, 1996; Rohács et al., 2003; D’Avanzo et al., 2010b), and Kir channel crystal structures have unambiguously identified the PIP2 binding site (primary site; Fig. 1 A) at the interface between the transmembrane domain (TMD) and the C-terminal domain (CTD; Hansen et al., 2011). Although the CTD is quite closely apposed to the TMD in the PIP2-bound Kir2.2 (PDB no. 3SPI) structure, the domain is displaced from the membrane by ∼6 Å in the Apo-Kir2.2 (PDB no. 3JYC) structure, suggesting that pulling the CTD toward the membrane provides the mechanistic link between PIP2 binding and Kir channel activation (Enkvetchakul and Nichols, 2003; Tao et al., 2009; D’Avanzo et al., 2010a; Hansen et al., 2011).
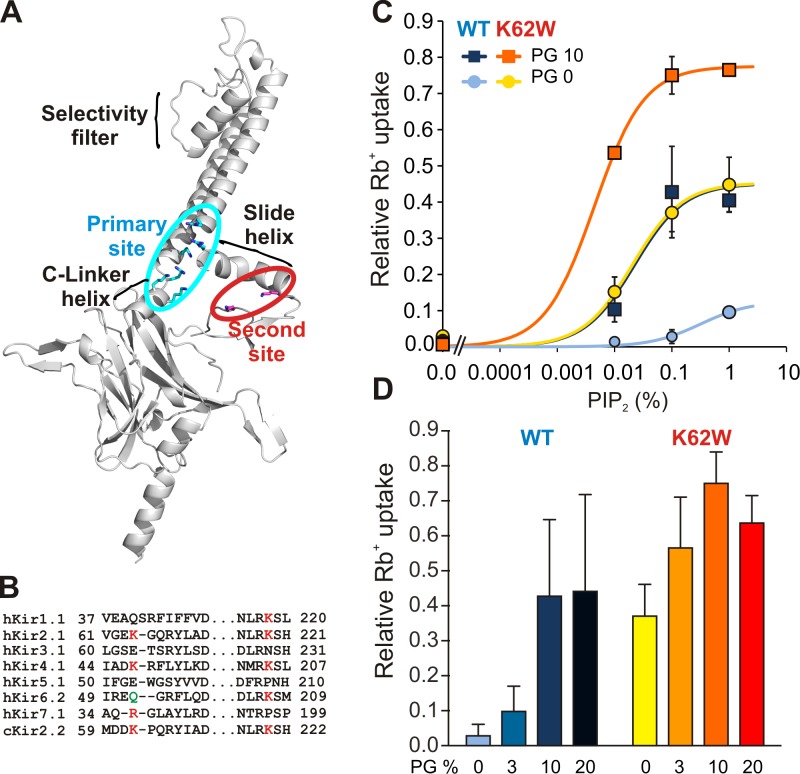
Figure 1.
Increased PIP2 sensitivity but decreased PL− sensitivity in K62W channels. (A) Ribbon diagram of Kir2.2 monomer structure (3SPI). Key functional parts of the protein are labeled, with residues comprising the primary and second sites shown in blue and red sticks, respectively. (B) Sequence alignments of selected regions of Kir subfamily members. Residues important for secondary PL− interaction are shown in red. Q52 of Kir6.2 shown in green causes gain-of-function if mutated to Arg. (C) 86Rb+ uptake versus PIP2 concentration for reconstituted chicken Kir2.2 WT and K62W mutant channels, in the presence of 0 or 10% POPG lipids (mean ± SE, n = 3). The line is the best fit of the one-site binding model in each case. (D) The same experiment was performed at constant 0.1% PIP2, with increasing POPG concentrations as indicated (mean ± SE, n = 3).
In addition to PIP2, bulk anionic phospholipids (PL−) are required for Kir2 channel gating, allosterically increasing PIP2 sensitivity by 10–100-fold and thereby making Kir2 channels active at physiological levels of PIP2 (Cheng et al., 2011). In silico docking studies identify an additional PL− binding site (second site), generated primarily by a lysine residue in the N-terminal end of the slide helix (K64 in human Kir2.1, K62 in chicken Kir2.2; Fig. 1, A and B; Lee et al., 2013). Given the relatively high level of PL− (>15% of all lipids) that is typically present in plasma membrane inner leaflets (van Meer et al., 2008; Ingólfsson et al., 2014) and the nonspecific character of PL− activation of Kir2 channels (Cheng et al., 2011), the second site interaction is likely to be consistently present in cell membranes. The mutation K64C in human Kir2.1 results in significant loss of PL− sensitivity, and reduced channel activity (Lee et al., 2013). However, modification of the cysteine with a long hydrophobic moiety generates high PIP2 sensitivity, even in the absence of PL−, suggesting that tethering of this site to the membrane inner leaflet induces the formation of the high-affinity primary PIP2 site and channel activation. Here, we performed further functional and structural characterization of the second site. We first show that mutation of the key residue to a membrane-associating tryptophan can fulfill the second site requirement to generate high PIP2 sensitivity. We then use crystallographic analysis of this mutant channel to show how the second site interaction with the membrane changes the channel structure and leads to formation of the high-affinity PIP2 binding site.
MATERIALS AND METHODS
Cloning, expression, and purification
The chicken Kir2.2 DNA plasmid was a generous gift from R. MacKinnon (The Rockefeller University, New York, NY). To use commercially available anti-flag resin (Sigma-Aldrich) for protein purification, the C-terminal 1D4-tagged sequence in cKir2.2 (Tao et al., 2009) was replaced with a Flag sequence. A single point mutation (K62W) was introduced using QuikChange site-directed mutagenesis kits (Agilent Technologies) and verified by sequencing. K62W mutant channels were expressed in Pichia pastoris cells and purified with 100 mM n-decyl-β-d-maltoside (DM; Anatrace), as described previously (Tao et al., 2009). In brief, frozen P. pastoris cells were broken using a model MM301 mixer mill (Retsch, Inc.; 5 × 3.0 min at 30 cps) and solubilized in lysis buffer (100 mM DM, 50 mM Tris, pH 7.5, 150 mM KCl, 1 mM EDTA, and 2 mM DTT) at room temperature for 1 h with stirring. Cell lysate was centrifuged at 30,000 g for 30 min at 10°C. About 750 µl of resin per 10 g of cells was added to the supernatant. Binding was performed at 4°C for >1.5 h with gentle rotation. The resin was washed with 20 column volumes of wash buffer (4 mM DM, 50 mM Tris, pH 7.5, 150 mM KCl, 1 mM EDTA, and 2 mM DTT), and protein was eluted by wash buffer with 200 µg/ml Flag peptide, over the course of 2–3 h. PreScission Protease (PPX) was added at a 1:20 ratio, and cleavage was performed for 12 h at 4°C with gentle rotation. The protein was concentrated and further purified on a Superdex 200 column (GE Healthcare) equilibrated in SEC buffer (20 mM Tris, pH 7.5, 150 mM KCl, 1 mM EDTA, 4 mM DM, and 20 mM DTT). Peak fractions were combined and concentrated to 5 mg/ml for crystallization experiments.
Rb+ uptake assay
Channel activity was assessed by 86Rb+ uptake into proteoliposomes. 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG) lipids were dissolved (10 mg/ml) in buffer A (450 mM KCl, 10 mM HEPES, 4 mM NMG, and 0.5 mM EGTA, pH 7.4) with 35 mM CHAPS. Porcine brain PIP2 was solubilized (2 mg/ml) in 8 mg/ml POPE solution. 1 mg of lipid mixture in 100 µl was incubated at room temperature for 2 h, and 3 µg of protein was added and incubated for another 20 min. The lipid–protein mixture was passed through partly dehydrated G-50 beads preequilibrated with buffer A to form proteoliposomes. Proteoliposomes were passed through partly dehydrated G-50 beads preequilibrated by buffer B (400 mM Sorbitol, 10 mM HEPES, 4 mM NMDG, and 0.5 mM EGTA, pH 7.4) to remove external KCl. 200 µl of buffer B containing 86Rb+ (PerkinElmer) at ∼4.5 nCi was added at time 0. After 10 min, 80-µl samples were harvested and passed through cation exchange beads to capture 86Rb+ in the external solution. The uptake was normalized to the maximum uptake in valinomycin after subtraction of leak uptake into protein-free liposomes. Porcine brain PI(4,5)P2, POPE, POPG, and lipids for crystallization were purchased from Avanti Polar Lipids, Inc.
Crystallization and structure determination
PIP2 was solubilized in 5 mM SEC buffer and added to the concentrated protein sample to 250 µM concentration 30 min before crystallization. Dioleoylglycerol pyrophosphate (DGPP/pyrophosphatidic acid [PPA]) and 1,2-didecanoyl-sn-glycero-3-phosphate (10:0 phosphatidic acid [PA]/PA) were solubilized at 60 mM in SEC buffer. PPA was added to protein samples at 3 mM and PA at 5 mM for crystallization experiments. Crystals were grown by hanging drop vapor diffusion by mixing 0.2 µl of protein and 0.2 µl of reservoir solutions, automated using CRYSTAL GRYPHON (Art Robbins Instruments). Apo-K62W crystals and crystals in the presence of PA or PPA grew in 100 mM tri-sodium citrate pH 5.8–7.0, 100 mM NaCl, and 20–26% PEG400. PIP2-bound K62W crystals grew in 50 mM HEPES, pH 6.3–6.8, 500 mM KCl, and 18–25% PEG400. All crystals grew at 20°C in 2–3 d and were cryoprotected by 30% (vol/vol) PEG400 and frozen in liquid nitrogen. X-ray diffraction data (2.0 Å for Apo, 2.8 Å for PIP2, 2.3 Å for PPA, and 2.0 Å for PA crystals) were collected at the Advanced Photon Source beamlines 24-ID-C and 24-ID-E at wavelength 0.9792 Å under liquid nitrogen stream. Phases were obtained for Apo-K62W crystals using the program MolRep (Vagin and Teplyakov, 2000) in the CCP4 suite (Winn et al., 2011) through molecular replacement with PIP2-bound cKir2.2 crystal structure (3SPI) as a search model. Iterative model building was performed in COOT (Emsley and Cowtan, 2004), and rounds of refinement were performed with REFMAC (Murshudov et al., 1997). The final model was obtained with R/Rfree of 0.199/0.227 and with 97.1% of residues in the most favored regions and none in the disallowed region in the Ramachandran plot. Phases for other crystals (PIP2-bound K62W, +PA, or +PPA) were determined with Apo-K62W structure as a search model, and COOT and REFMAC programs were used for iterative model building and refinement. The side chain orientation for residues such as Gln, His, and Asn was guided by the suggestions of MolProbity webserver (Chen et al., 2010). The final models were obtained with R/Rfree of 0.197/0.258 for PIP2, 0.194/0.221 for PA, and 0.205/0.235 for PPA crystals. There were no Ramachandran outliers for these three models, and 95.37% residues of PIP2, 96.53% of PA, and 97.27% of PPA models were in the most favored regions of the Ramachandran plot. Atomic coordinates and structure factors for the reported crystal structures have been deposited into the Protein Data Bank under accession nos. 5KUK (Apo-K62W) and 5KUM (PIP2-bound K62W).
Molecular dynamics (MD) simulations
MD simulations were performed using Gromacs software version 4.5.4 (Hess et al., 2008). 3 × 100-ns MD simulations were performed for both PIP2-bound K62W and WT (3SPI; Hansen et al., 2011) crystal structures. PIP2 was parameterized using the Hartree-Fock geometry optimization with the 6-31G* basis set (Frisch et al., 2009). Both structures were embedded in a lipid bilayer consisting of 592 1-Palmitoyl-2-oleoylphosphatidylcholine (POPC) lipids using the g_membed tool (Wolf et al., 2010) and solvated using the SPC/E water model (Berendsen et al., 1987). K+ and Cl− ions were randomly placed within the solvent to neutralize the system to obtain an ion concentration of 150 mM. The amber99sb force field (Hornak et al., 2006) was used for the protein. Berger lipid parameters were used for the POPC membrane (Berger et al., 1997; Cordomí et al., 2012). The corrected monovalent ion Lennard-Jones parameters for the amber force field were used as ion parameters (Joung and Cheatham, 2008). Cutoffs for electrostatic interactions were set to 1.0 nm. The particle-mesh Ewald method was used to treat long-range electrostatic interactions at every step (Darden et al., 1993). Lennard-Jones interactions were calculated with a cut off of 1.0 nm. To constrain bonds, we used the LINCS algorithm (Hess et al., 1997). To keep the simulation temperature constant, we used the Nosé-Hoover thermostat (Nosé, 1984; Hoover, 1985), coupling (τ = 0.2 ps) the protein, lipids (PIP2 and POPC), and solvent (water and ions) separately to a 310-K temperature bath. Pressure was kept constant using the Parrinello-Rahman barostat algorithm (Parrinello and Rahman, 1981) at 1 bar, using a coupling constant of 1 ps. Before each simulation, the protein atoms were restrained by a force constant of 1,000 kJ/mol/nm2 to their initial position, and 1,000 conjugate gradient energy-minimization steps were performed, followed by 5 ns of equilibrium simulation. Lipids, ions, and water were allowed to move freely. The root mean square deviation of simulated protein structures all converged to roughly 3.0 Å in the WT and 3.2 Å in the K62W mutant structure at around 20 ns (Fig. S2), indicating that the simulated systems were stable and at equilibrium.
Hydrogen bond analysis
Hydrogen bond analyses were performed using the Gromacs tool g_hbond. It defines a hydrogen bond with a distance cutoff of 3.5 Å and an angle cutoff of 30 degrees between donor-acceptor-hydrogen. Fig. 4 C shows means of the data over all four subunits over all three runs of PIP2-bound K62W versus 3SPI.
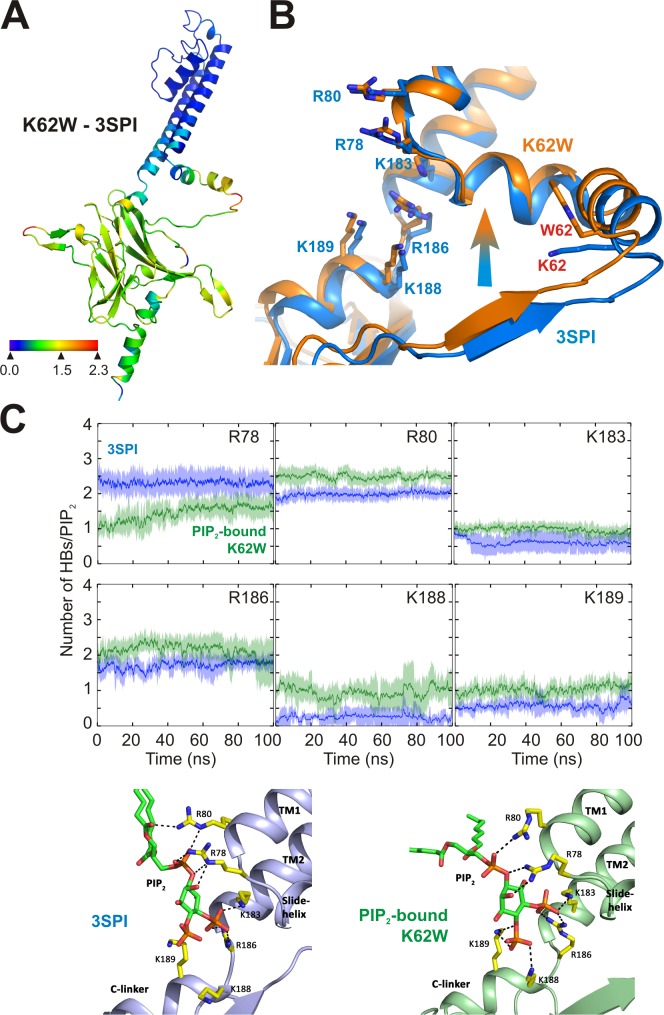
Figure 4.
Tighter tethering of the CTD to the membrane in K62W channels. (A) Ribbon diagrams, colored to indicate the conformational displacement perpendicular to the membrane plane (along the z axis) between the Apo-K62W and PIP2-bound WT (3SPI) structures. Positive distances indicate that the corresponding residues in the first structure are closer to the membrane plane. (B) Ribbon diagram showing overlaid structures of Apo-K62W (orange) and PIP2-bound WT (blue). The residues directly interacting with PIP2 are shown in sticks and labeled in blue, and residue 62 for bulk anionic lipid binding is shown in sticks and labeled in red. (C) Three independent MD simulations were performed for 100 ns, with PIP2-bound WT (3SPI) and K62W mutant structures. After 5-ns equilibration, hydrogen bond formation between PIP2 and neighboring basic residues (Arg78, Arg80, Lys 183, Arg186, Lys188, and Arg189) was assessed, based on the distance (3.5 Å) between donor and acceptor atoms and D-A-H angle (<30o). The results are means ± SD, three independent simulations of 100 ns in each case and of four tetramers in each case. (D) Snapshots showing details of the PIP2-binding site in the PIP2-bound WT (3SPI; left) and K62W (right) structures after 100 ns. Hydrogen bonds between PIP2 and the neighboring residues (yellow sticks) are shown as black dashes.
Online supplemental material
Fig. S1 shows comparison of backbone structures of WT and K62W cKir2.2 in the absence or presence of PIP2. Fig. S2 shows the time course of MD simulations of PIP2-bound WT (3SPI) and K62W mutant protein crystal structures. Fig. S3 shows electron density maps at the primary site, generated for crystals obtained with added PA or PPA lipids. Fig. S4 shows conformational changes perpendicular to membrane surface induced by Gβγ binding in GIRK2. Videos 1 and 2 illustrate positive fo-fc electron density at the primary site for Apo-K62W and PIP2-bound K62W, respectively. Table S1 provides crystallographic data and refinement statistics. Online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.201611616/DC1.
RESULTS
Trp substitution at the second site induces the synergistic PL− effect
Mutation of the key second site residue 64 in hKir2.1 from lysine to cysteine results in loss of PL− sensitivity, but modification of the cysteine with decyl-MTS tethers the site directly to the membrane inner leaflet, resulting in high-affinity PIP2 interaction with the primary site and high sensitivity of the channel to PIP2 (Lee et al., 2013). Aromatic tryptophan residues can also act to tether proteins at the membrane–water interface (Killian and von Heijne, 2000), so as a further test of the proposed functional role of the second site, we introduced a Trp mutation (K62W) at this site into cKir2.2 (Fig. 1 B) and then examined PL− sensitivity (Fig. 1, C and D). Like WT hKir2.1 and cKir2.2 channels, K62W mutant channels were inactive in the absence of PIP2. However, unlike WT channels, K62W channels exhibited high PIP2 sensitivity in even the complete absence of PL−: in zero POPG, K62W channel activity and apparent Kd for PIP2 were equivalent to those for the WT channel at saturating 10% POPG (Fig. 1 C). In the presence of a physiological level of PIP2 (0.1%), WT channels were inactive in the absence of POPG and markedly activated by POPG (∼10-fold), but K62W channels were highly active in the absence of POPG and only marginally more activated (<2-fold) by addition of POPG (Fig. 1 D). These results show that the K62W mutation essentially endows the same synergistic effect on PIP2 sensitivity and channel activation as PL−, even in the absence of PL− in the membrane.
Structures of cKir2.2 K62W with or without PIP2 are almost identical
Given that the mutation K62W, like chemical tethering of the K64C mutation in hKir2.1 (Lee et al., 2013), generates a similar PIP2 sensitizing effect to PL− binding, we crystallized cKir2.2[K62W] (Tao et al., 2009; Hansen et al., 2011) in the presence and absence of PIP2 to directly observe structural consequences. 2.0-Å and 2.8-Å resolution x-ray diffraction data were obtained for Apo- and PIP2-bound K62W crystals, respectively (Table S1). Phases were obtained by molecular replacement using 3SPI as a search model (Hansen et al., 2011), and structural models were built and refined to an Rfree of 0.227 for Apo- and 0.258 for PIP2-bound K62W. In the WT cKir2.2 structure obtained in the absence of PIP2 at the primary site (3JYC), the CTD is displaced ∼6 Å from the TMD (Fig. S1; Tao et al., 2009; Hansen et al., 2011). In marked contrast, the two final structural models for cKir2.2[K62W] are indistinguishable from each other, the CTD being tightly tethered to the TMD in both the presence and absence of PIP2 at the primary site (Fig. 2, A and B).
Figure 2.
Apo- and PIP2-bound K62W crystal structures. (A) K62W mutant channel structures determined without (Apo-, orange; PDB no. 5KUK) or with PIP2 (PIP2-bound, red; PDB no. 5KUM) are shown in ribbon diagram (PIP2 shown as space-filling balls). (B) Ribbon diagrams, colored to indicate the conformational displacement perpendicular to the membrane plane (along the z axis) between the two structures. The distance changes along the z axis at every Cα were computed after alignment of the two crystal structures at selectivity filter backbone atoms (residue 143–148). (C) I177 and M181 residues in the bundle crossing region are shown as space-filling spheres, for Apo (left) and PIP2 bound (right).
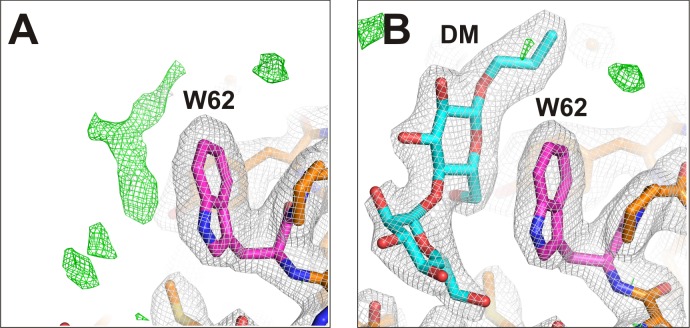
Knock-in Trp directly interacts with a detergent head group
Given the conformational and functional consequences of the K62W mutation, the electron density around residue 62 was examined closely (Fig. 3). Additional positive density that is evident very near the tryptophan side chain in both Apo- and PIP2-bound K62W crystals (Fig. 3 A) is well fit by a maltoside head group (Fig. 3 B). This is consistent with the introduced tryptophan directly interacting with decyl-maltoside during purification, as the detergent molecule replaces the native membrane lipid.
Figure 3.
K62W structure at the second PL− site. (A and B) The Apo-K62W structural model was generated by molecular replacement with 3SPI residue 62 as Trp (A), and final electron density and a model with a decylmaltoside detergent head group are shown (B). In each case, 2fo-fc electron density at 2.0-Å resolution, contoured at 1σ, is shown in gray. Positive fo-fc electron density at 2.0-Å resolution, contoured at 3σ, is shown in green. Pink indicates carbon atoms of the residue mutated; orange indicates carbon atoms of the wild-type residues; cyan indicates carbon atoms of the ligand (DM); blue indicates nitrogen atoms; red indicates oxygen atoms.
Tighter membrane tethering in K62W maintains the high-affinity primary site structure
Comparison of the Apo-WT and Apo-K62W structures indicates a global translation of the cytoplasmic domain along the z axis (i.e., perpendicular to the membrane) by ∼6 Å toward the membrane in the latter (Fig. S1). Even comparing with the PIP2-bound WT (3SPI) structure, there is ∼1-Å movement toward the membrane in the K62W structure (Fig. 4 A). The whole CTD is displaced toward the membrane, but this upward movement is greatest in the region of the second site. Induction of the high-affinity primary PIP2-binding site in the K62W structure, even in the absence of PIP2 (Fig. 4 B), provides a compelling structural mechanism for the positive allosteric effect of PL− binding on PIP2 sensitivity, i.e., that pulling of the cytoplasmic domain toward the membrane at the PL− site induces or facilitates formation of the high-affinity PIP2 site. In addition, the tighter apposition of the CTD to the membrane plane in the K62W structure brings several positively charged residues even closer to the primary site than in the PIP2-bound (3SPI) WT structure. MD simulations of PIP2-bound WT (3SPI) and PIP2-bound K62W crystal structures show that PIP2 is bound through more extensive hydrogen bonding to these residues in the K62W mutant channel (Fig. 4 C). In particular, at least one hydrogen bond was persistently present between PIP2 and K188 and K189 only in PIP2-bound K62W MD simulations. Previous studies have indicated that mutation of these residues to glutamine causes complete loss of function for Kir2.1 channels (Lopes et al., 2002; D’Avanzo et al., 2013). These results indicate that the second site interaction with the membrane strengthens interaction of PIP2 at the primary site.
The “bundle crossing” gate is still closed in K62W structures
At the narrowest region of the pore, the distance between the closest side chain carbon atoms (between Cα atoms in parentheses) of Ile 177 is 5.9 (11.43) Å for the Apo- and 6.0 (11.45) Å for PIP2-bound K62W channels, similar to 6.3 (11.07) Å in the PIP2-bound WT channel (Fig. 2 C; Hansen et al., 2011). Thus, even when both PIP2 and PL− interactions are satisfied, channels are still formally closed at the inner helix bundle crossing, as is also seen in Kir3.2 (GIRK2) channel structures in the presence of all relevant agonists (Whorton and MacKinnon, 2011, 2013).
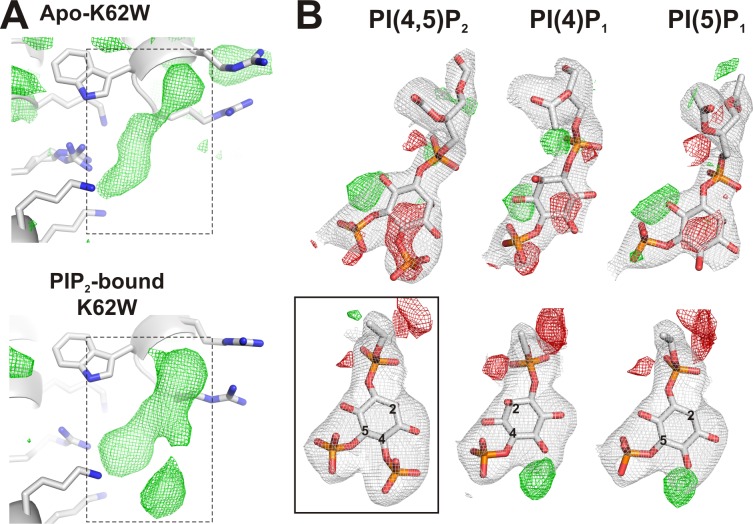
An unidentified molecule is present at the primary site in Apo-K62W crystals
In the absence of PIP2 in crystallization conditions, strong positive electron density is still detected at the primary site in Apo-K62W crystals. This raises the possibility that PIP2 or other phospholipid is copurified from the P. pastoris membrane. However, comparison of electron density maps (Fig. 5 A and Videos 1 and 2) for Apo-K62W and PIP2-bound K62W crystals before introduction of PIP2 in the refinement reveals obvious differences between the contour shapes of the unmodeled electron densities in these crystals, suggesting that the extra density in Apo-K62W crystal does not arise from copurified PIP2. As a further test of whether the copurified molecule is PIP2 or other phosphoinositides, electron density maps were recalculated with these lipids modeled at the primary site. Introduction of PIP2 fully accounts for the positive electron density in PIP2-bound K62W crystals (Fig. 5 B, boxed), but substantial mismatches result when PI(4,5)P2 or the monophosphoinositides PI(4)P or PI(5)P are modeled into the Apo-K62W structure (Fig. 5 B), in particular near the C4 and C6 positions, indicating that the presumably copurified molecule from P. pastoris is neither PIP2 nor monophosphoinositide.
Figure 5.
Positive density at the primary site in Apo-K62W. (A) Electron density at the primary site with no ligands modeled for Apo-K62W (top) or PIP2-bound K62W (bottom). Only fo-fc electron density calculated at 2.8-Å resolution and contoured at 2.5σ (green) is shown for clarity. The dashed boxes indicate the space corresponding to electron densities shown in B. Blue indicates nitrogen atoms. (B) Electron density recalculated after refinement in the presence of putative ligands, PI(4,5)P2, PI(4)P1, or PI(5)P1 (shown as sticks). The solid box indicates the correct assignment. 2fo-fc electron density at 2.8-Å resolution for Apo-K62W and PIP2-bound K62W, respectively, contoured at 1σ, is shown in gray. fo-fc electron density at 2.8-Å resolution contoured at 3σ and −3σ is shown in green and red, respectively. Orange indicates phosphate atoms.
At this juncture, we cannot differentiate whether this molecule is an opportunistic occupant of or catalyst to formation of the primary site. It is likely to be strongly negatively charged, like PIP2, to sit stably in the groove surrounded by basic residues. Our previous docking simulations on WT channels indicated that all negatively charged lipids favor binding to the primary site over the second site, with PA showing the most stable binding of singly negatively charged PL−s (Lee et al., 2013). In addition to PIP2, another strongly negatively charged PPA was resolved at the primary site in WT cKir2.2 channels (Hansen et al., 2011). Hence, we further examined whether such a lipid might be, or might displace, the copurifying P. pastoris molecule by crystallizing K62W proteins in the presence of high concentrations of PA (5 mM) or PPA (3 mM). Electron density contours at the primary site in these crystals were essentially identical to those in Apo-K62W crystals (Fig. S3, A and B) and inconsistent with the presence of PPA (3SPC; Fig. S3 D). Thus, this unidentified molecule bound more stably than PA or PPA but less stably than PIP2.
DISCUSSION
A refined model for lipid gating of Kir2 channels
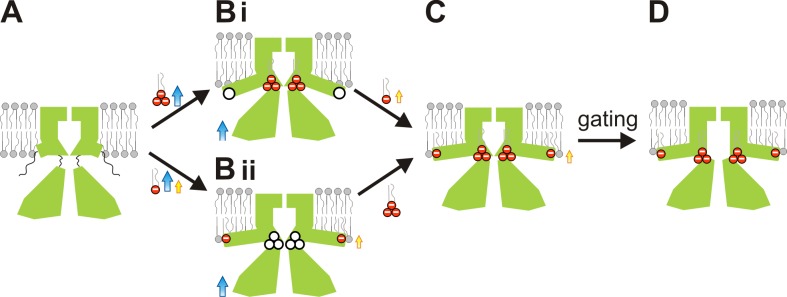
Combined with previous analyses (Tao et al., 2009; Hansen et al., 2011; Whorton and MacKinnon, 2011, 2013), our findings suggest a refined model for the role of the two lipid-binding sites in controlling Kir2 channel gating. In the absence of any negatively charged lipids, the CTD of Kir2 channels is disengaged from the TMD, with the C-linker and slide helix region unstructured (Tao et al., 2009); both the primary and second sites are amorphous (Fig. 6 A). Binding of PIP2 to the primary site (Fig. 6 B, top) or of bulk PL− to the second site (Fig. 6 B, bottom) pulls the CTD toward the membrane, thereby inducing the other binding site. Subsequent binding of PL− at the second site (Fig. 6 C, top) or of PIP2 at the primary site (Fig. 6 C, bottom) then establishes the state from which the channel can readily open (Fig. 6 D), presumably by splaying of the inner helices and widening of the bundle crossing, as hinted at in other crystal structures (Jiang et al., 2002; McCusker et al., 2012; Linder et al., 2013; Whorton and MacKinnon, 2013). Because the second site interaction requirement is likely to be constitutively met in most cellular membranes, the gating transition is likely to occur primarily through the bottom pathway in the model (i.e., Fig. 6, A→Bii→C→D).
Figure 6.
Lipid-dependent gating of Kir2 channels. (A) In the absence of any PL−s, the two binding sites are unstructured and the CTD is displaced away from the membrane. (B) Binding of PIP2 at the primary site or of PL− binding at the second site initiates upward movement of the CTD toward the membrane and induces the formation of the other site (white circles). (C) Subsequent binding to the other site places the channel in a preactivated state (D), from which opening is favored.
Tryptophan mimicry of the second site PL− interaction
Preferential positioning of aromatic residues, particularly tryptophan (Trp) and tyrosine (Tyr), at the membrane interface is apparent in many membrane protein crystal structures (Wallin et al., 1997). Trp is energetically favored for residing at the zwitterionic membrane interface by ∼3 kcal/mol compared with the positively charged Lys residue (Wimley and White, 1996). A sophisticated study involving organic molecules that are similar to the Trp side chain structure and solid state nuclear magnetic resonance methods indicates that stable Trp interaction at the membrane interface results mainly from aromaticity: π electrons interact with the electrostatically complex interface environment while the rigid flat shape of the aromatic ring disfavors deeper penetration into the hydrophobic core of the membrane (Yau et al., 1998). As a consequence, Trp inserted in the middle of a hydrophobic peptide can make the otherwise integral membrane peptide remain at the interface (Braun and von Heijne, 1999). Such strong Trp propensity to reside at the membrane interface provides a compelling explanation for how the K62W mutation effectively tethers the PL− site at the membrane surface, whereas the WT K62 residue is displaced from the membrane in the absence of electrostatic interaction with PL−. Thus, there is a lack of interaction of the native Lys residue in the WT Kir2.2 protein but strong interaction of Trp in the K62W mutant with the membrane lipid, replaced by interaction with the detergent head group in the crystal structures (Fig. 3).
Convergent allosteric mechanisms for PIP2-mediated Kir channel family gating
Although comparable studies have not yet been performed on purified proteins from other members of the Kir channel family, the key positively charged residues in the secondary PL− interaction site (K64 and K219 in human Kir2.1; K62 and K220 in chicken Kir2.2) are not highly conserved among Kir subfamilies (Fig. 1 B). This suggests that regulation by PL− specifically may not be a universal regulator for all Kir subfamilies, but the structural mechanism of cytoplasmic domain tethering for induction of PIP2 activation might be. Consistent with this idea, it is notable that those Kir subfamily members that exhibit low conservation at the PL− site tend to have other regulatory ligands that interact with the same region. Kir6 and Kir3 subfamilies use auxiliary regulatory protein subunits, sulfonylurea receptor and Gβγ, respectively. Kir5.1, in which neither of these residues is conserved, only generates active channels as an obligatory heterotetramer with Kir4 subunits, in which both PL−-interacting residues are conserved (Hibino et al., 2010). It is worth noting that the second site structurally overlaps with binding sites for other allosteric ligands, including Na+ (Ho and Murrell-Lagnado, 1999; Whorton and MacKinnon, 2011) and alcohol (Aryal et al., 2009), as well as Gβγ (Yokogawa et al., 2011; Whorton and MacKinnon, 2013) and Gαi/o (Mase et al., 2012), that have been identified in Kir3 channel structures. Moreover, structural changes along the membrane normal induced by Gβγ binding in Kir3 channels appear qualitatively similar to those induced by K62W in Kir2 channels (Fig. S4), consistent with a common structural basis for these disparate structural effectors.
Interestingly, residue Q52 in Kir6.2 aligns with K62 of Kir2.1. A naturally occurring positive charge substitution (Q52R) at this position, causally linked to human neonatal diabetes, results in a dramatic increase in open state stability and reduction of ATP inhibition (Proks et al., 2004; Koster et al., 2005). A recent study indicates that this residue may physically interact with the sulfonylurea receptor subunit to affect ATP sensitivity of the channels (Pratt et al., 2012), providing further consistency with the idea that the secondary site is a common allosteric regulatory site in Kir channels. The present study provides a physical model for the positive allosteric regulation of PIP2 sensitivity in Kir2 channels by PL− binding, but the convergent location of secondary allosteric sites in other Kir channels is consistent with a common physical basis, i.e., apposition of the slide helix to the membrane, for regulation of PIP2 sensitivity throughout the Kir channel family.
Supplementary Material
ACKNOWLEDGMENTS
The authors are very grateful to Dr. Roderick MacKinnon for providing the chicken Kir2.2 DNA construct that we used for P. pastoris expression and crystallization. We thank the staffs at APS beamlines 24-ID-C/E, especially Dr. Kay Perry for assistance with data collection and Dr. Zengqin Deng for help with crystallization.
This work was supported by National Institutes of Health grant HL54171 (to C.G. Nichols), American Heart Association fellowship 15POST22390016 (to S.-J. Lee), Center for the Investigation and Membrane Excitability Diseases Pilot and Feasibility grant CIMED-15-01 (to P. Yuan), and Austrian Science Fund grants W1232 (to E.-M. Zangerl-Plessl and A. Stary-Weinzinger) and I-2101-B26 (to A. Stary-Weinzinger). The computational simulations were carried out using the Vienna Scientific Cluster (VSC).
The authors declare no competing financial interests.
Author contributions: S.-J. Lee and C.G. Nichols conceived of the project. S.-J. Lee, S. Heyman, and F. Ren carried out experiments. P. Yuan supervised the crystallography and analyses. The molecular modeling was carried out by E.-M. Zangerl-Plessl and A. Stary-Weinzinger. C.G. Nichols, S.-J. Lee, and F. Ren analyzed the data. C.G. Nichols and S.-J. Lee wrote the paper, which was edited by F. Ren, P. Yuan, and A. Stary-Weinzinger.
Richard W. Aldrich served as editor.
Footnotes
Abbreviations used:
- CTD
- C-terminal domain
- MD
- molecular dynamics
- PA
- phosphatidic acid
- PIP2
- phosphatidylinositol-4,5-bisphosphate
- PPA
- pyrophosphatidic acid
- TMD
- transmembrane domain
References
- Aryal P., Dvir H., Choe S., and Slesinger P.A.. 2009. A discrete alcohol pocket involved in GIRK channel activation. Nat. Neurosci. 12:988–995. 10.1038/nn.2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen H.J.C., Grigera J.R., and Straatsma T.P.. 1987. The missing term in effective pair potentials. J. Phys. Chem. 91:6269–6271. 10.1021/j100308a038 [DOI] [Google Scholar]
- Berger O., Edholm O., and Jähnig F.. 1997. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 72:2002–2013. 10.1016/S0006-3495(97)78845-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun P., and von Heijne G.. 1999. The aromatic residues Trp and Phe have different effects on the positioning of a transmembrane helix in the microsomal membrane. Biochemistry. 38:9778–9782. 10.1021/bi990923a [DOI] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B. III, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., and Richardson D.C.. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66:12–21. 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W.W.L., D’Avanzo N., Doyle D.A., and Nichols C.G.. 2011. Dual-mode phospholipid regulation of human inward rectifying potassium channels. Biophys. J. 100:620–628. 10.1016/j.bpj.2010.12.3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordomí A., Caltabiano G., and Pardo L.. 2012. Membrane protein simulations using AMBER force field and Berger lipid parameters. J. Chem. Theory Comput. 8:948–958. 10.1021/ct200491c [DOI] [PubMed] [Google Scholar]
- D’Avanzo N., Cheng W.W., Wang S., Enkvetchakul D., and Nichols C.G.. 2010a Lipids driving protein structure? Evolutionary adaptations in Kir channels. Channels (Austin). 4:139–141. 10.4161/chan.4.3.12129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avanzo N., Cheng W.W., Xia X., Dong L., Savitsky P., Nichols C.G., and Doyle D.A.. 2010b Expression and purification of recombinant human inward rectifier K+ (KCNJ) channels in Saccharomyces cerevisiae. Protein Expr. Purif. 71:115–121. 10.1016/j.pep.2010.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avanzo N., Lee S.J., Cheng W.W., and Nichols C.G.. 2013. Energetics and location of phosphoinositide binding in human Kir2.1 channels. J. Biol. Chem. 288:16726–16737. 10.1074/jbc.M113.452540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T., York D., and Pedersen L.. 1993. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089 10.1063/1.464397 [DOI] [Google Scholar]
- Emsley P., and Cowtan K.. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Enkvetchakul D., and Nichols C.G.. 2003. Gating mechanism of KATP channels: function fits form. J. Gen. Physiol. 122:471–480. 10.1085/jgp.200308878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Mennucci B., Petersson G.A., et al. 2009. Gaussian 09, Revision A.02. Gaussian Inc., Wallingford, CT. Available at: http://www.gaussian.com/g_prod/g09.htm.
- Hansen S.B., Tao X., and MacKinnon R.. 2011. Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature. 477:495–498. 10.1038/nature10370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B., Bekker H., Berendsen H.J.C., and Fraaije J.G.E.M.. 1997. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463–1472. [DOI] [Google Scholar]
- Hess B., Kutzner C., van der Spoel D., and Lindahl E.. 2008. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4:435–447. 10.1021/ct700301q [DOI] [PubMed] [Google Scholar]
- Hibino H., Inanobe A., Furutani K., Murakami S., Findlay I., and Kurachi Y.. 2010. Inwardly rectifying potassium channels: their structure, function, and physiological roles. Physiol. Rev. 90:291–366. 10.1152/physrev.00021.2009 [DOI] [PubMed] [Google Scholar]
- Hilgemann D.W., and Ball R.. 1996. Regulation of cardiac Na+,Ca2+ exchange and KATP potassium channels by PIP2. Science. 273:956–959. 10.1126/science.273.5277.956 [DOI] [PubMed] [Google Scholar]
- Ho I.H., and Murrell-Lagnado R.D.. 1999. Molecular determinants for sodium-dependent activation of G protein-gated K+ channels. J. Biol. Chem. 274:8639–8648. 10.1074/jbc.274.13.8639 [DOI] [PubMed] [Google Scholar]
- Hoover W.G. 1985. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 31:1695–1697. 10.1103/PhysRevA.31.1695 [DOI] [PubMed] [Google Scholar]
- Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., and Simmerling C.. 2006. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 65:712–725. 10.1002/prot.21123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingólfsson H.I., Melo M.N., van Eerden F.J., Arnarez C., Lopez C.A., Wassenaar T.A., Periole X., de Vries A.H., Tieleman D.P., and Marrink S.J.. 2014. Lipid organization of the plasma membrane. J. Am. Chem. Soc. 136:14554–14559. 10.1021/ja507832e [DOI] [PubMed] [Google Scholar]
- Jiang Y., Lee A., Chen J., Cadene M., Chait B.T., and MacKinnon R.. 2002. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 417:515–522. 10.1038/417515a [DOI] [PubMed] [Google Scholar]
- Joung I.S., and Cheatham T.E. III. 2008. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B. 112:9020–9041. 10.1021/jp8001614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian J.A., and von Heijne G.. 2000. How proteins adapt to a membrane-water interface. Trends Biochem. Sci. 25:429–434. 10.1016/S0968-0004(00)01626-1 [DOI] [PubMed] [Google Scholar]
- Koster J.C., Remedi M.S., Dao C., and Nichols C.G.. 2005. ATP and sulfonylurea sensitivity of mutant ATP-sensitive K+ channels in neonatal diabetes: implications for pharmacogenomic therapy. Diabetes. 54:2645–2654. 10.2337/diabetes.54.9.2645 [DOI] [PubMed] [Google Scholar]
- Lee S.-J., Wang S., Borschel W., Heyman S., Gyore J., and Nichols C.G.. 2013. Secondary anionic phospholipid binding site and gating mechanism in Kir2.1 inward rectifier channels. Nat. Commun. 4:2786 10.1038/ncomms3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder T., de Groot B.L., and Stary-Weinzinger A.. 2013. Probing the energy landscape of activation gating of the bacterial potassium channel KcsA. PLOS Comput. Biol. 9:e1003058 10.1371/journal.pcbi.1003058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes C.M., Zhang H., Rohacs T., Jin T., Yang J., and Logothetis D.E.. 2002. Alterations in conserved Kir channel-PIP2 interactions underlie channelopathies. Neuron. 34:933–944. 10.1016/S0896-6273(02)00725-0 [DOI] [PubMed] [Google Scholar]
- Mase Y., Yokogawa M., Osawa M., and Shimada I.. 2012. Structural basis for modulation of gating property of G protein-gated inwardly rectifying potassium ion channel (GIRK) by i/o-family G protein α subunit (Gαi/o). J. Biol. Chem. 287:19537–19549. 10.1074/jbc.M112.353888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker E.C., Bagnéris C., Naylor C.E., Cole A.R., D’Avanzo N., Nichols C.G., and Wallace B.A.. 2012. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat. Commun. 3:1102 10.1038/ncomms2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., and Dodson E.J.. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255. 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- Nichols C.G., and Lopatin A.N.. 1997. Inward rectifier potassium channels. Annu. Rev. Physiol. 59:171–191. 10.1146/annurev.physiol.59.1.171 [DOI] [PubMed] [Google Scholar]
- Nosé S. 1984. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 81:511. [Google Scholar]
- Parrinello M., and Rahman A.. 1981. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 52:7182 10.1063/1.328693 [DOI] [Google Scholar]
- Pratt E.B., Zhou Q., Gay J.W., and Shyng S.L.. 2012. Engineered interaction between SUR1 and Kir6.2 that enhances ATP sensitivity in KATP channels. J. Gen. Physiol. 140:175–187. 10.1085/jgp.201210803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proks P., Antcliff J.F., Lippiat J., Gloyn A.L., Hattersley A.T., and Ashcroft F.M.. 2004. Molecular basis of Kir6.2 mutations associated with neonatal diabetes or neonatal diabetes plus neurological features. Proc. Natl. Acad. Sci. USA. 101:17539–17544. 10.1073/pnas.0404756101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohács T., Lopes C.M., Jin T., Ramdya P.P., Molnár Z., and Logothetis D.E.. 2003. Specificity of activation by phosphoinositides determines lipid regulation of Kir channels. Proc. Natl. Acad. Sci. USA. 100:745–750. 10.1073/pnas.0236364100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X., Avalos J.L., Chen J., and MacKinnon R.. 2009. Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 A resolution. Science. 326:1668–1674. 10.1126/science.1180310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A., and Teplyakov A.. 2000. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 56:1622–1624. 10.1107/S0907444900013780 [DOI] [PubMed] [Google Scholar]
- van Meer G., Voelker D.R., and Feigenson G.W.. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9:112–124. 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin E., Tsukihara T., Yoshikawa S., von Heijne G., and Elofsson A.. 1997. Architecture of helix bundle membrane proteins: an analysis of cytochrome c oxidase from bovine mitochondria. Protein Sci. 6:808–815. 10.1002/pro.5560060407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorton M.R., and MacKinnon R.. 2011. Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell. 147:199–208. 10.1016/j.cell.2011.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorton M.R., and MacKinnon R.. 2013. X-ray structure of the mammalian GIRK2–βγ G-protein complex. Nature. 498:190–197. 10.1038/nature12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimley W.C., and White S.H.. 1996. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3:842–848. 10.1038/nsb1096-842 [DOI] [PubMed] [Google Scholar]
- Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67:235–242. 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M.G., Hoefling M., Aponte-Santamaría C., Grubmüller H., and Groenhof G.. 2010. g_membed: Efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation. J. Comput. Chem. 31:2169–2174. 10.1002/jcc.21507 [DOI] [PubMed] [Google Scholar]
- Yau W.M., Wimley W.C., Gawrisch K., and White S.H.. 1998. The preference of tryptophan for membrane interfaces. Biochemistry. 37:14713–14718. 10.1021/bi980809c [DOI] [PubMed] [Google Scholar]
- Yokogawa M., Osawa M., Takeuchi K., Mase Y., and Shimada I.. 2011. NMR analyses of the Gβγ binding and conformational rearrangements of the cytoplasmic pore of G protein-activated inwardly rectifying potassium channel 1 (GIRK1). J. Biol. Chem. 286:2215–2223. 10.1074/jbc.M110.160754 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.