Abstract
HLA molecules play an important role for immunoreactivity in allogeneic hematopoietic stem cell transplantation. To elucidate the effect of specific HLA alleles on acute graft-versus-host disease, we conducted a retrospective analysis using 6967 Japanese patients transplanted with T-cell-replete marrow from an unrelated donor. Using unbiased searches of patient and donor HLA alleles, patient and/or donor HLA-B*51:01 (patient: HR, 1.37, P<0.001; donor: HR, 1.35, P<0.001) and patient HLA-C*14:02 (HR, 1.35, P<0.001) were significantly associated with an increased risk of severe acute graft-versus-host disease. The finding that donor HLA-C*14:02 was not associated with severe acute graft-versus-host disease prompted us to elucidate the relation of these high-risk HLA alleles with patient and donor HLA-C allele mismatches. In comparison to HLA-C allele match, patient mismatched HLA-C*14:02 showed the highest risk of severe acute graft-versus-host disease (HR, 3.61, P<0.001) and transplant-related mortality (HR, 2.53, P<0.001) among all patient mismatched HLA-C alleles. Although patient HLA-C*14:02 and donor HLA-C*15:02 mismatch was usually KIR2DL-ligand mismatch in the graft-versus-host direction, the risk of patient mismatched HLA-C*14:02 for severe acute graft-versus-host disease was obvious regardless of KIR2DL-ligand matching. The effect of patient and/or donor HLA-B*51:01 on acute graft-versus-host disease was attributed not only to strong linkage disequilibrium of HLA-C*14:02 and -B*51:01, but also to the effect of HLA-B*51:01 itself. With regard to clinical implications, patient mismatched HLA-C*14:02 proved to be a potent risk factor for severe acute graft-versus-host disease and mortality, and should be considered a non-permissive HLA-C mismatch in donor selection for unrelated donor hematopoietic stem cell transplantation.
Introduction
Hematopoietic stem cell transplantation (HSCT) from an unrelated (UR) donor is now an established curative therapy for hematologic malignancies and other hematologic or immunological disorders. Human leukocyte antigen (HLA) molecules play an important role in the cellular discrimination of ‘self’ and ‘non-self’ in allogeneic HSCT, and the critical role of patient and donor human HLA matching status on graft rejection, graft-versus-host disease (GvHD), and survival after UR-HSCT has been well defined. The risk of transplant-related immunological events may vary according to patient-donor mismatching HLA locus,1–4 mismatching specific HLA epitopes,5,6 and specific mismatch combinations.7 Furthermore, several studies have demonstrated that specific individual HLA genotypes may also affect transplant outcome,8–12 although the results are not always consistent.
The human major histocompatibility (MHC) region is the most gene-dense region of the human genome, containing more than 150 protein coding genes at approximately 4 Mb. Many of the HLA gene products have coordinated roles in immune function.13 The MHC region has been recognized as one of the most important genetic regions associated with many kinds of human disease, including autoimmune diseases, infections, and cancers.14 Linkage disequilibrium (LD) is the non-random association of alleles at different loci, and the long-range LD blocks observed in MHC haplotypes15 induce genetic fixity in this region. Our previous study revealed that the three common Japanese HLA haplotypes were highly conserved among unrelated individuals up to 8 Mb, and suggested that these haplotypes might contribute to the occurrence of acute GvHD in HLA-identical UR-HSCT.16 We therefore hypothesized that the immunological response following UR-HSCT depends, not only on patient-donor HLA allele matching, but also on specific individual HLA alleles themselves and their linked HLA region.
Here we analyzed the effect of specific HLA alleles on acute GvHD using a large UR-HSCT cohort with extensive HLA-allele data. Grades III–IV acute GvHD was the clinical end point in this study, as this is considered to represent a severe or life-threatening immunological event after allo-HSCT. In unbiased screening of patient and donor HLA alleles, patient and/or donor HLA-B*51:01 and patient HLA-C*14:02 were significantly associated with a higher risk of severe acute GvHD. The effects of these alleles on acute GvHD were attributed to the specific patient mismatched HLA-C allele and its genetically linked HLA-B allele.
Methods
Study population
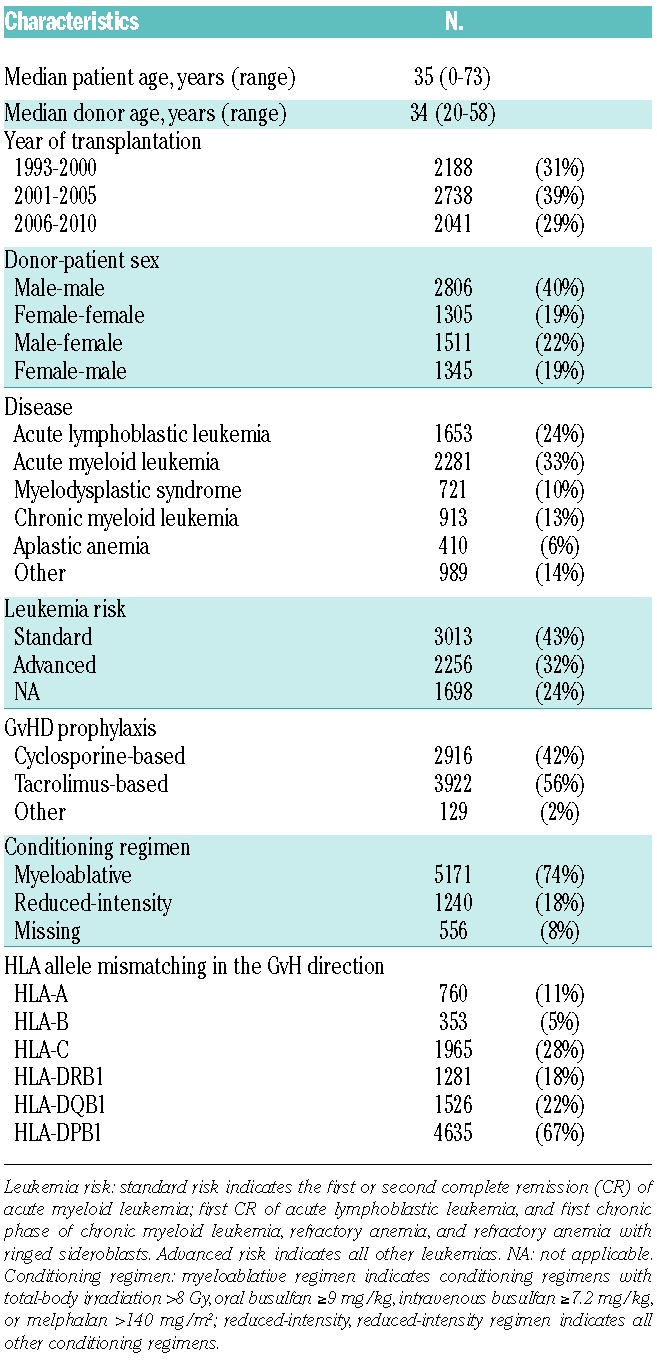
A total of 6967 unrelated donor transplant pairs from the Japan Marrow Donor Program (JMDP) database satisfied the following criteria and were included in the analysis: 1) transplanted from a serologically HLA-A, -B, and -DR antigen-matched donor between January 1993 and December 2010; 2) transplantation pairs were re-typed for HLA-A, -B, -C, -DRB1, -DQB1 and -DPB1 alleles; 3) transplanted non-T-cell-depleted marrow without in vivo use of anti-thymocyte globulin (ATG) for GvHD prophylaxis; 4) first transplantation; 5) Japanese ethnicity; and 6) survival for more than seven days after transplantation. Patients’ and donors’ characteristics are shown in Table 1. A final clinical survey of the patients was completed by September 2012. Informed consent was obtained from patients and donors in accordance with the Declaration of Helsinki, and approval of the study was obtained from the Institutional Review Board of Aichi Cancer Center, Fujita Health University and the JMDP.
Table 1.
Characteristics of patients and donors.
HLA typing and definitions
All donor-patient pairs were retrospectively genotyped for HLA-A, -C, -B, -DRB1, -DQB1, and -DPB1 alleles at the field 1 and field 2 level using either or both the polymerase chain reaction sequence-specific oligonucleotide and PCR-Sequencing Based Typing methods, as described elsewhere.4 HLA allele mismatch in the GvH direction among donor-patient pairs was defined as the patient’s alleles not being shared by the donor. Natural killer cell immunoglobulin-like receptor (KIR) ligand specificity of the HLA-C antigen was determined according to the amino acid residues of the HLA-C allele. C1 ligand specificity consists of Asn80 and C2 ligand specificity consists of Lys80. KIR ligand mismatch in the GvH direction was defined as the donor’s KIR ligand for HLA-C not being shared by the patient’s ligand.17
Biostatistical methods
The study evaluated the impact of specific HLA alleles with a frequency of more than 5% on the outcome of grades III–IV acute GvHD. Outcomes were compared among patient- and donor-specific HLA allele-positive and -negative groups using multivariable competing risk regression analysis,18 adjusted for clinical factors and HLA allele matching (Table 1). We included separate variables for HLA-A,-C,-B,-DRB1,-DQB1 and -DPB1 allele mismatches in the GvH direction.
To analyze the effect of patient mismatched HLA-C allele, those pairs matched for 1 HLA-C allele and mismatched for another HLA-C allele were extracted. The risk of each patient mismatched HLA-C allele on grades III–IV acute GvHD was compared with the HLA-C allele match. The influences of the level of expression of the patient mismatched HLA-C allotype were assessed as described previously.19
The effects of HLA-C allele mismatch combinations were also evaluated using the pairs matched for 1 HLA-C allele and mismatched for another HLA-C allele, and the risk of each HLA-C mismatch combination of grades III–IV acute GvHD was compared with the HLA-C allele match.
Multivariable competing risk regression analyses18 were conducted to evaluate the impact on acute GvHD and transplant-related mortality. A Cox’s proportional hazards regression model was used to evaluate the impact on overall survival (OS).20
A detailed description of the statistical methods is available in the Online Supplementary Appendix.
Results
Identification of HLA alleles associated with grades III–IV acute GvHD
The number of HLA alleles with a frequency more than 5% in each locus was as follows: HLA-A 7, -C 8, -B 8, -DRB1 7, -DQB1 8, and -DPB1 5. P<0.00116 was considered statistically significant (Bonferroni correction). Among 43 HLA alleles with a frequency more than 5%, the only alleles significantly associated with an increased risk of grades III–IV acute GvHD were patient and donor HLA-B*51:01 (patient: HR, 1.37; 95% confidence interval [CI], 1.19–1.59; P<0.001; donor: HR, 1.35; 95%CI: 1.17–1.56; P<0.001) and patient HLA-C*14:02 (HR, 1.35; 95%CI: 1.15–1.58; P<0.001) (Table 2 and Online Supplementary Table S1). These HLA alleles were also associated with a higher risk of mortality (patient HLA-B*51:01: HR, 1.18; 95%CI: 1.07–1.29; P<0.001; donor HLA-B*51:01: HR, 1.15; 95%CI: 1.05–1.26; P=0.001; patient HLA-C*14:02: HR, 1.18; 95%CI: 1.07–1.30; P=0.001).
Table 2.
Effect of HLA-C*14:02, -B*51:01 and their haplotypes on acute graft-versus-host disease (GvHD) and mortality.
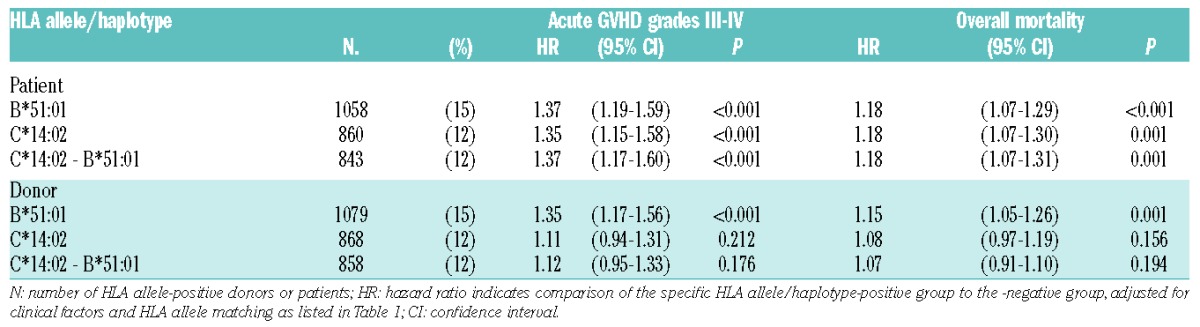
HLA-C*14:02 and -B*51:01 were in strong linkage disequilibrium
Since patient and/or donor HLA-B*51:01, patient HLA-C*14:01 were associated with a higher risk of severe acute GvHD, the linkage between these alleles was examined. Firstly, because almost all patients with HLA-B*51:01 (1053 of 1058) received transplants from donors with HLA-B*51:01, we were unable to determine which patient or donor HLA-B*51:01 contributed to increasing the risk of acute GvHD. Secondly, HLA-B*51:01 demonstrated strong positive LD with HLA-C*14:02 among Japanese.21 In the present analysis, 98% of patients with HLA-C*14:02 (843 of 860) were HLA-B*51:01-positive; therefore, the patient HLA-C*14:02-B*51:01 haplotype showed a similar effect in increasing the risk of grades III–IV acute GvHD (HR, 1.37; 95%CI: 1.17–1.60; P<0.001) and mortality (HR, 1.18; 95%CI: 1.07–1.31; P<0.001) as patient HLA-C*14:02 (Table 2). Among HLA-B*51:01-positive patients (n=1058), 843 patients (80%) had HLA-C*14:02, while 215 patients had HLA-C alleles other than HLA-C*14:02. We analyzed the effect of HLA-B*51:01 on acute GvHD in patients without HLA-C*14:02 to ascertain whether HLA-B*51:01 itself has an increased risk of acute GvHD irrespective of HLA-C*14:02. Patient HLA-B*51:01 was significantly associated with an increased risk of grades III–IV GvHD in the subgroup of excluded patients with HLA-C*14:02 (n=6197) (HR, 1.34; 95%CI: 1.01–1.79; P=0.046) (Table 3).
Table 3.
Subgroup analysis of the effect of patient HLA-B*51:01 and -C*14:02 on grades III–IV acute GvHD.
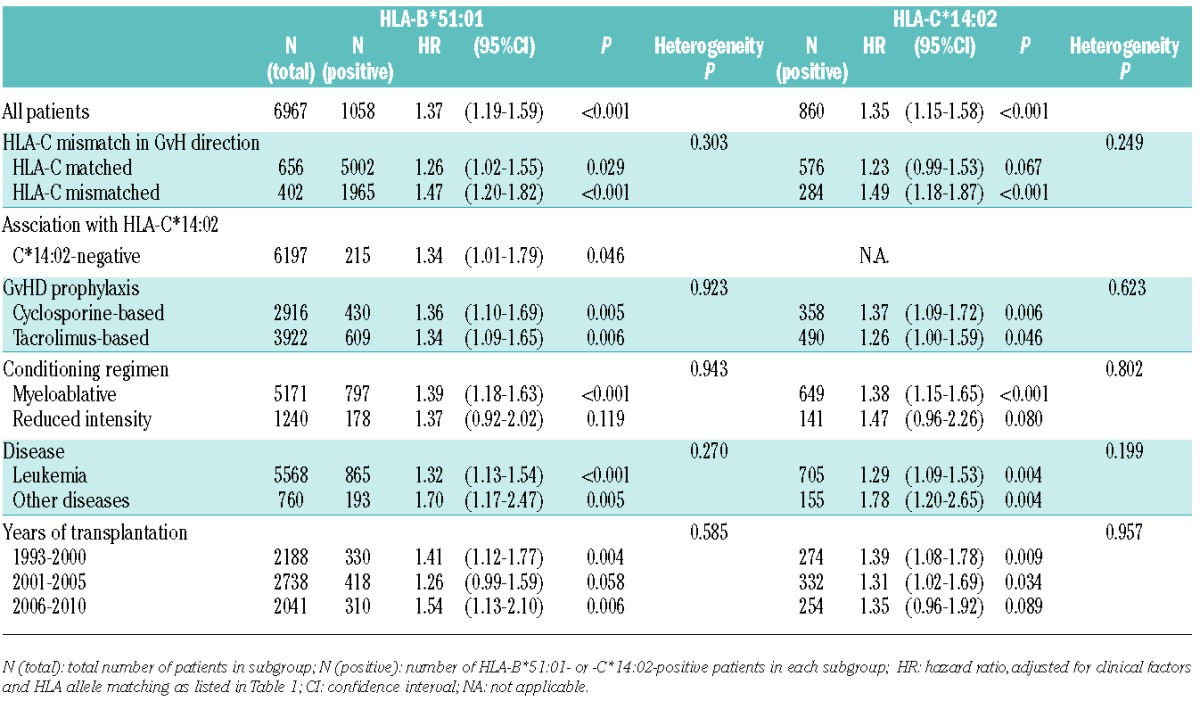
Increasing risk effect of HLA-C*14:02 and -B*51:01 on acute GvHD was prominent in HLA-C mismatched transplant
In contrast to the increasing risk effect of patient HLA-C*14:02 on grades III–IV acute GvHD, the effect of donor HLA-C*14:02 was not significant (HR, 1.11; 95%CI: 0.94–1.31; P=0.212) (Table 2). We assumed that the difference in the impact of HLA-C*14:02 on acute GvHD between patients and donors was attributable to HLA-C allele mismatch between patient and donor, and accordingly performed subgroup analysis of patient groups stratified by HLA-C matching status (Table 3). The association between patient HLA-C*14:02 and risk of grades III–IV acute GvHD was more prominent in pairs with HLA-C allele mismatch in the GvH direction (n=1965) (HR, 1.49; 95%CI: 1.18–1.87; P<0.001) than pairs with HLA-C match in the GvH direction (n=5002) (HR, 1.23; 95%CI: 0.99–1.53; P=0.067). Similarly, the effect of HLA-B*51:01 on acute GvHD was more obvious in pairs with HLA-C allele mismatch in the GvH direction (HR, 1.47; 95%CI: 1.20–1.82; P<0.001) than in matched pairs (HR, 1.26; 95%CI: 1.02–1.55; P=0.029).
We also performed subgroup analysis of patient groups stratified by various clinical factors (i.e. GvHD prophylaxis, conditioning regimen, disease, year of transplantation) (Table 3). There were no statistically significant heterogeneities in the effects of patient HLA-B*51:01 and HLA-C*14:02 in these subgroups, indicating the absence of interactions between the risk of this HLA allele and these clinical factors.
Effect of patient mismatched HLA-C alleles on acute GvHD
The effects of patient HLA-B*51:01 and -C*14:02 on grades III–IV acute GvHD were more prominent in HLA-C allele mismatched pairs than matched pairs, which prompted us to examine the relationship between patient-donor HLA-C allele mismatch and the effect of these alleles on acute GvHD.
First, we examined the effect of patient mismatched HLA alleles on grades III–IV acute GvHD (Figure 1), as these are HLA targets in the GvH direction. The patients transplanted from donors with matched for 1 HLA-C allele and mismatched for another HLA-C allele (n=1795) were compared with HLA-C matched transplant (n=4825). Mismatched alleles having fewer than 20 pairs were grouped together as “Others”. Median HR of patient mismatched HLA-C alleles was 1.56 compared with HLA-C matched transplants. Interestingly, patient mismatched HLA-C*14:02 showed a strikingly high risk of grades III–IV acute GvHD among all patient mismatched HLA-C alleles (HR, 3.61; 95%CI: 2.71–4.82; P<0.001). Cumulative incidences of grades III–IV acute GvHD in patients with patient HLA-C*14:02 mismatch, other patient HLA-C allele mismatch, and HLA-C match were 41.6% (95%CI: 32.6–50.3), 23.0% (95%CI: 21.0–25.0), and 13.6% (95%CI: 12.6–14.6) (Figure 2A). Patient HLA-C*14:02 mismatched transplant also showed the highest risk of transplant-related mortality (HR, 2.53; 95%CI: 1.92–3.34; P<0.001) and overall mortality (HR, 1.91; 95%CI: 1.52–2.42; P<0.001) among all patient mismatched HLA-C alleles (Online Supplementary Figures S1 and S2). Overall survival in patients with patient HLA-C*14:02 mismatch, with other patient HLA-C allele mismatch, and with HLA-C match were 35.5% (95%CI: 26.7–44.3), 46.1% (95%CI: 43.6–48.6) and 51.4% (95%CI: 49.9–52.9) (Figure 2B).
Figure 1.
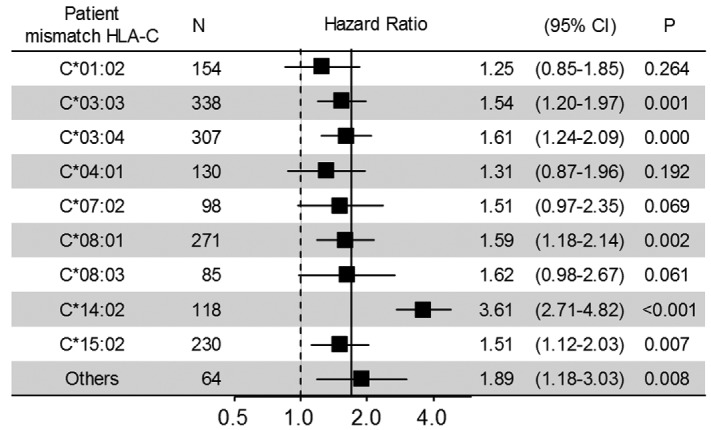
Impact of patient mismatched HLA-C allele on grades III–IV acute GvHD. Results of multivariable competing risk regression analysis for the effect of patient mismatched HLA-C allele on grades III–IV acute GvHD. The hazard ratio (HR) of each mismatched HLA-C allele was estimated by comparison to HLA-C match (n=4825). The solid vertical line at 1.69 indicates the HR of overall HLA-C mismatch in the GvH direction. Others contain patient mismatched HLA-C alleles with fewer than 20 patients, as follows: C*01:03, C*03:14, C*03:23, C*05:01, C*07:04, C*12:02, C*12:03 and HLA-C*14:03.
Figure 2.
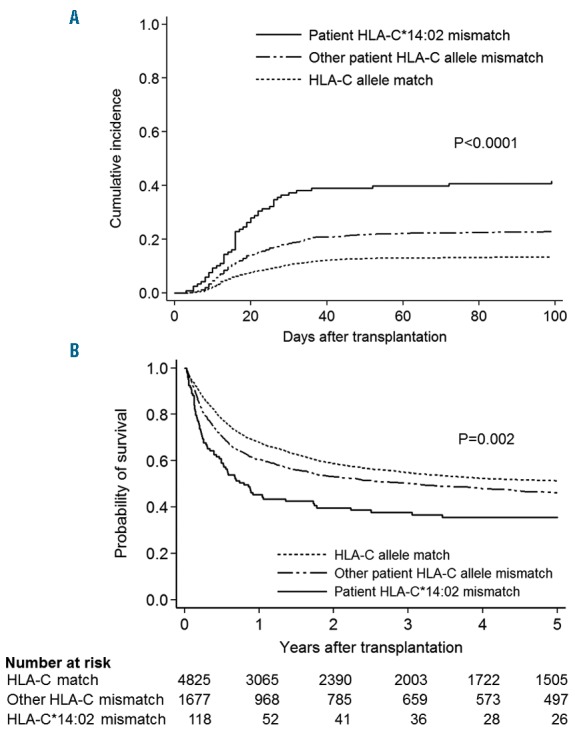
Acute graft-versus-host disease (GvHD) and survival curve by patient mismatched HLA-C. Patients were divided into three groups as follows: patient HLA-C*14:02 mismatch (n=118), other patient HLA-C allele mismatch (n=1677), and HLA-C allele match (4825). (A) Cumulative incidences of grades III–IV and (B) Kaplan-Meier curve of survival are shown. P-value indicates comparison between patient HLA-C*14:02 mismatch and other patient HLA-C mismatch.
A recent report showed that the expression level of a patient’s mismatched HLA-C allotype affects severe acute GvHD and mortality after unrelated hematopoietic stem cell transplantation.19 We analyzed whether the expression levels of patient mismatched HLA-C allotype influence grades III–IV acute GvHD. In comparison with patient mismatched HLA-C*07, HLA-C*14 was associated with a significantly higher risk of grades III–IV acute GvHD (HR, 2.14; 95%CI: 1.27–3.60; P=0.004) and higher transplant-related mortality (HR, 1.82; 95%CI: 1.15–2.86; P=0.01), whereas there were no significant differences between other HLA-C allotypes and HLA-C*07 (Online Supplementary Table S2). An increasing expression level of patient mismatched HLA-C was not associated with either the risk of grades III–IV acute GvHD (P=0.228) or transplant-related mortality (P=0.108) (Online Supplementary Figure 3A and B).
Impact of HLA-C allele mismatch combinations on acute GvHD
The previous JMDP study showed that specific donor-patient allele mismatch combinations were associated with severe acute GvHD.6 We analyzed the association between the impact of patient HLA-C*14:02 itself and that of donor-patient HLA-C allele mismatch combinations on grades III–IV acute GvHD. There were 33 HLA-C allele mismatch combinations with more than 15 donor-patient pairs, and the mismatch combinations having fewer than 15 pairs were grouped together as “Others”. HRs of HLA-C allele mismatch combinations for grades III–IV acute GvHD in comparison to HLA-C allele match are shown in the Online Supplementary Table S3. Among 33 HLA-C mismatch combinations, there were three combinations having a patient HLA-C allele of -C*14:02, and these three combinations showed higher HRs than the median HR for all mismatch combinations (median HR=1.75), as follows: donor C*15:02 and patient C*14:02 (HR, 4.39; 95%CI: 2.98–6.45; P<0.001), donor C*03:04 and patient C*14:02 (HR, 2.66; 95%CI: 1.43–4.93; P=0.002), and donor HLA-C*01:02 and patient C*14:02 (HR, 4.01; 95%CI: 2.19–7.35; P<0.001).
Relation of KIR2DL ligand mismatch and high-risk HLA-C allele mismatch combination
As described above, the donor C*15:02 and patient C*14:02 mismatch combination was one of the highest risk HLA-C allele mismatch combinations for grades III–IV acute GvHD. Our previous JMDP study demonstrated that KIR2DL-L mismatch in the GvH direction (KIR2DL-L-MM-G) was associated with an increased risk of severe acute GvHD and mortality in T-cell-replete UR-HSCT.22
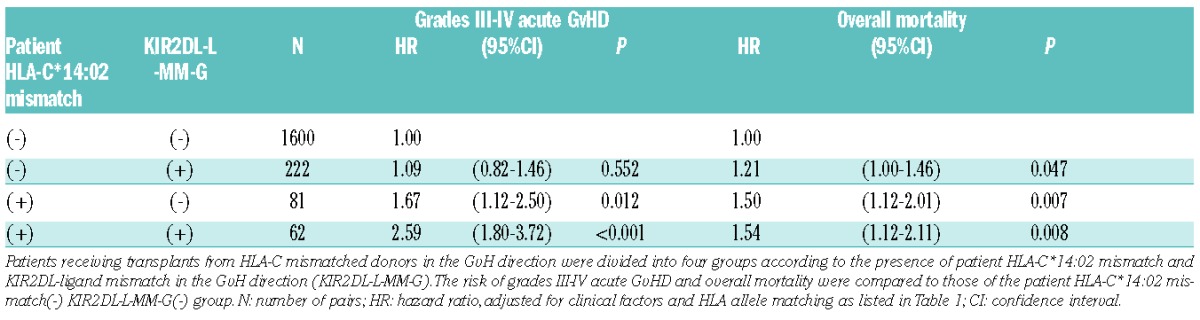
We examined the impact of patient-donor KIR2DL ligand mismatch on the risk of acute GvHD related to this HLA-C mismatch combination. In our Japanese cohort, the frequencies of KIR2DL-ligand HLA-C1 and -C2 were 93% and 7%, respectively, and the majority (91%) of KIR2DL-L-MM-G was the combination of patient HLA-C1/C1 and donor HLA-C1/C2. HLA-C*14:02 belongs to HLA-C1, while HLA-C*15:02 belongs to HLA-C2; therefore, donor HLA-C*15:02 and patient HLA-C*14:02 mismatch combination is usually KIR2DL-L-MM-G. In fact, 92% of pairs (60 of 65) with the donor C*15:02 and patient C*14:02 mismatch combination were KIR2DL-L-MM-G, and they accounted for approximately 21% of all pairs with KIR2DL-L-MM-G (n=284). KIR2DL-L-MM-G among patients transplanted from donors with HLA-C allele mismatch in the GvH direction was associated with a significantly higher risk of grades III–IV acute GvHD (HR, 1.34; 95%CI: 1.05–1.69; P=0.015). To clarify the effect of patient HLA-C*14:02-mismatch on grades III–IV acute GvHD in relation to KIR2DL-L-MM-G, patients transplanted from HLA-C mismatched donors were divided into four groups (Table 4). Both the patient HLA-C*14:02-mismatch(+)KIR-2DL-L-MM-G(+) group (n=62) and patient HLA-C*14:02-mismatch(+)KIR-2DL-L-MM-G(−) group (n=81) showed a significantly higher risk of acute GvHD (HR, 2.59; P<0.001 and HR, 1.67; P=0.012, respectively) and overall mortality (HR, 1.54; P=0.008 and HR, 1.50; P=0.007, respectively) than the patient HLA-C*14:02-mismatch(−)KIR-2DL-L-MM-G(−) group (n=1600). On the other hand, the patient HLA-C*14:02-mismatch(−)KIR-2DL-L-MM-G(+) group (n=222) showed no significant difference in grades III–IV acute GvHD (HR, 1.09; P=0.552), in spite of the higher trend in overall mortality (HR, 1.21; P=0.047) than in the HLA-C*14:02-mismatch(−)KIR-2DL-L-MM-G(−) group. Among 62 pairs in the patient HLA-C*14:02-mismatch(+)KIR-2DL-L-MM-G(+) group, 60 pairs had the mismatch combination of patient C*14:02 and donor C*15:02.
Table 4.
Effect of patient HLA-C*14:02 mismatch on severe acute graft-versus-host disease (GvHD) and mortality in relation to KIR2DL-L-MM-G
Discussion
In this study, we examined the influence of patient and donor HLA alleles on severe acute GvHD after UR-HSCT using 6967 unrelated Japanese pairs. Given the importance of GvHD prophylaxis to acute GvHD in allogeneic HSCT, we aimed to elucidate the biological effects of HLA alleles on acute GvHD by selecting patients who underwent transplantation from non-T-cell-depleted bone marrow donors without ATG.
We demonstrated that patient and/or donor HLA-B*51:01 and patient HLA-C*14:02 were significantly associated with an increased risk of grades III–IV acute GvHD. Our finding that patient HLA-C*14:02 was associated with an increased risk of severe acute GvHD, whereas donor HLA-C*14:02 was not, prompted us to investigate further the association of these high-risk HLA-alleles with patient-donor HLA-C allele mismatches. Results showed that patient mismatched HLA-C*14:02 was the key factor in the risk of severe acute GvHD. Petersdorf et al. reported that increasing expression of patient mismatched HLA-C was significantly associated with an increased risk of grades III–IV acute GvHD in UR-HSCT.19 In their report, the expression level of HLA-C*14 was highest among 14 HLA-C allotypes, and was associated with a strikingly high risk of acute GvHD and non-relapse mortality compared to other HLA-C allotypes. In our present study, patient mismatched HLA-C*14:02 also showed the highest risk of severe acute GvHD, transplant-related mortality, and overall mortality among all the patient mismatched HLA-C alleles. Although we did not demonstrate correlations between the expression levels of patient mismatched HLA-C alleles and transplant outcomes in our study, an extraordinarily high risk of patient mismatched HLA-C*14:02 for severe acute GvHD has been seen in other ethnic cohorts, indicating that this particular HLA-C*14:02 could play a very important role in immunoreactivity in allo-HSCT.
In our analysis of the association between donor-patient HLA-C allele mismatch combinations and acute GvHD, each of the three mismatch combinations with patient HLA-C*14:02 showed a higher HR than the other HLA-C mismatch combinations. In the previous JMDP study,6 the donor-C*15:02 and patient C*14:02 mismatch was identified as a non-permissive mismatch combination for severe acute GvHD, and the other two mismatches (donor-C*03:04 and patient-C*14:02, donor-C*01:02 and patient-C*14:02) were also associated with a higher risk of severe acute GvHD. Our present results are, therefore, consistent with those of this previous study.
KIR2DL-L-MM-G was reported to be significantly associated with a higher risk of severe acute GvHD than KIR2DL-L-match in the previous JMDP study.22 In our present JMDP study, KIR2DL-L-MM-G was consistently associated with a higher risk of grades III–IV acute GvHD even after restriction of the analysis population to pairs with HLA-C mismatch in the GvH direction. KIR2DL-L mismatching was first reported to be associated with improved survival and reduced relapse without acute GvHD in HLA mismatched T-cell-depleted and CD34+− selected haploidentical transplantation.23 Subsequent studies examining the effect of KIR2DL-L mismatch on transplant outcome in UR-HSCT showed inconsistent results, presumably because of differences in patient population, conditioning regimen, GvHD prophylaxis and sample size.24–29 Petersdorf et al. demonstrated that the negative effect of non-relapse mortality was evident in the high expression of patient mismatched HLA-C allotype (C*01 and C*14) compared to low expression (C*03 and C*07) among KIR2DL-L mismatched pairs, but not among KIR2DL-L matched pairs, suggesting that the KIR2DL-L mismatching effect was influenced by the expression levels of patient mismatched HLA-C alleles.19 In our cohort, patient HLA-C*14:02 and donor HLA-C*15:02 mismatch, which was eventually classified as KIR2DL-L-MM-G, showed the highest frequency among mismatch combinations with patient HLA-C*14:02. Furthermore, the negative effect of patient mismatched HLA-C*14:02 on grades III–IV acute GvHD and survival was also evident in KIR2DL-L matched pairs. These findings indicate that patient mismatched HLA-C*14:02 is a critical factor in severe acute GvHD and survival regardless of KIR2DL-L matching status. Therefore, we assume that the higher risk of grades III–IV acute GvHD for KIR2DL-L-MM-G in the JMDP cohort was attributable to this particular mismatch combination of patient HLA-C*14:02 and donor -C*15:02.
A recent study using Japanese UR-HSCT data in the Transplant Registry Management Program demonstrated that the adverse impact of high-risk HLA allele mismatch combinations identified in a previous JMDP study was significant in the early transplant years (1993–2001), but became less significant in later transplant years (after 2002).30 We analyzed the effect of patient HLA-C*14:02 and HLA-C*51:01 on grades III–IV acute GvHD and overall mortality (Online Supplementary Table S4) and the effect of patient mismatched HLA-C on transplant outcomes (Online Supplementary Figure S4) in the patients transplanted from 2002 through 2010. The adverse effect of patient HLA-C*14:02, -B*51:01, and patient mismatched HLA-C*14:02 on grades III–IV acute GvHD and mortality were obvious during this period. These results suggest that the biological impact of these HLA alleles on transplant outcomes should be considered even in current transplantation conditions.
Since HLA-C*14:02 is strongly linked to HLA-B*51:01 among Japanese, it is possible that the significantly negative effect of patient HLA-B*51:01 on severe acute GvHD was detected as a consequence of the effect of patient mismatched HLA-C*14:02. However, patient HLA-B*51:01 was also associated with a higher risk of severe acute GvHD in subgroups of patients without HLA-C*14:02 and of HLA-C matched pairs. We therefore assume that HLA-B*51:01 itself plays a role in the development or exacerbation of acute GvHD. As for the genetic association between HLA-B51 and autoimmune or inflammatory diseases, an association with Behçet’s disease has been shown in numerous ancestry groups and populations.31 Behçet’s disease is an inflammatory disease characterized by recurrent oral aphthous ulcers and numerous potentially systemic manifestations.32 Although the pathogenic mechanisms related to HLA-B51 still need to be clarified, several possible mechanisms for a genetic linkage to HLA-B51 have been suggested. HLA-B*51:01 binds a potentially large pool of peptides with relatively low affinity, and it has been speculated that inefficient tolerance to HLA-B*51:01-binding self-peptides might be a predisposing factor for the development of Behçet’s disease.33 Recently, HLA-B*51:01 and HLA-C*14:02 were reported to be susceptibility alleles among Japanese for Crohn’s disease, a subtype of inflammatory bowel mucosal disease.34 Since the linkage between HLA-C*14:02 and -B*51:01 in other ethnic groups is weaker than in Japanese,35 further extensive studies are required to elucidate the immunological mechanisms of the genetic association between HLA-B*51:01and acute GvHD in other ethnic groups.
In conclusion, using unbiased searches of patient and donor HLA alleles in the JMDP database, we found that patient mismatched HLA-C*14:02 was the most potent risk factor of severe acute GvHD and mortality, and that this was eventually the most highly expressed HLA-C allele, as previously reported, and might evoke strong allogeneic immune reactions. Furthermore, an increased risk of patients and/or donor HLA-B*51:01 for severe acute GvHD was attributed not only to the genetic linkage of HLA-C*14:02 and -B*51:01 in the Japanese population, but also to HLA-B*51:01 itself.
With regard to clinical implications, patient mismatched HLA-C*14:02 should be considered a non-permissive mismatch in UR-HSCT. Studies should investigate whether consideration of this particular high-risk HLA mismatch can be extended to other graft sources and donors in allo-HSCT, such as cord blood and HLA-mismatch-related transplantation.
Acknowledgments
The authors thank the staff members of the transplantation centers and donor centers, the Japan Marrow Donor Program office, and the Japanese Data Center for Hematopoietic Cell Transplantation for their generous co-operation.
Footnotes
Funding
This work was supported by grants from Practical Research for Allergic Disease and Immunology (Research on Technology of Medical Transplantation), the Japan Agency for Medical Research and Development, and the Japanese Ministries of Health, Labor and Welfare (H23-Immunology-010 and H26-Immunology-106) and Education, Culture, Sports, Science and Technology (MEXT KAKENHI Grant Number 22133011 and JSPS KAKENHI Grant Number 26461455).
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/101/4/491
References
- 1.Sasazuki T, Juji T, Morishima Y, et al. Effect of matching of class I HLA alleles on clinical outcome after transplantation of hematopoietic stem cells from an unrelated donor. Japan Marrow Donor Program. N Engl J Med. 1998;339(17):1177–1185. [DOI] [PubMed] [Google Scholar]
- 2.Morishima Y, Sasazuki T, Inoko H, et al. The clinical significance of human leukocyte antigen (HLA) allele compatibility in patients receiving a marrow transplant from serologically HLA-A, HLA-B, and HLA-DR matched unrelated donors. Blood. 2002;99(11):4200–4206. [DOI] [PubMed] [Google Scholar]
- 3.Lee SJ, Klein J, Haagenson M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110(13):4576–4583. [DOI] [PubMed] [Google Scholar]
- 4.Morishima Y, Kashiwase K, Matsuo K, et al. Biological significance of HLA locus matching in unrelated donor bone marrow transplantation. Blood. 2015;125(7):1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrara GB, Bacigalupo A, Lamparelli T, et al. Bone marrow transplantation from unrelated donors: the impact of mismatches with substitutions at position 116 of the human leukocyte antigen class I heavy chain. Blood. 2001;98(10):3150–3155. [DOI] [PubMed] [Google Scholar]
- 6.Kawase T, Morishima Y, Matsuo K, et al. High-risk HLA allele mismatch combinations responsible for severe acute graft-versus-host disease and implication for its molecular mechanism. Blood. 2007; 110(7):2235–2241. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez-Vina MA, Wang T, Lee SJ, et al. Identification of a permissible HLA mismatch in hematopoietic stem cell transplantation. Blood. 2014;123(8):1270–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin PJ, Gooley T, Anasetti C, Petersdorf EW, Hansen JA. HLAs and risk of acute graft-vs.-host disease after marrow transplantation from an HLA-identical sibling. Biol Blood Marrow Transplant. 1998; 4(3):128–133. [DOI] [PubMed] [Google Scholar]
- 9.Battiwalla M, Hahn T, Radovic M, et al. Human leukocyte antigen (HLA) DR15 is associated with reduced incidence of acute GVHD in HLA-matched allogeneic transplantation but does not impact chronic GVHD incidence. Blood. 2006;107(5):1970–1973. [DOI] [PubMed] [Google Scholar]
- 10.Stern M, Passweg J, Tiercy JM, et al. Human leukocyte antigen DR15 is associated with reduced relapse rate and improved survival after human leukocyte antigen-identical sibling hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2006;12(11):1169–1175. [DOI] [PubMed] [Google Scholar]
- 11.Battiwalla M, Ellis K, Li P, et al. HLA DR15 antigen status does not impact graft-versus-host disease or survival in HLA-matched sibling transplantation for hematologic malignancies. Biol Blood Marrow Transplant. 2012;18(8):1302–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khouri IF, Bassett R, Poindexter N, et al. Nonmyeloablative allogeneic stem cell transplantation in relapsed/refractory chronic lymphocytic leukemia: long-term follow-up, prognostic factors, and effect of human leukocyte histocompatibility antigen subtype on outcome. Cancer. 2011; 117(20):4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet. 2009;54(1):15–39. [DOI] [PubMed] [Google Scholar]
- 14.Trowsdale J. The MHC, disease and selection. Immunol Lett. 2011;137(1–2):1–8. [DOI] [PubMed] [Google Scholar]
- 15.Yunis EJ, Larsen CE, Fernandez-Vina M, et al. Inheritable variable sizes of DNA stretches in the human MHC: conserved extended haplotypes and their fragments or blocks. Tissue Antigens. 2003;62(1):1–20. [DOI] [PubMed] [Google Scholar]
- 16.Morishima S, Ogawa S, Matsubara A, et al. Impact of highly conserved HLA haplotype on acute graft-versus-host disease. Blood. 2010;115(23):4664–4670. [DOI] [PubMed] [Google Scholar]
- 17.Ruggeri L, Capanni M, Casucci M, et al. Role of natural killer cell alloreactivity in HLA-mismatched hematopoietic stem cell transplantation. Blood. 1999;94(1):333–339. [PubMed] [Google Scholar]
- 18.Fine JPGR. A proportional hazards model for subdistribution of a competing risk. J Am Stat Assoc. 1999;94:456–509. [Google Scholar]
- 19.Petersdorf EW, Gooley TA, Malkki M, et al. HLA-C expression levels define permissible mismatches in hematopoietic cell transplantation. Blood. 2014;124(26):3996–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cox D. Regression model and life tables. J R Stat Soc B. 1972;34(2):187–200. [Google Scholar]
- 21.Ikeda N, Kojima H, Nishikawa M, et al. Determination of HLA-A, -C, -B, -DRB1 allele and haplotype frequency in Japanese population based on family study. Tissue Antigens. 2015;85(4):252–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morishima Y, Yabe T, Matsuo K, et al. Effects of HLA allele and killer immunoglobulin-like receptor ligand matching on clinical outcome in leukemia patients undergoing transplantation with T-cell-replete marrow from an unrelated donor. Biol Blood Marrow Transplant. 2007;13(3):315–328. [DOI] [PubMed] [Google Scholar]
- 23.Ruggeri L, Capanni M, Urbani E, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295(5562):2097–2100. [DOI] [PubMed] [Google Scholar]
- 24.Davies SM, Ruggieri L, DeFor T, et al. Evaluation of KIR ligand incompatibility in mismatched unrelated donor hematopoietic transplants. Killer immunoglobulin-like receptor. Blood. 2002;100(10):3825–3827. [DOI] [PubMed] [Google Scholar]
- 25.Giebel S, Locatelli F, Lamparelli T, et al. Survival advantage with KIR ligand incompatibility in hematopoietic stem cell transplantation from unrelated donors. Blood. 2003;102(3):814–819. [DOI] [PubMed] [Google Scholar]
- 26.Bornhauser M, Schwerdtfeger R, Martin H, Frank KH, Theuser C, Ehninger G. Role of KIR ligand incompatibility in hematopoietic stem cell transplantation using unrelated donors. Blood. 2004;103(7):2860–2861. [DOI] [PubMed] [Google Scholar]
- 27.Schaffer M, Malmberg KJ, Ringden O, Ljunggren HG, Remberger M. Increased infection-related mortality in KIR-ligand-mismatched unrelated allogeneic hematopoietic stem-cell transplantation. Transplantation. 2004;78(7):1081–1085. [DOI] [PubMed] [Google Scholar]
- 28.Beelen DW, Ottinger HD, Ferencik S, et al. Genotypic inhibitory killer immunoglobulin-like receptor ligand incompatibility enhances the long-term antileukemic effect of unmodified allogeneic hematopoietic stem cell transplantation in patients with myeloid leukemias. Blood. 2005; 105(6):2594–2600. [DOI] [PubMed] [Google Scholar]
- 29.Farag SS, Bacigalupo A, Eapen M, et al. The effect of KIR ligand incompatibility on the outcome of unrelated donor transplantation: a report from the center for international blood and marrow transplant research, the European blood and marrow transplant registry, and the Dutch registry. Biol Blood Marrow Transplant. 2006; 12(8):876–884. [DOI] [PubMed] [Google Scholar]
- 30.Kanda Y, Kanda J, Atsuta Y, et al. Changes in the clinical impact of high-risk human leukocyte antigen allele mismatch combinations on the outcome of unrelated bone marrow transplantation. Biol Blood Marrow Transplant. 2014;20(4):526–535. [DOI] [PubMed] [Google Scholar]
- 31.Maldini C, Lavalley MP, Cheminant M, de Menthon M, Mahr A. Relationships of HLA-B51 or B5 genotype with Behcet’s disease clinical characteristics: systematic review and meta-analyses of observational studies. Rheumatology. 2012;51(5):887–900. [DOI] [PubMed] [Google Scholar]
- 32.Direskeneli H. Behcet’s disease: infectious aetiology, new autoantigens, and HLA-B51. Ann Rheum Dis. 2001;60(11):996–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gebreselassie D, Spiegel H, Vukmanovic S. Sampling of major histocompatibility complex class I-associated peptidome suggests relatively looser global association of HLA-B*5101 with peptides. Hum Immunol. 2006;67(11):894–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oryoji D, Hisamatsu T, Tsuchiya K, et al. Associations of HLA class I alleles in Japanese patients with Crohn’s disease. Genes Immun. 2015;16(1):54–56. [DOI] [PubMed] [Google Scholar]
- 35.Maiers M, Gragert L, Klitz W. High-resolution HLA alleles and haplotypes in the United States population. Hum Immunol. 2007;68(9):779–788. [DOI] [PubMed] [Google Scholar]


