Abscission is the last step of cytokinesis, allowing the physical separation of daughter cells at the end of cell division. It has been considered a cell autonomous process, yet Jungas et al. report that Ephrin/Eph signaling controls the completion of abscission.
Abstract
Cytokinesis is the last step of cell division, culminating in the physical separation of daughter cells at the end of mitosis. Cytokinesis is a tightly regulated process that until recently was mostly viewed as a cell-autonomous event. Here, we investigated the role of Ephrin/Eph signaling, a well-known local cell-to-cell communication pathway, in cell division. We show that activation of Eph signaling in vitro leads to multinucleation and polyploidy, and we demonstrate that this is caused by alteration of the ultimate step of cytokinesis, abscission. Control of abscission requires Eph kinase activity, and Src and citron kinase (CitK) are downstream effectors in the Eph-induced signal transduction cascade. CitK is phosphorylated on tyrosines in neural progenitors in vivo, and Src kinase directly phosphorylates CitK. We have identified the specific tyrosine residues of CitK that are phosphorylated and show that tyrosine phosphorylation of CitK impairs cytokinesis. Finally, we show that, similar to CitK, Ephrin/Eph signaling controls neuronal ploidy in the developing neocortex. Our study indicates that CitK integrates intracellular and extracellular signals provided by the local environment to coordinate completion of cytokinesis.
Introduction
Cytokinesis is the last step of cell division, allowing physical separation of the two daughter cells and faithful partitioning of genetic and cytoplasmic material (Green et al., 2012; Mierzwa and Gerlich, 2014). Tight control of cytokinesis completion is essential because cytokinesis failure has been associated with carcinogenesis (Sagona and Stenmark, 2010), but also because incomplete cytokinesis is an evolutionarily conserved physiological event required for development and homeostasis of several tissues harboring polyploid cells (Davoli and de Lange, 2011; Haglund et al., 2011). Cytokinesis begins with the assembly of an equatorial contractile ring whose constriction allows the formation of a narrow intercellular bridge (ICB) between the nascent daughter cells. Physical separation of the two daughter cells, also called abscission, ends cytokinesis. Completion of abscission requires the coordination of several cellular functions such as membrane trafficking, lipid turnover, cytoskeletal rearrangements, and orderly recruitment of molecular complexes to the midbody (Agromayor and Martin-Serrano, 2013; Mierzwa and Gerlich, 2014; Cauvin and Echard, 2015). One of the master regulators of cytokinesis is the small GTPase RhoA, whose accumulation at the equatorial cell cortex is the first event of cytokinesis, participating in specifying the position and promoting assembly and contraction of the actomyosin ring (Bement et al., 2005). After cleavage furrow ingression, active RhoA participates in the stabilization of the ICB by recruiting other proteins to the midbody, including Anillin and citron kinase (CitK; Madaule et al., 1998; Hickson and O’Farrell, 2008). At late stages of cytokinesis, the cytoskeleton is cleared from the ICB; disassembly of actin filaments requires inactivation of RhoA and changes in lipid composition of the plasma membrane (Emoto et al., 2005; Saurin et al., 2008; Dambournet et al., 2011), whereas microtubule severing is accomplished by spastins (Connell et al., 2009). Lastly, components of the ESCRT (endosomal sorting complexes required for transport) complex are recruited to the ICB, and membrane abscission ensues (Morita et al., 2007).
A key player in the maintenance of RhoA at the midbody is CitK, a protein that itself localizes to the midbody (Madaule et al., 1998; Naim et al., 2004; Bassi et al., 2011, 2013; Gai et al., 2011). Although CitK was first thought to be important for contraction of the equatorial actomyosin ring via phosphorylation of MLC2, it has more recently emerged that CitK is in fact dispensable for these steps and that its role is primarily to act as a scaffold protein during late cytokinesis and abscission (Naim et al., 2004; Gai et al., 2011; Serres et al., 2012; Watanabe et al., 2013). Indeed, CitK loss of function causes abscission defects with frequent reopening of the ICB, causing multinucleation (Echard et al., 2004; Gai et al., 2011; Watanabe et al., 2013). In mammals, CitK is strictly required for cytokinesis of a limited number of cell types, including neural progenitors of the developing neocortex. Accordingly, loss of CitK impairs cytokinesis of these cells, leading to an increase in the number of binucleated and polyploid neurons as well as neuronal cell death (Di Cunto et al., 2000; Sarkisian et al., 2002; LoTurco et al., 2003; Sgro et al., 2016).
In solid organs, dividing cells are part of tissues, and recent studies suggest that in addition to intracellular events, successful cytokinesis requires coordination with extracellular processes (Herszterg et al., 2014; Le Bras and Le Borgne, 2014). For instance, cooperation between dividing cells and their neighbors is necessary to remodel cell adhesion during cytokinesis while maintaining tissue cohesiveness (Founounou et al., 2013; Guillot and Lecuit, 2013; Herszterg et al., 2013). Ephrin/Eph signaling is a local communication pathway that regulates adhesion between neighboring cells and plays an important role in development but also in tumor dissemination and stem cell homeostasis (Gucciardo et al., 2014; Laussu et al., 2014; Kania and Klein, 2016). Here, we investigated the role of Ephrin/Eph signaling in cell division. We show that activation of EphB2 leads to a defect in abscission that correlates with multinucleation and polyploidy. In vivo, loss of Ephrin/Eph signaling decreases neuronal polyploidy in the developing neocortex. We identified Src kinase (Src) and CitK as downstream effectors of the pathway. We show that CitK is phosphorylated on tyrosines in vivo, and using mass spectrometry (MS), we have identified the specific tyrosine residues of CitK that are directly phosphorylated by Src. Finally, we provide evidence that tyrosine phosphorylation of CitK modulates its interaction with RhoA and is detrimental to cytokinesis.
Results
Activation of EphB2 forward signaling induces abscission defects
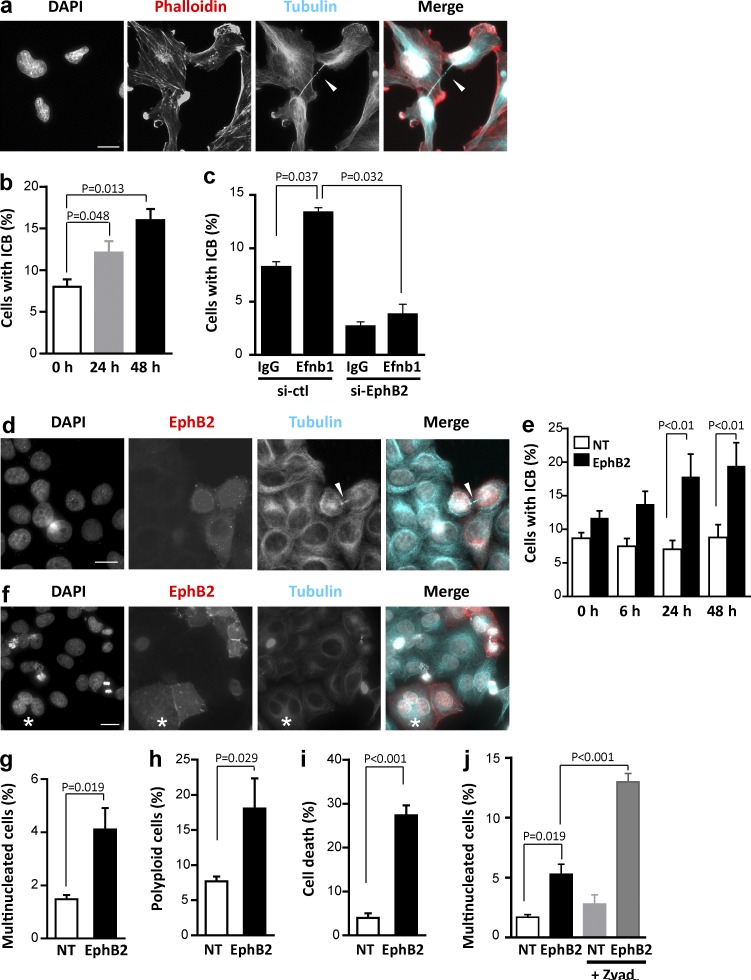
To test whether Ephrin/Eph signaling regulates cell division, we first used U251 cells, a human glioma cell line that expresses several Eph receptors endogenously (Fig. S1 a; Nakada et al., 2004). Stimulation of Eph forward signaling in U251 cells using bath application of clustered Efnb1-Fc recombinant protein led to a time-dependent increase in the proportion of daughter cells connected by an ICB (Fig. 1, a and b). Because EphB2 is the member of the family that is most highly expressed in U251 (Fig. S1 a), we next tested whether this receptor was responsible for this phenotype by silencing EphB2 in U251 cells. Depletion of endogenous EphB2 (Fig. S1 b), with or without stimulation, led to a significant decrease in the fraction of cells connected by an ICB compared with cells treated with a control siRNA (Fig. 1 c), indicating that EphB2 plays a role in persistence of the ICB in U251 cells. Next, we transfected EphB2 in HeLa cells, which express very little to no detectable levels of EphB receptors (Fig. S1 a), and compared transfected and nontransfected (NT) cells. As in U251 cells, stimulation of forward signaling led to an increase in the proportion of EphB2+ cells connected by an ICB, whereas the proportion of NT cells connected by an ICB did not change over time (Fig. 1, d and e). These results suggest that activation of EphB2 forward signaling leads to an abscission defect. One of the main consequences of abscission defects is the generation of multinucleated and/or polyploid cells (Lacroix and Maddox, 2012). Consistent with this, the number of multinucleated and polyploid cells more than doubled in EphB2+ cells compared with NT cells after stimulation of forward signaling (Fig. 1, f–h). Abscission failure, when not resolved by reopening of the bridge (leading to multinucleation and polyploidy), can lead to cell death (Joshi et al., 2011), and we observed that stimulation of Eph forward signaling led to a sharp increase in cell death after 24 h (Fig. 1 i). To find out whether this increase in cell death was a consequence of abscission defect, we inhibited apoptosis before activation of Eph forward signaling and monitored multinucleation. In these conditions, the fraction of multinucleated cells further increased after activation of Eph forward signaling (Fig. 1 j), supporting the notion that a fraction of EphB2+ cells undergoes cell death as a consequence of abscission failure. This was confirmed by time-lapse video microscopy, indicating that several stimulated EphB2+ HeLa cells underwent cell death after entering telophase (Video 1). Of note, EphB2 has also been reported to induce cell death via cytokinesis-independent mechanisms (Kandouz et al., 2010), and these other mechanisms also likely contribute to the high fraction of dead cells observed after EphB2 activation (Fig. 1 i).
Figure 1.
EphB2 forward signaling induces persistence of the ICB and multinucleation. (a) U251 cells were stimulated with Efnb1-Fc for 24 h and processed for DNA staining (gray, DAPI), actin staining (red, phalloidin), and immunostaining to detect β-tubulin (blue). Arrowhead indicates an ICB between daughter cells. (b) Quantification of the proportion of nascent daughter cells connected by an ICB after Efnb1-Fc stimulation for the indicated times. (c) U251 cells were transfected with control siRNAs or siRNAs targeting EphB2 and mock stimulated or stimulated with Efnb1-Fc. The proportion of U251 cells connected by an ICB was quantified as described in Methods. (d) HeLa cells transiently transfected with EphB2 and stimulated with Efnb1-Fc for 24 h were processed for DNA staining (gray, DAPI) and immunostained to detect EphB2 (red) and β-tubulin (blue). Arrowhead indicates an ICB between EphB2+ daughter cells. (e) Quantification of the proportion of NT (white bars) and EphB2-transfected (EphB2, black bars) cells connected by an ICB after Efnb1-Fc stimulation for the indicated time. (f) HeLa cells transiently transfected with EphB2 and stimulated with Efnb1-Fc for 24 h were stained as in d. Asterisk indicates a multinucleated cell. (g) Proportion of multinucleated cells in NT (white bars) and EphB2-transfected (EphB2, black bars) populations. (h and i) NT and EphB2-transfected (EphB2) HeLa cells were stimulated with Efnb1-Fc for 24 h and fixed, and DNA was stained using propidium iodide. DNA content of EphB2− (white bars) and EphB2+ (black bars) cells was analyzed by flow cytometry to quantify the proportion of polyploid cells (>4N DNA content; h) and dead cells (sub-G1 DNA content; i). (j) NT and EphB2-transfected (EphB2) HeLa cells were untreated or treated with Zvad and stimulated with Efnb1-Fc for 24 h, and the fraction of multinucleated cells was quantified. Error bars correspond to SEM. Statistical p-value is indicated when significant. Bars, 10 µm.
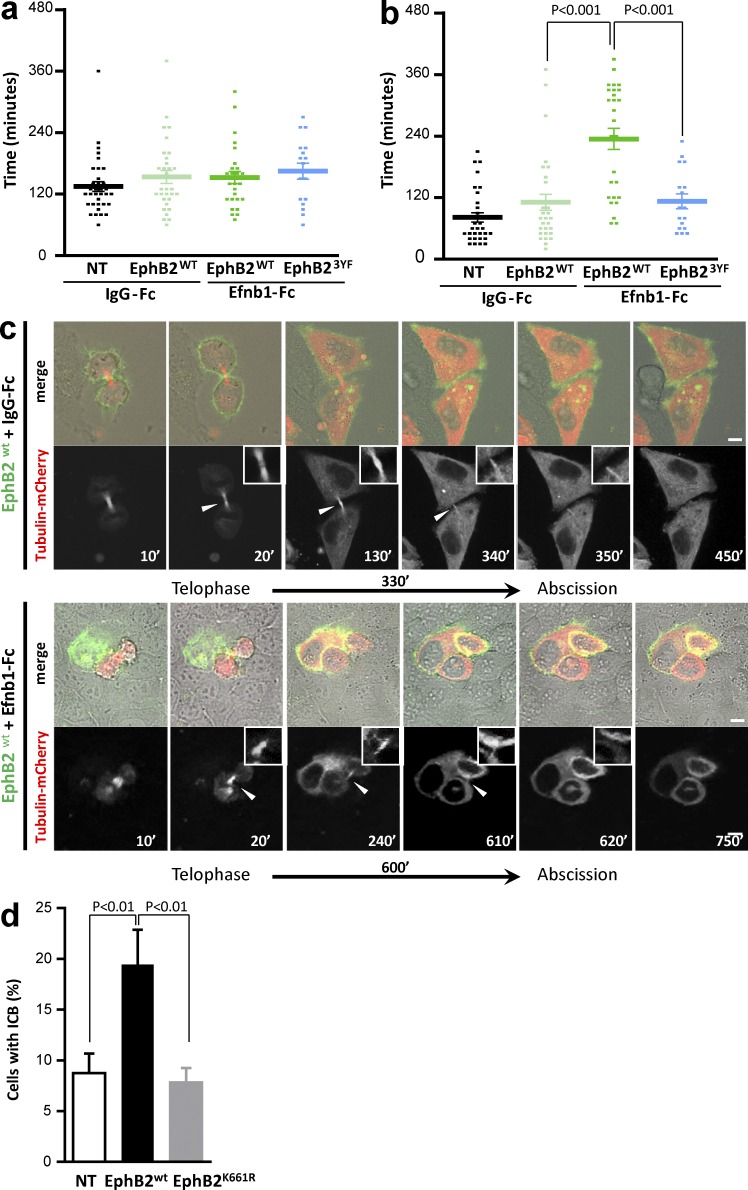
To better characterize EphB2-induced abscission defects, we recorded cell division events by time-lapse video microscopy of NT cells and cells expressing wild-type EphB2 (EphB2WT), either mock stimulated or stimulated with Efnb1-Fc to activate forward signaling (Fig. 2, a–c; and Fig. S2 a). Frame-by-frame analyses of these recordings showed that although activation of Eph forward signaling did not modify the progression from anaphase to late telophase (Fig. 2 a), the progression from late telophase to disappearance of the ICB was significantly delayed (Fig. 2 b) compared with control conditions. In extreme cases, daughter cells remained connected throughout the 8-h recording (Fig. S2 a, bottom). We confirmed persistence of the ICB in stimulated EphB2+ HeLa cells by performing high-resolution live imaging on cells cotransfected with EphB2-GFP and tubulin-mCherry, which allowed a direct visualization of the ICB in EphB2+ cells (Fig. 2 c). Altogether, these results demonstrate that activation of EphB2 forward signaling causes defects at late stages of cytokinesis ranging from abscission delay to complete failure of physical separation of daughter cells, resulting in multinucleation or cell death.
Figure 2.
EphB2-induced abscission defect is kinase dependent. (a and b) HeLa cells were left untransfected (NT) or were transfected with EphB2WT-GFP (EphB2WT) or EphB23YF-GFP (EphB23YF) for 48 h and synchronized, and cell division was recorded with an Incucyte microscope, in conditions of mock treatment (IgG-Fc) or Efnb1-Fc treatment. (a) Duration of the progression from anaphase to telophase in IgG-Fc– or Efnb1-Fc–treated cells. (b) Duration of the progression from telophase to disappearance of the ICB in IgG-Fc– or Efnb1-Fc–treated cells. (c) Still images from confocal recordings of dividing cells in EphB2WT-GFP cells expressing tubulin-mCherry mock treated or treated with Efnb1-Fc (as indicated). The top rows show fluorescence and phase contrast overlay, and the bottom rows show the tubulin-mCherry fluorescence to highlight the ICB (arrowhead). Insets are higher-magnification images of the ICB. (d) HeLa cells either NT or transfected with EphB2WT or EphB2K661R were treated with Efnb1-Fc. The proportion of cells connected by an ICB after 24 h of treatment was quantified in each condition. Error bars correspond to SEM. Statistical p-value is indicated when significant. Bars, 10 µm.
Ephs are receptor tyrosine kinases (RTKs) that exert many of their biological functions via their kinase activity (Lisabeth et al., 2013). To find out whether abscission defects were dependent on the kinase activity of EphB2, we transfected kinase-dead mutants of EphB2 (EphB2K661R and EphB23YF) in HeLa cells (Fig. S2, b–d) and monitored their ability to induce abscission defects. First, the progression from late telophase to abscission of daughter cells was not delayed in cells expressing EphB23YF, and the duration of that phase of cytokinesis was comparable to that of control cells (Fig. 2, a and b). Second, unlike cells expressing EphB2WT, cells expressing EphB2K661R did not exhibit persistence of the ICB upon incubation with Efnb1-Fc (Fig. 2 d). These results indicate that EphB2-mediated regulation of abscission requires the kinase activity of EphB2.
CitK is a target of EphB2 signaling
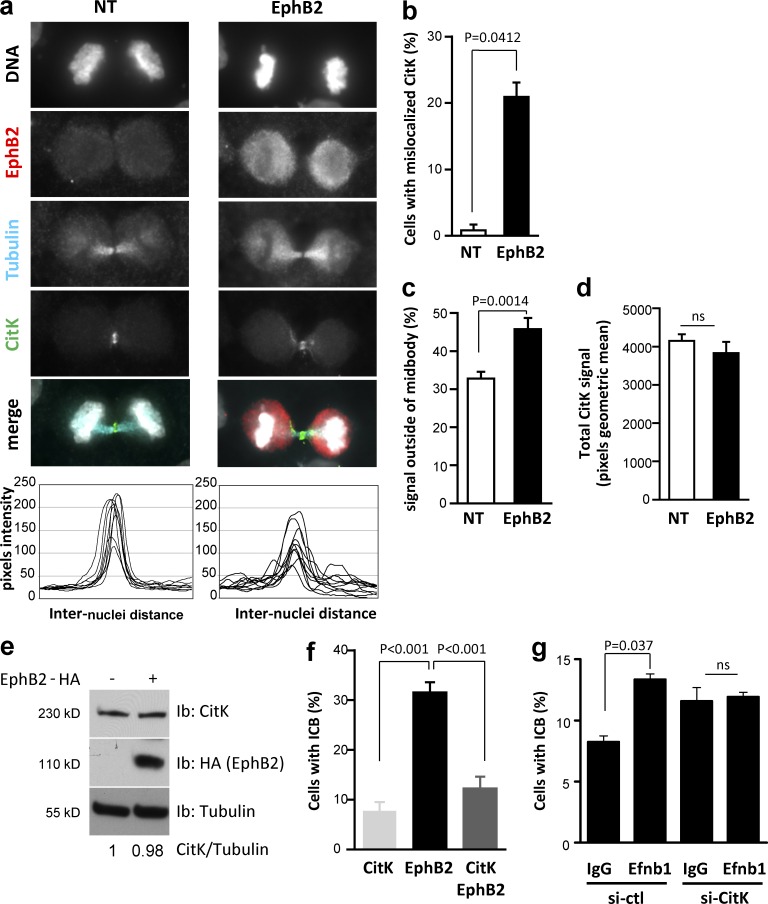
To characterize the mechanisms by which EphB2 forward signaling impairs abscission, we monitored the distribution of several proteins known to localize to the midbody and participate in abscission, including CitK, Anillin, and MgcRacGAP. Among them, only CitK exhibited altered localization in the ICB after EphB2 activation (Fig. 3 a and Fig. S3). After stimulation of forward signaling, a significant fraction of EphB2+ cells in telophase exhibited abnormal distribution of CitK in the ICB (no longer restricted to the midbody; Fig. 3, a–c). Indeed, endogenous CitK exhibited a broader distribution (Fig. 3, a–c), with no change of protein levels assessed by either quantification of total CitK signal intensity on confocal images (Fig. 3 d) or immunoblot (Fig. 3 e). CitK is well known for its role in controlling cytokinesis, and both gain and loss of CitK lead to multinucleation, indicating that its activity at the midbody must be tightly controlled for normal progression through cytokinesis. To test whether EphB2-induced abscission defects are caused by the deregulation of CitK, we performed gain- and loss-of-function experiments. Overexpression of full-length CitK (CitK FL) in stimulated EphB2+ HeLa cells decreased the fraction of cells still connected with an ICB (Fig. 3 f). Furthermore, activation of EphB2 in U251 cells depleted of endogenous CitK did not increase the fraction of cells connected by ICBs (Figs. 3 g and S1). Of note, depletion of CitK in U251 cells and HeLa cells induced a strong increase in the fraction of multinucleated cells (Fig. S1). Altogether, these results suggest that CitK is an effector of EphB2 in the control of abscission.
Figure 3.
Functional interaction between EphB2 and CitK in abscission. (a) HeLa cells untransfected (NT) or transfected with EphB2 and stimulated with Efnb1-Fc were stained for DNA (gray, DAPI) and immunostained to detect EphB2 (red), β-tubulin (blue), and CitK (green). The bottom panels represent the distribution of the CitK signal in the ICB for n = 10 cells for each condition. (b) Proportion of stimulated NT (n = 79) and EphB2-transfected (n = 59) cells in telophase with abnormal distribution of CitK in the ICB. (c) Proportion of CitK signal outside of the midbody in stimulated NT (n = 10) and EphB2-transfected (n = 10) cells. (d) Quantification of total CitK signal in stimulated NT (n = 10) and EphB2-transfected (n = 10) cells. (e) Protein lysates from HeLa cells untransfected (−) or transfected with HA-tagged EphB2 (+) and stimulated with Efnb1-Fc were immunoblotted to detect EphB2-HA, endogenous CitK, and tubulin. (f) HeLa cells were transiently transfected with CitK FL, EphB2, or EphB2+CitK and stimulated with Efnb1-Fc for 24 h. The proportion of daughter cells connected by an ICB was quantified in each condition. (g) U251 cells were transfected with control siRNAs or siRNAs targeting CitK and mock stimulated or stimulated with Efnb1-Fc. The proportion of U251 cells connected by an ICB was quantified. These experiments were performed at the same time as the experiments presented in Fig. 1 c; thus the controls (si-ctl) are the same. Error bars correspond to SEM. Statistical p-value is indicated when significant. ns, nonsignificant. Bars, 10 µm.
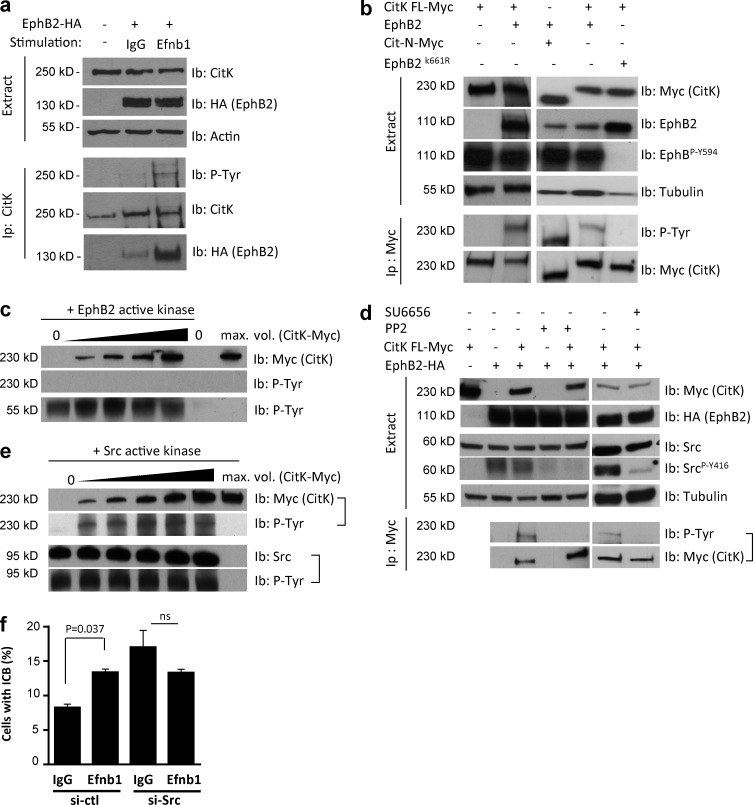
As described earlier, EphB2-induced abscission defects were dependent on the tyrosine kinase activity of EphB2, raising the possibility that CitK might be phosphorylated by EphB2. Indeed, immunoprecipitation of endogenous CitK revealed that it is phosphorylated on tyrosines after stimulation of forward signaling in HeLa cells (Fig. 4 a). Furthermore, upon EphB2 stimulation, the EphB2 receptor coimmunoprecipitated with endogenous CitK (Fig. 4 a). Similarly, immunoprecipitation of overexpressed CitK revealed that both CitK FL and the isoform of CitK lacking its kinase domain (Cit-N) were phosphorylated on tyrosines in EphB2+ stimulated cells (Fig. 4 b), whereas tyrosine phosphorylation of CitK FL was not detected in stimulated cells expressing EphB2K661R (Fig. 4 b). To assess whether CitK is a direct substrate of EphB2, we performed in vitro kinase assays with purified recombinant EphB2 kinase domain and CitK FL. No phosphorylation on tyrosine was detected on CitK by immunoblotting with the P-Tyr antibody, even at the highest amount of CitK tested (Fig. 4 c), whereas detection of phosphorylated recombinant EphB2 kinase validated the kinase assay (Fig. 4 c, bottom). It has been shown previously that Src-family kinases are activated downstream of Eph receptors in some cellular contexts (Gucciardo et al., 2014). Accordingly, Src was activated in EphB2+ cells stimulated with Efnb1-Fc, as detected by immunoblotting with an antibody against activated Src (SrcP-Y416; Fig. 4 d, extract). As expected, pretreatment of EphB2+ cells with two different Src inhibitors, PP2 and SU6656, before activation of Eph forward signaling prevented Src activation (Fig. 4 d, extract). Notably, PP2 and SU6656 treatments also abolished tyrosine phosphorylation of immunoprecipitated CitK in EphB2-stimulated cells (Fig. 4 d, Ip Myc), suggesting that CitK may be phosphorylated by Src. To test this hypothesis, we performed in vitro kinase assays with recombinant active Src kinase and purified CitK FL as substrate. Immunoblotting with antiphosphotyrosine antibodies revealed that purified CitK was indeed phosphorylated in the presence of recombinant Src kinase (Fig. 4 e). To confirm the involvement of Src in the EphB2-induced abscission defect, we depleted endogenous Src in U251 cells, which induced persistence of the ICB and multinucleation in U251 cells (Figs. 4 f and S1 d), consistent with previously published results reporting a requirement for Src activity before cleavage furrow ingression (Kasahara et al., 2007). Importantly, in Src-depleted cells, activation of EphB2 did not lead to a further increase in the fraction of cells connected with an ICB (Fig. 4 f), suggesting that Src is required for the EphB2-induced abscission defect. Altogether, these results indicate that EphB2 forward signaling activates Src kinase, which phosphorylates CitK on tyrosines and impairs abscission.
Figure 4.
CitK is tyrosine phosphorylated by Src in response to EphB2 signaling. (a) HeLa cells were transfected with EphB2-HA and either mock stimulated or stimulated with Efnb1-Fc. Protein lysates (top, Extract) were immunoblotted with antibodies against CitK and HA. Actin was used as loading control. Endogenous CitK was immunoprecipitated (bottom, Ip) and immunoblotted with P-Tyr, CitK, and HA antibodies. (b) HEK-293 cells were cotransfected with EphB2 and either myc-tagged CitK FL or myc-tagged Cit-N and stimulated with Efnb1-Fc for 20 min. Protein lysates (top, Extract) were immunoblotted with antibodies against Myc, EphB2, and P-EphB (EphBY594). Tubulin was used as a loading control. Exogenous CitK was immunoprecipitated (bottom, Ip) with an anti-Myc antibody and immunoblotted with Myc and P-Tyr antibodies. (c) Increasing amounts of purified His-Myc–tagged CitK produced in HEK-293 cells were incubated with recombinant EphB2 kinase domain in the presence of ATP. Negative controls (two lanes on the right) are samples without EphB2 recombinant kinase. Samples were analyzed by immunoblotting with antibodies against Myc and P-Tyr (top). No tyrosine phosphorylation on CitK was detected. Autophosphorylation of recombinant EphB2 kinase (bottom) indicates that the kinase was active. (d) HEK-293 cells were cotransfected with HA-tagged EphB2 and Myc-tagged CitK FL as indicated and either mock treated or pretreated with PP2 or SU6656 before stimulation with Efnb1-Fc for 20 min. Protein lysates (Extract, top) were immunoblotted with antibodies against Myc, HA, Src, and activated Src (SrcP-Y416), as indicated. Tubulin was used as loading control. Exogenous CitK was immunoprecipitated using an anti-Myc antibody and immunoblotted using P-Tyr and anti-Myc antibodies (Ip, bottom). (e) Increasing amounts of purified His-Myc–tagged CitK were incubated with recombinant Src in the presence of ATP. The negative control (right lane) is a sample without recombinant Src. Samples were analyzed by immunoblotting with Myc, Src, and P-Tyr antibodies. Purified CitK is phosphorylated on tyrosines in the presence of recombinant Src. Bottom panels show that recombinant Src kinase is autophosphorylated. (f) U251 cells were transfected with control siRNAs or siRNAs targeting Src and mock stimulated or stimulated with Efnb1-Fc. The proportion of U251 cells connected by an ICB was quantified. These experiments were performed at the same time as the experiments presented in Fig. 1 c; thus the controls (si-ctl) are the same. Error bars correspond to SEM. Statistical p-value is indicated when significant. ns, nonsignificant.
Because other RTKs are known to activate Src, we asked whether phosphorylation of CitK and control of abscission was specific for EphB2 or could be generalized. We tested four other RTKs known to activate Src (Fig. S4) for their ability to induce persistence of the ICB in both U251 and HeLa cells. Among them, only activation of EGFR led to persistence of the ICB (Fig. S4, b and d). However, EGFR has been shown previously to control cytokinesis by Src-independent mechanisms, including activation of RhoA via ERK (Tong et al., 2016) and phosphorylation of MLC-2 (Jiang et al., 2014). Thus, these results show that control of cytokinesis via activation of Src and phosphorylation of CitK is not a generic response to RTK activation but rather a specific feature of EphB2, possibly caused by the ability of activated EphB2 to interact with CitK.
Tyrosine phosphorylation of CitK impairs abscission
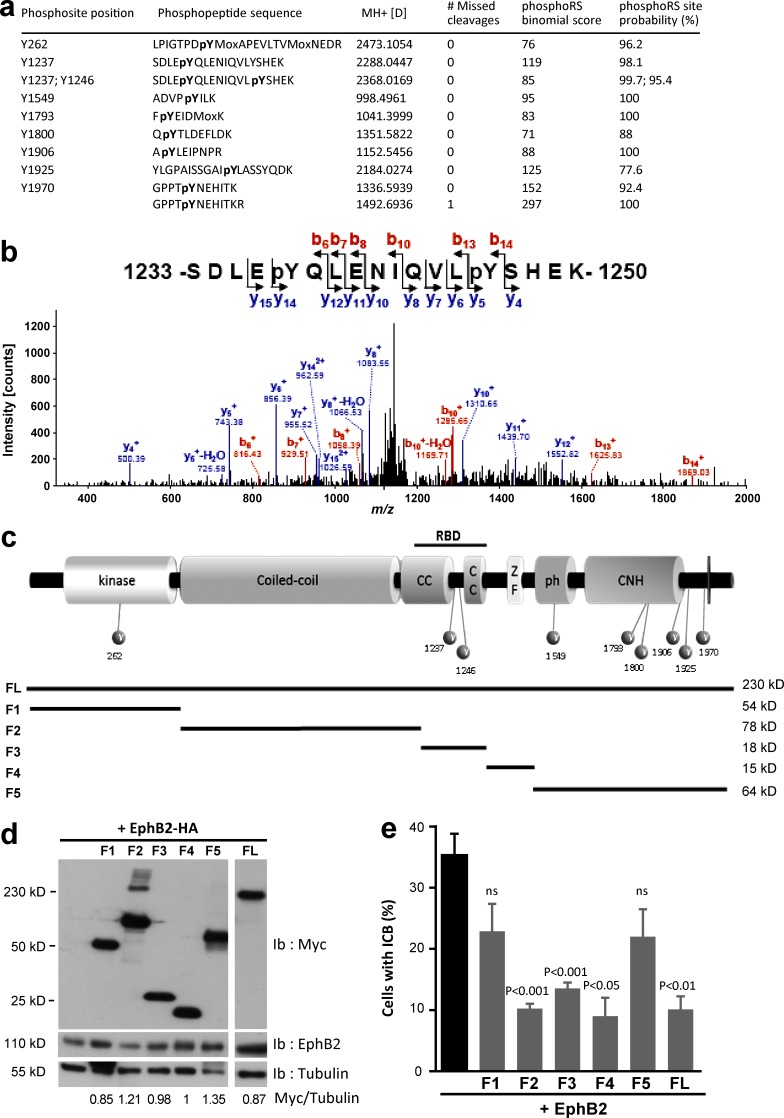
CitK is a large multidomain protein that contains several putative serine, threonine, and tyrosine phosphorylation sites. To identify the specific tyrosine residues of CitK that are phosphorylated by Src, we used in vitro kinase assays described earlier and nano–liquid chromatography-tandem MS (nanoLC-MS/MS) analysis. We identified nine tyrosines on CitK that are directly phosphorylated by Src among the 47 tyrosines present in the amino acid sequence (Fig. 5, a and b; and Table S2). Phosphorylated tyrosines were distributed along the entire length of the protein, some of them matching to key domains (Fig. 5 c). To assess the relevance of these modifications for cytokinesis, we first expressed individual domains of CitK and tested their ability to rescue the abscission defect induced by EphB2 activation (Fig. 5, d and e). Unlike CitK FL, CitK fragments corresponding to the serine/threonine kinase domain (F1) and the PH+CNH domains (F5) did not significantly rescue the persistence of the ICB between daughter cells (Fig. 5 e). On the other hand, expression of CitK fragments corresponding to the coiled-coil domain, Rho-binding domain (RBD), and Zinc finger domain (F2, F3, and F4, respectively) rescued the EphB2-induced ICB persistence phenotype (Fig. 5 e). These data are consistent with previously published results demonstrating that CitK mediates the transition from constriction to abscission via its coiled-coiled domain (Watanabe et al., 2013). Rescue by the Zinc finger/C1 domain was unexpected based on the published literature; however, it is possible that expression of this lipid–protein interaction domain may help tether endogenous CitK to the midbody and plasma membrane (as recently found for MgcRacGAP; Lekomtsev et al., 2012), thus partially rescuing EphB2-induced abscission defects.
Figure 5.
Identification of phosphorylated tyrosines on CitK. (a) Tyrosine-phosphorylated peptides identified on CitK by nanoLC-MS/MS. (b) The CID MS/MS spectrum of the diphosphorylated peptide, 1233-SDLEpYQLENIQVLpYSHEK-1250 (doubly charged precursor ion, MH2+, at m/z 2 1184.5121) displays a series of y- and b-ions, indicating that Y1237 and Y1246 are phosphorylated. pY, phosphorylated tyrosine residues. (c) Schematic drawing illustrating the distribution of Src-phosphorylated tyrosines identified by nanoLC-MS/MS analysis on CitK and the F1–F5 fragments of CitK. (d) HEK-293 cells were cotransfected with HA-tagged EphB2 and full-length myc-tagged CitK (FL) or the indicated myc-tagged CitK fragments (F1 to F5). Protein lysates were immunoblotted for Myc and EphB2. Tubulin was used as loading control (the upper band in the F1 sample corresponds to leftover signal from the Myc immunoblot). Ratio of signal intensity is provided below. (e) HeLa cells were transfected with EphB2 alone or with CitK FL or the indicated CitK fragments (F1 to F5). Cells were stimulated with Efnb1-Fc, and the proportion of cells connected by an ICB at 24 h of treatment was quantified in each condition. Statistical significance was calculated by comparison to the EphB2-alone condition (black bar). Error bars correspond to SEM. Statistical p-value is indicated when significant. ns, nonsignificant.
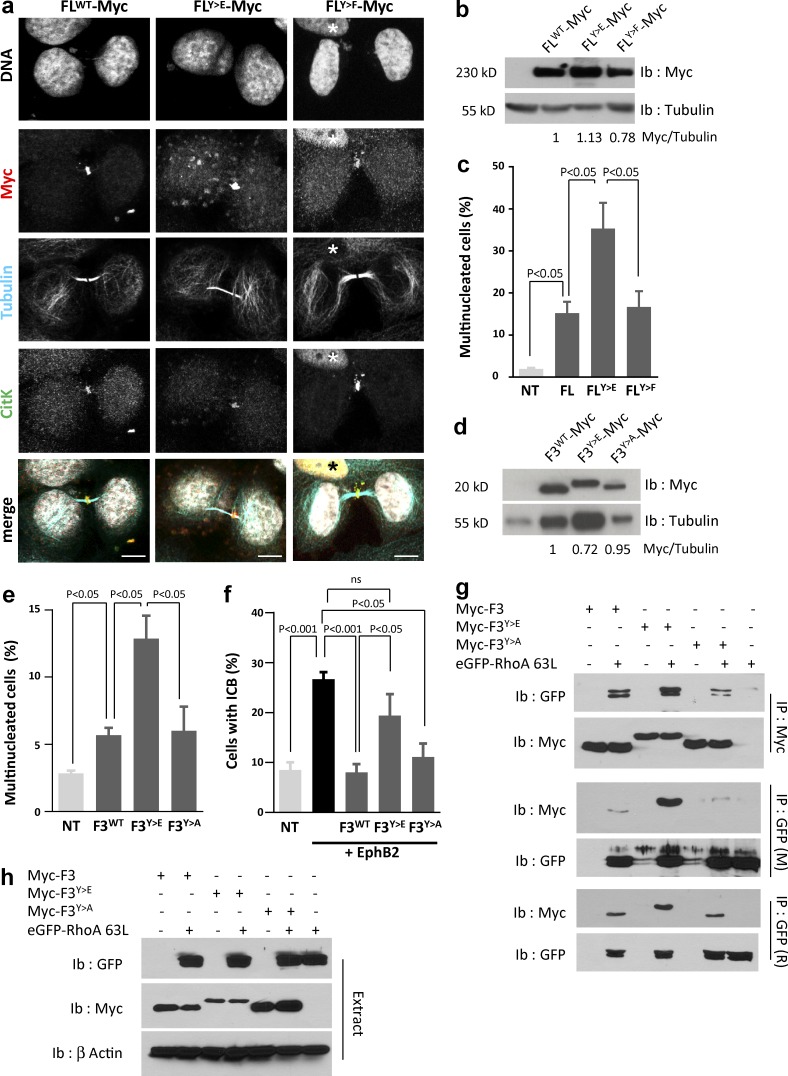
Based on these results, we narrowed down the list of phosphorylated tyrosines to be tested functionally to tyrosines 1237 and 1246, located in the RBD of CitK, and postulated that phosphorylation on those tyrosine residues may negatively impact CitK function during abscission. Both tyrosines were mutated into either phospho-mimetic (Y to E) or unphosphorylatable (Y to F) residues in CitK FL, and we verified that these mutations did not preclude localization of CitK to the midbody (Fig. 6 a). Next, we functionally tested the effect of these mutations on cytokinesis. Expression of the phosphomimetic FLY>E protein caused a marked increase in the number of multinucleated cells compared with WT or unphosphorylatable FLY>F proteins (Fig. 6, b and c). Although less potent than CitK FL mutants, mutated versions of the F3 fragment (RBD) of CitK produced similar results, and expression of the phosphomimetic RBD mutant was sufficient to cause multinucleation compared with WT or unphosphorylatable RBD (Fig. 6, d and e). Finally, we tested whether the mutant F3 fragments could rescue the phenotypes induced by EphB2 forward signaling. Although both F3WT and unphosphorylatable F3Y>A fragments prevented EphB2-induced ICB persistence, expression of the phosphomimetic F3Y>E fragment did not rescue ICB maintenance induced by EphB2 (Fig. 6 f). These results clearly show that phosphorylation of CitK on tyrosines 1237 and 1246 prevents the completion of cytokinesis. One possible mechanism by which phosphorylation of these two tyrosine residues may affect CitK function during cytokinesis is by modulating its association with its partners. The RBD of CitK mediates its interaction with Rho proteins (Madaule et al., 1998); we therefore assessed the interaction between RhoA, a major effector of cytokinesis, and the mutated forms of CitK RBD. Immunoprecipitation of either myc-tagged F3 fragments or GFP-tagged RhoA63L (a constitutively activated form of RhoA) revealed a consistently increased association between F3Y>E and RhoA63L compared with F3WT and F3Y>A (Fig. 6, g and h). Altogether, these results suggest that phosphorylation of tyrosines 1237 and 1246 within the RBD of CitK stabilizes the interaction of CitK with active RhoA.
Figure 6.
Tyrosine phosphorylation of CitK is detrimental to abscission. (a) HeLa cells were transfected with Myc-tagged CitK FL, phosphomimetic CitK (FLY>E), or unphosphorylatable CitK (FLY>F), fixed, and immunostained to detect Myc (red), tubulin (cyan), or CitK (green). Nuclei were stained with DAPI (gray). Nuclear localization of CitK (asterisks) corresponds to interphase cells. Bars, 10 µm. (b and c) HeLa cells were untransfected (NT) or transfected for 48 h with Myc-tagged CitK FL, phosphomimetic CitK (FLY>E), or unphosphorylatable CitK (FLY>F). (b) Protein extracts were immunoblotted with anti-myc and -tubulin antibodies. Ratio of signal intensity is provided below. (c) The proportion of multinucleated cells was quantified in each condition. (d and e) HeLa cells were untransfected (NT) or transfected for 48 h with Myc-tagged CitK-F3 (F3WT), phosphomimetic CitK-F3 (F3Y>E), or unphosphorylatable CitK-F3 (F3Y>A) fragments. (d) Protein extracts were immunoblotted with anti-myc and -tubulin antibodies. Ratio of signal intensities is indicated below. (e) The proportion of multinucleated cells was quantified in each condition. (f) HeLa cells were untransfected (NT) or cotransfected for 48 h with EphB2 alone (black bar) or Myc-tagged CitK-F3WT, phospho-mimetic CitK-F3Y>E, or unphosphorylatable CitK-F3Y>A fragments. Cells were stimulated with Efnb1-Fc for 24 h, and the proportion of cells connected by an ICB was quantified in each condition. (g and h) HEK-293 cells were transfected with Myc-tagged CitK-F3WT (F3), phospho-mimetic CitK-F3Y>E (F3Y>E), or unphosphorylatable CitK-F3Y>A (F3Y>A) and an activated form of RhoA tagged with GFP (eGFP-RhoA63L) as indicated. (g) CitK fragments were immunoprecipitated (top, Ip Myc) with an anti-Myc antibody and immunoblotted with GFP and Myc antibodies. RhoA was immunoprecipitated (middle and bottom, Ip GFP) with anti-GFP antibodies of different species and immunoblotted with GFP and Myc antibodies. (h) Protein lysates (Extract) were probed with antibodies to detect GFP and Myc. Actin was used as loading control. Western blots are representative of at least two independent experiments. Error bars correspond to SEM. Statistical p-value is indicated when significant. ns, nonsignificant.
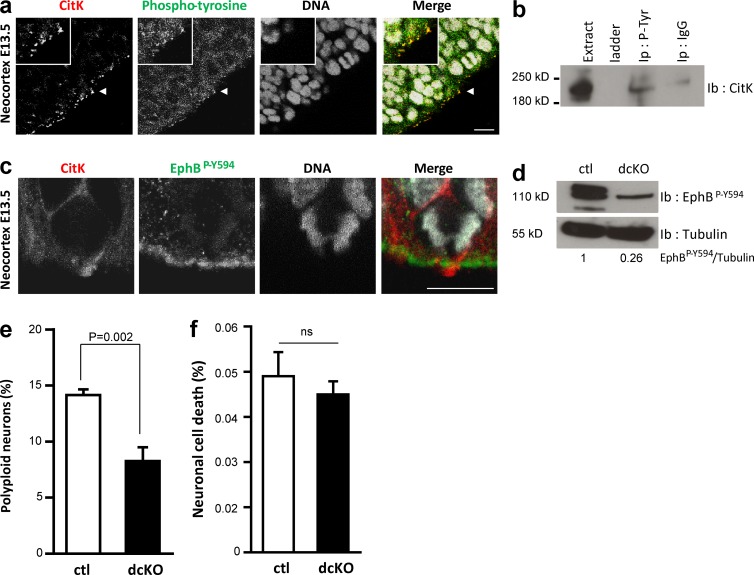
As mentioned previously, CitK is required to control cytokinesis of neural progenitors in the developing neocortex, and loss of CitK leads to an increase in the number of polyploid neurons (Di Cunto et al., 2000; Sarkisian et al., 2002; Sgro et al., 2016). We showed previously that EphB2 is expressed in neural progenitors (Arvanitis et al., 2013) and thus decided to test whether cytokinesis of neural progenitors may be controlled by Ephrin/Eph signaling. We focused our analyses on the neocortex at embryonic day 13.5 (E13.5), because at this developmental stage the majority of cells in the neocortex are neural progenitors undergoing active proliferation, with mitosis of apical neural progenitors taking place at the ventricular surface of the neocortex (Taverna et al., 2014). As reported previously, CitK is present at the ventricular surface of the neocortex, its punctate distribution reflecting its enrichment in midbodies and midbody remnants (Di Cunto et al., 2000; Chang et al., 2010). Coimmunostaining of paraffin sections with P-Tyr and CitK antibodies indicated a partial colocalization between CitK and tyrosine-phosphorylated proteins at the ventricular surface of the neocortex (Fig. 7 a), suggesting the putative tyrosine phosphorylation of CitK in neural progenitors. To confirm this, we performed immunoprecipitations from E13.5 cortices using the P-Tyr antibody. Immunoblotting with the CitK antibody revealed that endogenous CitK was immunoprecipitated with the P-Tyr antibody (Fig. 7 b) indicating that it is phosphorylated on tyrosine in vivo. Interestingly, the phosphorylated form of EphB receptors was markedly enriched at the ventricular surface of the neocortex (Fig. 7 c), and coimmunostaining showed that P-EphB2 and CitK are both present and in close proximity at the apical surface of dividing neural progenitors (Fig. 7 c). To test whether Ephrin/Eph signaling controls cytokinesis in vivo, we generated mice in which Ephrin/Eph signaling is specifically abrogated in neural progenitors and quantified neuronal ploidy as performed previously in CitK mutants (Di Cunto et al., 2000). To avoid potential functional compensation between Eph receptors, we generated mice deficient for the two ephrinB ligands that are expressed in neural progenitors using the Cre-lox system (Efnb1loxlox; Efnb2loxlox; Nestin-Cre). As expected, phosphorylation of EphBs was strongly decreased in the neocortex of these mutants (Fig. 7 d). Cytometry analyses indicated that loss of Ephrin/Eph signaling in neural progenitors led to a reduction in the fraction of polyploid neurons present at P0 in the neocortex of ephrin mutants compared with age-matched controls (Fig. 7 e and Fig. S5). Importantly, cell death was not affected in the neocortex of ephrin mutants (Fig. 7 f).
Figure 7.
Ephrin/Eph signaling controls neuronal polyploidy in the developing neocortex. (a) Paraffin sections of E13.5 embryos at the level of the neocortex were immunostained for CitK (red) and proteins phosphorylated on tyrosines (green). Nuclei were stained with DAPI (gray). Arrowheads indicate regions of colocalization between CitK and phosphotyrosines. Bar, 20 µm. (b) Protein lysates from E13.5 neocortex were used to immunoprecipitate tyrosine-phosphorylated proteins (Ip P-Tyr). Anti–human IgG was used as a control (Ip IgG). Whole lysate (Extract) and immunoprecipitated proteins were immunoblotted with a CitK antibody. Western blots are representative of two independent biological replicates. (c) High magnification of a dividing neural progenitor in the neocortex immunostained for CitK (red) and EphBP-Y594 (green). DNA is stained with DAPI (gray). Bar, 20 µm. (d) Protein lysates extracted from the neocortex of E13.5 control (ctl; Efnb1loxlox; Efnb2loxlox) and double mutant (dcKO, Efnb1loxlox; Efnb2loxlox; Nestin-Cre) embryos were immunoblotted with the EphBP-Y594 antibody. Tubulin was used as a loading control. Ratio of signal intensity is provided below. (e) Neuronal ploidy was quantified by flow cytometry in cortical neurons from control (ctl) and double conditional mutant (dcKO) P0 pups. (f) Neuronal cell death was quantified by flow cytometry in cortical neurons from control (ctl) and double-mutant (dcKO) P0 pups. Error bars correspond to SEM. Statistical p-value is indicated when significant. ns, nonsignificant.
Discussion
Our results show that activation of EphB2 forward signaling affects the completion of abscission. EphB2 activates Src, which phosphorylates CitK within its RBD, on tyrosines 1237 and 1246. Although it was shown previously that Src and CitK colocalize at the ingression furrow and midbody during cytokinesis (Shafikhani et al., 2008) and that Src kinase activity plays a role in cytokinesis (Ng et al., 2005; Kasahara et al., 2007; Soeda et al., 2013), the direct phosphorylation of CitK by Src has never been described. Two independent large-scale proteomic analyses of adult mouse brain have reported phosphorylation of CitK on tyrosine 1237, demonstrating that this exact modification is physiological (Ballif et al., 2008; Wiśniewski et al., 2010). Interestingly, an interaction between Citron-N (the neuronal variant that lacks the kinase domain) and Src has been recently reported in mouse brain to play a role in dendritic spine and synapse formation (Repetto et al., 2014), although it is unclear whether this interaction was direct or mediated via binding of Citron-N to other partners, such as the Src inhibitory protein p140Cap/SRCIN1 (Repetto et al., 2014). In light of our results, this Citron-N–Src interaction may reflect the fact that Citron-N is also a substrate of Src in postmitotic adult neurons.
Loss of CitK in the developing brain was previously reported to increase the fraction of polyploid neurons, and this was attributed to its role in cytokinesis (Di Cunto et al., 2000; Sarkisian et al., 2002). We show here that loss of Ephrin/Eph signaling in neural progenitors induces an opposite phenotype: a decrease in the number of polyploid neurons. This is a striking observation in the context of Ephrin/Eph signaling because the role of this pathway in vivo is generally associated with migration or adhesion processes rather than DNA content variation or cell division. It is now recognized that in physiological conditions, a substantial fraction of neurons of the postnatal mammalian cerebral cortex present DNA content variations, including polyploidy, and this feature is invoked as a basis for neuronal diversity (Muotri and Gage, 2006; Bushman and Chun, 2013) or as a means to meet higher metabolic demands (López-Sánchez et al., 2011). The mechanisms by which these somatic variations in DNA content arise remain poorly characterized, and our results suggest that incomplete cytokinesis of neural progenitors could contribute to this process. Regardless of the mechanism, our observation that physiological neuronal polyploidy is genetically controlled indicates that it is not caused by random cell division errors. Of note, another receptor tyrosine kinase, p75NTR, has also been implicated in controlling neuronal ploidy in the neocortex, but no mechanistic description was reported (López-Sánchez and Frade, 2013).
Tyrosine phosphorylation of CitK correlates with its mislocalization from the midbody, abscission delay, and cytokinesis defects (persistence of the ICB, multinucleation, and polyploidy). Overexpression of wild-type CitK or domains of CitK involved in protein–protein interactions is sufficient to compensate for alterations of endogenous CitK function, presumably because a significant fraction of overexpressed CitK remains unphosphorylated. Consistent with this idea, a phosphomimetic CitK RBD is unable to rescue EphB2-induced abscission defects. Furthermore, expression of phosphomimetic CitK is sufficient to cause cytokinesis defects regardless of EphB2 activation status, demonstrating that tyrosine phosphorylation of CitK modulates its function in abscission. Both loss and gain of CitK have been reported previously to promote multinucleation (Echard et al., 2004; Gai et al., 2011; Watanabe et al., 2013), indicating that tight control of CitK function is required for the completion of cytokinesis. Tyrosine phosphorylation appears to act as such a regulatory switch, allowing the fine tuning of the progression through abscission.
As mentioned earlier, during cytokinesis CitK plays an important role in controlling the orderly recruitment of key effectors at the midbody, including RhoA, Anillin, MKLP1, and KIF14 (Gruneberg et al., 2006; Gai et al., 2011; Bassi et al., 2013; Watanabe et al., 2013), and a recent study showed that phosphorylation of CitK by aurora kinase controls stereotyped organization of the midbody (McKenzie et al., 2016). We show here that tyrosine phosphorylation of the CitK RBD promotes its interaction with activated RhoA. Whether this disrupts the timing of recruitment of CitK to the contractile ring and whether it prevents the timely inactivation of RhoA at the midbody to allow completion of cytokinesis remains to be explored (Saurin et al., 2008; Gai et al., 2011; Watanabe et al., 2013). Indeed, RhoA must be inactivated at late stages of cytokinesis to allow for disassembly of actin filaments (Saurin et al., 2008; Manukyan et al., 2015). For instance, Anillin was reported to recruit p190RhoGAP to the midbody, and p190RhoGAP depletion results in abscission failure because of accumulation of active RhoA with similar rates to those observed in this study (Manukyan et al., 2015). Likewise, the recruitment of PKCε/14-3-3 to the midbody ring where RhoA and CitK localize is needed to extinguish RhoA activity at the midbody, and PKCε inhibition results in elevated RhoA activity and abscission failure (Saurin et al., 2008). Local control of membrane lipid composition, such as phosphatidyl ethanolamine and PI(4,5)P2, around the midbody is also crucial in cytokinesis and abscission, likely allowing the timely recruitment of various effectors of these processes, and the PI(4)P 5-kinase PIP5Kβ, a RhoA effector, and the PI(4,5)P2 5-phosphatase OCRL both play a critical role in the completion of cytokinesis (Emoto et al., 2005; Dambournet et al., 2011). From our results, it appears likely that sequential tyrosine phosphorylation and dephosphorylation of CitK is required to allow its activation and inactivation required for the completion of abscission, possibly by modulating RhoA activation levels at the midbody (Gai et al., 2011). Indeed, an attractive possibility is that activation of the EphB2/Src pathway leads to sustained tyrosine phosphorylation of CitK, promoting the CitK–RhoA interaction and maintaining elevated RhoA activity at the midbody, thus preventing abscission. The CitK–RhoA interaction could possibly prevent access of a RhoGAP to inactivate RhoA. In this context, it would be interesting to identify potential tyrosine phosphatases implicated in the completion of abscission, and PTPN13 appears to be a possible candidate (Hagemann et al., 2013). Finally, we cannot rule out the possibility that activation of Eph forward signaling and phosphorylation of CitK could perturb its recently described midbody microtubule-stabilizing function (Sgro et al., 2016), or the function of the molecular machinery required for membrane splitting on each side of the midbody, including components of the endosomal sorting complex required for transport–III (Mierzwa and Gerlich, 2014).
In conclusion, our study uncovers a novel role for Ephrin/Eph signaling in controlling cell division and neuronal polyploidy. In addition, it provides the first functional characterization of tyrosine phosphorylation events on CitK in cytokinesis. More generally, our findings support the concept that the local environment plays an important role in pacing abscission and suggest that CitK could be an integrator of extrinsic and intrinsic information during cytokinesis.
Materials and methods
Plasmids
Plasmids coding for EphB2 (pcDNA3.1-HA-EphB2 coding for WT mouse EphB2 and pcDNA3.1-HA-K661R coding for kinase-dead mouse EphB2) were obtained from T. Pawson (Samuel Lunenfeld Institute, Toronto, ON, Canada). The CitK cDNA was provided by F. Di Cunto (University of Turin,Turin, Italy). E. Pasquale (Burnham Institute for Medical Research, La Jolla, CA) provided a mutant cDNA for EphB2. The pcDNA3-tubulin:mCherry plasmid was a gift from A. Merdes (Center for Integrative Biology, Toulouse, France). The EphA3 cDNA (#23911; Addgene) was a gift from W. Hahn (Broad Institute, Boston, MA) and was subcloned by In-fusion recombination (Takara Bio Inc.) into a pcDNA3.1 Hygro vector. The pEGFP-N1-TrkB plasmid (#32500; Addgene) was a gift from R. Segal (Dana Farber Cancer Institute, Boston, MA). Plasmids coding for peGFP-RhoA and for CitK, pcDNA3.1Hygro-Myc-CRIK_FL, CRIK_F1, CRIK_F2, CRIK_F3, CRIK_F4, CRIK_F5, and pcDNA6-Myc-His-CRIK_FL were described previously (Besson et al., 2004; Serres et al., 2012).
Mutagenesis to produce CitK F3 phosphomimetic (F3Y>E) and unphosphorylatable (F3Y>A) coding plasmids was performed using the gBlocks technology (idtDNA) and the pcDNA3.1Hygro-Myc-CRIK_F3 plasmid as template. gBlocks were synthesized with the coding sequence for oriented cloning with KpnI and XhoI restriction enzymes flanking the CRIK_F3 coding sequence, modified for Tyr 1237 and 1246. CRIK_F3 coding sequence was excised from the pcDNA3.1-Myc-CRIK_F3 plasmid using those restriction enzymes and replaced by the gBlocks fragments using Rapid DNA ligation kit (11635379-001; Roche). CitK FL phosphomimetic (FLY>E) and unphosphorylatable (FLY>F) mutants were generated by site-directed mutagenesis using primers carrying the mutations and the Phusion Taq polymerase and DpnI enzyme (Thermo Fisher Scientific). All plasmids were validated by sequencing before plasmid amplification.
Antibodies
Anti–α/β-tubulin (rabbit, #2148, immunofluorescence [IF] 1:50), anti-phosphotyrosine (mouse, 4G10, IF 1:200, Western blot [WB] 1:2,000; #9411), anti-Src (rabbit, WB 1:1,000; #2108), and anti–phospho Src Tyr416 (rabbit, WB 1:1,000; #2108) were purchased from Cell Signaling Technology. Phalloidin (IF 1:200; ab176753) and anti–phospho EphB Y594 (rabbit, IF 1:200, WB 1:1,000; ab61791) were purchased from Abcam. Anti-CitK (goat, IF 1:200, WB 1:1,000; sc-1848), anti-Crik (mouse, BD, WB 1/500, 3 µg per immunoprecipitation [IP]), anti-cMyc (mouse, 9E10, IF 1:100, WB 1:1,000, coimmunoprecipitation [coIP] 1:200; sc-40), anti-cMyc (rabbit, A14, WB 1:1,000, 3 µg per IP; sc-789), anti-GFP (rabbit, FL, WB 1/1,000, 3 µg per IP; sc-8334), and anti-GFP (mouse, B2, WB 1/1,000, 3 µg per IP; sc-9996) were purchased from Santa Cruz Biotechnology, Inc. Anti-HA (mouse, IF 1:500, WB 1:2,000; 16B12) was purchased from Covance. Anti-EphB2 (goat, IF 1:50, WB 1:500; AF467) was purchased from R&D Systems. Anti–α-tubulin (mouse, IF 1:50; clone AA4.3) was purchased from DSHB. Anti–α-tubulin (mouse, DM1.a, WB 1:1,000) and anti–β-actin (mouse, WB 1:10,000; AC74) were purchased from Sigma-Aldrich. C-terminal Anti–Anillin (IF 1:200; GTX 107742) and anti-RacGAP1 (IF 1:200; GTX 113320) were purchased from GeneTex. Anti-NeuN (mouse, cytometry 1:100; MAB377) was purchased from EMD Millipore. Secondary antibodies coupled to Alexa or Cyanine (IF 1:250) or coupled to HRP (WB 1:5,000) were obtained from Jackson ImmunoResearch Laboratories, Inc. DAPI (D8417) was obtained from Sigma-Aldrich. For WB of brains, immunoprecipitation mouse TrueBlot (Rockland Immunochemicals; 1:1,000 dilution) was used for detection.
Mice
Efnb1loxlox, Efnb2loxlox, and Nestin-Cre mouse lines have been described (Tronche et al., 1999; Davy et al., 2004; Grunwald et al., 2004). To generate compound mutants, Efnb1loxlox; Efnb2loxlox females were bred with Nestin-Cre; Efnb1Ylox; Efnb2loxlox males. Neonates were individually genotyped by PCR. Animal procedures were preapproved by the appropriate Animal Care Committee (MP/07/21/04/11).
Cell lines and treatments
Human glioma U251, human cervix carcinoma HeLa, and human embryonic kidney HEK-293T cell lines were cultured in DMEM-glutamax (61965-059; Invitrogen) supplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin (15140-122; Invitrogen), and 1 mM sodium pyruvate (11530-396; Invitrogen) at 37°C in 5% CO2 in a humidified incubator. Stimulation of forward signaling was obtained by adding preclustered recombinant proteins to the culture medium. Preclustering was realized by mixing 0.1 µg/ml goat anti–human IgG (#G-101-C; R&D Systems) with 1 µg/ml recombinant human IgG1-Fc (#110-HG; R&D Systems) for control or recombinant mouse Efnb1-Fc chimera (#473-EB; R&D Systems) for effective stimulation, for 1 h at 37°C in culture medium. Cells overexpressing EphA3 were stimulated with clustered Efna5-Fc chimera (#374-EA). Cells overexpressing TrkB receptors were stimulated with 50 ng/ml hBDNF (B3795; Sigma-Aldrich). To stimulate endogenous EGF receptors and FGF receptors, cells were stimulated with 50 ng/ml human EGF (F0291; Sigma-Aldrich) and 20 ng/ml human FGF (9644; Sigma-Aldrich), respectively. To inhibit Src activity, cells were incubated with 5 µM inhibitor PP2 (P0042; Sigma-Aldrich) or SU6656 (S9692; Sigma-Aldrich) 1 h before stimulation, and inhibitors were maintained until the end of the experiments.
Transfections
Cells were plated 1 day before transfection at a density of 5 × 104 cells/ml on glass coverslips placed at the bottom of a 12-well plate or at a density of 2 × 105 cells/ml in six-well plates and 100-mm-diameter plates. HeLa cells were transfected using Fugene HD (E2311; Roche) according to the manufacturer’s instructions, in OptiMEM (51985-026; Invitrogen) using 0.5 µg plasmid for 12-well plates, 1 µg plasmid for six-well plates, and 5–10 µg for 100-mm-diameter plates. HEK-293T cells were transfected with the calcium phosphate procedure using 5 µg plasmid in 1 ml phosphate calcium solution for a 100-mm-diameter plate. For cotransfections, total amounts of DNA were identical to those described earlier, with a 1:3 ratio for Eph coding plasmid and cotransfected plasmid, respectively.
Si-RNAs (SMARTpool-ON TARGET Plus) consisting of siRNA targeting human CitK (L-004613-00), siRNA targeting human SRC (L-003175-00), siRNA targeting human EphB2 (L-003122-00), and siRNA control (D-001810-10-05) were purchased form GE Healthcare. Transfection was performed using the Lipofectamine RNAiMAX technology in a forward protocol and 50 µm siRNA duplex per well in a 35-mm Petri dish. Validation for gene extinction was done 48 h after transfection.
Immunofluorescence and flow cytometry
For immunofluorescence, cells were grown and treated as described earlier on glass coverslips and fixed at RT with 2% paraformaldehyde (15710; Electron Microscopy Sciences) for 20 min, permeabilized, and blocked with PBTA solution (0.5% BSA, 0.5% FBS, and 0.1% Triton X-100 in PBS) for 30 min. Primary and secondary antibodies diluted in PBTA were incubated for 2 h or 45 min, respectively, at RT in a humidified atmosphere and rinsed three times for 1 min each in successive baths of PBS. Coverslips were mounted on glass slides (Superfrost) using mounting medium (4.8% wt/vol Mowiol and 12% wt/vol glycerol in 50 mM Tris, pH 8.5).
For flow cytometry, cells were grown in six-well plates and collected using trypsin-EDTA 0.25% (Invitrogen) and mechanical dissociation by pipetting. Approximately 1 × 106 cells in 1.2 ml ice-cold PBS were fixed by slowly adding 3 ml cold 100% ethanol while vortexing to obtain a 70% final concentration. The incubation procedure with primary and secondary antibodies was as described for immunofluorescence, with centrifugation at 2,200 rpm for 10 min at each step. Finally, cells were resuspended in 500 µl PI/RNase solution (550825; BD) for 15 min in the dark. Acquisition was performed on a FACSCalibur (BD) with a minimum of 1 × 104 cells analyzed with CellQuestPro software (BD).
For analysis of neuronal ploidy, cortices of P0 pups from distinct litters were collected in ice-cold PBS-2% BSA and mechanically dissociated, and cells were sieved through a 40-µm nylon mesh (542-040; Greiner). Cells were washed with ice-cold PBS and fixed as described earlier in 70% ethanol. Incubation procedures with primary (NeuN) and secondary antibodies were as described earlier. Cells were resuspended in 500 µl PI/RNase (550825; BD) solution for 15 min in the dark. Acquisition was performed on a FACSCalibur (BD) with 2 × 105 cells analyzed with CellQuestPro software (BD). Debris and doublets were excluded using morphometric parameters and pulse area parameters from PI emission, respectively. To set the threshold of NeuN-positive cells, control samples were cells incubated with secondary antibody and PI only. Neuronal DNA content was estimated by analyzing PI emission intensity of NeuN-positive cells only. Five and three embryos were analyzed for controls (WT and Efnb1lox; Efnb2loxlox) and mutants (Nes-Cre; Efnb1lox; Efnb2loxlox), respectively.
Live imaging and microscopy
For live imaging on Incucyte, HeLa cells were plated at a density of 2 × 104 cells per well in ImageLock 24-well plates (#4365; Essen BioScience) and transfected with the indicated plasmids the next day. 24 h after transfection, cells were synchronized in prophase using the CDK1 inhibitor Ro-3306 (SLM0569; Sigma-Aldrich) at 10 µM for 18 h and then washed three times with 1 ml prewarmed inhibitor-free medium. For stimulation, 1 ml culture medium containing preclustered ligands (described earlier) was added, and plates were immediately loaded into the Incucyte incubator (FLR; Essen BioScience) for live imaging acquisition at 37°C in 5% CO2 in a humidified atmosphere. Software (IncuCyte 2011A) was calibrated as follows: three fields per well, one frame every 10 min for 8 h, with autofocus and autocalibration for green fluorescence. The time taken for plate loading and software calibration was estimated at ∼10 min on the first experiment and was included in subsequent experiments before starting acquisition. Cells were considered transfected when GFP was detected at both the beginning and the end of the acquisition sequence. Phase-contrast time-lapse images were manually analyzed (frame by frame) to monitor the presence or absence of the ICB between nascent cells. The ICB was visualized based on its refringence, and its time of disappearance was defined as the first frame in which no refringent structure between nascent daughter cells could be visualized.
High-resolution live imaging was performed on a 710 confocal microscope (25×/0.8 APO, PMT; ZEISS) equipped with a thermostat chamber with CO2 atmosphere control. HeLa cells were plated at a density of 2 × 104 cells per well in six-well glass-bottom plates and cotransfected as described earlier with EphB2-GFP and tubulin-mCherry plasmids. Synchronization and stimulation were performed as described earlier. Acquisition was performed with the Zen software (ZEISS) calibrated as follows: seven fields per well, one frame every 10 min for 12 h, 15 z-stacks of 2 µm.
Other microscope acquisitions were performed on either an SP5 DM600B confocal microscope (40×/1.3 APO; 20×/0.7 APO, PMT; Leica Biosystems) or an Eclipse 80i (20×/0.5 Nikon; DXM1200C camera; Nikon). Observation was performed using Type-F mineral oil (1153859; Leica Biosystems). Cell counting and pixel quantification were performed with the use of ImageJ software. On fixed cells, the ICB was defined as a thin tubulin-positive structure connecting two cells.
Image analyses of CitK subcellular distribution in the ICB
To calculate total CitK signal, the geometric mean intensity of pixels present in a zone of fixed size centered on the dark zone (absence of tubulin signal) of the ICB was collected from images of cells in telophase (n = 10). The sum (S) of the geometric mean intensity of all pixels represents the total amount of CitK in the measured area. To quantify the distribution of CitK, the measurement zone was subdivided in three parts (left, middle, and right), and S was calculated for each part (Sl, Sm, and Sr). The ratio of Sl and Sr versus Sm represents CitK distribution in the ICB; the more this ratio increases, the less CitK is restricted to the midbody.
Protein extraction, immunoprecipitation, and Western blot
For protein extraction, cells were scraped in an ice-cold PBS solution, pelleted by centrifugation at 1,500 rpm for 5 min, and washed once in a large volume of ice-cold PBS. Pellets were suspended in cold protein lysis buffer (50 mM Hepes, pH 7.5, containing 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 0.1% Tween 20, 10% glycerol, 1% NP-40, and 0.5 mM DTT) supplemented with anti-protease (11836170-001, complete; Roche) and anti-phosphatase (04906834-001, PhoStop; Roche) for 1 h on ice. Protein lysates were then vortexed, sonicated, and centrifuged at 13,000 rpm for 10 min. Cleared lysates were either used for Western blot analyses or processed for coIP assays. CoIP was performed overnight with primary antibodies, and 20 µl prewashed protein A beads (Sigma-Aldrich) were added to each tube for the last 45 min. Beads were washed three times in protein lysis buffer, and samples were denatured by boiling in loading buffer (4× 100 mM Tris-HCL, pH 6.8, 8% SDS, 40% glycerol, 4% β-mercaptoethanol, and bromophenol blue) before loading and electrophoresis on a 4–20% SDS-PAGE gel (456-1094; Bio-Rad Laboratories). Proteins were transferred onto a nitrocellulose membrane (11998905; GE Healthcare), which was blocked for 30 min and incubated with primary antibody in 5% nonfat dry milk in TBS-T (20 mM Tris base, 150 mM NaCl, and 0.05% Tween 20 adjusted to pH 7.6 with 1 M HCl) overnight at 4°C. Milk was replaced with 5% BSA (04-100-811-C; Euromedex) for detection of phosphorylated epitopes. Western blots presented are representative of at least three independent experiments.
For immunoprecipitation from brain tissues, cortices from E13.5 embryos were microdissected and pooled in 1 ml protein lysis buffer. Protein solubilization was performed as described earlier. For immunoprecipitation, protein lysates were precleared for 45 min at 4°C with protein A agarose beads to remove endogenous IgG and decrease nonspecific protein binding. Beads were removed, and cleared lysates were used for immunoprecipitation as described earlier.
In vitro kinase assay
For the in vitro kinase assay, HEK-293T cells were transfected with pcDNA6_CRIK_FL_Myc-Histidine–tagged plasmid for 48 h in 100-mm Petri plates. The cell monolayer was collected by scraping in ice-cold PBS and washed once, and the pellet was resuspended in 2 ml of 50 mM Hepes, pH 7.5, containing 250 mM NaCl, 10% glycerol, 0.5% NP-40, and 10 mM imidazole supplemented with antiprotease and phosphatase inhibitors for 1 h on ice, and then vortexed, sonicated, and centrifuged at 13,000 rpm for 10 min. 300 µl Talon beads (#635636; Takara Bio Inc.) were added to the cleared lysate for 90 min on a rocker at 4°C. Beads were centrifuged for 3 min at 3,000 rpm at 4°C and washed three times in 1 ml of 50 mM Hepes, pH 7.5, containing 300 mM NaCl, 0.05% Tween20, and 20 mM imidazole. CitK histidine-tagged protein was eluted by adding 50 µl washing buffer supplemented with 400 mM imidazole for 5 min on ice and vortexing. Indicated volumes of eluate were incubated with 50 ng EphB2 kinase (SRP-0346; Sigma-Aldrich) or 100 ng Src kinase (S1076; Sigma-Aldrich) in 20 µl kinase buffer (25 mM Hepes, pH 7.5, containing 2.5 mM MgCl2, 4 mM MnCl2, protease and phosphatase inhibitors, and 200 µM ATP) for 15 min at 37°C. Controls were included in the experiment by replacing the eluate, ATP, or kinases with buffer in the reaction mixture. The reaction was stopped by denaturation in loading buffer (4× 100 mM Tris-HCl, pH 6.8, 8% SDS, 40% glycerol, 4% β-mercaptoethanol, and bromophenol blue) and boiling for 10 min. Samples were then loaded and treated as described in the Western blotting section.
In-gel tryptic digestion and nanoLC-MS/MS analysis
For MS analysis, his-tagged CitK was purified as described earlier and processed for kinase assay. Samples were reduced for 30 min at 55°C, by adding 1× Laemmli sample buffer containing 25 mM DTT and alkylated in 90 mM iodoacetamide for 30 min in the dark at RT. Equal volumes of samples were separated on 4–20% SDS-PAGE. After protein separation, the acrylamide gel was stained using PhastGel Blue R (Sigma-Aldrich) according to the manufacturer’s instructions. Bands at expected size (∼230 kD) were excised and subjected to in-gel tryptic digestion using modified porcine trypsin (Promega) at 20 ng/µl as previously described (Shevchenko et al., 1996). The dried peptide extracts obtained were dissolved in 14 µl of 0.05% trifluoroacetic acid in 2% acetonitrile and analyzed by online nanoLC using an UltiMate 3000 RSLCnano LC system (Thermo Fisher Scientific) coupled to an ETD-enabled Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific) for an alternative decision tree–driven collision-induced dissociation (CID)/electron transfer dissociation (ETD) fragmentation acquisition (Swaney et al., 2008). 5 µl peptide extract was loaded on a 300-µm ID × 5 mm PepMap C18 precolumn (Thermo Fisher Scientific) at 20 µl/min in 2% acetonitrile and 0.05% trifluoroacetic acid. After 3-min desalting, peptides were separated online on a 75-µm ID × 15 cm C18 column (packed in-house with Reprosil C18-AQ Pur 3 µm resin; Dr. Maisch; Proxeon Biosystems). The flow rate was set at 300 nl/min. Peptides were eluted using a 5–50% linear gradient of solvent B in 60 min (solvent A was 0.2% formic acid in water and solvent B was 0.2% formic acid in 80% acetonitrile). Survey scans of peptide precursors from 300 to 2,000 m/z were performed at 120K resolution (at 200 m/z). Most intense ions per survey scan were selected at 2 m/z with the quadrupole for subsequent data-dependent decision tree–based CID/ETD fragmentation, and the resulting fragments were analyzed by a rapid-scan MS analysis in the ion trap. The settings for the data-dependent decision tree–based CID/ETD method were as follows: ETD was performed instead of CID if charge state was 3 and m/z was <650, or if the charge state was 4 and the m/z was <900, or if the charge state was 5 and the m/z was <950. ETD was performed for all precursor ions with charge states >5. The dynamic exclusion duration was set to 60 s with a 10-ppm tolerance around the selected precursor and its isotopes. Monoisotopic precursor selection was turned on. The instrument was run in top speed mode with 3-s cycles, meaning the instrument would continuously perform MS2 events until the list of nonexcluded precursors diminished to zero or 3 s, whichever was shorter. For internal calibration, the 445.120025 ion was used as lock mass.
Database search and data analysis
Peak lists extraction from Xcalibur raw files were automatically performed using Proteome Discoverer software (version 1.4.0.288; Thermo Fisher Scientific). The following parameters were set for creation of the peak lists: parent ions in the mass range 300–5,000 and no grouping of MS/MS scans. The nonfragment filter was used to simplify ETD spectra with the following settings: the precursor peak was removed within a 4-D window, charged reduced precursors were removed within a 2-D window, and neutral losses from charged reduced precursors were removed within a 2D window (the maximum neutral loss mass was set to 120 D). Peak lists were searched against the SwissProt mouse database implemented with the His-tagged CitK sequence and using Mascot software (version 2.3.01; Matrix Science). Cysteine carbamidomethylation was set as a fixed modification and methionine oxidation and serine/threonine/tyrosine phosphorylations as variable modifications. Up to three missed trypsin cleavages were allowed. Mass tolerances in MS and MS/MS were set to 10 ppm and 0.6 D, respectively. Peptide spectral matches were validated using percolator based on q-values at a 5% false discovery rate (Brosch et al., 2009). With Proteome Discoverer, peptide identifications were grouped into proteins according to the law of parsimony and filtered to 5% false discovery rate. The phosphorylation site localization of identified phosphopeptides was performed with phosphoRS algorithm 3.1 (Taus et al., 2011) implemented in Proteome Discoverer. Phosphopeptides and phosphoresidue localization were validated by a phosphoRS binomial score >50 and a phosphoRS site probability >75%. Phosphorylated site identification was confirmed by manual interpretation of corresponding MS/MS data.
Quantitative PCR
RNA was extracted from cell pellets using TRIreagent according to the manufacturer’s instructions. 1 µg RNA was used for reverse transcription. Genomic DNA was degraded with 1 µl DNase (RQ1; Roche) for 20 min at 37°C in 20 µl RNase/DNase-free water (W4502; Sigma-Aldrich), and the reaction was stopped by adding 1 µl stop solution under heat inactivation at 65°C for 10 min. 2 µl dNTPs (10 mM; Promega) and 2 µl oligodTs (100 mM, idtDNA) were added for 5 min at 65°C, then 8 µl of 5× buffer, 2 µl RNasin (N2511; Roche), and 4 µl of 100 mM DTT (Promega) were added for 2 min at 42°C. The mix was divided into equal volumes in a reverse-transcriptase–negative control tube with addition of 1 µl water and in a reverse-transcriptase–positive tube with 1 µl superscript enzyme (Invitrogen) and placed at 42°C for 1 h. The reaction was stopped at 70°C for 15 min, and cDNAs were diluted (10-, 100-, and 1,000-fold) and processed for quantitative PCR in triplicate for each dilution. 10 µl diluted cDNA was mixed with 10 µl premix Evagreen (BTIU31019; VWR International) containing 1 µM of each primer, and the PCR program was run for 35 cycles on a MyiQ BioRad thermocycler. mRNA relative expression levels were calculated using the 2-ΔΔCts method. Primers are listed in Table S1.
Statistical analysis
For experiments involving a single pair of conditions, statistical significance between the two sets of data were analyzed with a two-tailed, unpaired Student’s t test with Prism5 (GraphPad software). For datasets containing more than two samples, one-way analysis of variance with a classical Bonferroni multiple comparison posttest was used to determine adjusted p-values. Sample sizes of sufficient power were chosen on the basis of similar published research and were confirmed statistically by appropriate tests. Each experiment was performed at least three times, and 200–1,000 cells were counted for each condition of each experiment. Statistically significant differences are reported at P < 0.05, P < 0.01, P < 0.001, or exact value mentioned on the graphs.
Online supplemental material
Fig. S1 shows validation of siRNAs. Fig. S2 shows still images from Incucyte recordings. Fig. S3 shows immunofluorescence staining for Anillin and MgcRacGAP. Fig. S4 shows the effect of activation of other RTKs on abscission and phosphorylation of CitK. Fig. S5 shows cytometry parameters used for the analysis of neuronal polyploidy. Video 1 shows a stimulated EphB2+ HeLa cell undergoing cell death after entering telophase. Table S1 shows primer sequences used. Table S2 shows the mass spectrometry dataset. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201602057/DC1.
Supplementary Material
Acknowledgments
The EphB2-GFP, EphB2-3YF-GFP, HA-EphB2WT, and HA-EphB2K661R constructs were gifts from Dr. Tony Pawson. The CitK cDNA was provided by Dr. Ferdinando Di Cunto. We are grateful to Dr. Elena Pasquale and Dr. Andreas Merdes for sharing plasmids. FACS analyses were performed at the Centre de Biologie Intégrative platform, and images were acquired in the Toulouse Rio Imaging facility with the help of Brice Ronsin and Stéphanie Bosch for live imaging. We thank Maxime Bertrand for technical help and our laboratory colleagues for critical reading of the manuscript.
This work was supported by core funding from the Centre National de la Recherche Scientifique, a grant from Fondation ARC pour la Recherche sur le Cancer to A. Davy (SFI20101201607), an Agence Nationale de la Recherche grant to A. Davy (ANR-15-CE13-0010-01), and in part by the French Ministry of Research with the Investissement d’Avenir Infrastructures Nationales en Biologie et Santé program (Proteomics French Infrastructure project, ANR-10-INBS-08). A. Besson is supported by grants from the Fondation ARC pour la Recherche sur le Cancer and Ligue Contre le Cancer.
The authors declare no competing financial interests.
Author contributions: T. Jungas designed, carried out, and analyzed experiments. R.T. Perchey and M. Fawal performed and analyzed experiments. C. Callot generated various mutant forms of CitK FL and established the protocol for CitK purification. C. Froment and O. Burlet-Schiltz performed and analyzed MS experiments. A. Besson performed experiments, co-supervised the project, and participated in writing the manuscript. A. Davy wrote the manuscript and supervised the project.
Footnotes
Abbreviations used:
- CID
- collision-induced dissociation
- CitK
- citron kinase
- coIP
- coimmunoprecipitation
- E
- embryonic day
- ETD
- electron transfer dissociation
- FL
- full-length
- ICB
- intercellular bridge
- IF
- immunofluorescence
- IP
- immunoprecipitation
- LC-MS/MS
- liquid chromatography-tandem MS
- MS
- mass spectrometry
- NT
- nontransfected
- RBD
- Rho-binding domain
- RTK
- receptor tyrosine kinase
- WB
- Western blot
- WT
- wild-type
References
- Agromayor M., and Martin-Serrano J.. 2013. Knowing when to cut and run: Mechanisms that control cytokinetic abscission. Trends Cell Biol. 23:433–441. 10.1016/j.tcb.2013.04.006 [DOI] [PubMed] [Google Scholar]
- Arvanitis D.N., Béhar A., Tryoen-Tóth P., Bush J.O., Jungas T., Vitale N., and Davy A.. 2013. Ephrin B1 maintains apical adhesion of neural progenitors. Development. 140:2082–2092. 10.1242/dev.088203 [DOI] [PubMed] [Google Scholar]
- Ballif B.A., Carey G.R., Sunyaev S.R., and Gygi S.P.. 2008. Large-scale identification and evolution indexing of tyrosine phosphorylation sites from murine brain. J. Proteome Res. 7:311–318. 10.1021/pr0701254 [DOI] [PubMed] [Google Scholar]
- Bassi Z.I., Verbrugghe K.J., Capalbo L., Gregory S., Montembault E., Glover D.M., and D’Avino P.P.. 2011. Sticky/citron kinase maintains proper RhoA localization at the cleavage site during cytokinesis. J. Cell Biol. 195:595–603. 10.1083/jcb.201105136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi Z.I., Audusseau M., Riparbelli M.G., Callaini G., and D’Avino P.P.. 2013. Citron kinase controls a molecular network required for midbody formation in cytokinesis. Proc. Natl. Acad. Sci. USA. 110:9782–9787. 10.1073/pnas.1301328110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement W.M., Benink H.A., and von Dassow G.. 2005. A microtubule-dependent zone of active RhoA during cleavage plane specification. J. Cell Biol. 170:91–101. 10.1083/jcb.200501131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson A., Gurian-West M., Schmidt A., Hall A., and Roberts J.M.. 2004. p27Kip1 modulates cell migration through the regulation of RhoA activation. Genes Dev. 18:862–876. 10.1101/gad.1185504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch M., Yu L., Hubbard T., and Choudhary J.. 2009. Accurate and sensitive peptide identification with Mascot Percolator. J. Proteome Res. 8:3176–3181. 10.1021/pr800982s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman D.M., and Chun J.. 2013. The genomically mosaic brain: Aneuploidy and more in neural diversity and disease. Semin. Cell Dev. Biol. 24:357–369. 10.1016/j.semcdb.2013.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauvin C., and Echard A.. 2015. Phosphoinositides: Lipids with informative heads and mastermind functions in cell division. Biochim. Biophys. Acta. 1851:832–843. 10.1016/j.bbalip.2014.10.013 [DOI] [PubMed] [Google Scholar]
- Chang Y., Klezovitch O., Walikonis R.S., Vasioukhin V., and LoTurco J.J.. 2010. Discs large 5 is required for polarization of citron kinase in mitotic neural precursors. Cell Cycle. 9:1990–1997. 10.4161/cc.9.10.11730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell J.W., Lindon C., Luzio J.P., and Reid E.. 2009. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 10:42–56. 10.1111/j.1600-0854.2008.00847.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambournet D., Machicoane M., Chesneau L., Sachse M., Rocancourt M., El Marjou A., Formstecher E., Salomon R., Goud B., and Echard A.. 2011. Rab35 GTPase and OCRL phosphatase remodel lipids and F-actin for successful cytokinesis. Nat. Cell Biol. 13:981–988. 10.1038/ncb2279 [DOI] [PubMed] [Google Scholar]
- Davoli T., and de Lange T.. 2011. The causes and consequences of polyploidy in normal development and cancer. Annu. Rev. Cell Dev. Biol. 27:585–610. 10.1146/annurev-cellbio-092910-154234 [DOI] [PubMed] [Google Scholar]
- Davy A., Aubin J., and Soriano P.. 2004. Ephrin-B1 forward and reverse signaling are required during mouse development. Genes Dev. 18:572–583. 10.1101/gad.1171704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cunto F., Imarisio S., Hirsch E., Broccoli V., Bulfone A., Migheli A., Atzori C., Turco E., Triolo R., Dotto G.P., et al. 2000. Defective neurogenesis in citron kinase knockout mice by altered cytokinesis and massive apoptosis. Neuron. 28:115–127. 10.1016/S0896-6273(00)00090-8 [DOI] [PubMed] [Google Scholar]
- Echard A., Hickson G.R., Foley E., and O’Farrell P.H.. 2004. Terminal cytokinesis events uncovered after an RNAi screen. Curr. Biol. 14:1685–1693. 10.1016/j.cub.2004.08.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto K., Inadome H., Kanaho Y., Narumiya S., and Umeda M.. 2005. Local change in phospholipid composition at the cleavage furrow is essential for completion of cytokinesis. J. Biol. Chem. 280:37901–37907. 10.1074/jbc.M504282200 [DOI] [PubMed] [Google Scholar]
- Founounou N., Loyer N., and Le Borgne R.. 2013. Septins regulate the contractility of the actomyosin ring to enable adherens junction remodeling during cytokinesis of epithelial cells. Dev. Cell. 24:242–255. 10.1016/j.devcel.2013.01.008 [DOI] [PubMed] [Google Scholar]
- Gai M., Camera P., Dema A., Bianchi F., Berto G., Scarpa E., Germena G., and Di Cunto F.. 2011. Citron kinase controls abscission through RhoA and anillin. Mol. Biol. Cell. 22:3768–3778. 10.1091/mbc.E10-12-0952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R.A., Paluch E., and Oegema K.. 2012. Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 28:29–58. 10.1146/annurev-cellbio-101011-155718 [DOI] [PubMed] [Google Scholar]
- Gruneberg U., Neef R., Li X., Chan E.H., Chalamalasetty R.B., Nigg E.A., and Barr F.A.. 2006. KIF14 and citron kinase act together to promote efficient cytokinesis. J. Cell Biol. 172:363–372. 10.1083/jcb.200511061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald I.C., Korte M., Adelmann G., Plueck A., Kullander K., Adams R.H., Frotscher M., Bonhoeffer T., and Klein R.. 2004. Hippocampal plasticity requires postsynaptic ephrinBs. Nat. Neurosci. 7:33–40. 10.1038/nn1164 [DOI] [PubMed] [Google Scholar]
- Gucciardo E., Sugiyama N., and Lehti K.. 2014. Eph- and ephrin-dependent mechanisms in tumor and stem cell dynamics. Cell. Mol. Life Sci. 71:3685–3710. 10.1007/s00018-014-1633-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot C., and Lecuit T.. 2013. Adhesion disengagement uncouples intrinsic and extrinsic forces to drive cytokinesis in epithelial tissues. Dev. Cell. 24:227–241. 10.1016/j.devcel.2013.01.010 [DOI] [PubMed] [Google Scholar]
- Hagemann N., Ackermann N., Christmann J., Brier S., Yu F., and Erdmann K.S.. 2013. The serologically defined colon cancer antigen-3 interacts with the protein tyrosine phosphatase PTPN13 and is involved in the regulation of cytokinesis. Oncogene. 32:4602–4613. 10.1038/onc.2012.485 [DOI] [PubMed] [Google Scholar]
- Haglund K., Nezis I.P., and Stenmark H.. 2011. Structure and functions of stable intercellular bridges formed by incomplete cytokinesis during development. Commun. Integr. Biol. 4:1–9. 10.4161/cib.13550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herszterg S., Leibfried A., Bosveld F., Martin C., and Bellaiche Y.. 2013. Interplay between the dividing cell and its neighbors regulates adherens junction formation during cytokinesis in epithelial tissue. Dev. Cell. 24:256–270. 10.1016/j.devcel.2012.11.019 [DOI] [PubMed] [Google Scholar]
- Herszterg S., Pinheiro D., and Bellaïche Y.. 2014. A multicellular view of cytokinesis in epithelial tissue. Trends Cell Biol. 24:285–293. 10.1016/j.tcb.2013.11.009 [DOI] [PubMed] [Google Scholar]
- Hickson G.R., and O’Farrell P.H.. 2008. Rho-dependent control of anillin behavior during cytokinesis. J. Cell Biol. 180:285–294. 10.1083/jcb.200709005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Wang Y., Wang T., Hawke D.H., Zheng Y., Li X., Zhou Q., Majumder S., Bi E., Liu D.X., et al. 2014. PKM2 phosphorylates MLC2 and regulates cytokinesis of tumour cells. Nat. Commun. 5:5566 10.1038/ncomms6566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S., Braithwaite A.W., Robinson P.J., and Chircop M.. 2011. Dynamin inhibitors induce caspase-mediated apoptosis following cytokinesis failure in human cancer cells and this is blocked by Bcl-2 overexpression. Mol. Cancer. 10:78 10.1186/1476-4598-10-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandouz M., Haidara K., Zhao J., Brisson M.L., and Batist G.. 2010. The EphB2 tumor suppressor induces autophagic cell death via concomitant activation of the ERK1/2 and PI3K pathways. Cell Cycle. 9:398–407. 10.4161/cc.9.2.10505 [DOI] [PubMed] [Google Scholar]
- Kania A., and Klein R.. 2016. Mechanisms of ephrin-Eph signalling in development, physiology and disease. Nat. Rev. Mol. Cell Biol. 17:240–256. 10.1038/nrm.2015.16 [DOI] [PubMed] [Google Scholar]
- Kasahara K., Nakayama Y., Nakazato Y., Ikeda K., Kuga T., and Yamaguchi N.. 2007. Src signaling regulates completion of abscission in cytokinesis through ERK/MAPK activation at the midbody. J. Biol. Chem. 282:5327–5339. 10.1074/jbc.M608396200 [DOI] [PubMed] [Google Scholar]
- Lacroix B., and Maddox A.S.. 2012. Cytokinesis, ploidy and aneuploidy. J. Pathol. 226:338–351. 10.1002/path.3013 [DOI] [PubMed] [Google Scholar]
- Laussu J., Khuong A., Gautrais J., and Davy A.. 2014. Beyond boundaries—Eph:ephrin signaling in neurogenesis. Cell Adhes. Migr. 8:349–359. 10.4161/19336918.2014.969990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bras S., and Le Borgne R.. 2014. Epithelial cell division—multiplying without losing touch. J. Cell Sci. 127:5127–5137. 10.1242/jcs.151472 [DOI] [PubMed] [Google Scholar]
- Lekomtsev S., Su K.C., Pye V.E., Blight K., Sundaramoorthy S., Takaki T., Collinson L.M., Cherepanov P., Divecha N., and Petronczki M.. 2012. Centralspindlin links the mitotic spindle to the plasma membrane during cytokinesis. Nature. 492:276–279. 10.1038/nature11773 [DOI] [PubMed] [Google Scholar]
- Lisabeth E.M., Falivelli G., and Pasquale E.B.. 2013. Eph receptor signaling and ephrins. Cold Spring Harb. Perspect. Biol. 5:a009159 10.1101/cshperspect.a009159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Sánchez N., and Frade J.M.. 2013. Genetic evidence for p75NTR-dependent tetraploidy in cortical projection neurons from adult mice. J. Neurosci. 33:7488–7500. 10.1523/JNEUROSCI.3849-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Sánchez N., Ovejero-Benito M.C., Borreguero L., and Frade J.M.. 2011. Control of neuronal ploidy during vertebrate development. Results Probl. Cell Differ. 53:547–563. 10.1007/978-3-642-19065-0_22 [DOI] [PubMed] [Google Scholar]
- LoTurco J.J., Sarkisian M.R., Cosker L., and Bai J.. 2003. Citron kinase is a regulator of mitosis and neurogenic cytokinesis in the neocortical ventricular zone. Cereb. Cortex. 13:588–591. 10.1093/cercor/13.6.588 [DOI] [PubMed] [Google Scholar]
- Madaule P., Eda M., Watanabe N., Fujisawa K., Matsuoka T., Bito H., Ishizaki T., and Narumiya S.. 1998. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature. 394:491–494. 10.1038/28873 [DOI] [PubMed] [Google Scholar]
- Manukyan A., Ludwig K., Sanchez-Manchinelly S., Parsons S.J., and Stukenberg P.T.. 2015. A complex of p190RhoGAP-A and anillin modulates RhoA-GTP and the cytokinetic furrow in human cells. J. Cell Sci. 128:50–60. 10.1242/jcs.151647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie C., Bassi Z.I., Debski J., Gottardo M., Callaini G., Dadlez M., and D’Avino P.P.. 2016. Cross-regulation between Aurora B and Citron kinase controls midbody architecture in cytokinesis. Open Biol. 6:6 10.1098/rsob.160019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzwa B., and Gerlich D.W.. 2014. Cytokinetic abscission: Molecular mechanisms and temporal control. Dev. Cell. 31:525–538. 10.1016/j.devcel.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Morita E., Sandrin V., Chung H.Y., Morham S.G., Gygi S.P., Rodesch C.K., and Sundquist W.I.. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26:4215–4227. 10.1038/sj.emboj.7601850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri A.R., and Gage F.H.. 2006. Generation of neuronal variability and complexity. Nature. 441:1087–1093. 10.1038/nature04959 [DOI] [PubMed] [Google Scholar]
- Naim V., Imarisio S., Di Cunto F., Gatti M., and Bonaccorsi S.. 2004. Drosophila citron kinase is required for the final steps of cytokinesis. Mol. Biol. Cell. 15:5053–5063. 10.1091/mbc.E04-06-0536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada M., Niska J.A., Miyamori H., McDonough W.S., Wu J., Sato H., and Berens M.E.. 2004. The phosphorylation of EphB2 receptor regulates migration and invasion of human glioma cells. Cancer Res. 64:3179–3185. 10.1158/0008-5472.CAN-03-3667 [DOI] [PubMed] [Google Scholar]
- Ng M.M., Chang F., and Burgess D.R.. 2005. Movement of membrane domains and requirement of membrane signaling molecules for cytokinesis. Dev. Cell. 9:781–790. 10.1016/j.devcel.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Repetto D., Camera P., Melani R., Morello N., Russo I., Calcagno E., Tomasoni R., Bianchi F., Berto G., Giustetto M., et al. 2014. p140Cap regulates memory and synaptic plasticity through Src-mediated and citron-N-mediated actin reorganization. J. Neurosci. 34:1542–1553. 10.1523/JNEUROSCI.2341-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagona A.P., and Stenmark H.. 2010. Cytokinesis and cancer. FEBS Lett. 584:2652–2661. 10.1016/j.febslet.2010.03.044 [DOI] [PubMed] [Google Scholar]
- Sarkisian M.R., Li W., Di Cunto F., D’Mello S.R., and LoTurco J.J.. 2002. Citron-kinase, a protein essential to cytokinesis in neuronal progenitors, is deleted in the flathead mutant rat. J. Neurosci. 22:RC217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin A.T., Durgan J., Cameron A.J., Faisal A., Marber M.S., and Parker P.J.. 2008. The regulated assembly of a PKCε complex controls the completion of cytokinesis. Nat. Cell Biol. 10:891–901. 10.1038/ncb1749 [DOI] [PubMed] [Google Scholar]
- Serres M.P., Kossatz U., Chi Y., Roberts J.M., Malek N.P., and Besson A.. 2012. p27(Kip1) controls cytokinesis via the regulation of citron kinase activation. J. Clin. Invest. 122:844–858. 10.1172/JCI60376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgro F., Bianchi F.T., Falcone M., Pallavicini G., Gai M., Chiotto A.M., Berto G.E., Turco E., Chang Y.J., Huttner W.B., and Di Cunto F.. 2016. Tissue-specific control of midbody microtubule stability by citron kinase through modulation of TUBB3 phosphorylation. Cell Death Differ. 23:801–813. 10.1038/cdd.2015.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafikhani S.H., Mostov K., and Engel J.. 2008. Focal adhesion components are essential for mammalian cell cytokinesis. Cell Cycle. 7:2868–2876. 10.4161/cc.7.18.6674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., and Mann M.. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850–858. 10.1021/ac950914h [DOI] [PubMed] [Google Scholar]
- Soeda S., Nakayama Y., Honda T., Aoki A., Tamura N., Abe K., Fukumoto Y., and Yamaguchi N.. 2013. v-Src causes delocalization of Mklp1, Aurora B, and INCENP from the spindle midzone during cytokinesis failure. Exp. Cell Res. 319:1382–1397. 10.1016/j.yexcr.2013.02.023 [DOI] [PubMed] [Google Scholar]
- Swaney D.L., McAlister G.C., and Coon J.J.. 2008. Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat. Methods. 5:959–964. 10.1038/nmeth.1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taus T., Köcher T., Pichler P., Paschke C., Schmidt A., Henrich C., and Mechtler K.. 2011. Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10:5354–5362. 10.1021/pr200611n [DOI] [PubMed] [Google Scholar]
- Taverna E., Götz M., and Huttner W.B.. 2014. The cell biology of neurogenesis: Toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell Dev. Biol. 30:465–502. 10.1146/annurev-cellbio-101011-155801 [DOI] [PubMed] [Google Scholar]
- Tong J., Li L., Ballermann B., and Wang Z.. 2016. Phosphorylation and activation of RhoA by ERK in response to epidermal growth factor stimulation. PLoS One. 11:e0147103 10.1371/journal.pone.0147103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F., Kellendonk C., Kretz O., Gass P., Anlag K., Orban P.C., Bock R., Klein R., and Schütz G.. 1999. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 23:99–103. 10.1038/12703 [DOI] [PubMed] [Google Scholar]
- Watanabe S., De Zan T., Ishizaki T., and Narumiya S.. 2013. Citron kinase mediates transition from constriction to abscission through its coiled-coil domain. J. Cell Sci. 126:1773–1784. 10.1242/jcs.116608 [DOI] [PubMed] [Google Scholar]
- Wiśniewski J.R., Nagaraj N., Zougman A., Gnad F., and Mann M.. 2010. Brain phosphoproteome obtained by a FASP-based method reveals plasma membrane protein topology. J. Proteome Res. 9:3280–3289. 10.1021/pr1002214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.