How CRISPR Cas9–guide RNA complexes navigate the nucleus and interrogate the genome is not well understood. Ma et al. track these complexes in live cells and find that mutations in the guide seed region significantly reduced the complex’s target residence time, with a commensurate impairment of cleavage.
Abstract
The bacterial CRISPR-Cas9 system has been repurposed for genome engineering, transcription modulation, and chromosome imaging in eukaryotic cells. However, the nuclear dynamics of clustered regularly interspaced short palindromic repeats (CRISPR)–associated protein 9 (Cas9) guide RNAs and target interrogation are not well defined in living cells. Here, we deployed a dual-color CRISPR system to directly measure the stability of both Cas9 and guide RNA. We found that Cas9 is essential for guide RNA stability and that the nuclear Cas9–guide RNA complex levels limit the targeting efficiency. Fluorescence recovery after photobleaching measurements revealed that single mismatches in the guide RNA seed sequence reduce the target residence time from >3 h to as low as <2 min in a nucleotide identity- and position-dependent manner. We further show that the duration of target residence correlates with cleavage activity. These results reveal that CRISPR discriminates between genuine versus mismatched targets for genome editing via radical alterations in residence time.
Introduction
The clustered regularly interspaced short palindromic repeats (CRISPR) bacterial acquired immunity system, as redesigned for application in eukaryotic cells, has become a powerful tool for genome editing (Jinek et al., 2012; Cong et al., 2013; Mali et al., 2013), gene expression modulation (Qi et al., 2013), and genomic DNA visualization (Chen et al., 2013; Ma et al., 2015, 2016). The CRISPR-associated protein 9 (Cas9)–guide RNA complex initially locates a protospacer adjacent motif (PAM) sequence and a vicinal 10–12 nt seed sequence for complementarity, both of which are essential for DNA targeting and cleavage (Jinek et al., 2014; Sternberg et al., 2014; Jiang et al., 2015; Richardson et al., 2016). Biochemical studies suggest that structural reorganization of the Cas9–guide RNA complex and R-loop formation at the target are essential for DNA cleavage (Szczelkun et al., 2014; Rutkauskas et al., 2015; Sternberg et al., 2015; Jiang et al., 2016). Single molecule tracking in living cells revealed that the Cas9–guide RNA complex searches DNA by 3D diffusion with a dwell time of <1 s at off-target sites and established a lower limit of ∼5 min for on-target residence time (Knight et al., 2015).
However, the true on-target dwell time of Cas9–guide RNA and its influence on CRISPR activity in living cells remain unknown. Here, we have investigated the intranuclear assembly and target-binding dynamics of the Cas9–guide RNA complex in live cells and have found that target residence time highly correlates with cleavage activity.
Results
Visualization of Cas9, guide RNA, and DNA target in living cells
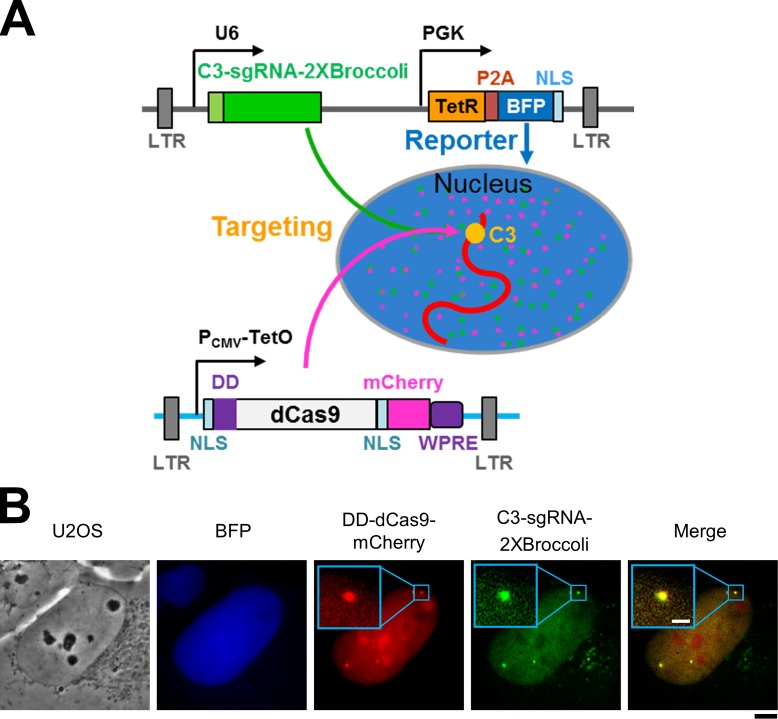
In previous studies, either nuclease-dead Cas9 (dCas9) from Streptococcus pyogenes was tagged with fluorescent proteins for live cell tracking (Chen et al., 2013; Ma et al., 2015), or RNA aptamers were inserted into the guide RNA to recruit fluorescent protein-fused binding partners for visualization of DNA targets (Ma et al., 2016). In this study, we directly labeled both dCas9 and the guide RNA to track their assembly and target-site interrogation. The system we designed is diagrammed in Fig. 1 A, and a detailed description of its molecular components and strategic operation is given in the figure legend. The guide RNA carries the aptamer “Broccoli” (Filonov et al., 2014) so that one can convert it to a fluorescent RNA by adding a specific small molecule to the medium, which, after cellular uptake, forms a conjugate with the guide RNA’s aptamer that elicits the small molecule’s fluorescence (see also Fig. S1). The guide-Broccoli RNA is constitutively expressed via the U6 promoter, and its expression is reported by the coexpression of blue fluorescent protein (BFP) downstream via a constitutive phosphoglycerol kinase promoter. In parallel, the system employs a ligand-tunable destabilization domain (DD; Banaszynski et al., 2006) in addition to the Tet-On system to tightly control the expression of dCas9-mCherry. As the genomic target for interrogation by Cas9–guide RNA, we chose a repeated sequence uniquely situated in the subtelomeric region of the long arm of chromosome 3 (C3; Ma et al., 2016), which facilitated visualization of the complex at the target with a favorable ratio of foci signal/nuclear background. As shown in Fig. 1 B, DD-dCas9-mCherry and the C3-targeting guide RNA–2XBroccoli were colocalized on distinct foci (C3).
Figure 1.
Multicolor labeling system for CRISPR nuclear dynamics. (A) Diagram of the CRISPR dynamics tracking system. dCas9 was fused to a DD at the N terminus and mCherry at the C terminus as well as two nuclear localization signals (NLSs) and a posttranscriptional regulatory element of woodchuck hepatitis virus. Lentiviral long terminal repeats were used for integration, and expression was under the control of the cytomegalovirus (CMV) promoter with the 2× TetO2 tetracycline operator. Expression of DD-dCas9-mCherry was induced by the addition of doxycycline (Dox), and any of the protein resulting from leaky repression was subjected to degradation by ubiquitinylation-proteasome system because of the presence of the DD. The C3–guide RNA was tagged by a pair of the Broccoli RNA aptamer and was constitutively produced from the U6 promoter but became fluorescent only upon addition of the cell-permeant small molecule DFHBI-1T. This same plasmid contained the tetracycline repressor (TetR) and BFP fused with an NLS via a 2A self-cleaving peptide (P2A) linker constitutively expressed via the PGK promoter. Stable cells were generated for constitutively expressed C3–guide RNA and controllable DD-dCas9-mCherry to study how their level impacts the DNA target (C3) recognition. BFP is used for a reporter for C3–guide RNA expression. The chromosomal target is a repeated sequence uniquely situated at a subtelomeric site on the long arm of C3 (Fig. 3 A and detailed in Materials and methods). (B) Simultaneous visualization of dCas9, guide RNA, and the C3 target in live cells. Localization of BFP reporter (blue), DD-dCas9-mCherry (red), and C3–guide RNA–2XBroccoli (green) stably expressed in a U2OS-derived cell line. Bars: (black) 5 µm; (white) 1 µm.
Cas9 is indispensable for guide RNA stability in living cells
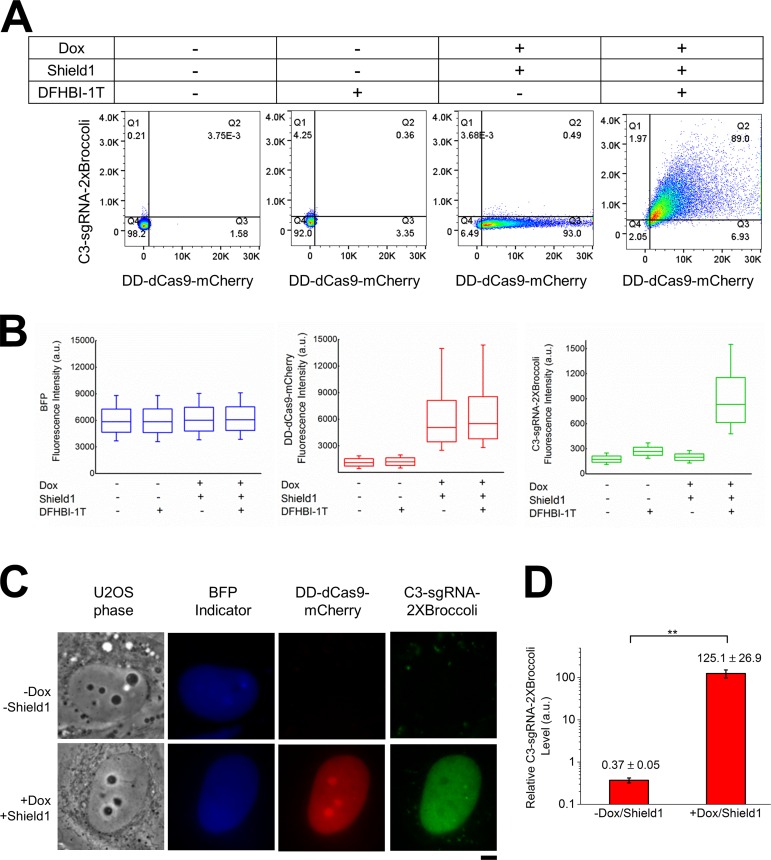
To directly measure the Cas9 and guide RNA levels in living cells, we used FACS (Fig. 2 A). The results showed that the DD-dCas9-mCherry level was tightly controlled by the addition of doxycycline (Dox) and Shield1, whereas C3–guide RNA–2XBroccoli was not fluorescent until the cells were exposed to DFHBI-1T. Intriguingly, in the absence of DD-dCas9-mCherry, the level of fluorescent guide RNA was too low to be detected by either FACS or microscopy (Fig. 2, A and C) under similar BFP expression (Fig. 2, B and C), indicating that the guide RNA is extremely unstable in the absence of dCas9, which was confirmed by RT-PCR analysis (Fig. 2 D).
Figure 2.
Cas9 is required to stabilize guide RNA in living cells. (A) FACS analysis of guide RNA–2XBroccoli in presence or absence of dCas9 and DFHBI-1T. The cellular level of BFP reporter, C3–guide RNA–2XBroccoli, and induced DD-dCas9-mCherry (48 h of induction) were measured in the presence of 5 µM DFHBI-1T for FACS analysis. Data in all panels are representative of experiments performed at least three times. (B) Box-and-whisker plots show the expression levels of BFP (blue), DD-dCas9-mCherry (red), and C3–guide RNA–2XBroccoli (green) in the indicated conditions. Box spans from first to last quartiles. Whiskers represent 10/90 percentiles, and middle lines represent the position of the mean value of the data distribution. (C) Images of C3–guide RNA–2X Broccoli were captured in presence and absence of dCas9. The U2OS-derived cell line stably expressing BFP indicator (blue) and C3–guide RNA–2XBroccoli (green) was imaged before or after induction of DD-dCas9-mCherry (red) by Dox with Shield1, and 10 µM DFHBI-1T was added before imaging. Bar, 5 µm. Data in all panels are representative of experiments performed at least three times. (D) RT-PCR analysis. RNA was extracted from the stable cells line in which dCas9 had or had not been induced for the previous 48 h and subjected to RT-PCR as detailed in the Materials and methods. Data are presented as means ± SD (n = 4), and statistical significance is calculated by Student’s t test: **, P < 0.005.
To investigate the stability of the dCas9–guide RNA complex, cell lines expressing C3–guide RNA–2XBroccoli and DD-dCas9-mCherry were treated with actinomycin D at a concentration that inhibits transcription by all three RNA polymerases. Approximately 50% of the guide RNA disappeared within 15 min, and the remainder declined over the subsequent 4 hours, whereas both BFP and Cas9 levels remained stable, confirming their expression and that of the guide RNA (Fig. 3, A and B). In additional experiments, we examined the effect of guide RNA–2XBroccoli with shorter guide sequences (i.e., 11, 6, or 0 nt in length) on the stability of the dCas9–guide RNA complex. As shown in Fig. 3 C (middle), stability decreased with the shorter guide RNA lengths.
Figure 3.
Stability of the Cas9–guide RNA complex in live cells. (A) The U2OS-derived cell line stably expressing BFP indicator, C3–guide RNA–2XBroccoli and with DD-dCas9-mCherry having been induced for the previous 48 h, was incubated with actinomycin D (10 µg/ml) for the indicated times, and cells were harvested at each time point. To measure the level of the guide RNA-2XBroccoli, 5 µM DFHBI-1T was added before FACS analysis. (B) Plots of the fluorescence levels of BFP reporter, DD-dCas9-mCherry, or C3–guide RNA–2XBroccoli in the left, middle, and right, respectively. Box spans from first to last quartiles. Whiskers represent 10/90 percentiles, and middle lines represent the mean value of the data distribution. Data in panels are representative of experiments performed at least three times. (C) Stably transfected cells with guide RNA–2XBroccoli of 11,6, or 0 nt in length of guide RNAs were used to measure the stability of the dCas9–guide RNA complex, as reflected by the guide RNA–2XBroccoli levels.
Nuclear guide RNA concentration is a limiting factor for efficient DNA targeting
To understand how the stability of the guide RNA affects DNA interrogation, we compared the guide RNA containing a full-length tracrRNA (trans-activating CRISPR RNA) component (+85 nt) to guide RNAs with truncated tracrRNA components (+54 or +67 nt), which had been shown to be less stable (Hsu et al., 2013; Mekler et al., 2016). Reduced nuclear guide RNA–Broccoli signal and reduced foci signals were evident with guide RNAs with truncated tracrRNA components compared with the full-length one (Fig. 4 A). The expression levels of BFP and dCas9-mCherry were comparable in each condition, indicating that Cas9–guide RNA complexes with truncated tracrRNAs are less stable and result in inefficient DNA targeting. The level of nuclear C3–guide RNA–Broccoli, presumably reflecting the assembled Cas9–guide RNA complex, positively correlated with target binding as shown by foci brightness (Fig. 4 B and Fig S2). Hence, DNA interrogation by Cas9–guide RNA is limited by the guide RNA stability and assembly with Cas9 and is driven by the concentration of the assembled Cas9–guide RNA complex. Interestingly, with the two shorter guide RNAs, we observed a nucleolar localization of Cas9 (Fig. 4 A), raising the possibility that some dCas9 remains unassembled with these shorter guide RNAs and enters nucleoli where there is a high concentration of hundreds of small noncoding RNA species (Jorjani et al., 2016).
Figure 4.
The effect of nuclear guide RNA level on target DNA interrogation. (A) U2OS cells were cotransfected with dCas9-mCherry and plasmids encoding C3–guide RNA–Broccoli-54 (54-nt truncated tracrRNA component), C3–guide RNA–Broccoli-67 (67-nt truncated tracrRNA component), or C3–guide RNA–Broccoli-85 (85 nt full tracrRNA component), each plasmid containing the BFP reporter; all cells were exposed to 5 µM DFHBI-1T to visualize Broccoli. Data in all panels are representative of experiments performed at least three times. (B) The effect of nuclear C3–guide RNA–2XBroccoli expression level on the foci brightness. Bars: (black) 5 µm; (white) 1 µm. Data in all panels are representative of experiments performed at least three times.
Target residence time determines guide RNA targeting efficiency
To understand how CRISPR complex interrogates true or mismatched target, we then used FRAP of GFP-dCas9 to measure the on-target residence time (Fig. 5, A and B). The residence time and the off-rate of the dCas9/C3-11–guide RNA complex on the C3 target were estimated to be 206 ± 4.5 min and 2.9 ± 0.1 × 10−4 s−1 (Fig. 5 C and Fig. S3). Complexes with mismatched guide RNAs at the seed sequence all displayed shorter residence times from one third to one hundredth on the target in an identity- and position-dependent manner (Fig. 5 C and Fig. S3). These results, obtained in live cells, show that even a single mismatch between the guide RNA and the target DNA significantly reduces the dwell time of the dCas9–guide RNA complex on the target. A mismatched guide RNA that had shorter target residence was also reflected in reduced overall foci brightness (Fig. 5, D and E). Purine-to-purine mismatches at position -5 away from the PAM previously were shown to result in loss of cleavage activities in all 46 guide RNAs examined (Doench et al., 2016). Here, we show that -5 (C) to G or A mutations, resulting in G to G or G to A mismatches, resulted in reduction of residence times to 7.4 ± 3.6 min or 1.4 ± 0.6 min, respectively (Fig. 5C). These results reveal that mismatches on guide RNA seed sequence reduce target residence time in an identity- and position-dependent manner, but not with respect to their proximity to the PAM site, at least as compared with the mismatches at the -5 or -3 positions.
Figure 5.
Mismatched guide RNA reduce targeting efficiency by lessening residence time. (A) Schematic of C3-specific repeat locus. FRAP analysis was used to estimate the target residence time of guide RNAs carrying single mismatches (red and underlined) at the -3 or -5 positions of the seed region. The PAM sequence in the target is shown in orange. (B) FRAP images of Cas9/C3-11–guide RNA on the target. The bleached spot is indicated by the arrow. Bar, 5 µm. (C) Recovery times of dCas9-3XGFP at the target as a function of each C3-11 guide RNA indicated. Inset table: the recovery times (τ) and dissociation rates (koff) were estimated by fitting the FRAP curves as detailed in Materials and methods. *The values represent the mean with standard errors in the plot from three independent experiments. (D) Targeting efficiency with mismatches on seed sequences. C3-11 guide RNAs carrying a single mutation at position -3 or -5 were expressed in U2OS cells along with dCas9-3XGFP, and the foci brightness was examined under similar nuclear fluorescent backgrounds. Bar, 5 µm. Data in all panels are representative of experiments performed at least three times. (E) The relative foci brightness of guide RNA mutants (n = 28 for 11wt, 23 for 11m3.1, 20 for 11m3.2, 20 for 11m3.3, 23 for 11m5.1, 30 for 11m5.2, and 22 for 11m5.3). Box spans from first to last quartiles. Whiskers represent 10/90 percentiles, and middle lines represent the mean position.
The correlation between shortened residence time and lower target-binding efficiency was also observed when the guide RNA length truncated from 11 to 8 nt, and the residence time decreased from 206.0 ± 4.6 min to 25.3 ± 7.3 min (Fig. S4). This finding aligns with recent studies that guide RNAs 12 and 20 nt in length were comparable in transcription activation, whereas 1 of 8 nt had almost negligible activation (Kiani et al., 2015) because the residence time of transcription factors on the promoters has been shown to be correlated with transcription activation (Lickwar et al., 2012).
CRISPR cleavage activity is correlated with residence time
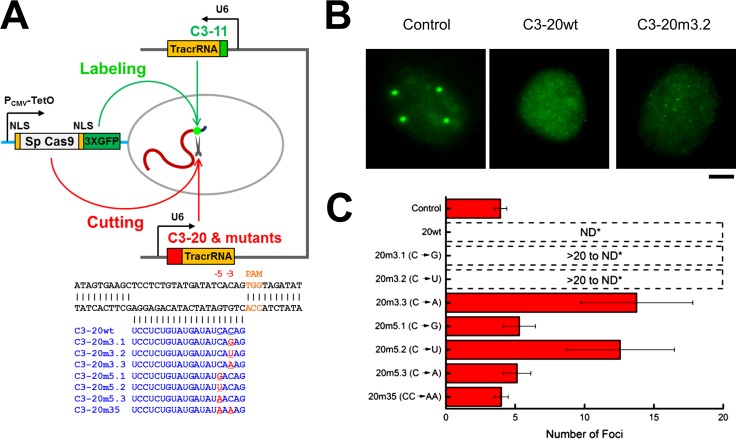
We then asked how the residence time on the target correlates with CRISPR activity. To directly visualize cleavage activity, target foci were labeled using pairs of guide RNAs, and loss of foci brightness caused by cleavage was monitored (Fig. 6 A). Truncated (11 nt; C3-11 for labeling only; Ma et al., 2016) and full-length (20 nt; C3-20 as cleavage competent) guide RNAs were simultaneously used together with nuclease-active Sp Cas9-3XGFP. Because the target is a repeated sequence, cleavage will be manifest by signal diminution and dispersal. Mutations in the C3-20 guide RNAs are predicted to result in variable degrees of cleavage. Intermediate degrees of cleavage will result in an increase in the number of foci with lower intensity, whereas higher levels of cleavage will result in loss of detectable foci altogether. When only C3-11 guide RNA (labeling competent, cleavage noncompetent) was used, four distinct target foci were detected, whereas no distinct foci were observed when the C3-20 guide RNA was coexpressed with C3-11 (Fig. 6, B and C). Mismatched guide RNAs that displayed shorter target residence times (Fig. 5 C) displayed less cleavage activity (Fig. 6, B and C; Fig. S5). The degree to which cleavage activity was impaired by each of the mismatched guide RNAs aligns with previously reported studies (Hsu et al., 2013). These results suggest that radical alterations of residence time occur when there are mismatches in the seed sequence, implying that in living cells, the Cas9–guide RNA complex acutely detects a true target DNA before executing the cleavage step.
Figure 6.
CRISPR cleavage activity decreases with residence time. (A) Schematic of strategy for direct visualization of cleavage activities. C3-11 guide RNA was used for labeling, and C3-20 or variants thereof with single or double mutations (red and underlined) at position -3 or -5 adjacent to the PAM site were cloned into the same plasmid (as denoted by the black bracket between them in the diagram) Concurrently, nuclease-active Cas9 fused to 3XGFP was used for simultaneously labeling and cleaving the C3 target. (B) U2OS cells were cotransfected with guide RNAs and Cas9-3XGFP and then fixed after 24 h and imaged by collecting z-stack images to capture all foci in each nucleus examined. Foci (control, noncleaved target), no foci (C3-20wt, highly cleaved target), or dispersed punctae (C3-20m3.2, cleaved target) are shown in representative images. Bar, 5 µm. (C) The mean number of either foci or dispersed punctae observed with the control (wild-type) guide RNA or mutants are plotted; ND denotes not detectable as fluorescence above the nuclear background. Each histogram bar represents the mean foci numbers (n = 16 for control, 14 for 20wt, 14 for 20m3.1, 15 for 20m3.2, 15 for 20m3.3, 12 for 20m5.1, 11 for 20m5.2, 10 for 20m5.3, and 11 for 20m35).
Discussion
In the type II CRISPR-Cas system, Cas9 and guide RNAs assemble into a ribonucleoprotein complex and act as a single-turnover enzyme to cleave target DNA highly efficiently in vitro (Jinek et al., 2012; Sternberg et al., 2015). However, the cell’s microenvironment of Cas9, guide RNA, and the target might alter target recognition efficiency (Farasat and Salis, 2016; Fu et al., 2016). Here, we examined Cas9 and guide RNA’s intracellular location, level, and stability in living cells and their effects on RNP assembly for target recognition. We found that guide RNAs are extremely unstable in the absence of Cas9 or when the guide RNA has a truncated tracrRNA component. In the presence of Cas9, the guide RNA displayed two nuclear populations with half-lives of ∼15 min and ∼2 h, respectively (Fig. 3). We speculate that the former represents Cas9–guide RNA complexes roaming the nucleus, and the latter represents ones that are stabilized by DNA binding, either to targets or nontargets. The fact that the guide RNA is so unstable in the absence of Cas9 emphasizes the importance of expressing it at stoichiometric levels with respect to Cas9 in gene-editing studies. Conversely, the nucleolar localization of Cas9–guide RNA complexes under some conditions, as we have observed (Fig. 4 A) and also reported previously (Chen et al., 2013), suggests that attention must be paid in gene-editing endeavors to the possible sequestration of CRISPR machinery in this RNA-rich nuclear body.
It has not been well understood how the Cas9–guide RNA complex reaches targets within structurally organized chromatin environments. The genome contains a vast number of off-target binding sites for any given guide RNA (Kuscu et al., 2014; Wu et al., 2014), and these would be expected to attenuate free intranuclear diffusion of the Cas9–guide RNA complex, thus slowing the kinetics of the search process for the bona fide target (Farasat and Salis, 2016). These model studies suggested that the on-target cleavage activity would be increased and off-target binding would be substantially reduced when the genome size increases from the ∼50,000 base pairs of λ phage to ∼3 billion base pairs in the human genome. We find that increased nuclear Cas9 and guide RNA concentrations significantly enhance DNA targeting. Moreover, our finding that guide RNAs are extremely unstable in the absence of Cas9 means that achieving high and equal levels of both nuclear Cas9 and guide RNA will be essential for CRISPR efficiency, whether for editing, gene regulation, or chromosomal locus labeling.
In the type I CRISPR-Cas system, in vitro kinetics studies have shown that target recognition occurs through directional R-loop zipping from the PAM and that intermediate R-loops stall at mutations and collapse in a PAM proximity–dependent manner, resulting in shorter residence times at targets with mismatches closer to the PAM (Rutkauskas et al., 2015). Here, our in vivo measurements, with a type II CRISPR-Cas9 system, revealed that on-target residence time radically changes with single mismatches in the guide RNA seed sequence in a base identity- and position-dependent manner, but independent of PAM proximity. Specifically, the transversion mutation (C to A) reduced the residence time to a greater extent than the transition mutation (C to U), which agrees with the reported higher residual CRISPR activity of seed transition mutations than transversions (Hsu et al., 2013; Tsai et al., 2015). Moreover, we found that the distance of a mismatch from the PAM is not directly correlated with residence time and cleavage activity. We found that C to G or A mismatches -5 away from the PAM were more detrimental than ones at position -3.
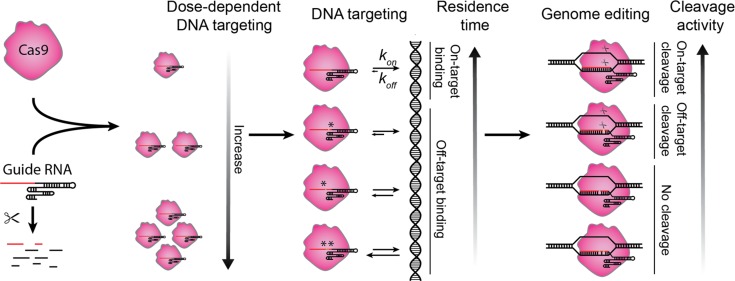
In summary, our measurements of target residence times revealed the critical negative impact of certain mismatches in the seed sequence and their inverse correlation with cleavage activity (Fig. 7). Measurements of target residence time for mismatches at each position can thus be used in the rational design of optimal guide RNAs. It will be intriguing to examine whether new Cas orthologues (Hou et al., 2013; Ran et al., 2015; Zetsche et al., 2015) or S. pyogenes Cas9 variants with increased specificity (Kleinstiver et al., 2016; Slaymaker et al., 2016) sensitize target discrimination by altering residence times.
Figure 7.
A working model of CRISPR nuclear dynamics. Schematic of CRISPR nuclear dynamics and target interrogation in living cells. Guide RNA is rapidly degraded if not assembled with Cas9, and the Cas9–guide RNA complex interrogates DNA in a concentration-dependent manner; guide RNA and target mismatches reduce residence time in an identity- and position-dependent manner, and the degree of changes determines the cleavage activity.
Materials and methods
Plasmid construction
The expression vector for dCas9-mCherry, 3XGFP from S. pyogenes was described previously, originally constructed from pHAGE-TO-DEST (Ma et al., 2015), and a DD was inserted into the N-terminal region. The guide RNA expression vector was based on the pLKO.1 lentiviral expression system (Addgene), in which TetR-P2A-BFP (Addgene) was inserted right after the phosphoglycerate kinase (PGK) promoter, with sequences coding for the desired guide RNA, guide RNA–1XBroccoli, or guide RNA–2XBroccoli inserted immediately after the U6 promoter. To place C3-20 and C3-11 guide RNAs in the same plasmid, DNA sequences for the U6 promoter-C3-11 guide RNA were amplified by PCR and cloned downstream of the U6–guide RNA cassette, and then C3-20 or the other guide RNA mutants were cloned into the same vector. The rapid guide RNA expression plasmid construction protocol was described previously (Ma et al., 2015). Details on the Cas9s and guide RNAs used in this study are given in Table S1.
Cell culture and transfection
Human osteosarcoma U2OS cells (ATCC) were cultured on 35-mm glass-bottom dishes (MatTek Corporation) at 37°C in DMEM (Thermo Fisher Scientific) containing high glucose and supplemented with 10% (vol/vol) FBS. For transfection, typically 200 ng of dCas9-mCherry or dCas9-3xGFP plasmid DNA and 1 µg of total guide RNA plasmid DNA indicated were cotransfected using Lipofectamine 2000 (Thermo Fisher Scientific), and the cells were incubated for another 24–48 h before imaging. For actinomycin D (Thermo Fisher Scientific) treatment, cells were seeded in 6-well plates in medium containing 2 µg/ml doxycycline (Sigma-Aldrich) and 0.5 µM Shield1 (Takara Bio Inc.). After 2 d, actinomycin D (Sigma-Aldrich) was added at 10 µg/ml at the indicated times before FACS analysis. For Fig. 6 B and Fig. S5, cells were transfected for 24 h and fixed in 4% formaldehyde for 10 min before capturing images.
Lentivirus production and transduction
HEK293T cells were maintained in Iscove’s Modified Dulbecco’s Medium (Thermo Fisher Scientific) containing high glucose and supplemented with 1% GlutaMAX (Thermo Fisher Scientific), 10% FBS (Hycolne FBS; Thermo Fisher Scientific), and 1% penicillin/streptomycin (Thermo Fisher Scientific). 24 h before transfection, ∼5 × 105 cells were seeded in 6-well plates. For each well, 0.5 µg of pCMV-dR8.2 dvpr (Addgene) and 0.3 µg of pCMV-VSV-G (Addgene), each constructed to carry HIV long terminal repeats, and 1.5 µg of plasmid containing the gene of interest were cotransfected by using TransIT transfection reagent (Mirus) according to the manufacturer’s instructions. After 48 h, the virus was collected by filtration through a 0.45-µm polyvinylidene fluoride filter (Pall Laboratory). The virus was immediately used or stored at -80°C. For lentiviral transduction, U2OS cells maintained as described in Cell culture and transfection were transduced by spinfection in 6-well plates with lentiviral supernatant for 2 d and ∼2 × 105 cells were combined with 1 ml lentiviral supernatant and centrifuged for 30 min at 1,200 g.
Flow cytometry
Cells expressing the desired fluorescent Cas9 and/or guide RNA were selected by fluorescence-activated sorting (FACSAria cell sorter; BD) or analyzed on an LSR II cytometer (BD). All data were processed with FlowJo software (Tree Star). Both the FACSAria cell sorter and LSR II cytometer were equipped with 405-, 488-, and 561-nm excitation lasers, and the emission signals were detected by using filters at 450/50 nm (wavelength/bandwidth) for BFP, 530/30 nm for Broccoli or GFP, and 610/20 nm for mCherry. Single cells were sorted onto individual wells in 96-well plates containing 1% GlutaMAX, 20% FBS, and 1% penicillin/streptomycin in chilled DMEM medium. For the Broccoli signal analysis, 5 µM DFHBI-1T was added before FACS.
FRAP and fluorescence microscopy
A microscope (DMIRB; Leica Biosystems) was equipped with an EMCCD camera (iXon-897D; Andor Technology), mounted with a 2× magnification adapter and 100× oil objective lens (NA 1.4), resulting in a total 200× magnification equal to a pixel size of 80 nm in the image. The microscope stage incubation chamber was maintained at 37°C. A custom-built FRAP module was inserted between the fluorescent lamp and the microscope body, with a motorized translating mirror (Leica Biosystems) used to switch between imaging and FRAP mode. The FRAP module used a 488-nm laser (Obis; Coherent) set to 20 mW output power and created a diffraction limited spot of roughly 800 nm in diameter. The laser was fiber coupled (Pointsource; Qioptiq) and collimated out of an FC connector using a 30 mm focal length lens and projected into the sample with a 200 mm focal length lens and the microscope’s excitation tube lens and objective (Leica Biosystems). The FRAP module was controlled through a custom-built mechatronic assembly consisting of a fast linear actuator (Everest), DC motor driver (MCP Technology Systems), custom electrical circuitry, and 3D-printed adapters. The guide RNA used was 11 nt in length to achieve the necessary high signal/noise ratio for live-cell imaging as compared with one of 20 nt in length (Ma et al., 2016). Our use of a guide RNA with an 11-nt seed was also based on the facts that a guide RNA with a 10-nt seed sequence forms a stable Cas9–guide RNA–DNA target ternary complex in vitro (Richardson et al., 2016) and that a guide RNA with a 12-nt seed region determined the Cas9–guide RNA targeting specificity (Larson et al., 2013). The C3 target and its immediate periphery were bleached for 5 s, and the postbleach images were acquired up to 4 h as necessitated by the recovery times being observed in experiments with the various guide RNAs. GFP was excited with an excitation filter at 470/28 nm (Chroma Technology Corp.), and its emission was collected using an emission filter at 512/23 nm (Chroma Technology Corp.). Imaging data were acquired by MetaMorph acquisition software (Molecular Devices). Thresholds were set on the basis of the ratios between nuclear focal signals to background nucleoplasmic fluorescence. For fixed cells in Fig. 5, cells were imaged by using a custom-built, single-molecule, real-time microscope as previously described (Grünwald and Singer, 2010; Ma et al., 2016). In brief, imaging was performed on a custom-built, dual-channel setup housing a 150× 1.45 NA oil immersion objective (Olympus) combined with 200-mm focal length tube lenses (LAO-200, cvi; Melles Griot). This produces an effective magnification of 167× and a 95.8-nm pixel. The emission is split in the primary beam path onto three electron-multiplying charge-coupled devices (Andor Technology): iXon3-897E, iXon-897D, and iXonUltra-897U. Emission filters (Semrock) are 460/60 for BFP, 534/20 for mNeongreen, and 600/37 for mCherry. Excitation of fluorescent proteins was with a 405-nm and 515-nm diode laser (Obis, Coherent) and a solid-state 561-nm laser (SE; Cobolt), and the intensity and on/off switching were controlled by an acousto-optic tunable filter (AA Opto-Electronics). In Fig. 6 B and Fig. S5, the step size in z-stacks was 160 nm, and the exposure time was 150 ms. Each z-stack series was 31 frames, and each image was obtained by a maximum projection of a z-stack series using ImageJ.
Image processing
The time series FRAP images were registered and analyzed by Fiji and then plotted using Origin 9.0 software (OriginLab). The images were first corrected for cellular movements by using the StackReg plugin (Thévenaz et al., 1998). All fluorescence intensities were corrected by background subtraction. Double-bleach correction was performed by using the fluorescence level from the whole nucleus as the indication of photobleaching. The first post bleach image was set to time 0, and the prebleach image was set to intensity 1. We fitted the FRAP curves based on the Diffusion-Uncoupled model (McNally, 2008) with a minor modification: where t is time, A1 is a constant to represent the highest recovery level, Φapp is the apparent bleached fraction, and koff is the dissociation rate.
Chromosome-specific repeats
Mining of chromosome-specific repeats was described previously (Ma et al., 2015). The chromosome 3–specific locus is situated in the subtelomeric region q29, having the identifier Chr 3: 195199022–195233876 in the human reference genome hg19 at the University of California Santa Cruz genome browser (http://genome.ucsc.edu).
RT-PCR analysis
Cells were treated as described in Fig. 2 D, and RNA was extracted with an RNeasy Plus Mini Kit (QIAGEN) and then subjected to RT-PCR using the following primers and probe (Integrated DNA Technologies) for C3–guide RNA–2XBroccoli: Forward primer: 5′-TGGGCTCTAGCAAGTTCAAATAA-3′; complementary to nt 55–78 of the RNA; Probe: 5′-ACTTGAGACGGTCGGGGTCCAGATA-3′; complementary to nt 94–118 of the RNA; Reverse primer: 5′-CCCACACTCTACTCGACAAGATA-3′; complementary to 144–122 nt of the RNA.
Online supplemental material
Figs. S1–S5 include the sequence and structure of C3–guide RNA–2XBroccoli, target interrogation related to the intranuclear Cas9 and guide RNA levels, target residence times from FRAP analysis, effects on residence time of shorter guide RNAs, and images of live-cell cleavage assay. Table S1 lists all guide RNA sequences used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201604115/DC1.
Supplementary Material
Acknowledgments
We thank Rita Strack and Samie Jaffrey for the Broccoli aptamer. We thank Aviva Joseph for sharing her lentiviral transduction protocol, Bonginkhosi Vilakati for performing the RT-PCR analyses, and Ankit Gupta, Mehmet Fatih Bolukbasi, Yu-Chieh Chung, and Feng He for valuable discussions. We are also grateful to Erik Sontheimer and Scot Wolfe for critical input and comments on the manuscript.
This work was supported in part by the Vitold Arnett Professorship Fund, National Institutes of Health grant U01 DA-040588-01 and Amyotrophic Lateral Sclerosis Association grant 16-LGCA-300 (H. Ma and T. Pederson), and National Institutes of Health grants 5 U01 EB021238-02 to D. Grunwald and R01 GM102515 to S. Zhang.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- BFP
- blue fluorescent protein
- C3
- chromosome 3
- Cas9
- CRISPR-associated protein 9
- CRISPR
- clustered regularly interspaced short palindromic repeats
- dCas9
- nuclease-dead Cas9
- DD
- destabilization domain
- Dox
- doxycycline
- PAM
- protospacer adjacent motif
- tracrRNA
- trans-activating CRISPR RNA
References
- Banaszynski L.A., Chen L.C., Maynard-Smith L.A., Ooi A.G., and Wandless T.J.. 2006. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 126:995–1004. 10.1016/j.cell.2006.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Gilbert L.A., Cimini B.A., Schnitzbauer J., Zhang W., Li G.W., Park J., Blackburn E.H., Weissman J.S., Qi L.S., and Huang B.. 2013. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 155:1479–1491. 10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., and Zhang F.. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science. 339:819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R., et al. . 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34:184–191. 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farasat I., and Salis H.M.. 2016. A biophysical model of CRISPR/Cas9 activity for rational design of genome editing and gene regulation. PLOS Comput. Biol. 12:e1004724 10.1371/journal.pcbi.1004724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filonov G.S., Moon J.D., Svensen N., and Jaffrey S.R.. 2014. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc. 136:16299–16308. 10.1021/ja508478x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu B.X.H., St Onge R.P., Fire A.Z., and Smith J.D.. 2016. Distinct patterns of Cas9 mismatch tolerance in vitro and in vivo. Nucleic Acids Res. 44:5365–5377. 10.1093/nar/gkw417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünwald D., and Singer R.H.. 2010. In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature. 467:604–607. 10.1038/nature09438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z., Zhang Y., Propson N.E., Howden S.E., Chu L.F., Sontheimer E.J., and Thomson J.A.. 2013. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl. Acad. Sci. USA. 110:15644–15649. 10.1073/pnas.1313587110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., et al. . 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31:827–832. 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Zhou K., Ma L., Gressel S., and Doudna J.A.. 2015. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 348:1477–1481. 10.1126/science.aab1452 [DOI] [PubMed] [Google Scholar]
- Jiang F., Taylor D.W., Chen J.S., Kornfeld J.E., Zhou K., Thompson A.J., Nogales E., and Doudna J.A.. 2016. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 351:867–871. 10.1126/science.aad8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., and Charpentier E.. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337:816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Jiang F., Taylor D.W., Sternberg S.H., Kaya E., Ma E., Anders C., Hauer M., Zhou K., Lin S., et al. . 2014. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 343:1247997 10.1126/science.1247997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorjani H., Kehr S., Jedlinski D.J., Gumienny R., Hertel J., Stadler P.F., Zavolan M., and Gruber A.R.. 2016. An updated human snoRNAome. Nucleic Acids Res. 44:5068–5082. 10.1093/nar/gkw386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiani S., Chavez A., Tuttle M., Hall R.N., Chari R., Ter-Ovanesyan D., Qian J., Pruitt B.W., Beal J., Vora S., et al. . 2015. Cas9 gRNA engineering for genome editing, activation and repression. Nat. Methods. 12:1051–1054. 10.1038/nmeth.3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., and Joung J.K.. 2016. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 529:490–495. 10.1038/nature16526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S.C., Xie L., Deng W., Guglielmi B., Witkowsky L.B., Bosanac L., Zhang E.T., El Beheiry M., Masson J.B., Dahan M., et al. . 2015. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science. 350:823–826. 10.1126/science.aac6572 [DOI] [PubMed] [Google Scholar]
- Kuscu C., Arslan S., Singh R., Thorpe J., and Adli M.. 2014. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 32:677–683. 10.1038/nbt.2916 [DOI] [PubMed] [Google Scholar]
- Larson M.H., Gilbert L.A., Wang X., Lim W.A., Weissman J.S., and Qi L.S.. 2013. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 8:2180–2196. 10.1038/nprot.2013.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickwar C.R., Mueller F., Hanlon S.E., McNally J.G., and Lieb J.D.. 2012. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature. 484:251–255. 10.1038/nature10985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Naseri A., Reyes-Gutierrez P., Wolfe S.A., Zhang S., and Pederson T.. 2015. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc. Natl. Acad. Sci. USA. 112:3002–3007. 10.1073/pnas.1420024112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Tu L.C., Naseri A., Huisman M., Zhang S., Grunwald D., and Pederson T.. 2016. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat. Biotechnol. 34:528–530. 10.1038/nbt.3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., and Church G.M.. 2013. RNA-guided human genome engineering via Cas9. Science. 339:823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally J.G. 2008. Quantitative FRAP in analysis of molecular binding dynamics in vivo. Methods Cell Biol. 85:329–351. 10.1016/S0091-679X(08)85014-5 [DOI] [PubMed] [Google Scholar]
- Mekler V., Minakhin L., Semenova E., Kuznedelov K., and Severinov K.. 2016. Kinetics of the CRISPR-Cas9 effector complex assembly and the role of 3′-terminal segment of guide RNA. Nucleic Acids Res. 44:2837–2845. 10.1093/nar/gkw138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., and Lim W.A.. 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 152:1173–1183. 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S., et al. . 2015. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 520:186–191. 10.1038/nature14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., and Corn J.E.. 2016. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 34:339–344. 10.1038/nbt.3481 [DOI] [PubMed] [Google Scholar]
- Rutkauskas M., Sinkunas T., Songailiene I., Tikhomirova M.S., Siksnys V., and Seidel R.. 2015. Directional R-loop formation by the CRISPR-Cas surveillance complex cascade provides efficient off-target site rejection. Cell Reports. 10:1534–1543. 10.1016/j.celrep.2015.01.067 [DOI] [PubMed] [Google Scholar]
- Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., and Zhang F.. 2016. Rationally engineered Cas9 nucleases with improved specificity. Science. 351:84–88. 10.1126/science.aad5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg S.H., Redding S., Jinek M., Greene E.C., and Doudna J.A.. 2014. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 507:62–67. 10.1038/nature13011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg S.H., LaFrance B., Kaplan M., and Doudna J.A.. 2015. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 527:110–113. 10.1038/nature15544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczelkun M.D., Tikhomirova M.S., Sinkunas T., Gasiunas G., Karvelis T., Pschera P., Siksnys V., and Seidel R.. 2014. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc. Natl. Acad. Sci. USA. 111:9798–9803. 10.1073/pnas.1402597111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenaz P., Ruttimann U.E., and Unser M.. 1998. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7:27–41. 10.1109/83.650848 [DOI] [PubMed] [Google Scholar]
- Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P., et al. . 2015. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33:187–197. 10.1038/nbt.3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Scott D.A., Kriz A.J., Chiu A.C., Hsu P.D., Dadon D.B., Cheng A.W., Trevino A.E., Konermann S., Chen S., et al. . 2014. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 32:670–676. 10.1038/nbt.2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A., et al. . 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 163:759–771. 10.1016/j.cell.2015.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.