Abstract
The persistence of latent HIV proviruses in long-lived CD4+ T cells despite antiretroviral therapy (ART)1–3 is a major obstacle to viral eradication4–6. Because current candidate latency-reversing agents (LRAs) induce HIV transcription but fail to clear these cellular reservoirs,7–8 new approaches for killing these reactivated latent HIV reservoir cells are urgently needed. HIV latency depends upon transcriptional quiescence of the integrated provirus and circumvention of immune defense mechanisms4–6,9. These defenses include cell-intrinsic innate responses that use pattern-recognition receptors (PRR) to detect viral pathogens and subsequently induce apoptosis of the infected cell10. Retinoic acid-inducible gene I (RIG-I) forms one class of pattern-recognition receptors that mediates apoptosis and elimination of infected cells after recognition of viral RNA11–14. Here we show that acitretin, an FDA-approved retinoic-acid derivative, enhances RIG-I signaling ex vivo, increases HIV transcription, and induces preferential apoptosis of HIV-infected cells. These effects are abrogated by RIG-I knockdown. Acitretin also decreases proviral DNA levels in CD4+ T cells from HIV-infected subjects on suppressive ART, an effect amplified by combination with suberoylanilide hydroxamic acid (SAHA), a histone deacetylase inhibitor. Pharmacologic enhancement of an innate cellular defense network could provide a means to eliminate reactivated cells in the latent HIV reservoir.
Despite suppression of HIV replication by ART, HIV persists as latent proviruses in CD4+T cells, enabling viral rebound when ART is interrupted1–3. Latent proviruses produce little HIV RNA and protein expression and allow immune evasion. Ideal LRAs would reactivate latent proviruses without provoking broad T cell activation, enabling host cell killing by either viral cytopathic or immune responses. This is the basis for "shock-and-kill" therapeutic approach to eliminate the latent reservoir4–9. However, in clinical studies, SAHA used alone did not deplete reservoir cells even though it induced viral expression7. Moreover, when therapy is delayed, the latent reservoir contains mutations in the virus allowing for immune escape from cytotoxic T-lymphocytes8. Consequently, alternative killing strategies are needed9.
Cell-intrinsic innate immune defenses against viral infection involve pathogen detection through pattern recognition receptors (PRRs). Retinoic-acid-inducible gene I (RIG-I) is a cytosolic PRR10–15 that can sense HIV-RNA16 and activate innate antiviral signaling leading to the death of infected cells17. However, HIV infection impedes RIG-I signaling by sequestration and degradation18,19,and RIG-I expression is lower in HIV patients20. RIG-I recognizes full-length HIV RNA more efficiently than short viral transcripts16 that predominate during latent infection2–6 thereby limiting RIG-I activity in patients on suppressive ART.
Retinoic acid (RA) induces RIG-I expression11 potentially arming innate immune defense21,22. RA also activates p300 acetyl transferase23–24, and could lead to stimulation of HIV transcription6. Acitretin is an FDA-approved RA derivative that has been used to treat HIV patients with psoriasis and is well-tolerated25,26. We hypothesized that acitretin could be used to increase HIV transcription and activate the RIG-I pathway to promote the preferential death of reactivated reservoir cells.
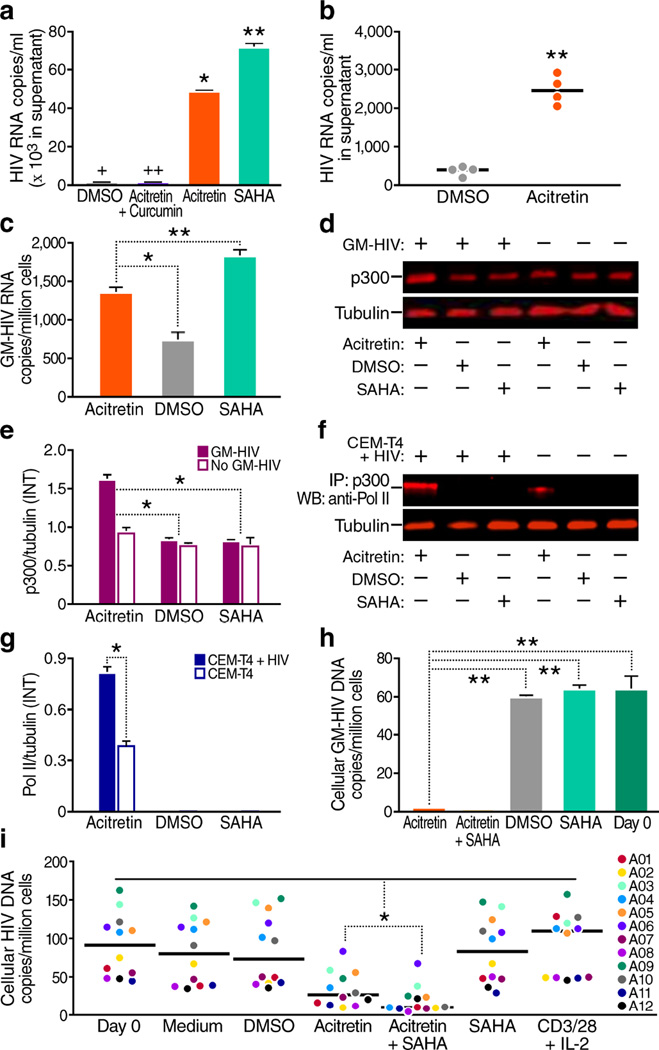
First, we assessed the effects of acitretin on viral expression from a latent HIV infected human T cell line (ACH-2)27. Treatment of ACH-2 with acitretin or SAHA for 72 hours increased HIV transcription (Fig. 1a). The effects of acitretin were partially blocked by curcumin, an inhibitor of p30028 (Fig. 1a). Acitretin also activated HIV RNA expression in latently infected CD4+T-cells from HIV+ subjects on suppressive ART (Fig. 1b).
Figure 1.
Acitretin induces HIV expression and reduces cellular HIV-DNA in CD4+ T cells. (a) HIV copy number in the supernatant of a latent HIV T-cell line (ACH-2) after 72 h of treatment. Both acitretin (P < 0.05) and SAHA significantly increased HIV transcription (*P < 0.01), and curcumin (an inhibitor of p300) limited acitretin induction of HIV. (+HIV ‘copy number, 486.6 ± 5.9’), (++HIV ‘copy number, 379.6 ± 17.8’). (b) HIV-RNA copy number in supernatants from cultures of CD4+ T cells from four aviremic HIV+ subjects on ART at day 6. Acitretin significantly increased HIV transcription (P < 0.01 versus. DMSO control). (c) Cellular GM-HIV-RNA copies/million cells after 24 h of the indicated treatment of infected primary CD4+ T cells. Both acitretin and SAHA increased GM-HIV transcription to a greater extent than DMSO. The increase was greater with SAHA than acitretin (P < 0.01). (d) Immunoblot analysis of p300 and tubulin proteins from both GM-HIV-infected and uninfected CD4+ T cells (from the same donor) after 48 h of treatment. (e) The ratio of mean value intensities (INT) for p300 and tubulin from panel (d, n = 4) confirming significantly higher expression of p300 in infected cells treated with acitretin than in cells treated with DMSO or SAHA. (f) Immunoblot analysis of co-immunoprecipitation of protein extracts of CEM-T4 cells with or without latent GFP-HIV virus using antibody against p300 and western blot for RNA pol II after 48 h of treatment. Association of p300 with RNA pol II is enhanced by acitretin. (g) The ratio of RNA pol II to tubulin from (f, n = 4) is greatest with acitretin treatment of cells with GFP-HIV. (h) GM-HIV-DNA content in cellular DNA after 72 h of treatment. GM-HIV-DNA was significantly lower after treatment with acitretin or acitretin plus SAHA than with SAHA or DMSO (P < 0.001). GM-HIV-DNA was not detectable despite testing of cellular DNA from two million cells after treatment with acitretin plus SAHA. (i) HIV-DNA concentrations at day 7 of treatment in CD4+ T cells from HIV+ subjects on ART (n = 12). Both acitretin and acitretin plus SAHA significantly lowered HIV-DNA concentrations in cells from all 12 HIV+ subjects (P < 0.05 compared to treatment with DMSO, SAHA, medium, or anti-CD3 and anti-CD28 antibodies beads plus IL-2 (CD3/28+IL-2). HIV-DNA concentrations were significantly lower after treatment with acitretin plus SAHA than after treatment with acitretin alone (P < 0.05). Values represent mean ± s.e.m. of duplicate samples from HIV+ subjects (b,i), and triplicate samples from the ACH-2 (a) and GM-HIV infection model(c, h) from three independent experiments. A student's t-Test was used to compare experimental conditions (a, b, c, e, g, h, i); *P<0.05; **P<0.01.
Induction of HIV transcription alone may not clear HIV reservoirs4–9. Since acitretin induces RIG-I, and RIG-I signaling can eliminate virally infected cells10–15, we tested the ability of acitretin to promote clearance of HIV-infected cells in a primary T-cell model29. We used a single round viral construct GM-HIV containing a mutated gag gene (Supplementary Fig. 1) to infect unstimulated CD4+T-cells from healthy donors by spinoculation29,30 then treated cells with acitretin, SAHA, or DMSO. One day after treatment, both acitretin and SAHA induced HIV-RNA expression (Fig. 1c). Next, we examined whether the induction of HIV-RNA by acitretin was accompanied by p300 induction. Indeed, 48 hours after acitretin treatment, p300 expression was increased in infected with GM-HIV more than in uninfected cells (Fig. 1d,e) and enhancement of p300-association with RNA Pol II (Fig. 1f,g) was greater in HIV-infected CEM-T4 cells (a human lymphoblastoid T-cell line)14, than in uninfected cells. Furthermore, after 72 hours of treatment, acitretin significantly reduced cellular GM-HIV-DNA levels measured by real time PCR (Fig. 1h).
We next tested whether acitretin reduces HIV-DNA levels in samples from HIV+ subjects on ART. Treatment of CD4+T-cells from twelve ART-suppressed HIV+ subjects (Supplementary Table 1) with acitretin or acitretin plus SAHA for 7 days reduced HIV-DNA levels significantly more than treatment with DMSO, SAHA, or anti-CD3/anti-CD28 beads (Fig. 1i). The reduction was greatest when acitretin was combined with SAHA. This reduction in HIV-DNA concentration by acitretin was not due to expansion of uninfected cells (Supplementary Fig. 2). Thus, acitretin facilitates the reduction of HIV-DNA levels in CD4+T-cells from HIV+ subjects in vitro, an effect not seen with any candidate LRAs studied to date.
In other models, RIG-I limits viral infection by inducing apoptosis of infected cells13–15. To determine whether acitretin induces apoptosis in HIV-infected cells, we measured annexin-V staining and caspase-3 activity in GM-HIV-infected and uninfected CD4+T-cells after 72 hours of treatment. Acitretin significantly increased the percentage of annexin-V+ cells (Fig. 2a) and caspase-3 activity in the infected cells (Fig. 2b), consistent with reports that activation of RIG-I induces caspase-3-mediated apoptosis13.
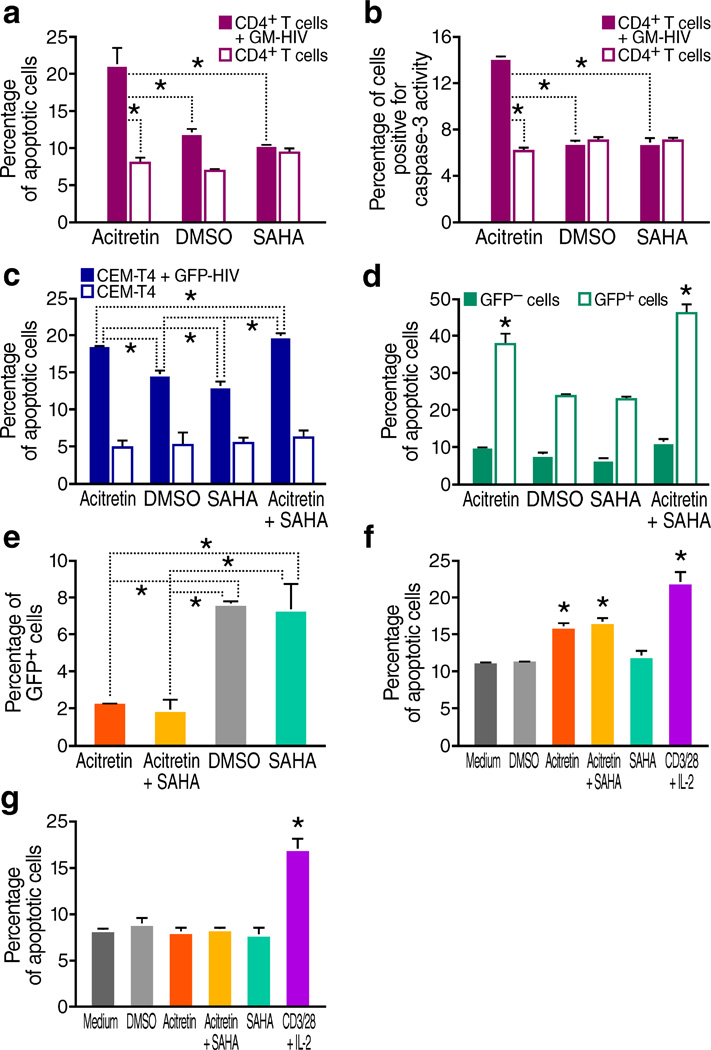
Figure 2.
Acitretin preferentially induces apoptosis in HIV-infected cells. (a) Percentage of apoptotic cells determined by annexin V staining of GM-HIV-infected or mock-infected CD4+Tcells after 72 h of treatment. The percentage of apoptotic cells was significantly greater in the presence of acitretin than with SAHA or DMSO in GM-HIV-infected but not uninfected CD4+T cells (P < 0.05). (b) Percentage of cells expressing active caspase-3 determined by flow cytometry assessed 72 h of treatment with DMSO, SAHA or acitretin. Caspase-3 activity was preferentially increased in GM-HIV infected CD4+T cells treated with acitretin (P<0.05). (c) Percentage of apoptotic cells determined by annexin V staining of GFP-HIV-infected and uninfected CEM-T4 cells. Acitretin and acitretin plus SAHA increased apoptosis at 48 h in GFP-HIV-infected cells to a significantly greater extent than SAHA or DMSO alone (P <0.05) although both of these agents produced higher than expected levels of cell death in infected cells compared to uninfected cells. No differences were detected in uninfected CEM-T4 cells. (d) Percentage of apoptotic cells determined by annexin V staining of infected cells from (c) gated for the presence or absence of expression of GFP encoded by the reporter virus. Acitretin and acitretin plus SAHA increased apoptosis of GFP-positive cells to a significantly greater degree than SAHA or DMSO, while no significant increases were induced by these agents in GFP negative cells (P < 0.05). (e) Percentage of GFP-positive cells after 7 d of treatment, as determined by flow cytometry of infected cells from (c). Both acitretin and acitretin plus SAHA reduced the number of GFP-HIV-positive cells significantly more (P < 0.05) than treatment with SAHA or DMSO. (f) Average percentage of apoptotic cells determined by annexin V staining of CD4+ T cells from HIV+ subjects on ART (n = 12). After 7 d of treatment with acitretin and acitretin plus SAHA, the frequency of apoptotic cells was significantly increased in patient CD4+ T cells compared to DMSO, SAHA, or medium (P < 0.05). Apoptosis was also significantly increased following treatment with anti-CD3 and anti-CD28 antibodies beads plus IL-2 (CD3/28+IL-2) (P < 0.05). (g) Mean percentage of apoptotic cells determined by annexin V staining in cultures of CD4+ T cells from four healthy control subjects. Neither acitretin, nor acitretin plus SAHA induced higher levels of apoptosis than DMSO, SAHA, or medium at day 7 in these normal cells while increased apoptosis was observed when these normal cells were treated with anti-CD3 and anti-CD28 antibodies beads plus IL-2(CD3/28+IL-2) (P < 0.05). Values represent mean ± s.e.m. of duplicate samples from experiments with cells from 12 HIV+ subjects (f), values represent mean ± s.e.m. of triplicate samples for (a,b,c,d,e) from three independent experiments, and values represent mean ± s.e.m.of triplicate samples for (g) from four healthy donor CD4 T cells experiments. A student's t-Test was used to compare experimental conditions (a–g); **P < 0.01, *P < 0.05.
To more directly assess whether acitretin induces apoptosis preferentially in HIV-infected cells and to test for potential synergistic activity with SAHA31, cells were infected with GFP-HIV29, and infected and uninfected cells were treated with acitretin, SAHA, or both. At 48 hours, acitretin and acitretin plus SAHA resulted in significantly higher levels of apoptosis in the GFP-positive cells than in the total cells or the GFP-negative cells (Fig. 2c,d), indicating preferential induction of apoptosis in GFP-HIV infected cells. Furthermore, GFP-positive cells became significantly less frequent after treatment with acitretin or acitretin plus SAHA compared to SAHA or DMSO (Fig. 2e). These results are consistent with preferential induction of apoptosis in acitretin or acitretin + SAHA treated HIV-expressing cells, leading to their elimination.
Next, we tested whether acitretin induces apoptosis in CD4+T-cells from HIV+ subjects on suppressive ART. Acitretin and acitretin plus SAHA each stimulated significantly higher levels of apoptosis than treatment with DMSO, SAHA, or medium. No difference was seen in uninfected cells from healthy controls. Meanwhile treatment with anti-CD3/anti-CD28+IL2 led to high levels of apoptosis in cells from both HIV+ and healthy control participants (Fig. 2f,g). In contrast to anti-CD3/anti-CD28, acitretin did not upregulate the activation markers CD69 and HLA-DR on CD4+T-cells (Supplementary Fig. 2h). Kinetic studies showed that the induction of RNA expression occurred later with acitretin than with SAHA, and the acitretin-induced apoptosis was greater for any given HIV RNA level and time point compared to SAHA or DMSO (Supplementary Fig. 3). These results support the conclusion that acitretin preferentially reduces HIV containing cells by inducing apoptosis.
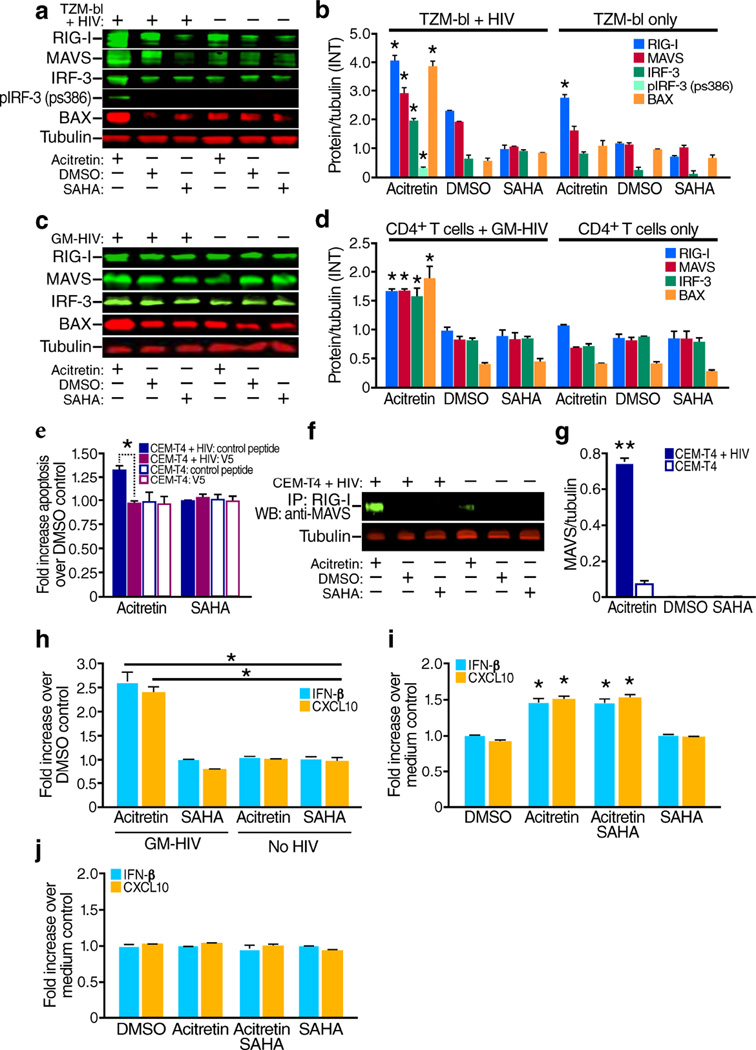
We examined whether depletion of cells harboring HIV-DNA and induction of apoptosis by acitretin was dependent on RIG-I signaling. Binding of viral RNA to RIG-I leads to a conformational change in RIG-I32 that enables binding to the mitochondrial antiviral signaling protein (MAVS)33 and phosphorylation of interferon-regulatory factor 3 (IRF3) at serine 38634–37 culminating in type I interferon (IFN) induction and caspase-3 activation10–15.To assess acetritin effects on RIG-I expression and signaling, we immunoblotted extracts from HIV infected and uninfected TZM-bl cells following 48 hours of treatment of acitretin, SAHA or DMSO with antibodies against RIG-I pathway proteins. Acitretin induces RIG-I in both infected and uninfected cells (Fig. 3a,b). However only in HIV-infected cells did acitretin significantly enhance MAVS, IRF3 and phosphoserine386-IRF3 levels. Similar patterns were seen in latent GM-HIV infected CD4+T-cells (Fig. 3c,d). Acitretin treatment significantly increases MAVS binding to RIG-I33 in latently HIV infected CEM-T4 cells compared to uninfected cells (Fig. 3f,g). These observations suggest the initial induction of RIG-I and viral RNA by acitretin and subsequent RIG-I sensing of viral RNA12–15,32 promotes RIG-I association with MAVS38. RIG-I-induced apoptosis in virus-infected cells depends on the interaction of IRF-3 with BCL-2-associated X protein (BAX), triggering caspase-mediated apoptosis12–13. In the presence of acitretin, BAX expression was significantly increased, and pretreatment with a BAX inhibitor(V5) peptide39, but not a control peptide, diminished acitretin-induced apoptosis(Fig. 3a–e). These findings support a role for BAX-signaling in acitretin-induced apoptosis12–13.
Figure 3.
Acitretin increases expression of RIG-I signaling pathway proteins including MAVS, IRF3, p-IRF3, and BAX, and increases production of IFN-β, and CXCL10 in cells infected with HIV. (a) Representative immunoblot analysis of RIG-I, MAVS, IRF3, p-IRF3 (antibody detecting phosphorylation at Ser 386), BAX, and tubulin at 48 h following treatment of HIV infected or uninfected TZM-bl cells with acitretin, DMSO or SAHA. (b) The ratio of mean value intensities (INT) for each protein and tubulin for the immunoblot bands in (a) combined with results from three additional independent experiments (n = 4). (c) Representative immunoblot analysis of RIG-I, MAVS, IRF3, BAX. and tubulin at 48 h after acitretin treatment of GM- HIV-infected and uninfected CD4+ T cells with acitretin, DMSO or SAHA. (d) The ratio of mean value intensities (INT) of each protein and tubulin for the immunoblot bands in panel (c) combined with results from three additional independent experiments (n = 4). RIG-I, MAVS, IRF3,p-IRF3 and BAX significant increased treated with acitretin compare to SAHA or MDSO control (P<0.05). RIG-I was also increased by acitretin in uninfected TZM-bl cells and reached significant (P<0.05)(b), but in uninfected CD4+ T cells from the same donors this increase did not reach statistical significance when compared to the DMSO control(d). (e) Fold increases in annexin V positive, apoptotic cells in uninfected or HIV-infected CEM-T4 cells treated with acitretin or SAHA (versus DMSO) in the presence of the V5 peptide (a BAX inhibitor) or a control peptide. Note significant decreases in apoptosis production by the V5 peptide (a BAX inhibitor) in acitretin treated infected cells (P<0.05) but not in the other conditions. (f, g) Immunoblot analysis of co-immunoprecipitation of protein extracts of CEM-T4 cells with or without latent GFP-HIV virus using antibody against RIG-I followed by immunoblotting for MAVS after 48 h of treatment with acitretin, DMSO, or SAHA. Note increased association of RIG-I with MAVS in infected cells treated with acitretin. (g) The association of MAVS with RIG-I following acitretin treatment in (f, n = 4) was higher in GFP-HIV infected cells than in uninfected cells. (h) Fold increase over DMSO control in type 1 interferon (IFN-β) and CXCL10 expression in supernatants from GM-HIV-infected, and uninfected CD4+ T cells 72 h after treatment. Acitretin increased IFN-β and CXCL10 levels to a greater extent than SAHA (P < 0.05) in infected but not uninfected cells. (i) Fold increase over medium control of IRF3-induced type 1 interferon (IFN-β) and CXCL10 expression in supernatants of cultures of CD4+ T cells from HIV+ subjects on ART (n = 12) on day 7 of treatment. Both acitretin and acitretin plus SAHA significantly induced IFN-β and CXCL10 (P < 0.05 vs. SAHA and DMSO). (j) No differences in IFN-β and CXCL10 expression occurred in healthy control CD4+ T cells treated with DMSO, acitretin, SAHA or acitretin plus SAHA. All values represent mean ± s.e.m. of duplicate samples from HIV+ subjects(i), and values represent mean ± s.e.m. of triplicate samples for (e,h) from three independent experiments, and values represent mean ± s.e.m.of triplicate samples for (j) from four healthy donor CD4 T cells experiments. A student's t-Test was used to compare experimental conditions (b, d, e, g, h, i, j); **P < 0.01, *P < 0.05.
Next, we measured type I IFN and CXCL10 chemokine production,18,33–37 following 72 hours of acitretin treatment in GM-HIV-infected and uninfected primary CD4+T-cells. Acitretin, but not SAHA significantly increased IFN-β and CXCL10 secretion in infected, but not in uninfected cells (Fig. 3h). Similarly, IFN-β and CXCL10 levels in day 7 supernatants of T-cell cultures from ART-suppressed HIV+ subjects were higher after treatment with acitretin or acitretin plus SAHA compared to DMSO or SAHA alone (Fig. 3i). In contrast, no increased IFN-β or CXCL10 production was detected in acitretin or acitretin plus SAHA-treated cells from healthy volunteers (Fig. 3j). Acitretin produced dose-related changes in both apoptosis and IFN-β or CXCL10 production in infected cells (Supplementary Fig. 4). Together, these results indicate that acitretin stimulates preferential RIG-I-mediated signaling in HIV-infected cells leading not only to apoptosis but also to type 1 IFN and chemokine production. The release of interferon and subsequent action of interferon stimulated genes might promote the death of some uninfected bystander as well as infected cells explaining why the total number of dying cells is greater than the number of latently infected cells present (Fig. 2a–c,f).
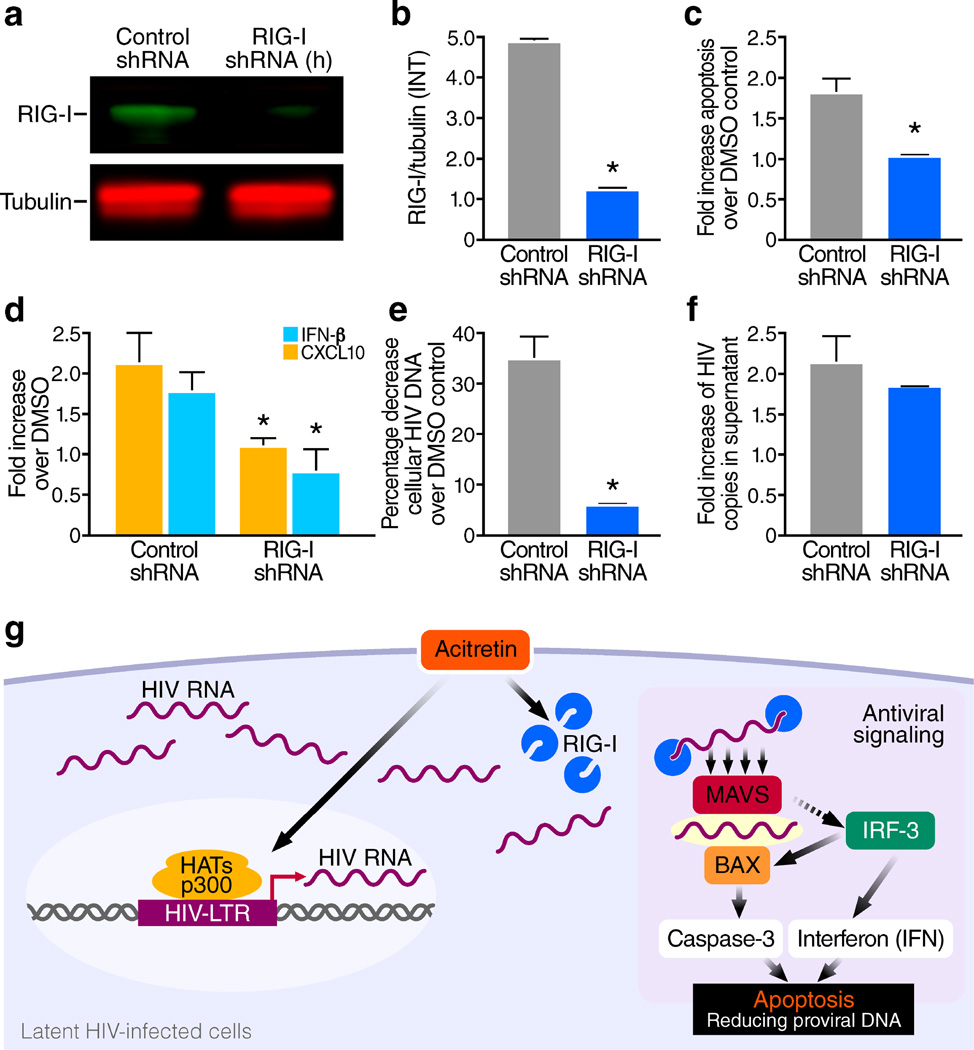
Finally, to establish that the effects of acitretin treatment are mediated through RIG-I, shRNA was used to inhibit expression of RIG-I in CEM-T4 cells. RIG-I knockdown abrogated the induction of apoptosis, production of CXCL10 and IFN-β and reduction in HIV-DNA by acitretin treatment, but had no impact on the release of HIV virions in this latent infection model29 (Fig. 4a–f). These findings demonstrate the essential role of RIG-I in the observed acitretin effects. Further studies are needed to define the nature of HIV-transcripts that bind and activate RIG-I10.
Figure 4.
Short hairpin RNA (shRNA) knockdown of RIG-I expression markedly inhibits acitretin enhancement of apoptosis, induction of IFN-β and CXCL10 production and preferential depletion of HIV-DNA+ cells. CEM T4 cells were transfected with either RIG-I-shRNA plasmid DNA, or control-shRNA (scrambled shRNA) plasmid DNA, cultured for 72 h and then selected in the presence of puromycin (10µg/ml) for 7 d. Next, these cells were infected with HIV-NL43 and maintained for 7 d with antiviral drugs as previously described for the in vitro latency model. (a) Immunoblot analysis of RIG-I and tubulin after the puromycin selection, (b) the ratio of mean value intensities (INT) of RIG-I to tubulin from (a, n = 4), RIG-I shRNA transfection significantly reduced RIG-I expression (P<0.05).(c) Knockdown of RIG-I significantly diminished (P<0.05) acitretin induced apoptosis at day 7 post treatment compared to the scrambled shRNA control, and (d) significantly reduced (P<0.05) acitretin induced CXCL10 and IFN-β release and (e) significantly impaired acitretin-induced decreases in HIV-DNA levels (P<0.05). (f) Knockdown of RIG-I expression conversely had no significant impact on acitretin-stimulated release of HIV-RNA into supernatant at day 7, the fold increase of HIV copies in supernatant are from acitretin compared to DMSO control in both conditions. All values represent mean ± s.e.m. of triplicate samples (b,c,d,e,f) from three independent experiments. A student's t-Test was used to compare experimental conditions (b–f); **P < 0.01, *P < 0.05.
(g) Model depicting the preferential enhancement of RIG-I signaling by acitretin in latently infected HIV cells. Acitretin increases both RIG-I and HIV-RNA expression in the cytoplasm of latent HIV-infected cells; recognition of HIV-RNA by RIG-I initiates antiviral signaling. RIG-I–RIG-I interactions produce conformational changes that permit binding to MAVS, which leads in turn promotes IRF-3 activation. Activated IRF-3 activates two pathways: one inducing type I interferon (IFN) production and another resulting in the binding of BAX and activation of caspase-3-dependent apoptosis in the virally infected cell. These two pathways work together to protect the host against further spread of the pathogen. LTR, long terminal repeat.
In summary, our data support a model (Fig. 4g) whereby acitretin, at clinically achievable concentrations40 increases HIV transcription, induces RIG-I expression, and augments RIG-I signaling culminating in the preferential apoptotic death of the reactivated reservoir cells. Addition of LRAs like SAHA further enhances these effects of acitretin. Thus, one possible approach to induce the death of reservoir cells could be to use combinations of LRAs and acitretin to preferentially “shock and kill” CD4+ T-cells harboring HIV. This approach would likely eliminate cells producing both infectious and non-infectious forms of HIV.
Online Methods
Cell lines
CEM-T4 cells (HIV-negative, human CD4+, lymphoblastic cell line), TZM-bl cells (HIV-negative, HeLa cell line with luciferase, beta-galactosidase reporters under control of the HIV promoter), and ACH2 cells (latent HIV T cell line derived from human T cells) were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH (courtesy of Dr. J.P. Jacobs, Dr. John C. Kappes, Dr. Xiaoyun, Wu, and Dr. Thomas Folks, respectively).
Media and reagents
All T cells and T cell lines were cultured in RPMI 1600 supplemented with penicillin (50 U/ml), streptomycin (50 µg/ml), L-glutamine (2 mM) (all from Life Technologies, Grand Island, NY), and 10% heat-inactivated fetal calf serum (FCS) (from Sigma, St. Louis, MO). TZM-bl cells were cultured in DMEM supplemented with the same additives as above. During cell culture post treatment or activation, 1 µM indinavir (IDV), 10 µM nevirapine (NVP), and 600 nM raltegravir (RAL) (NIH AIDS Reagent Program) were included to prevent spreading HIV infection. Working stocks of acitretin and SAHA were dissolved in DMSO (all from Sigma, St. Louis, MO). anti-CD3 and anti-CD28 antibodies beads (CD3/28) (Life Technologies, Grand Island, NY) were used at 1 bead per cell with human IL2 (Chiron, Emeryville, CA) at 10U/ml.
Cell culture and drug treatment
Cell treatment for both primary T-cells and cell lines was performed by resuspending cells at 0.5 million cells per ml in complete RPMI with antiretroviral drugs as indicated above. TZM-bl cells were used by plating approximately 2×105 cells/ well of a 24 well tissue culture plate the day prior to infection. This results in approximately 75% cell confluence at time of infection and further testing. In general, test drugs were used at the following final concentrations: SAHA (350nM); acitretin (5 µM); equivalent DMSO (final DMSO concentration did not exceed 0.05%) input for negative control. Dose-response studies used acitretin varying from 1 to 25 µM using the DMSO (Final DMSO concentration did not exceed 0.25%) equivalent to the 25 µM of acitretin comparator. Cells were maintained in the same drug concentration throughout the duration of experiments ranging from 3 to 7days. Most assays employed acitretin at a fixed concentration of 5 µM (1650ng/ml). RAs are very lipophilic, so while serum concentrations peak at 500 to 700ng/ml, tissue concentrations measured in patients on established therapy reach levels of 16,000 ng/ml or approximately 50 µM40. Thus the concentrations used in the current studies are clinically achievable.
Generation of GM-HIV for single-round HIV infection
We excised the gag region from pNL4-3 (NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, courtesy of Dr. Malcolm Martin) by restriction digest with BssHII (711) and SpeI (1507) subcloning this region into the pcDNA3.1 TOPO TA vector (Life Technologies, Grand Island, NY). Using the Quikchange II XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla CA), we mutated a region of gag from amino acids 1404 to 1432 (Supplementary Fig. 1 a). The sequence-verified mutated gag was re-cloned into pNL4-3 to make pGM-HIV. To generate GM-HIV capable of only a single round of infection, we co-transfected 293T cells with the pGM-HIV clone and a plasmid expressing wild-type gag (Supplementary Fig. 1 a), only pGM-HIV plus gag expressing vector can produce p24 into supernatant (Supplementary Fig. 1 b). To confirm infectivity of GM-HIV, we first infected 0.5×106 TZM-bl cells with 1 ng of p24 supernatant from pGM-HIV plus gag expression vector, the same volume of supernatant from pGM-HIV plus empty vector, and 1 ng of p24 of HIV-1(NL-4-3) as a positive control. After three days, HIV infectivity was measured by Bright-GloTM Luciferase assay system (Promega) and expressed as relative light units (RLU). Only supernatant from pGM-HIV plus gag expressing vector, and HIV-1(NL4-3) were able to infect TZM-bl cells (Supplementary Fig. 1 c). Next we infected PHA stimulated 1×10^6 CD4+T cells with 5ng of p24 in supernatant from pGM-HIV plus gag expressing vector, or an equal volume of supernatant from pGM-HIV plus empty vector by spinoculation at 2000 g for 2 hours29. The cells were washed with RPMI three times immediately after infection, and once the next day to remove all residual inoculum. Subsequently, the cells were cultured in RPMI with IL-2 (10u/ml), then GM-HIV DNA and RNA concentrations were measured in cellular RNA and DNA extracts by real-time PCR at day1 and day7. GM-HIV was only measurable in cells infected with supernatant from pGM-HIV plus gag expression vector (Supplementary Fig. 1 d).
HIV+ participants on ART
All HIV-positive participants were on combination ART and had undetectable plasma viral loads (<50 copies mL−1) for at least 1 year (median 5 years) (Supplementary Table 1). They were recruited from SF VAMC Infectious Diseases Clinic to meet the predetermined number of study participants. The study was approved by the UCSF Committee on Human Research and the SF VAMC Human Subject Research subcommittee and all research participants gave written informed consent.
Isolation and culture of CD4 T cells from HIV-infected participants and healthy donors
Peripheral blood mononuclear cells were purified from whole blood (HIV+, on ART) or PBMC eluted from leukoreduction filters (healthy donors) by density centrifugation. CD4 T lymphocytes were enriched by negative selection with the EasySep Human CD4 T-cell enrichment kit (Stemcell, Vancouver, BC). The purity of CD4 T cells was assessed by flow cytometry and was typically >95%. Cells were rested overnight before additional use.
In vitro infection of primary CD4+ Tcells with GM-HIV for a primary T-cell model of latent HIV infection29
Healthy donor CD4 T cells enriched by negative selection were cultured overnight, then treated according to the method of Lassen29by first infecting the unstimulated, resting CD4 T cells with GM-HIV (5 ng p24/1×106 cells) by spinoculation at 2000 g for 2 hours. Uninfected controls underwent mock spinoculation. The cells were washed with RPMI three times immediately after infection, and once the next day to remove all residual inoculum. Following spinoculation of unstimulated CD4+Tcells, subsequently, the cells were cultured in RPMI for 10 days, integrated, latent proviral infection is established29. To confirm that the cells carried latent proviruses, we stimulated the cells with either PHA plus IL-2 or anti-CD3/28-beads plus IL-2 for 48 hours then measured cellular GM-HIV RNA. Both PHA plus IL-2 (PHA+IL-2) and anti-CD3 and anti-CD28 antibodies beads plus IL-2(CD3/28+IL-2) treatment resulted in significantly increased cellular GM-HIV RNA compared to medium control (Supplementary Figure. 1 e).
Generation of latent GFP-HIV-infected CEM-T4 cells
A GFP reporter HIV construct (GFP-HIV)29 was transfected into 293T cells to produce GFP-HIV virions. CEM-T4 cells (1 × 106) were infected with 1000 pg of p24 for 5 hours, and unbound virus was removed by washing the cells three times with RPMI. Subsequently, the cells were cultured in medium with Saquinavir (5 µM) for 10 days to prevent spreading infection and allow for attrition of productively infected cells29. To study whether the cells have latent proviruses, we stimulated the cells with 5µM acitretin, 350nM SAHA, or 500nM prostratin for 72 hours, then harvested supernatant to measure the p24 production. Acitretin, SAHA, and prostratin all significantly increased p24 production compared to medium control, demonstrating that the cells contained proviruses that respond to LRA stimulation (Supplementary Fig. 3 a).
Assays for Apoptosis
The following assays were performed in duplicate with the EasyCyte6HT-2L flow cytometer (Millipore, Billerica, MA) according to the manufacturer’s instructions: (1) Guava ViaCount assay (Millipore, Billerica, MA) for absolute total cell counts and percentages of apoptotic cells. (2) Alexa Fluor 488 annexin V/Dead Cell Apoptosis Kit (Life Technologies, Grand Island, NY) to detect apoptosis for all experiments except those using GFP-HIV virus (50,000 cells per sample at each time point) (Fig. 1 and Fig. 3). (3) CellEvent Caspase-3/7 Green Flow Cytometry Assay Kit (Life Technologies, Grand Island, NY) to detect caspase-3 and caspase-7 activity (50,000 cells per sample) (Fig. 1). (4) Alexa Fluor 647 Annexin V (Life Technologies, Grand Island, NY) to detect apoptosis in GFP-HIV-infected CEM-T4 cells (100,000 cells per sample) (Fig. 2), since GFP would interfere with Alexa Fluor 488 annexin V apoptosis and caspase 3/7-green flow cytometry assays.
Assay with BAX inhibitor-V5
Both GFP-HIV-infected and un-infected CEM-T4 cells prepared as described above under “Generation of latent GFP-HIV-infected CEM-T4 cells” were incubated with 200µM of either BAX-inhibitor-V5 or BAX-Inhibiting Peptide Negative Control (Millipore, Billerica, MA) for three hours, then treated with the following drugs (final concentrations): SAHA (350nM); Acitretin (5 µM); equivalent DMSO input. After treatment for 24 hours39, the cells were stained with Alexa Fluor 647 Annexin V (Life Technologies, Grand Island, NY) to detect apoptosis (100,000 cells per sample) using the EasyCyte6HT-2L flow cytometer (Millipore, Billerica, MA) according to the manufacturer’s instructions.
ELISA assays for cytokine
Supernatants from GM-HIV-infected and uninfected CD4 T-cell cultures were collected 72 hours after drug treatment and from HIV-positive or healthy donor CD4 T cells 7 days after drug treatment. The supernatants were analyzed with the following ELISAs: (1) Verikine Human IFN-β kit (BPL Assay Science, Piscataway, NJ), and (2) Human CXCL10/IP-10 kit (R&D Systems, Minneapolis, MN). Assays were done according to the manufacturers’ protocols.
Immunoblot analysis (Western blots)
Cellular protein was extracted with the Mammalian Cell Lysis Kit (Sigma, St. Louis, MO), separated on a 4–12% Bis-Tris gel (Life Technologies, Grand Island, NY), and transferred to an Immobilon-FL-PVDF membrane (Millipore, Billerica, MA). The membrane was blocked with Odyssey Blocking buffer and incubated overnight with 1:200 dilution of primary antibodies against RIG-I, p300, MAVS, IRF3, BAX, Pol-II, tubulin (all from Santa Cruz Biotechnology, Santa Cruz, CA, USA), or p-IFR3 (phospho-Ser 386) (GeneTex, Irvine, CA). The membranes were then incubated with 1:15000 dilution of secondary antibodies: IRDye 800CW (green) for RIG-I, MAVS, and IRF3, p-IRF3(ps386) and IRDye680 (red) for BAX, p300, Pol-II,and tubulin (all from LI-COR, Lincoln, NE). Finally, the western blot membranes were analyzed with the Odyssey CLx Infrared Imaging System (LI-COR, Lincoln, NE). Protein expression is represented as mean band intensity (INT) normalized to results for tubulin.
Co-immunoprecipitation assays (IP)with RIG-I or P300
30×106 of each cell sample were washed twice with cold PBS, and lysed using the Mammalian Cell Lysis kit (Sigma, MO). The lysates were centrifuged at 4°C for 10 minutes at 12,000× g to pellet cellular debris, and the clarified lysate was transferred to a fresh pre-chilled tube. 20µl of lysates from each sample was saved for tubulin western blot to determine the amount of protein in the lysates. The remaining lysates were precleared by incubation with protein A/Sepharose for 1 hour at 4°C, subjected to immunoprecipitation with 2µg of anti-RIG-I polyclonal antibody (C-15), or 2µg of anti-p300 polyclonal antibody (C-20) overnight at 4°C, then treated with 50µl of protein G/Sepharose for 2 hours. The immunoprecipitates were washed 10 times with 1ml of cold wash buffer (1% NP-40, 1% triton-100, 50mM Tris-Cl(pH7.5), 1mM EDTA, 1mM EGTA, 0.27M Sucrose, 10mM Na4P2O7, 50mM NaF,1mM NaVO4, 0.1% 1-mercaptoethanol,1mM PMSF, 0.25M NaCl), then eluted from the protein G/Sepharose with 50µl of elution Buffer (1M Glycine 0.25M NaCl, pH2.8). The eluate was neutralized with 2µl of 1M Tris-Cl (pH9.5), then subjected to10% SDS-PAGE gel electrophoresis, and transferred to nitrocellulose membranes. Membranes were treated with blocking buffer (Odyssey blocking buffer) for one hour then incubated overnight in blocking buffer with a either 1:200 dilution of anti-MAVS antibody (T-20) for RIG-I-immunoprecipitation or 1:200 dilution of anti RNA polymerase II (Pol II) antibody(F-12)) for p300-immunoprecipitation at 4°C. Detection was performed using 1:15000 dilution of secondary antibodies: IRDye 800CW (green) for MAVS, and IRDye680 (red) for Pol II for two hours. The bands were analyzed using Odyssey Infrared Imaging System (LI-COR, Nebraska).
Preparation and evaluation of cells with attenuated RIG-I expression
CEM-T4 cells were transfected with either RIG-I-short hairpin RNA (shRNA) (h) plasmid (sc-61480-SH), or control-shRNA plasmid (sc-108083) (both from Santa Cruz Biotechnology, Santa Cruz, CA) using FuGENE HD transfection reagent (Roche, Indianapolis, IN) according to the manufacturers’ protocols. 72 hours after transfection, puromycin at 10µg/ml (Santa Cruz Biotechnology, Santa Cruz, CA) was added to select for transfected cells for 7 days. Immunoblot analysis of protein prepared from both shRNA and control-plasmid transfected cells was performed to confirm RIG-I expression knockdown using anti-RIG-I (C-15). RIG-I-knockdown and control cells were infected with 1000 pg of p24 of HIV NL4-3 for 5 hours, and unbound virus was removed by washing the cells three times with RPMI. Subsequently, the cells were cultured in medium with Saquinavir (5 µM) for 7 days to prevent spreading infection and to permit a reversion to latency29.
RIG-I-shRNA and control plasmid-transfected cells were then each treated with acitretin (5 µM) or the equivalent DMSO input (negative control) and maintained with antiretroviral drug cocktail of 1 µM indinavir (IDV), 10 µM nevirapine (NVP), and 600 nM raltegravir (RAL) for 7 days.
Measurement of intracellular HIV-1 RNA, DNA and HIV-1 RNA in supernatant
Total cellular RNA was extracted with the Rneasy Mini Kit (Qiagen, Valencia, CA). Genomic DNA was extracted with the DNA blood mini kit (Qiagen, Valencia, CA). RNA and DNA concentrations were measured with an ND-1000 Spectrophotometer (NanoDrop, Wilmington, DE). Supernatant HIV-1 RNA was extracted with the QIAamp Viral RNA mini kit (Qiagen, Valencia, CA). HIV RNA copy numbers were measured with a locked nucleic acid (LNA) Taqman assay. Briefly, we performed a single-tube, one-step assay in a total volume of 25 µl using the TaqMan RNA-to-CT-1STEP KIT (Life Technologies, Grand Island, NY), primers G19-2-F-7Y (AGCAGCYATGCAAATGTTA) and G-20-R (AGAGAACCAAGGGGAAGTGA) (400 nM each), and a dual-labeled fluorescent LNA probe (6-FAM/+C+C+ATCA+A+T+G+A+G+G+A/BHQ1) (200 nM); “+” in front of nucleotides indicates an LNA. Cycling conditions were 50°C for 30 min, 95°C for 5 min, and then 50 cycles of 95°C for 15 sec and 59°C for 1 min.
HIV DNA copy numbers were assessed with the same primer/probe combination and the TaqMan Gene Expression Master mix (Life Technologies, Grand Island, NY). Results were expressed as averaged HIV-1 RNA or DNA copies per million CD4 equivalents for cellular nucleic acids and as RNA copies/ml for supernatant HIV RNA concentration. The limit of detection for all samples was 10 copies per million cells.
For quantification of GM-HIV RNA and DNA, the same primers and methods were used but with the following probe: 6-VIC/ AACGAAGTCAGTCGACCACACTCACTCTCAC/ BHQ1.
Statistical analyses
ACH-2 cell, CEM-T4 cell, and GM-HIV-infected CD4 T-cell assays were run in triplicate and repeated at least three independent experiments. Experiments with primary CD4+ T cells were run in triplicate and repeated with at least four different donors. Experiments with unstimulated CD4+ T cells from HIV+ subjects were run in duplicate. All immunoblot analysis was repeated from four independent experiments. Microsoft Excel 2008 was used for statistical analyses. Data are presented as means ± s.e.m. Significance was assessed by student's t-Test; P<0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank the study participants without whom this research could not be performed, Drs. Steve Deeks, Huldrych Gunthard, Carolina Lopez and Hiroyu Hatano for helpful comments and support, and Dr. Mai Vu for assistance with participant recruitment. We thank John C.W. Carroll for graphics arts, Stephen Ordway for editorial assistance and Sharon Wilcox for administrative assistance. This work was supported by the National Institutes of Health [grants 1R21AI104445-01A1 (PL), R56 AI116342 and R21 AI116218 (JW)], the Department of Veterans Affairs Merit Review Award 5101 BX001048 (JW), the UCSF-Gladstone Center for AIDS Research Virology Core P30AI027763 (WCG, JW), U19 AI096113 (WCG) and research supported as part of the amfAR Institute for HIV Cure Research with grant number 109301(WG, JW, and PL).
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
Author Contributions
P.L. contributed to design the research, to perform the experiments, to interpret the data, and to write the paper. P.K. contributed to assist with experiments, to interpret the data and to help writing the paper. H.W.L. contributed to recruit HIV subjects for the study. P.K. and S.A.Y. contributed to assist with experiments. D.V.H. and W.C.G. contributed to provide key suggestions, and to interpret the data. W.C.G. also contributed to write the paper. J.K.W. contributed to recruit HIV subjects for the study, to interpret the data, and to write the paper.
References
- 1.Wong JK, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 2.Finzi D, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 3.Chun TW, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deeks SG, Lewin SR, Havlir DV. The end of AIDS: HIV infection as a chronic disease. Lancet. 2013;382:1525–1533. doi: 10.1016/S0140-6736(13)61809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Passaes CP, Saez-Cirion A. HIV cure research: advances and prospects. Virology. 2014;454–455:340–352. doi: 10.1016/j.virol.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 6.Ruelas DS, Greene WC. An integrated overview of HIV-1 latency. Cell. 2013;155:519–529. doi: 10.1016/j.cell.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Archin NM, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng K, et al. Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature. 2015;517:381–385. doi: 10.1038/nature14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Badley AD, Sainski A, Wightman F, Lewin SR. Altering cell death pathways as an approach to cure HIV infection. Cell Death Dis. 2013;4:e718. doi: 10.1038/cddis.2013.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kell AM, Gale M., Jr RIG-I in RNA virus recognition. Virology. 2015;479–480:110–121. doi: 10.1016/j.virol.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang SL, Shyu RY, Yeh MY, Jiang SY. Cloning and characterization of a novel retinoid-inducible gene 1(RIG1) deriving from human gastric cancer cells. Mol Cell Endocrinol. 2000;159:15–24. doi: 10.1016/s0303-7207(99)00207-5. [DOI] [PubMed] [Google Scholar]
- 12.Goubau D, et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5'-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chattopadhyay S, et al. Viral apoptosis is induced by IRF-3-mediated activation of Bax. EMBO J. 2010;29:1762–1773. doi: 10.1038/emboj.2010.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broz P, Monack DM. Newly described pattern recognition receptors team up against intracellular pathogens. Nat Rev Immunol. 2013;13:551–565. doi: 10.1038/nri3479. [DOI] [PubMed] [Google Scholar]
- 15.Schlee M. Master sensors of pathogenic RNA - RIG-I like receptors. Immunobiology. 2013;218:1322–1335. doi: 10.1016/j.imbio.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berg RK, et al. Genomic HIV-RNA induces innate immune responses through RIG-I-dependent sensing of secondary-structured RNA. PLoS One. 2012;7:e29291. doi: 10.1371/journal.pone.0029291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Wang X, Li J, Zhou Y, Ho W. RIG-I activation inhibits HIV replication in macrophages. J Leukoc Biol. 2013;94:337–341. doi: 10.1189/jlb.0313158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solis M, et al. RIG-I-mediated antiviral signaling is inhibited in HIV-1 infection by a protease-mediated sequestration of RIG-I. J Virol. 2011;85:1224–1236. doi: 10.1128/JVI.01635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doehle BP, Hladik F, McNevin JP, McElrath MJ, Gale M., Jr Human immunodeficiency virus type 1 mediates global disruption of innate antiviral signaling and immune defenses within infected cells. J Virol. 2009;83:10395–10405. doi: 10.1128/JVI.00849-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Britto AM, et al. Expression levels of the innate response gene RIG-I and its regulators RNF125 and TRIM25 in HIV-1-infected adult and pediatric individuals. AIDS. 2013;27:1879–1885. doi: 10.1097/QAD.0b013e328361cfbf. [DOI] [PubMed] [Google Scholar]
- 21.Cassani B, Villablanca EJ, De Calisto J, Wang S, Mora JR. Vitamin A and immune regulation: role of retinoic acid in gut-associated dendritic cell education, immune protection and tolerance. Mol Aspects Med. 2012;33:63–76. doi: 10.1016/j.mam.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raverdeau M, Mills KH. Modulation of T-cell and innate immune responses by retinoic Acid. J Immunol. 2014;192:2953–2958. doi: 10.4049/jimmunol.1303245. [DOI] [PubMed] [Google Scholar]
- 23.Dietze EC, et al. CBP/p300 induction is required for retinoic acid sensitivity in human mammary cells. Biochem Biophys Res Commun. 2003;302:841–848. doi: 10.1016/s0006-291x(03)00266-3. [DOI] [PubMed] [Google Scholar]
- 24.Kawasaki H, et al. Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature. 1998;393:284–289. doi: 10.1038/30538. [DOI] [PubMed] [Google Scholar]
- 25.Ortiz NE, Nijhawan RI, Weinberg JM. Acitretin. Dermatol Ther. 2013;26:390–399. doi: 10.1111/dth.12086. [DOI] [PubMed] [Google Scholar]
- 26.Buccheri L, Katchen BR, Karter AJ, Cohen SR. Acitretin therapy is effective for psoriasis associated with human immunodeficiency virus infection. Arch Dermatol. 1997;133:711–715. [PubMed] [Google Scholar]
- 27.Clouse KA, et al. Monokine regulation of human immunodeficiency virus-1 expression in a chronically infected human T-cell clone. J Immunol. 1989;142:431–438. [PubMed] [Google Scholar]
- 28.Marcu MG, et al. Curcumin is an inhibitor of p300 histone acetylatransferase. Med Chem. 2006;2:169–174. doi: 10.2174/157340606776056133. [DOI] [PubMed] [Google Scholar]
- 29.Lassen KG, Hebbeler AM, Bhattacharyya D, Lobritz MA, Greene WC. A flexible model of HIV-1 latency permitting evaluation of many primary CD4+T-cell reservoirs. PLoS One. 2012;7:e30176. doi: 10.1371/journal.pone.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pace MJ, et al. Directly infected resting CD4+T-cells can produce HIV Gag without spreading infection in a model of HIV latency. PLoS Pathog. 2012;8:e1002818. doi: 10.1371/journal.ppat.1002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med. 2014;20:425–429. doi: 10.1038/nm.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peisley A, Wu B, Xu H, Chen ZJ, Hur S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509:110–114. doi: 10.1038/nature13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 34.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 35.Sumpter R, et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samanta M, Iwakiri D, Kanda T, Imaizumi T, Takada K. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 2006;25:4207–4214. doi: 10.1038/sj.emboj.7601314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rehwinkel J. Exposing viruses: RNA patterns sensed by RIG-I-like receptors. J Clin Immunol. 2010;30:491–495. doi: 10.1007/s10875-010-9384-7. [DOI] [PubMed] [Google Scholar]
- 38.Wu B, et al. Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol Cell. 2014;55:511–523. doi: 10.1016/j.molcel.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshida T, et al. Matsuyama S.Bax-inhibiting peptide derived from mouse and rat Ku70. Biochem Biophys Res Commun. 2004;321:961–966. doi: 10.1016/j.bbrc.2004.07.054. [DOI] [PubMed] [Google Scholar]
- 40.Wiegand UW, Chou RC. Pharmacokinetics of acitretin and etretinate. J Am Acad Dermatol. 1998;39:S25–S33. doi: 10.1016/s0190-9622(98)70441-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.